Abstract
High-throughput technologies for genomics, transcriptomics, proteomics, and metabolomics, and integrative analysis of these data, enable new, systems-level insights into disease pathogenesis. Mitochondrial diseases are an excellent target for hypothesis-generating omics approaches, as the disease group is mechanistically exceptionally complex. Although the genetic background in mitochondrial diseases is in either the nuclear or the mitochondrial genome, the typical downstream effect is dysfunction of the mitochondrial respiratory chain. However, the clinical manifestations show unprecedented variability, including either systemic or tissue-specific effects across multiple organ systems, with mild to severe symptoms, and occurring at any age. So far, the omics approaches have provided mechanistic understanding of tissue-specificity and potential treatment options for mitochondrial diseases, such as metabolome remodeling. However, no curative treatments exist, suggesting that novel approaches are needed. In this Review, we discuss omics approaches and discoveries with the potential to elucidate mechanisms of and therapies for mitochondrial diseases.
Introduction
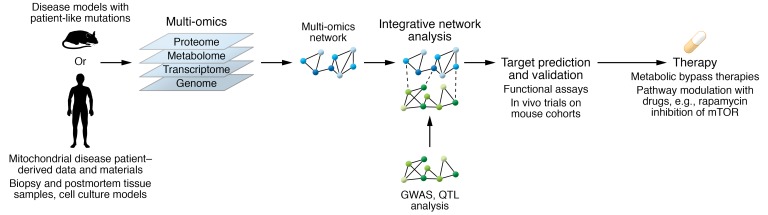
The high-throughput omics technologies, based on next-generation sequencing and mass spectrometry, have considerably advanced in recent years, allowing molecular physiology studies of whole-tissue and organismal homeostasis in exceptional depth. The technological improvements include simplification of sample preparation workflows (1), enhanced instrument sensitivity (2), and upgraded computational analysis methods (3). In studies of mitochondrial biology, omics data have been especially beneficial in identification of relevant genes, metabolites, and proteins (4, 5). Moreover, use of genomics, transcriptomics, proteomics, and metabolomics data has provided valuable insights into pathogenic pathways involved in mitochondrial diseases, including molecular stress responses and major remodeling of the metabolome and proteome (6–10). Integration of different levels of these data (multi-omics) facilitates robust insights into complex molecular functional mechanisms by reinforcing complementary evidence from multiple levels (11). As mitochondrial functions are involved at multiple levels of regulatory pathways, the multi-omics approaches are expected to be of extensive value for elucidating the pathogenic pathways in mitochondrial diseases, a disease group extending to almost all fields of medicine (Figure 1).
Figure 1. From mouse models and patient-derived samples to therapy development.
Mitochondrial diseases are modeled in mouse knockouts of mitochondrial proteins or in mouse models that recapitulate patient mutations in conserved homologous amino acids. Patient samples are derived from biopsies, postmortem tissue, and induced pluripotent stem cells. Integrative analysis of genomics, transcriptomics, proteomics, and metabolomics data allows the prediction and validation of targets.
Mitochondrial diseases are the most common group of inherited metabolic diseases, with exceptional clinical variability (12). Primary mitochondrial diseases, which arise from pathogenic mutations in the nuclear or mitochondrial genome and affect a variety of mitochondrial functions, have an estimated prevalence of 1:2000 to 1:5000 and can manifest at any age (13). Furthermore, secondary mitochondrial diseases include, for instance, neurodegenerative disorders, such as Parkinson’s disease, and mitochondrial metabolism also may contribute to cancer progression (14). The reasons why mitochondrial disorders show tissue-specific manifestations in almost any organ system are still poorly understood. Sometimes a mitochondrial disease manifests in a newborn immediately after birth with, e.g., a multisystem disease or an isolated cardiomyopathy with severe lactic acidosis, leading to death within a few days. Sometimes a healthy teenager develops acute, severe epilepsy and liver dysfunction and progresses to a severe neurodegenerative disease (12). Despite the scarcity of curative options, preliminary evidence suggests that sometimes interventions modifying a critical target in metabolic pathways may ameliorate the disease, indicating the vast importance of molecular studies to reveal such targets (15, 16). For these studies, omics analyses have become a valuable tool, facilitating a wide-angle view on the complex mechanisms of tissue homeostasis.
Mitochondria are best known as the cellular power plants; via the process of oxidative phosphorylation (OXPHOS), they catabolize nutrients to provide cells with fuel, adenosine triphosphate (ATP). Mitochondrial DNA (mtDNA) encodes 13 proteins, which form the core components of the respiratory chain enzymes and two subunits of ATPase, as well as the transfer RNAs and ribosomal RNAs that mitochondrial ribosomes require to synthesize these proteins. However, the mitochondrial proteome mostly consists of approximately 1500 nuclear-encoded proteins that are translated in the cytoplasm and translocated into the mitochondria through elaborate and complex import machinery (17). These proteins are assembled with mtDNA-encoded OXPHOS-enzyme subunits to form functional enzyme complexes, involving coordinated insertion of metal cofactors. Lipid synthesis and catabolism is dependent upon functional interactions between mitochondria and the peroxisomes and on physical contact sites between mitochondria and the endoplasmic reticulum (17). Furthermore, these events respond to nutrient status and stress signals, adding to the complexity of mitochondrial physiology. The metabolic activity of a given cell type tends to correlate with the number of mitochondria (18–20), being high for instance in cardiac muscle, nerve cells, sensory organs, skeletal muscle, and the liver. However, mitochondrial diseases rarely affect all these tissues, and can be restricted even to one tissue, indicating that deficient ATP synthesis does not solely explain tissue-specific disease manifestations. In addition to catabolic nutrient breakdown, mitochondria are versatile players in anabolic cellular functions, including biosynthetic one-carbon cycle, iron-sulfur cluster synthesis, and cellular stress responses (17). Mitochondria also contribute to apoptosis as well as calcium storage and signaling between cells (17, 21). The mitochondrial contributions to anabolic biosynthesis pathways are an intriguing mechanism to explain the tissue-specific disease manifestations (8, 9, 17).
Because of the variable manifestations, diagnosis of mitochondrial disorders is challenging. The development of next-generation sequencing technologies has been especially valuable, as a known genetic defect can be identified in up to 40% to 60% of the patients (22). The genetic diagnosis provides means for counseling and sometimes directs treatment choices, and it also is the first step toward understanding the disease mechanisms. To facilitate the discovery of novel mechanisms contributing to the tissue-specific manifestations, a combination of multifaceted molecular and physiological analyses of carefully designed disease models and patient materials is needed.
In this Review, we describe how research related to human mitochondrial physiology and diseases has evolved in recent years, fueled by the increasing use of omics data. We discuss the prospects of omics analyses for understanding the mechanisms of tissue-specificity of mitochondrial disorders and discovering clinically relevant biomarkers, with special focus on metabolic biomarkers.
Animal models of mitochondrial function and disease
Owing to the complexity of mitochondrial functions, precise mouse models for mitochondrial diseases are invaluable tools for omics approaches. Tissue-specificity of metabolic disorders is very likely associated with the metabolic needs of the tissue at a specific point in time — and only sensitive omics can reveal such pathways. Therefore, animal models that faithfully replicate human disease mutations and manifestations are especially valuable, combined with human samples to study the relevance of the identified pathways in humans (Figure 1).
Omics analyses of animal models have provided completely novel — even unexpected — views of pathogenic pathways. For instance, metabolomics analyses have suggested that mitochondrial defects modify not only organellar functions but also whole cellular flux of metabolic pathways and affect nuclear transcription and even nuclear genome maintenance (23). A recent study using transcriptomics, metabolomics, and in vivo imaging showed that the affected tissue can send hormone-like signals to the whole body; for example, in the case of mitochondrial muscle disease, FGF21 endocrine signaling rewires the metabolism of the brain (24). These studies highlight the importance of a global view of changes in the metabolome. Proteomics and transcriptomics can also provide complementary views.
To date, only a few animal models with primary mtDNA mutations exist (25, 26). However, mtDNA mutagenesis and mtDNA depletion have been generated by modification of mtDNA maintenance proteins encoded by the nucleus (27–29), and approximately 50 knockout mouse lines have been described for mitochondrial proteins encoded by the nuclear genome (29, 30). Knockout mice are excellent mechanistic models, but do not often serve as good disease models: even the most severe mitochondrial disease models often retain a low level of expression of the mutant protein, as full knockouts are embryonically lethal (with some exceptions) (31). Furthermore, tissues have crosstalk and send secreted factors to modulate organismal physiology. Therefore, tissue-specific postnatal conditional knockout strategies that lead to death of the targeted cell are non-optimal to assess disease physiology. Importantly, transgenic/knockin mice harboring homologous patient mutations have shown their value: the proteins of interest are well conserved in species, and the findings and physiology have been replicated remarkably well in human patient samples (8, 27, 32). However, to date, few of these models have been analyzed by integrative multi-omics techniques.
Multi-omics analyses reveal pathways in mitochondrial disease models.
Recent studies have provided valuable examples of how multi-omics approaches can discover specific metabolic pathways and metabolites mediating mitochondrial dysfunction (summarized in Table 1). Nikkanen et al. analyzed the cell transcriptome and metabolome in two mouse models with recessive and dominant mutations in the mtDNA helicase Twinkle (encoded by Twnk), and in human patients carrying the same mutations. They identified a major remodeling of the anabolic folate-driven one-carbon cycle specifically in the affected tissues in mice, driving one-carbon units for purine and glutathione synthesis, suggesting therapy targets in the folate cycle as a means of treatment (8). A similar finding was reported by Bao et al. in human cells depleted for another gene essential for mtDNA maintenance, mtDNA polymerase-γ (encoded by Polg) (9). Kühl et al. analyzed the cellular transcriptome and mitochondrial proteome of five different conditional knockout mouse models of genes essential for mtDNA expression in mouse heart tissue (Twnk, Tfam, Polrmt, Mterf4, and Lrpprc) (10) and similarly found remodeling of the one-carbon cycle, substantially reduced coenzyme Q (CoQ) levels, and decreased levels of multiple mitochondrial CoQ biosynthesis enzymes (10). CoQ functions as an electron carrier from complexes I and II to complex III in the inner mitochondrial membrane and is a redox-active compound with antioxidant characteristics (33). Thus, these omics studies propose controlled therapy trials with CoQ derivatives for patients with mtDNA maintenance defects.
Table 1. In vivo and in vitro model studies using multi-omics approach.
Identifying biomarkers and targets in mitochondrial disease models.
A milestone achievement of multi-omics approaches in mitochondrial research was the identification of novel regulators of the mitochondrial integrated stress response (ISRmt), a multifaceted tissue-specific response activated upon mitochondrial stress, such as in mitochondrial muscle myopathy patients (7, 8, 24, 34). Although some components of it overlap with the mitochondrial unfolded protein response (UPRmt), described mostly in studies of Caenorhabditis elegans (35), the key transcription factors and downstream targets differ (8–10, 24). The cellular response signature of ISRmt comprises changes in gene expression mediated by activating transcription factor 4 (ATF4), ATF5, and ATF3, resulting in de novo synthesis of the metabolic cytokines FGF21 and GDF15 and remodeling of the one-carbon and folate metabolisms and thus leading to increased serine and nucleotide pools. All these processes are collectively mediated by the upstream kinase, called mechanistic target of rapamycin complex 1 (mTORC1), and, strikingly, inhibition of mTORC1 with rapamycin reverses these molecular defects and skeletal muscle tissue-level pathology in a mouse model of mitochondrial muscle myopathy (34, 36). FGF21 and GDF15 are secreted from the affected muscle to circulation and also show high specificity to mitochondrial disorders in humans, serving as sensitive and specific blood biomarkers especially for muscle-manifesting mitochondrial diseases (24, 37, 38).
Another comprehensive multi-omics study provided a global transcriptome, proteome, and metabolome analysis of ISRmt in human HeLa cells elicited by treating the cells with drugs that block mitochondrial translation, import, membrane potential, or OXPHOS (39). In addition to identifying ATF4 as the key regulator of the mitochondrial stress–activated transcriptional program in cultured cells, the authors also described a global decrease in mitochondrial ribosomal protein levels and mitochondrial translation and a global increase in amino acid biosynthesis as cellular response markers of ISRmt. Importantly, interrogating gene expression databases for humans (40) and mice (BXD strain; refs. 41, 42) to identify ISRmt-induced genes revealed tight clustering of mitochondrial stress genes across multiple tissues in both humans and mice, highlighting the relevance of ISRmt processes identified in mouse models to human mitochondrial disease physiology.
Another mitochondrial myopathy, the muscle-specific and rapidly progressive mtDNA depletion syndrome, appears in infants with mutations in nuclear-encoded thymidine kinase 2 (TK2), encoding an enzyme that localizes to the mitochondria and is required in deoxyribose nucleoside triphosphate synthesis during mtDNA replication (43). Siegmund et al. found rapamycin to increase survival of a mouse model with a mutant Tk2 and identified tissue-specific metabolic changes based on large-scale transcriptomics analysis of brain and liver and metabolomics analysis of brain, liver, and plasma (44). However, although rapamycin holds a promising avenue for treatment of some mitochondrial disease patients, it would require long-term treatment. Moreover, its side effects, including immunosuppression, make it a non-optimal treatment choice (45).
Integrating complex data from multiple reference strains and models.
A particularly useful approach to explore genetic and environmental factors underlying complex systems, such as metabolism, is the analysis of omics data generated from a large number of different mouse strains, such as the Collaborative Cross mouse genetic reference population (42). Although this approach has not been used for mitochondrial disease per se, it has yielded interesting new information about aspects of mitochondrial regulation in other physiological and disease settings. For example, the integrative analysis of liver transcriptomes and whole genome sequences of approximately 100 different mouse strains revealed mitochondrial dysfunction and novel metabolic genes (Pkl2 and Chchd6) underlying nonalcoholic fatty liver disease (46). In another study, combined analysis of transcriptomic and proteomic data sets overlaid with genome sequences from 40 different mouse strains characterized coregulated genes of the mitochondrial UPRmt and further identified novel genes implicated in glucose homeostasis and diabetes (47). Looking forward, investigations of omics data sets generated using the mouse genetic reference population after treatment with different kinds of metabolic stressors (nutritional, genetic, or drug-induced) hold remarkable potential to discover novel mitochondrial pathways relevant to human disease.
Novel omics methods that reach beyond merely quantifying levels of transcripts and metabolites, reporting on posttranslational modifications and signaling pathway activation, are continuously being developed (48, 49). For example, the methyl cycle is dependent on the folate cycle, which is remodeled by mtDNA maintenance defects; integrative approaches to study genome-wide methylation and its cellular effects in disease models would be invaluable. Another important future development lies in the invention of new omics methods that enable analysis of activities in fine subcellular localizations: in mitochondria, but also in other organelles that intimately interact with the mitochondria, such as the endoplasmic reticulum, lysosomes, and peroxisomes. The recent generation and use of the transgenic MITO-Tag mouse line, which expresses a mitochondria-localized epitope tag that can be used for immunoisolation of mitochondria, are notable advancements toward solving the lack of cellular and subcellular resolution (50). Finally, the combined use of CRISPR-based genetic screens with deep sequencing and other omics methods provides powerful ways of discovering new genes and pathways essential for cellular health under mitochondrial stress. Notable examples of such approaches have uncovered genes that are essential for OXPHOS (51) and for survival upon inhibition of the respiratory chain (5).
Substantial challenges still lie ahead, not necessarily in our ability to generate omics data, but more in developing appropriate data analysis methods and biologically insightful computational models that would provide candidate therapeutic targets for intervention. However, progress achieved in the last decade in discovering novel metabolic pathways and therapeutic targets by the use of animal models and omics analyses holds great promise for future progress in development of treatment options for patients. The field is ready to start utilizing the data from model systems and translate the results to well-designed treatment studies.
Different approaches for multi-omics data integration
As reviewed in the previous section, in vivo and in vitro disease model studies have used multiple omics levels to gain novel biological insights into mitochondrial function and disease (summarized in Table 1). The majority of these studies have investigated more than two levels of omics, most commonly the underlying genome with the transcriptome and, further, either with the proteome and/or the metabolome. These data levels can be studied either separately with cross-comparison between the different omics results or in a more integrative fashion.
In general, integrative approaches can be broadly classified into supervised and unsupervised approaches (3). The supervised approaches typically use multiple omics levels to train a model with labeled training data to predict a phenotype — for instance, to distinguish a disease phenotype from control. The unsupervised approaches characteristically aim to integrate different omics levels by clustering the omics features, such as expression levels of genes/proteins/metabolites or samples, including network-based methods. In this section, we focus on the commonly used unsupervised approaches used in the studies presented in Table 1.
In order to gain insight into the genetic contributions to the regulation of molecular pathways and phenotypes, quantitative trait loci (QTL) analysis is commonly used. The associations of genetic loci or intervals with expression profiles of transcriptome, methylome, proteome, or metabolome are determined to define eQTLs, meQTLs, pQTLs, or mQTLs, respectively (Figure 2). The underlying mutations in mitochondrial diseases have been generated in animal models with known genotype information, allowing QTL analysis when combined with expression data. For instance, the GeneNetwork database contains whole-genome sequence data or high-density genotype information for the whole BXD family of mouse strains and their parents along with expression data for the key cell types, tissues, and organs (52). For the human genome, the Genotype-Tissue Expression Project (GTEx) is a large initiative that collects eQTL information on multiple human tissues (40). Williams et al. analyzed eQTLs, pQTLs, and mQTLs in BXD mice to identify genomic variants associated with gene, protein, or metabolite expression levels and thereby influencing cellular processes and overall mitochondrial function and metabolism (53). Their analysis led to a number of interesting findings, such as mapping of mitochondrial respiratory chain supercomplex assembly to the Cox7a2l gene.
Figure 2. Multi-omics data analysis workflow using integrative analysis approaches to identify a set of multibiomarkers for downstream validation.
Integrative approaches commonly include clustering and association analysis of the omics features that can be inspected in networks. Enrichment and mapping of candidate multibiomarkers to known canonical pathways can be used to interpret the findings. Metabolic data with or without accompanying transcriptome-level data can be analyzed by GEMs that combine data from various sources to represent the metabolism of a given biological system. The illustration of genome-scale metabolic models is adapted with permission from Cell Systems (105).
QTLs can be studied either in cis, where a genetic variant affects expression of nearby genes, gene products, or methylated gene regions, or in trans, where the variant’s effect ranges further away in the same chromosome or in other chromosomes. For example, a trans-eQTL analysis in the BXD mouse genetic reference population was used to identify the mitochondrial protein fumarate hydratase 1 (FH1) in the control of ATF4 activity, which is proposed to contribute to mitochondrial stress responses (24, 39).
Networks have become popular tools to present all associations of a system, such as all QTL associations (54). A network is represented as a graph where molecules and their relationships are depicted as nodes and edges, respectively. Nodes in a network are typically connected based on various association or similarity measures, enabling also combination of different types of data. For instance, QTL results can be combined with protein-protein interactions (PPIs), which can be used to build networks together with expression or association data. Wang et al. applied a hidden Markov random field model to integrate QTLs with PPI information to improve prediction accuracy of disease-related genes (55). Kogelman et al. first built a coexpression network of QTL genes and subsequently incorporated known PPI information to prioritize the QTL results (56).
Overall, networks provide convenient means for data integration (Figure 2). For example, Wu et al. used robust Spearman correlation measures to construct pairwise protein association networks and compared those with the corresponding transcript association networks in order to detect regulatory genes in the UPRmt pathway (47). The weighted gene coexpression network analysis (WGCNA) (57) has become a widely used approach to integrate genomics, transcriptomics, and more recently metabolomics data. WGCNA groups genes/proteins/metabolites into modules based on coexpression patterns, considering also the network topology to reduce spurious connections. The modules have often been shown to be enriched for similar functions (58). While WGCNA has been used for time-resolved data, it has been shown that group averaging of individual-level expression measurements may worsen the reproducibility of the results; hence, an approach based on individual-level networks has been suggested to provide more reproducible results (59).
While most pairwise methods are not able to identify the directions of regulatory interactions, Bayesian networks consist of directed acyclic graphs, and hence they can represent causal regulatory relationships among nodes, e.g., genes or proteins or metabolites (60). However, the identification of a network topology of a Bayesian network becomes challenging with respect to the number of nodes and connecting edges. In order to apply a Bayesian approach to large-scale network reconstruction, several methods have been developed, including approximation methods for network topology identification as well as methods reducing the search space (60, 61).
Identifying and interpreting metabolomic multibiomarkers using integrated multi-omics
Metabolomics aims to capture and quantify small-molecule metabolites (<1500 Da) detected within a given biological system. The metabolome can be considered the downstream product of upstream cellular biological regulatory processes, with contributions from underlying layers of the genome, epigenome, transcriptome, and proteome as well as environmental factors ultimately linking genotype to phenotype (62). In the following sections, we describe approaches that use multilayered omics data to identify and interpret metabolomic multibiomarkers.
Targeted versus global omics approaches.
Approaches to study genomes, transcriptomes, proteomes, and metabolomes can be either targeted or global. For instance, targeted metabolomics aims to measure either the absolute concentrations (quantitative analysis) or relative intensities (semiquantitative analysis) of a predefined set of metabolites based on prior knowledge about the given biological system. In contrast, global metabolomics aims to measure all the small-molecule metabolites in a cell, fluid, tissue compartment, or organism in a semiquantitative manner.
Both targeted and global approaches can be applied to determine metabolic fingerprints, i.e., subsets of the metabolome that have potential to distinguish phenotypes (63). Such fingerprints can be used as multibiomarkers for different disease phenotypes (6), as well as for patient stratification (64), for drug discovery (65), and to monitor disease progression, treatment effect, and/or toxicity (66). Khan et al. recently demonstrated this use in wild-type mice and those with mtDNA deletions, showing that rapamycin shifted the metabolome considerably in the skeletal muscle. After treatment, the metabolomes of control and diseased mice were overlapping with each other and not with the original untreated cohorts, revealing specific rewiring of the metabolome by rapamycin (34).
Common to all omics data analysis workflows is the computationally intensive data preprocessing, followed by reduction of data complexity and interpretation of the results. Especially in the context of metabolomics, the data preprocessing tends to be more straightforward for the targeted approach than for the global approach. For the global approach, the preprocessing step of noise reduction is especially critical, as noise signals introduced by contaminants and artifacts need to be filtered from the biological signals. While the two most commonly used tools for noise reduction of metabolomics data, MZmine 2 (67) and XCMS (68), perform relatively well, a recent study showed that there is still room for improvement to achieve high-quality data for downstream analyses (69).
Extracting functional insights from exceptionally complex data.
Omics data, including metabolomics data, are typically extremely complex, and “top-down” methods consist most commonly of comparative analysis, enrichment analysis, and pathway mapping. Comparative analysis to determine differential abundance of metabolites between two groups of samples still often uses t test, although there is evidence (70) that tools including limma (71) or ROTS (72), which were originally developed for other omics data, can be beneficial to metabolomics studies.
Commonly used approaches to gain functional insights into metabolic fingerprints use overrepresentation or enrichment analysis (73), with or without quantitative data. These approaches rely on known annotations in public databases. The discovered set of metabolites, such as a metabolic fingerprint, is compared with publicly available, functionally annotated metabolite data sets, such as metabolic pathways (74). The aim is to identify whether certain pathways contain a significantly larger number of fingerprint metabolites than expected by chance (Figure 2). The enrichment analyses most commonly use Fisher’s exact test or the hypergeometric distribution, but many other enrichment methods have been applied, such as Kolmogorov-Smirnov or Wilcoxon statistical tests (75, 76). While the first enrichment analysis tools for omics data were designed for gene expression studies, in recent years, several tools have been developed specifically for metabolomics data, such as MetaboAnalyst 4.0 (77) and IMPaLA (78). A recent comparison of the most commonly used tools for metabolite enrichment analysis showed that, in general, the different tools yielded consistent results (79).
The widely used metabolic pathway databases and metabolite data collections include the Human Metabolome Database (HMDB) (80), the Kyoto Encyclopedia of Genes and Genomes (KEGG) (81), the PubChem database (82), the Chemical Entities of Biological Interest (ChEBI) (83), the Metabolite and Chemical Entity Database METLIN (84), and the human metabolic reconstruction Recon 2.2 (85) (and the most recent version, Recon3D; ref. 86), PubChem being the largest.
Challenges in these types of knowledge-based approaches relate most commonly to the quality and completeness of the annotation databases (87, 88). This is particularly a problem with metabolite disease sets, which have not yet reached the level of completeness that would allow disease-based enrichment analysis (79). However, there have been recent efforts to improve metabolite disease sets. For instance, the number of compounds with disease links in the newest version of HMDB has increased by 77% in comparison with the previous version (80). The recently published MetSigDis database contains 6849 curated relationships between 2420 metabolites and 129 diseases across eight species, including human (89).
GEMs incorporate omics data sets into metabolic modeling.
Genome-scale metabolic models (GEMs) are network-based models that aim to capture the chemical reactions and metabolites that represent the metabolism of a given biological system (90, 91). The involved gene-protein-reaction associations are determined on the basis of various sources, including databases, literature, and experimental measurements. Different omics and other measurement data can be used in the modeling, including transcriptomics data, metabolic constraints, localization information, and thermodynamic measurements (Figure 2). Recent efforts have reconstructed human metabolic models with all known metabolic reactions, Recon3D (86) being one of the most comprehensive models. Recon3D encompasses 4140 unique metabolites, 12,890 protein structures, 13,543 reactions, and 3288 associated genes. The metabolic reactions and their metabolites are distributed over nine cellular compartments, mitochondria being one of them.
In multicellular organisms, like humans, metabolic reactions that are activated in disease settings vary across different cells and tissues, and hence a general model needs to be tailored to construct cell type–specific metabolic models. These context-specific models aim to shed light on the underlying molecular basis of the metabolic phenotype and can be used to predict regulatory mechanisms. Recently, several algorithms have been developed to extract context-specific models. However, many factors, including algorithms’ parameter settings, influence the model content and predictive accuracy (92). An evaluation of six algorithms to construct cell line– and tissue-specific metabolic models concluded that, although the choice of the parameters affected the model content, the largest variability to the output came from the selected algorithm (92).
MitoCore is a GEM of human central metabolism with two compartments, representing the cytosolic and the mitochondrial side. It encompasses 74 metabolites, 324 reactions, and 83 transport steps between mitochondrion and cytosol. It incorporates both normal and abnormal physiological conditions, including hypoxia and mitochondrial diseases. The authors of MitoCore compared the model with the human metabolic reconstruction Recon 2 and found only partial overlap, mainly because of the different partitioning of metabolism between the cytosol and the mitochondrial matrix (93). This indicates the importance of resolution and context in measuring metabolites; as resolution and context-specificity improve, GEMs become increasingly comprehensive and accurate. However, already in their current state, GEMs have been useful in exploring and gaining new insights into metabolomics (94). Considering modeling of large-scale metabolic data and its complexity, machine learning methods have proved useful in different steps of metabolism model building, including parameter determination and model optimization. Several machine learning approaches for metabolism modeling have recently been developed (95).
Limitations in metabolomics data integration and interpretation.
Associations between different omics layers are typically complex. Although transcriptome-level changes are often reflected in the proteome, discordances exist. Furthermore, a large variety of post-transcriptional and posttranslational modifications modulate protein functions. Still, these different omics approaches often point to changes in similar key pathways, which can then be specifically tested with traditional cell biological and protein biochemical means (8–10). It is also noteworthy that metabolite detection with aforementioned techniques only captures products or substrates of an enzymatic reaction (96): the steady-state levels of specific metabolites, which can reflect their increased use or decreased production. Integrative omics can bring light to dynamics, as construction of pathways (e.g., high substrate, low enzyme protein levels indicating a putative block or bottleneck in the pathway) may give insight into usage versus synthesis. Use of stable isotope–labeled metabolites (such as 13C-glucose or glutamine) and analysis of their carbon flux to downstream pathways enable direct interpretation of pathway dynamics (97). Recently, computational approaches have been developed to incorporate experimental flux profiling to build differential flux maps at a genome scale and context-specifically (98). These examples indicate that interpretation of data requires multidisciplinary approaches and development of computational approaches specifically addressing the challenges (99).
While mass spectrometry–based (MS-based) detection of metabolites is more sensitive than the NMR-based approach, the diversity of metabolites with respect to their chemical properties remains a technical challenge also with MS-based detection (100). Furthermore, the approach may be (a) targeted, in which case a specific preselected set of metabolites is analyzed, in a semiquantitative manner, or (b) untargeted, giving hundreds, even thousands of compounds, the nature of which is interpreted by their mass. As a result, the coverage of the total metabolome may be limited, and key metabolites may be missed. To overcome these limitations, progress has been made in improving the MS instrumentation and sample handling strategies, as well as metabolite identification and pathway analyses (101).
Conclusions and future directions
Thanks to the continuously improving omics technology, research on mitochondrial diseases is poised to be transformed. A growing body of studies on animal disease models highlights how different layers of omics data and their integrative analyses can help to elucidate novel molecular mechanisms of mitochondrial function and disease. For instance, functional genetic screens coupled with deep sequencing enable identification of new genes essential for specific bioenergetic or metabolic aspects of mitochondrial function, which would otherwise be exceedingly difficult and costly to predict by a series of conventional experiments.
With tissue-specificity postulated to underlie the observed heterogeneity of clinical manifestations of mitochondrial disorders, efforts have been undertaken to identify which factors determine the tissue-specificity of mitochondrial diseases. However, a large number of constitutive or cell type–specific knockouts for mitochondrial proteins remain to be investigated with respect to global changes in their tissue-specific gene expression and metabolome. Further, these analyses can be coupled with well-designed treatment studies with metabolic supplementation, drugs, or environmental stressors that alter cellular metabolism. Ultimately, the study of the genome, transcriptome, proteome, and metabolome of mitochondrial disease patient cohorts is anticipated to reveal human disease–specific perturbations in gene expression and metabolism and uncover the range of genetic and environmental contributions to metabolic regulation. Finally, longitudinal studies of patient groups allow characterization of disease stage–specific clinical markers that can be used for follow-up of disease progression and therapy response.
The development of measurement technologies and computational tools, including machine learning approaches, will undoubtedly facilitate new lines of research. For instance, early progression of cellular stress can be investigated by cell type–specific morphological studies using still images or time-lapse videos (102, 103). Recently, a machine learning–based mitochondrial imaging approach was developed (102). Moreover, the technological advances in single-cell technologies allow improved resolution of cell mechanisms that will further extend the understanding of mitochondrial disease mechanisms (101, 104).
Acknowledgments
LLE reports grants from the European Research Council (677943); the European Union’s Horizon 2020 research and innovation programme (675395); the Academy of Finland (296801, 304995, 310561, and 313343); the Juvenile Diabetes Research Foundation (2-2013-32); Tekes, the Finnish Funding Agency for Innovation (1877/31/2016); and the Sigrid Juselius Foundation. Support from the University of Turku, Åbo Akademi University, Biocenter Finland, and ELIXIR Finland Node is acknowledged. AS thanks the following funding sources: the Academy of Finland (307592, 303349, and 307431), the Sigrid Juselius Foundation, the Finnish Foundation for Cardiovascular Research (180072), and the University of Helsinki.
Version 1. 01/02/2020
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(1):20–28.https://doi.org/10.1172/JCI129202.
Contributor Information
Sofia Khan, Email: sofia.khan@utu.fi.
Gulayse Ince-Dunn, Email: gulayse.dunn@helsinki.fi.
Anu Suomalainen, Email: anu.wartiovaara@helsinki.fi.
Laura L. Elo, Email: laura.elo@utu.fi.
References
- 1.Cui H, et al. Comprehensive next-generation sequence analyses of the entire mitochondrial genome reveal new insights into the molecular diagnosis of mitochondrial DNA disorders. Genet Med. 2013;15(5):388–394. doi: 10.1038/gim.2012.144. [DOI] [PubMed] [Google Scholar]
- 2.Palmfeldt J, Bross P. Proteomics of human mitochondria. Mitochondrion. 2017;33:2–14. doi: 10.1016/j.mito.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front Genet. 2017;8:84. doi: 10.3389/fgene.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arroyo JD, et al. A genome-wide CRISPR death screen identifies genes essential for oxidative phosphorylation. Cell Metab. 2016;24(6):875–885. doi: 10.1016/j.cmet.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buzkova J, et al. Metabolomes of mitochondrial diseases and inclusion body myositis patients: treatment targets and biomarkers. EMBO Mol Med. 2018;10(12):e9091. doi: 10.15252/emmm.201809091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyynismaa H, et al. Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet. 2010;19(20):3948–3958. doi: 10.1093/hmg/ddq310. [DOI] [PubMed] [Google Scholar]
- 8.Nikkanen J, et al. Mitochondrial DNA replication defects disturb cellular dNTP pools and remodel one-carbon metabolism. Cell Metab. 2016;23(4):635–648. doi: 10.1016/j.cmet.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 9.Bao XR, et al. Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife. 2016;5:e10575. doi: 10.7554/eLife.10575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kühl I, et al. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. Elife. 2017;6:e30952. doi: 10.7554/eLife.30952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman GS, et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- 13.Gorman GS, et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol. 2015;77(5):753–759. doi: 10.1002/ana.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5):e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan NA, et al. Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol Med. 2014;6(6):721–731. doi: 10.1002/emmm.201403943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domínguez-González C, et al. Deoxynucleoside therapy for thymidine kinase 2–deficient myopathy. Ann Neurol. 2019;86(2):293–303. doi: 10.1002/ana.25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suomalainen A, Battersby BJ. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cell Biol. 2018;19(2):77–92. doi: 10.1038/nrm.2017.66. [DOI] [PubMed] [Google Scholar]
- 18.Fernández-Vizarra E, Enríquez JA, Pérez-Martos A, Montoya J, Fernández-Silva P. Tissue-specific differences in mitochondrial activity and biogenesis. Mitochondrion. 2011;11(1):207–213. doi: 10.1016/j.mito.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Mishra P, Chan DC. Metabolic regulation of mitochondrial dynamics. J Cell Biol. 2016;212(4):379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melser S, Lavie J, Bénard G. Mitochondrial degradation and energy metabolism. Biochim Biophys Acta. 2015;1853(10 pt B):2812–2821. doi: 10.1016/j.bbamcr.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maldonado EM, Taha F, Rahman J, Rahman S. Systems biology approaches toward understanding primary mitochondrial diseases. Front Genet. 2019;10:19. doi: 10.3389/fgene.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hämäläinen RH, et al. Defects in mtDNA replication challenge nuclear genome stability through nucleotide depletion and provide a unifying mechanism for mouse progerias. Nat Metab. doi: 10.1038/s42255-019-0120-1. doi: 10.1038/s42255-019-0120-1. [published online ahead of print October 7, 2019]. [DOI] [PubMed] [Google Scholar]
- 24.Forsström S, et al. Fibroblast growth factor 21 drives dynamics of local and systemic stress responses in mitochondrial myopathy with mtDNA deletions. Cell Metab. doi: 10.1016/j.cmet.2019.08.019. doi: 10.1016/j.cmet.2019.08.019. [published online ahead of print September 9, 2019]. [DOI] [PubMed] [Google Scholar]
- 25.Kauppila JHK, et al. A phenotype-driven approach to generate mouse models with pathogenic mtDNA mutations causing mitochondrial disease. Cell Rep. 2016;16(11):2980–2990. doi: 10.1016/j.celrep.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue K, et al. Generation of mice with mitochondrial dysfunction by introducing mouse mtDNA carrying a deletion into zygotes. Nat Genet. 2000;26(2):176–181. doi: 10.1038/82826. [DOI] [PubMed] [Google Scholar]
- 27.Tyynismaa H, et al. Mutant mitochondrial helicase Twinkle causes multiple mtDNA deletions and a late-onset mitochondrial disease in mice. Proc Natl Acad Sci U S A. 2005;102(49):17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trifunovic A, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 29.Tyynismaa H, Suomalainen A. Mouse models of mitochondrial DNA defects and their relevance for human disease. EMBO Rep. 2009;10(2):137–143. doi: 10.1038/embor.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruzzenente B, Rötig A, Metodiev MD. Mouse models for mitochondrial diseases. Hum Mol Genet. 2016;25(R2):R115–R122. doi: 10.1093/hmg/ddw176. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X, et al. Progressive loss of mitochondrial DNA in thymidine kinase 2-deficient mice. Hum Mol Genet. 2008;17(15):2329–2335. doi: 10.1093/hmg/ddn133. [DOI] [PubMed] [Google Scholar]
- 32.Akman HO, et al. Thymidine kinase 2 (H126N) knockin mice show the essential role of balanced deoxynucleotide pools for mitochondrial DNA maintenance. Hum Mol Genet. 2008;17(16):2433–2440. doi: 10.1093/hmg/ddn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Littarru GP, Tiano L. Bioenergetic and antioxidant properties of coenzyme Q10: recent developments. Mol Biotechnol. 2007;37(1):31–37. doi: 10.1007/s12033-007-0052-y. [DOI] [PubMed] [Google Scholar]
- 34.Khan NA, et al. mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 2017;26(2):419–428.e5. doi: 10.1016/j.cmet.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol. 2014;217(pt 1):137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351(6274):728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suomalainen A, et al. FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: a diagnostic study. Lancet Neurol. 2011;10(9):806–818. doi: 10.1016/S1474-4422(11)70155-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lehtonen JM, et al. FGF21 is a biomarker for mitochondrial translation and mtDNA maintenance disorders. Neurology. 2016;87(22):2290–2299. doi: 10.1212/WNL.0000000000003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quirós PM, et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol. 2017;216(7):2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.GTEx Consortium. et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andreux PA, et al. Systems genetics of metabolism: the use of the BXD murine reference panel for multiscalar integration of traits. Cell. 2012;150(6):1287–1299. doi: 10.1016/j.cell.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collaborative Cross Consortium. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190(2):389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saada A, Shaag A, Mandel H, Nevo Y, Eriksson S, Elpeleg O. Mutant mitochondrial thymidine kinase in mitochondrial DNA depletion myopathy. Nat Genet. 2001;29(3):342–344. doi: 10.1038/ng751. [DOI] [PubMed] [Google Scholar]
- 44.Siegmund SE, et al. Low-dose rapamycin extends lifespan in a mouse model of mtDNA depletion syndrome. Hum Mol Genet. 2017;26(23):4588–4605. doi: 10.1093/hmg/ddx341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Kim SG, Blenis J. Rapamycin: one drug, many effects. Cell Metab. 2014;19(3):373–379. doi: 10.1016/j.cmet.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chella Krishnan K, et al. Integration of multi-omics data from mouse diversity panel highlights mitochondrial dysfunction in non-alcoholic fatty liver disease. Cell Syst. 2018;6(1):103–115.e7. doi: 10.1016/j.cels.2017.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, et al. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell. 2014;158(6):1415–1430. doi: 10.1016/j.cell.2014.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Archer TC, et al. Proteomics, post-translational modifications, and integrative analyses reveal molecular heterogeneity within medulloblastoma subgroups. Cancer Cell. 2018;34(3):396–410.e8. doi: 10.1016/j.ccell.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzawa M, Noguchi K, Nishi K, Kozuka-Hata H, Oyama M, Ui-Tei K. Comprehensive identification of nuclear and cytoplasmic TNRC6A-associating proteins. J Mol Biol. 2017;429(21):3319–3333. doi: 10.1016/j.jmb.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 50.Bayraktar EC, et al. MITO-Tag Mice enable rapid isolation and multimodal profiling of mitochondria from specific cell types in vivo. Proc Natl Acad Sci U S A. 2019;116(1):303–312. doi: 10.1073/pnas.1816656115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell. 2015;162(3):540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulligan MK, Mozhui K, Prins P, Williams RW. GeneNetwork: a toolbox for systems genetics. Methods Mol Biol. 2017;1488:75–120. doi: 10.1007/978-1-4939-6427-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams EG, et al. Systems proteomics of liver mitochondria function. Science. 2016;352(6291):aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fagny M, et al. Exploring regulation in tissues with eQTL networks. Proc Natl Acad Sci U S A. 2017;114(37):E7841–E7850. doi: 10.1073/pnas.1707375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Zheng J, Wang Z, Li H, Deng M. Inferring gene-disease association by an integrative analysis of eQTL genome-wide association study and protein-protein interaction data. Hum Hered. 2018;83(3):117–129. doi: 10.1159/000489761. [DOI] [PubMed] [Google Scholar]
- 56.Kogelman LJ, et al. An integrative systems genetics approach reveals potential causal genes and pathways related to obesity. Genome Med. 2015;7:105. doi: 10.1186/s13073-015-0229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:article 17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 58.Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: state of the art. J Biopharm Stat. 2010;20(2):281–300. doi: 10.1080/10543400903572753. [DOI] [PubMed] [Google Scholar]
- 59.Elo LL, Schwikowski B. Analysis of time-resolved gene expression measurements across individuals. PLoS One. 2013;8(12):e82340. doi: 10.1371/journal.pone.0082340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xing L, Guo M, Liu X, Wang C, Wang L, Zhang Y. An improved Bayesian network method for reconstructing gene regulatory network based on candidate auto selection. BMC Genomics. 2017;18(S9):844. doi: 10.1186/s12864-017-4228-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. doi: 10.1155/2018/1591878. Beretta S, Castelli M, Gonçalves I, Henriques R, Ramazzotti D. Learning the structure of bayesian networks: a quantitative assessment of the effect of different algorithmic schemes. Complexity . [DOI]
- 62.Fiehn O. Metabolomics — the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1–2):155–171. [PubMed] [Google Scholar]
- 63.Roede JR, Park Y, Li S, Strobel FH, Jones DP. Detailed mitochondrial phenotyping by high resolution metabolomics. PLoS One. 2012;7(3):e33020. doi: 10.1371/journal.pone.0033020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sreekumar A, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457(7231):910–914. doi: 10.1038/nature07762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 65.Dubuis S, Ortmayr K, Zampieri M. A framework for large-scale metabolome drug profiling links coenzyme A metabolism to the toxicity of anti-cancer drug dichloroacetate. Commun Biol. 2018;1:101. doi: 10.1038/s42003-018-0111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mina SG, Alaybeyoglu B, Murphy WL, Thomson JA, Stokes CL, Cirit M. Assessment of drug-induced toxicity biomarkers in the brain microphysiological system (MPS) using targeted and untargeted molecular profiling. Front Big Data. 2019;2:23. doi: 10.3389/fdata.2019.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11(1):395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78(3):779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 69.Myers OD, Sumner SJ, Li S, Barnes S, Du X. Detailed investigation and comparison of the XCMS and MZmine 2 chromatogram construction and chromatographic peak detection methods for preprocessing mass spectrometry metabolomics data. Anal Chem. 2017;89(17):8689–8695. doi: 10.1021/acs.analchem.7b01069. [DOI] [PubMed] [Google Scholar]
- 70.Myint L, Kleensang A, Zhao L, Hartung T, Hansen KD. Joint bounding of peaks across samples improves differential analysis in mass spectrometry-based metabolomics. Anal Chem. 2017;89(6):3517–3523. doi: 10.1021/acs.analchem.6b04719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suomi T, Seyednasrollah F, Jaakkola MK, Faux T, Elo LL. ROTS: an R package for reproducibility-optimized statistical testing. PLoS Comput Biol. 2017;13(5):e1005562. doi: 10.1371/journal.pcbi.1005562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chagoyen M, Pazos F. MBRole: enrichment analysis of metabolomic data. Bioinformatics. 2011;27(5):730–731. doi: 10.1093/bioinformatics/btr001. [DOI] [PubMed] [Google Scholar]
- 74.Xia J, Wishart DS. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38(Web Server issue):W71–W77. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21(9):1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 76.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chong J, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamburov A, Cavill R, Ebbels TMD, Herwig R, Keun HC. Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics. 2011;27(20):2917–2918. doi: 10.1093/bioinformatics/btr499. [DOI] [PubMed] [Google Scholar]
- 79.Marco-Ramell A, et al. Evaluation and comparison of bioinformatic tools for the enrichment analysis of metabolomics data. BMC Bioinformatics. 2018;19(1):1. doi: 10.1186/s12859-017-2006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wishart DS, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim S, et al. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hastings J, et al. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44(D1):D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guijas C, et al. METLIN: a technology platform for identifying knowns and unknowns. Anal Chem. 2018;90(5):3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swainston N, et al. Recon 2.2: from reconstruction to model of human metabolism. Metabolomics. 2016;12:109. doi: 10.1007/s11306-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brunk E, et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36(3):272–281. doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Altman T, Travers M, Kothari A, Caspi R, Karp PD. A systematic comparison of the MetaCyc and KEGG pathway databases. BMC Bioinformatics. 2013;14(1):112. doi: 10.1186/1471-2105-14-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frainay C, et al. Mind the gap: mapping mass spectral databases in genome-scale metabolic networks reveals poorly covered areas. Metabolites. 2018;8(3):51. doi: 10.3390/metabo8030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng L, et al. MetSigDis: a manually curated resource for the metabolic signatures of diseases. Brief Bioinformatics. 2019;20(1):203–209. doi: 10.1093/bib/bbx103. [DOI] [PubMed] [Google Scholar]
- 90.Gu C, Kim GB, Kim WJ, Kim HU, Lee SY. Current status and applications of genome-scale metabolic models. Genome Biol. 2019;20(1):121. doi: 10.1186/s13059-019-1730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Brien EJ, Monk JM, Palsson BO. Using genome-scale models to predict biological capabilities. Cell. 2015;161(5):971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Opdam S, Richelle A, Kellman B, Li S, Zielinski DC, Lewis NE. A systematic evaluation of methods for tailoring genome-scale metabolic models. Cell Syst. 2017;4(3):318–329.e6. doi: 10.1016/j.cels.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith AC, Eyassu F, Mazat JP, Robinson AJ. MitoCore: a curated constraint-based model for simulating human central metabolism. BMC Syst Biol. 2017;11(1):114. doi: 10.1186/s12918-017-0500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boczonadi V, et al. Mitochondrial oxodicarboxylate carrier deficiency is associated with mitochondrial DNA depletion and spinal muscular atrophy–like disease. Genet Med. 2018;20(10):1224–1235. doi: 10.1038/gim.2017.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cuperlovic-Culf M. Machine learning methods for analysis of metabolic data and metabolic pathway modeling. Metabolites. 2018;8(1):4. doi: 10.3390/metabo8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Mol Cell. 2015;58(4):699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buescher JM, et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Basler G, Fernie AR, Nikoloski Z. Advances in metabolic flux analysis toward genome-scale profiling of higher organisms. Biosci Rep. 2018;38(6):BSR20170224. doi: 10.1042/BSR20170224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cavill R, Jennen D, Kleinjans J, Briedé JJ. Transcriptomic and metabolomic data integration. Brief Bioinform. 2016;17(5):891–901. doi: 10.1093/bib/bbv090. [DOI] [PubMed] [Google Scholar]
- 100.Kapoore RV, Vaidyanathan S. Towards quantitative mass spectrometry-based metabolomics in microbial and mammalian systems. Philos Trans A Math Phys Eng Sci. 2016;374(2079):20150363. doi: 10.1098/rsta.2015.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duncan KD, Fyrestam J, Lanekoff I. Advances in mass spectrometry based single-cell metabolomics. Analyst. 2019;144(3):782–793. doi: 10.1039/C8AN01581C. [DOI] [PubMed] [Google Scholar]
- 102.Zahedi A, et al. Deep analysis of mitochondria and cell health using machine learning. Sci Rep. 2018;8(1):16354. doi: 10.1038/s41598-018-34455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harwig MC, et al. Methods for imaging mammalian mitochondrial morphology: a prospective on MitoGraph. Anal Biochem. 2018;552:81–99. doi: 10.1016/j.ab.2018.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ludwig LS, et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell. 2019;176(6):1325–1339.e22. doi: 10.1016/j.cell.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yilmaz LS, Walhout AJ. A Caenorhabditis elegans genome-scale metabolic network model. Cell Syst. 2016;2(5):297–311. doi: 10.1016/j.cels.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]