Abstract
Mitochondrial dysfunction or loss is evident in neurodegenerative diseases. Furthermore, mitochondrial DNA (mtDNA) mutations associated with NADH dehydrogenase subunits and nuclear gene mutations that affect mitochondrial function result in optic neuropathies. In this issue of the JCI, Del Dotto et al. and Piro-Mégy et al. identify heterozygous mutations in nuclear-encoded mitochondrial single-strand binding protein 1 (SSBP1) in patients with apparently dominant optic neuropathy with or without extraocular phenotypes. Both research groups reported similar mitochondrial findings in response to SSBP1 mutations. However, the specific SSBP1 mitochondria–associated function in retinal ganglion cells (RGCs) and the resulting optic nerve remains unclear. We suggest that high expression of SSBP1 during RGC differentiation is critical for mtDNA maintenance to produce appropriate optic nerve connectivity and that SSBP1 mutations in dominant optic atrophy patients do not permit stable binding to N6-methyldeoxyadenosine on the heavy strand involved with replication, leading to disruptions of mtDNA and, eventually, optic nerve dysfunction.
Mitochondrial dysfunction in optic neuropathies
Optic neuropathies are a heterogenous group of inherited diseases affecting the optic nerve. Axons of retinal ganglion cells (RGCs) form the optic nerve, which transmits visual information from the retina to the visual cortex. Defects in RGC function are associated with vision impairment in glaucoma, Leber optic atrophy (OMIM 535000), and other neuropathies. Leber hereditary optic neuropathy (LHON) was the first reported mitochondrial DNA (mtDNA) disease (1), with male patients being two to five times more likely to develop blindness than females. Color vision is severely affected in LHON from early stages, and visual acuity can deteriorate to 20/200 (legal blindness) or only light perception. Some LHON patients show extraocular phenotypes, such as cardiac conduction abnormalities, dystonia, and multiple sclerosis–like illness. Autosomal dominant optic atrophies (DOAs) are caused by mutations in nuclear-encoded OPA genes that affect diverse mitochondrial functions. DOAs present a much earlier age of onset than LHON, yet most patients report only mild to medium reduction in visual acuity, dyschromatopsia, and/or abnormalities in visual evoked potentials. Additional clinical phenotypes in DOA may include hearing loss, peripheral neuropathy, myopathy, ataxia, and chronic progressive external ophthalmoplegia.
Maintenance of healthy mitochondria requires recurring fission and fusion for repairing damaged components and is especially critical in neurons with extensive axonal/dendritic processes because of high local demands for energy and of metabolites (2). Nuclear genes provide almost all of the necessary mitochondrial proteins, whereas the mitochondrial genome encodes only 37 genes that are critical for survival. LHON is exclusively caused by mutations in the mitochondrial genes encoding NADH dehydrogenase subunits of complex 1; therefore, it exhibits a maternal inheritance pattern. LHON mutations exhibit low penetrance and varying prognosis, indicating the need for additional factors to produce clinical manifestations. The most well-studied nuclear OPA gene encodes intramitochondrial Dynamin GTPase OPA1 (OMIM 165500), which participates in maintenance of mitochondrial genome integrity, replication and distribution of mtDNA, mitochondrial fusion, and energy metabolism (3). Mitonuclear crosstalk is essential for numerous cellular processes, yet how changes in mtDNA and nuclear OPA mutations largely affect RGCs and the optic nerve is unclear.
Nuclear protein influencing mtDNA replication/repair
In this issue of the JCI, two reports, Del Dotto et al. and Piro-Mégy et al. (4, 5), add further mystery to the continuing saga of the mitonuclear tango. Both groups identified a total of six different mutations in a nuclear gene encoding mitochondrial single-strand binding protein (SSBP1) in multiple families with autosomal dominant optic neuropathy (with and without extraocular phenotypes). The two research groups revealed different genetic trajectories, yet both reached the same conclusion and presented similar biochemical findings (using patient fibroblasts). Key points include a marked depletion of mtDNA with impaired replication rate, defective mitochondrial structure and respiration, modeling of SSBP1 predicting the effect of the mutations, and the absence of SSBP1 tetramers (4, 5). Del Dotto and colleagues were also able to reproduce some of the disease-associated phenotypes in a zebrafish ssbp1-knockout model generated by CRISPR-Cas9 (4). Another recent report further validates SSBP1 as the bona fide cause of DOA (6).
SSBP1 is part of the mtDNA replication, maintenance, and repair machinery, together with the mitochondrial polymerase (POLγ), mtDNA helicase (TWINKLE), and mtRNA polymerase (POLRMT) (7, 8). mtDNA replication is proposed to initiate at the origin of the heavy (H) strand, and DNA synthesis then proceeds to produce a new H-strand (9). SSBP1 binds as a tetramer to the displaced parental H-strand to protect it against nucleases, prevent secondary structure formation, and stimulate TWINKLE’s helicase activity as well as increase the processivity of POLγ (10, 11). Retrograde translocation of mitochondrial SSBP1 into the nucleus as a response to proteotoxic stresses helps in cell survival and maintenance of mitochondrial membrane potential by augmenting heat shock factor HSF1 (12). Functional relevance of SSB proteins has been suggested in DNA damage response (13).
Mitochondrial dysfunction as well as single-strand breaks have commonly and independently been associated with neurodegeneration (14–16). High penetrance of optic neuropathies in patients with SSBP1 mutations raises a number of questions, the primary one being, why are only RGCs and the optic nerve, and not other neurons, affected? Is mitochondrial maintenance and damage response machinery different in RGCs? Are specific subtypes of RGCs more affected? Why is there no involvement of other CNS neurons even though dysregulation of mitochondrial biogenesis and mitochondrial DNA depletion have been observed in Parkinson’s neurons (17, 18)?
A model suggesting how SSBP1 mutations affect mitochondrial function
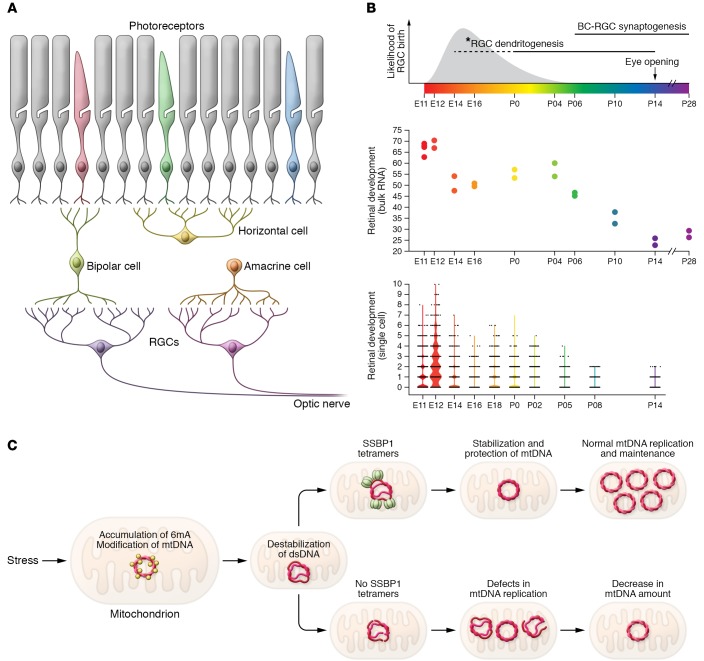
The mammalian retina has five major types of neurons, and RGCs are among the first cells born during development (Figure 1A). High expression of SSBP1 is evident at E12 at the time of RGC birth in both bulk and single-cell RNA-Seq data, with lower transcript levels observed in mature retina (Figure 1B and refs. 19, 20). We therefore hypothesize that SSBP1 is needed for mitochondrial biogenesis during early RGC differentiation. Notably, mitochondria enter RGC dendrites before branching patterns are established, and the dendritic mitochondria gradually become enriched (>15-fold) near branch points (21). Thus, RGCs appear to require mitochondria in order to maintain the synaptic landscape. Without healthy mitochondria, the ability of RGCs to transmit visual information might be compromised, likely leading to the scotomas that patients report. Disruption of mitochondrial fusion in Opa1+/– mice is reported to result in shortened mitochondria, which accumulate in proximal RGC dendrites; and the number of excitatory synapses and branches of RGC dendrites decline with age (22), further highlighting the importance of mitochondrial fusion for dendritic and synaptic integrity.
Figure 1. SSBP1 mutations and mtDNA replication/repair in the retina.
(A) Schematic of retinal architecture and time course of neuronal birth in mouse retina. (B) SSBP1 gene expression in the developing mouse retina by bulk RNA-Seq (upper panel) and single-cell RNA-Seq. Plots were generated from publicly available data on gene expression profiles (19, 20). *RGC dendritogenesis period (shown by dashed line) is based on gene profiles and not anatomy. BC, bipolar cell. (C) A model of mitochondrial dysfunction/loss by SSBP1 mutations.
To explain how SSBP1 mutations affect mitochondrial function, especially in developing RGCs, we propose a model shown in Figure 1C, as follows. Genome-wide mapping studies have uncovered asymmetric enrichment of N6-methyldeoxyadenosine (6mA) clusters on the H-strand of mtDNA, and 6mA could destabilize double-stranded DNA, promoting the recruitment of SSBP1 (23). Increased accumulation of 6mA disrupts oxidative phosphorylation (23) and might lead to impaired response to stress and probably neuropsychiatric disorders (24). We hypothesize that the harsh environment in the mitochondria results in enhanced accumulation of 6mA on the H-strand of mtDNA and that mutant SSBP1 protein is unable to form stable tetramers, leaving it vulnerable to damage. Increased accumulation of 6mA, combined with mutant SSBP1, could result in errors (including depletion) during mtDNA replication and/or repair, eventually leading to mitochondrial dysfunction or loss. Future studies directed at protecting single-stranded DNA for error-free mtDNA repair or replacement of the damaged H-strand DNA would help in maintaining healthy mitochondria and designing a treatment of optic atrophies.
Acknowledgments
The authors are supported by the Intramural Research Program of the National Eye Institute (ZIAEY000450 and ZIAEY000546). We thank Andrew Smith, Anupam Mondal, and Partha Dey for helpful comments and Ben Fadl for assistance with the single cell panel in Figure 1.
Version 1. 11/18/2019
Electronic publication
Version 2. 01/02/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(1):62–64. https://doi.org/10.1172/JCI132532.
See the related article at Dominant mutations in mtDNA maintenance gene SSBP1 cause optic atrophy and foveopathy.
See the related article at SSBP1 mutations cause mtDNA depletion underlying a complex optic atrophy disorder.
Contributor Information
Lina Zelinger, Email: lina.zelinger@nih.gov.
Anand Swaroop, Email: swaroopa@nei.nih.gov.
References
- 1.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242(4884):1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 2.Misgeld T, Schwarz TL. Mitostasis in neurons: maintaining mitochondria in an extended cellular architecture. Neuron. 2017;96(3):651–666. doi: 10.1016/j.neuron.2017.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elachouri G, et al. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21(1):12–20. doi: 10.1101/gr.108696.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Del Dotto V, et al. SSBP1 mutations cause mtDNA depletion underlying a complex optic atrophy disorder. J Clin Invest. 2020;130(1):108–125. doi: 10.1172/JCI128514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piro-Mégy C, et al. Dominant mutations in mtDNA maintenance gene SSBP1 cause optic atrophy and foveopathy. J Clin Invest. 2020;130(1):143–156. doi: 10.1172/JCI128513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jurkute N, et al. SSBP1 mutations in dominant optic atrophy with variable retinal degeneration. Ann Neurol. 2019;86(3):368–383. doi: 10.1002/ana.25550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. Reconstitution of a minimal mtDNA replisome in vitro. EMBO J. 2004;23(12):2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wanrooij S, Fusté JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc Natl Acad Sci U S A. 2008;105(32):11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 10.Farr CL, Wang Y, Kaguni LS. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein. Template-primer DNA binding and initiation and elongation of DNA strand synthesis. J Biol Chem. 1999;274(21):14779–14785. doi: 10.1074/jbc.274.21.14779. [DOI] [PubMed] [Google Scholar]
- 11.Korhonen JA, Gaspari M, Falkenberg M. TWINKLE Has 5’ -> 3’ DNA helicase activity and is specifically stimulated by mitochondrial single-stranded DNA-binding protein. J Biol Chem. 2003;278(49):48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 12.Tan K, Fujimoto M, Takii R, Takaki E, Hayashida N, Nakai A. Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nat Commun. 2015;6:6580. doi: 10.1038/ncomms7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croft LV, et al. Human single-stranded DNA binding protein 1 (hSSB1, OBFC2B), a critical component of the DNA damage response. Semin Cell Dev Biol. 2019;86:121–128. doi: 10.1016/j.semcdb.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 14.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 15.Wright AF, et al. Lifespan and mitochondrial control of neurodegeneration. Nat Genet. 2004;36(11):1153–1158. doi: 10.1038/ng1448. [DOI] [PubMed] [Google Scholar]
- 16.Jeppesen DK, Bohr VA, Stevnsner T. DNA repair deficiency in neurodegeneration. Prog Neurobiol. 2011;94(2):166–200. doi: 10.1016/j.pneurobio.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grünewald A, Rygiel KA, Hepplewhite PD, Morris CM, Picard M, Turnbull DM. Mitochondrial DNA depletion in respiratory chain-deficient parkinson disease neurons. Ann Neurol. 2016;79(3):366–378. doi: 10.1002/ana.24571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal A, Drerup CM. Axonal transport and mitochondrial function in neurons. Front Cell Neurosci. 2019;13:373. doi: 10.3389/fncel.2019.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks MJ, et al. Improved retinal organoid differentiation by modulating signaling pathways revealed by comparative transcriptome analyses with development in vivo. Stem Cell Rep. doi: 10.1016/j.stemcr.2019.09.009. doi: 10.1016/j.stemcr.2019.09.009. [published online ahead of print October 8, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clark BS, et al. Single-Cell RNA-Seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron. 2019;102(6):1111–1126.e5. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faits MC, Zhang C, Soto F, Kerschensteiner D. Dendritic mitochondria reach stable positions during circuit development. Elife. 2016;5:e11583. doi: 10.7554/eLife.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams PA, Piechota M, von Ruhland C, Taylor E, Morgan JE, Votruba M. Opa1 is essential for retinal ganglion cell synaptic architecture and connectivity. Brain. 2012;135(Pt 2):493–505. doi: 10.1093/brain/awr330. [DOI] [PubMed] [Google Scholar]
- 23.Koh CWQ, et al. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018;46(22):11659–11670. doi: 10.1093/nar/gky1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao B, et al. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun. 2017;8(1):1122. doi: 10.1038/s41467-017-01195-y. [DOI] [PMC free article] [PubMed] [Google Scholar]