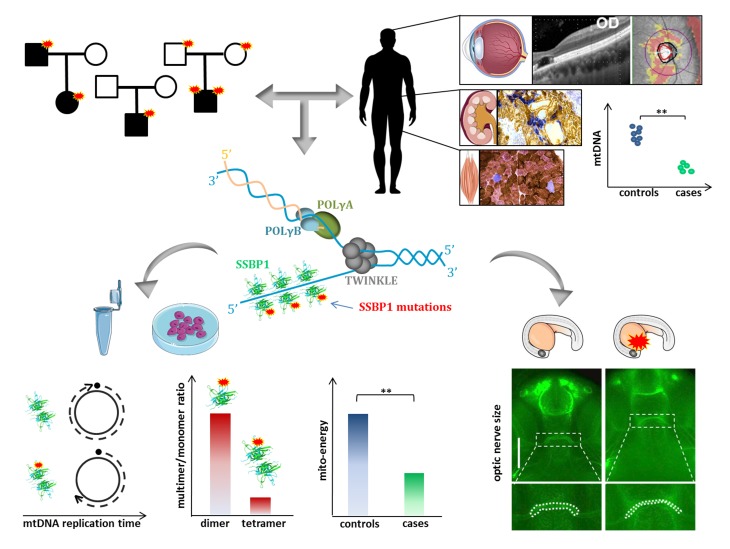
Abstract
Inherited optic neuropathies include complex phenotypes, mostly driven by mitochondrial dysfunction. We report an optic atrophy spectrum disorder, including retinal macular dystrophy and kidney insufficiency leading to transplantation, associated with mitochondrial DNA (mtDNA) depletion without accumulation of multiple deletions. By whole-exome sequencing, we identified mutations affecting the mitochondrial single-strand binding protein (SSBP1) in 4 families with dominant and 1 with recessive inheritance. We show that SSBP1 mutations in patient-derived fibroblasts variably affect the amount of SSBP1 protein and alter multimer formation, but not the binding to ssDNA. SSBP1 mutations impaired mtDNA, nucleoids, and 7S-DNA amounts as well as mtDNA replication, affecting replisome machinery. The variable mtDNA depletion in cells was reflected in severity of mitochondrial dysfunction, including respiratory efficiency, OXPHOS subunits, and complex amount and assembly. mtDNA depletion and cytochrome c oxidase–negative cells were found ex vivo in biopsies of affected tissues, such as kidney and skeletal muscle. Reduced efficiency of mtDNA replication was also reproduced in vitro, confirming the pathogenic mechanism. Furthermore, ssbp1 suppression in zebrafish induced signs of nephropathy and reduced optic nerve size, the latter phenotype complemented by WT mRNA but not by SSBP1 mutant transcripts. This previously unrecognized disease of mtDNA maintenance implicates SSBP1 mutations as a cause of human pathology.
Keywords: Genetics, Ophthalmology
Keywords: Bioenergetics, Genetic diseases, Mitochondria
Introduction
The expanding genetic landscape of inherited optic neuropathies has highlighted mitochondrial dysfunction as a major driver of this pathology (1, 2). Overall, the genetic defects leading to optic atrophy range from mitochondrial DNA (mtDNA) point mutations in Leber hereditary optic neuropathy (LHON) (3) to dominant and recessive mutations affecting a cluster of nuclear genes implicated in mitochondrial dynamics (4). These include OPA1, whose protein product is necessary for fusing the inner mitochondrial membrane (5, 6), MFN2, for fusion of the outer mitochondrial membrane (7), DNM1l (8), OPA3 (9), and SLC25A46 (10), involved in mitochondrial fission. In addition to optic neuropathy, mutations in several of these genes have also been hallmarked by broader clinical phenotypes defined as “plus,” associated with mtDNA instability, as characterized by secondary accumulation of multiple deletions in postmitotic tissues, such as skeletal muscle and brain (11–13). In patients, mtDNA multiple deletions are phenotypically reflected by ocular myopathy with chronic progressive external ophthalmoplegia (CPEO) and ptosis, in association or not with more widespread brain involvement, including parkinsonism and dementia (14, 15).
Originally, CPEO and ptosis with mtDNA multiple deletions were noted for their remarkable association of Mendelian inheritance and secondary mtDNA instability (16). The genes associated with this initial group of mitochondrial disorders were all implicated in mitochondrial replisome, such as the mitochondrial polymerase (POLG1 and POLG2), the helicase Twinkle (TWNK), other genes instrumental to mtDNA replication (RRM2B, RNaseH1, DNA2, MGME1), and genes implicated in nucleotide availability and balance (SLC25A4, TYMP, TK2, DGOUK, MPV17) (17).
We are now aware that allelic mutations in all these genes, respectively implicated in mitochondrial dynamics, replisome, and nucleotide metabolism, may either affect so profoundly mtDNA replication that the major outcome is depletion of mitogenomes with fatal infantile encephalomyopathies or induce slow somatic accumulation of mtDNA multiple deletions with various syndromes of adult life dominated by CPEO (18).
However, one key factor implicated in mtDNA replication has been missing: mitochondrial single-strand binding protein 1 (SSBP1), whose cDNA cloned in 1993 (19) and whose gene was mapped to 7q34 in 1995 (20). Despite being a good candidate for mtDNA maintenance disorders, it was not pathogenically associated with any human disease. SSBP1 has been shown to coat the displaced, parental H-strand during mtDNA synthesis, a critical function according to the strand displacement mode of mtDNA replication (21).
Here, we report the identification of a spectrum of phenotypes associated with SSBP1 mutations and mtDNA depletion transmitted as autosomal dominant and recessive traits that ranged from isolated optic atrophy to additional clinical features, including retinal macular dystrophy, sensorineural deafness, mitochondrial myopathy, and kidney failure necessitating transplantation.
Results
Exome sequencing identifies dominant and recessive mutations in SSBP1
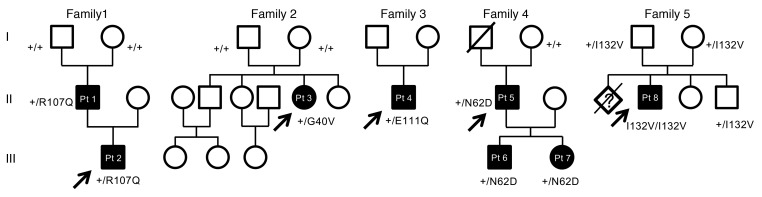
SSBP1 mutations and their segregation in 5 unrelated families.
Two unrelated families from Italy and the US with the common feature of congenital or early onset optic atrophy, negative for the most frequent causes, underwent trio whole exome sequencing (WES) independently in different centers (Supplemental Methods and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI128514DS1).
De novo mutations in SSBP1 were identified in both families, which we connected through GeneMatcher (22). In the Italian family (family 1 in Figure 1), we identified a heterozygous missense mutation NM 003143.2: c.320G>A (p.R107Q) (Supplemental Table 2), which arose de novo in the father and was transmitted to his affected child. The US proband (family 2 in Figure 1) carried a de novo heterozygous missense mutation, c.119G>T (p.G40V) (Supplemental Table 2).
Figure 1. Pedigrees of the 5 families carrying SSBP1 mutations.
Affected individuals (black circles/squares) present with variable combinations of optic atrophy with clinical phenotypes, including retinal dystrophy, kidney insufficiency, and mitochondrial myopathy, among others. All mutations segregate consistently with the disease phenotype.
Based on these findings, a total of 135 Italian probands with optic atrophy of unknown genetic origin were screened for SSBP1 mutations. In 2 unrelated individuals, we found additional heterozygous missense mutations in SSBP1: c.331G>C (p.E111Q) in family 3 and c.184A>G (p.N62D) in family 4 (Figure 1 and Supplemental Table 2). No members of family 3 were available for segregation. In family 4, the heterozygous mutation segregated in both proband’s offspring, whereas it was absent in his mother (the only parent available for testing). The father died in his 70s, without any report of visual impairment. The segregation was therefore compatible with a de novo event in the proband.
In a fifth family from Austria with a single proband (family 5 in Figure 1) presenting with a largely overlapping phenotype, WES identified a homozygous mutation in SSBP1, c.394A>G (p.I132V) (Supplemental Table 2). Parental consanguinity was not reported, and estimation of genomic inbreeding using WES data failed to reveal excess of homozygosity (Supplemental Table 3). Homozygosity for p.I132V can be explained as a founder mutation because the parents were from a remote mountain area in Austria.
Clinical presentation of affected individuals.
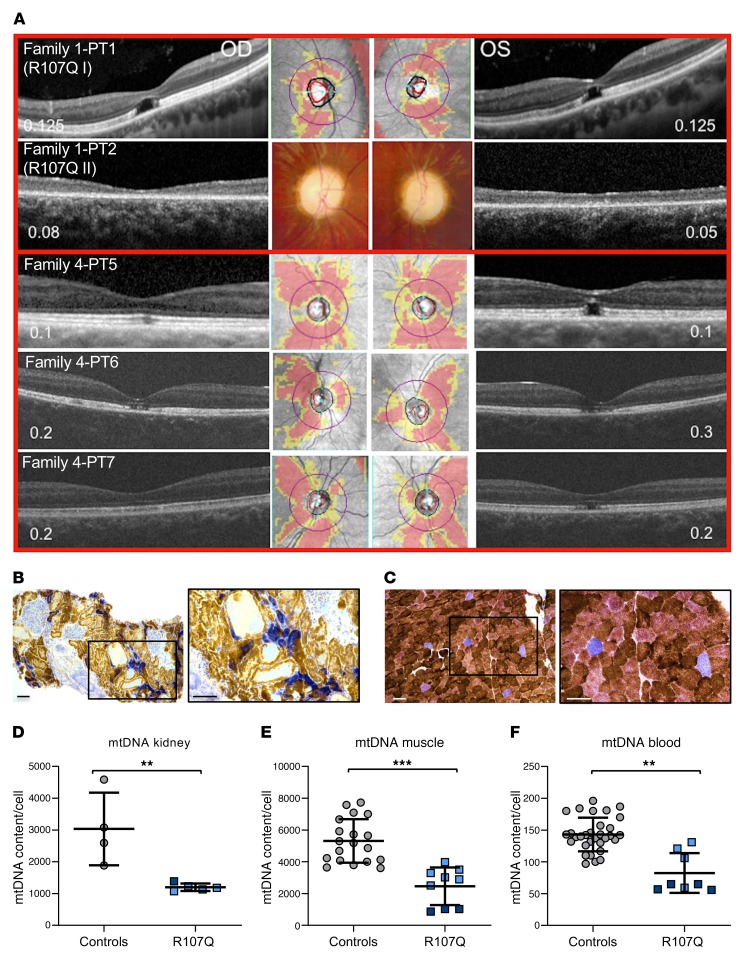
Family 1 included 2 probands, a father and son, who presented with childhood onset optic atrophy, retinal macular dystrophy, sensorineural deafness, and nephropathy, which in the child ultimately led to kidney transplantation (Figure 2, A–C, Supplemental Figure 1, and Supplemental Methods). Muscle and kidney biopsies from both patients revealed histoenzymatic features compatible with mitochondrial dysfunction, such as cytochrome c oxidase–negative (COX-negative) cells (Figure 2, B and C, and Supplemental Figure 1B). The mtDNA molecular analysis revealed partial depletion of copy number in both tissues (Figure 2, D and E). Blood-derived cells were also mtDNA depleted, similarly to kidney and muscle (Figure 2F). However, both long range and digital droplet PCR failed to identify and quantify mtDNA-deleted molecules in kidney, muscle, blood, and urinary sediment cells (Supplemental Figure 2, A–D). A slight reduction of 7S DNA, the third strand of the mtDNA displacement loop (D-loop) was also noted (Supplemental Figure 2, E–H). Thus, muscle and kidney histoenzymatic analysis, as well as mtDNA investigations, were suggestive of mitochondrial dysfunction as pathogenic mechanism.
Figure 2. OCT, muscle and kidney histopathology, and tissue mtDNA quantification of patients carrying SSBP1 mutations.
(A) Macular and optic nerve OCT and visual acuity (numbers within each panel expressed as decimals) of patients from families 1 and 4. Family1-PT1 patient shows a complete foveal cavitation characterized by the absence of inner segment/outer segment and outer segment/RPE junctions. Family1-PT2 patient shows diffuse atrophy of the photoreceptors’ layers. Family 4-PT5 patient shows incomplete (OD) and complete (OS) foveal cavitation. Family 4-PT6 patient shows incomplete foveal cavitation characterized by partial disruption of inner/outer segment and outer segment/RPE junctions. Family 4-PT7 patient shows mild rarefaction of the hyperreflectivity of the inner segment/outer segment junction (OD) and the incomplete form of foveal cavitation (OS). Optic disc atrophy is documented by the diffuse retinal nerve fiber layer thinning in the OCT deviation map of the optic nerve or in the color optic nerve photograph (middle, only for patient family 1-PT2). (B) Kidney and (C) muscle histochemistry of proband PT1 of family 1 (R107Q I). COX/SDH staining reveals COX-negative cells in the tubular components of kidney parenchyma and in sporadic muscle fibers. Boxes on merged images correspond to magnified insets. Scale bars: 100 μm. (D–F) mtDNA copy numbers from different tissues of healthy individuals (controls) and the PT1 and PT2 patients from family 1: proband (R107Q I, light blue) and his son (R107Q II, dark blue). (D) Data are shown as mean ± SD of 4 controls and patients (1 experiment for R107Q I and 3 experiments for R107Q II). (E) Data are shown as mean ± SD of 19 controls and patients (2 biopsies of R107Q I analyzed in 6 experiments and 3 experiments for R107Q II). (F) Data are shown as mean ± SD of 31 controls and R107Q patients (2 blood samples for patients, analyzed in 4 experiments). A reduction of mtDNA content was observed in patient tissues. **P < 0.01; ***P < 0.001, Student’s 2-tailed t test.
Family 2 included a single proband presenting a similar phenotype, with childhood-onset severe optic atrophy and progressive retinal degeneration exhibiting a cone-rod dystrophy (CORD) phenotype (Figure 3). Despite relatively good foveal preservation, visual acuity was severely reduced due to the severity of the optic atrophy. In association with these eye findings, this patient also exhibited progressive nephropathy requiring transplantation and sensorineural hearing loss. No functional or histological studies were available for this patient.
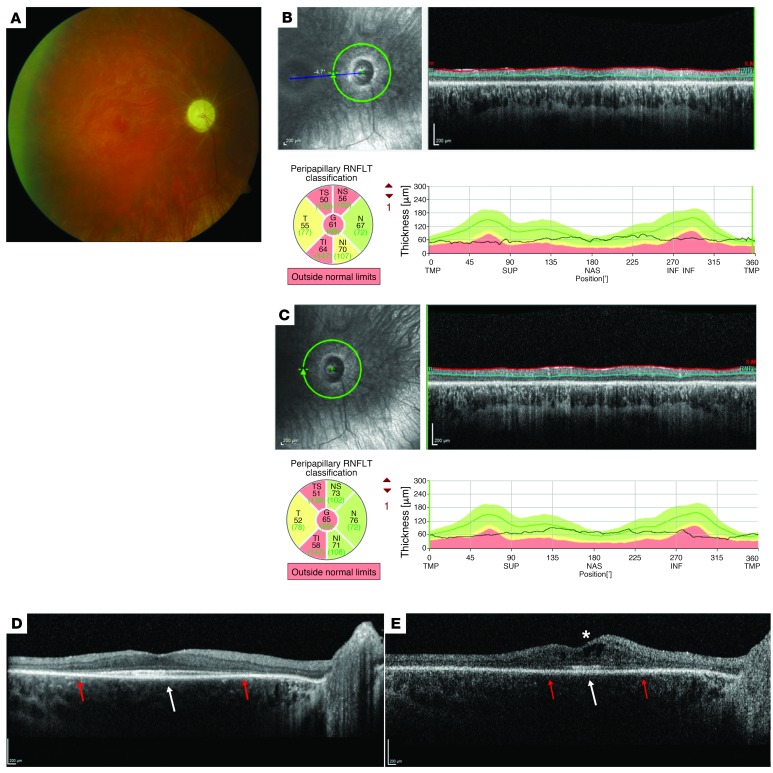
Figure 3. Ophthalmologic phenotype associated with the p.G40V SSBP1 mutation.
In all panels, the right eye (OD) is illustrated as representative of both eyes. Disease expression in this proband ascertained at Duke (PT3) was symmetric. (A) Ophthalmoscopy showed diffuse optic nerve pallor and blunted foveal reflexes, but enhanced vitreoretinal interface reflexes and marked vasculature attenuation with ghost vessel–like appearance. (B) RNFL OCT scan obtained at the age of 17 years, 10 months: the average RNFL thickness was only approximately 60–65 μm in each eye. (C) At age 23 years, there was no significant change in the extent of the RNFL loss. (D) The macular OCT obtained at the age of 17 years, 10 months, shows mild thinning of all retinal layers and marked loss of the EZ (red arrows), with foveal sparing and presence of subfoveal hyperreflectivity at the EZ/RPE interface (white arrow). (E) Followup macular OCT at age 23 years showed significant increase in the thinning of all retinal layers and further contraction of EZ residue (red arrows), but persistent subfoveal hyperreflectivity (white arrow). Hyporeflective cystic spaces consistent with macular edema had developed at this age as well (white asterisk) Scale bars 200 µm.
The sporadic patient of family 3 was in his 70s and presented with isolated optic atrophy and no retinal changes in the macula. The proband of family 4 had 2 affected offspring with neuroophthalmological assessment revealing features virtually identical to those of family 1 in all 3 individuals (Figure 2A). The mtDNA analysis in blood-derived cells of patients from families 3 and 4 revealed a copy number in the lower end of the control range, suggesting a tendency to reduction (Supplemental Figure 2J).
The proband of family 5, with homozygous p.I132V, initially developed blindness due to retinal dystrophy and deafness. This clinical picture was later complicated by hypertrophic cardiomyopathy, nephropathy, ataxia,and growth retardation. Muscle biopsy revealed COX-negative fibers; biochemical studies documented a combined deficiency of complexes I and III, whereas citrate synthase (CS) was elevated.
SSBP1 mutations frequency and in silico prediction of deleteriousness.
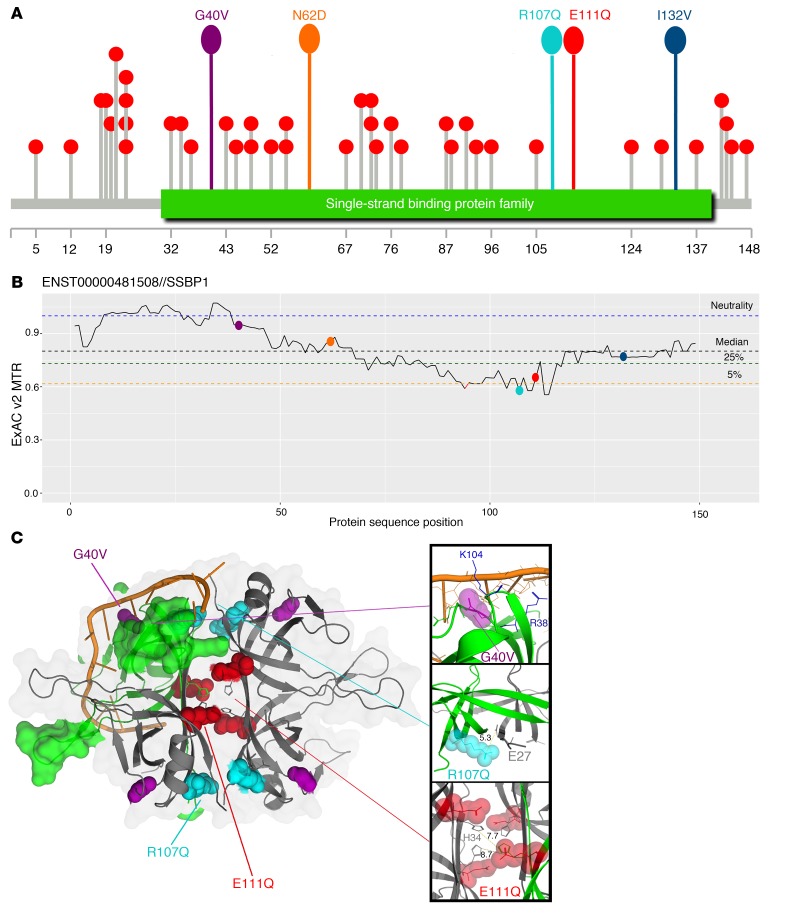
All dominant mutations were novel based on the variant database gnomAD, version 2.0.2. The recessive mutation was reported in only 2 heterozygous alleles in gnomAD and was absent in the homozygous state. All missense mutations were evolutionarily conserved (phyloP100way scores ranging 6.2 to 8.9), with high potential for deleteriousness according to Combined Annotation Dependent Depletion (CADD Phred scores ranging from 21.8 to 29.4) (Supplemental Table 2). All mutations mapped within the single-strand binding domain (Figure 4A), and 2 of them (p.R107Q and p.E111Q) affected residues under the strongest purifying selection relative to SSBP1 according to missense tolerance ratio (MTR), having MTRs within the fifth percentile of most missense depleted regions of the gene (Figure 4B). Mutations p.G40V, p.R107Q, and p.E111Q are predicted to disrupt molecular function according to in silico protein structure modeling by VIPUR (https://github.com/EvanBaugh/VIPUR) (Supplemental Methods, Supplemental Table 4, and ref. 23), although with seemingly distinct deleterious effects (Figure 4C). The p.G40V is predicted to have an unfavorable backbone conformation and appears to disrupt the interaction of SSBP1 with ssDNA by destabilizing the nearby nucleotide-binding residues. Conversely, p.E111Q and p.R107Q are predicted to affect SSBP1 oligomerization by disruption of stabilizing salt bridges of E111 and R107 with H34 and E27, respectively. Both p.N62D and p.I132V are not predicted to be grossly disruptive. However, p.N62D occurs at dimer interface and in close spatial proximity to R107 and introduces a negative charge that may interfere with dimerization. Finally, p.I132V is assumed to be tolerated mainly due to incomplete site conservation; however, it still has a high structural disruption score that suggests destabilizing potential. Notably, all 3 mutations with a disruptive prediction appear to act through distinct mechanisms: p.G40V damaging ssDNA binding, p.R111Q disrupting tetramer assembly, and p.R107Q disrupting both dimerization and tetramerization.
Figure 4. Distribution of SSBP1 mutations and protein in silico model.
(A) Lollipop (61) diagram of ultrarare population and patients’ variants along the protein: variants with ≤ 2 gnomAD alleles are represented by gray sticks with red circles on top, while patients’ variants sticks and circles are uniformly colored. The green box represents the SSB domain. (B) MTR diagram for SSBP1 and location of patients’ variants: MTR viewer, version 0.3 (62), was used with window size 31 on ENST00000481508 transcript. MTR is plotted against SSBP1 sequence, and locations of variants are represented with dots using the same color code as in A. Dotted lines represent neutrality (blue) or different percentiles — black (median), green (25th), yellow (5th) — of most missense depleted gene regions. (C) Structural model of the SSBP1 homotetramer (from PDB code 3ULL) with aligned ssDNA (from structural alignment to 3ULP): the 3 positions carrying the most deleterious predictions are highlighted on WT homotetramer with same color code as in A and B. Upper inset: Gly40 occurs close to the approximate ssDNA binding site, not directly contacting DNA, but forming a highly constrained loop coordinating DNA-contacting residues Arg38 and Lys104. Middle inset: Arg107 occurs on the outer surface of the homotetramer at both homodimeric and homotetrameric interfaces. It is spatially close to Glu27 (5.3°A away) and likely forms a stabilizing salt -bridge across the dimer interface. Lower inset: Glu111 occurs directly in the tetrameric interface and potentially forms a stabilizing salt bridge with His34, although the available model does not clearly indicate the monomer this interaction occurs with (both His34 residues on opposing dimers are spatially close to Glu111, 7.7°A and 8.7°A, respectively).
Analysis of SSBP1 in patient-derived mutant fibroblasts
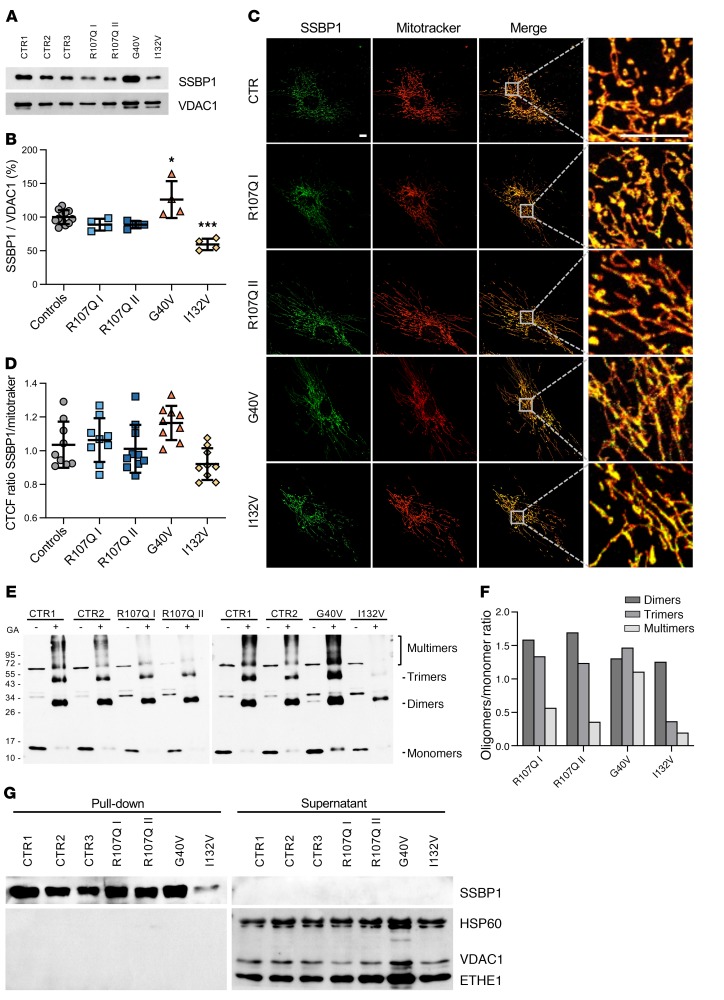
To assess the functional impact of SSBP1 mutations on protein levels, we performed Western blot analysis on mitochondria isolated from primary fibroblasts of 4 patients (both patients from family 1, probands from families 2 and 5) and from controls. Quantification of SSBP1 relative to the loading control VDAC1 indicated that abundance of p.R107Q mutant was comparable to that of controls, while p.G40V showed a significantly increased level of about 25% and p.I132V mutant a significant decrease of 39 % protein level instead (Figure 5, A and B). Immunofluorescence experiments evaluating colocalization of SSBP1 with MitoTracker red revealed a trend, congruent with Western blot analysis, toward an increase in the mutant p.G40V protein and decrease in the p.I132V (Figure 5, C and D).
Figure 5. Effect of SSBP1 mutations on protein stability, oligomerization, and ssDNA binding in fibroblasts.
(A) Western blot analysis of SSBP1 expression levels on isolated mitochondria; VDAC1 was used as a loading control. A representative blot out of 4 independent experiments is shown. (B) Densitometric analysis of 4 independent Western blot experiments shows an increase and a reduction of SSBP1 levels in G40V and I132V cells, respectively. All values (mean ± SD) are normalized to control cells. n = 12 (controls) and 4 (mutants). *P < 0.05; ***P < 0.001. Statistical significance was determined using 1-way ANOVA with Tukey’s correction.(C) Representative confocal images of fibroblasts labeled with anti-SSBP1 antibody (green) and MitoTracker red (red). Boxes on merged images correspond to magnified insets at right of each panel. Scale bar: 10 μm. (D) Quantification of SSBP1-MitoTracker colocalization, expressed as corrected total cell fluorescence (CTCF) ratio on 9 images per group. Data are represented as mean ± SD. (E) SSBP1 oligomerization analysis performed on the same samples used in A. GA: 0.1% GA. The presence of monomeric (molecular weight around 15 kDa), dimeric (molecular weight around 30 kDa), trimeric (molecular weight around 45 kDa), and multimeric (molecular weights > 60 kDa) forms are indicated. The protein amount utilized for the different samples was previously determined for Western blot analysis in Figure 3A. (F) Densitometric analysis of E shows that p.R107Q and p.I132V mutations, but not p.G40V, interfere with SSBP1 multimerization. All values represent the ratio between each oligomer amount in the presence of GA and monomers without GA. (G) SSBP1-ssDNA binding assay performed on isolated mitochondria shows that SSBP1 mutants were able to bind ssDNA. Streptavidin-agarose beads were used to precipitate biotinylated ssDNA together with associated proteins. Supernatants and pull-down fractions were run on a SDS-PAGE and immunoblotted with anti-SSBP1, anti-VDAC1, anti-HSP60, and anti-ETHE1 antibodies. A representative blot out of 3 is shown. *P < 0.05; ***P < 0.001. Statistical significance was determined using 1-way ANOVA with Tukey’s correction.
To monitor the effects of the mutations on homooligomerization of SSBP1, we performed a protein crosslinking experiment with isolated mitochondria treated with 0.1% glutaraldehyde (GA) to induce protein crosslinking or left untreated. Lysates were then separated on a denaturing SDS-polyacrylamide gel and the SSBP1 monomer, dimer, trimer, and multimer were detected by Western blot. In the absence of GA (–) the majority of SSBP1 was in the monomeric form (molecular weight of approximately 15 kDa), whereas in the presence of GA (+), some SSBP1 oligomers were crosslinked (Figure 5E). The relative levels of the oligomeric crosslinked products (GA+) and the monomeric form (GA–) were determined by densitometry and expressed as a ratio, where the ratio for controls was set equal to 1 (Figure 5F). The p.R107Q and p.G40V mutations induced the accumulation of dimeric and trimeric forms, while the detection of SSBP1 tetramers and multimers was severely reduced in the case of p.R107Q, but not affected by p.G40V. In the cell line with p.I132V mutation, we hardly detected any trimeric and multimeric products. These results suggest that p.R107Q and p.I132V mutations interfere with SSBP1 multimerization.
Next, we tested binding of WT and affected proteins to ssDNA. We performed an in vitro pull-down assay by incubating mitochondrial lysate with biotinylated ssDNA. We found that the SSBP1 antibody detected the protein only in the pull-down fraction both in controls and patients (Figure 5G). No protein was observed in the supernatant, indicating that WT and SSBP1 mutants were able to bind ssDNA. Moreover, anti-HSP60, anti-VDAC, and anti-ETHE1 antibodies were able to detect the corresponding proteins exclusively in the supernatant, but not in the pull-down fraction. These results indicated that only the SSBP1-ssDNA complex was precipitated specifically. Since we were wondering how the complex SSBP1(p.I132V mutation)-ssDNA could be precipitated despite only small amounts of tetramers being detected, we expressed the mutant protein in E. coli and tested its ability to form tetramers. Size exclusion chromatography of WT and p.I132V mutation demonstrated in both cases a stable tetramer (Supplemental Figure 3A). However, as observed by differential scanning fluorimetry, the p.I132V mutation has a somewhat lower thermostability than WT SSBP1, indicative of mild alterations to the physical properties of the mutation, though both proteins melt well above physiologically relevant temperatures (Supplemental Figure 3B). Together, these experiments suggest that the different mutations did not prevent binding of SSBP1 to ssDNA under these experimental conditions.
Analysis of mutant fibroblasts reveals depletion of mtDNA and nucleoids with altered dynamics of mitogenomes repopulation and impaired in vitro replication
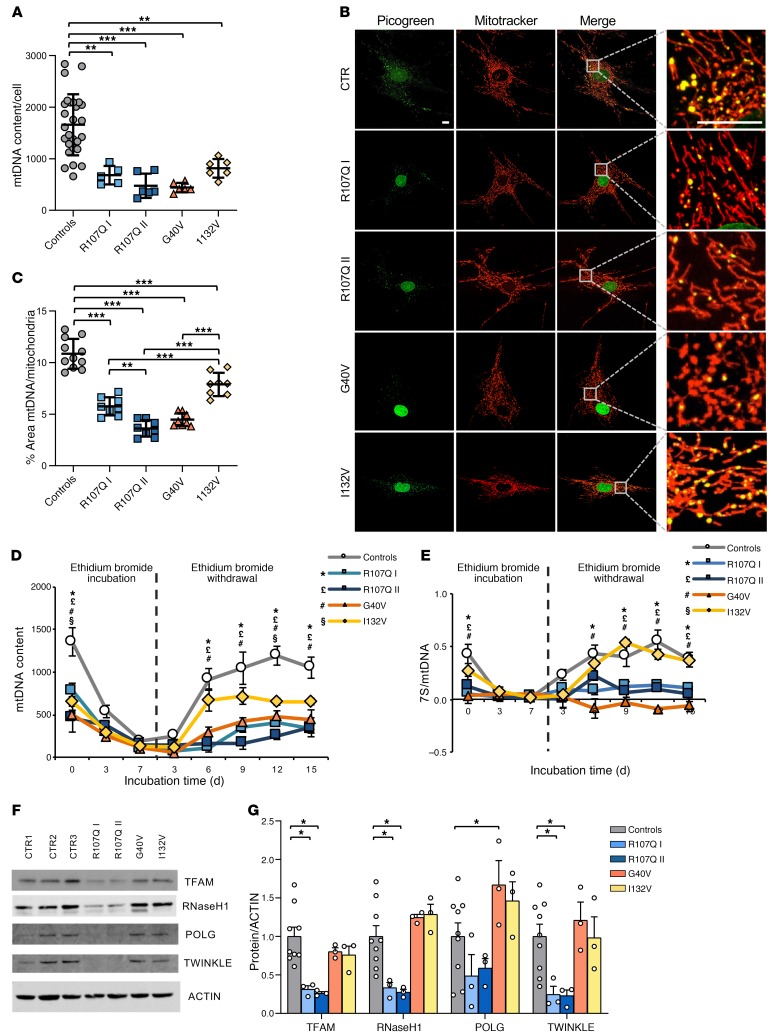
Based on the partial mtDNA depletion observed in patient-derived tissues (Figure 2, D–F), we investigated mtDNA maintenance in SSBP1 mutant fibroblasts by quantifying nucleoid and mtDNA copy numbers compared with those of controls. In all 4 patient cell lines, we found significantly reduced mtDNA content, ranging from 54% to 78% depletion compared with that of controls (Figure 6A). The p.I132V mutation appeared the less severe in terms of mtDNA depletion, whereas p.R107Q II and p.G40V were the most severe (Figure 6A). This result was matched by nucleoid quantification, as assessed by PicoGreen/MitoTracker red combined staining, showing a significant reduction of nucleoids, which were particularly prominent in p.R107Q II and p.G40V cells (Figure 6, B and C).
Figure 6. Effect of SSBP1 mutation on mtDNA amount, nucleoids, and the dynamics of genome repopulation in fibroblasts.
(A) mtDNA copy number quantification reveals a depletion in all mutant cells. Data are shown as mean ± SD of controls (n = 21) and mutants (n = 6) except for R107Q I (n = 5). **P < 0.01; ***P < 0.001. (B) Representative live confocal images of fibroblasts labeled with PicoGreen (green) and MitoTracker red (red). Boxes on merged images correspond to magnified insets at right of each panel. Scale bars: 10 μm. (C) Quantification of nucleoids, expressed as the ratios between the percentage of the area occupied by nucleoids (PicoGreen, nuclei excluded) and the area occupied by mitochondria (MitoTracker), shows a significant reduction of nucleoid numbers in all mutant cell lines. Data are shown as mean ± SD of controls (n = 11) and mutants (n = 9). **P < 0.01; ***P < 0.001. (D) mtDNA repopulation after depletion by EtBr in fibroblasts. mtDNA content is shown as mean ± SEM of controls (n = 7) and mutant cells (n = 3). A severe effect is observed for p.R107Q and p.G40V cells. *P < 0.05, R107Q I cells versus controls; £P < 0.05, R107Q II cells versus controls; #P < 0.05, G40V cells versus controls; §P < 0.05, I132V cells versus controls. (E) Quantification of 7S in the same samples analyzed in D reveals a marked reduction in p.R107Q and G40V cells. Data are shown as mean ± SEM. *P < 0.05, R107Q I cells versus controls; £P < 0.05, R107Q II cells versus controls; #P < 0.05, G40V cells versus controls; §P < 0.05, I132V cells versus controls. (F) Western blot of TFAM, RNaseH1, POLγ,and TWINKLE expression levels; ACTIN was used as a loading control. One representative blot out of 3 is shown. (G) Densitometric analysis of data shown in F shows a reduction of some of the replisoma proteins in p.R107Q cells. All values (means ± SEM) are normalized to the control cells. *P < 0.05. Statistical significance was determined using 1-way ANOVA (A, D, and E), 2-way ANOVA (G) with Dunnett’s correction, or 1-way ANOVA with Tukey’s correction (C).
To assess the global efficiency of mtDNA replication, we next performed a depletion/repopulation experiment, in which cells were mtDNA depleted by 7 days of exposure to low concentrations of ethidium bromide (EtBr), followed by its withdrawal and mtDNA repopulation in 15 days. Each mutant cell line started from a lower mtDNA content at point 0, reached a profound depletion similar to that of controls at day 7, and resumed mtDNA replication with different efficiencies (Figure 6D). The most severe effects were observed for p.R107Q and p.G40V mutations, whereas the homozygous p.I132V mutation was associated with milder outcomes. Considering the mtDNA amount at point 0 as 100 %, it is notable that only p.R107Q was significantly slower in mitogenomes repopulation (Supplemental Figure 4A). At the last time point of this experiment, all cell lines regained approximately the original levels of mtDNA copy number, with the exception of R107Q I.
The same samples were also quantified for 7S DNA (Figure 6E). Control fibroblasts at time 0 had ratio of 7S DNA/mtDNA of approximately 0.43. This ratio matched perfectly the changes of mtDNA copy number during the EtBr experiment in controls, with a similar pattern for the mild p.I132V mutation (ratio 0.28 at time 0). In contrast, p.R107Q and p.G40V mutations showed a low amount of 7S DNA (0.09 and 0.03 at time 0, respectively), which remained mostly unchanged during the experiment. No deletions were observed at times 0 and 15 (data not shown).
To assess the possible presence of low levels of mtDNA heteroplasmic mutations, which might expand after the depletion/repopulation experiment, mtDNA deep sequencing (mean coverage 7412X) was carried out at point 0 and at 15 days after EtBr withdrawal. Overall, mutant cells at time 0 had a significantly higher number of heteroplasmic variants, considering heteroplasmy within 20% of total copy number, but the load of heteroplasmy did not differ compared with that of controls and was not substantially changed by the bottleneck due to EtBr treatment at day 15 (Supplemental Figure 4, B–E). The mtDNA complete sequence of all cell lines and blood circulating cells for the remaining cases allowed for reconstruction of the haplotype of each individual enrolled in this study and their phylogenetic relationship (Supplemental Data File). The only notable variant was observed in the case R107Q II (Pt 2 in family 1), who presented a C insertion at position 57 (57insC) within the OH region that we believe to be novel.
Next, we quantified the levels of TFAM and other components of the mtDNA replisome (Figure 6, F and G). Western blot analysis revealed a significant reduction of TFAM, RNaseH1, and TWINKLE in both p.R107Q mutant cells, with only a modest decrease of POLγ. In contrast, we did not observe any protein reduction in the other 2 mutant cell lines, with a significant increase of POLγ in p.G40V mutant. Overall, these results suggest a severe effect of p.R107Q and p.G40V mutations on mtDNA maintenance, replication, and 7S DNA amounts in fibroblasts. The p.I132V mutation was associated with only a baseline mtDNA reduction, confirming its milder nature.
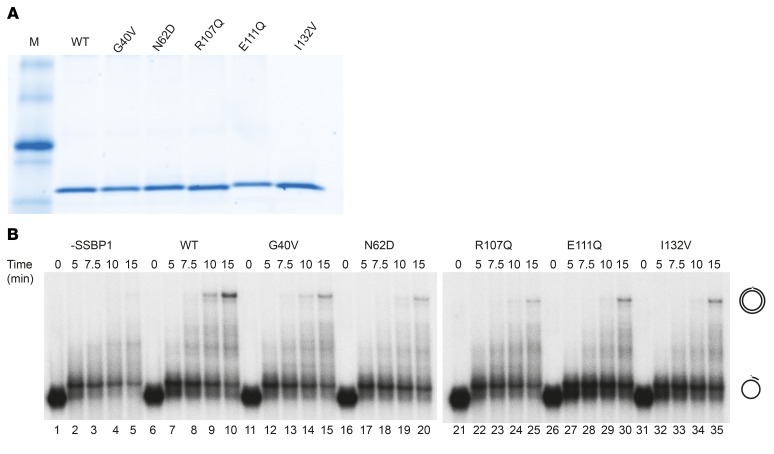
SSBP1 is known to stimulate the activity of POLγ (24); thus, we decided to investigate in vitro whether the identified mutations affected this ability. To this end, we purified WT SSBP1 and mutant derivatives thereof in recombinant form and analyzed them using SDS-PAGE to confirm purity (Figure 7A). We next monitored the ability of SSBP1 to stimulate DNA synthesis on a circular, ssDNA template of about 3000 nt in the presence of POLγ. The primer needed to initiate DNA synthesis was a 32P-labeled oligonucleotide (50 nt) that had been annealed to ssDNA substrate. We used saturating amounts of SSBP1 that could cover the single-stranded template completely (1 SSBP1 tetramer was calculated to bind 60 nt ssDNA). All mutations were able to support full-length DNA synthesis (Figure 7B, compare lanes 1–5, no SSBP1 added, with 11–35), but were all less efficient than the WT protein (Figure 7B, lanes 6–10). In conclusion, all SSBP1 mutations had reduced ability to stimulate POLγ-dependent DNA synthesis in vitro.
Figure 7. Effect of mutant SSBP1 on DNA replication in vitro.
(A) Ten pmoles of each purified SSBP1 version were separated on SDS-PAGE. M, marker. (B) DNA polymerase assay performed on a primed single-stranded circular DNA shows that all tested mutants displayed a lower stimulatory effect on DNA synthesis than the WT SSBP1.
mtDNA depletion reflects on bioenergetics of SSBP1 mutant fibroblasts
Considering the mtDNA depletion in all patient fibroblasts, we next characterized the impact of SSBP1 mutations on bioenergetics. Oxygen consumption rate (OCR) analysis showed a severe respiratory deficit in both p.R107Q mutant cells and a partial defect in p.G40V, whereas no differences were found for p.I132V mutation (Figure 8, A and B). Furthermore, all mutants, with the exception of p.I132V, showed a significant shift toward glycolysis, as indicated by decreased OCR/ECAR ratios (Supplemental Figure 4F).
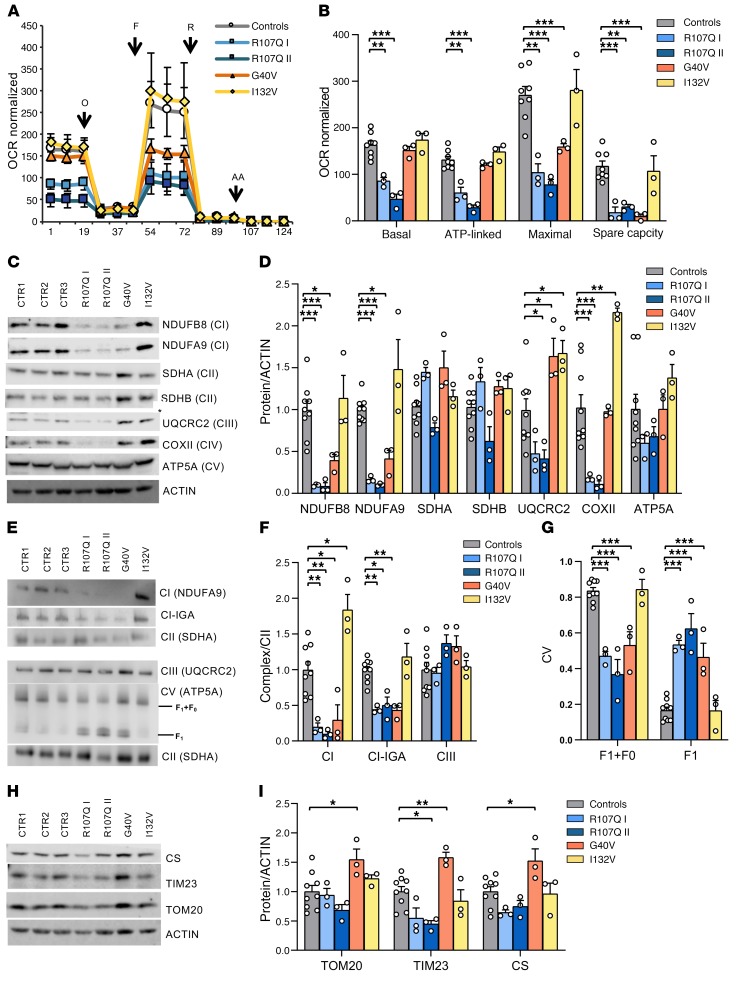
Figure 8. Energetic profile of SSBP1 mutated fibroblasts.
(A) OCR of fibroblasts, expressed as pmoles O2/min normalized for protein content, under basal conditions and after injection of oligomycin (O), carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP; F), rotenone (R) and antimycin A (AA). Data are shown as mean ± SEM of control (n = 7) and mutant cells (n = 3). (B) Basal, ATP-linked, maximal respiration, and spare respiratory capacity were calculated from OCR traces and are reported in the graph as mean ± SEM. OCR experiments show a severe reduction of respiratory capacity in p.R107Q and a partial defect in p.G40V mutants. (C) Western blot of OXPHOS subunit expression levels; ACTIN was used as a loading control. One representative experiment out of 3 is shown. (D) Densitometric analysis of data shown in C shows a variable reduction of OXPHOS subunits in p.R107Q and p.G40V cells. Data, normalized to the control cells, are shown as mean ± SEM of 3 independent experiments. (E) Analysis of complexes assembly was carried out in digitonin-treated mitoplasts resolved by CN and BN-PAGE, as described in Methods. SDHA (CII) was used as a loading control. One representative experiment out of 3 is shown. (F) Densitometric analysis of CI and CIII complexes. Data are shown as mean ± SEM of 3 independent experiments. (G) Densitometric analysis of CV complex, showing an increase of F1 subunit not assembled in R107Q and G40V fibroblasts. (H) Western blot of CS, TIM23, and TOM20; ACTIN was used as a loading control. One representative experiment out of 3 is shown. (I) Densitometric analysis of the mitochondrial mass proteins. Data, normalized to the control cells, are shown as mean ± SEM of 3 independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, 1-way (G) or 2-way ANOVA (B, D, F, and I) with Dunnett’s test.
We also performed Western blot analysis of representative subunits of OXPHOS complexes (Figure 8, C and D). A marked decrease in the amount of NDUFB8 and NDUFA9 (CI), and COX II (CIV) characterized p.R107Q cells, with a slight reduction of UQCRC2 (CIII), which was statistically significant only in R107Q-II. The p.G40V mutation was associated with a milder but significant reduction of CI subunits and an increase of UQCRC2. Concordant with OCR results, p.I132V cells did not show reduction of OXPHOS subunits, but rather an increase of UQCRC2 and COX II. To confirm these data, we quantified the amount of assembled complexes by Blue-Native PAGE (Figure 8E). All patient cell lines, with the exception of p.I132V, exhibited a significant reduction of CI, both by Western blot and in-gel activity (Figure 8F), and a partially disassembled complex V (CV), as shown by the appearance of 2 bands at lower molecular weight corresponding to free F1 (Figure 8G). No alteration of CIII amounts was observed (Figure 8F). Finally, the analysis of supercomplexes assembly showed similar results, with reduction of supercomplexes containing complex I (I + III2, I + III2 + IV, and I + III2 + IVn) and increase of isolated CIII2, not assembled with CI, in fibroblasts carrying p.R107Q and p.G40V mutations. Again, the incomplete assembly of CV was detected in these 3 cell lines (Supplemental Figure 4G).
To assess whether mtDNA depletion was paralleled by a reduction of mitochondrial mass, we quantified CS, TIM23, and TOM20 levels (Figure 8, H and I). This analysis revealed a slight reduction and an increase of mitochondrial mass in R107Q-II and G40V mutations, respectively.
These results indicate that the SSBP mutations p.R107Q and p.G40V cause an energetic defect driven by CI reduction, a defect that was less severe in the latter mutation, probably due to the compensatory increment of mitochondrial mass. In contrast, fibroblasts carrying p.I132V, despite having mtDNA depletion, did not display a detectable energetic defect.
To evaluate whether the phenotypic defect due to the homozygous recessive mutation could be rescued by the WT protein, we integrated by lentiviral transduction the WT SSBP1 in a control and in the p.I132V cell lines. The SSBP1 complementation, confirmed by Western blot (Supplemental Figure 5, A and B), rescued the mtDNA copy number and induced a nonsignificant increase in respiration in p.I132V cells (Supplemental Figure 5, C–E).
Disruption of ssbp1 in zebrafish results in optic nerve atrophy and mitochondrial depletion
SSBP1 has an established role in mitochondrial biogenesis, and previous RNA in situ hybridization studies have documented ubiquitous expression in zebrafish from 0 to 2.5 days post fertilization (dpf) (25). To establish relevance of SSBP1 disruption to the optic atrophy phenotype of patients, we generated zebrafish models. We performed reciprocal BLAST searches and identified a single ortholog in the zebrafish genome (72 % identity; 87 % similarity for human [NP_003134.1] versus zebrafish [NP_001017806.1] protein). Next, we generated and characterized an efficient sgRNA-targeting ssbp1 exon 4 (Supplemental Figure 6A); we injected 50 pg sgRNA with 100 pg of Cas9 protein into 1-cell stage embryos, performed heteroduplex analysis, and estimated an average mosaicism of approximately 94 % in F0 mutant embryos (Supplemental Figure 6, B and C). At 2 dpf, F0 ssbp1 mutants displayed no overt morphological abnormalities compared with controls (Supplemental Figure 6D).
We and others have reported previously that the zebrafish is a tractable model for evaluating optic nerve ultrastructure (26, 27). To assess optic nerve integrity, we conducted immunostaining of whole-mount F0 mutant embryos fixed at 2 dpf and stained with anti-acetylated tubulin antibody (investigator masked to experimental condition). We acquired ventral images of whole-mount embryos using fluorescence microscopy and measured the area of the optic nerve chiasm at a consistent position framed bilaterally by the notochord (Figure 9A and Supplemental Figure 6E). We found that F0 mutants displayed an optic nerve phenotype indistinguishable from that of uninjected controls or batches injected with an equivalent dose of sgRNA alone (Supplemental Figure 6, E and F). Additionally, we performed quantitative reverse-transcriptase PCR (qRT-PCR) on RNA harvested from F0 ssbp1 embryos at 2 dpf (Supplemental Figure 6G), but consistent with our optic nerve phenotyping data, we did not find any significant differences between mutants and controls.
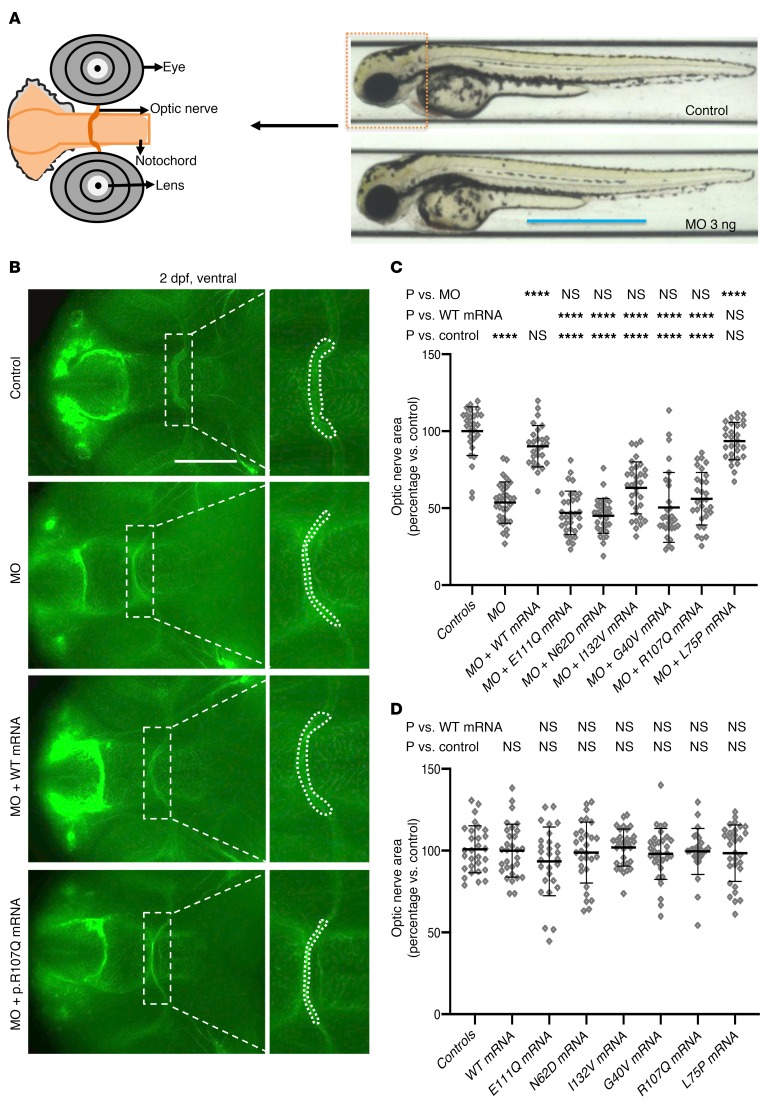
Figure 9. Optic nerve phenotypes and in vivo complementation studies in zebrafish ssbp1 models.
(A) Left, schematic representation of the ventral view of a 2 dpf embryo showing the optic chiasm framed by the notochord as demarcated by acetylated tubulin staining. Right, representative lateral view of 2 dpf larvae (control and ssbp1 morphant) with the anterior region outlined by a dashed box. Scale bar: 800 μm. (B) Representative ventral images of whole-mount 2 dpf embryos stained with anti-acetylated tubulin antibody to mark the optic nerve. Dashed boxes correspond to magnified insets at right of each panel. Region measured is indicated in inset with a white dashed line outlining the area of the optic chiasm. Scale bar: 50 μm. Original magnification, ×2 (insets). (C) Quantification of the optic chiasm area in embryo batches (as indicated in A). WT mRNA ameliorated MO phenotype significantly, while mRNA harboring patient mutations failed to rescue MO phenotypes. L75P is a negative control variant (rs78598246, 5 homozygotes in gnomAD; accessed January 2019). Three biological replicates. (D) Quantification of the optic chiasm area in embryo batches after overexpression of mRNA; optic nerve phenotypes were not affected. ****P < 0.0001, ANOVA with Tukey’s multiple comparisons test. n = 26–35 embryos/batch; 3 biological replicates gave similar results. Error bars represent SEM.
We hypothesized that the apparent lack of early optic nerve phenotype in F0 mutants could be due to the presence of maternal ssbp1, and we aged animals for evaluation at later time points. By 3 weeks after fertilization, we saw marked growth restriction and lethality in F0 larvae compared with controls. We speculated that the F0 mutant lethality could be due to the depletion of mitochondria over time. To explore this possibility, we injected 50 pg sgRNA plus 100 pg Cas9 protein into the Tg(XlEef1a1:mlsEGFP) transgenic zebrafish line, which targets GFP to the mitochondria with the COXVIII mitochondrial localization signal (mls). We aged F0 mutants to 15 dpf, obtained lateral fluorescent images, and quantified GFP intensity in the lens of live anesthetized embryos. Consistent with our hypothesis, we found a significant reduction of green signal in F0 mutants compared with controls or larvae injected with sgRNA alone (Supplemental Figure 6, H and I).
We next sought to evaluate the effects of sspb1 loss at an earlier developmental time point by using a translation blocking (tb) morpholino (MO) to suppress ssbp1 transcript (both maternal and embryonic mRNA; Supplemental Figure 7A). To determine the efficiency of ssbp1 tb MO, we performed immunoblotting on total protein extracted from pools of embryos at 2 dpf; we observed depletion of ssbp1 protein in morphants reduced to approximately 5% of protein levels in controls (Supplemental Figure 7, B and C). Next, we tested a dose curve by injecting tb-MO in 1- to 4-cell stage embryos at 3 different doses (1 ng, 2 ng, and 3 ng). Immunostaining and quantification of optic nerve chiasm area at 2 dpf showed a dose-dependent and significant reduction in optic nerve size for each dose tested in comparison with controls (Supplemental Figure 7, D and E). To determine the specificity of the tb-MO phenotype, we coinjected WT human SSBP1 mRNA with MO and observed a significant amelioration of the optic nerve phenotype to a level nearly indistinguishable from that of controls (Figure 9, A–C, and Supplemental Figure 8, A and B). We also matured larvae to 4 dpf, a developmental time point by which the zebrafish pronephros is formed (28, 29). We noted a dose-dependent cardiac, yolk sac, and periorbital edema suggestive of nephropathy (ref. 30 and Supplemental Figure 7, F and G). However, other phenotypes, such as abnormal otolith formation (a proxy for the mammalian ear), relevant to our SSBP1 human cohort were indistinguishable in ssbp1 morphants versus controls (Supplemental Figure 9A). Additionally, we did not detect differences in mlsEGFP quantity in morphants on our mitochondrial transgenic reporter at 2 dpf, possibly due to detection thresholds at this early developmental stage (Supplemental Figure 9, A and B). Finally, we quantified mtDNA content in ssbp1 morphants and controls using a quantitative PCR (qPCR) assay to monitor mtDNA amount relative to nuclear DNA (nDNA) (using mt-nd1 and polg1 primers, respectively; 2 dpf embryo pools of 30 embryos each) and did not detect significant differences between either experimental group (Supplemental Figure 9C).
In vivo complementation is a sensitive and specific approach for testing pathogenicity of nonsynonymous mutations in the context of optic nerve phenotypes (26). To test the effect of the missense mutations identified in affected individuals (including those studied in fibroblasts as well as the other 2 mutations found in families 3 and 4), we coinjected MO with SSBP1 mutation-bearing mRNAs and compared their rescue efficiency with that of WT SSBP1 message or MO alone. For each of the p.G40V, p.N62D, p.R107Q, p.E111Q, and p.I132V encoding mRNAs, we detected no significant differences between MO alone and mutant mRNA rescue experiments, suggesting that they are functional null mutations (Figure 9, A–C, and Supplemental Figure 8, A and B). Consistent with the in vitro studies, we observed a milder effect for p.I132V, the sole mutation that segregated in a recessive inheritance pattern. Although p.I132V mRNA rescued optic nerve chiasm area less efficiently than WT mRNA, the mean optic nerve area was improved by 18% in p.I132V rescue batches compared with MO alone (P = 0.0148, 2-tailed unpaired t test). However, this difference did not reach statistical significance when corrected for multiple testing (Figure 9, A–C, and Supplemental Figure 8, A and B). Further, SSBP1 complementation with mRNA harboring a common variant (p.L75P: 5 homozygotes in gnomAD; MAF 1.776e-3) rescued similarly to WT mRNA, supporting the specificity of our assay (Figure 9, A–C, and Supplemental Figure 8, A and B). Injection of mRNA encoding any of the 5 patients’ mutations, p.L75P, or WT SSBP1 yielded no apparent optic nerve phenotype compared with controls (Figure 9D and Supplemental Figure 8C). Moreover, titration of WT and p.R107Q mRNAs did not show any significant phenotype (Supplemental Figure 8D), suggesting that dominant-negative effect of these mutations is unlikely.
Together, our data indicate that MO-based zebrafish models of ssbp1 suppression recapitulate optic nerve atrophy observed in individuals with dominant and recessive SSBP1 mutations. Furthermore, our in vivo complementation data suggest that missense mutations in cases confer a loss of function, supporting a loss-of-function model of disease when cellular levels of mitochondria fall below a critical dosage threshold.
Discussion
We report SSBP1 mutations associated with an optic atrophy spectrum disorder, including retinal dystrophy, kidney insufficiency requiring transplantation, sensorineural deafness, and mitochondrial myopathy with mtDNA depletion. SSBP1 mutations impaired mtDNA maintenance and replication, as demonstrated in cells and in vitro. Reduced mtDNA copies were reflected in a variable phenotype of impaired OXPHOS, either in vitro in the study of fibroblasts or ex vivo in biopsies of affected tissues, such as kidney and skeletal muscle. In zebrafish, loss of ssbp1 was shown to affect optic nerve development and induce signs of nephropathy. All mutants failed to rescue the optic nerve phenotype, suggesting that dominant mutations induced loss of function, whereas the recessive behaved as a hypomorph.
From the genotype-phenotype standpoint, a few features deserve a comment. To date, almost all mtDNA-depletion disorders are fatal infantile syndromes (17), whereas we describe an adult disease dominated by optic atrophy with pure partial mtDNA depletion, without coexisting multiple deletions. A further example of adult phenotype with prevalent mtDNA depletion is the spectrum disorder associated with recessive mutations in MPV17, ranging from severe epathocerebral encephalopathy to adult neurohepathopathy, recurrent in Navajo Indians (31, 32). In contrast, mitochondrial neurogastrointestinal-encephalopathy (MNGIE), another adult disease with mtDNA depletion, also combines multiple deletions and CPEO/ptosis (33). Remarkably, none of our patients displayed CPEO or ptosis, which is usually the hallmark of mtDNA multiple deletion accumulation, such as in MNGIE and DOA “plus” phenotypes (11, 12, 14, 15, 17), or of sporadic mtDNA single deletion, such as in Kearns-Sayre syndrome (KSS) (34). Thus, these SSBP1 mutations selectively impinge on efficiency of mtDNA replication, apparently without affecting its fidelity. Interestingly, optic atrophy most probably is a congenital or childhood-onset reduction of axons that remains stable in adulthood, frequently combined with a prevailing cone retinal degenerative phenotype that worsens over time. Most patients exhibited a foveal cone photoreceptor ellipsoid zone (EZ) loss, visible as a foveal hyporeflective gap of EZ and retinal pigmented epithelium (RPE) layers (Figure 2A). This resembles the known cavitation lesions seen in congenital disorders, such as achromatopsia (35, 36) and blue cone monochromacy (37), as well as in degenerative entities, such as KCNV2-related retinopathy (38). A progressive cone-predominant disease expression configuring a CORD phenotype was seen in the family 2 (p.G40V) proband, lacking the foveal cavitation, but with a unique hyperreflectivity that also persisted as the retinal disease progressed. These neuroretinal features have not been previously reported in mtDNA depletion syndromes, although a CORD phenotype was found in KSS (34), whereas kidney involvement was observed since the first study on infantile syndromes with mtDNA depletion (39). Besides the genotype-phenotype variability with different SSBP1 mutations, a different severity was also observed within family 1 (p.R107Q). Dominant disorders frequently display incomplete penetrance, which may also reflect on phenotype expressivity. Nuclear background may obviously play a role, as well as mtDNA haplotype. Our complete mtDNA sequence only revealed an insertion affecting the OH region in the younger proband with severe nephropathy. This could affect mtDNA replication efficiency, potentially worsening the defect due to the SSBP1 mutation. Specific experiments should be designed to confirm this hypothesis.
We documented causal association of SSBP1 mutations with the disease, demonstrating, both in vivo and in vitro, that all patients’ missense SSBP1 mutations are pathogenic. First, suppression of ssbp1 transcript in zebrafish induced reduction of optic nerve chiasm size and depletion of mitochondria numbers, possibly also affecting kidney function. The optic nerve phenotype, the constant clinical feature in all patients, was fully rescued by WT or by the common polymorphism p.L75P mRNA, but not by mutant SSBP1 mRNA, confirming their deleteriousness. These experiments clearly support loss of function for all dominant mutations, whereas the recessive mutation p.I132V, improving the optic nerve chiasm size less effectively as compared with WT, revealed its possible hypomorphic nature, which results in a disease outcome only when in homozygous state.
Second, cellular and in vitro studies clearly documented that all SSBP1 mutations hamper mtDNA replication, as was also evident by mtDNA copy number assessment in patients’ tissues, including skeletal muscle, kidney, and blood. In fact, both the depletion/repopulation experiment after EtBr in mutant fibroblasts and the in vitro synthesis experiment confirmed that SSBP1 mutations cause reduced efficiency of mtDNA replication. Indeed, the in vitro assay showed that, even though the mutants could support full-length DNA synthesis, there was a delay as compared with the reactions with WT SSBP1, indicating either lower affinity to the DNA or to the POLγ holoenzyme. Regardless of the reason, the mutants seemed to have lost some of their stimulatory effect, possibly giving a molecular explanation of the disease phenotype, i.e., decreasing POLγ activity.
Notably, all patients’ mutations were missense changes, while we found no protein-truncating or splice-site alterations. This suggests that SSBP1 dosage reduction is not the molecular mechanism underlying disease. Consistently, we did not observe reduction in protein products by Western blot, except for the recessive p.I132V. In large public population databases such as gnomAD, a few ultrarare potential dosage-affecting SSBP1 alleles (Supplemental Table 5) suggest that monoallelic haploinsufficiency may be tolerated or be implicated in overlapping disease phenotypes as possibly hypomorphic alleles. An example of this is the start-loss c.3G>A variant proposed to act as modifier for penetrance of the m.1555A>G mtDNA deafness mutation, associated with mtDNA depletion and multiple deletions limited to skeletal muscle (40).
SSBP1 is a small protein, and much of its surface is involved in binding interactions with DNA, itself, and other replisome proteins. The pull-down experiments in fibroblasts failed to reveal a defect in ssDNA binding, also for the p.G40V, which was predicted to disrupt this interaction in silico. This apparent contradiction may be explained by the fact that the pull-down experiment is not quantitative and cannot measure the dynamic of SSBP1-ssDNA interaction. Conversely, the crosslinking experiment showed that p.R107Q hampers SSBP1 oligomerization, as predicted in silico for this mutation and its neighboring p.E111Q. Although p.N62D is predicted to be neutral, in vivo studies support it as a pathogenic mutation and its spatial proximity to p.R107Q suggests that the 2 mutations share the same mechanism. The recessive p.I132V was not predicted to be deleterious; however, its destabilizing potential, evidenced by lower thermostability, reduction of mutant SSBP1 oligomers, complementation studies in fibroblasts, and persistence of optic nerve atrophy in MO+p.I132V zebrafish, all argue in favor of its pathogenic potential.
Despite all mutant fibroblasts being characterized by a decreased mtDNA amount, only p.R107Q cells presented a reduced TFAM level and replisome proteins, according to its most severe effect on mtDNA replication. Considering that TFAM stabilizes mtDNA by packing a single mitogenome into nucleoids (41, 42), we would have rather expected that all mutants displayed a similar trend of reduced TFAM levels. Furthermore, we also observed a gradual worsening of mitochondrial energetic functions based on the different mutations: no energetic alteration on p.I132V, a partial respiratory defect driven by CI reduction in p.G40V, and a very severe energetic deficit in p.R107Q. We can speculate either that this latter mutation severely affects the stability of the replication proteins and consequently affects bioenergetic efficiency, or that p.G40V and p.I132V cells may have particularly efficient compensatory activation of mitochondrial biogenesis. Our results support this last hypothesis. Indeed, we found that p.G40V fibroblasts presented an increase of mitochondrial mass, a mechanism observed also in other mitochondrial diseases, such as LHON (43). Remarkably, a mechanism of increased transcription efficiency in association with mtDNA depletion was already observed in the livers of the Mpv17 KO mouse model (44). Thus, it is not surprising that these mutant cells had milder or no respiratory defect, also considering that fibroblasts are not the target tissue of the pathology and may display only a very mild energetic defect, as previously reported (45, 46). The SSBP1-associated disease, in fact, displayed a clear tissue specificity in patients with an inconstant clinical expression in kidney, for example. Tissue-specific sensitivity to little variations in mtDNA depletion and compensatory response may determine clinical expression. In the same line, the failure to detect a clear mtDNA depletion in 2 dpf zebrafish embryos might just be due to the early stage at which this experiment was done and, again, to the tissue-specific reduction of the mtDNA copy number.
SSBP1 is required for efficient initiation and elongation of mtDNA replication (47, 48). A majority of all mtDNA replication events are prematurely terminated at the end of the D-loop, forming 7S DNA. The relative levels of abortive mtDNA replication appear to be a regulated event. When more mtDNA is required, 7S DNA synthesis is reduced in favor of complete mtDNA replication (49, 50). The drop in 7S DNA levels associated with mutations in SSBP1 is therefore most likely a compensatory effect. Even if all 3 tested mutations (p.G40V, p.R107Q, and p.I132V) negatively regulate mitochondrial genome stability, only dominant mutations induce a severe reduction of 7S abundance. The milder reduction of 7S DNA associated with p.I132V may be compatible with its hypomorphic nature. Alterations of 7S DNA levels have also been observed in other mitochondrial disorders caused by impaired mtDNA replication (51–53).
In conclusion, we add SSBP1 to the list of genes implicated in mtDNA maintenance human diseases. Our current findings will open many different avenues of further investigation, ultimately contributing to a better understanding of mtDNA replication, enlarging the landscape of phenotypic expression of mitochondrial diseases in humans.
Methods
Cells and culture conditions.
Skin fibroblasts were derived from 7 healthy donors, 2 related patients with the p.R107Q mutation, 1 patient with the p.G40V mutation, and 1 patient with the p.I132V mutation. Fibroblasts were grown in DMEM medium supplemented with 10% FBS (FBS, Gibco, Life Technologies), 2 × 10-3 mol/L l–glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin in an incubator with a humidified atmosphere of 5% CO2 at 37°C. The mtDNA depletion/repopulation experiment was performed as described (43). Briefly, cells were grown in glucose medium supplemented 0.05 mg/ml uridine and 50 ng/ml EtBr to induce mtDNA depletion. After 7 days, EtBr was removed from the medium and cells were propagated until the 15th day.
mtDNA content and quantification of deletions and 7S DNA.
Quantification of mtDNA copy number relative to nDNA was performed for fibroblasts, and muscle, kidney, and blood tissues, as described (54).
Quantification of mtDNA deletion and 7S DNA was performed on fibroblasts and muscle, kidney, urine, and blood tissues. Briefly, the absolute quantification of mitochondrial genome deletions was based on duplex amplification in droplet digital PCR (ddPCR) with specific probes in MT-ND4 (major arc) and MT-ND1 (control region) adapted from published qPCR methods (55) and expressed as ratio ND4/ND1.
The quantification of 7S DNA was performed as previously described (51) and adapted to ddPCR (Bio-Rad). The 7S DNA was expressed as ratio –1 (mean ± SD) of mtDNA+7S DNA over mtDNA using the primer pairs (a + b1) and (a + b2), respectively, where b1 is inside the 7S region, amplifying both mtDNA and 7S DNA, and b2 is outside, amplifying only mtDNA.
Oligomerization assay.
SSBP1 oligomerization was carried out as described (56). Briefly, 10 μg of mitochondria isolated from controls and patients’ primary fibroblasts, as described (57), were treated or not with crosslinking agent (GA) at a final concentration of 0.1%. The reaction was quenched after 10 minutes with 100 × 10–3 mol/L PBS/glycine. Monomers or multimers were detected by SDS-PAGE with anti-SSBP1 antibody. Densitometric analysis was carried out using ImageJ software (NIH) and was expressed as crosslinked oligomers to monomers ratio.
Pull-down assay.
Pull-down assay was carried out as described (58, 59) with some modifications. Briefly, 10 μg of isolated mitochondria were solubilized with 1% dodecyl-maltoside (DDM) in binding buffer (20 mM HEPES pH 7.4, 50 × 10–3 mol/L NaCl, 10 × 10–3 mol/L MgCl2, 10–3 mol/L CaCl2, 8 × 10–3 mol/L DTT, 0.1 mg/ml BSA, 10% glycerol, 0.02% Tween 20, 1× protease inhibitor cocktail) for 30 minutes on ice; 20 μg of biotinylated ssDNA (biotin-5′-GGACTATTTATTCAATATATTTAAGAACTAATTCCAGCTGAGCGCCGG) (60) was added and incubated on a wheel shaker for 30 minutes at room temperature. To each reaction, 50 μL of streptavidin-agarose beads (Sigma-Aldrich) was added and incubated for 30 minutes at room temperature. Beads were pelleted at 600 g for 1 minute, and supernatants were collected and precipitated as described below, while pellets were washed 10 times with binding buffer and finally resuspended in 2× Laemmli buffer. Supernatants were precipitated adding 1 volume of 20% trichloroacetic acid (TCA), washed with cold acetone, air-dried, and resuspended in 1× Laemmli buffer. Pulled down and supernatant fractions were analyzed by immunoblotting using anti-SSBP1, anti-VDAC1, anti-HSP60, and anti-ETHE1 antibodies.
Expression and purification of recombinant proteins.
Recombinant baculoviruses encoding POLγB and POLγA were expressed in Sf9 insect cells. These recombinant proteins lacked the N-terminal mitochondrial targeting sequence and carried a carboxy-terminal 6′ His-tag. The proteins were purified over HIS-Select Nickel Affinity Gel (Sigma-Aldrich) and HiTrap Heparin HP (GE Healthcare), followed by HiTrap SP HP or HiTrap Q HP columns (GE Healthcare), depending on the net electrical charge of the protein. SSBP1 lacking the mitochondrial targeting sequence/MTS (aa 1–16) and containing an N-terminal 6′ His-tag was expressed in E. coli cells and purified over HIS-Select Nickel Affinity Gel (Sigma-Aldrich) and HiTrap Heparin HP (GE Healthcare), followed by HiTrap SP. Patient point mutations were introduced using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies) and verified by sequencing before being expressed and purified as for WT SSBP1.
Second-strand synthesis assay.
A 32P-labelled 50 nt long oligonucleotide (5′-GTGGCACTTTTCGGGGAAATGTGCGCGGAACCCCTATTTGTTTATTTTTC-3′) was annealed to single-stranded pBluescript SK II (–). DNA synthesis assays were performed using a 10 fmol template, 200 fmol of POLγ holoenzyme, and 500 fmol (as tetramer) of WT or mutant SSBP1 in 25 × 10–3 mol/L Tris-HCl (pH 7.8), 10–3 mol/L TCEP, 10 × 10-3 mol/L MgCl2, 0.1 mg/ml BSA, and 10 μM (each) dNTPs. Reactions were incubated at 37°C for the indicated times and stopped by the addition of 6 μl stop buffer (90 mM EDTA, 6% SDS, 30% glycerol, 0.25% bromophenol blue, and 0.25% xylene cyanol). Samples were separated on a 0.8% agarose gel and visualized by autoradiography.
Additional information.
WES and mtDNA sequencing methods appear in Supplemental Methods. Genome data were deposited at the European Genome-phenome Archive (EGA), which is hosted at the European Bioinformatics Institute (EBI) and the Centre for Genomic Regulation (CRG) (EGAS00001003850 and EGAS00001004003). Additionally, standard methods for fluorescence microscopy, OCR, Western blot, assessment of OXPHOS complexes and respiratory supercomplexes assembly, functional SSBP1 complementation, and all methods related to zebrafish experiments are detailed in the Supplemental Methods.
Study approval.
The local ethical committee at each center approved the study as follows: University of Bologna’s Comitato Etico di Area Vasta Emilia Centro della Regione Emilia-Romagna (CE-AVEC) 211/2018/SPER/AUSLBO for families 1, 3, and 4; Duke University’s Health System Institutional Review Board for Clinical Investigators (DUHS IRB, FWA #00009025) protocol #32301: Genomic Study of Medical, Developmental, or Congenital Problems of Unknown Etiology, for family 2; and the Ethical Committee of the University of Innsbruck (Ethikkommission der Medizinischen Universität Innsbruk AN2014-0090 335/4.7) for family 5. All patients and healthy donors or their parents or guardians gave informed consent.
Statistics.
GraphPad Prism for Windows (GraphPad Software) was used for statistical analyses. For patient tissues and fibroblast experiments, comparison of 2 groups used unpaired Student’s 2-tailed t test and comparisons among multiple groups used 1-way or 2-way ANOVA with Tukey’s or Dunnett’s multiple comparisons tests. For zebrafish experiments, unpaired Student’s 2-tailed t test and ANOVA with Tukey’s multiple comparisons test were used. For all analyses, differences were considered significant at P ≤ 0.05.
Author contributions
VDD, FU, IDM, PM, MG, AM, LC, FP, FT, BM, ZS, CP, MAG, WCC, IC, CZ, RK, MA, MP, KK, and NS carried out the experiments. CLM, PB, MC, MLV, RL, EB, RC, FE, FDC, and VC performed clinical investigation of families 1, 3, and 4. VS, JS, SN, MED, and AI performed clinical investigation of the proband from family 2. SB, SW, and WS performed clinical investigation of the proband from family 5. VDD, FU, IDM, PM, MG, AM, LC, CP, EED, and EHB analyzed and interpreted the data. VDD, FU, IDM, PM, EED, AI, and WCC contributed to study design and reviewed and revised the manuscript. VC, HP, NK, VT, TP, WCC, MS, and MF designed and supervised the study, acquired funding, and wrote the manuscript.
Supplementary Material
Acknowledgments
This study was supported by Ricerca Corrente funding (AM, LC, FT, FP, MC, and VC) and by grant GR-2016-02361449 to LC, both from the Italian Ministry of Health; by the German BMBF and Horizon2020 through E-Rare project GENOMIT (01GM1603 to HP and FWF-I 2741-B26) and the German Network for Mitochondrial Disorders (mitoNET 01GM1906B to HP); and by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (ES065078 to WCC and MAG). Financial support from the Mariani Foundation, Milan, and of Mitocon, Italy (grant no. 2018-01 to VT) are acknowledged. CP is supported by a fellowship from Associazione Luigi Comini Onlus, Italy. FU and KK were funded by an International Research Support Initiative Program fellowship from the Higher Education Commission of Pakistan. We thank T. Khan for technical assistance with zebrafish experiments. Family 2 was evaluated through the Duke Genome Sequencing Clinic, supported by the Duke University Health System, and partially funded by UCB Celltech. We thank Federico Sadun for referring patients from family 1 and Rosanna Carroccia for technical help with the kidney biopsy of PT1.
Version 1. 09/24/2019
In-Press Preview
Version 2. 11/18/2019
Electronic publication
Version 3. 01/02/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(1):108–125.https://doi.org/10.1172/JCI128514.
See the related Commentary at SSBP1 faux pas in mitonuclear tango causes optic neuropathy.
Contributor Information
Valentina Del Dotto, Email: vale.deldotto@gmail.com.
Farid Ullah, Email: faridullahhu@gmail.com.
Ivano Di Meo, Email: ivano.dimeo@istituto-besta.it.
Pamela Magini, Email: pamela.magini@unibo.it.
Mirjana Gusic, Email: mirjana.gusic@helmholtz-muenchen.de.
Alessandra Maresca, Email: alessandramaresca@gmail.com.
Leonardo Caporali, Email: leonardo.caporali@gmail.com.
Flavia Palombo, Email: flavia.palombo@unito.it.
Francesca Tagliavini, Email: tagliavini.fra@gmail.com.
Bertil Macao, Email: bertil.macao@medkem.gu.se.
Zsolt Szilagyi, Email: zsolt.szilagyi@medkem.gu.se.
Margaret A. Gustafson, Email: margaret.gustafson@nih.gov.
Kamal Khan, Email: kamal.khan@duke.edu.
Chiara La Morgia, Email: chiara.lamorgia@unibo.it.
Piero Barboni, Email: p.barboni@studiodazeglio.it.
Michele Carbonelli, Email: dmcarbonelli@gmail.com.
Maria Lucia Valentino, Email: marialucia.valentino@unibo.it.
Rocco Liguori, Email: rocco.liguori@unibo.it.
Vandana Shashi, Email: vandana.shashi@duke.edu.
Shashi Nagaraj, Email: shashi.nagaraj@duke.edu.
Mays El-Dairi, Email: mays.el-dairi@duke.edu.
Alessandro Iannaccone, Email: alessandro.iannaccone@duke.edu.
Ioana Cutcutache, Email: ioana.cutcutache@ucb.com.
Enrico Bertini, Email: enricosilvio.bertini@opbg.net.
Rosalba Carrozzo, Email: rosalba.carrozzo@opbg.net.
Francesco Emma, Email: francesco.emma@opbg.net.
Francesca Diomedi-Camassei, Email: francesca.diomedi@opbg.net.
Claudia Zanna, Email: claudia.zanna@unibo.it.
Martin Armstrong, Email: Martin.Armstrong@ucb.com.
Nicholas Stong, Email: nestong@gmail.com.
Sylvia Boesch, Email: sylvia.boesch@i-med.ac.at.
Robert Kopajtich, Email: robert.kopajtich@helmholtz-muenchen.de.
Wolfgang Sperl, Email: w.sperl@salk.at.
William C. Copeland, Email: copelan1@niehs.nih.gov.
Marco Seri, Email: marco.seri@unibo.it.
Maria Falkenberg, Email: maria.falkenberg@medkem.gu.se.
Holger Prokisch, Email: prokisch@helmholtz-muenchen.de.
Nicholas Katsanis, Email: katsanis@cellbio.duke.edu.
Valeria Tiranti, Email: valeria.tiranti@istituto-besta.it.
Tommaso Pippucci, Email: tommaso.pippucci@gmail.com.
Valerio Carelli, Email: valerio.carelli@unibo.it.
References
- 1.Carelli V, La Morgia C, Ross-Cisneros FN, Sadun AA. Optic neuropathies: the tip of the neurodegeneration iceberg. Hum Mol Genet. 2017;26(R2):R139–R150. doi: 10.1093/hmg/ddx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu-Wai-Man P, Votruba M, Burté F, La Morgia C, Barboni P, Carelli V. A neurodegenerative perspective on mitochondrial optic neuropathies. Acta Neuropathol. 2016;132(6):789–806. doi: 10.1007/s00401-016-1625-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242(4884):1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 4.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 5.Alexander C, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26(2):211–215. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 6.Delettre C, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26(2):207–210. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 7.Züchner S, et al. Axonal neuropathy with optic atrophy is caused by mutations in mitofusin 2. Ann Neurol. 2006;59(2):276–281. doi: 10.1002/ana.20797. [DOI] [PubMed] [Google Scholar]
- 8.Gerber S, et al. Mutations in DNM1L, as in OPA1, result in dominant optic atrophy despite opposite effects on mitochondrial fusion and fission. Brain. 2017;140(10):2586–2596. doi: 10.1093/brain/awx219. [DOI] [PubMed] [Google Scholar]
- 9.Reynier P, et al. OPA3 gene mutations responsible for autosomal dominant optic atrophy and cataract. J Med Genet. 2004;41(9):e110. doi: 10.1136/jmg.2003.016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrams AJ, et al. Mutations in SLC25A46, encoding a UGO1-like protein, cause an optic atrophy spectrum disorder. Nat Genet. 2015;47(8):926–932. doi: 10.1038/ng.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amati-Bonneau P, et al. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain. 2008;131(Pt 2):338–351. doi: 10.1093/brain/awm298. [DOI] [PubMed] [Google Scholar]
- 12.Hudson G, et al. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain. 2008;131(Pt 2):329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- 13.Rouzier C, et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain. 2012;135(Pt 1):23–34. doi: 10.1093/brain/awr323. [DOI] [PubMed] [Google Scholar]
- 14.Yu-Wai-Man P, et al. Multi-system neurological disease is common in patients with OPA1 mutations. Brain. 2010;133(Pt3):771–786. doi: 10.1093/brain/awq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carelli V, et al. Syndromic parkinsonism and dementia associated with OPA1 missense mutations. Ann Neurol. 2015;78(1):21–38. doi: 10.1002/ana.24410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. An autosomal dominant disorder with multiple deletions of mitochondrial DNA starting at the D-loop region. Nature. 1989;339(6222):309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 17.Viscomi C, Zeviani M. MtDNA-maintenance defects: syndromes and genes. J Inherit Metab Dis. 2017;40(4):587–599. doi: 10.1007/s10545-017-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiMauro S, Schon EA, Carelli V, Hirano M. The clinical maze of mitochondrial neurology. Nat Rev Neurol. 2013;9(8):429–444. doi: 10.1038/nrneurol.2013.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiranti V, Rocchi M, DiDonato S, Zeviani M. Cloning of human and rat cDNAs encoding the mitochondrial single-stranded DNA-binding protein (SSB) Gene. 1993;126(2):219–225. doi: 10.1016/0378-1119(93)90370-I. [DOI] [PubMed] [Google Scholar]
- 20.Tiranti V, et al. Chromosomal localization of mitochondrial transcription factor A (TCF6), single-stranded DNA-binding protein (SSBP), and endonuclease G (ENDOG), three human housekeeping genes involved in mitochondrial biogenesis. Genomics. 1995;25(2):559–564. doi: 10.1016/0888-7543(95)80058-T. [DOI] [PubMed] [Google Scholar]
- 21.Miralles Fusté J, et al. In vivo occupancy of mitochondrial single-stranded DNA binding protein supports the strand displacement mode of DNA replication. PLoS Genet. 2014;10(12):e1004832. doi: 10.1371/journal.pgen.1004832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36(10):928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baugh EH, et al. Robust classification of protein variation using structural modelling and large-scale data integration. Nucleic Acids Res. 2016;44(6):2501–2513. doi: 10.1093/nar/gkw120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farr CL, Wang Y, Kaguni LS. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. Template-primer DNA binding and initiation and elongation of DNA strand synthesis. J Biol Chem. 1999;274(21):14779–14785. doi: 10.1074/jbc.274.21.14779. [DOI] [PubMed] [Google Scholar]
- 25. Thisse B, Thisse C. Fast release clones: a high throughput expression analysis. ZFIN Direct Data Submission. http://zfin.org Accessed October 23, 2019.
- 26.Carnes MU, et al. Discovery and functional annotation of SIX6 variants in primary open-angle glaucoma. PLoS Genet. 2014;10(5):e1004372. doi: 10.1371/journal.pgen.1004372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samuel A, Rubinstein AM, Azar TT, Ben-Moshe Livne Z, Kim SH, Inbal A. Six3 regulates optic nerve development via multiple mechanisms. Sci Rep. 2016;6:20267. doi: 10.1038/srep20267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drummond IA, Davidson AJ. Zebrafish kidney development. Methods Cell Biol. 2016;134:391–429. doi: 10.1016/bs.mcb.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 29.Hentschel DM, et al. Rapid screening of glomerular slit diaphragm integrity in larval zebrafish. Am J Physiol Renal Physiol. 2007;293(5):F1746–F1750. doi: 10.1152/ajprenal.00009.2007. [DOI] [PubMed] [Google Scholar]
- 30.Anderson BR, et al. In vivo modeling implicates APOL1 in nephropathy: evidence for dominant negative effects and epistasis under anemic stress. PLoS Genet. 2015;11(7):e1005349. doi: 10.1371/journal.pgen.1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spinazzola A, et al. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38(5):570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 32.Karadimas CL, et al. Navajo neurohepatopathy is caused by a mutation in the MPV17 gene. Am J Hum Genet. 2006;79(3):544–548. doi: 10.1086/506913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giordano C, et al. Gastrointestinal dysmotility in mitochondrial neurogastrointestinal encephalomyopathy is caused by mitochondrial DNA depletion. Am J Pathol. 2008;173(4):1120–1128. doi: 10.2353/ajpath.2008.080252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carelli V, et al. Mitochondrial Ophthalmology. In: DiMauro S, Hirano M, Schon EA, eds. Mitochondrial Medicine. Abington, Oxon, United Kingdom: Informa Healthcare; 2006:105–142. [Google Scholar]
- 35.Genead MA, et al. Photoreceptor structure and function in patients with congenital achromatopsia. Invest Ophthalmol Vis Sci. 2011;52(10):7298–7308. doi: 10.1167/iovs.11-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sundaram V, et al. Retinal structure and function in achromatopsia: implications for gene therapy. Ophthalmology. 2014;121(1):234–245. doi: 10.1016/j.ophtha.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo X, et al. Blue cone monochromacy: visual function and efficacy outcome measures for clinical trials. PLoS ONE. 2015;10(4):e0125700. doi: 10.1371/journal.pone.0125700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sergouniotis PI, Holder GE, Robson AG, Michaelides M, Webster AR, Moore AT. High-resolution optical coherence tomography imaging in KCNV2 retinopathy. Br J Ophthalmol. 2012;96(2):213–217. doi: 10.1136/bjo.2011.203638. [DOI] [PubMed] [Google Scholar]
- 39.Moraes CT, et al. mtDNA depletion with variable tissue expression: a novel genetic abnormality in mitochondrial diseases. Am J Hum Genet. 1991;48(3):492–501. [PMC free article] [PubMed] [Google Scholar]
- 40.Kullar PJ, et al. Heterozygous SSBP1 start loss mutation co-segregates with hearing loss and the m.1555A>G mtDNA variant in a large multigenerational family. Brain. 2018;141(1):55–62. doi: 10.1093/brain/awx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci U S A. 2011;108(33):13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kukat C, et al. Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci U S A. 2015;112(36):11288–11293. doi: 10.1073/pnas.1512131112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giordano C, et al. Efficient mitochondrial biogenesis drives incomplete penetrance in Leber’s hereditary optic neuropathy. Brain. 2014;137(Pt 2):335–353. doi: 10.1093/brain/awt343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viscomi C, et al. Early-onset liver mtDNA depletion and late-onset proteinuric nephropathy in Mpv17 knockout mice. Hum Mol Genet. 2009;18(1):12–26. doi: 10.1093/hmg/ddn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zanna C, et al. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131(Pt 2):352–367. doi: 10.1093/brain/awm335. [DOI] [PubMed] [Google Scholar]
- 46.Del Dotto V, et al. Deciphering OPA1 mutations pathogenicity by combined analysis of human, mouse and yeast cell models. Biochim Biophys Acta Mol Basis Dis. 2018;1864(10):3496–3514. doi: 10.1016/j.bbadis.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Posse V, et al. RNase H1 directs origin-specific initiation of DNA replication in human mitochondria. PLoS Genet. 2019;15(1):e1007781. doi: 10.1371/journal.pgen.1007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23(12):2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milenkovic D, et al. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum Mol Genet. 2013;22(10):1983–1993. doi: 10.1093/hmg/ddt051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jemt E, et al. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015;43(19):9262–9275. doi: 10.1093/nar/gkv804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antes A, et al. Differential regulation of full-length genome and a single-stranded 7S DNA along the cell cycle in human mitochondria. Nucleic Acids Res. 2010;38(19):6466–6476. doi: 10.1093/nar/gkq493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reyes A, et al. RNASEH1 mutations impair mtDNA replication and cause adult-onset mitochondrial encephalomyopathy. Am J Hum Genet. 2015;97(1):186–193. doi: 10.1016/j.ajhg.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kornblum C, et al. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat Genet. 2013;45(2):214–219. doi: 10.1038/ng.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mussini C, et al. Effect of treatment interruption monitored by CD4 cell count on mitochondrial DNA content in HIV-infected patients: a prospective study. AIDS. 2005;19(15):1627–1633. doi: 10.1097/01.aids.0000186019.47297.0d. [DOI] [PubMed] [Google Scholar]
- 55.Krishnan KJ, Bender A, Taylor RW, Turnbull DM. A multiplex real-time PCR method to detect and quantify mitochondrial DNA deletions in individual cells. Anal Biochem. 2007;370(1):127–129. doi: 10.1016/j.ab.2007.06.024. [DOI] [PubMed] [Google Scholar]
- 56.Hjerpe R, et al. Oligomerization conditions Mdm2-mediated efficient p53 polyubiquitylation but not its proteasomal degradation. Int J Biochem Cell Biol. 2010;42(5):725–735. doi: 10.1016/j.biocel.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 57.Fernández-Vizarra E, López-Pérez MJ, Enriquez JA. Isolation of biogenetically competent mitochondria from mammalian tissues and cultured cells. Methods. 2002;26(4):292–297. doi: 10.1016/S1046-2023(02)00034-8. [DOI] [PubMed] [Google Scholar]
- 58.Lyonnais S, et al. The human mitochondrial transcription factor A is a versatile G-quadruplex binding protein. Sci Rep. 2017;7:43992. doi: 10.1038/srep43992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu B, et al. Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol Cell. 2013;49(1):121–132. doi: 10.1016/j.molcel.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ciesielski GL, Rosado-Ruiz FA, Kaguni LS. Purification and comparative assay of human mitochondrial single-stranded DNA-binding protein. Methods Mol Biol. 2016;1351:211–222. doi: 10.1007/978-1-4939-3040-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jay JJ, Brouwer C. Lollipops in the clinic: information dense mutation plots for precision medicine. PLoS ONE. 2016;11(8):e0160519. doi: 10.1371/journal.pone.0160519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traynelis J, et al. Optimizing genomic medicine in epilepsy through a gene-customized approach to missense variant interpretation. Genome Res. 2017;27(10):1715–1729. doi: 10.1101/gr.226589.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.