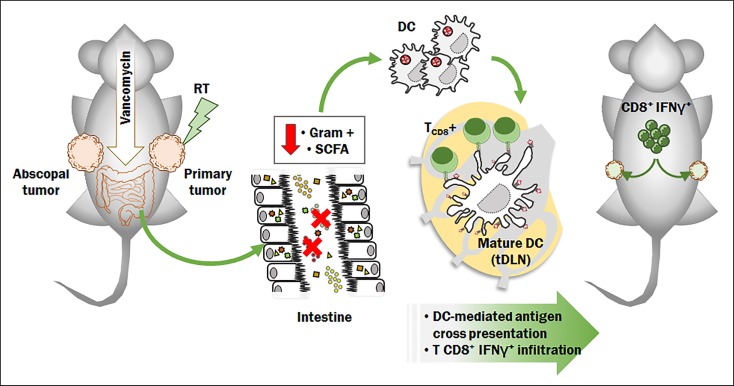
Abstract
Alterations in gut microbiota impact the pathophysiology of several diseases, including cancer. Radiotherapy (RT), an established curative and palliative cancer treatment, exerts potent immune modulatory effects, inducing tumor-associated antigen (TAA) cross-priming with antitumor CD8+ T cell elicitation and abscopal effects. We tested whether the gut microbiota modulates antitumor immune response following RT distal to the gut. Vancomycin, an antibiotic that acts mainly on gram-positive bacteria and is restricted to the gut, potentiated the RT-induced antitumor immune response and tumor growth inhibition. This synergy was dependent on TAA cross presentation to cytolytic CD8+ T cells and on IFN-γ. Notably, butyrate, a metabolite produced by the vancomycin-depleted gut bacteria, abrogated the vancomycin effect. In conclusion, depletion of vancomycin-sensitive bacteria enhances the antitumor activity of RT, which has important clinical ramifications.
Keywords: Immunology, Oncology
Keywords: Antigen presentation, Dendritic cells, Radiation therapy
Introduction
Radiotherapy (RT) is a well-established antitumor treatment, with more than 50% of newly diagnosed cancer patients with solid tumors receiving RT at some point during their treatment, usually in combination with chemotherapy (1). High-dose, hypofractionated radiotherapy was initially introduced for intracranial tumors as stereotactic radiosurgery and was later extended to extracranial sites as stereotactic body RT (SBRT) (8–30 Gy/fraction) (2).
Hypofractionated, high-dose RT can generate immunogenic cell death (ICD) and elicits adaptive antitumor immunity through tumor-associated antigen (TAA) cross-priming (3–5). Tumor cells succumbing to ICD stimulate antigen presenting cells (APCs), such as dendritic cells (DCs), to efficiently engulf tumor antigens and cross-present them to CD8+ cytotoxic T lymphocytes (6, 7). This process culminates in the generation of a specific immune response capable of impacting distant nonirradiated tumors, known as the abscopal effect (8).
Combination of RT with immunotherapy has recently garnered considerable clinical interest due to its potential for transforming RT from a modality used to treat localized disease to a modality used to treat systemic malignancies (9). However, several issues concerning dosage, timing, patient selection, and toxicity need to be resolved (10–13). Moreover, combinations of immunotherapy with other modalities are curative in only a fraction of patients, raising the question of whether important regulators of RT-mediated antitumor immune responses are yet to be discovered.
The intestinal microbiota plays a critical role in secondary and tertiary lymphoid structure: germ-free mice that lack microbiota have smaller Peyers patches and a reduced number of CD4+ T cells and IgA-producing plasma cells. Indeed, early studies have identified significantly impaired host immune responses to pathogens in mice raised under germ-free conditions (14). Moreover, a direct correlation exists between the presence of specific bacteria in the gut microbiota and T cell development and differentiation (15, 16).
Since the gut microbiota impacts the function of T cells and other immune cell subsets, both within the gut-associated lymphoid tissue (GALT) and beyond, microbiota-derived signals have been shown to have broad roles in the regulation of several diseases and in their treatments, including cancer and its response to chemotherapy (17–20). Therefore, we hypothesized that perturbation of the gut microbiota can modulate the antitumor effects of ablative RT and influence antigen-specific antitumor responses. In line with this hypothesis, we recently demonstrated that the gut microbiota impacts the antitumor effects of adoptive cell therapy (ACT) (21).
In the present manuscript we describe the impact of the gut microbiota composition on hypofractionated RT and demonstrate that the microbiota can be altered to improve RT efficacy. We selected vancomycin, a glycopeptide antibiotic that acts mainly on gram-positive bacteria, as an agent to specifically perturb the gut microbiota, because vancomycin is very poorly absorbed after oral administration and does not enter the systemic circulation (22, 23).
Using both a melanoma model (24, 25) and a HPV E6/7-expressing lung and cervical cancer model (26), we found that the antitumor effects elicited by RT in tumor-bearing mice are positively impacted by treatment with vancomycin. Notably, this effect was strictly dependent on a functioning immune system and was abrogated by the intake in trans of sodium butyrate, a key metabolite of gram-positive bacteria. Our results suggest that alterations in the gram-positive gut bacterial community can elicit tumor microenvironment remodeling, mediate increased antigen presentation in draining lymph nodes, and improve RT antitumor efficacy. Since vancomycin is a widely used clinical agent with a relatively safe profile, these findings raise the potential for utilizing this antibiotic to enhance the effects of RT in patients with cancer.
Results
Oral vancomycin treatment enhances the direct and abscopal antitumor effects of hypofractionated RT in preclinical melanoma and lung/cervical tumor models.
Given the role of the gut microbiota in modulating immune cells that are also known to be involved in the response to RT, we examined whether the microbiota-regulated systemic immune response contributes to the RT-mediated anticancer immune response. The effects of oral vancomycin treatment are localized and impact the gut microbiota directly without any known systemic effects (21–23).
Vancomycin (mostly targeting gram-positive bacteria) or a neomycin/metronidazole (Neo/Met) regimen (mostly targeting gram-negative bacteria) was administered orally in C57/Bl6 mice (27). The following day, the mice were challenged subcutaneously with B16-OVA or tissue culture number 1 (TC-1) tumor cells. Each cell line was injected on both flanks of each mouse, and when tumors were approximately 50 mm3 (10–11 days), RT was delivered on 1 tumor on each mouse with a 21 Gy single fraction dose using an XRAD320iX irradiator (28), while taking precautions to avoid irradiating any areas of the gut by shielding nonirradiated areas.
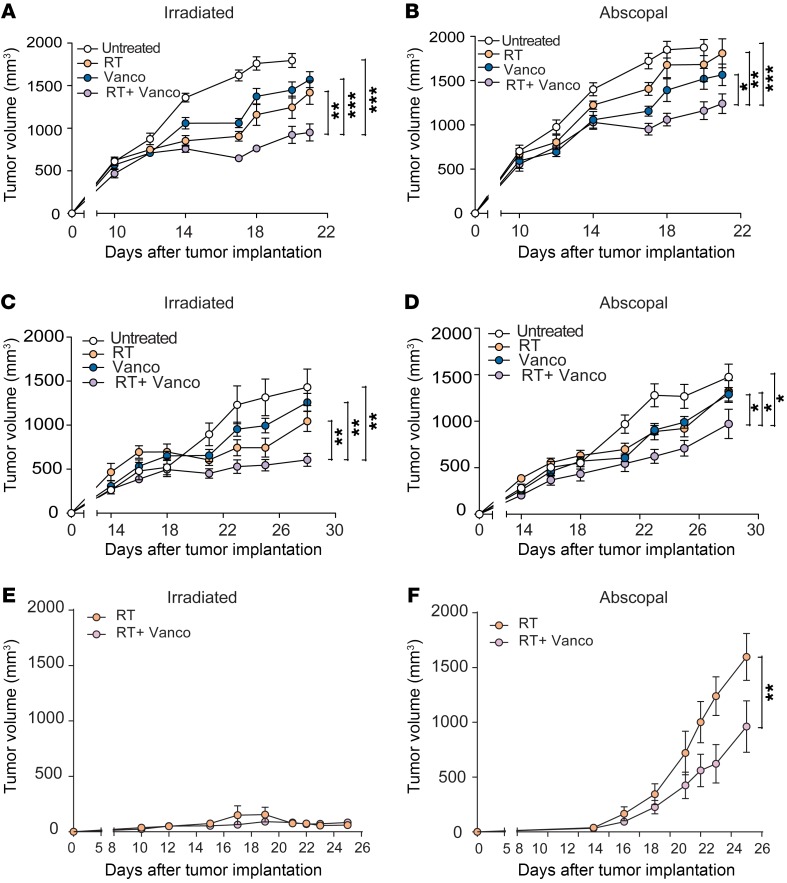
Previously published work has shown that in breast and colorectal cancers, cross-priming can be affected in a dose-dependent manner due to the activation of TREX1 nucleases at doses above 8 Gy (13). Because we used melanoma and lung cancer, we chose to test 2 RT modalities implemented in clinic (29–31) relevant to such mechanism: one consisting of 3 × 8 Gy fractionated RT and a single dose of 21 Gy. As demonstrated in Supplemental Figure 1 (supplemental material available online with this article; https://doi.org/10.1172/JCI124332DS1), the 2 treatments produced similar tumor control in this model after 3 weeks. Therefore, in subsequent experiments with this tumor model, we maintained the 1 × 21 Gy regimen (Supplemental Figure 1). We observed in both the B16-OVA (Figure 1A, and Supplemental Figure 2) and TC-1 (Figure 1C) tumor models that the addition of vancomycin with RT as a combination therapy produced antitumor effects that were greater than the antitumor effects mediated by either vancomycin alone or RT alone as single modalities (P < 0.001, P < 0.001, P = 0.0018 when the RT+vancomycin group was compared with untreated, vancomycin, and RT respectively). In contrast, preadministration of gram-negative targeted antibiotics (i.e., Neo/Met) did not augment the antitumor effects of RT (Supplemental Figure 3). The nonirradiated (abscopal) tumors in RT+vancomycin combination-treated mice cohorts in the B16-OVA (Figure 1B,) and TC-1 (Figure 1D) models also experienced delayed growth compared with control untreated and RT-alone-treated cohorts. The combined antitumor effects for RT+vancomycin was consistent in B16-OVA-bearing mice sourced from a different vendor (Jackson laboratories) (Supplemental Figure 4). Of note, when the 21-Gy dose of RT was delivered directly to B16-OVA tumors sized less than 50 mm3, RT controlled primary tumor growth with or without vancomycin (Figure 1E), which is consistent with our previous results. However, in the abscopal tumor, growth was significantly decreased with the addition of vancomycin to RT compared with RT alone (P < 0.01) (Figure 1F). Cumulatively, these results demonstrate that vancomycin enhances RT-mediated antitumor effects both locally at the tumor site and systemically via abscopal effects.
Figure 1. Oral vancomycin treatment enhances the direct and abscopal antitumor effects of hypofractionated RT in a preclinical melanoma and lung/cervical tumor model.
Shown are tumor volumes from control (untreated), vancomycin treatment alone (Vanco), RT treatment alone (RT), or vancomycin plus RT combination treatment (RT+Vanco) on irradiated tumors derived from the B16-OVA melanoma model (A) or from the TC-1 lung/cervical cancer model (C) and abscopal tumors (B and D, respectively). (E) B16-OVA tumor volumes of mice treated with RT alone, or RT and vancomycin (F) nonirradiated (abscopal) B16-OVA tumor volumes from mice treated with RT alone, or RT and vancomycin. n = 5 to 14 mice per group. Data are representative of at least 2 independent experiments. Mean ± SEM are shown. Statistical significance was assessed by 2-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001.
The effects of vancomycin are abrogated in CD8-depleted animals.
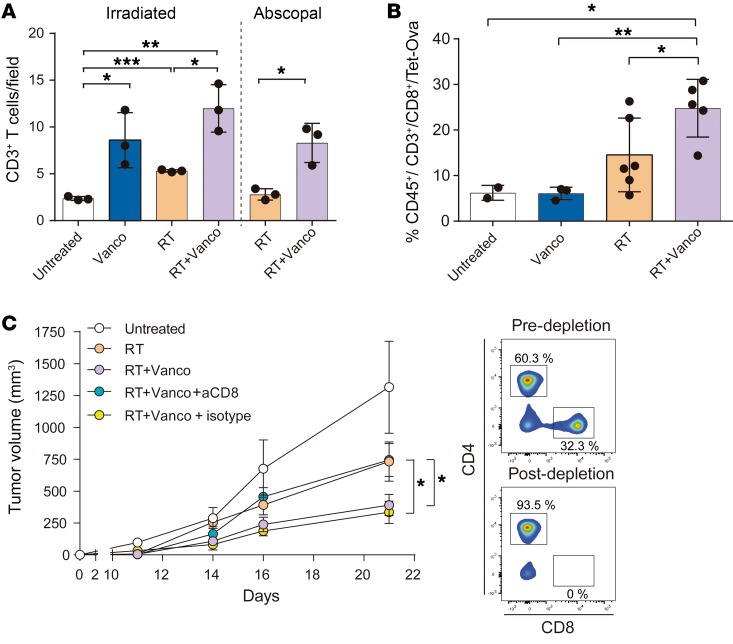
Since T cells mediate both local and systemic antitumor immune response after RT (32, 33), we initially tested the overall presence of infiltrating CD3+ T cells within tumor sections from both irradiated and abscopal B16-OVA tumors by immunohistochemistry. We found a measurable CD3+ T cell infiltrate within B16-OVA primary tumors that were treated with vancomycin alone, RT alone, or with the combination of RT+vancomycin (P < 0.05, P < 0.01, and P < 0.001 respectively) (Figure 2A). B16-OVA abscopal tumors treated with RT alone or with the combination of RT+vancomycin also had measurable infiltrating CD3+ T cells, and CD3+ T cell infiltration was significantly higher following RT+vancomycin combination treatment compared with RT treatment alone (P < 0.05) (Figure 2A).
Figure 2. The effects of vancomycin are abrogated in CD8-depleted mice.
(A) Quantification of CD3+ cell infiltration by immunohistochemistry of B16-OVA–derived primary tumor sections from untreated, vancomycin treatment alone (Vanco), RT treatment alone (RT), or vancomycin plus RT combination treatment (RT+Vanco), and of abscopal tumor sections from RT abscopal or RT+Vanco abscopal combination treated. Mean ± SEM are shown. Statistical significance was assessed by Tukey’s test. (B) Primary tumors from each treatment group were digested and individual cells were analyzed by flow cytometry to determine the percentage of OVA-specific CD8+ T cells (CD45+/CD3+/CD8+/TET-OVA). Mean ± SEM are shown. Statistical significance was assessed by Tukey’s test. (C) Tumor growth rates in CD8-depleted mice. n = 5 to 10 mice per group. Mean ± SEM are shown. Statistical significance was assessed by 2-way ANOVA. Data are representative of at least 2 independent experiments. *P < 0.05, **P < 0.001, ***P < 0.01.
In addition to analyzing the numbers of infiltrating CD3+ T cells, we also characterized the antigen specificity of the tumor infiltrating CD8+ T cells by using OVA MHC1 tetramer. Tumors from mice treated with vancomycin alone, RT alone, or the combination of RT+vancomycin were digested, and individual cells were assayed for tetramer staining by flow cytometry. The results showed that tumors from mice treated with the combination of RT+vancomycin had a significantly higher number of OVA-specific CD8+ T cells (i.e., percentage of CD45+/CD3+/CD8+/TET-OVA cells) compared with control untreated tumors, vancomycin alone–treated tumors, and RT alone–treated tumors (P = 0.0117, P = 0.0028, and P = 0.0472, respectively) (Figure 2B). Therefore, vancomycin treatment increased the number of RT-generated tumor cytolytic CD8+ T cells within the tumor. Because T regulatory cells (Tregs) have been reported to be influenced by microbiota composition (15, 34) and because Tregs influence RT antitumor effects (35, 36), we evaluated the impact of the vancomycin treatment on Tregs in the tumor. In our model, the number of intratumoral Tregs were only minimally affected by vancomycin (Supplemental Figure 5).
To determine whether a causal link exists between the increase in CD8+ T cell infiltration and the improved antitumor response in mice receiving RT+vancomycin combination treatment, we selectively depleted CD8+ T cells by pretreating the mice with an anti-CD8 monoclonal antibody (37). Depletion of CD8+ cells prior to the combination treatment of RT+vancomycin abrogated the antitumor effects of the combination treatment (Figure 2C, Supplemental Figure 6), demonstrating that the CD8+ T cell population is required for mediating the observed antitumor effects (P = 0.0120). Taken together, the results of these experiments demonstrate that vancomycin enhances the systemic antitumor effects mediated by RT via a mechanism that requires a cytolytic CD8+ T cell population.
Antitumor effects of RT+vancomycin combination treatment are IFN-γ dependent.
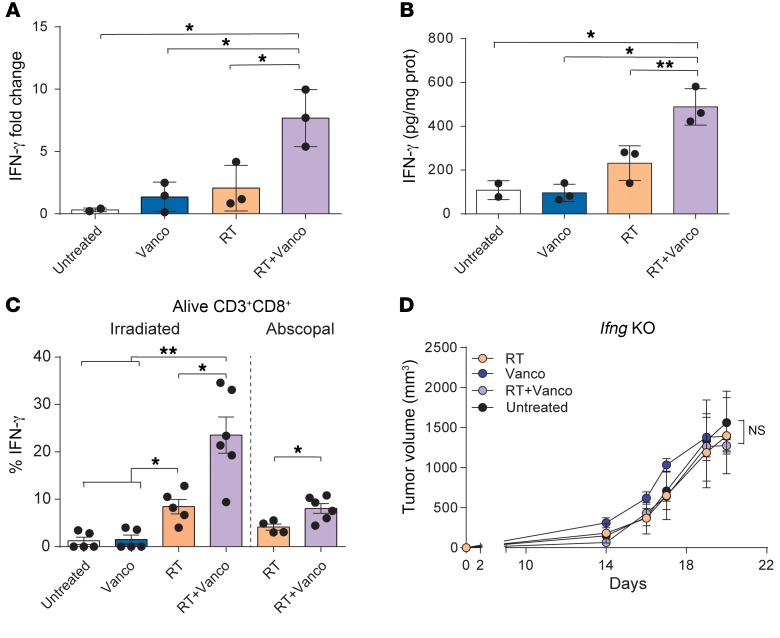
Several cytokines contribute to the priming of the effector functions of cytotoxic T cells, and IFN-γ is known to play a critical role in both differentiation and effector functions of CD8+ cytolytic T cells in the antitumor immune response (38, 39). To determine whether IFN-γ is involved in the antitumor effects of RT+vancomycin combination treatment in the tumor microenvironment, we measured intratumoral expression of IFN-γ in B16-OVA tumors at 5 days after radiation. We found that Ifng mRNA expression levels were significantly increased in the RT+vancomycin combination treatment group compared with treatment with either vancomycin alone or RT alone (P = 0.0131 and P = 0.0293, respectively) (Figure 3A). Previously, we reported that vancomycin in combination with T cell therapy induces the expansion of CD8+ DCs and affects the level of IL-12; in turn, IL-12 stimulates the production of IFN-γ and promotes both Th1 differentiation and CTL activity (21, 40). Hence, we assessed the levels of such cytokine in the tumors of treated mice. In agreement with the IFN-γ results, we found increased levels of Il12 expression (Supplemental Figure 7). In parallel, we found that IFN-γ protein expression levels were significantly increased in the RT+vancomycin combination treatment group (P = 0.0028 and P = 0.0179, respectively, for vancomycin and RT) (Figure 3B). IFN-γ is produced by T cells and is a signature of the Th1 phenotype (41). To assess the functionality of the antigen-specific tumor–infiltrating CD8+ T cells, we dissociated tumor cell suspensions, incubated with Kb-OVA (SIINFEKL) peptide overnight, and measured intracellular IFN-γ within the CD8+ T cell fraction. Tumors treated with RT+vancomycin combination had a significantly increased number of IFN-γ+ CD8+ T cells compared with either of the single treatments (P < 0.05 for both comparisons) (Figure 3C). To further test the role of IFN-γ in the antitumor response, we used Ifng knockout mice. We found that in B16-OVA tumor–challenged Ifng knockout mice, the enhancement of the radiation effects by vancomycin was ablated (P = 0.5601 and P = 0.0911 compared with RT and vancomycin, respectively) (Figure 3D). These results indicate that vancomycin remodels the tumor microenvironment and increases the functionality of tumor-infiltrating OVA-specific CD8+ T cells, and that IFN-γ is required to augment the RT-induced immune effect against the tumor.
Figure 3. Antitumor effects of vancomycin plus RT combination treatment are IFN-γ dependent.
(A) Ifng mRNA expression levels in B16-OVA primary tumors collected at 5 days after radiation (21 Gy). (B) ELISA analysis of IFN-γ protein expression levels in tumor lysates 5 days after radiation (21 Gy). (C) Intracellular IFN-γ expression in infiltrating T cells after overnight Kb-OVA peptide stimulation. Mean ± SEM are shown. Statistical significance was assessed by Tukey’s test. (D) The antitumor effects of vancomycin+RT combination treatment are lost in Ifng-KO mice. n = 5 to 10 mice per group. Mean ± SEM are shown. Statistical significance was assessed by 2-way ANOVA. Data are representative of at least 2 independent experiments. *P < 0.05, **P < 0.01.
Vancomycin treatment increases local TAA cross-presentation and antigen recognition in tumors.
Recent studies have shown that appropriate dosing and scheduling of high-dose RT can lead to increased T cell priming and T cell–dependent tumor regression (28). We therefore reasoned that enhanced antigen presentation may be a driver of enhanced CD8+ T cell activity.
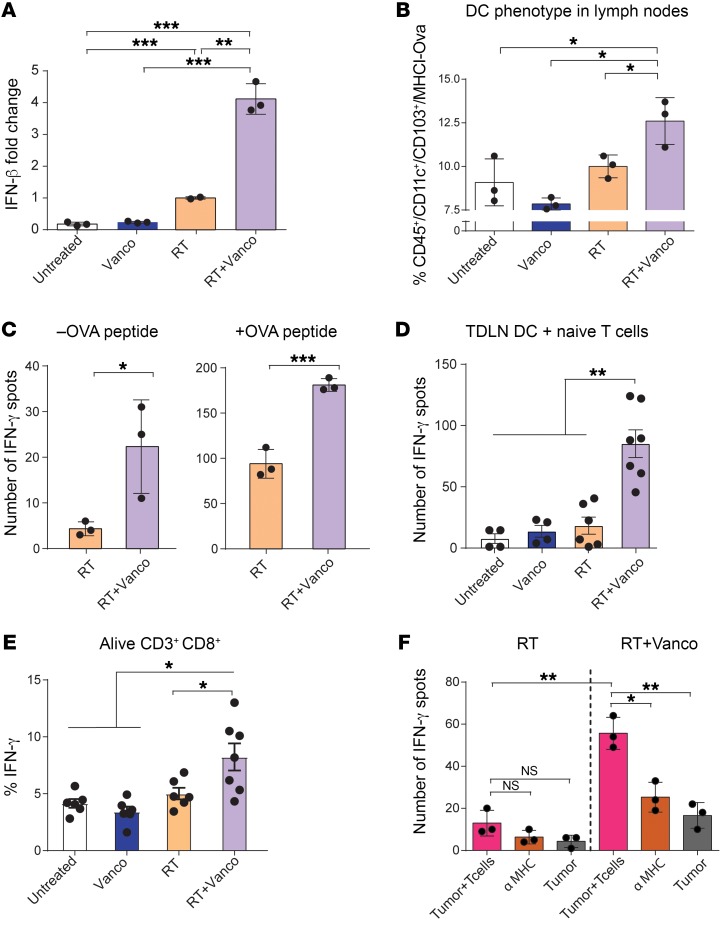
Hypofractionated RT activates the cross-priming capacity of tumor-infiltrating DCs, with IFN-β serving as the master regulator of tumor antigen cross-presentation (42). We examined the expression of Ifnb1 in tumor-draining lymph nodes (TDLNs) and tumors after RT of B16-OVA tumors. Concordantly, vancomycin treatment synergistically enhanced Ifnb1 gene expression compared with RT alone in TDLNs (P = 0.0032) (Figure 4A) and in the tumor (Supplemental Figure 8).
Figure 4. Vancomycin treatment increases local TAA cross-presentation and antigen recognition in tumors.
(A) Ifnb1 mRNA expression levels in TDLNs 1 day after irradiation. (B) Anti-MHC1 (Kb)-SL8 OVA peptide staining on tumor-infiltrating CD11c+ CD103+ DCs 5 days after RT. (C) IFN-γ ELISPOT assay plated with TDLN single-cell suspension and OT1 T cells (1:5 TDLN cells/T cells) in absence (left) or presence (right) of OVA peptide. Cells were harvested 5 days after RT treatment. (D) Coculture of purified CD11c+ DCs from TDLNs from each treatment group were incubated overnight with naive OT1 T cells in an IFN-γ ELISPOT assay (1:10 DCs/T cells). (E) Percentage of IFN-γ expression from overnight OT1 cells cocultured with DCs from mice treated with each therapeutic approach. (F) B16-OVA tumors from mice treated with RT alone or with the RT and vancomycin (RT+VANCO) combination treatment were dissociated and plated with OT1 cells in an IFN-γ ELISPOT plate for 24 hours. n = 5 to 10 mice per group. Data are representative of at least 2 independent experiments. Mean ± SEM are shown. Statistical significance was assessed by Tukey’s test. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we directly assessed the level of antigen presentation in MHC class I by staining the TDLN cell suspension with an antibody specific for the MHC1 (Kb)-SL8 OVA peptide, and found a significant increase in DCs from the RT+vancomycin combination treatment group (P = 0.0396) when compared with RT (Figure 4B and Supplemental Figure 9). To confirm an increase in functional antigen presentation in TDLNs of treated animals, we assessed the production of IFN-γ by coculturing OT1 T cells with APCs from treated mice. Given that, in vivo, the priming of reactive T cells occurs mostly in the lymph nodes, and in order to maintain the complexity of the lymphoid tissue in vitro, we used a whole TDLN cell suspension as a source for APCs. Specifically, we incubated a cell suspension of TDLNs from B16-OVA tumors coming from mice treated with the RT+vancomycin combination or with RT alone, in the absence or presence of OVA peptide together with OT1 T cells in an IFN-γ ELISPOT assay (Figure 4C). In order to confirm that all the IFN-γ signals in the ELISPOT were produced by OT1 T cells, TDLNs were also seeded alone without any significant number of spots detected. However, when cocultured with OT1 T cells, we observed a significant increase in the number of IFN-γ–producing cells with TDLNs derived from the mice treated with RT+vancomycin combination compared with TDLNs from mice treated with RT alone (P = 0.0398 and P = 0.0010 for assays in absence or presence of OVA peptide, respectively) (Figure 4C). Next, to confirm priming of naive OVA-specific T cells by CD11c+ DCs, we purified the CD11c+ population from the TDLNs and set up an IFN-γ ELISPOT assay where we cocultured CD11c+ DCs from TDLNs from each treatment group with naive OT1 CD3+ T cells. Our results demonstrated that vancomycin in combination with RT enhances the local antigen presentation functionality of CD11c+ DCs (Figure 4D). Moreover, using intracellular staining, we demonstrated that purified CD11c+ cells isolated from TDLNs from the RT+vancomycin group (but not the other groups) specifically increase IFN-γ production in CD8+ T cells from unprimed (naive) OT1 cells (i.e., percentage of IFN-γ–producing CD8+ T cells) (P<0.05) (Figure 4E).
Finally, we dissociated B16-OVA tumors from treated mice and coincubated the cells with OT1 OVA-specific CD8+T cells purified from OVA transgenic OT1 mice and assayed for IFN-γ–producing cells by ELISPOT. The combination treatment with RT+vancomycin augmented IFN-γ secretion compared with RT treatment alone (P = 0.0016) (Figure 4F). Importantly, MHC1 blocking antibodies significantly reduced IFN-γ–producing cells, demonstrating MHC1-dependent activation of CD8+ T cells (P = 0.0073) (Figure 4F). Similar results were observed with the TC-1 tumor model (Supplemental Figure 10). Collectively, our data indicate that vancomycin enhances local antigen presentation, promoting increased tumor recognition by antigen-specific T cells.
Short-chain fatty acids impair APC activity in vitro and abrogate vancomycin-enhanced RT antitumor activity in vivo.
The gut microbiota aids host digestion and generates a large repertoire of metabolites, including short-chain fatty acids (SCFAs) that constitute the major products of bacterial fermentation (43). SCFAs are saturated aliphatic organic acids that contain between 1–6 carbon atoms of which acetate (2 carbon atoms [C2]), propionate (3 carbon atoms [C3]), and butyrate (4 carbon atoms [C4]) represent 95% of total SCFAs present in the intestine (44, 45). Interestingly, SCFAs can directly modulate cytokine production and impact DC functions, including IL-12 transcription and secretion (46).
Given the central role of DCs in T cell priming and the role of CD8+ T cells in RT+vancomycin combination, we examined the potential contributions of C3 and C4 in DC APC function in vitro.
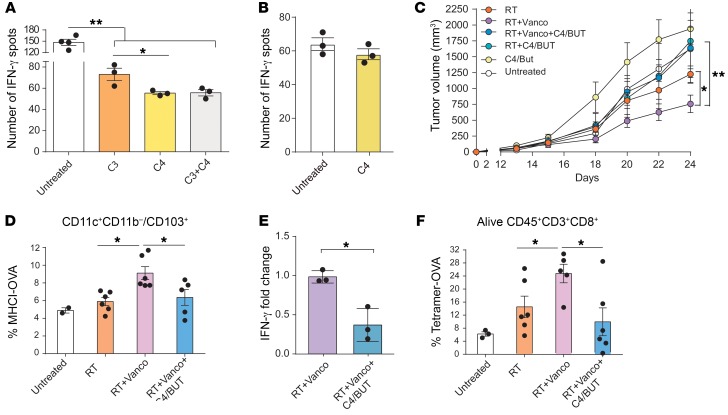
For this, we generated bone marrow DCs (bmDCs) by culturing bone marrow cells in GM-CSF/IL-4 for 7 days and adding C3 and/or C4 at 100 μM during OVA protein pulsing. The DCs pulsed with the full-length OVA protein were then cultured with T cells taken from OT1 mice in an IFN-γ ELISPOT assay. Our results demonstrate that the APC function of DCs was inhibited by C4 alone, as shown by measurements of decreased IFN-γ–secreting cells in bmDCs treated with C4 relative to untreated control. Interestingly, C3-treated DCs also exhibited a partial inhibition of antigen presentation when used alone but did not potentiate the inhibition induced by C4 alone (P = 0.0134) (Figure 5A). Therefore, we focused on the role of C4 in subsequent experiments. Next, we phenotypically characterized bmDCs exposed to SCFAs. We found that although overall CD11c+ population was not affected, their activation state, measured by CD86 expression, was reduced by the presence of SFCA (Supplemental Figure 11). To ensure that SCFAs were affecting primarily DCs and not T cells, we evaluated the direct impact of C4 on purified T cells by stimulating overnight T cells purified from splenocytes of healthy mice with αCD3/αCD28 antibodies in the presence or absence of C4. The presence of C4 during αCD3/αCD28 stimulation resulted in no significant effects on IFN-γ production by T cells (Figure 5B). In line with previously published results, these data confirm the impact of SCFAs on DC phenotype and inhibition of antigen presentation.
Figure 5. SCFAs impair APC activity in vitro and abrogate vancomycin-enhanced RT antitumor activity in vivo.
(A) Bone marrow–derived DCs (bmDCs) were either untreated, treated with 100 μM butyrate (C4), or treated with 100 μM propionate (C3) together with OVA protein (100 μg/mL) for 24 hours before they were plated to an ELISPOT plate with T cells from OT1 mice. (B) Purified T cells were in vitro–stimulated with aCD3/aCD28 in the presence or absence of 100 μM C4/butyrate in a IFN-γ ELISPOT plate. (C) In vivo effects on tumor growth in irradiated mice treated with vancomycin-containing drinking water with or without C4. Mean ± SEM are shown. Statistical significance was assessed by 2-way ANOVA. (D) Ifng mRNA expression levels in tumors from irradiated mice treated with vancomycin-containing drinking water with and without C4/butyrate. Mean ± SEM are shown. Statistical significance was assessed by Tukey’s test. (E) Flow cytometry analysis of OVA-presenting DCs and (F) CD3+CD8+Tet-OVA+ cell subsets. Mean ± SEM are shown. Statistical significance was assessed by Tukey’s test. n = 5 to 10 mice per group. Data are representative of at least 2 independent experiments. *P < 0.05, **P < 0.01.
We then tested whether C4/butyrate can negatively impact the synergy of RT+vancomycin combination therapy in vivo by supplying sodium butyrate to the drinking water of mice at the same time they were being treated with vancomycin. One day later, the mice were challenged with B16-OVA, and irradiated (21 Gy) when their tumors reached approximately 50 mm3. In agreement with our previous results, vancomycin enhanced the tumor-inhibitory effects of RT (P = 0.0307), but the addition of C4 abrogated this effect (P = not significant) (Figure 5C). To determine whether the effects of the C4 in vivo correlate with the observed impact in vitro, we enumerated OVA-presenting DCs in lymph nodes of mice and found a significant decrease of this population in mice treated with butyrate (Figure 5D). Moreover, we analyzed the mRNA expression levels of Ifng and Il12b in the tumors by real-time quantitative PCR (qPCR). Notably, the impact of vancomycin on Ifng and Il12b levels was reduced when C4 was included in the treatment regimen (Figure 5E and Supplemental Figure 12). Finally, the addition of butyrate to RT+vancomycin treatment reduced the percentage of intratumoral, antigen-specific T cells compared with RT+vancomycin (Figure 5F). Collectively, our results indicate that C4/butyrate produced by intestinal bacteria impairs the activity of APCs in vitro and interferes with the cross-priming activity and abrogates vancomycin-enhanced RT-mediated antitumor effects in vivo.
Vancomycin treatment alters short-chain fatty acid concentration and bacterial community composition.
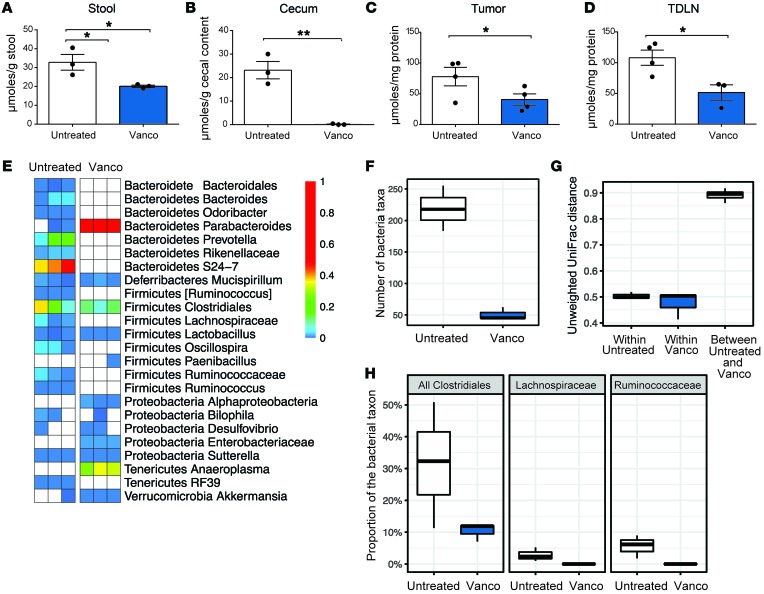
To determine whether a causal relationship exists between butyrate production and vancomycin treatment, we investigated whether vancomycin treatment decreased the levels of SCFAs in mouse feces and altered the proportion of bacteria producing SCFAs in the gut microbiota. Cecum contents and stools from untreated mice, mice treated with vancomycin alone, and mice treated with the combination of RT+vancomycin were collected, and C4 levels were analyzed. Vancomycin treatment alone or RT+vancomycin combination treatment significantly decreased the concentration of C4 relative to untreated controls (Figure 6, A and B).
Figure 6. Vancomycin treatment alters SCFA concentration and bacterial community composition.
(A) Concentration of C4 SCFAs in stool, (B) cecal contents, (C) tumor, and (D) TDLNs. Mean ± SEM are shown. Statistical significance was assessed by Tukey’s test. *P < 0.05, **P < 0.01. (E) Heatmap of bacterial taxon abundance in vancomycin-treated mice. White squares represent taxa not observed in the sample; n = 3 per group. (F) Bacterial community diversity as determined by 16S rRNA marker gene sequencing. (G) Dissimilarity of bacterial communities in stool from control and vancomycin-exposed mice. (H) Relative abundance of taxa known to contain butyrate-producing bacterial species. Data are representative of at least 2 independent experiments.
Since the SCFA concentrations in the blood and tissues have been reported to be substantially lower than in intestinal content (47), we examined the effects of vancomycin in both the TDLNs and the tumors by collecting tissue from tumor-bearing animals that were either untreated (control) or treated with vancomycin. We observed that in both tumor and TDLN tissues, vancomycin significantly decreased the amount of available C4 (Figure 6, C and D).
The bacterial population in the cecum has been reported to be more abundant in Firmicutes, the phylum of SCFA-producing gut bacteria, relative to feces (48). Therefore, we carried out 16S rRNA marker gene sequencing and confirmed that the levels of gram-positive SCFA-producing bacteria were substantially reduced following treatment with vancomycin alone. Fecal samples from mice treated with 0.5 g/L oral vancomycin were sequenced and compared with samples from untreated control mice. As expected, we found that vancomycin treatment induced major alterations in the bacterial community composition, including elimination of the majority of gram-positive taxa together with some impact on gram-negative taxa (Figure 6E). As a result, the number of bacterial taxa found in the vancomycin-treated group was 4 times lower than that of the untreated controls (P = 0.01) (Figure 6F). The composition of the bacterial community was altered consistently by vancomycin treatment, as indicated by an analysis of unweighted UniFrac distance, which is a measure of community dissimilarity between samples. Sample distances within the control and vancomycin treatment groups were drastically lower than the between-group distances (P < 0.001) (Figure 6G).
Vancomycin treatment eliminated the 2 major families of short-chain fatty acid–producing Clostridia: Ruminococcaceae and Lachnospiraceae (P < 0.01 for both families) (Figure 6H). Because some bacteria remained under the parent order of Ruminococcaceae and Lachnospiraceae after vancomycin treatment, we aligned the 16S sequence reads that were assigned to Clostridiales to a database of all characterized bacterial species and examined SCFA products attributed to the closest matching bacterial species using a 90% sequence similarity cut off. Based on our analyses, we determined that the vancomycin treatment group showed no evidence of remaining butyrate-producing bacteria from the Clostridiales order. Our data demonstrate that vancomycin treatment preferentially targets the gram-positive bacterial populations, including butyrate-producing bacteria, and decreases SCFA concentrations in fecal and tissues samples.
Discussion
In this study, we describe the impact of gut microbiota composition on hypofractionated RT delivered to sites distal to the gut, and demonstrate that the gut microbiota can be modulated to improve RT-mediated antitumor responses. Using both a B16-OVA melanoma model and a TC-1 lung/cervical cancer model (expressing HPV E6/7), we report that vancomycin pretreatment enhanced the antitumor effects of RT in tumor-bearing mice, whereas pretreatment with a neomycin and metronidazole (Neo/Met) regimen had no such effect. The improving effects of vancomycin in irradiated mice were confirmed using mice sourced from a second animal vendor, proving that our observation is not limited to a specific microbiota baseline. We also showed that the RT-enhancing effects of vancomycin treatment are IFN-γ– and CD8-dependent, and that vancomycin treatment increases antigen presentation by CD11c+ DCs in the TDLNs of RT-treated mice. Furthermore, we report that butyrate/C4-producing bacteria are eliminated by vancomycin treatment and that C4-mediated impairment of APC function abrogates vancomycin-enhanced RT antitumor activity. Collectively, our data suggest a new mechanism in which the vancomycin-mediated increase in RT-mediated efficacy involves the elimination of bacteria-producing SCFAs, which is accompanied by tumor microenvironment remodeling together with increased antigen presentation and cytotoxic T cell infiltration in the tumor.
Although an increase in the total number of T cells infiltrating the tumor in the vancomycin-alone group was observed, the OVA T cell–specific response was, unexpectedly, not improved in the same group, suggesting that in nontreated tumors, cross presentation may increase the recognition of other TAAs (49, 50).
Although the interplay between the intestinal microbiota and the immune system was originally described some time ago, it has recently regained momentum (51–55). Our findings using 2 distinct preclinical cancer models showed that the use of a specific antibiotic such as vancomycin can augment the antitumor effects of RT. Importantly, vancomycin is retained locally within the gut when administered orally (56), strongly suggesting that our observed phenotype is due to the local interactions between the gut microbiota and the immune system that results in long-range systemic effects. Previously, Viaud et al. (20) studied the impact of antibiotic treatment on the antitumor immune effects of cyclophosphamide. In contrast to our data, that study demonstrated that vancomycin administration abrogated the response to cyclophosphamide therapy. Interestingly, the study by Viaud et al. also showed that the antitumor immune response to cyclophosphamide therapy was mediated by the specific phenotype of Th17 cells (20). In contrast, our data show that the enhanced antitumor effects of RT+vancomycin combination treatment is dependent on IFN-γ and cytotoxic T cells. Moreover, Ivanov et al. (14) demonstrated that depletion of certain gram-positive bacteria, including SFB, influenced the local and systemic Th17 development, suggesting that there are different mechanisms involved in the 2 models.
Another notable difference between the results presented here and previous studies is that the delivery of hypofractionated localized RT largely avoids any direct effects on the gut, whereas chemotherapy, being systemic in nature, may also impact the gut microbiome, which can be a confounding factor in such comparisons. As demonstrated by previous work in chemotherapy and systemic-based RT, therapies such as total body irradiation (TBI) have a direct effect on gut microbiota and damage to the gut epithelial barrier, with possible transient intraperitoneal extraversion and inflammation, which can in turn influence tumor response (20, 57, 58). In our studies, RT is locally delivered to the tumor, avoiding direct damage to the gut epithelium, which parallels most clinical modalities involving radiotherapy.
One important aspect to consider when dealing with antiimmune responses to radiotherapy is the radiation dose and fractionation scheme. Vanpouille-Box et al. (13) recently showed that DNA exonuclease TREX1 is induced by radiation doses above 12–18 Gy and attenuates their immunogenicity by degrading DNA that accumulates in the cytosol upon radiation. Interestingly, in our studies we did not observe significant differences between 8 × 3-Gy and 1 × 21-Gy regimens. While we did not analyze TREX1 activation in our study, it is possible that these differences are due to the distinct tumor types used in the 2 studies (breast carcinoma vs. melanoma) as well as possible differences in the gut microbiota, as indicated by our data.
Recently, our group demonstrated in preclinical models that gut microbiota influence the impact of ACT, which is in line with the present results that vancomycin treatment increases the antitumor effects of ACT (21). This increase in the antitumor effects of ACT, in agreement with our current data in RT, also involved DCs, which, in the case of ACT, sustained systemic adoptively transferred antitumor T cells through IL-12. Moreover, in patients undergoing allogeneic hematopoietic cell transplantation, administration of oral vancomycin also increased IL-12 levels (21).
This complexity in immune response and microbiome diversity is also evident in recent human and mouse studies that demonstrate an impact of specific bacteria on the antitumor effects of treatments with CTLA-4 or PD1/PD-L1 blockade (51, 54, 59–61). Elimination of bacterial species by broad-spectrum antibiotics or fecal transfer from immunotherapy-resistant patients to germ-free mice eliminated the response to immune checkpoint therapy. These findings and our data suggest that different bacterial species may be redundant in their impact on the immune system by the presence of possibly overlapping metabolites, with different possible outcomes for tumor growth (60).
Although it is established that some of the “off-target” antitumor effects of RT are immune-mediated (42), the mechanism(s) by which RT+vancomycin combination treatment mediates the antitumor effects has not been investigated.
Dietary fiber (e.g., complex carbohydrates) can be digested and subsequently fermented in the colon by gut microbes, primarily Clostridia (gram-positive bacteria), generating SCFAs such as C4, C3, and C2 (62). These SCFAs activate the G protein–coupled receptors GPR41, GPR43, GPR109a, and Olfr78, which are widely expressed in a variety of cell types, including gut enteroendocrine cells and immune cells (63–65). Specifically, SCFAs were shown to have a direct impact on the functionality of macrophages and DCs. Propionate was shown to affect mouse DCs and macrophage biology in the bone marrow and to impair the ability of DCs to promote Th2 cell effector function in the lungs (66). Butyrate and propionate treatment of human DCs significantly reduced LPS-induced Il6 mRNA and Il12 gene expression and enhanced leukocyte trafficking, and SCFAs strongly reduced the release of several proinflammatory chemokines (46). In agreement, our data demonstrate that butyric acid impacted expression of costimulatory molecules and antigen presentation of bmDCs, but did not directly decrease IFN-γ secretion by T cells. Although DCs can also modulate T cells in a butyrate-dependent manner (67), to our knowledge, the direct impact on antigen presentation in the context of RT has not been investigated. Finally, butyrate administrated in vivo abrogated the increase of antigen presenting cells in TDLNs and decreased tumor levels of IFN-γ and IL-12. Similarly, the adjuvant antitumor effects of vancomycin on RT were lost in animals fed with butyrate. SCFAs produced by colonic Clostridia were also shown to induce the differentiation of colonic Treg cells in mice (68). However, in our model, we did not observe significant changes when we investigated this population in the tumor.
In conclusion, our data suggest that the response to antitumor therapies in mice is mediated by a complex interplay between microbes and immune system interactions. Consistent results with antibiotics demonstrate the effectiveness of this treatment to modulate the gut composition. However, the range of results observed in our study and in previous studies emphasizes the need to carefully characterize gut microbial composition when studying host-microbial interactions in cancer immunotherapy. Based on our findings, we propose the use of gut modulation in a patient-specific manner in order to translate local antitumor effects of RT into a systemic response that can target metastatic disease.
Methods
Mouse strains and cell lines.
Six-to-eight-week-old C57BL/6 female mice were purchased from Envigo and Jackson Laboratories. Six-to-eight-week-old Ifng-KO female (strain B6.129S7-Ifngtm1Ts/J) mice were purchased from Jackson Laboratories. OT1 mice were provided by Jackson Laboratories (strain C57BL/6-Tg(TcraTcrb)1100Mjb/J)). Since gut microbiota play an important role in the design of these experiments, the mice were maintained under special conditions upon delivery; only one cage was placed inside the hood, and both glove replacement and decontamination using chlorine dioxide–based sterilant (clydox-S) were carried out between cages, to avoid any bacterial exchange. Autoclaved food and individual autoclaved water bottles were used in order to protect against any contamination. Furthermore, to ensure microbiota homogeneity between wild-type and Ifng-KO when cages were changed, a mix of mouse feces coming from all used cages was added into the clean unused cages to normalize the animal’s bacterial flora. This process was discontinued after tumor challenge. The mice were randomly assigned to different experimental groups before tumor challenge. TC-1 cells were propagated in 5% CO2 at 37°C and cultured in RPMI 1640 medium with 2 mM l-glutamine (Corning, catalog 25005CI) supplemented with 10% FBS (Gibco, lot 1456821), adjusted to contain 1.1 g/L sodium pyruvate and 10 mM HEPES (Corning, catalog 25060CI), supplemented with 2 mM nonessential amino acids (Gibco, catalog 11150050), 100 U/mL penicillin, and 100 mg/mL streptomycin (Corning, catalog 30002CI). The murine melanoma tumor model B16 cell line engineered to express ovalbumin (B16-OVA) was grown in DMEM (Gibco, catalog 10566) supplemented with 10% FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin. G418 (500 μg/mL) was used for positive selection of B16-OVA–transfected cells. Both B16 and TC-1 cell cultures (ATCC) were tested to detect mycoplasma contamination.
In vitro dendritic cell generation.
After bone marrow cells were harvested from the mouse femur, the resulting cells were seeded in 6-well plates and incubated with granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4 at concentrations of 10 ng/mL for 7 days. Media containing each treatment were replaced every 2 days. On day 7, cells were collected and adjusted to 1 × 106 cells/mL and incubated with 1 μg/ml LPS (Invivogen, catalog tlrl-eblps) overnight (69).
In vitro SCFA treatment.
BMDCs were treated with 100 μM sodium butyrate (MilliporeSigma, catalog B5887), 100 μM sodium propionate (MilliporeSigma, catalog P1880), or a combination of both for 24 hours (46), and pulsed with OVA peptide. After LPS and LPS+SCFA incubations, the cells were detached and placed in an ELISPOT plate with T cells from the OT1 mice. CD3+ T cells were isolated from spleens of OT1 mice using the Miltenyi Pan T cell isolation kit (catalog 130-095-130). Purified T cells (1 × 105) were seeded in triplicate in a 96-well ELISPOT plate and stimulated overnight with αCD3 (clone 145-2C11, BioLegend) / αCD28 (clone 37.51, Invitrogen) in the presence or absence of 100 μM sodium butyrate. Part of the bmDCs were collected, stained, and analyzed by flow cytometry.
ELISPOT.
Ninety-six well multiscreen immobilon-P (MAIP) filtration plates (Millipore, catalog MAIPS4510) were coated overnight with a 2.5 μg/mL solution of rat anti-mouse IFN-γ (BD Biosciences, catalog 551216) in sterilized PBS. The assay was carried out using R10 medium containing RPMI with HEPES and glutamine, 10% FBS, 100 U/mL penicillin, 100 mg/mL streptomycin, and 55 μM 2-mercaptoethanol (Gibco, catalog 21985023). The plates were washed 3 times with sterilized PBS and blocked with R10 for 2 hours before cells were plated. Lymph node cells (0.5 × 106 cells/well), bmDCs (1 × 104 cells/well and 2 × 104 cells/well), and tumor cells (0.1 × 106 cells/well) were incubated in triplicate for 20 hours at 37°C with 0.1 × 106 T cells from OT1 mice with or without 1 μg/mL OVA peptide. bmDCs from C57BL/6J were pulsed with OVA protein (100 μg/mL) and LPS (10 ng/mL) during the final 24 hours. After the incubation, the plates were washed 6 times with PBS containing 0.05% Tween-20 (Bio-Rad). This process was repeated before the plates were incubated with anti-mouse biotin-conjugated anti–IFN-γ antibody (clone, BD Biosciences, catalog 5 54410) for 3 hours at room temperature, followed by incubation with streptavidin-alkaline phosphatase conjugate (BD Biosciences, catalog 55065) for 30 minutes at 37°C. Finally, the plates were washed 3 times with washing buffer and 3 times with PBS and then developed by adding nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Pierce, catalog 34042). The spots were measured using an automated ELISPOT reader (Autoimmun Diagnostika GmbH).
Tumor challenge.
Six-to-eight-week-old C57BL/6 mice (Jackson Laboratories or Envigo) were injected subcutaneously in both legs with 1 × 105 TC-1 or B16-OVA cells per mouse in 100 μL PBS. The nonirradiated (abscopal) tumor was injected 3 days after the primary one. Once tumors were detectable, development was monitored every other day by caliper measurements.
Antibiotic and sodium butyrate administration.
Mice received drinking water containing vancomycin (0.5 g/L, MilliporeSigma, catalog V2002) or neomycin (1 g/L, MilliporeSigma, catalog N1876), plus metronidazole (1 g/L, MilliporeSigma, catalog M3761) (Neo/Met) ad libitum beginning 1 day before the tumor challenge. In the butyrate-containing experiments, sodium butyrate (5 g/L) was included in the water at the same time and in combination with vancomycin. All the reagents were purchased from Sigma-Aldrich. Neo/Met treatment was supplemented with 1.5 g/L of a dextrose, maltodextrin, and sucralose mix. No significant differences in water consumption were observed among groups.
Irradiation.
Tumors were irradiated with 21 Gy when tumor volumes were approximately 50 mm3, using an XRAD320iX, an x-ray system capable of delivering a precise radiation dosage to small animals such as mice. The process included shielding the gut area from the irradiation with lead, in order to avoid direct perturbations in the gut microbiome. Irradiation of TDLNs was also avoided in order to allow T cell priming. The Department of Radiation Oncology houses an X-Rad 320IX Biological X-Ray Irradiator (Precision X-Ray) in the Smilow Center for Translational Research. The irradiator is calibrated for absolute dose using the AAPM TG-61 protocol for kV x-ray beam dosimetry (70). The calibrations are performed with a 0.6-cc volume Exradin Model A12 Farmer chamber (Standard Imaging) and paired Welhoffer Dose 1 (IBA Dosimetry) electrometer, both with the National Institute of Standards and Technology (NIST) and Accredited Dosimetry Calibration Laboratory (ADCL) certificates. Regular film and optically stimulated luminescent dosimeter (OSLD) measurements were performed to monitor irradiator output.
In vivo CD8 depleting antibody treatment.
Mice received 3 doses (once a week) of 200 μg/mouse of either anti-mouse monoclonal CD8-blocking antibody (InVivoMAb anti-CD8α YTS 169.4, BioXCell, catalog BE0117) or rat IgG2b isotype control (InVivoMAb LTF-2, BioXCell, catalog BE0090) by intraperitoneal injection (37).
Cell isolation and purification.
Tumors were digested using 600 U/mL of collagenase type IV (Gibco, catalog 17104019) resuspended in HBSS (Gibco, catalog 14175-079). DCs were isolated from tumors and lymph nodes using CD11c MicroBead UltraPure (Miltenyi Biotec, catalog 130-108-338) according to the manufacturer’s instructions. TDLNs and spleens were digested with collagenase D (2 mg/mL, Sigma-Aldrich, catalog 11088858001) in HBSS. T cells were isolated from OT1 mouse splenocytes using the mouse Pan T Cell Isolation Kit II (Miltenyi Biotec, catalog 130-095-130) according to the manufacturer’s instructions.
Coculture assay.
Tumors and draining lymph nodes from treated mice were collected and dissociated. Tumor cells (1 × 105) were incubated for 24 hours with 1 × 105 T cells from OT1 mice or from E6/7 immunized mice in IFN-γ ELISPOT plates (71). CD11c+ cells isolated from TDLN cell suspension were cocultured with purified naive CD3+ T cells from OT1 mice in a ratio 1:10 and incubated overnight. OT1 T cells were then collected, stained, and analyzed by flow cytometry.
Flow cytometry.
Cells were subjected to up to 8-parameter flow cytometry on a FACS Canto flow cytometer using BD FACS Diva software (BD Biosciences) and data were analyzed using FlowJo version X (Tree Star). Briefly, intracellular staining to detect T cell polarization was performed from isolated purified CD3+ T cells. Cells were washed, stained with surface bodies, fixed, permeabilized, and incubated with intracellular antibodies using the FOXP3/Transcription Factor Staining Buffer Set from eBioscience (catalog 00-5523-00). The following monoclonal antibodies against mouse markers were used to phenotype the T cells: anti-CD45 (clone 30-F11, Invitrogen) anti-CD3 (clone 17A2, BD Pharmingen), anti-CD25 (clone 145-2C11, BD Pharmingen), anti-CD4 (clone RM4-5, BL), and anti-CD8 (clone 53-6.7, BioLegend). iTAg Tetramer/PE–H-2 Kb OVA (SIINFEKL) (MBL International) was used to detect specifically infiltrating OVA T cells. LIVE/DEAD Fixable Aqua Dead Cell Stain (Life Technologies) was used to gate living cells. Intracellular staining was carried out using anti–IFN-γ (clone XMG1.2, eBioscience) and anti-FOXP3 (clone FJK-16s, Invitrogen) after cells were stimulated by incubating 1 × 106 cells with 5 μg/mL OVA peptide (257-264, GenScript, catalog RP10611) overnight or PMA (25 ng/mL), ionomycin (1 μg/mL) and Golgi stop for 6 hours.
DC phenotyping was carried out using the following monoclonal antibodies against mouse markers: MHC2 (clone M5/114.15.2, BioLegend), CD11c (clone N418, eBioscience), CD11b (clone M1/70, BioLegend), anti-CD86 (clone GL-1, BioLegend) and CD45 (clone 104, BioLegend). Anti–H-2kb bound to SIINFEKL (clone 25-D1.16, BL) was used because of its specific reaction with ovalbumin-derived peptide SIINFEKL ((Kb)-SL8 OVA peptide) bound to H-2Kb of MHC class I, avoiding both unbound H-2Kb and H-2Kb bound with an irrelevant peptide.
Immunohistochemical tumor analyses.
Tumors were embedded in OCT (VWR International LLC, catalog 4583) or immediately snap frozen in dry ice. Sections (6-μm thick) were stained for mouse CD3 (CD3 Early T-Cell Marker, rabbit monoclonal antibody, Thermo Fisher Scientific, catalog RM-9107-S0) with hematoxylin as a counterstain. Images of the slides were taken using a Nikon Eclipse Ti microscope. Investigators performed a blind assessment of the IHC sections.
Cytokines analyzed by ELISA.
Mouse IFN-γ levels were measured from supernatants using a DuoSet ELISA mouse IFN-γ kit (R&D Systems, catalog DY485) according to manufacturer’s instructions. Protein extractions from tumors were carried out using T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific, catalog 78510), and the resultant lysates were used to analyze IFN-γ levels with the Mouse IFN-γ Quantikine ELISA Kit (R&D Systems, catalog MIF00). The results were normalized to the total protein levels measured by BCA assay.
Gene expression analysis.
The relative quantification of the expression levels of selected genes was carried out by real-time reverse transcription PCR (qPCR). Total RNA from B16-OVA tumors and from TDLNs was extracted using TRIzol reagent (Invitrogen, catalog 15596018) according to the manufacturer’s instructions. The quality of the RNA samples was determined by visualization of intact 18S and 28S RNA bands following agarose gel electrophoresis with ethidium bromide staining. Two micrograms of total RNA were reverse transcribed with random primers using the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, catalog 4368814). Twenty-five nanograms of cDNA were used in each qPCR reaction with a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). TaqMan gene expression assays were used to quantify expression levels of the following target genes 1 day after radiation: Infb1 (Mm00439546_s1); and 3–5 days after radiation: Ifng (Mm01168134_m1), Il12b (Mm01288989). Gapdh (Mm99999915_g1) was used as housekeeping gene.
Analysis of SCFAs in fecal samples.
Stools and fecal contents were collected from mice and stored frozen in sterile tubes until analysis. Fecal samples were homogenized in volatile free fatty acid mix (5 μL/mg stool, Sigma-Aldrich) and centrifuged (13,000g for 5 minutes). The supernatant was filtered using 1.2 μm, 0.65 μm, and 0.22 μm filter plates (Millipore, catalog MSBVN1210, MSDVN6510, and MSGVN2210, respectively). The filtrate was loaded into total recovery vials (Waters, catalog 186007197C) for analysis. SCFAs were quantified using a Waters Acquity uPLC System with a Photodiode Array Detector and an autosampler (192 sample capacity). Samples were analyzed on a HSS T3 1.8 μm 2.1 × 150 mm column. The flow rate was 0.25 mL/min, the injection volume was 5 μL, the column temperature was 40°C, the sample temperature was 4°C, and the run time was 25 minutes per sample. Eluent A was 100 mM sodium phosphate monobasic, pH 2.5; eluent B was methanol; the weak needle wash was 0.1% formic acid in water; the strong needle wash was 0.1% formic acid in acetonitrile; and the seal wash was 10% acetonitrile in water. The gradient was 100% eluent A for 5 minutes, gradient to 70% eluent B from 5–22 minutes, and then 100% eluent A for 3 minutes. The photodiode array was set to read absorbance at 215 nm with 4.8 nm resolution. Samples were quantified against standard curves of at least 5 points run in triplicate. Standard curves were run at the beginning and end of each metabolomics run. Quality control checks (blanks and standards) were run every 8 samples. Results were rejected if the standards deviated by more or less than 5%. Concentrations in the samples were calculated as the measured concentration minus the concentration of the solvent (volatile free fatty acid mix, 10 mM); the range of detection was at least 1–100 μmol/g stool. Analysis was performed by the Microbial Culture & Metabolomics Core, PennCHOP Microbiome Program.
Analysis of SCFAs in TDLNs and tumor samples by liquid chromatography high-resolution mass spectrometry (LC-HRMS).
SCFA analysis was conducted using a 2-hydrazinoquinoline derivatization and LC-HRMS modified from ref. 72. Tumor tissues or lymph nodes were placed in 0.4 mL 80% methanol/water (from –80°C) spiked with 20 μL internal standard mix (1000 ng [13C4]-β-hydroxy-butyrate and 1000 ng [13C22H3]-acetate). The tissues and lymph nodes in metabolic solution were homogenized using a Bullet Blender 24 AU (Model BB24-AU) for 5 minutes at power 12 at 4°C. For serum SCFA, 0.2 mL 80% methanol/water (from –80°C) and 20 μL internal standard mix were added to 10 μL serum. Samples were then placed on ice for 20 minutes to allow for metabolite extractions from tissues into the methanol solution. The tubes were hand vortexed for 5 seconds each, then spun down for 5 minutes at 20,400g, and the clear supernatants were moved to a clean Eppendorf tube. The protein pellets from the precipitate were used for protein quantification by a Bradford assay (Thermo Fisher Scientific). Supernatant was evaporated to dryness under nitrogen gas, then 50 μL derivatization solution (2.5 μM 2-hydrazinoquinoline and 2 μM 2,2-dipyridyl disulfide in acetonitrile) was added followed by 50 μL 2 μM triphenylphosphine in acetonitrile. Derivatization was done for 15 minutes in a water bath at 60°C. The mixture was placed on ice for 5 minutes to cool, followed by an addition of 100 μL water and vortex mixing to stop the reaction. A quantity of 100 μL of this reaction mixture was transferred to HPLC vials. A quantity of 10 μL was injected on a Ultimate 3000 UHPLC with a Waters Acquity UPLC BEH C18 column (1.0 × 150 mm, 1.7 μm) using a reversed phase gradient with solvent A of 2 mM ammonium acetate in water with 0.05% acetic acid to solvent B 2 mM ammonium acetate in 95% acetonitrile/5% water with 0.05% acetic acid at 0.08 mL/min flow rate at 55°C. The LC was coupled to a Q Exactive Plus (Thermo Fisher Scientific) with a heated ESI source operating in positive ion mode alternating full-scan and MS/MS. The [M+H]+ ion of each analyte and its internal standard was quantified with a 5 ppm window in the full-scan acquisition, with peak confirmation by MS/MS using the product ion derived from the HQ derivative (143 and 160 m/z).
16S rRNA marker gene sequencing.
Mouse fecal samples were collected and frozen at –80°C. DNA was extracted using the DNA Stool Kit from Qiagen, according to the manufacturer’s instructions. PCR amplification of the 16S rRNA marker gene was carried out using Phusion High-Fidelity DNA Polymerase (New England Biolabs). The primers targeted the V4-V6 regions of the 16S gene (5′-TGYCAGCMGCCGCGGTA-3′, 5′-TCACGRCACGAGCTGACG-3′), and contained 10 bp DNA barcodes used to identify samples in sequencing. Products were purified using AMPure XP from Beckman Coulter. The DNA was sequenced on a Roche 454 GS FLX instrument. DNA sequence data were processed using the QIIME pipeline version 1.8 using default parameters unless indicated (73). We required an exact match to the expected barcode and primer sequence. We removed low-quality sequence reads with more than 2 ambiguous base calls, or if the sequence length was less than 200 bp. Operational taxonomic units were selected with UCLUST (74) and taxonomic assignments were generated using the default consensus assignment method in QIIME. Representative sequences were aligned with PyNAST (75) and used to construct a phylogenetic tree with FastTree (76). UniFrac distances (77) were computed between each pair of samples using QIIME. Nonparametric statistical tests (Mann-Whitney, Kruskal-Wallis) were used to compare the relative abundance of taxa and within-group UniFrac distances. Sequencing data is deposited in the NCBI Sequence Read Archive (SRA) under accession number PRJNA565607.
Statistics.
Sample sizes were chosen on the basis of pilot experiments and on our experience with similar experiments. Two-tailed Student’s t tests were used to compare data sets where indicated. Two-way ANOVA was used to compare tumor progression. The relative abundance of bacterial taxa was compared using a 2-sided t test after log transformation. Sample dissimilarity between groups was evaluated using permutational multivariate ANOVA (PERMANOVA). Survival over time was analyzed using the Kaplan-Meier estimator. A P value less than 0.05 was considered significant. Correction for false discovery rate was used to correct for multiple comparisons.
Study approval.
All animal studies were approved by the IACUC and the University Laboratory Animal Resources at the University of Pennsylvania. Mice were treated in accordance with University of Pennsylvania guidelines.
Author contributions
MUH performed the experiments for the rebuttal and validated the initial experiments, analyzed the data, and helped draft the manuscript for submission after revision. SR initiated the project, performed the initial experiments, analyzed the data, and drafted the manuscript. SB assisted with all the experiments, analyzed the data, and helped draft the manuscript. LGDG performed the experiments and analyzed the data regarding SCFA experiments. IV, S Pustylnikov, S Pierini, and RPL helped perform the experiments and analyze the data. KB performed microbiota bioinformatics analyses and gave suggestions for the manuscript. FB gave suggestions for the manuscript and helped with critical advice on how to correctly perform the microbiome studies. IAB, CAM, and NWS performed and helped with SCFA mass spectroscopy experiments and analysis. CK advised on experimental design, helped with critical advice on radiation protocols, gave suggestions and reviewed and edited the manuscript. AF conceived the project, designed the study, supervised project planning and execution, and wrote the manuscript. The order of the co–first authors was assigned based on the relative contributions of those individuals.
Supplementary Material
Acknowledgments
This work was supported by NIH 1R01CA219871-01A1.
Version 1. 12/09/2019
Electronic publication
Version 2. 01/02/2020
Print issue publication
Footnotes
SR’s present address is: Adaptimmune LLC, Philadelphia, Pennsylvania, USA.
LGDG’s present address is: Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
KB’s present address is: Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2020;130(1):466–479.https://doi.org/10.1172/JCI124332.
Contributor Information
Mireia Uribe-Herranz, Email: mireiau@upenn.edu.
Stavros Rafail, Email: psil@hotmail.com.
Silvia Beghi, Email: silvia.beghi@pennmedicine.upenn.edu.
Luis Gil-de-Gómez, Email: luisgdegomez@hotmail.com.
Ioannis Verginadis, Email: vioannis@mail.med.upenn.edu.
Kyle Bittinger, Email: kylebittinger@gmail.com.
Sergey Pustylnikov, Email: sergei.pustylnikov@uphs.upenn.edu.
Stefano Pierini, Email: Stefano.Pierini@uphs.upenn.edu.
Renzo Perales-Linares, Email: renzo.peraleslinares@pennmedicine.upenn.edu.
Frederic Bushman, Email: bushman@mail.med.upenn.edu.
Constantinos Koumenis, Email: koumenis@xrt.upenn.edu.
Andrea Facciabene, Email: facciabe@pennmedicine.upenn.edu.
References
- 1.Jaffray DA. Image-guided radiotherapy: from current concept to future perspectives. Nat Rev Clin Oncol. 2012;9(12):688–699. doi: 10.1038/nrclinonc.2012.194. [DOI] [PubMed] [Google Scholar]
- 2.Park C, Papiez L, Zhang S, Story M, Timmerman RD. Universal survival curve and single fraction equivalent dose: useful tools in understanding potency of ablative radiotherapy. Int J Radiat Oncol Biol Phys. 2008;70(3):847–852. doi: 10.1016/j.ijrobp.2007.10.059. [DOI] [PubMed] [Google Scholar]
- 3.Apetoh L, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13(9):1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 4.Golden EB, Frances D, Pellicciotta I, Demaria S, Helen Barcellos-Hoff M, Formenti SC. Radiation fosters dose-dependent and chemotherapy-induced immunogenic cell death. Oncoimmunology. 2014;3:e28518. doi: 10.4161/onci.28518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demaria S, Coleman CN, Formenti SC. Radiotherapy: Changing the game in immunotherapy. Trends Cancer. 2016;2(6):286–294. doi: 10.1016/j.trecan.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kepp O, et al. Molecular determinants of immunogenic cell death elicited by anticancer chemotherapy. Cancer Metastasis Rev. 2011;30(1):61–69. doi: 10.1007/s10555-011-9273-4. [DOI] [PubMed] [Google Scholar]
- 7.Derer A, Deloch L, Rubner Y, Fietkau R, Frey B, Gaipl US. Radio-immunotherapy-induced immunogenic cancer cells as basis for induction of systemic anti-tumor immune responses — pre-clinical evidence and ongoing clinical applications. Front Immunol. 2015;6:505. doi: 10.3389/fimmu.2015.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demaria S, et al. Ionizing radiation inhibition of distant untreated tumors (abscopal effect) is immune mediated. Int J Radiat Oncol Biol Phys. 2004;58(3):862–870. doi: 10.1016/j.ijrobp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 9.Shevtsov M, Sato H, Multhoff G, Shibata A. Novel approaches to improve the efficacy of immuno-radiotherapy. Front Oncol. 2019;9:156. doi: 10.3389/fonc.2019.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reynders K, Illidge T, Siva S, Chang JY, De Ruysscher D. The abscopal effect of local radiotherapy: using immunotherapy to make a rare event clinically relevant. Cancer Treat Rev. 2015;41(6):503–510. doi: 10.1016/j.ctrv.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wattenberg MM, Fahim A, Ahmed MM, Hodge JW. Unlocking the combination: potentiation of radiation-induced antitumor responses with immunotherapy. Radiat Res. 2014;182(2):126–138. doi: 10.1667/RR13374.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Twyman-Saint Victor C, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanpouille-Box C, et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat Commun. 2017;8:15618. doi: 10.1038/ncomms15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dzutsev A, Goldszmid RS, Viaud S, Zitvogel L, Trinchieri G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur J Immunol. 2015;45(1):17–31. doi: 10.1002/eji.201444972. [DOI] [PubMed] [Google Scholar]
- 20.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uribe-Herranz M, et al. Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI Insight. 2018;3(4):94952. doi: 10.1172/jci.insight.94952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moellering RC. Pharmacokinetics of vancomycin. J Antimicrob Chemother. 1984;14 Suppl D:43–52. doi: 10.1093/jac/14.suppl_d.43. [DOI] [PubMed] [Google Scholar]
- 23.Isaac S, et al. Short- and long-term effects of oral vancomycin on the human intestinal microbiota. J Antimicrob Chemother. 2017;72(1):128–136. doi: 10.1093/jac/dkw383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Netland PA, Zetter BR. Metastatic potential of B16 melanoma cells after in vitro selection for organ-specific adherence. J Cell Biol. 1985;101(3):720–724. doi: 10.1083/jcb.101.3.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poste G, Doll J, Hart IR, Fidler IJ. In vitro selection of murine B16 melanoma variants with enhanced tissue-invasive properties. Cancer Res. 1980;40(5):1636–1644. [PubMed] [Google Scholar]
- 26.Lin KY, et al. Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 1996;56(1):21–26. [PubMed] [Google Scholar]
- 27.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3(2):148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, et al. Therapeutic effects of ablative radiation on local tumor require CD8+ T cells: changing strategies for cancer treatment. Blood. 2009;114(3):589–595. doi: 10.1182/blood-2009-02-206870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Twyman-Saint Victor C, et al. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 2015;520(7547):373–377. doi: 10.1038/nature14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maity A, et al. A phase I trial of pembrolizumab with hypofractionated radiotherapy in patients with metastatic solid tumours. Br J Cancer. 2018;119(10):1200–1207. doi: 10.1038/s41416-018-0281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beli I, et al. Hypofractionated radiotherapy in non small cell lung cancer: a review of the current literature. Rev Recent Clin Trials. 2010;5(2):103–111. doi: 10.2174/157488710791233608. [DOI] [PubMed] [Google Scholar]
- 32.Deng L, et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41(5):843–852. doi: 10.1016/j.immuni.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta A, et al. Radiotherapy promotes tumor-specific effector CD8+ T cells via dendritic cell activation. J Immunol. 2012;189(2):558–566. doi: 10.4049/jimmunol.1200563. [DOI] [PubMed] [Google Scholar]
- 34.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 35.Oweida A, et al. Resistance to radiotherapy and PD-L1 blockade is mediated by TIM-3 upregulation and regulatory T-cell infiltration. Clin Cancer Res. 2018;24(21):5368–5380. doi: 10.1158/1078-0432.CCR-18-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muroyama Y, et al. Stereotactic radiotherapy increases functionally suppressive regulatory T cells in the tumor microenvironment. Cancer Immunol Res. 2017;5(11):992–1004. doi: 10.1158/2326-6066.CIR-17-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pastille E, et al. Transient ablation of regulatory T cells improves antitumor immunity in colitis-associated colon cancer. Cancer Res. 2014;74(16):4258–4269. doi: 10.1158/0008-5472.CAN-13-3065. [DOI] [PubMed] [Google Scholar]
- 38.Trapani JA, Smyth MJ. Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol. 2002;2(10):735–747. doi: 10.1038/nri911. [DOI] [PubMed] [Google Scholar]
- 39.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3(2):133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 40.Mehrotra PT, Wu D, Crim JA, Mostowski HS, Siegel JP. Effects of IL-12 on the generation of cytotoxic activity in human CD8+ T lymphocytes. J Immunol. 1993;151(5):2444–2452. [PubMed] [Google Scholar]
- 41.Kennedy R, Celis E. Multiple roles for CD4+ T cells in anti-tumor immune responses. Immunol Rev. 2008;222:129–44. doi: 10.1111/j.1600-065X.2008.00616.x. [DOI] [PubMed] [Google Scholar]
- 42.Burnette BC, et al. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. 2011;71(7):2488–2496. doi: 10.1158/0008-5472.CAN-10-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibson GR, Willems A, Reading S, Collins MD. Fermentation of non-digestible oligosaccharides by human colonic bacteria. Proc Nutr Soc. 1996;55(3):899–912. doi: 10.1079/PNS19960087. [DOI] [PubMed] [Google Scholar]
- 44.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nastasi C, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci Rep. 2015;5:16148. doi: 10.1038/srep16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang X, et al. Bidirectional interactions between indomethacin and the murine intestinal microbiota. eLife. 2015;4:e08973. doi: 10.7554/eLife.08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Charette M, Marabelle A, Houot R. Turning tumour cells into antigen presenting cells: The next step to improve cancer immunotherapy? Eur J Cancer. 2016;68:134–147. doi: 10.1016/j.ejca.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 50.Overwijk WW, Restifo NP. B16 as a mouse model for human melanoma. Curr Protoc Immunol. 2001;Chapter 20:Unit 20.1. doi: 10.1002/0471142735.im2001s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gopalakrishnan V, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and anticancer immunosurveillance. Cell. 2016;165(2):276–287. doi: 10.1016/j.cell.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Pitt JM, et al. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity. 2016;44(6):1255–1269. doi: 10.1016/j.immuni.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Routy B, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 55.Zitvogel L, Daillère R, Roberti MP, Routy B, Kroemer G. Anticancer effects of the microbiome and its products. Nat Rev Microbiol. 2017;15(8):465–478. doi: 10.1038/nrmicro.2017.44. [DOI] [PubMed] [Google Scholar]
- 56.Rice LB. Antimicrobial resistance in gram-positive bacteria. Am J Infect Control. 2006;34(5 Suppl 1):S11-9; discussion S64-73. doi: 10.1016/j.ajic.2006.05.220. [DOI] [PubMed] [Google Scholar]
- 57.Paulos CM, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest. 2007;117(8):2197–2204. doi: 10.1172/JCI32205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bowers JS, et al. Dendritic cells in irradiated mice trigger the functional plasticity and antitumor activity of adoptively transferred Tc17 cells via IL12 signaling. Clin Cancer Res. 2015;21(11):2546–2557. doi: 10.1158/1078-0432.CCR-14-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vétizou M, Daillère R, Zitvogel L. [The role of intestinal microbiota in the response to anti-tumor therapies] Med Sci (Paris) 2016;32(11):974–982. doi: 10.1051/medsci/20163211013. [DOI] [PubMed] [Google Scholar]
- 60.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matson V, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–108. doi: 10.1126/science.aao3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 63.Samuel BS, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105(43):16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ang Z, Ding JL. GPR41 and GPR43 in Obesity and Inflammation - Protective or Causative? Front Immunol. 2016;7:28. doi: 10.3389/fimmu.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pluznick JL, et al. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA. 2013;110(11):4410–4415. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 67.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 69.Pierini S, et al. A tumor mitochondria vaccine protects against experimental renal cell carcinoma. J Immunol. 2015;195(8):4020–4027. doi: 10.4049/jimmunol.1500281. [DOI] [PubMed] [Google Scholar]
- 70.Ma CM, et al. AAPM protocol for 40-300 kV x-ray beam dosimetry in radiotherapy and radiobiology. Med Phys. 2001;28(6):868–893. doi: 10.1118/1.1374247. [DOI] [PubMed] [Google Scholar]
- 71.Facciabene A, et al. Local endothelial complement activation reverses endothelial quiescence, enabling t-cell homing, and tumor control during t-cell immunotherapy. Oncoimmunology. 2017;6(9):e1326442. doi: 10.1080/2162402X.2017.1326442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu Y, Yao D, Chen C. 2-Hydrazinoquinoline as a derivatization agent for LC-MS-based metabolomic investigation of diabetic ketoacidosis. Metabolites. 2013;3(4):993–1010. doi: 10.3390/metabo3040993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 75.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26(2):266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PloS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71(12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.