Abstract
N-3 docosapentaenoic acid–derived resolvin D5 (RvD5n-3 DPA) is diurnally regulated in peripheral blood and exerts tissue-protective actions during inflammatory arthritis. Here, using an orphan GPCR screening approach coupled with functional readouts, we investigated the receptor(s) involved in mediating the leukocyte-directed actions of RvD5n-3 DPA and identified GPR101 as the top candidate. RvD5n-3 DPA bound to GPR101 with high selectivity and stereospecificity, as demonstrated by a calculated KD of approximately 6.9 nM. In macrophages, GPR101 knockdown limited the ability of RvD5n-3 DPA to upregulate cyclic adenosine monophosphate, phagocytosis of bacteria, and efferocytosis. Inhibition of this receptor in mouse and human leukocytes abrogated the pro-resolving actions of RvD5n-3 DPA, including the regulation of bacterial phagocytosis in neutrophils. Knockdown of the receptor in vivo reversed the protective actions of RvD5n-3 DPA in limiting joint and gut inflammation during inflammatory arthritis. Administration of RvD5n-3 DPA during E. coli–initiated inflammation regulated neutrophil trafficking to the site of inflammation, increased bacterial phagocytosis by neutrophils and macrophages, and accelerated the resolution of infectious inflammation. These in vivo protective actions of RvD5n-3 DPA were limited when Gpr101 was knocked down. Together, our findings demonstrate a fundamental role for GPR101 in mediating the leukocyte-directed actions of RvD5n-3 DPA.
Keywords: Infectious disease, Inflammation
Keywords: Arthritis, Bacterial infections, G protein–coupled receptors
Introduction
Development of the inflammatory response was a fundamental step in the evolution of multicellular organisms, given that this mechanism is vital in warding off invading pathogens and in coordinating reparative and regenerative mechanisms (1, 2). Leukocytes play a central role in the clearance of pathogens, in which, for example, neutrophils and macrophages are tasked with the uptake and killing of bacteria (3, 4). These cells are also important in coordinating the termination of acute inflammation as well as initiating tissue repair and regeneration (4–6). Central to the protective actions exerted by leukocytes is a novel genus of mediators termed specialized pro-resolving mediators (SPMs) (4, 7, 8). These autacoids are produced through the enzymatic conversion of essential fatty acids, which yields molecules with a defined stereochemistry and potent biological properties (4, 7, 8).
Recent studies demonstrate that in addition to arachidonic acid, eicosapentaenoic acid, and docosahexaenoic acid (DHA), n-3 docosapentaenoic acid (n-3 DPA) is also a substrate for conversion to novel, structurally distinct bioactive mediators (9, 10). One of these novel mediators, n-3 DPA–derived resolvin D5 (RvD5n-3 DPA) (7S,17S-dihydroxy-8E,10Z,13Z,15E,19Z-docosapentaenoic acid), is diurnally regulated in human peripheral blood, and loss of its production, together with that of the related mediators RvD1n-3 DPA (7S,8R,17S-trihydroxy-9E,11E,13Z,15E,19Z-docosapentaenoic acid) and RvD2n-3 DPA (7S,16,17S-trihydroxy-8,10Z,12,14,19Z- docosapentaenoic acid), in patients with cardiovascular disease is linked with increased peripheral blood neutrophil, monocyte, and platelet activation (9). RvD5n-3 DPA also exerts tissue-protective actions by regulating mouse leukocyte trafficking and responses in vivo, as well as neutrophil, monocyte, and macrophage responses in vitro (9). We recently uncovered a homeostatic role for RvD5n-3 DPA in regulating intestinal epithelial barrier function, where decreased levels of this mediator during inflammatory arthritis are linked with increased intestinal barrier permeability and joint inflammation (11). Treatment of arthritic mice with RvD5n-3 DPA restored barrier function, reduced leukocyte trafficking into the arthritic joints, and limited joint inflammation (11).
The biological actions of lipid mediators are stereospecific, suggesting that they activate cognate receptors to evoke downstream protective activities (12). Studies investigating the identity of these proteins demonstrate that lipid mediators, including the classic eicosanoids and SPMs, activate a class of receptors known as GPCRs (12, 13). In this context, the first pro-resolving mediator receptor identified was the lipoxin A4 receptor, also referred to as formyl peptide receptor 2 (ALX) or FPR2 (14). This receptor was later found to also mediate the biological actions of DHA-derived resolvin D1 (7S,8R,17S-trihydroxy-4Z,9E,11E,13Z,15E,19Z-docosahexaenoic acid) as well as the pro-resolving protein annexin A1 (15, 16). Recent studies have uncovered a role for the orphan receptors GPR32 and GPR18 in mediating the biological actions of pro-resolving mediators, with RvD1 and RvD3 (4S,11R,17S-trihydroxy-5Z,7E,9E,13Z,15E,19Z-docosahexaenoic acid) activating GPR32 (16, 17) and RvD2 activating GPR18 (18). Of note, the identity of the receptor mediating the biological actions of RvD5n-3 DPA is not known.
In the present study, we used an integrated approach to establish the identity of the GPCR that mediates the pro-resolving actions of RvD5n-3 DPA on leukocytes. Results from these experiments identified GPR101 as a candidate receptor for RvD5n-3 DPA. Flow cytometric analysis demonstrated that GPR101 was expressed on both human and mouse peripheral blood leukocytes. Using radiolabeled RvD5n-3 DPA, we observed specific binding of this mediator to GPR101 with an affinity that was within its bioactive range. Knockdown of Gpr101 in mice led to an abrogation of the protective actions of RvD5n-3 DPA in vivo by limiting its ability to regulate inflammatory arthritis and infectious inflammation.
Results
Identification of candidate receptors for RvD5n-3 DPA.
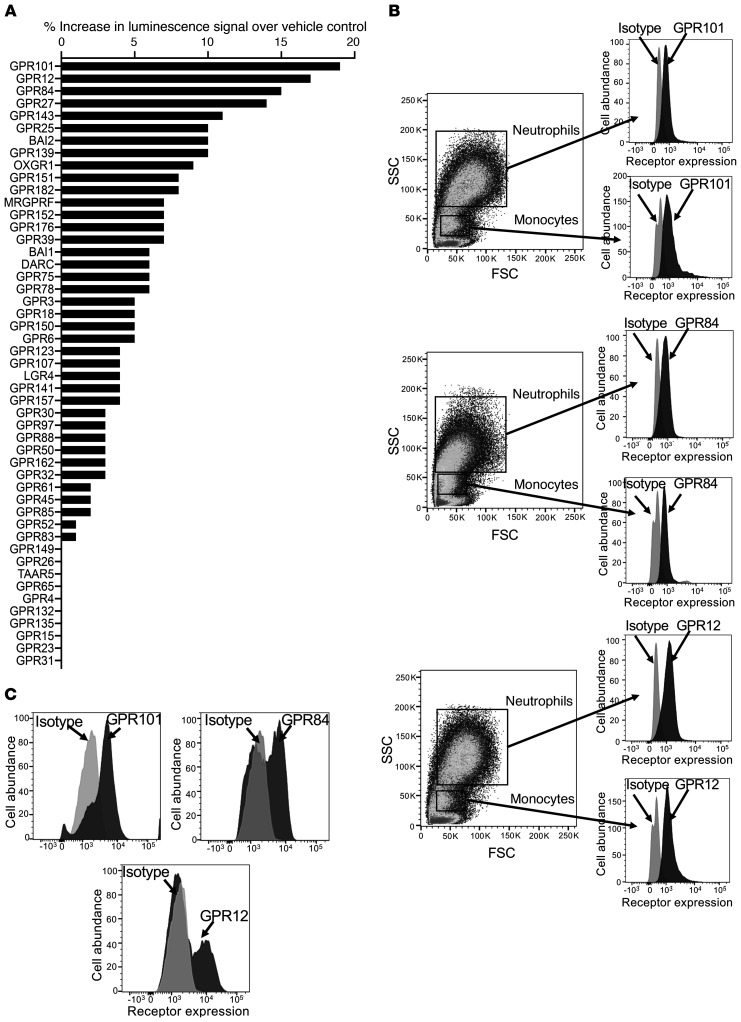
In order to establish whether the biological actions of RvD5n-3 DPA were mediated by a GPCR, we screened a panel of orphan GPCRs to determine whether RvD5n-3 DPA showed agonistic activity toward these receptors, using 10 nM RvD5n-3 DPA and assessing increases in luminescence as a readout for receptor activation. Here, we found that the strongest agonistic signals elicited by this pro-resolving mediator were with GPR101, GPR12, and GPR84 (Figure 1A), with values approximately 15%–20% above the control value. Given that RvD5n-3 DPA regulates the biological actions of monocyte-derived macrophages and peripheral blood leukocytes (9, 10), we next investigated the expression of these 3 receptors on circulating human neutrophils and monocytes and identified all 3 receptors (Figure 1B). Moreover, human monocyte–derived macrophages also expressed all 3 receptors on their cell surface (Figure 1C).
Figure 1. RvD5n-3 DPA receptor candidates are expressed on human leukocytes.
(A) Activation of orphan receptors by RvD5n-3 DPA (10 nM). Results represent the percentage increase in luminescence signal over vehicle control. (B and C) Expression of the top 3 candidate receptors on human (B) peripheral blood leukocytes and (C) macrophages. Results are representative of 4 donors. FSC, forward scatter; SSC, side scatter.
RvD5n-3 DPA stereospecifically activates GPR101.
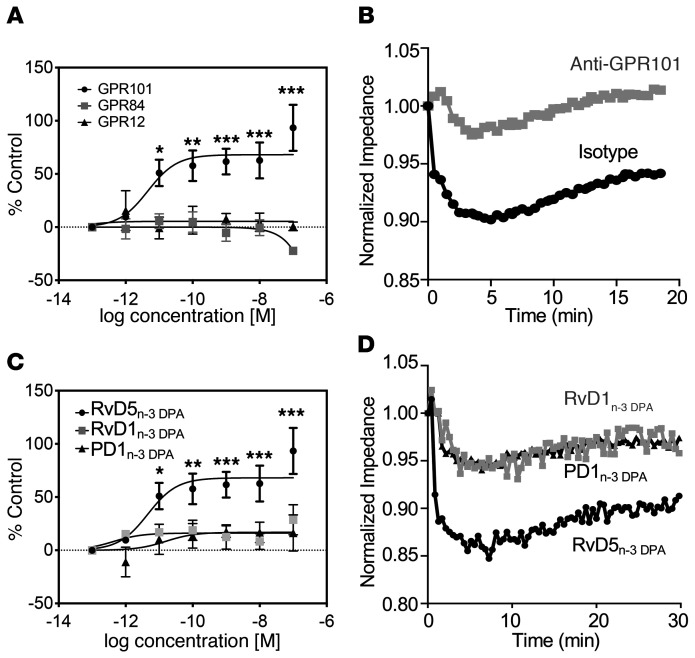
To establish the role of these receptors in mediating the biological actions of RvD5n-3 DPA, we evaluated the ability of this ligand to activate each of these 3 receptors using a β-arrestin–based ligand receptor interaction screening system, which enabled the construction of full dose-response curves (19). In these settings, RvD5n-3 DPA increased chemiluminescence in a concentration-dependent manner in cells overexpressing GPR101, with a calculated EC50 of 4.6 × 10–12 M (Figure 2A). Of note, this increase in chemiluminescence was not observed in cells expressing either GPR12 or GPR84 (Figure 2A). Using the β-arrestin system, we also tested whether RvD5n-3 DPA activates the pro-resolving receptors GPR32 (also known as DRV1) and GPR18 (also known as DRV2). Here, we found that RvD5n-3 DPA displayed an affinity for GPR32/DRV1 comparable to that observed with RvD1, with an EC50 of approximately 1.4 × 10−11 M and approximately 1.5 × 10−12 M, respectively. Of note, RvD5n-3 DPA did not appear to activate GPR18/DRV2 at biologically relevant concentrations (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI131609DS1).
Figure 2. Activation of GPR101 by RvD5n-3 DPA.
(A) RvD5n-3 DPA was incubated at the indicated concentrations with CHO cells expressing human GPR101 (circles), GPR84 (squares), or GPR12 (triangles) coupled with the β-arrestin reporter system, and receptor activation was measured as an increase in luminescence signal. Results represent the mean ± SEM. n = 5–7 independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 versus the respective vehicle control group; 2-way ANOVA with Tukey’s post hoc multiple comparisons test. (B) CHO cells overexpressing GPR101 were incubated with either isotype control or anti-GPR101 antibody (30 minutes at room temperature) and then with 1 nM RvD5n-3 DPA, and impedance was measured over a 20-minute period using the xCELLigence DP system. Results are representative of 3 distinct experiments. (C) CHO cells expressing GPR101 coupled with the β-arrestin reporter system were incubated with the indicated concentrations of RvD5n-3 DPA, RvD1n-3 DPA, PD1n-3 DPA, or vehicle (PBS containing 0.01% ethanol), and receptor activation was measured as an increase in luminescence signal. Note that the same data is shown for the RvD5n-3 DPA trace as the GPR101 trace in A. Results represent the mean ± SEM. n = 5–7 independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 versus the vehicle control group; 2-way ANOVA with Tukey’s post hoc multiple comparisons test. (D) RvD5n-3 DPA, RvD1n-3 DPA, and PD1n-3 DPA (10 nM) were incubated with GPR101-expressing CHO cells, and impedance was measured over a 30-minute period using the xCELLigence DP system. Results are representative of 3 distinct experiments.
Given that GPCR activation leads to changes in both cell shape and size (20), we next used an impedance-based assay to further evaluate the interactions between RvD5n-3 DPA and GPR101. We found that incubation of RvD5n-3 DPA with GPR101-expressing CHO cells led to changes in impedance, an observation that was markedly reversed when these cells were preincubated with an anti-GPR101 antibody (Figure 2B).
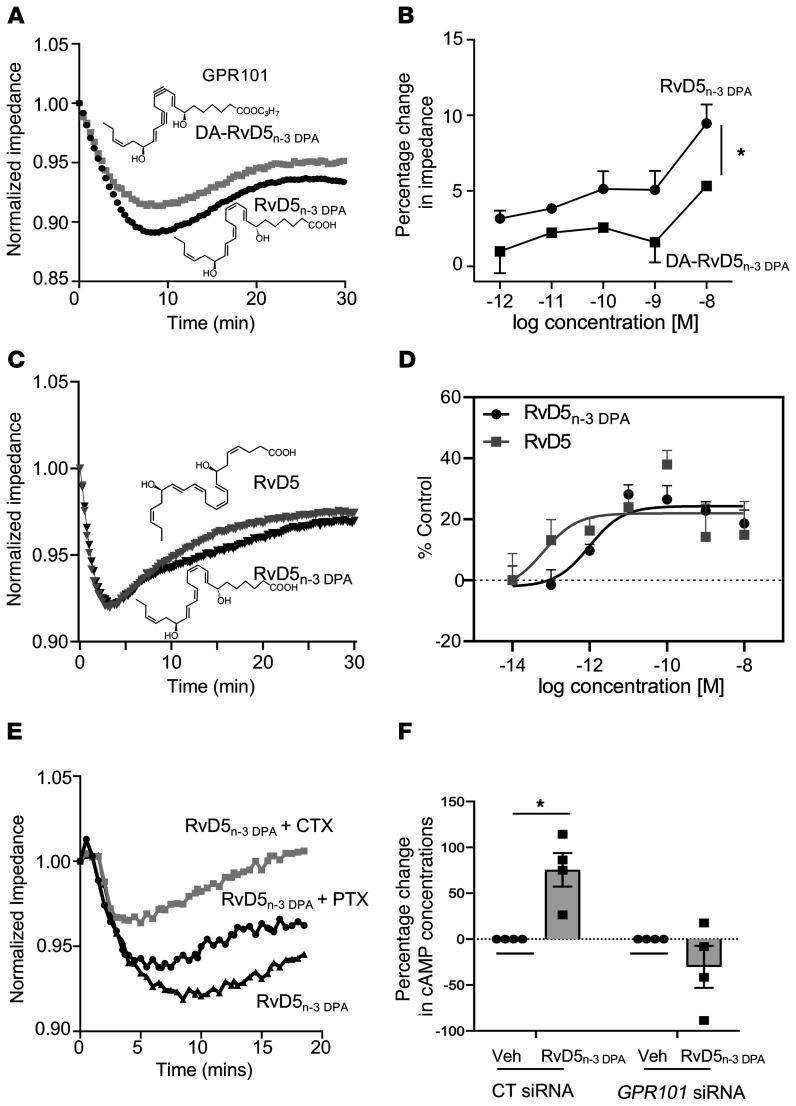
We next investigated the stereospecificity of the interaction between RvD5n-3 DPA and GPR101, assessing the capacity of related n-3 DPA–derived SPMs to activate this receptor. Incubation of GPR101-expressing cells with either RvD1n-3 DPA or PD1n-3 DPA (10R,17S-dihydroxy-7Z,11E,13E,15Z,19Z-docosapentaenoic acid) did not elicit notable increases in chemiluminescent signals (Figure 2C), indicating a degree of selectivity for RvD5n-3 DPA. We made similar observations when assessing impedance changes, in which addition of RvD1n-3 DPA or PD1n-3 DPA to GPR101-expressing cells only elicited marginal changes in impedance (Figure 2D). To investigate the stereospecific nature of these receptor-ligand interactions, we next tested the ability of isopropyl-(7S,8E,15E,17S,19Z)-7,17-dihydroxydocosa-8,15,19-trien-10,13-diynoate (DA-RvD5n-3 DPA), an analog of RvD5n-3 DPA, to elicit changes in impedance. We found that substituting the double bond in the middle portion of the analog for triple bond, which would increase the rigidity of the molecule, led to a significant decrease in the ability of the molecule to activate the receptor (Figure 3, A and B). We further evaluated the stereospecific nature of the interaction between RvD5n-3 DPA and GPR101 by assessing whether the DHA-derived RvD5 (7S,17S-dihydroxy-4Z,8E,10Z,13Z,15E,19Z-docosahexaenoic acid), which differs from the n-3 DPA congener by having an extra double bond on carbons 4 and 5, activates GPR101 to a similar degree. Using both impedance assays and the β-arrestin system, we found that RvD5 activated GPR101 to an extent similar to that seen with RvD5n-3 DPA, with a calculated EC50 of approximately 6.4 × 10−14 M and approximately 9.7 × 10−13 M, respectively (Figure 3, C and D).
Figure 3. RvD5n-3 DPA stereospecifically activates GPR101 and increases cAMP in human macrophages.
(A) GPR101-expressing CHO cells were incubated with RvD5n-3 DPA or DA-RvD5n-3 DPA (10 nM), and impedance was measured for 30 minutes. Results are representative of 3 distinct experiments. (B) GPR101-expressing CHO cells were incubated with the indicated concentrations of the ligands described in A, and changes in impedance from baseline values were determined at t = 10 minutes. Results represent the mean ± SEM. n = 3 from 3 distinct experiments. *P < 0.05; 1-way ANOVA with a Holm-Sidak post hoc multiple comparisons test. (C) GPR101-overexpressing CHO cells were incubated with RvD5n-3 DPA or DHA-derived RvD5 (1 nM), and cell impedance was measured over a 30-minute period using the xCELLigence DP system. Results are shown as the mean ± SEM (n = 4 in 3 independent experiments). (D) CHO cells expressing GPR101 coupled with the β-arrestin luminescence reporter system were incubated with the indicated concentrations of RvD5n-3 DPA, DHA-derived RvD5, or vehicle (cell-plating reagent containing 0.01% ethanol), and receptor activation was measured as an increase in luminescence signal. Results are shown as the mean ± SEM (n = 3 in 2 independent experiments). (E) GPR101-expressing CHO cells were incubated with CTX (1 μg/mL, 2 hours), PTX (1 μg/mL, 16 hours), or vehicle and then with RvD5n-3 DPA (10 nM), and impedance was measured over a 30-minute period. Results are representative of 3 distinct experiments. (F) Human monocyte–derived macrophages were incubated with either an siRNA against GPR101 or a control sequence (CT siRNA; 72 hours at 37°C) and then with RvD5n-3 DPA (10 nM) or vehicle (Veh) (PBS containing 0.01% ethanol) for 2 minutes, and cAMP concentrations were assessed. Results represent the mean ± SEM (n = 4 donors). *P < 0.05; 1-way ANOVA with Holm-Sidak post hoc multiple comparisons test.
We next tested the role of G proteins in mediating the signals generated by RvD5n-3 DPA via the activation of GPR101. For this purpose, we incubated cells with either cholera toxin (CTX) (Gαs-like protein inhibitor) or pertussis toxin (PTX) (Gαi-like protein inhibitor) prior to addition of RvD5n-3 DPA. Incubation of cells with CTX, but not PTX, markedly inhibited the changes in impedance elicited by RvD5n-3 DPA (Figure 3E). These results suggest that activation of GPR101 by RvD5n-3 DPA promotes the recruitment of Gαs-like proteins. Given that cAMP is the downstream second messenger of Gαs, we next investigated whether RvD5n-3 DPA upregulates cAMP in human monocyte–derived macrophages using an siRNA approach. Incubation of these cells with RvD5n-3 DPA led to an increase in cAMP concentrations (168 ± 56 pmol vs. 290 ± 98 pmol per 0.4 × 106 cells), an increase that was absent in cells transfected with an siRNA against GPR101 (Figure 3F).
Binding of 3H-RvD5n-3 DPA to GPR101.
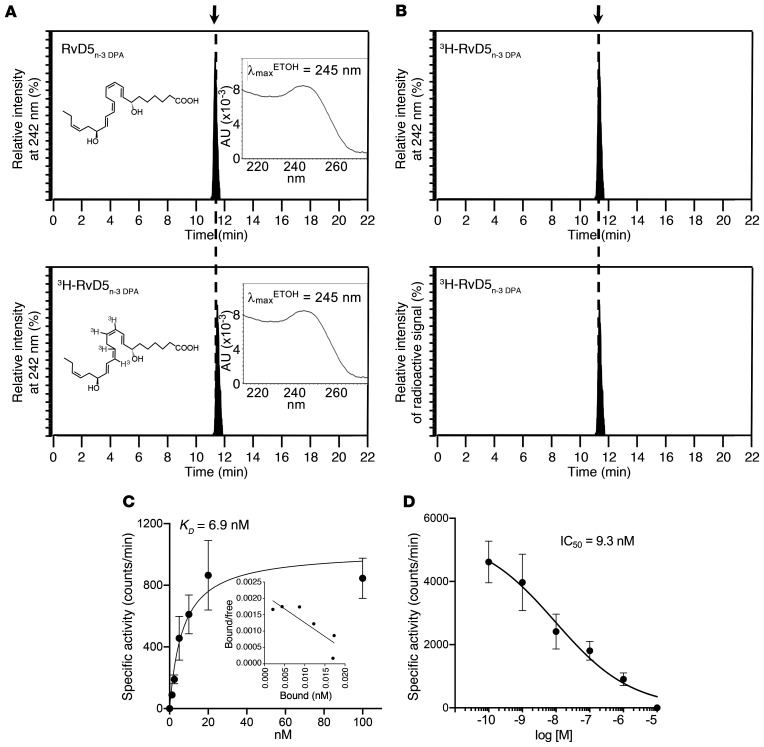
Next, we determined the affinity of the ligand-receptor interaction. For this purpose, we prepared tritium-labeled RvD5n-3 DPA by catalytic tritiation with a specific activity of approximately 40 Ci/mmol. The purity and integrity of the tritiated product were determined using reversed-phase ultraviolet HPLC (RP-UV-HPLC) to be approximately 95%, as demonstrated by essentially identical retention times between the authentic RvD5n-3 DPA and [10, 11, 13, 143H]-RvD5n-3 DPA as well as matching UV chromophores with an UV absorbance maximum in ethanol (λmaxETOH) of 245 nm (Figure 4A). Furthermore, quantification of radioactivity demonstrated that approximately 98% of the activity was associated with [10, 11, 13, 143H]-RvD5n-3 DPA, given that the peak in activity coincided with the eluting of the molecule (Figure 4B). We next conducted saturation binding studies to establish the KD for binding of RvD5n-3 DPA to GPR101. These experiments yielded a calculated KD of approximately 6.9 nM (Figure 4C). To determine the specificity of the RvD5n-3 DPA-GPR101 interaction, we next sought to establish the IC50 of the interaction. We found that RvD5n-3 DPA and [10, 11, 13, 143H]-RvD5n-3 DPA competed for GPR101 binding, with an IC50 of approximately 9.3 nM (Figure 4D). Together, these results demonstrate that the affinity of RvD5n-3 DPA for GPR101 was within the bioactive range for this mediator (9, 10).
Figure 4. Specific binding of [3H]-RvD5n-3 DPA to human GPR101.
(A and B) Characterization of [10, 11, 13, 14 3H]-RvD5n-3 DPA ([3H]-RvD5n-3 DPA). (A) RP-UV-HPLC chromatogram for RvD5n-3 DPA and [3H]-RvD5n-3 DPA. (B) RP-UV-HPLC chromatogram of [3H]-RvD5n-3 DPA and online radioactivity monitoring. (C) GPR101-overexpressing CHO cells (0.5 × 106 cells in 100 μL) were incubated with [3H]-RvD5n-3 DPA at the indicated concentrations in the presence or absence of 10 μM RvD5n-3 DPA (60 minutes at 4°C). Cell incubations were transferred to a vacuum manifold, unbound radioligand was removed, and activity was measured. Results represent the mean ± SEM (n = 4 from 2 distinct experiments). Inset shows a Scatchard plot. (D) To assess competition binding, GPR101-expressing CHO cells (0.5 × 106 cells in 100 μL) were incubated with 3 nM [3H]-RvD5n-3 DPA in the presence or absence of increasing concentrations of RvD5n-3 DPA for 60 minutes at 4°C.
GPR101 mediates the regulatory actions of RvD5n-3 DPA on macrophages.
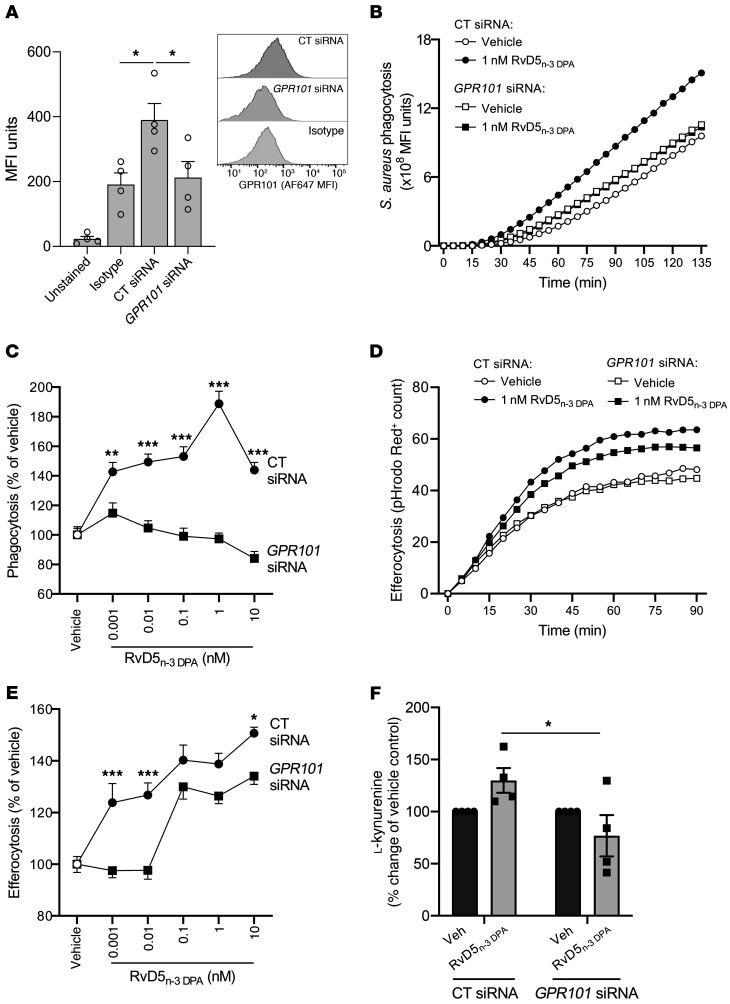
In order to test the functional role of GPR101 in mediating the pro-resolving actions of RvD5n-3 DPA in human monocyte–derived macrophages, we next knocked down the expression of this receptor using siRNA. Given the central role of macrophages in promoting the resolution of bacterial infections via the uptake and killing of pathogens in a process termed phagocytosis (4, 7, 18), we next investigated whether RvD5n-3 DPA regulates phagocytosis of bacterial particles in a GPR101-dependent manner. For this purpose, we used pHrodo Green–conjugated Staphylococcus aureus (S. aureus) bioparticles that become increasingly fluorescent as pH decreases, thus providing a readout of phagocytic uptake and subsequent phagolysosomal degradation of ingested material. Incubation of scrambled control siRNA–transfected macrophages with RvD5n-3 DPA led to a dose-dependent increase in phagocytosis. Transfection of macrophages with an siRNA against GPR101, which significantly reduced receptor expression, abrogated the ability of RvD5n-3 DPA to upregulate phagocytosis (Figure 5, A–C).
Figure 5. GPR101 mediates the protective actions of RvD5n-3 DPA on human macrophages.
(A) Human monocyte–derived macrophages were incubated with either an siRNA against GPR101 or a control sequence (CT siRNA) for 96 hours, and GPR101 expression was assessed using flow cytometry (n = 4 donors). (B) Cells were transfected as in A and then incubated with RvD5n-3 DPA (0.001–10 nM) or vehicle (RPMI-1640 containing 0.1% ethanol, 15 minutes, 37°C), after which (B and C) efferocytosis of pHrodo Red–conjugated apoptotic HL-60 cells and (D and E) phagocytosis of pHrodo Green–conjugated S. aureus bioparticles were measured using a Zeiss Celldiscoverer 7 high-content imager. B and D show the increase in signal over time for vehicle and 1 nM RvD5n-3 DPA groups, whereas C and E show the AUC for all tested concentrations. Results represent the mean ± SEM (n = 6 donors from 2 distinct experiments). *P < 0.05, **P < 0.01, and ***P < 0.001; 2-way ANOVA with Tukey’s post hoc multiple comparisons test. (F) Human monocyte–derived macrophages were incubated with either an siRNA against GPR101 or a control sequence and then with 10 nM RvD5n-3 DPA or vehicle (RPMI-1640 containing 0.1% ethanol; 2 hours at 37°C), and the expression of l-kynurenine was measured using LC-MS/MS. Results represent the mean ± SEM (n = 4 donors from 2 distinct experiments). *P < 0.05; Friedman’s test with Dunn’s post hoc multiple comparisons test.
Next, we tested the ability of these macrophages to take up apoptotic cells, a key pro-resolving action termed efferocytosis (4, 6, 12, 16, 21). Treatment of control siRNA–transfected macrophages with RvD5n-3 DPA led to a dose-dependent increase in efferocytosis of pHrodo Red–labeled apoptotic cells. Meanwhile, transfection of macrophages with an siRNA against GPR101 limited the ability of RvD5n-3 DPA to upregulate efferocytosis in a similar manner (Figure 5, D and E).
We recently found that RvD5n-3 DPA upregulates the activity of indoleamine 2,3-dioxygenase (IDO) in macrophages (11), which metabolizes tryptophan to l-kynurenine, a mechanism implicated in upregulation of the homeostatic IL-10 receptor (IL-10R) (22). Therefore, we tested the role of GPR101 in mediating the upregulation of l-kynurenine concentrations by monocyte-derived macrophages. In line with published findings, we observed that incubation of control siRNA–transfected macrophages with RvD5n-3 DPA led to an upregulation of l-kynurenine concentrations, whereas addition of this mediator to cells transfected with an siRNA against GPR101 only marginally regulated the concentrations of this metabolite (Figure 5F). Together, these findings demonstrate a nonredundant role for GPR101 in mediating the biological actions of RvD5n-3 DPA on human monocyte–derived macrophages.
GPR101 mediates the protective actions of RvD5n-3 DPA on neutrophils and monocytes.
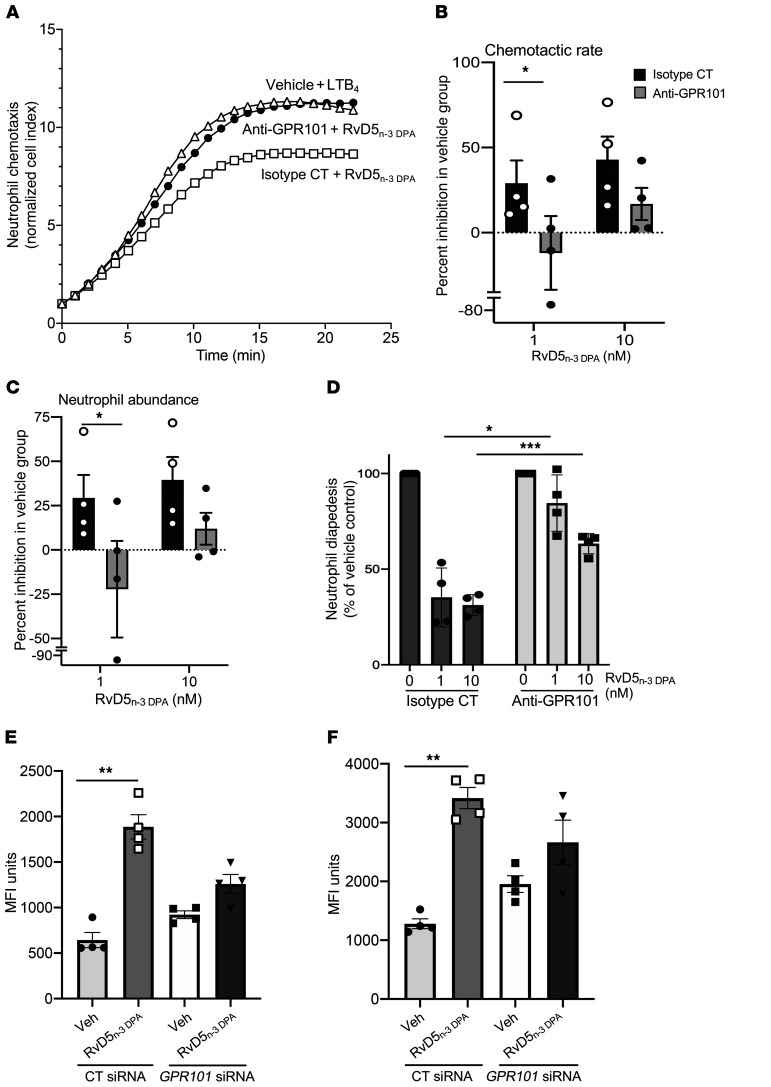
Since RvD5n-3 DPA regulates the biological actions of neutrophils (9, 10, 23) and GPR101 is expressed on these cells (Figure 1), we complemented the results produced with human monocyte–derived macrophages by determining the neutrophil-directed actions of RvD5n-3 DPA. Incubation of RvD5n-3 DPA with human neutrophils led to a significant and dose-dependent reduction in both the rate of neutrophil migration toward leukotriene B4 (LTB4) and the total number of cells migrated (Figure 6, A–C). In addition, RvD5n-3 DPA also reduced the number of cells that transmigrated through an activated endothelial cell monolayer under flow conditions (Figure 6D). Incubation of neutrophils to an antibody targeting GPR101 reversed the biological actions of RvD5n-3 DPA, increasing both neutrophil chemotaxis and transmigration (Figure 6, A–D).
Figure 6. GPR101 mediates the protective actions of RvD5n-3 DPA on neutrophils and monocytes.
(A–C) Human neutrophils were incubated with anti-GPR101 antibody or an isotype control antibody (15 minutes at room temperature) and then with RvD5n-3 DPA at the indicated concentrations or with vehicle (PBS containing 0.1% ethanol), and chemotaxis toward LTB4 (10 nM) was assessed using the xCELLigence DP system. (A) representative traces, (B) neutrophil chemotactic rate calculated from the slope of the curve, and (C) neutrophil chemotaxis calculated from the AUC of the traces shown in A. *P < 0.05 versus the indicated control group; Kruskal-Wallis test with Dunn’s post hoc multiple comparisons test. (D) Neutrophils were isolated and incubated as detailed above, and neutrophil–endothelial cell interactions were assessed after perfusing (0.1 Pa) neutrophils over an activated endothelial cell monolayer. Results represent the mean ± SEM (n = 4 donors from 3–4 distinct experiments). *P < 0.05 and ***P < 0.001 versus the indicated control group; 2-way ANOVA with Sidak’s post hoc test. (E and F) Mice were administered 9 μg siRNA against mouse Gpr101 or a scrambled control sequence. After 72 hours, blood was collected and cells incubated with RvD5n-3 DPA (10 nM) or vehicle (20 minutes at 37°C) and then with 5 × 107 CFU fluorescently labeled bacteria (30 minutes at 37°C), and phagocytosis in (E) neutrophils and (F) monocytes was assessed by flow cytometry. Results represent the mean ± SEM (n = 4 per group from 2 distinct experiments). **P < 0.01 versus the indicated control group; Kruskal-Wallis test with Dunn’s post hoc multiple comparisons test.
Since neutrophils are pivotal for the clearance of bacterial infections (3, 4), we next tested whether RvD5n-3 DPA, via GPR101, increases the ability of these cells to take up bacteria. Flow cytometric assessment of peripheral blood neutrophils and monocytes demonstrated that these cells expressed the murine ortholog of human GPR101 (Supplemental Figure 2). Incubation of whole blood from mice transfected with a control siRNA sequence with RvD5n-3 DPA led to a significant increase in bacterial phagocytosis by both neutrophils (643 ± 96 vs. 1886 ± 154 MFI units) and monocytes (1280 ± 97 vs. 3418 ± 208 MFI units). These actions were reversed when the mediator was added to peripheral blood collected from mice transfected with siRNA against Gpr101 that reduced the expression of the receptor on peripheral blood leukocytes (Figure 6, E and F, and Supplemental Figure 3). Together, these findings establish a role for GPR101 in mediating the biological actions of RvD5n-3 DPA on neutrophils and monocytes.
GPR101 mediates the antiarthritic actions of RvD5n-3 DPA.
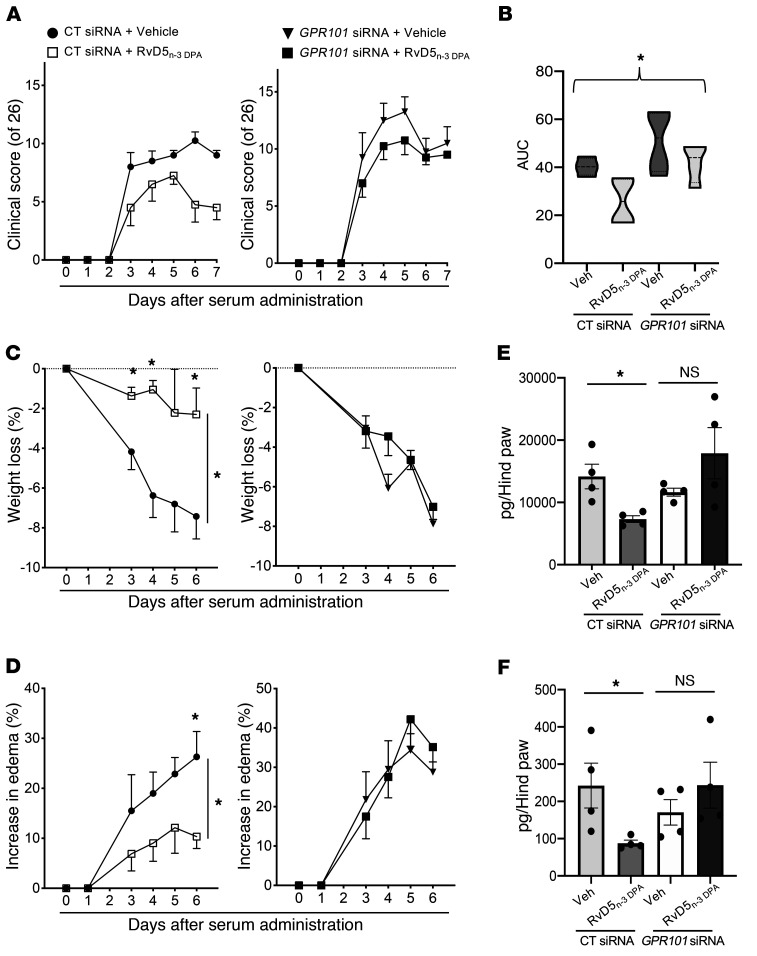
RvD5n-3 DPA displays potent antiarthritic actions that regulate both neutrophil and macrophage responses in inflammatory arthritis (11). Thus, we next investigated the role of GPR101 in mediating these protective actions. For this purpose, we treated mice with either an siRNA targeting mouse Gpr101 to knock down the expression of this receptor in vivo, or a control sequence. We then challenged the mice with arthritogenic serum from K/BxN mice, which initiates a polyarthritic disease driven by both monocytes and neutrophils (24). Assessment of clinical scores revealed that treatment of mice transfected with a control siRNA sequence with RvD5n-3 DPA led to a significant reduction in clinical scores when compared with vehicle-treated mice (5.5 ± 1.8 in RvD5n-3 DPA–treated mice vs. 9.0 ± 0.5 in mice given vehicle), whereas in mice transfected with an siRNA against Gpr101, this antiarthritic activity of RvD5n-3 DPA was lost (9.5 ± 0.3 in RvD5n-3 DPA –treated mice vs. 10.5 ± 1.7 in mice given vehicle) (Figure 7, A and B). Transfection of mice with an siRNA against Gpr101 diminished the ability of this mediator to protect against weight loss (a marker of disease severity), limit joint edema, and reduce joint concentrations of the inflammation-initiating prostaglandins and LTB4 (Figure 7, C–F).
Figure 7. Knockdown of GPR101 reverses the antiarthritic actions of RvD5n-3 DPA.
Mice were administered 9 μg siRNA targeting mouse Gpr101 or a scrambled control sequence. After 24 hours and 72 hours, mice were administered arthritogenic serum and then treated with RvD5n-3 DPA (150 ng/mouse) or vehicle (72 hours and 96 hours after siRNA administration). (A) Clinical scores, (B) AUC for clinical scores, (C) weight loss, and (D) edema were determined throughout the disease process. (E and F) On day 7, hind paws were harvested and eicosanoid concentrations determined using LC-MS/MS–based lipid mediator profiling. (E) Prostaglandin and (F) LTB4 metabolomic concentrations. Results represent the mean ± SEM (n = 4 mice per group). *P < 0.05 versus the vehicle-treated group; Kruskal-Wallis test with Dunn’s post hoc multiple comparisons test (B, E, and F) and 2-way ANOVA (C and D).
We recently found that inflammatory arthritis disrupts gut barrier function, which promotes joint inflammation (11). RvD5n-3 DPA administration reduced intestinal inflammation and eicosanoid concentrations and upregulated intestinal pro-resolving mediator levels (11). Therefore, in this study, we assessed whether GPR101 was central to these modulatory properties of RvD5n-3 DPA. Lipid mediator profiling of intestinal tissues demonstrated that in mice administered a control siRNA sequence, RvD5n-3 DPA reduced the concentrations of the inflammatory eicosanoids, including PGD2 (193.4 ± 38.9 in vehicle-treated mice vs. 95.0 ± 15.7 pg/50 mg tissue) and PGE2 (376.3 ± 58.3 in vehicle-treated mice vs. 177.8 ± 16.9 pg/50 mg tissue), and increased the concentrations of select SPMs, namely RvD4 (0.6 ± 0.24 in vehicle-treated-mice vs. 2.7 ± 0.3 pg/50 mg tissue) and 17R-RvD1 (0.0 ± 0.0 in vehicle-treated mice vs. 0.21 ± 0.05 pg/50 mg tissue). Of note, these protective actions were diminished in mice transfected with the siRNA against Gpr101 prior to RvD5n-3 DPA administration (Supplemental Figure 4).
RvD5n-3 DPA accelerates the resolution of bacterial infections via GPR101.
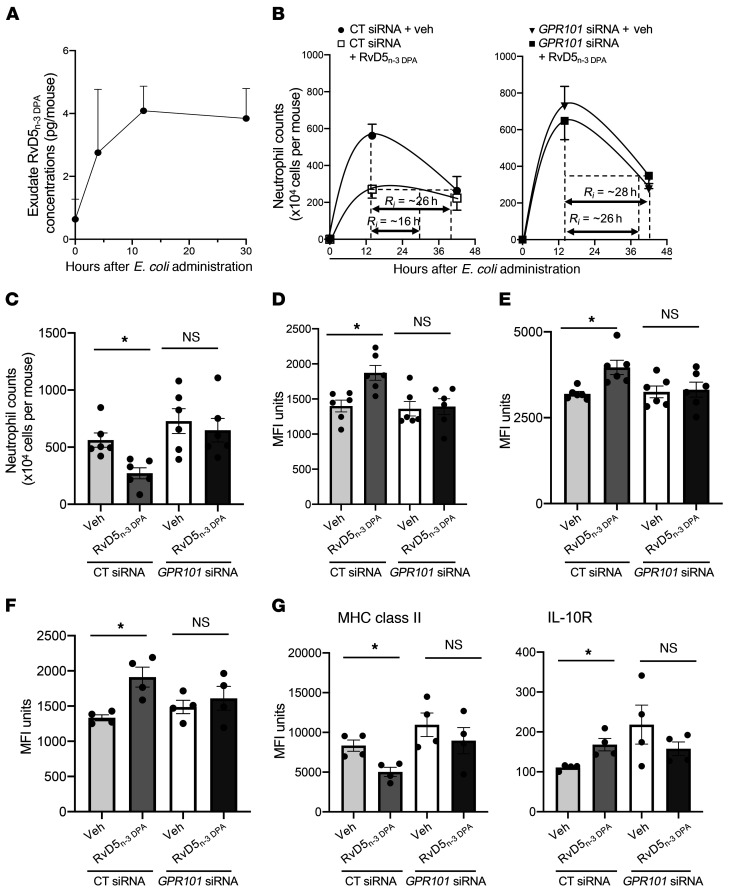
Having established that RvD5n-3 DPA increased the ability of monocytes, macrophages, and neutrophils to take up bacteria, we next questioned whether this mediator was produced during bacterial infections and whether its production was temporally regulated during self-limited infections. For this purpose, we inoculated mice with E. coli and assessed the temporal production of RvD5n-3 DPA using lipid mediator profiling. This analysis demonstrated that RvD5n-3 DPA was produced during bacterial infections, with its production reaching a maximum at the 12-hour interval and remaining sustained into the resolution phase of the inflammatory response (Figure 8A). We next tested whether the protective actions of RvD5n-3 DPA in promoting bacterial clearance as observed in vitro were retained in vivo and examined the role of GPR101 in mediating the observed regulatory actions. In mice given a control siRNA sequence, administration of RvD5n-3 DPA at a dose as low as 100 ng/mouse accelerated the resolution of E. coli–initiated inflammation, reducing the resolution interval (Ri) (i.e., the time it takes for neutrophil numbers to reduce from peak values to half maximum) from 26 hours to 16 hours (Figure 8B). This reduction in the resolution interval was linked with a significant reduction in the number of neutrophils recruited to the peritoneum (5.6 ± 0.9 × 106 in vehicle-treated mice vs. 2.7 ± 0.7 × 106 cells per mouse in RvD5n-3 DPA–treated mice) (Figure 8C); a significant upregulation in the ability of exudate neutrophils (1400 ± 118 MFI units in vehicle-treated mice vs. 1873 ± 150 MFI units in RvD5n-3 DPA–treated mice) and macrophages (3195 ± 94.7 MFI units in vehicle-treated mice vs. 3965 ± 292 MFI units in RvD5n-3 DPA–treated mice) to take up bacteria (Figure 8, D and E); and macrophage efferocytosis (Figure 8F). In these exudates, we also found a significant reduction in PGD2, PGE2 and the LTB4 metabolome, which encompasses the neutrophil chemoattractant LTB4 (Supplemental Figure 5).
Figure 8. Gpr101 knockdown limits the ability of RvD5n-3 DPA to activate host responses to promote the resolution of E. coli infections.
(A) Mice were inoculated with 105 CFU E. coli via i.p. injection and lavages collected at the indicated time intervals before (0 hours) or after inoculation. RvD5n-3 DPA concentrations were determined using lipid mediator profiling. Results represent the mean ± SEM (n = 3 mice per group). (B) Mice were administered 9 μg siRNA against mouse Gpr101 or a scrambled control sequence, and after 72 hours, they were administered RvD5n-3 DPA (100 ng/mouse) or vehicle control (PBS containing 0.1% ethanol) and then inoculated with 105 CFU E. coli via i.p. injection. (C) Fourteen-hour exudate neutrophil counts. Results represent the mean ± SEM (n = 6 mice per group from 2 distinct experiments). (D and E) Bacterial phagocytosis was determined in exudate (D) neutrophils and (E) macrophages at the 14-hour interval using flow cytometry. (F) Efferocytosis was determined at the 14-hour interval in CD64+F4/80+ macrophages using flow cytometry. (G) The expression of MHC class II and IL-10R was assessed in CD64+F4/80+ macrophages using flow cytometry. Results represent the mean ± SEM (n = 6 mice per group from 2 distinct experiments). *P < 0.05 versus the vehicle-treated group; Kruskal-Wallis test with Dunn’s post hoc multiple comparisons test (C–G).
Since macrophages are central players in both the termination and propagation of acute inflammation (4, 7, 18), we next investigated whether RvD5n-3 DPA regulates exudate macrophage phenotype during infectious inflammation. In exudate macrophages from mice given RvD5n-3 DPA and a control siRNA, we observed a decrease in the expression of MHC class II molecules (4863 ± 542 MFI units vs. 9136 ± 1536 MFI units in RvD5n-3 DPA–treated mice vs. vehicle-treated mice). We observed an increase in the expression of the fractalkine receptor CX3CR1 (4010 ± 198 MFI units vs. 3160 ± 148 MFI units in RvD5n-3 DPA–treated mice vs. vehicle-treated mice); the lipid mediator biosynthetic enzyme COX-2 (994 ± 70 MFI units vs. 1317 ± 106 MFI units in RvD5n-3 DPA–treated mice vs. vehicle-treated mice); and the IL-10R (195 ± 35 MFI units vs. 88 ± 20 MFI units in RvD5n-3 DPA–treated mice vs. vehicle-treated mice) (Figure 8G and Supplemental Figure 6), markers associated with a homeostatic and resolution macrophage phenotype (25, 26). Of note, transfection of mice with an siRNA against mouse Gpr101, which led to a reduction in neutrophil and macrophage receptor expression (Supplemental Figure 7), abolished the protective actions of RvD5n-3 DPA on both of these cell types (Figure 8, A–G, and Supplemental Figure 7). Together, these findings demonstrate that RvD5n-3 DPA activates host innate immune responses to promote the termination of infectious inflammation, actions that are mediated by GPR101.
Discussion
In the present study, using a screening approach, we identified GPR101 as a receptor for RvD5n-3 DPA. We found that RvD5n-3 DPA bound and activated GPR101 at a concentration that was commensurate with its biological actions. Inhibition of GPR101 using anti-GPR101 antibodies and siRNA demonstrated a role for this receptor in mediating the immunoregulatory actions of RvD5n-3 DPA in both human and mouse leukocytes. Knockdown of Gpr101 in vivo reversed the joint protective actions of RvD5n-3 DPA during inflammatory arthritis and the pro-resolving actions of this mediator during bacterial infections.
GPR101 is an X-linked receptor expressed on chromosome Xq26.3, the longer arm of the X chromosome. The GPR101 gene encodes a 508-aa deduced protein that shares approximately 30% sequence homology in the transmembrane regions with the α-1A-adrenergic receptor and the serotonin 5HT1A receptor (https://www.omim.org/entry/300393#1). In human brain tissues, two GPR101 transcript variants have been identified, one of 9.5 kb and the second of 4.2 kb, whereas in mouse brain, a 5.5-kb transcript was identified. The human GPR101 protein shares approximately 71% identity with the deduced mouse protein, which is predicted to be 511 aa in size. In the present study, we demonstrate that this receptor was expressed on human and mouse neutrophils, monocytes, and macrophages (Figure 1 and Supplemental Figures 2, 3, and 7). In humans, several mutations have been identified for this receptor that include the E308D mutation, which was identified in tumour DNA (27). A second mutation, the substitution of D366E, was identified in a patient with sporadic acromegaly (28). However, the functional impact of these mutations is not well characterized, given that the endogenous ligand(s) for this receptor were not known.
Pro-resolving mediators exert potent actions in the regulation of innate immune responses. These biological actions are mediated via activation of select GPCRs, to which these molecules bind with high stereospecificity and affinity (14–18). In the present study, using radiolabeled RvD5n-3 DPA, we found that this mediator bound to GPR101 with a KD of approximately 6.9 nM, which is comparable to its bioactive concentration range. We also found that RvD5n-3 DPA at concentrations as low as 0.1–10 nM was able to activate the receptor. This ability to activate intracellular signaling was found to be stereospecific, since neither RvD1n-3 DPA nor PD1n-3 DPA elicited marked changes in luminescence or impedance in GPR101-transfected CHO cells. Furthermore, subtle changes to the chemical backbone such as the substitution of the double bond at C10 and C13 for triple bond, which increases the rigidity of the molecule, also led to a significant (P < 0.5) decrease in its ability to activate the receptor.
Flow cytometric analysis revealed that GPR101 was expressed on both human and mouse neutrophils, monocytes, and macrophages. Leukocytes are involved in the initiation and termination of both sterile and inflammatory responses, with neutrophils, monocytes, and macrophages being the first responders (1, 2, 6, 7, 9, 18). SPMs, including RvD5n-3 DPA, have leukocyte-directed actions that regulate leukocyte trafficking to the site of inflammation (4, 8, 10, 18), promoting the uptake and killing of bacteria and enhancing the clearance of apoptotic cells and cellular debris. Results from the present studies demonstrate that knockdown of GPR101 limited the ability of human macrophages to take up both apoptotic cells and bacteria in response to RvD5n-3 DPA. Furthermore, inhibition of the receptor on human neutrophils using an anti-GPR101 antibody limited the biological actions of RvD5n-3 DPA, thereby reducing the ability of this mediator to promote the uptake and killing of bacteria, reversing its antichemotactic actions on neutrophils, and limiting its ability to reduced neutrophil diapedesis.
Rheumatoid arthritis is an autoimmune disease perpetuated by unremitting joint inflammation that results in bone destruction and joint deformation (29). Neutrophils and monocytes are implicated in the propagation of the effector phase of the disease. In a recent study by our group, we found that, in addition to joint inflammation, inflammatory arthritis also led to gut inflammation and weakening of the gut barrier, an observation that was linked with a reduction in intestinal levels of RvD5n-3 DPA (11). Furthermore, administration of RvD5n-3 DPA to arthritic mice reduced both joint and gut inflammation, restoring barrier function and reducing leukocyte infiltration into the paws (9). Here, we report that knockdown of GPR101 limited the protective actions of RvD5n-3 DPA in reducing joint and intestinal inflammation in experimental inflammatory arthritis. Knockdown of this receptor also blunted the ability of RvD5n-3 DPA to reduce the levels of inflammatory eicosanoids in the gut and joints of arthritic mice as well as promote the upregulation of SPMs in arthritic mice.
Bacterial infections result from an inability of the host immune response to contain invading pathogens (1). The prevailing approach to treating infections is to inhibit bacterial replication using antibiotics. The recent rise in antibiotic-resistant bacteria has spurred the search for novel approaches to treat infections. Studies investigating endogenous mechanisms promoting the resolution of bacterial infections demonstrate that SPMs reprogram the host immune response to positively regulate the uptake and killing of bacteria (4). Indeed, we found here that RvD5n-3 DPA promoted the uptake of bacteria by neutrophils, monocytes, and macrophages in a GPR101-dependent manner. Administration of RvD5n-3 DPA to mice also reduced the exudate concentrations of inflammatory eicosanoids, neutrophil infiltration into the site of inflammation, and the resolution interval from 26 hours to 16 hours. Of note, knockdown of GPR101 reversed these protective actions, underscoring the role of SPMs in reprogramming host immune responses during infection and the role of GPR101 in mediating the immunomodulatory actions of RvD5n-3 DPA during infections.
In summary, we have identified an immunoregulatory axis centered on GPR101/RvD5n-3 DPA that promotes the termination of both sterile and infectious inflammation. These findings demonstrate that GPR101 is the conveyor for the immune-regulatory properties of RvD5n-3 DPA and establish a role for this receptor in mediating the regulation of joint inflammation, both locally and remotely through the gut barrier, and in instructing the host to efficiently dispose of bacterial pathogens. Thus, these results provide new cues for harnessing the immune-modulatory actions of pro-resolving mediators by developing innovative therapeutic approaches based on the RvD5n-3 DPA/GPR101 axis.
Methods
GPCR screening
Using PathHunter β-arrestin enzyme fragment complementation (EFC) technology coupled with β-gal (DiscoveRx), we screened 49 orphan receptors. GPCR screening was performed by incubating 10 nM RvD5n-3 DPA or vehicle control (PBS in 0.01% ethanol) at 37°C for 90 minutes with the candidate GPCRs, and receptor activation was determined by measuring the luminescence signal. Negative controls were used to measure the potential constitutive activity in the absence of ligand. This custom screening was performed in duplicate, and the mean chemiluminescence value was used for analysis. The percentage of activity was calculated using the following formula: percentage of activity = 100% × (mean relative luminescence units [RLU] of the test sample – mean RLU of the vehicle control)/(mean RLU of the vehicle control).
GPCR–β-arrestin assays
To monitor downstream receptor activation, the β-arrestin PathHunter system (DiscoveRx) was used. CHO cells stably expressing human recombinant GPR12, GPR84, or GPR101 coupled to a β-arrestin reporter system were cultured in media containing 10% heat-inactivated FBS. GPCR and β-arrestin expression was maintained by dual selection in hygromycin B and geneticin according to the manufacturer’s instructions (DiscoveRx). For the experiments, single-use (eXpress) CHO cells stably expressing human recombinant GPR18 or GPR32 coupled to a β-arrestin reporter system were used. For all GPCR–β-arrestin assays, cells were plated in 96-well plates 48 hours before experiments using AssayComplete Cell Plating Reagent (DiscoveRx). Cells were incubated with the indicated concentrations of RvD5n-3 DPA (Vinresol, custom synthesis), PD1n-3 DPA, or RvD1n-3 DPA (provided by Trond Vidar Hansen, University of Oslo, Oslo, Norway) for 1 hour at 37°C, and receptor activation was determined by measuring chemiluminescence using the PathHunter Detection Kit (DiscoveRx).
xCELLigence RTCA DP system
Ligand-receptor interactions were assessed by monitoring impedance changes across single-cell monolayers using an xCELLigence DP System (ACEA Biosciences). GPR101-overexpressing CHO cells were plated on electronic microtiter plates (E-Plates). Test compounds were added to the chambers in serum-free medium, and real-time impedance changes were monitored (0–30 minutes at 37°C). For antibody incubations, anti-GPR101 antibody (clone G278, host: rabbit, Assay Biotechnology) or nonimmune rabbit IgG was incubated with cells on microtiter plates at 1:50 dilutions for 20 minutes before addition of the indicated mediators. In select experiments, GPR101-overexpressing CHO cells were incubated with CTX (1 μg/mL, 2 hours), PTX (1 μg/mL, 16 hours), or vehicle. Cells were then washed with serum-free HAM F-12 serum-free medium. RvD5n-3 DPA (10 nM) was added and impedance changes were assessed over a 30-minute period.
Preparation of [10, 11, 13, 143H]-RvD5n-3 DPA and radioligand binding
Total organic synthesis of [10, 11, 13,143H]-RvD5n-3 DPA was done by Vinresol (custom synthesis) via the tritiation of (7S,8E,15E,17S,19Z)-7,17-dihydroxydocosa-8,15,19-trien-10,13-diynoate. The integrity and activity of the radioligand was ascertained using reverse-phase UV-HPLC (RV-UV-HPLC) system (1100 Series, Agilent Technologies) equipped with an Eclipse Plus C18 Column (100 mm × 4.6 mm × 1.8 μm; Agilent Technologies) coupled with a diode array detector (DAD) (G1315B; Agilent Technologies). A gradient of methanol/water of 55:45 (vol/vol) was ramped to 63:37 (vol/vol) over a 22-minute period and then to 98:2 (vol/vol) for the next 8 minutes. The flow rate was maintained at 0.5 mL/min.
To assess the affinity of the radioligand for human GPR101, GPR101-overxpressing CHO cells were suspended in DPBS containing Ca2+ and Mg2+. For competition binding experiments, 0.5 × 106 cells/0.1 mL were incubated with approximately 3 nM [10, 11, 13,143H]-RvD5n-3 DPA (specific activity ~40 Ci/mmol) in the absence or presence of increasing concentrations of unlabeled RvD5n-3 DPA (1 nM–10 μM). Following incubation for 60 minutes at 4°C, cells were transferred to Whatman GF/C glass microfiber filters (Thermo Fisher Scientific), and unbound ligand was removed by washing twice with 4 mL ice-cold Dulbecco’s PBS (DPBS) using a vacuum manifold (MilliporeSigma). Bound radioactivity was then determined using a scintillation counter (Beckman Coulter). Nonspecific binding was determined in the presence of 10 μM unlabeled RvD5n-3 DPA.
For saturation binding experiments, 0.5 × 106 cells/0.1 mL cells were incubated with [10, 11, 13,143H]-RvD5n-3 DPA (1.25–100 nM), and then with or without 10 μM RvD5n-3 DPA for 60 minutes at 4°C. Cells were then transferred onto Whatman GF/C glass microfiber filters, and unbound ligand was removed by washing twice with 4 mL ice-cold DPBS using a vacuum manifold (MilliporeSigma). Bound radioactivity was determined using a scintillation counter (Beckman Coulter).
GPCR expression on primary cells
Human leukocyte GPCR expression.
Expression of GPR101, GPR12 (host: rabbit, lot AG11012912, catalog bs-13514R, Bioss) and GPR84 (host: mouse, lot 08248 WECz, catalog H00053831-A01, Abnova) was assessed in human peripheral blood monocytes, neutrophils, and macrophages. Nonspecific binding was quenched using FC blocking solution. Human cells were incubated with rabbit anti–human GPR101, rabbit anti–human GPR12, or mouse anti–human GPR84 (30 minutes at 4°C) and then with AF488-conjugated goat anti–rabbit IgG (Thermo Fisher Scientific) or AF488-conjugated goat anti–mouse IgG (Thermo Fisher Scientific) for 30 minutes at 4°C. Cells were then incubated with AF647-conjugated anti–human CD14 (clone HCD14, BioLegend) and APC-Cy7–conjugated anti–human CD16 (clone 3G8, BioLegend; for monocytes) antibodies (30 minutes at 4°C). For whole blood incubation, RBCs were lysed using a BD Pharmingen Red Blood Cell Lysis Kit following the manufacturer’s instructions, and fluorescence staining was evaluated using an LSR Fortessa Cell Analyzer (BD Biosciences) and FlowJo software, version 10 (Tree Star).
Mouse leukocyte GPCR expression.
Peripheral blood was collected via cardiac puncture in 3.2 % sodium citrate. Cells were then incubated with anti–mouse CD16/CD32 (clone 93, eBioscience; dilution 1:100, 20 minutes at 4°C in PBS containing 5% FCS staining solution). Peripheral blood cells were then incubated with AF700-conjugated anti–mouse Ly6G (clone 1A8, BioLegend; for neutrophils) and BV421-conjugated anti–mouse CD115 (clone AFS98, BioLegend; for monocytes) antibodies (30 minutes at 4°C), and receptor expression was evaluated as described above.
Targeted lipid mediator profiling
Paws and intestinal tissues were collected and immediately transferred to liquid nitrogen prior to homogenization in 1 mL ice-cold MeOH using a glass dounce. All samples for LC-MS/MS–based profiling were extracted using solid-phase extraction columns as described previously (30). Prior to sample extraction, deuterated internal standards, representing each region in the chromatographic analysis (500 pg each) were added to facilitate quantification. Samples were kept at –20°C for a minimum of 45 minutes to allow for protein precipitation. Supernatants were subjected to solid-phase extraction, and the methyl formate fraction was collected, brought to dryness, and suspended in phase (methanol/water, 1:1, vol/vol) for injection on a Shimadzu LC-20AD HPLC and a Shimadzu SIL-20AC autoinjector, paired with a QTrap 6500 Plus (Sciex). An Agilent Poroshell 120 EC-C18 column (100 mm × 4.6 mm × 2.7 μm) was kept at 50°C and the mediators eluted using a mobile phase consisting of methanol/water/acetic acid ratio of 20:80:0.01 (vol/vol/vol) that was ramped to 50:50:0.01 (vol/vol/vol) over 0.5 minutes and then to 80:20:0.01 (vol/vol/vol) from 2 minutes to 11 minutes, maintained until 14.5 minutes, and then rapidly ramped to 98:2:0.01 (vol/vol/vol) for the next 0.1 minutes. This was subsequently maintained at 98:2:0.01 (vol/vol/vol) for 5.4 minutes, and the flow rate was maintained at 0.5 mL/min. QTrap 6500 Plus was operated using a multiple reaction monitoring method as previously described (30). Each lipid mediator was identified using established criteria including matching of the retention time to synthetic or authentic standards and at least 6 diagnostic ions (30). Calibration curves were obtained for each mediator using lipid mediator mixtures at 0.78, 1.56, 3.12, 6.25, 12.5, 25, 50, 100, and 200 pg that gave linear calibration curves with r2 values of 0.98–0.99.
Animal experiments
E. coli peritonitis.
Six- to eight-week-old male C57BL/6 mice (Charles River Laboratories, the supplier fed the mice a lab diet containing essential fatty acids) were administered 9 μg Accell Control siRNA or Accell Mouse GPR101 siRNA SMARTpool (Dharmacon) via i.p. injection. After 3 days, mice were administered RvD5n-3 DPA (100 ng/mouse) or vehicle via i.p. injection immediately prior to i.p. injection of live E. coli (serotype O6:K2:H1, 105 CFU per mouse). At designated time intervals, the mice were euthanized, blood was collected via cardiac puncture in sodium citrate, and peritoneal exudates were collected in 4 mL PBS.
The cellular composition in the exudates was determined using Turk’s solution, light microscopy, and flow cytometry. For the latter, exudate cells were incubated with anti–mouse CD16/CD32 (clone 93, eBioscience; dilution 1:100, 20 minutes at 4°C in PBS containing 5% FCS staining solution), followed by incubation with phycoerythrin (PE) CD64 (FcγRI, clone X54-5/7.1, host mouse, BioLegend), Alexa Fluor 700 Ly-6G (clone 1A, host rat, BioLegend), Brilliant Violet 711 CD115 (CSF-1R) (clone AFS98, host rat, BioLegend), Brilliant Violet 510 CD43 (clone 1G10, host mouse, BD Biosciences), and APC/cyanine7 F4/80 (clone BM8, host rat, BioLegend) for 45 minutes on ice. To assess bacterial phagocytosis in peritoneal exudate leukocytes, exudate cells were incubated with anti–mouse CD16/CD32 (clone 93, eBioscience; dilution 1:100, 20 minutes at 4°C in PBS containing 5% FCS staining solution) and then with Alexa Fluor 700–conjugated anti–mouse Ly6G (for neutrophils), APC/Cy7-conjugated anti–mouse F4/80, and PE-conjugated anti–mouse CD64 antibodies (monocyte-derived macrophages; 30 minutes at 4°C in staining solution). Cells were then fixed and permeabilized using BD Perm/Wash Buffer (BD Biosciences) following the manufacturer’s instructions and incubated with FITC-conjugated anti–E. coli antibody (catalog GTX13626, GenTex; 1:50 dilution, 30 minutes at 4°C in BD Perm/Wash Buffer).
We assessed efferocytosis in infectious exudates by incubating exudate leukocytes with anti–mouse CD16 and anti–mouse CD32 and then with APC- and Cy7-conjugated anti–mouse F4/80 and PE-conjugated anti–mouse CD64 antibodies (30 minutes at 4°C in staining solution). Cells were then fixed and permeabilized using BD Perm/Wash Buffer (BD Biosciences) and incubated with FITC-conjugated anti–mouse Ly6G antibody (30 minutes at 4°C, in BD Perm/Wash Buffer).
In select experiments, resolution indices were calculated as described previously (17, 18), where the time difference for neutrophil numbers to go from maximum to half maximum was determined as the Ri. To assess the pro-resolving actions of the test products, these products were administered 12 hours after E. coli (105 CFU) inoculation, and resolution indices were assessed.
Inflammatory arthritis.
Ten-week-old male C57BL/6 mice (Charles River Laboratories, the supplier fed the mice a lab diet containing essential fatty acids) were administered 9 μg Accell Control siRNA or Accell Mouse GPR101 siRNA SmartPool (Dharmacon) via i.p. injection. After 24 hours, mice were i.p. administered 100 μl K/BxN serum (provided by Mauro Perretti and Dianne Cooper, Queen Mary University of London, London, United Kingdom) on days 0 and 2 to initiate inflammatory arthritis. Clinical scores were monitored daily using a 26-point arthritic scoring system (11). Mouse ankles, wrists, pads, and digits were inspected daily for swelling and redness. Blood, paws, and intestines were collected on day 8.
Mouse peripheral blood leukocyte phagocytosis.
Six- to eight-week-old male C57BL/6 mice (Charles River Laboratories; the supplier fed the mice a lab diet containing essential fatty acids) were i.p. administered 8 μg Accell Control siRNA or Accell Mouse GPR101 siRNA SMARTpool (Dharmacon). After 3 days, blood was collected via cardiac puncture and incubated with either RvD5n-3 DPA (10 nM) or vehicle (PBS containing 0.1% ethanol) for 20 minutes at 37°C, and then with 5 × 107 CFU fluorescently labeled E. coli for 60 minutes at 37°C. At the end of the incubations, RBCs were lysed using a BD Pharmingen Red Blood Cell Lysis Kit following the manufacturer’s instructions. The cells were then fixed and incubated with anti-CD16/CD32 antibody (20 minutes at 4°C) followed by AF700-conjugated anti–mouse Ly6G (clone 1A8, BioLegend; for neutrophils) and BV-421–conjugated anti–mouse CD115 (clone AFS98, BioLegend; for monocytes) antibodies (30 minutes at 4°C). Cells were then washed and staining evaluated using a BD Fortessa II with BD FACSDiva software, and the results were analyzed using FlowJo software, version 10.4 (Tree Star).
Blood collection from healthy volunteers
Venous peripheral blood was collected at the indicated intervals in sodium citrate (3.2%) from fasting volunteers who declared they had not taken NSAIDS for at least 14 days or consumed caffeine or alcohol for at least 24 hours or fatty fish for at least 48 hours.
Human neutrophil responses
Human neutrophil chemotaxis.
Human peripheral blood neutrophils were isolated from healthy volunteers as detailed above. Cells were incubated with either an antibody against GPR101 or an isotype control for 15 minutes at room temperature and then with RvD5n-3 DPA (0.1–10 nM) or vehicle control (PBS containing 0.01% ethanol) for 15 minutes at 37°C. Neutrophils were then transferred to the top chamber of a preloaded cell invasion and migration plate (CIM-Plate 16) on an xCELLigence DP system, and chemotaxis toward LTB4 (10 nM) was assessed in real time for 30 minutes.
Human neutrophil–endothelial cell interactions.
Human neutrophils, isolated as detailed above, were incubated with either an antibody against GPR101 or an isotype control for 15 minutes at room temperature and then with RvD5n-3 DPA (0.1–10 nM) or vehicle control (PBS containing 0.01% ethanol) for 15 minutes at 37°C. Cells were then perfused over TNF-α–activated human umbilical endothelial cells (10 ng/mL, 4 hours) at 0.1 PA for 8 minutes, 6 random 10-second fields were recorded per condition, and neutrophil–endothelial cell interactions were quantified as previously described (4).
Human macrophage responses
Human monocyte–derived macrophage efferocytosis.
PBMCs from healthy volunteers were purchased from the NHS Blood and Transplant Bank. PBMCs were isolated using Histopaque 1077 (MilliporeSigma) density centrifugation from blood cones. Macrophages were prepared using previously published protocols (31). Briefly, PBMCs were plated onto 10-cm tissue culture plates and incubated at 37°C for 30 minutes in PBS containing calcium and magnesium. Subsequently, cells were washed using PBS without calcium and magnesium, and adherent cells were incubated in RPMI 1640 containing 10% human serum and 20 ng/mL granulocyte macrophage–CSF (GM-CSF) for 7 days at 37°C in 5% CO2.
On day 7, monocyte-derived macrophages were then seeded onto 96-well plates at 4 × 104 cells per well and transfected with 1 μM Accell Human Control siRNA or Accell Anti–human GPR101 siRNA SMARTpool for 72 hours in serum-free Accell siRNA Delivery Medium (Dharmacon). Apoptotic target cells for efferocytosis were generated as follows: human promyelocytic HL-60 cells were seeded at 1 × 106 cells/mL onto 35-mm plates, irradiated with UV-C light (254 nm) for 15 minutes, and incubated at 37°C for 2 hours. Apoptosis induction was verified by flow cytometry using the APC Annexin V Apoptosis Detection Kit with PI (BioLegend), in which annexin V–positive and annexin V/PI–double-positive cells were considered to be apoptotic. Apoptotic HL-60 cells were washed with PBS and labeled with 1 μM pHrodo Red Succinimidyl Ester (Invitrogen, Thermo Fisher Scientific) in PBS for 30 minutes at room temperature. Macrophages were stained with CellBrite Blue (Biotium) for 1 hour at 37°C to visualize cell membranes, washed with RPMI-1640, and incubated with RvD5n-3 DPA (0.001–10 nM) or vehicle (RPMI-1640 containing 0.1% ethanol) for 15 minutes at 37°C. Apoptotic pHrodo Red–labeled HL-60 cells were added directly to the macrophages at a 3:1 ratio (apoptotic HL-60/macrophage), and the increase in pHrodo Red signal over time (representing apoptotic cell efferocytosis by macrophages) was quantified using a Zeiss Celldiscoverer 7 high-content imaging system.
Human monocyte–derived macrophage phagocytosis.
Monocyte-derived macrophages were seeded onto 96-well plates at 4 × 104 cells per well and transfected with 1 μM Accell Human Control siRNA or Accell Anti–human GPR101 siRNA SMARTpool for 72 hours in serum-free Accell siRNA Delivery Medium (Dharmacon). Macrophages were stained with Hoechst 33342 (Thermo Fisher Scientific) for 1 hour at 37°C to visualize cell nuclei, washed with RPMI-1640, and incubated with RvD5n-3 DPA (0.001–10 nM) or vehicle (RPMI-1640 containing 0.1% ethanol) for 15 minutes at 37°C. pHrodo Green–labeled S. aureus bioparticle conjugates (Invitrogen, Thermo Fisher Scientific) were opsonized by incubation in PBS containing 20% (v/v) human serum. Opsonized pHrodo Green–labeled S. aureus bioparticles were directly added to the macrophages at a final concentration of 5 μg per well, and the increase in pHrodo Green signal over time (representing phagocytosis of bacterial particles by macrophages) was quantified using a Zeiss Celldiscoverer 7 high-content imaging system.
cAMP measurements.
Human monocyte–derived macrophages were seeded onto 12-well plates at 5 × 105 cells per well and then transfected with 1 μM Accell Human Control siRNA or Accell Anti–human GPR101 siRNA SMARTpool for 72 hours in serum-free Accell siRNA Delivery Medium (Dharmacon). Cells were incubated with RvD5n-3 DPA (1 nM) or vehicle (PBS containing 0.01% ethanol) for 2 minutes, after which incubations were quenched with 5% Triton X-100, cells were homogenized, and cAMP levels were determined by ELISA following the manufacturer’s instruction (Elite cAMP ELISA Assay kit, eEnzyme).
Monocyte-derived macrophage l-kynurenine.
Macrophages were prepared as detailed above and then seeded onto 12-well plates at 2.5 × 105 cells per well and kept overnight in RPMI 1640 containing 10% FBS. Cells were then incubated with 10 nM RvD5n-3 DPA or vehicle. Two hours later, cells were collected using methanol containing deuterium-labeled choline and subjected to LC-MS/MS analysis as detailed previously (11) to determine l-kynurenine concentrations.
Statistical analysis
We performed all statistical analyses and data derivation using GraphPad Prism 8 (GraphPad Software) and Microsoft Excel software. Results presented in the figures are expressed as the mean ± SEM. Statistical differences between groups were determined as described in the figure legends. Briefly, Gaussian distribution of the data was tested using the Shapiro-Wilk normality test. For data following a normal distribution, statistical differences were tested using 1-way or 2-way ANOVA with a Holm-Sidak or Tukey’s post hoc multiple comparisons tests. For data not following a normal distribution, a Kruskal-Wallis test with Dunn’s post hoc multiple comparisons test was used. Sample sizes for each experiment were determined on the basis of the variability observed in prior experiments.
Study approval
Animal experiments were approved by the United Kingdom Home Office, London, United Kingdom (P998AB295) and adhered to the Guidance on the Operation of Animals, Scientific Procedures Act and to Laboratory Animal Science Association Guidelines (Guiding Principles on Good Practice for Animal Welfare and Ethical Review Bodies, Royal Society for the Prevention of Cruelty to Animals [RSPCA], 2015). Studies involving human primary cells were approved by the Queen Mary Research Ethics Committee, London, United Kingdom (QMREC 2014:61). All volunteers provided informed consent before blood donation. Healthy volunteers who donated blood gave written consent in accordance with a Queen Mary Research Ethics Committee (QMREC 2014:61) and Declaration of Helsinki principles.
Author contributions
JD conceived the overall research plan. MBF, DSK, AS, JS, KP, FP, and JD conducted the experiments and analyzed results. MBF and JD wrote the manuscript. All authors contributed to manuscript preparation. The order of the co–second authors was assigned on the basis of both experimental and intellectual contributions.
Supplementary Material
Acknowledgments
This work was supported by funding from the European Research Council under the European Union’s Horizon 2020 Research and Innovation Program (grant 677542, to JD); a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (grant 107613/Z/15/Z); the Barts Charity (grant MGU0343, to JD); a Marie Sklodowska Curie IF Fellowship under the European Union’s Horizon 2020 Research and Innovation Program (grant 746183); and the Barts Charity (grant MGU0439, to MBF). We also thank Jane Sosabowski, Roxana Kashani, and Julie Foster (Centre for Molecular Oncology, Queen Mary University of London) for assistance with radiolabel binding assays.
Version 1. 12/03/2019
Electronic publication
Version 2. 01/02/2020
Print issue publication
Version 3. 12/12/2022
Legend text for Figure 2C edited for clarity
Footnotes
Copyright: © 2020, Flak et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2020;130(1):359–373. https://doi.org/10.1172/JCI131609.
Contributor Information
Magdalena B. Flak, Email: m.b.flak@qmul.ac.uk.
Duco S. Koenis, Email: d.koenis@qmul.ac.uk.
Agua Sobrino, Email: a.sobrinogregorio@qmul.ac.uk.
Kimberly Pistorius, Email: k.c.d.f.pistorius@qmul.ac.uk.
Francesco Palmas, Email: f.palmas@qmul.ac.uk.
Jesmond Dalli, Email: j.dalli@qmul.ac.uk.
References
- 1.Majno G. The ancient riddle of sigma eta psi iota sigma (sepsis) J Infect Dis. 1991;163(5):937–945. doi: 10.1093/infdis/163.5.937. [DOI] [PubMed] [Google Scholar]
- 2. Malagoli D. The evolution of the immune system: conservation and diversification. Cambridge, Massachusetts, USA: Academic Press; 2016. [Google Scholar]
- 3.Scapini P, Cassatella MA. Location in the spleen dictates the function of murine neutrophils. J Exp Med. 2017;214(5):1207–1209. doi: 10.1084/jem.20170655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spite M, et al. Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature. 2009;461(7268):1287–1291. doi: 10.1038/nature08541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quiros M, et al. Macrophage-derived IL-10 mediates mucosal repair by epithelial WISP-1 signaling. J Clin Invest. 2017;127(9):3510–3520. doi: 10.1172/JCI90229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sumagin R, et al. Neutrophil interactions with epithelial-expressed ICAM-1 enhances intestinal mucosal wound healing. Mucosal Immunol. 2016;9(5):1151–1162. doi: 10.1038/mi.2015.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalli J, Serhan CN. Pro-resolving mediators in regulating and conferring macrophage function. Front Immunol. 2017;8:1400. doi: 10.3389/fimmu.2017.01400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hellmann J, et al. Biosynthesis of D-series resolvins in skin provides insights into their role in tissue repair. J Invest Dermatol. 2018;138(9):2051–2060. doi: 10.1016/j.jid.2018.03.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colas RA, et al. Impaired production and diurnal regulation of vascular RvDn-3 DPA increase systemic inflammation and cardiovascular disease. Circ Res. 2018;122(6):855–863. doi: 10.1161/CIRCRESAHA.117.312472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalli J, Colas RA, Serhan CN. Novel n-3 immunoresolvents: structures and actions. Sci Rep. 2013;3:1940. doi: 10.1038/srep01940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flak MB, Colas RA, Muñoz-Atienza E, Curtis MA, Dalli J, Pitzalis C. Inflammatory arthritis disrupts gut resolution mechanisms, promoting barrier breakdown by Porphyromonas gingivalis. JCI Insight. 2019;4(13):125191. doi: 10.1172/jci.insight.125191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalli J, Serhan CN. Identification and structure elucidation of the pro-resolving mediators provides novel leads for resolution pharmacology. Br J Pharmacol. 2019;176(8):1024–1037. doi: 10.1111/bph.14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugimoto Y, Narumiya S. Prostaglandin E receptors. J Biol Chem. 2007;282(16):11613–11617. doi: 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- 14.Fiore S, Romano M, Reardon EM, Serhan CN. Induction of functional lipoxin A4 receptors in HL-60 cells. Blood. 1993;81(12):3395–3403. [PubMed] [Google Scholar]
- 15.Perretti M, et al. Endogenous lipid- and peptide-derived anti-inflammatory pathways generated with glucocorticoid and aspirin treatment activate the lipoxin A4 receptor. Nat Med. 2002;8(11):1296–1302. doi: 10.1038/nm786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnamoorthy S, et al. Resolvin D1 binds human phagocytes with evidence for proresolving receptors. Proc Natl Acad Sci U S A. 2010;107(4):1660–1665. doi: 10.1073/pnas.0907342107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dalli J, et al. Resolvin D3 and aspirin-triggered resolvin D3 are potent immunoresolvents. Chem Biol. 2013;20(2):188–201. doi: 10.1016/j.chembiol.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang N, Dalli J, Colas RA, Serhan CN. Identification of resolvin D2 receptor mediating resolution of infections and organ protection. J Exp Med. 2015;212(8):1203–1217. doi: 10.1084/jem.20150225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson KR, Eglen RM. Beta galactosidase complementation: a cell-based luminescent assay platform for drug discovery. Assay Drug Dev Technol. 2007;5(1):137–144. doi: 10.1089/adt.2006.052. [DOI] [PubMed] [Google Scholar]
- 20.Peters MF, Scott CW. Evaluating cellular impedance assays for detection of GPCR pleiotropic signaling and functional selectivity. J Biomol Screen. 2009;14(3):246–255. doi: 10.1177/1087057108330115. [DOI] [PubMed] [Google Scholar]
- 21.Gerlach BD, et al. Resolvin D1 promotes the targeting and clearance of necroptotic cells. Cell Death Differ. doi: 10.1038/s41418-019-0370-1. [published online ahead of print June 20, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanis JM, et al. Tryptophan metabolite activation of the aryl hydrocarbon receptor regulates IL-10 receptor expression on intestinal epithelia. Mucosal Immunol. 2017;10(5):1133–1144. doi: 10.1038/mi.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gobbetti T, et al. Protectin D1n-3 DPA and resolvin D5n-3 DPA are effectors of intestinal protection. Proc Natl Acad Sci USA. 2017;114(15):3963–3968. doi: 10.1073/pnas.1617290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christensen AD, Haase C, Cook AD, Hamilton JA. K/BxN serum-transfer arthritis as a model for human inflammatory arthritis. Front Immunol. 2016;7:213. doi: 10.3389/fimmu.2016.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang W, et al. Neutrophils promote the development of reparative macrophages mediated by ROS to orchestrate liver repair. Nat Commun. 2019;10(1):1076. doi: 10.1038/s41467-019-09046-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zigmond E, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40(5):720–733. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Trivellin G, et al. Gigantism and acromegaly due to Xq26 microduplications and GPR101 mutation. N Engl J Med. 2014;371(25):2363–2374. doi: 10.1056/NEJMoa1408028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamenický P, Bouligand J, Chanson P. Gigantism, acromegaly, and GPR101 mutations. N Engl J Med. 2015;372(13):1264–1265. doi: 10.1056/NEJMc1500340. [DOI] [PubMed] [Google Scholar]
- 29.Pitzalis C, Kelly S, Humby F. New learnings on the pathophysiology of RA from synovial biopsies. Curr Opin Rheumatol. 2013;25(3):334–344. doi: 10.1097/BOR.0b013e32835fd8eb. [DOI] [PubMed] [Google Scholar]
- 30.Walker ME, Souza PR, Colas RA, Dalli J. 13-Series resolvins mediate the leukocyte-platelet actions of atorvastatin and pravastatin in inflammatory arthritis. FASEB J. 2017;31(8):3636–3648. doi: 10.1096/fj.201700268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalli J, Serhan CN. Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood. 2012;120(15):e60–e72. doi: 10.1182/blood-2012-04-423525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.