Abstract
Although most patients with type 1 diabetes (T1D) retain some functional insulin-producing islet β cells at the time of diagnosis, the rate of further β cell loss varies across individuals. It is not clear what drives this differential progression rate. CD8+ T cells have been implicated in the autoimmune destruction of β cells. Here, we addressed whether the phenotype and function of autoreactive CD8+ T cells influence disease progression. We identified islet-specific CD8+ T cells using high-content, single-cell mass cytometry in combination with peptide-loaded MHC tetramer staining. We applied a new analytical method, DISCOV-R, to characterize these rare subsets. Autoreactive T cells were phenotypically heterogeneous, and their phenotype differed by rate of disease progression. Activated islet-specific CD8+ memory T cells were prevalent in subjects with T1D who experienced rapid loss of C-peptide; in contrast, slow disease progression was associated with an exhaustion-like profile, with expression of multiple inhibitory receptors, limited cytokine production, and reduced proliferative capacity. This relationship between properties of autoreactive CD8+ T cells and the rate of T1D disease progression after onset make these phenotypes attractive putative biomarkers of disease trajectory and treatment response and reveal potential targets for therapeutic intervention.
Keywords: Autoimmunity, Immunology
Keywords: Autoimmune diseases, Diabetes, T cells
Introduction
Type 1 diabetes (T1D) is an organ-specific autoimmune disease that leads to the destruction of pancreatic islet β cells, resulting in glucose dysregulation and a life-long dependence on exogenous insulin treatment. This autoimmune process typically begins years prior to clinical diagnosis (stages 1–2) and involves humoral and cellular immune responses (1). Upon diagnosis (stage 3), most individuals with T1D retain some level of functioning β cells, as indicated by the presence of circulating C-peptide, a byproduct of endogenous insulin processing (2), and the preservation of these functional β cells is associated with fewer clinical complications (3). Yet, the rate of functional β cell loss following diagnosis varies among individuals (4, 5). A key question for the prediction and prevention of T1D and other autoimmune diseases is what factors contribute to the rate of disease progression.
The character of the immune response probably plays a role in the rate of functional β cell loss following T1D diagnosis, as treatment with immune-modulating therapies results in short-term preservation of β cell function in some individuals (6). CD8+ T cells in particular influence T1D susceptibility and progression as clearly shown in mouse models (7–10), and islet-specific CD8+ T cells are detectable in the peripheral blood of individuals with T1D (11–15). Within the pancreas, CD8+ T cells are the most abundant lymphocyte of insulitic islets (16), and both polyclonal and islet-specific CD8+ T cells are more prevalent in the pancreas of individuals with T1D than in at-risk or healthy controls (HCs) (15, 17–19). In addition, some clues have emerged from responder analyses of immunotherapy clinical trials that implicate a role for CD8+ T cells. Partial exhaustion and central memory signatures of CD8+ T cells define responders in clinical trials of anti-CD3 (teplizumab) (20–23), and the frequency of memory islet-specific CD8+ T cells in peripheral blood increases with treatment (24, 25). Thus, islet-specific CD8+ T cells probably play an important role in T1D progression and outcome. However, these autoantigen-targeting cells remain poorly defined due in part to the technical difficulty of identifying and extensively phenotyping rare, low-affinity autoreactive T cells.
Here, we took advantage of high-content, single-cell mass cytometry, or cytometry by time of flight (CyTOF), along with a combinatorial pooled peptide–loaded MHC tetramer (Tmr) staining approach (26) to identify and extensively phenotype antigen-specific CD8+ T cells. We also introduce DISCOV-R, an analytical solution for phenotypic classification of rare subpopulations. Leveraging these technologies and analytical tools in HCs and individuals with T1D allowed us to (a) assess the phenotypic heterogeneity of rare islet-specific cells in individual subjects, (b) define common phenotypes of islet-specific CD8+ T cells across subjects, and (c) relate islet-specific phenotypes to the disease progression rate. We found that islet-specific CD8+ T cells exhibited heterogeneous phenotypes in both HCs and subjects with T1D. The rate of disease progression in T1D subjects was linked to 2 shared phenotypes: an activated memory phenotype was more frequent among islet-specific cells of rapid progressors, whereas an exhaustion phenotype was more prevalent in slow progressors. The exhaustion phenotype was confirmed functionally and was not merely a consequence of more advanced age, disease state, or disease duration. Together, these data implicate the phenotype and function of autoreactive CD8+ T cells as key mechanisms underlying the rate of disease progression.
Results
In spite of the complex phenotypes and heterogeneity within autoreactive CD8+ T cells, approaches to analyze these rare T cell subsets in T1D typically utilize a small number of single-parameter values (11–15, 27). To address this issue, we generated a high-content CyTOF panel incorporating HLA class I tetramers to identify antigen-specific CD8+ T cells and additional markers of differentiation, activation, and exhaustion (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI126595DS1). Tmrs contained a pooled set of HLA-A*0201–restricted peptides derived from known islet-associated autoantigens (Supplemental Table 2). We tested an insulin peptide separately from other islet antigens, as the phenotype of reactive cells might be expected to differ in individuals with T1D exposed to exogenous insulin. For comparison, we also included Tmrs containing 2 epitopes associated with chronic viral infection (12). We identified antigen-specific CD8+ T cells using a modified combinatorial Tmr staining approach based on the method described by Newell et al. (26) (Supplemental Figure 1). In verifying Tmr staining by CyTOF, we found that the intensity of Tmr staining was generally greater for the virus-specific pool than for the islet-specific pool (Supplemental Figure 1D and Figure 1A) and that both the Tmr+ frequencies and the intensities of markers were highly reproducible (Supplemental Figure 2), as previously described with flow cytometric Tmr analysis (11–15).
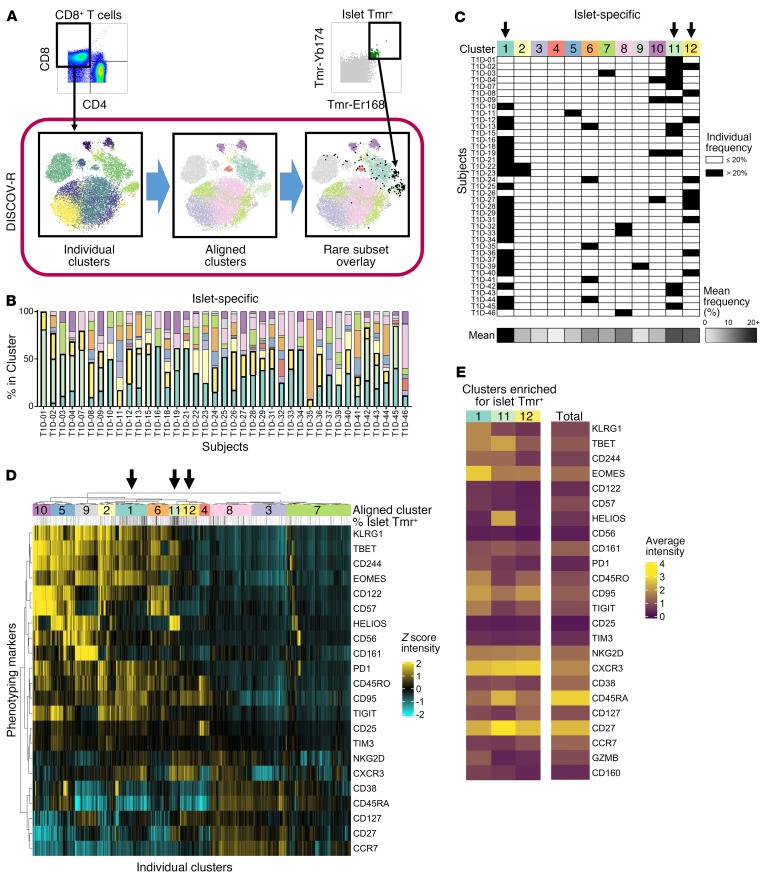
Figure 1. Islet-specific CD8+ T cells are dominated by three CXCR3+ memory phenotypes across subjects with T1D.
The DISCOV-R analysis method was applied to total CD8+ and islet-specific T cells from subjects with T1D (n = 46); the T cells had been assayed with the Tmr-CyTOF panel. (A) Schematic of the DISCOV-R method (see Methods and Supplemental Figure 3 for details) for 1 individual. (B and C) Distribution of islet-specific cells across the 12 aligned clusters for subjects with at least 5 Tmr+ cells (n = 39). (B) Data are displayed as a stacked bar graph for each subject, colored by cluster. The 3 clusters that are most dominant among islet-specific cells across subjects (clusters 1, 11, and 12) have heavy outlining and are stacked at the bottom. (C) Clusters containing more than 20% islet-specific cells for an individual are indicated in black. Arrows indicate clusters predominant in at least 25% of the samples; the detached bottom row indicates the mean frequency of cells within a cluster for all individuals on a scale from 0% (white) to 20% or higher (black). (D) Heatmap of Z scores using arcsinh-transformed expression of 22 consistent markers (rows) for all individual clusters (columns) from all T1D subjects (n = 46), grouped into 12 aligned clusters (annotated with numbers and colors). Negative Z scores (aqua) represent underexpression, and positive Z scores (yellow) represent overexpression of markers in an individual cluster compared with the mean of expression intensity on total CD8+ T cells within a subject. Frequency of islet-specific (Tmr+) cells within an individual cluster is annotated above (white = 0%, black = 20%+). (E) Heatmap of the mean absolute arcsinh-transformed expression of 24 markers for the 3 islet-specific clusters and total CD8+ T cells. Expression intensity ranges from 0 (dark purple) to 4+ (yellow).
We detected low numbers of autoantigen-specific events for Tmr+ cells analyzed by CyTOF in both HCs and individuals with T1D. We used a computational strategy called DISCOV-R (distribution analysis across clusters of a parent population overlaid with a rare subpopulation) (Figure 1A), in which total CD8+ T cells from each individual were clustered, in this case using Phenograph (28). Next, these individual clusters were aligned with CD8+ T cell clusters from other samples by hierarchical metaclustering to generate a common phenotypic landscape. Finally, Tmr+ cells were overlaid onto the CD8+ T cell landscapes for analysis of their distribution, as described in detail in Supplemental Figure 3. DISCOV-R facilitates direct comparisons of complex phenotypes between subjects while minimizing (a) skew introduced by disparate sample sizes, (b) sensitivity to outliers, and (c) homogenization resulting from the pooling of cells or subjects. This in turn enabled an unbiased assessment of the phenotypic distribution of rare, autoreactive cells both within and across subjects without masking individual heterogeneity.
Islet-specific CD8+ T cells are composed of 3 predominant CXCR3+ memory phenotypes.
For an extensive characterization of islet-specific CD8+ T cells, we applied our CyTOF panel and DISCOV-R to PBMCs from individuals with T1D (n = 46) (Table 1 and Supplemental Table 3). For characterization of the antigen-specific Tmr+ cell phenotype, we restricted analysis to samples that contained 5 or more Tmr+ cell events. We found heterogeneity of islet-specific CD8+ T cells within individual subjects and common phenotypes across subjects. Specifically, of the 12 shared phenotypes (clusters) we defined among total CD8+ T cells across all individuals, islet-specific Tmr+ cells were identified in more than 1 cluster for most subjects. However, no individual subject had more than 10 of the 12 clusters containing islet-specific Tmr+ cells (Figure 1B and Supplemental Figure 4). Three common clusters (labeled 1, 11, and 12 in Figure 1) contained the largest representation of islet-specific Tmr+ cells, accounting for greater than 20% of the islet-specific Tmr+ cells in more than 25% of the subjects (Figure 1C).
Table 1. Cohort demographics.
To define these 3 common islet-specific T cell phenotypes, we assessed the expression levels of phenotypic markers on all individual clusters of total CD8+ T cells (Figure 1D and Supplemental Figure 5) and the 3 aligned islet-specific clusters (Figure 1E). Cluster 11, which was dominant only among islet-specific cells (Supplemental Figure 6), had an activated transitional memory phenotype with high expression of HELIOS and CD27. Cluster 12 was also unique to islet-specific cells and had a transitional memory phenotype with high CD27 expression, but lacked HELIOS expression. Cluster 1 dominated among insulin- and virus-specific cells in addition to islet-specific cells (Supplemental Figure 6) and had a memory exhausted–like phenotype with high EOMES expression, intermediate TBET expression, and elevated expression of the multiple inhibitory receptors 2B4 (CD244), PD1, TIGIT, and CD160. All 3 islet-specific clusters were CXCR3+, consistent with previous reports (29). Thus, islet-specific CD8+ T cells are heterogeneous and dominated by 3 distinct CXCR3+ memory subsets: an exhaustion-like subset that was also dominant for insulin- and chronic virus–specific cells and 2 transitional memory phenotypes unique to islet-specific cells, 1 of which was HELIOS+.
Although inclusion of multiple antigen specificities within a pool of islet-specific Tmrs may account for the phenotypic heterogeneity, several lines of evidence argue against this. Antigen-specific cells identified by a single specificity (insulin) or only 2 pooled Tmrs (virus) occupied 2 or more prominent clusters in the majority of individuals (Supplemental Figure 6). Moreover, the number of predominant phenotypes of islet-specific cells from a given individual was not correlated with the number of positive islet antigen specificities determined by flow cytometry (Supplemental Figure 7). We performed cluster distribution analyses on all subjects with at least 5 Tmr+ events, resulting in exclusion of 7 of 46 subjects. Yet, when we only analyzed subjects with at least 25 Tmr+ events (n = 27), we found similar prominence of 3 clusters, suggesting that outliers were not skewing our results. Last, our finding of phenotypic heterogeneity within islet-specific cells is consistent with others’ recent reports (30, 31). Therefore, islet-specific CD8+ T cells were phenotypically heterogeneous at both the individual and population levels, and a core set of 3 predominant CXCR3+ memory phenotypes was conserved among islet-specific cells across subjects.
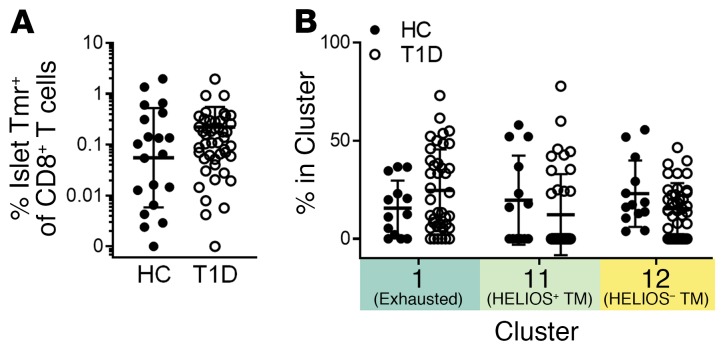
We also detected islet-specific CXCR3+ memory CD8+ T cells in PBMCs from nondiabetic HLA-A2+ HCs (Supplemental Figure 8). As illustrated in Figure 2, most HCs displayed similar frequencies and phenotypes of antigen-specific cells compared with the T1D subjects in this study (Supplemental Figures 6 and 8). In both HCs and individuals with T1D, the transitional memory phenotypes enriched for islet-specific cells (clusters 11 and 12) were not consistently represented among insulin- or chronic virus–specific cells, whereas the exhaustion phenotype (cluster 1) was prevalent in both virus- and insulin-specific cells. Thus, islet-specific cells are phenotypically heterogeneous and exhibit some unique phenotypes not consistently seen in other antigen-specific CD8+ T cells that are expected to have recurrent exposure to antigen.
Figure 2. Islet-specific CD8+ T cell frequency and phenotype do not differ between HCs and individuals with T1D.
HCs (n = 20) were assayed with our Tmr-CyTOF panel and included in the DISCOV-R analysis as in Figure 1. (A) Frequency of islet-specific (Tmr+) cells within total CD8+ T cells was assessed and compared for HCs (n = 20) and T1D subjects (n = 46) using a Mann-Whitney U test. (B) Frequency of islet-specific CD8+ T cells among the 3 common islet-specific clusters was assessed for HCs (n = 13) and individuals with T1D (n = 39) with 5 or more Tmr+ events using 2-way ANOVA with Sidak’s test for multiple comparisons. Data represent the mean ± SD. TM, transitional memory.
Activated memory and exhaustion phenotypes discriminate subjects with rapid and slow T1D progression.
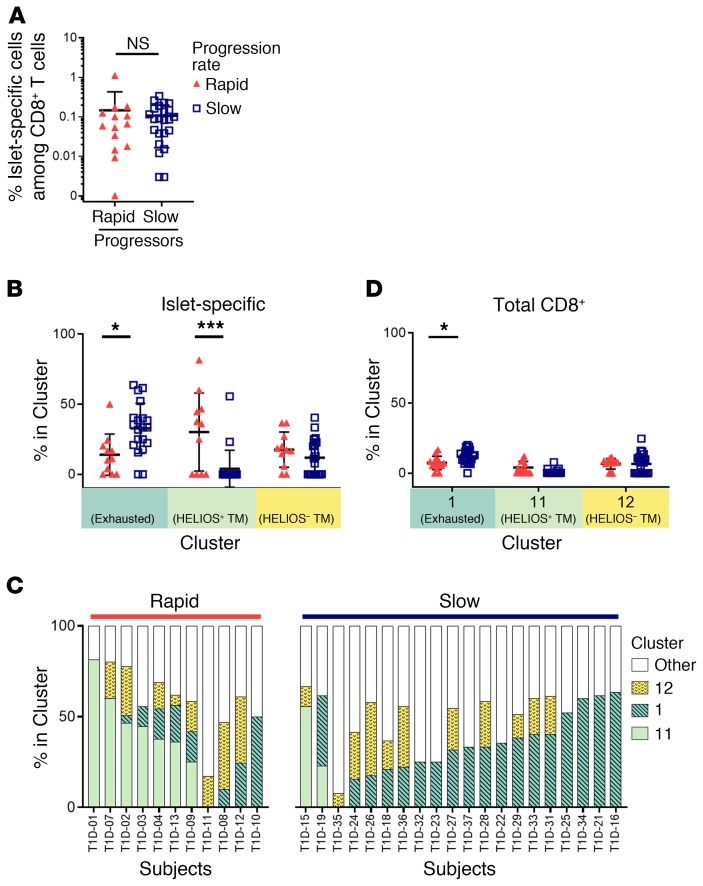
To address whether the phenotypes of autoreactive CD8+ T cells influence the rate of disease progression after onset in T1D, we stratified subjects by their rate of loss of β cell function. Subjects with rapid progression were less than 5 years from diagnosis but showed no detectable insulin secretion (<0.05 ng/mL) (C-peptide); subjects categorized as slow progressors were 5 or more years from diagnosis with >0.1 ng/mL C-peptide (Table 1 and Supplemental Table 3). We found no significant difference in the frequency of islet-specific CD8+ T cells between rapid and slow progressors (Figure 3A), even when outliers were excluded (data not shown); however, the proportion of exhausted (cluster 1) and HELIOS+ transitional memory (cluster 11) phenotypes among islet-specific cells differed significantly when comparing rapid and slow progressors, whereas the cluster 12 phenotype did not (Figure 3B). No other clusters differed among islet-specific cells between the rapid and slow progressors. On an individual basis, approximately half of the islet-specific CD8+ T cells in each cohort were of the common CXCR3+ memory phenotypes, and islet-specific cells from the majority of rapid progressors were enriched for cluster 11, whereas the majority of islet-specific cells of slow progressors were enriched for exhausted cluster 1 (Figure 3C). We also observed an increased frequency of the cluster 1 exhaustion phenotype in slow progressors among total CD8+ T cells (Figure 3D). These differences in the frequency of cluster 1 islet-specific cells were also seen when applying more stringent C-peptide cutoffs for rapid (<0.02 ng/mL, n = 9) and slow (>0.2 ng/mL, n = 12) rates of progression (P < 0.04, by 2-way ANOVA with Sidak’s test for multiple comparisons). A cutoff of 25% for the frequency of islet-specific cells residing in cluster 1 identified slow progressors with 70% sensitivity and 91% selectivity. Discrimination of disease progression by these phenotypes was particular to islet-specific cells, as we found no differences between their frequencies among virus- or insulin-specific CD8+ T cells (Supplemental Figure 9).
Figure 3. Phenotype, not frequency, of islet-specific CD8+ T cells is associated with the rate of disease progression in T1D.
The frequency of (A) islet-specific (Tmr+) cells within total CD8+ T cells was assessed for rapid (n = 14) and slow (n = 23) T1D progressors using a Mann-Whitney U test. The frequency of (B) islet Tmr+ or (D) total CD8+ T cells among the 3 common islet-specific clusters was assessed for rapid (n = 11, red solid triangles) and slow (n = 20, blue open squares) T1D progressors with 5 or more Tmr+ events using 2-way ANOVA with Sidak’s test for multiple comparisons. Data represent the mean ± SD. *P < 0.05 and ***P < 0.001. (C) Distribution of islet Tmr+ cells in clusters for individual samples; rapid progressors (n = 11) and slow progressors (n = 20) were organized by decreasing frequency of cluster 11 and increasing frequency of clusters 1 and 12 within each group.
Simplified gating for CXCR3, EOMES, and HELIOS, based on DISCOV-R clusters, approximated the phenotype of clusters 1, 11, and 12, with similar trends seen between rapid and slow progressors, though such comparisons were not consistently statistically significant (Supplemental Figure 10). Thus, multidimensional definitions beyond a few well-defined markers are required to fully distinguish these disease-associated phenotypes, which included an activated transitional memory phenotype linked to rapid progression (cluster 11) and an exhausted memory phenotype linked to slow progression (cluster 1), whereas none of the other clusters were associated with outcome.
The association between the exhaustion-like phenotype and T1D outcomes remains after accounting for disease duration and age.
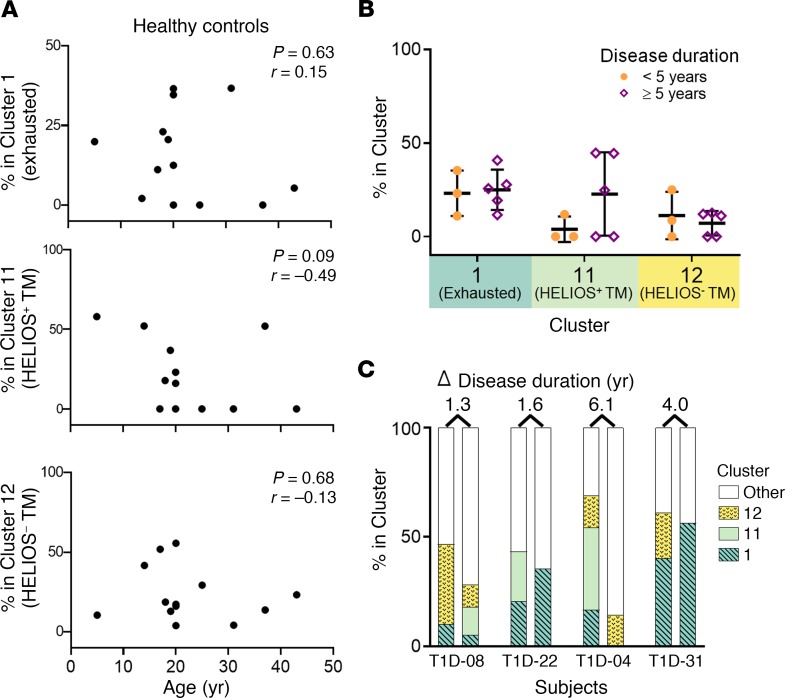
Factors beyond the rate of disease progression may contribute to the observed differential islet-specific CD8+ T cell phenotypes. Rapid and slow progressors were matched for sex and mean age at diagnosis. However, since slow progressors tended to be older in this cohort, and T cell exhaustion and memory are associated with more advanced age (32), we adjusted for age at sampling; yet, the differential phenotypes between rapid and slow progressors among islet-specific and total CD8+ T cells remained (Supplemental Figure 11). Furthermore, the 3 islet-specific phenotypes did not correlate with age in HCs (Figure 4A). Thus, exhaustion is not solely driven by the presence of disease or age but is instead related to disease outcome.
Figure 4. Age and disease duration do not determine islet-specific CD8+ T cell exhaustion.
The frequency of islet-specific phenotypes among islet-specific CD8+ T cells was assessed for subjects with 5 or more Tmr+ events. (A) Frequencies in HCs (n = 13) as a function of age based on DISCOV-R results from Figure 2. Statistical significance was determined by Spearman’s correlation. (B) Frequencies in T1D subjects who were not classified as rapid or slow progressors, grouped by disease duration (<5 years, n = 3, solid orange circles; ≥5 years, n = 5, open purple diamonds) on the basis of DISCOV-R results from Figure 1. A 2-way ANOVA with Sidak’s test for multiple comparisons revealed no statistically significant differences between the groups. Data represent the mean ± SD. (C) Frequencies in T1D subjects (n = 4) with samples drawn at 2 time points following disease onset, shown as paired, stacked bar graphs. The time points of the first draw were 3.2, 3.8, 4.8, and 5.5 years after disease onset, respectively.
By definition, slow and rapid progressors differed by their disease duration. Thus, we assayed an independent cohort of T1D subjects stratified by disease duration but not distinguished by their disease progression rate (Table 1 and Supplemental Table 3); we found no difference in the frequency of the 3 common islet-specific clusters by disease duration (Figure 4B). To further test the influence of disease duration on exhaustion cluster 1, we performed longitudinal analyses on a subset of subjects for whom the disease duration was relatively similar, yet the frequency of the exhausted islet-specific cells was divergent. We did not observe major fluctuations in exhausted cluster 1 frequencies in islet-specific CD8+ T cells over time, and those with a lower proportion of exhausted cells did not gain this phenotype with longer disease duration (Figure 4C). Together, these data indicate that islet-specific CD8+ T cell phenotypes differ by disease progression rate, and not age or disease duration.
Islet-specific CD8+ T cells from slow T1D progressors are functionally more exhausted.
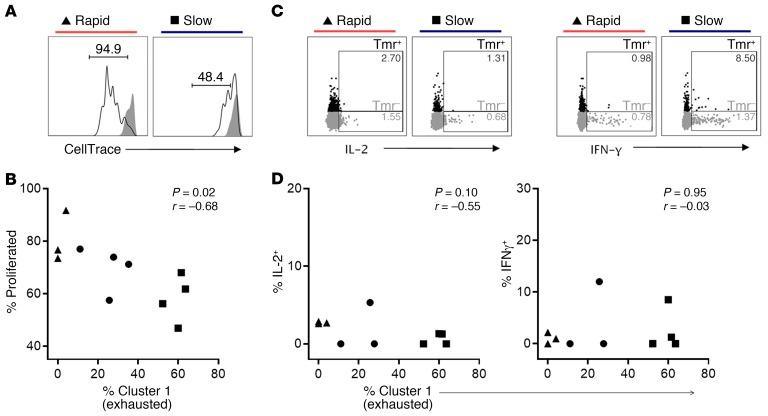
To confirm that cells in cluster 1 exhibited functional features of exhaustion, we tested the proliferation and cytokine production of islet-specific CD8+ T cells. Consistent with their more exhausted phenotype, islet-specific cells that were most dominated by cluster 1 were less proliferative in response to T cell receptor stimulation (Figure 5, A and B). By comparison, the frequencies of clusters 11 and 12 were positively correlated with proliferation (Supplemental Figure 12). Additionally, islet-specific cells maintained only a limited ability to produce the cytokines IL-2 and IFN-γ, irrespective of the abundance of cluster 1 (Figure 5, C and D). Taken together, these findings support an exhausted-like phenotype (33) of cluster 1, which dominates islet-specific cells from slow progressors, exhibits low production of cytokine, and is marked by elevated expression of multiple inhibitory receptors and reduced proliferative capacity compared with islet-specific cells from rapid progressors lacking cluster 1.
Figure 5. Islet-specific CD8+ T cells with an abundant cluster 1 (exhausted) phenotype are hypoproliferative and produce limited levels of the cytokines IL-2 and IFN-γ.
PBMCs from individuals with T1D (n = 11) with varying frequencies of cluster 1 among their islet-specific cells were stimulated with anti-CD3 plus anti-CD28. Cells were assayed by flow cytometry to identify islet-specific (Tmr+) CD8+ T cells (Supplemental Figure 13). Examples of gating for proliferation and cytokine production are shown for a rapid progressor (T1D-02) and a slow progressor (T1D-34) with low (4%) and high (60%) frequencies of cluster 1, respectively. (A) Representative examples of the frequency of proliferated cells on day 5 among stimulated (black line) islet Tmr+ cells as measured by CellTrace dye dilution, using unstimulated (solid gray) cells as a gating control. (B) Frequency of proliferated cells among islet Tmr+ cells after 5 days of stimulation, plotted against the frequency of cluster 1 determined by mass cytometry for each individual (n = 11). (C) Representative examples of IL-2 and IFN-γ production assessed at 6 hours among islet Tmr+ (black) or Tmr– CD8+ T cells (gray). (D) Frequency of IL-2+ and IFN-γ+ cells among islet Tmr+ cells after 6 hours of stimulation, plotted against the frequency of cluster 1 determined by mass cytometry for each individual (n = 10); no substantial cytokine production (<1%) was observed in the absence of stimulation. Statistical significance was determined by Spearman’s correlation. The difference in proliferation between islet-specific cells of rapid progressors (triangles, n = 3) and slow progressors (squares, n = 4) was not significant (P = 0.057), nor was cytokine production (P > 0.05) by Mann-Whitney U test.
Discussion
Although islet-autoreactive CD8+ T cells were present in both HCs and individuals with diabetes, we found that characteristic phenotypes of these cells reflect the rate of disease progression in T1D. Ample evidence indicates that islet-specific CD8+ T cells are a significant driver of β cell destruction in T1D (34). However, across numerous studies involving individuals with T1D, the frequencies of islet-specific CD8+ T cells in peripheral blood have been neither consistently altered nor strongly correlated with disease progression (11–15, 35). Here, through an unbiased and multidimensional approach, we demonstrate that islet-specific CD8+ T cells of HCs and T1D subjects comprise 3 dominant phenotypes that display characteristics of transitional memory or exhausted memory cells. Crucially, however, autoreactive CD8+ T cell phenotypes in T1D subjects predicted outcome after onset of disease, an activated transitional memory phenotype with high proliferative potential was associated with rapid progression, and a more functionally exhausted phenotype corresponded with slow disease progression. Detection of these phenotypes may be used to more precisely classify patients and to select therapies that promote the maintenance of β cell health.
The phenotype of autoreactive CD8+ T cells was not uniform. CD8+ T cells have various functions associated with distinct activation and differentiation states (36, 37). For example, following vaccination, viral antigen–specific cells exhibit a defined phenotype during the effector and memory phases of the immune response (38). Using the analytical tool DISCOV-R, we were able to define variable phenotypes of CD8+ T cells in multiple dimensions and subsequently assess the phenotypes of rare antigen-specific cells. In contrast to temporal vaccine-induced, virus-specific cells and antigen-elicited gluten-specific CD4+ T cells (39), we found that chronic and latent virus-specific CD8+ T cells had substantial phenotypic heterogeneity. Our findings were consistent with another report (26) and indicate the variable nature of the immune response to recurring antigen exposure (40). Importantly, we found that islet-specific cells were also phenotypically heterogeneous within an individual (30, 31), suggestive of variability in islet-specific cell immune history and thus having potential consequences for autoimmune disease progression.
Three predominant CXCR3+ memory phenotypes of islet-specific cells were common across both HCs and subjects with T1D. An exhaustion-like phenotype was broadly shared among chronic virus–, insulin-, and islet-specific CD8+ T cells, consistent with repeated antigen exposure in all 3 settings (41). However, there are several states of exhaustion (42). The islet-specific exhaustion cluster 1 differed from other clusters that also share features of exhaustion and were enriched in virus- and insulin-specific cells. Unlike islet-enriched exhaustion cluster 1, clusters 2, 6, and 10 lacked CXCR3, expressed late differentiation or senescence markers, and were less abundant in both islet-specific and total CD8+ T cells. These more terminally exhausted cells were more prominent among insulin-specific cells, even within the same individual. The CXCR3+ and less terminal phenotype of the islet-specific cluster 1 suggests a precursor exhausted population (42). Two other phenotypes we identified were unique to islet-specific cells (clusters 11 and 12) and not dominant among insulin- and virus-specific cells, indicating a different fate and function associated with exposure to native autoantigens from the pancreas as opposed to those seen for repeated viral antigen or abundant exogenous autoantigen as with insulin administration. These findings confirm and expand reports by other groups which have shown in a more focused manner that not all islet-specific cells exhibit the same differentiation state (19, 24, 27, 30, 31, 35, 43–45) and that the phenotype of islet-specific cells partially overlaps with chronic virus–specific cells (24, 30, 43, 45).
Despite differential expression of most markers, a unifying feature of the 3 islet-specific phenotypes was the high level of expression of CXCR3, whose ligand, CXCL10, is upregulated in the pancreas in T1D (46) and could therefore promote islet-specific T cell migration to that site. Although CXCR3 expression is greatest in these 3 islet-specific phenotypes, we also observed a CXCR3-expressing naive subset that dominated in polyclonal and virus-specific cells. Naive CD8+ T cells have been reported in the pancreas of subjects with recent-onset T1D (18). The potential for this subset to be recruited to the pancreas along with autoreactive cells has important implications, because the presence of nonspecific bystander CD8+ T cells in the pancreas in a mouse model of diabetes was found to be associated with attenuated effector functions of islet-specific cells and protection from disease (47). Thus, the phenotype and function of the migrating cells that are not autoreactive may also contribute to disease progression.
Autoimmune disease progression may be modulated by functional responses of CD8+ T cells. For example, inflammatory CD8+ T cells are associated with disease severity in systemic lupus erythematosus (SLE) (48) and multiple sclerosis (49). By contrast, increased CD8+ T cell exhaustion is associated with a beneficial response to therapy in recent-onset T1D (20, 50) as well as slower disease progression or a better prognosis in Crohn’s disease, SLE, and vasculitis (51, 52). Comparisons of phenotypes of autoreactive CD8+ T cells in health and disease have yielded mixed results. Some groups have found increased memory and CD57+ cells in individuals with T1D compared with HCs (27, 43), whereas others found no difference (15, 19). We found that the comprehensively defined islet-specific phenotypes, though variable across individuals, did not significantly differ by disease status and, indeed, were also present in HCs. Using established markers of CD8+ T cell differentiation, Yeo et al. described a positive correlation between changes in C-peptide and changes in effector memory CD57+ β cell–specific CD8+ T cells among young individuals who were newly diagnosed with T1D, implicating antigen load as a driver of differentiation and peripheral migration of this T cell subset (53). However, the relationship of autoreactive CD8+ T cell function and rate of progression in established T1D remains relatively unexplored. Here, we found no differences in the frequency of total islet-specific CD8+ T cells between rapid and slow progressors, but T1D subjects with rapid disease progression had a significantly greater proportion of islet-specific CD8+ T cells with a HELIOS+ transitional memory phenotype that is consistent with activation and proliferation (54) as well as exacerbated autoimmunity (55). Transitional memory cells are highly proliferative and polyfunctional, and this subset is transiently expanded in acute but not chronic HIV infection (56), suggestive of a more aggressive disease state. For the first time to our knowledge, we clearly associate this islet-specific transitional memory phenotype with a rapid rate of disease progression, opening the possibility for selective targeting of these cells therapeutically in patients with established T1D.
Individuals with T1D with slow disease progression showed enrichment for islet-specific cells with functional features of exhaustion, regardless of disease duration and after accounting for age. T cell exhaustion plays opposing roles; it is deleterious in tumor and chronic viral infection (42, 57) but, as recently appreciated, beneficial in autoimmunity (58). Interestingly, we also observed the association of the exhaustion phenotype with slow disease progression among the polyclonal CD8+ T cell population within the same subjects, which suggests that intrinsic factors may promote exhaustion of islet-specific and total CD8+ T cells in slow progressors. However, we observed that the phenotype is present in HCs and that not all specificities reflect this bias. Virus- and insulin-specific exhausted cells were not preferentially increased in slow progressors, indicating an additional role for antigen exposure. In chronic viral infections and cancer, it is known that exposure to antigen in the absence of costimulation leads to exhaustion, typically within a few weeks (33). Our findings suggest that increasing the exhaustion phenotype could have a beneficial effect on outcome. Indeed, in T1D, therapeutic manipulation of the T cell receptor alone correlated with a more exhausted-like CD8+ phenotype in responders to treatment (20, 25).
Whether exhaustion among islet-specific cells of slow progressors precedes disease or is merely a consequence of time and disease duration has important implications for disease mechanisms, monitoring, and therapy. Here, we found that the exhausted-like islet-specific phenotype was clearly not transient. Instead, we show in longitudinal analyses of a subset of subjects with a range of exhausted cells that exhaustion is relatively maintained over time within an individual. Also, the exhaustion phenotype was found at a similar range of frequencies in HCs. Stability of the exhausted phenotype has also been demonstrated in the setting of long-term, chronic HIV infection (59), consistent with preserved epigenetic and transcriptional programming (33, 42, 60). This contrasts with the dynamic changes in exhaustion features associated with disease severity in rheumatoid arthritis (61). Future longitudinal assessments of phenotype, function, and epigenetics across a range of disease stages will help further dissect mechanisms underlying this apparent stability. These results also indicate that early intervention to augment islet-specific T cell exhaustion may prevent or delay further disease progression.
In summary, using high-dimensional mass cytometry with a new analytical method, DISCOV-R, we revealed phenotypic heterogeneity among circulating autoreactive CD8+ T cells in HCs and individuals with T1D. We linked an activated memory phenotype with rapid disease progression after T1D onset and an exhausted phenotype with slow disease progression. Future studies may address whether this reflects islet-specific CD8+ T cell composition in the pancreas. Although our current studies focused on 4 well-defined islet antigens, many new and altered islet-specific epitopes have been discovered recently (62). Determining whether these new specificities express similar phenotypic markers may elucidate the role of specific antigens and their associated phenotypes in the T1D disease course. The finding that islet-specific CD8+ T cells from slow progressors were enriched for an exhausted phenotype indicates that therapies that augment and establish their exhaustion could be effective in preserving residual β cell function after onset and makes the phenotype an attractive putative biomarker to predict disease trajectory and monitor therapeutic efficacy.
Methods
Study design and samples.
On average, one-third of individuals diagnosed with T1D will lose detectable C-peptide within 5 years of disease onset (<0.017 nmol/L, or 0.05 ng/mL) (5, 63). Conversely, approximately one-third of individuals will retain C-peptide at levels 0.1 ng/mL or higher (0.03 nmol/L) 5 years after diagnosis (64). In this study, we sought to assess these ends of the disease spectrum cross-sectionally, using 5 years after onset and these C-peptide levels as cutoffs for distinguishing the rate of disease progression. Using our newly developed 35-parameter CyTOF panel with pooled Tmrs (Supplemental Table 2) in combination with 24 phenotyping markers (Supplemental Table 1) and applying a new analytical method for phenotyping rare subpopulations, DISCOV-R, we characterized antigen-specific CD8+ T cells from cryopreserved PBMCs from 20 HLA-A2+ HCs and 46 HLA-A2+ T1D subjects, a subset of which was stratified by rapid (n = 14, <0.05 ng/mL C-peptide within 5 years of diagnosis) and slow (n = 23, >0.1 ng/mL C-peptide at 5 or more years into disease) rate of disease progression following diagnosis. Because age at onset is a known predictor of disease progression, these groups were matched for this feature (Table 1 and Supplemental Table 3). All assays were run and analyzed in a blinded manner, and staining batches included an internal control of a single HLA-A2– individual with or without a consistent low frequency of spiked preproinsulin- or CMV-specific clone that was used to set gates for Tmr+ cells.
Peptide-MHC Tmr generation.
A biotinylated monomer of HLA-A2 (2 mg/mL) loaded with peptides (Supplemental Table 2) chosen for their demonstrated presence in T1D (12) was obtained from the NIH Tetramer Core Facility. Metal-conjugated avidin (0.5 mg/mL) was provided by Fluidigm prior to commercial availability; premium-grade phycoerythrin-conjugated (PE-conjugated) streptavidin (1 mg/mL) was obtained from Thermo Fisher Scientific. Each monomer was diluted in PBS, and then multimerized by 6 additions (each followed by a 10-minute incubation at 4°C) of 1/36th molar equivalents of 1 avidin, such that the final preparation contained a monomer at 0.16 mg/mL (2.4 mM) with a 6:1 molar ratio to avidin in 1% BSA, which was stored at 4°C for up to 1 month.
CyTOF staining, acquisition, and subset identification.
Thawed cryopreserved PBMCs (1.5 × 106 to 2.5 × 106 per stain) were first stained for viability using 100 μL cisplatin (100 μM, Enzo Life Sciences) in PBS for 1 minute at room temperature (RT), followed by quenching and washing with protein-containing media. Cells were then pretreated with 250 μL dasatinib (50 nM, LC Laboratories) for 8–10 minutes at 37°C and washed prior to staining with 50 μL solution containing 1 μL of each Tmr (Supplemental Table 2) in running buffer (RB) (PBS with 0.5% BSA and 2 mM EDTA) for 15 minutes at 37°C. Without washing, 50 μL 2× surface antibody cocktail (Supplemental Table 1) in RB was added, and the sample was incubated for an additional 30 minutes at 4°C. Samples were washed, fixed using the Maxpar Nuclear Antigen Staining Buffer (Fluidigm) for 20 minutes at RT, and stained with an intracellular antibody cocktail in Staining Perm Buffer (Fluidigm) for 30 minutes at 4°C. Samples were then washed, resuspended with 125 nM MaxPar Intercalator-Ir (Fluidigm) in Fix and Perm Buffer (Fluidigm), and stored at 4°C overnight or for up to 1 week prior to acquisition.
On the day of acquisition, cells were washed and resuspended in cold ultrapure water containing one-fifth EQ Four Element Calibration Beads (Fluidigm) by volume to a cell concentration of less than 0.5 × 106/mL. Samples were acquired at a rate of 300–500 events per second on a CyTOF 1.5 with upgrades (Fluidigm), running CyTOF Software, version 6.0.626 and using a Super Sampler system (Victorian Airship & Scientific Apparatus). Files were converted to flow cytometry standard (FCS) format and then randomized and normalized for EQ bead intensity using CyTOF software. FlowJo software, version 10.4, was used to manually gate and export the FCS files for CD8+ T cell and Tmr+ populations (Supplemental Figure 1), guided by control samples.
DISCOV-R computational analyses.
The FCS files prepared above were analyzed using custom R scripts (https://github.com/BenaroyaResearch/Wiedeman_Long_DISCOV-R) based on the flowCore (65), Rtsne (66–68), and cytofkit (28, 69–71) packages. Tmr+ events were concatenated to total CD8+ T cell FCS files for each sample prior to arcsinh transformation using the following parameters: a = 0 and b = 1/5. Subsequently, t-distributed stochastic neighbor embedding (t-SNE) analysis and Rphenograph clustering were performed for each sample using the following 22 phenotypic markers: KLRG1, HELIOS, TIM3, CD25, PD1, CCR7, CD45RO, CD57, CD45RA, CD38, CD127, TIGIT, CD27, CD161, TBET, CD95, NKG2D, CD122, EOMES, CXCR3, CD244, and CD56; granzyme B and CD160 intensities were less reproducible (>20% coefficient of variation), and were excluded from clustering. To align phenotypically similar clusters across individuals, hierarchical metaclustering of individual Phenograph clusters with higher than 1% frequency was performed across individuals using the Z score values of expression compared with each subject’s total CD8+ T cells, with Euclidean distance and Ward’s method used to assess phenotypic similarity. Heatmaps were generated in R using the ComplexHeatmap package (72). The dendrogram was cut into 12 segments using “cutree” from the stats R package (73), and the resulting cluster assignments were applied to the individual samples. To summarize the absolute expression values for each cluster, a weighted average of arcsinh-transformed intensities was calculated on the basis of the proportion of a subject’s total CD8+ T cells represented within a given cluster. This approach reduced bias due to varying sampling depths and PhenoGraph under-/overclustering for an individual. Tmr+ cells were overlaid on the t-SNE plot using the ggplot2 package (74) in R. Total CD8+ and Tmr+ T cells were counted for each cluster in each subject, and their frequency was calculated for samples with at least 5 Tmr+ events. Clusters were considered dominant if they contained more than 20% of the Tmr+ T cells, as this cutoff required more than 1 cell to be present, and the false-positive rate (proportion of Tmr+ T cells when randomly assigned to a cluster exceeded the observed value) dropped off considerably at this threshold. In our validation of the DISCOV-R method, we found that individual clustering and cluster alignment were both highly reproducible (<25% CV for clusters of at least 3% frequency), as was the number of dominant islet-specific clusters present in an individual (mean 2 ± 1).
Flow cytometric assays of cytokine production and proliferation.
Thawed cryopreserved PBMCs were cultured at 37°C with and without stimulation with plate-bound anti-CD3 (OKT3, 1 μg/mL, BioLegend) and soluble anti-CD28 (CD28.2, 2 μg/mL, BioLegend). For the cytokine production assay, cells were cultured for 6 hours, and brefeldin A and monensin (BioLegend) were each added at 1× for the last 4 hours. For the proliferation assay, cells were loaded with CellTrace Violet (Invitrogen, Thermo Fisher Scientific), according to the manufacturer’s instructions, prior to culturing for 5 days. Following culturing, cells were harvested; stained for viability using Zombie NIR (BioLegend) according to the manufacturer’s instructions; stained with pooled islet-specific Tmrs, as in the CyTOF staining, acquisition, and subset identification section; and subsequently stained with the surface antibodies anti-CD14-BUV737 (M5E2), anti–CD19-FITC (HIB19), anti–CD56-PE-Cy7 (NCAM16.2), anti-CD3-BUV395 (UCHT1), anti–CD4-BV605 (RPA-T4), and anti-CD8-BV786 (RPA-T8) in Brilliant Stain Buffer (all from BD Biosciences) to identify CD8+ T cells (Supplemental Figure 13). For the cytokine production assay, cells were further fixed and permeabilized using the Foxp3 Transcription Factor Staining Buffer Set (Invitrogen, Thermo Fisher Scientific) and stained with anti–IL-2-BB700 (MQ1-17H12), anti–TNF-α-BV650 (MAb11), and anti–IFN-γ-BV421 (B27) (all from BD Biosciences). Cells were acquired on an LSRFortessa (BD Biosciences) and analyzed using FlowJo software.
Statistics.
Beyond that which was described in DISCOV-R computational analyses, GraphPad Prism (version 7.05, GraphPad Software) was used to generate graphs and to perform either a Mann-Whitney U test to compare antigen-specific cell frequency or a 2-way ANOVA with Sidak’s test for multiple comparisons of the frequencies of the 3 islet-specific clusters between 2 groups. Spearman’s correlation was used for assessment of bivariate data. All statistical tests were 2 sided.
Study approval.
All samples were collected from the BRI’s Immune Mediated Disease Registry and Repository. Written informed consent was obtained from all subjects according to protocols approved by the BRI’s institutional review board (protocol number IRB07109).
Author contributions
AEW and SAL conceptualized the study. CS, CJG, ES, and GTN provided input on sample selection and clinical interpretation. EAJ gave guidance on Tmr staining methodology. AEW, GB, and AMK conducted the experiments. VSM, MGR, HAD, SP, BH, MJD and PSL performed computational and statistical analysis and provided consultation. AEW and SAL wrote the manuscript. All authors reviewed and edited the manuscript. SAL obtained funding and supervised the study.
Supplementary Material
Acknowledgments
Development of the CyTOF panel was supported by the ITN and sponsored by the National Institute of Allergy and Infectious Diseases (NIAID) subaward (to SAL) under award number UM1AI109565 (NIH grant, awarded to GTN). Funding from the JDRF (3-SRA-2014-315-M-R, to EAJ and SAL) supported all remaining components of the study. We are grateful to T.S. Nguyen and the Diabetes Clinical Research team for expert management of clinical samples and associated data; A. Hocking, K. Cerosaletti, B. Khor, J.L. Blanchfield, R. LaFond, and J. Calise for review, feedback, and editing of the manuscript; and Fluidigm for providing metal-conjugated avidin.
Version 1. 12/09/2019
Electronic publication
Version 2. 01/02/2020
Print issue publication
Footnotes
Conflict of interest: CJG has consulted for Bristol-Myers Squibb.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(1):480–490.https://doi.org/10.1172/JCI126595.
Contributor Information
Alice E. Wiedeman, Email: AWiedeman@benaroyaresearch.org.
Virginia S. Muir, Email: VMuir@benaroyaresearch.org.
Mario G. Rosasco, Email: MRosasco@benaroyaresearch.org.
Hannah A. DeBerg, Email: HDeBerg@benaroyaresearch.org.
Bertrand Haas, Email: bertrand.haas@gmail.com.
Matthew J. Dufort, Email: Mdufort@benaroyaresearch.org.
Cate Speake, Email: CSpeake@benaroyaresearch.org.
Elisavet Serti, Email: eserti@immunetolerance.org.
Gabriele Blahnik, Email: grblahnik@gmail.com.
Anna M. Kus, Email: akus@benaroyaresearch.org.
Eddie A. James, Email: ejames@benaroyaresearch.org.
S. Alice Long, Email: along@benaroyaresearch.org.
References
- 1.Insel RA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care. 2015;38(10):1964–1974. doi: 10.2337/dc15-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziegler AG, Nepom GT. Prediction and pathogenesis in type 1 diabetes. Immunity. 2010;32(4):468–478. doi: 10.1016/j.immuni.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leighton E, Sainsbury CA, Jones GC. A practical review of C-peptide testing in diabetes. Diabetes Ther. 2017;8(3):475–487. doi: 10.1007/s13300-017-0265-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenbaum CJ, et al. Fall in C-peptide during first 2 years from diagnosis: evidence of at least two distinct phases from composite type 1 diabetes TrialNet data. Diabetes. 2012;61(8):2066–2073. doi: 10.2337/db11-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis AK, et al. Prevalence of detectable C-peptide according to age at diagnosis and duration of type 1 diabetes. Diabetes Care. 2015;38(3):476–481. doi: 10.2337/dc14-1952. [DOI] [PubMed] [Google Scholar]
- 6.Ehlers MR. Immune interventions to preserve β cell function in type 1 diabetes. J Investig Med. 2016;64(1):7–13. doi: 10.1097/JIM.0000000000000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serreze DV, Leiter EH, Christianson GJ, Greiner D, Roopenian DC. Major histocompatibility complex class I-deficient NOD-B2mnull mice are diabetes and insulitis resistant. Diabetes. 1994;43(3):505–509. doi: 10.2337/diab.43.3.505. [DOI] [PubMed] [Google Scholar]
- 8.Wicker LS, et al. Beta 2-microglobulin-deficient NOD mice do not develop insulitis or diabetes. Diabetes. 1994;43(3):500–504. doi: 10.2337/diab.43.3.500. [DOI] [PubMed] [Google Scholar]
- 9.Wong FS, Visintin I, Wen L, Flavell RA, Janeway CA. CD8 T cell clones from young nonobese diabetic (NOD) islets can transfer rapid onset of diabetes in NOD mice in the absence of CD4 cells. J Exp Med. 1996;183(1):67–76. doi: 10.1084/jem.183.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiLorenzo TP, et al. Major histocompatibility complex class I-restricted T cells are required for all but the end stages of diabetes development in nonobese diabetic mice and use a prevalent T cell receptor alpha chain gene rearrangement. Proc Natl Acad Sci U S A. 1998;95(21):12538–12543. doi: 10.1073/pnas.95.21.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinuzzi E, et al. The frequency and immunodominance of islet-specific CD8+ T-cell responses change after type 1 diabetes diagnosis and treatment. Diabetes. 2008;57(5):1312–1320. doi: 10.2337/db07-1594. [DOI] [PubMed] [Google Scholar]
- 12.Velthuis JH, et al. Simultaneous detection of circulating autoreactive CD8+ T-cells specific for different islet cell-associated epitopes using combinatorial MHC multimers. Diabetes. 2010;59(7):1721–1730. doi: 10.2337/db09-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James EA, et al. Combinatorial detection of autoreactive CD8+ T cells with HLA-A2 multimers: a multi-centre study by the Immunology of Diabetes Society T Cell Workshop. Diabetologia. 2018;61(3):658–670. doi: 10.1007/s00125-017-4508-8. [DOI] [PubMed] [Google Scholar]
- 14.Sarikonda G, et al. CD8 T-cell reactivity to islet antigens is unique to type 1 while CD4 T-cell reactivity exists in both type 1 and type 2 diabetes. J Autoimmun. 2014;50:77–82. doi: 10.1016/j.jaut.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Culina S, et al. Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci Immunol. 2018;3(20):eaao4013. doi: 10.1126/sciimmunol.aao4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol. 2009;155(2):173–181. doi: 10.1111/j.1365-2249.2008.03860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YJ, et al. Multiplexed in situ imaging mass cytometry analysis of the human endocrine pancreas and immune system in type 1 diabetes. Cell Metab. 2019;29(3):769–783.e4. doi: 10.1016/j.cmet.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damond N, et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 2019;29(3):755–768.e5. doi: 10.1016/j.cmet.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Duque S, et al. Conventional and neo-antigenic peptides presented by β cells are targeted by circulating naïve CD8+ T cells in type 1 diabetic and healthy donors. Cell Metab. 2018;28(6):946–960.e6. doi: 10.1016/j.cmet.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Long SA, et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci Immunol. 2016;1(5):eaai7793. doi: 10.1126/sciimmunol.aai7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tooley JE, et al. Changes in T-cell subsets identify responders to FcR-nonbinding anti-CD3 mAb (teplizumab) in patients with type 1 diabetes. Eur J Immunol. 2016;46(1):230–241. doi: 10.1002/eji.201545708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herold KC, et al. Teplizumab treatment may improve C-peptide responses in participants with type 1 diabetes after the new-onset period: a randomised controlled trial. Diabetologia. 2013;56(2):391–400. doi: 10.1007/s00125-012-2753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herold KC, et al. An anti-CD3 antibody, teplizumab, in relatives at risk for type 1 diabetes. N Engl J Med. 2019;381(7):603–613. doi: 10.1056/NEJMoa1902226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cernea S, Herold KC. Monitoring of antigen-specific CD8 T cells in patients with type 1 diabetes treated with antiCD3 monoclonal antibodies. Clin Immunol. 2010;134(2):121–129. doi: 10.1016/j.clim.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alhadj Ali M, et al. Metabolic and immune effects of immunotherapy with proinsulin peptide in human new-onset type 1 diabetes. Sci Transl Med. 2017;9(402):eaaf7779. doi: 10.1126/scitranslmed.aaf7779. [DOI] [PubMed] [Google Scholar]
- 26.Newell EW, Sigal N, Nair N, Kidd BA, Greenberg HB, Davis MM. Combinatorial tetramer staining and mass cytometry analysis facilitate T-cell epitope mapping and characterization. Nat Biotechnol. 2013;31(7):623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vignali D, et al. Detection and characterization of CD8+ autoreactive memory stem T cells in patients with type 1 diabetes. Diabetes. 2018;67(5):936–945. doi: 10.2337/db17-1390. [DOI] [PubMed] [Google Scholar]
- 28.Levine JH, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(1):184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garyu JW, et al. Characterization of diabetogenic CD8+ T cells: immune therapy with metabolic blockade. J Biol Chem. 2016;291(21):11230–11240. doi: 10.1074/jbc.M115.713362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laban S, et al. Heterogeneity of circulating CD8 T-cells specific to islet, neo-antigen and virus in patients with type 1 diabetes mellitus. PLoS ONE. 2018;13(8):e0200818. doi: 10.1371/journal.pone.0200818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogura H, et al. Identification and analysis of islet antigen-specific CD8+ T cells with T cell libraries. J Immunol. 2018;201(6):1662–1670. doi: 10.4049/jimmunol.1800267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song Y, et al. T-cell immunoglobulin and ITIM domain contributes to CD8+ T-cell immunosenescence. Aging Cell. 2018;17(2):e12716. doi: 10.1111/acel.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faustman DL, Davis M. The primacy of CD8 T lymphocytes in type 1 diabetes and implications for therapies. J Mol Med. 2009;87(12):1173–1178. doi: 10.1007/s00109-009-0516-6. [DOI] [PubMed] [Google Scholar]
- 35.Paul M, et al. Pathophysiological characteristics of preproinsulin-specific CD8+ T cells in subjects with juvenile-onset and adult-onset type 1 diabetes: A 1-year follow-up study. Pediatr Diabetes. 2018;19(1):68–79. doi: 10.1111/pedi.12536. [DOI] [PubMed] [Google Scholar]
- 36.Klebanoff CA, Gattinoni L, Restifo NP. Sorting through subsets: which T-cell populations mediate highly effective adoptive immunotherapy? J Immunother. 2012;35(9):651–660. doi: 10.1097/CJI.0b013e31827806e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henning AN, Roychoudhuri R, Restifo NP. Epigenetic control of CD8+ T cell differentiation. Nat Rev Immunol. 2018;18(5):340–356. doi: 10.1038/nri.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wrammert J, Miller J, Akondy R, Ahmed R. Human immune memory to yellow fever and smallpox vaccination. J Clin Immunol. 2009;29(2):151–157. doi: 10.1007/s10875-008-9267-3. [DOI] [PubMed] [Google Scholar]
- 39.Christophersen A, et al. Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat Med. 2019;25(5):734–737. doi: 10.1038/s41591-019-0403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 41.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 42.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 43.Skowera A, et al. β-cell-specific CD8 T cell phenotype in type 1 diabetes reflects chronic autoantigen exposure. Diabetes. 2015;64(3):916–925. doi: 10.2337/db14-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sachdeva N, et al. Preproinsulin specific CD8+ T cells in subjects with latent autoimmune diabetes show lower frequency and different pathophysiological characteristics than those with type 1 diabetes. Clin Immunol. 2015;157(1):78–90. doi: 10.1016/j.clim.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 45.Luce S, et al. Single insulin-specific CD8+ T cells show characteristic gene expression profiles in human type 1 diabetes. Diabetes. 2011;60(12):3289–3299. doi: 10.2337/db11-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonelli A, Ferrari SM, Corrado A, Ferrannini E, Fallahi P. CXCR3, CXCL10 and type 1 diabetes. Cytokine Growth Factor Rev. 2014;25(1):57–65. doi: 10.1016/j.cytogfr.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 47.Christoffersson G, Chodaczek G, Ratliff SS, Coppieters K, von Herrath MG. Suppression of diabetes by accumulation of non-islet-specific CD8+ effector T cells in pancreatic islets. Sci Immunol. 2018;3(21):eaam6533. doi: 10.1126/sciimmunol.aam6533. [DOI] [PubMed] [Google Scholar]
- 48.Blanco P, Pitard V, Viallard JF, Taupin JL, Pellegrin JL, Moreau JF. Increase in activated CD8+ T lymphocytes expressing perforin and granzyme B correlates with disease activity in patients with systemic lupus erythematosus. Arthritis Rheum. 2005;52(1):201–211. doi: 10.1002/art.20745. [DOI] [PubMed] [Google Scholar]
- 49.Malmeström C, et al. Relapses in multiple sclerosis are associated with increased CD8+ T-cell mediated cytotoxicity in CSF. J Neuroimmunol. 2008;196(1-2):159–165. doi: 10.1016/j.jneuroim.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Long SA, et al. Remodeling T cell compartments during anti-CD3 immunotherapy of type 1 diabetes. Cell Immunol. 2017;319:3–9. doi: 10.1016/j.cellimm.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKinney EF, et al. A CD8+ T cell transcription signature predicts prognosis in autoimmune disease. Nat Med. 2010;16(5):586–591, 1p following 591. doi: 10.1038/nm.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG. T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature. 2015;523(7562):612–616. doi: 10.1038/nature14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeo L, et al. Autoreactive T effector memory differentiation mirrors β cell function in type 1 diabetes. J Clin Invest. 2018;128(8):3460–3474. doi: 10.1172/JCI120555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akimova T, Beier UH, Wang L, Levine MH, Hancock WW. Helios expression is a marker of T cell activation and proliferation. PLoS ONE. 2011;6(8):e24226. doi: 10.1371/journal.pone.0024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Digre A, et al. Overexpression of heparanase enhances T lymphocyte activities and intensifies the inflammatory response in a model of murine rheumatoid arthritis. Sci Rep. 2017;7:46229. doi: 10.1038/srep46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breton G, et al. Programmed death-1 is a marker for abnormal distribution of naive/memory T cell subsets in HIV-1 infection. J Immunol. 2013;191(5):2194–2204. doi: 10.4049/jimmunol.1200646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linsley PS, Long SA. Enforcing the checkpoints: harnessing T-cell exhaustion for therapy of T1D. Curr Opin Endocrinol Diabetes Obes. 2019;26(4):213–218. doi: 10.1097/MED.0000000000000488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McKinney EF, Smith KG. T cell exhaustion and immune-mediated disease-the potential for therapeutic exhaustion. Curr Opin Immunol. 2016;43:74–80. doi: 10.1016/j.coi.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 59.Buggert M, et al. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014;10(7):e1004251. doi: 10.1371/journal.ppat.1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seo H, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc Natl Acad Sci U S A. 2019;116(25):12410–12415. doi: 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Onofrio LI, et al. Inhibitory receptor expression on T cells as a marker of disease activity and target to regulate effector cellular responses in rheumatoid arthritis. Arthritis & rheumatology (Hoboken, NJ) 2018;70(9):1429–1439. doi: 10.1002/art.40521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McGinty JW, Marré ML, Bajzik V, Piganelli JD, James EA. T cell epitopes and post-translationally modified epitopes in type 1 diabetes. Curr Diab Rep. 2015;15(11):90. doi: 10.1007/s11892-015-0657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hao W, Gitelman S, DiMeglio LA, Boulware D, Greenbaum CJ, Type 1 Diabetes TrialNet Study Group Fall in C-peptide during first 4 years from diagnosis of type 1 diabetes: variable relation to age, HbA1c, and insulin dose. Diabetes Care. 2016;39(10):1664–1670. doi: 10.2337/dc16-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nordwall M, Ludvigsson J. Clinical manifestations and beta cell function in Swedish diabetic children have remained unchanged during the last 25 years. Diabetes Metab Res Rev. 2008;24(6):472–479. doi: 10.1002/dmrr.871. [DOI] [PubMed] [Google Scholar]
- 65.Hahne F, et al. flowCore: a Bioconductor package for high throughput flow cytometry. BMC Bioinformatics. 2009;10:106. doi: 10.1186/1471-2105-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Maaten LJP, Hinton GE. Visualizing high-dimensional data using t-SNE. Journal of Machine Learning Research. 2008;9(Nov):2579–2605. [Google Scholar]
- 67.van der Maaten L. Accelerating t-SNE using tree-based algorithms. Journal of Machine Learning Research. 2014;15(Oct):3221–3245. [Google Scholar]
- 68. Krijthe JH. R wrapper for van der Maaten’s Barnes-Hut implementation of t-distributed stochastic neighbor embedding. https://github.com/jkrijthe/Rtsne Accessed November 7, 2019.
- 69.Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J. Cytofkit: a Bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput Biol. 2016;12(9):e1005112. doi: 10.1371/journal.pcbi.1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Becher B, et al. High-dimensional analysis of the murine myeloid cell system. Nat Immunol. 2014;15(12):1181–1189. doi: 10.1038/ni.3006. [DOI] [PubMed] [Google Scholar]
- 71.Wong MT, et al. Mapping the diversity of follicular helper T cells in human blood and tonsils using high-dimensional mass cytometry analysis. Cell Rep. 2015;11(11):1822–1833. doi: 10.1016/j.celrep.2015.05.022. [DOI] [PubMed] [Google Scholar]
- 72.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 73. Becker RA, Chambers JM, Wilks AR. The New S Language: a programming environment for data analysis and graphics. Pacific Grove, CA: Wadsworth & Brooks/Cole; 1988. [Google Scholar]
- 74. Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.