Abstract
Cellular membranes undergo remodeling during many cellular processes including endocytosis, cytoskeletal protrusion, and organelle biogenesis. During these events, specialized proteins sense and amplify fluctuations in membrane curvature to create stably curved architectures. Amphiphysin1 is a multi-domain protein containing an N-terminal crescent-shaped BAR (Bin/Amphiphysin/Rvs) domain and a C-terminal domain that is largely disordered. When studied in isolation, the BAR domain of Amphiphysin1 senses membrane curvature and generates membrane tubules. However, the disordered domain has been largely overlooked in these studies. Interestingly, our recent work has demonstrated that the disordered domain is capable of substantially amplifying the membrane remodeling ability of the BAR domain. However, the physical mechanisms responsible for these effects are presently unclear. Here we elucidated the functional role of the disordered domain by gradually truncating it, creating a family of mutant proteins, each of which contained the BAR domain and a fraction of the disordered domain. Using quantitative fluorescence and electron microscopy, our results indicate that the disordered domain contributes to membrane remodeling by making it more difficult for the protein to bind to and assemble on flat membrane surfaces. Specifically, we found that the disordered domain began to significantly impact membrane remodeling when its projected area exceeded that of the BAR domain. Once this threshold was crossed, steric interactions with the membrane surface and with neighboring disordered domains gave rise to increased curvature sensing and membrane vesiculation, respectively. These findings provide insight into the synergy between structured and disordered domains, each of which play important biophysical roles in membrane remodeling.
INTRODUCTION
Deforming membranes such that they take on high curvatures is essential for many cellular processes, such as formation of trafficking vesicles,1, 2 viral budding,3 and cytokinesis.4 However, membranes are inherently resistant to deformation.5 Therefore, cells must employ proteins to sense and generate membrane curvature.3, 6, 7 Curvature sensing and curvature generation have been primarily attributed to protein domains with specific structural features. Some examples include wedge-like amphipathic helices and crescent-shaped BAR (Bin/Amphiphysin/Rvs) domains. Amphipathic helices sense membrane curvature by inserting into membrane packing defects, which are highly abundant on curved membrane surfaces.8 In contrast, crescent-shaped BAR domains match the curvature of highly curved membrane surfaces, resulting in stronger binding and curvature sensing.9 BAR domains have also been shown to generate membrane tubules by assembling into cylindrical scaffolds on membrane surfaces,10–12 forcing the underlying membrane to adopt the cylindrical geometry of the scaffold.13
Recently, we have demonstrated that intrinsically disordered proteins (IDPs) are also capable of sensing14, 15 and generating16, 17 membrane curvature. Disordered domains lack substantial regions of secondary structure, behaving more like random polymers than stably folded protein domains.18, 19 These polymer-like properties contribute to the ability of disordered proteins to sense and generate membrane curvature. In particular, disordered domains sample a variety of conformations, giving them a high degree of conformational entropy. When a disordered domain is tethered to a membrane surface, the membrane surface imposes a geometric constraint that substantially reduces the conformational entropy of the disordered domain. However, as the surface curves away from the protein and adopts convex curvature, the geometric constraint is relieved and the conformational entropy of the disordered domain increases. This increase in entropy favors binding of disordered domains to curved surfaces, rather than flat surfaces.14, 15 As more proteins continue to bind and become crowded on a membrane surface, steric pressure is generated between the proteins. This steric pressure can be alleviated by bending of the membrane away from the crowded proteins, increasing the average distance between them.20, 21 Importantly, the expanded conformations of disordered proteins make them efficient generators of membrane curvature.16, 17 Specifically, disordered domains have substantially larger hydrodynamic radii in comparison to stably folded proteins of equivalent molecular weight.19, 22
Interestingly, disordered domains are often coupled with structured domains within the same protein molecule. This coupling is particularly common in membrane trafficking pathways, where nearly 40% of proteins contain substantial regions of intrinsic disorder.23–25 As discussed above, the curvature sensing and generation properties of the structured domains have been frequently studied.9, 26 In contrast, the disordered domains have typically been neglected in these studies, based on the common assumption that curvature sensing and curvature generation require specific structural motifs. Therefore, with the recent discoveries of biophysical roles for disordered domains,14–17 it has become increasingly important to reevaluate the mechanisms responsible for membrane remodeling in trafficking pathways.
Amphiphysin1, a protein involved in the late stages of clathrin coated pit assembly, provides an interesting model protein for examining curvature sensing and generation, as it contains both a structured BAR domain and a substantial disordered domain.9, 27 Previous work has demonstrated that low concentrations of Amphiphysin1’s BAR domain are capable of sensing membrane curvature,9 while higher concentrations assemble into tubular scaffolds10–12 that drive the membrane to take on a tubular geometry.13 Interestingly, when the full-length Amphiphysin1 protein is used in similar studies, curvature sensitivity increases substantially,14 suggesting that the disordered domains make a significant contribution to the sensing process. Higher coverage of the membrane surface by full-length Amphiphysin1 results in membrane vesiculation,28 a process that is more energetically demanding than forming membrane tubules.29 Here membrane vesiculation has been hypothesized to arise from the propensity of assembled BAR scaffolds to locally concentrate bulky disordered domains, driving the local accumulation of steric pressure.28
The ability of the structured BAR domain and the disordered domain of Amphyphysin1 to collaborate to drive membrane remodeling suggests that other proteins that couple structured curvature sensors with disordered domains may function similarly. Many examples of this coupling are found in membrane trafficking pathways24, 25 and cytokinesis.30, 31 However, the disordered domains within these proteins vary greatly in length from less than 50 amino acids to more than 1000 amino acids.24 Therefore, it is important to assess how long the disordered amino acid chain needs to be before it can be expected to make a substantial contribution to membrane curvature sensing and generation.
To address this question, we investigated curvature sensing and membrane vesiculation as a function of the length of the disordered domain. In these studies we examined a set of proteins consisting of constructs for which the C-termini were successively truncated.. To examine these biophysical phenomena, we utilized quantitative fluorescence microscopy to observe protein partitioning and membrane remodeling, as well as electron microscopy to directly observe membrane deformation. Our results revealed that curvature sensitivity and membrane vesiculation capacity are successively reduced as the IDP domain is progressively shortened. Further, we found that the disordered domain made a substantial contribution to sensing and driving membrane curvature whenever its expected hydrodynamic radius significantly exceeded that of the BAR domain. For curvature sensing, the significance of crossing this size threshold is likely that the disordered domain begins to experience the conformational constraint associated with the underlying membrane surface. In contrast, for the process of curvature generation, a disordered domain that is larger than the BAR domain likely enables steric interactions between neighboring disordered domains, fueling the steric pressure that ultimately drives membrane vesiculation. Collectively these findings provide useful biophysical insights into the potential coupling between structured and disordered domains involved in membrane remodeling.
MATERIALS AND METHODS
Materials
DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), DOPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine), and PI(4,5)P2 (L-α-phosphatidylinositol-4,5-bisphosphate; Brain, Porcine) were purchased from Avanti Polar Lipids, Inc. Oregon Green-DHPE (Oregon Green 488 1,2,-dihexadecanoyl-sn-glycero-3-phosphoethanolamine) was purchased from Thermo Fisher Scientific. DP-EG10-biotin (dipalmitoyl-decaethylene-glycol-biotin) was provided by Darryl Sasaki from Sandia National Laboratories, Livermore, CA.32 MOPS (3-(N-morpholino)propanesulfonic acid), HEPES (4–2(2-hydroxymethyl)-1-piparazineethanesulphonic acid), Tris (tris(hydroxymethyl)aminomethane) hydrochloride, Sodium Chloride, DTT (dithiothreitol), IPTG (isopropyl-β-D-thiogalactopyranoside), β-mercaptoethanol, thrombin protease, HRV-3C (human rhinovirus-3c) protease, Triton X-100, Neutravidin, Glycerol, and Zeba spin desalting columns (7K MWCO, 5 mL) were purchased from Thermo Fisher Scientific. EGTA (ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid), EDTA (ethylenediaminetetraacetic acid), TCEP (Tris(2-carboxyethyl) phosphine hydrochloride), PMSF (phenylmethanesulfonyl fluoride), PLL (poly-L-lysine), EDTA-free protease inhibitor tablets, p-aminobenzamidine-agarose resin, and ATTO594 NHS-ester were purchased from Sigma-Aldrich. Amine reactive PEG (mPEG-succinimidyl Valerate, 5kDa) and PEG-biotin (biotin-PEG SVA, 5 kDa) were purchased from Laysan Bio, Inc. Glutathione Sepharose 4B was purchased from GE Healthcare. All reagents were used without additional purification.
Plasmids
Bacterial expression vectors for N-BAR and Amph-FL were prepared as described previously. Briefly, the pGex6P vector containing full-length human Amphiphysin1 (residues 2–695) was provided by the laboratory of Tobias Baumgart (University of Pennsylvania). The N-BAR domain of human Amphiphysin1 (residues 2–242) was cloned into the pGex4T2 vector using EcoRI and BamHI restriction sites. To generate the Amph-1/3 construct, the Amph-FL plasmid was PCR amplified using primers that introduced a premature stop codon (TAG) in place of the codon for glutamine at residue 370 (Table 1). For Amph-2/3, PCR primers introduced a premature stop codon (TAG) in place of the codon for alanine at residue 497 (Table 1).
Table 1.
Sequences of primers used for generating Amph-1/3 and Amph-2/3 plasmids. Stop codons are highlighted in red.
| Protein | Primer Sequence (5’ to 3’) |
|---|---|
| Amph-1/3 | Forward: GTGACACCTGCAGGTTCTGCTTAGGTGACCCACTCACCCATGTCT Reverse: AGACATGGGTGAGTGGGTCACCTAAGCAGAACCTGCAGGTGTCAC |
| Amph-2/3 | Forward: GAGGAAGCAGAGGCGGAGAAGTAGACTGTCCCTGCCGGGGAAGGA Reverse: TCCTTCCCCGGCAGGGACAGTCTACTTCTCCGCCTCTGCTTCCTC |
Protein Purification
All proteins were expressed as fusion proteins with an N-terminal GST tag, which was later cleaved. Proteins were expressed in BL21 E. coli cells following induction with 1 mM IPTG at 30°C for 6–8 hours. Bacteria was then harvested, pelleted, and resuspended into a lysis buffer that contained 500 mM Tris-HCl, 5 mM EDTA, 10 mM β-mercaptoethanol, 5% glycerol, 1 mM PMSF, 1% v/v Triton X-100, and 1x Roche protease inhibitor cocktail (pH=8.0). Cells were then lysed using probe sonication. Proteins were purified by incubating bacterial lysate with glutathione resin. After extensive washing of the glutathione columns, proteins were cleaved directly from the resin by incubating with soluble HRV-3C or thrombin proteases overnight at 4°C. Excess HRV-3C, which contained a GST tag, was removed by passage through another glutathione agarose column. Thrombin was removed using p-aminobenzamidine-agarose resin. Proteins were concentrated using EMD Millipore Amicon centrifugal filter units. Proteins were exchanged into buffer containing 25 mM HEPES, 150 mM NaCl, and 5 mM TCEP (pH = 7.4) using Zeba Spin Desalting columns. Proteins were stored as small aliquots at −80°C.
Protein Labeling
Proteins used for curvature sensing experiments were fluorescently labeled with ATTO-594. ATTO-594 NHS-ester was dissolved in DMSO to create a 10 mM stock, which was stored in small aliquots at −80°C. N-BAR, Amph-1/3, -2/3, and –FL were labeled in a buffer consisting of 25 mM HEPES, 150 mM NaCl, and 5 mM TCEP (pH = 7.4). Protein concentrations varied from 10–40 μM. ATTO-594 stock was combined with protein such that the stoichiometric ratio of dye:protein was 2:1. The total amount of DMSO present never exceeded 1 v/v%. ATTO-594 NHS-ester reacted with primary amines within the proteins. The reactions were allowed to proceed for 30 minutes at room temperature. Afterward, unreacted dye was removed using Centri-Spin size exclusion columns (Princeton Separations). Protein and dye concentrations were measured using UV-Vis spectroscopy. The labeling ratios for all proteins varied from 0.5 to 1.5 dyes per protein. The dye ratios used helped ensure that the binding abilities of the proteins were impacted minimally. Labeled proteins were then stored as small aliquots at −80°C.
Vesicle Preparation
DOPC, DOPS, DP-EG10-biotin, and Oregon Green-DHPE stocks were stored in chloroform at −80°C. PI(4,5)P2 stocks were stored in a mixture containing 66% v/v chloroform, 30% v/v methanol, and 3% v/v water, also at −80°C. Lipid stocks were brought to room temperature and combined such that the final molar percentages were 76% DOPC, 15% DOPS, 5% PI(4,5)P2, 2% DP-EG10-biotin, and 2% Oregon Green DHPE. Additional methanol was added to the combined lipid mixture to bring the final methanol concentration to 30% v/v. The mixture was dried under a stream of nitrogen gas, then stored under vacuum for at least 12 hours to ensure solvent removal. The resulting lipid film was then resuspended in buffer containing 20 mM MOPS, 150 mM sodium chloride, 0.5 mM EGTA , and 0.5 mM EDTA (pH = 7.4). A mixture of sonicated and extruded vesicles were used for curvature sensing experiments. Extruded vesicles alone were used for fission experiments. For sonication, lipid suspensions were sonicated in an ice bath using a probe tip sonicator (Branson Ultrasonics). For extrusion, lipid suspensions were extruded through either 100 nm or 200 nm polycarbonate membranes (Whatman plc). Vesicles were prepared at concentrations ranging from 100–200 μM and diluted as necessary.
Curvature Sensing Experiments
All curvature sensing experiments were carried out in buffer containing 20 mM MOPS, 150 mM sodium chloride, 0.5 mM EGTA, and 0.5 mM EDTA (pH = 7.4). Imaging wells consisted of 0.8 mm thick silicone gaskets (Grace Bio-labs) adhered to no. 1.5 glass coverslips. Wells were passivated with buffer solution containing biotinylated PEG as described previously.14 After 20 minutes of incubation, excess biotinylated PEG was rinsed from the wells. Neutravidin was added at a concentration of 0.2 mg/mL and allowed to incubate for 10 minutes. Excess neutravidin was then rinsed from the wells using the MOPS buffer. Wells were then rinsed with a MOPS buffer solution containing 50 nM sonicated vesicles and 500 nM extruded vesicles (100 nm extruded). After 10 minutes, excess vesicles were rinsed from the wells and protein was added at specified concentrations. Protein was incubated for at least 20 minutes prior to imaging. Wells were stored in sealed containers to prevent evaporation.
Tethering of Vesicles after Exposure to Proteins
All fluorescence-based membrane remodeling assays were carried out in buffer containing 20 mM MOPS, 150 mM sodium chloride, 0.5 mM EGTA, 0.5 mM EDTA, and 5 mM TCEP (pH = 7.4). Experiments were performed by combining extruded vesicles (200 nm extruded) at a concentration of 10 μM with unlabeled proteins at specified concentrations in the aforementioned MOPS buffer. Vesicles and protein were then incubated at 37°C for 30 minutes. During this incubation time, imaging wells were prepared as outlined in the “Curvature Sensing Experiments” section. After excess neutravidin was rinsed from the wells and protein-vesicle incubation was complete, protein and vesicle mixtures were added to the wells and allowed to tether for 10 minutes at room temperature. Afterward, wells were rinsed to remove untethered vesicles.
Fluorescence Microscopy
A spinning disc confocal microscope (Zeiss Axio Observer Z1 with Yokogawa CSU-X1M) was used to visualize tethered vesicles and membrane-bound proteins. Green and Red fluorescent samples were excited with lasers having wavelengths of 488 nm and 562 nm, respectively. Emission filters were centered at 525 nm and 629 nm with widths of 50 nm and 62 nm, respectively. The microscope was equipped with a 100× 1.4 N.A. oil immersion objective, as well as a cooled (−70°C) EMCCD iXon3 897 camera (Andor Technology).
Image Processing
Tethered vesicles were imaged using a perpendicular “z-stack” method.14 Images stacks were acquired in the direction perpendicular to the surface of the glass cover slip. The image with the highest mean fluorescence intensity was chosen as the plane of focus. Vesicles appeared as diffraction-limited puncta. These puncta were fit to two dimensional Gaussians using the publicly-available particle detection program, cmeAnalysis.33 Gaussian amplitudes corresponded to the brightness of each vesicle. Only puncta with amplitudes significantly above the local fluorescence background were included in our analysis. To further ensure that puncta in the lipid channel were above the noise threshold of our images, we only included puncta that persisted in the same location throughout three consecutive frames. In the presence of fluorescently labeled protein, amplitudes of Gaussian fits were obtained in both the lipid and protein fluorescent channels. The program used an algorithm to ensure that the centroids of puncta in the lipid and protein channels were spatially colocalized within a specified search radius. This search radius was equal to three times the standard deviation of the Gaussian fit.
Determination of Vesicle Diameter and Number of Membrane-bound Proteins
Vesicle diameter was calibrated using a method developed initially by the Stamou research group8, 34, 35 and previously used in our research group.14, 16 The vesicle diameter distribution was measured using dynamic light scattering and the vesicle fluorescence intensity distribution was measured using the aforementioned image processing technique. We used the means of these two distributions to compute an approximate scaling factor between fluorescence intensity and vesicle surface area. This scaling factor was then used to convert vesicle fluorescence intensities to vesicle diameters.
In the presence of fluorescently-labeled protein, the number of proteins bound to the surface of each tethered vesicle was estimated using a fluorescence calibration experiment. First we measured the average fluorescent emission from a single ATTO-594 labeled protein. In these calibration measurements, fluorescently labeled proteins were adsorbed to glass cover slips and appeared as diffraction-limited puncta. Importantly, proteins that were used for these calibration measurements were in stoichiometric excess to dye molecules, ensuring that very few proteins contained multiple fluorophores. The fluorescence intensity distributions of these puncta were acquired using the publicly-available particle detection program, cmeAnalysis.33 These fluorescence intensity distributions contained one prominent peak, which corresponded to mean brightness of a single ATTO-594 molecule. Once the single molecule brightness value was determined, we used the same imaging conditions to measure the brightness of proteins that colocalized with tethered vesicles. Dividing these values by the single molecule brightness value yielded an estimate of the number of membrane-bound proteins per vesicle.
Transmission Electron Microscopy (TEM)
Extruded vesicles (200 nm extruded) and protein were combined at 100 μM and 5 μM, respectively, and allowed to incubate for 30 minutes at 37°C. Vesicles and protein were incubated in buffer containing 20 mM MOPS, 150 mM sodium chloride, 0.5 mM EGTA, 0.5 mM EDTA, and 5 mM TCEP (pH = 7.4). Next, 5 μL of the protein-lipid mixture was placed onto glow-discharged, 300 square mesh, carbon-coated grid and stained with 2% uranyl acetate (Electron Microscopy Sciences). Samples were then imaged on a Tecnai Spirit BioTwin T12 electron microscope (Tecnai). Vesicles diameters were measured using ImageJ software.
RESULTS AND DISCUSSION
The effect of disordered domain length on membrane curvature sensing.
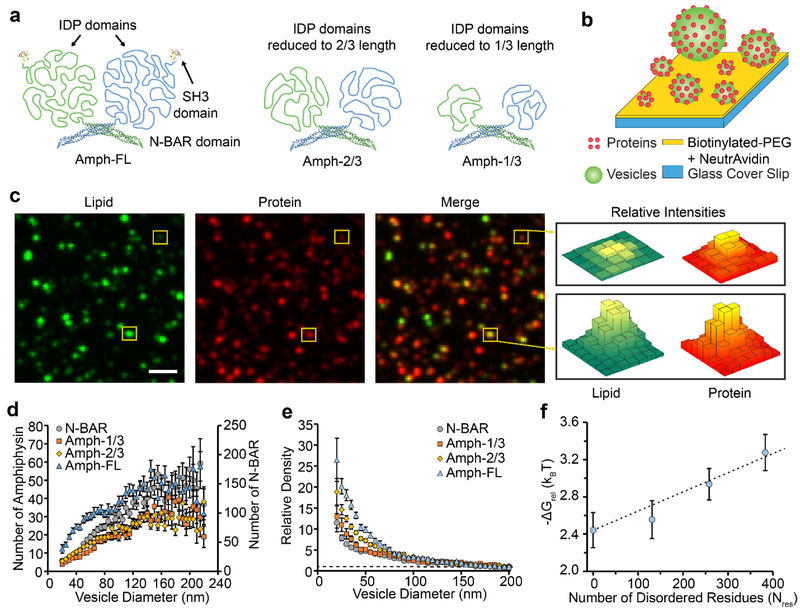
Amphiphysin1 contains an N-terminal BAR domain, an intrinsically disordered protein (IDP) domain, and a C-terminal SH3 domain. The BAR domain facilitates membrane binding.9 When the BAR domain was removed, the IDP domain exhibited no measureable binding to lipid vesicles, Supplemental Figure S1. Therefore, we can conclude that the C-terminal domain does not contain any membrane-binding motifs. The protein forms a native dimer as shown in Figure 1a.9 We examined curvature sensing by Amphiphysin1 (human, residues 2–695) as a function of IDP domain length by generating three truncation mutants. In total we examined four different proteins: full-length Amphiphysin1 (Amph-FL; residues 2–695), Amph-2/3 (residues 2–496), Amph-1/3 (residues 2–369), and N-BAR (residues 2–242), Figure 1a. Amph-1/3 and Amph-2/3 contained the N-BAR domain and approximately the first 1/3 and 2/3 of the disordered domain, respectively. N-BAR (residues 2–242) lacked the disordered domain entirely. Using polymer scaling approximations from measurements that were performed previously,14 we estimate that the total projected area of the individual disordered domains to be 75 nm2, 50 nm2, and 25 nm2 for Amph-FL, Amph-2/3, and Amph-1/3, respectively. For comparison, the projected area of the N-BAR monomer is approximately 23 nm2, which represents half of the projected area of the N-BAR dimer in the published crystal structures.36
Figure 1: Amphiphysin1 curvature sensitivity decreases as the disordered domains are truncated.
(a) Structure of full-length Amphiphysin1 and its truncation mutants. (b) Schematic of the assay used for tethering vesicles and measuring curvature sensitivity. (c) Lipid, protein, and merged fluorescent images of vesicles exposed to 2 nM Amph-FL. Yellow boxes indicate vesicles with different fluorescence intensities, which correspond to vesicles with different diameters. (d) The average number of proteins bound to each vesicle as a function of vesicle diameter for populations of vesicles exposed to either N-BAR, Amph-1/3, Amph-2/3, or Amph-FL. (e) The relative density of Amph-FL and its truncation mutants as a function of vesicle diameter. The dashed line corresponds to Relative Density = 1. (f) ΔGrel plotted as a function of the number of residues in the IDP domain, Nres. The dashed line represents the scaling expected based on polymer theory. All vesicles were composed of 76% DOPC, 15% DOPS, 5% PI(4,5)P2, 2% Oregon Green-DHPE, and 2% DP-EG10-biotin. Scale bar in c represents 2 μm. Data in d-e is presented as a 5 nm-increment moving average of the raw data (>1000 total data points, see Supplemental Figure S2). Each error bar represents the standard error of the mean within each bin. Each of these bins contains from 15–272 data points acquired cumulatively from three independent replicates. In d-e, N-BAR and Amph-1/3 concentrations were 5 nM, while Amph-2/3 and Amph-FL concentrations were 2 nM. Concentrations were varied to ensure that all proteins occupied the vesicles at comparable membrane coverages, Supplemental Figure S2. Error bars in f represent the propagated error from the corresponding error in e.
To measure the sensitivity of these proteins to membrane curvature, we utilized the tethered vesicle assay depicted in Figure 1b. Here biotin-NeutrAvidin interactions were used to tether vesicles with diameters ranging from 20 to 250 nm to glass cover slips passivated with polyethylene glycol (PEG), as described previously.8, 14 This range of vesicle diameter encompasses many biologically relevant structures, including trafficking vesicles.3 Binding of Amphiphysin1 to membranes is enhanced in the presence of anionic lipids due to electrostatic attraction between the negatively-charged membrane and positive charges on the concave surface of the N-BAR domain.9, 37 Therefore, we facilitated membrane binding by incorporating the anionic lipids DOPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine) and PI(4,5)P2 (L-α-phosphatidylinositol-4,5-bisphosphate) into our vesicles at molar percentages of 15% and 5%, respectively. After tethering vesicles to glass cover slips, we incubated vesicles with protein-containing solutions and visualized protein binding using fluorescence microscopy.
Vesicles were fluorescently labeled using Oregon Green DHPE (Oregon Green 488 1,2,-dihexadecanoyl-sn-glycero-3-phosphoethanolamine). Proteins were fluorescently labeled using ATTO-594, which was conjugated covalently to primary amines within each protein. Using the tethered-vesicle assay, we monitored partitioning of Amph-FL and its truncation mutants between vesicles of different diameters. Vesicles appeared as diffraction-limited puncta, as shown in Figure 1c. In these images, tethered vesicles were incubated with Amph-FL. Here the ability of the protein to sense curvature is evident upon visual inspection, consistent with our recent report.14 Specifically, as indicated by the boxed puncta, the ratio of the protein fluorescence intensity to lipid fluorescence intensity appeared to generally increase as vesicle diameter decreased. Therefore, in merged images, larger vesicles appeared more green while smaller vesicles appeared more red, Figure 1c.
To quantitatively compare curvature sensitivities of Amph-FL and its truncation mutants, we measured fluorescence intensities of colocalized puncta within the lipid and protein fluorescence channels to determine the diameter of each vesicle and the number of proteins bound to each vesicle. The vesicle diameter was determined by comparing the mean Oregon Green DHPE intensity for tethered vesicles to the mean diameter obtained from dynamic light scattering, as outlined in the methods section. The number of proteins bound to each vesicle was determined by dividing the fluorescence intensity in the protein channel by the calibrated fluorescence intensity of a single ATTO 594-labeled protein, as detailed in the methods section. For each protein examined, the number of bound proteins increased monotonically as vesicle diameter increased, Figure 1d. This general trend is expected since larger vesicle surface areas correspond to larger capacities for protein binding.
If a protein binds preferentially to membranes of high curvature, then the density of membrane-bound protein should increase as vesicle diameter decreases. Here density is defined as the number of membrane-bound proteins per vesicle surface area. Therefore, to evaluate the curvature sensitivity of Amph-FL and its truncation mutants, we calculated relative density as a function of vesicle diameter, Figure 1e. Relative density is defined as the protein density value for a particular vesicle diameter normalized by the protein density value for vesicles having a reference diameter of 200 nm. In previous works, we have utilized proteins that do not sense membrane curvature as negative controls in the tethered-vesicle assay.14, 15 For these proteins, relative density remained nearly constant and did not change substantially as vesicle diameter decreased. In contrast, the relative density of N-BAR increased 11-fold as vesicle diameter decreased from 200 nm to 20 nm, while the relative density of Amph-FL increased 26-fold over the same range of vesicle diameter. These values are similar to those in our recent report14, where we speculated that the disordered domains contributed to curvature sensing through an entropic mechanism. Specifically, the membrane surface is expected to impose a steric constraints on the disordered chains, which can be partially alleviated by binding of the protein to a highly curved membrane surface.
As the disordered domain becomes shorter, we would expect its conformational entropy to decrease, in accordance with the principles of polymer physics38. Therefore, if the contribution of the disordered domains to membrane curvature sensing arises primarily from an entropic mechanism, then truncating the disordered domain should reduce the sensitivity of protein binding to membrane curvature. In line with this prediction, the truncation mutants exhibited lesser degrees of curvature sensitivity compared to the full-length protein. Specifically, as vesicle diameter decreased from 200 nm to 20 nm, Amph-2/3 displayed a 19-fold increase in relative density, while Amph-1/3 displayed a 13-fold increase in relative density.
This trend of decreasing curvature sensing with decreasing disordered domain length can be predicted based on the principles of polymer physics. Specifically, if we model the disordered domain as a freely-joined chain, then conformational entropy should increase linearly with the number of residues, Nres.14, 39 Since enthalpy should be approximately the same for all configurations, free energy can be approximated as the product of temperature and entropy. Therefore, free energy should also increase linearly as Nres increases.
If we assume that protein binding to the membrane surface is a random, Boltzmann distributed process, then the relative decrease in free energy as a protein moves from a flatter to a more highly curved membrane surface, ΔGrel, can be described using Equation 1.39
| (1) |
Using this equation, ΔGrel can be estimated based on our experimental measurements of the relative density of membrane bound proteins as a function of vesicle diameter. Specifically, Figure 1f plots estimates of the change in free energy as a function of Nres for protein binding to a vesicle of 20 nm diameter relative to a vesicle of 200 nm diameter. Here it is evident that ΔGrel increases approximately linearly with Nres, as expected for entropic curvature sensing by a freely-joined chain.14 The intercept with the vertical axis is set by the ΔGrel value of the N-BAR domain alone, for which Nres is zero. Interestingly, for Amph-1/3 protein, the value of ΔGrel derived from the experimental measurements deviates somewhat from the theoretical trend. The deviation could arise from the small degree of contact between the shortened disordered domain and the membrane surface. Specifically, the first third of the disordered domain is expected to have a hydrodynamic radius that only slightly exceeds the dimensions of the BAR domain. Therefore, the membrane surface may not place a substantial constraint on the conformational entropy of this relatively short disordered domain. Another possible explanation arises from the distribution of charged residues within Amphiphysin’s disordered domain, as listed in Table 2. In particular, the density of negatively charged residues has its lowest value, 4%, within the first third of the domain, substantially smaller than the overall charge density within the full-length disordered domain, 12%. We have recently shown that a high net charge can substantially increase the ability of a disordered domain to sense membrane curvature, owing to electrostatic repulsion from the membrane surface, which itself has a substantially negative surface potential.15 This electrostatic mechanism would be expected to contribute less significantly to curvature sensing by the Amph-1/3 protein than for the Amph-2/3 and Amph-FL, owing to the non-uniform distribution of negatively charged residues within the disordered domain, Table 2. This disparate contribution could also help to explain the deviation of the data from the Amph-1/3 protein from the trend established by the other data.
Table 2.
Properties of the disordered domains in full-length Amphiphysin1 and its truncation mutants at pH 7.4 based on sequence information.
| Protein | Number of disordered residues | Expected net charge in disordered domain at pH 7.4 | Charge density of disordered domain (net charge per residue) |
|---|---|---|---|
| Amph-1/3 | 131 | −5 | −0.04 |
| Amph-2/3 | 258 | −24 | −0.09 |
| Amph-FL | 383 | −47 | −0.12 |
Collectively these results confirm that reducing the length of Amphiphysin’s disordered domain sequentially reduces the ability of the protein to sense membrane curvature. This finding is consistent with the entropic and electrostatic mechanisms of curvature sensing by disordered proteins, which we have recently reported.15 As protein concentration on the membrane surfaces increases, we would expect a transition from curvature sensing to curvature generation.40 Therefore it is reasonable that the length of the disordered domain may also impact membrane deformation by Amphiphysin1. This relationship is investigated in the next section.
Impact of disordered domain length on membrane vesiculation.
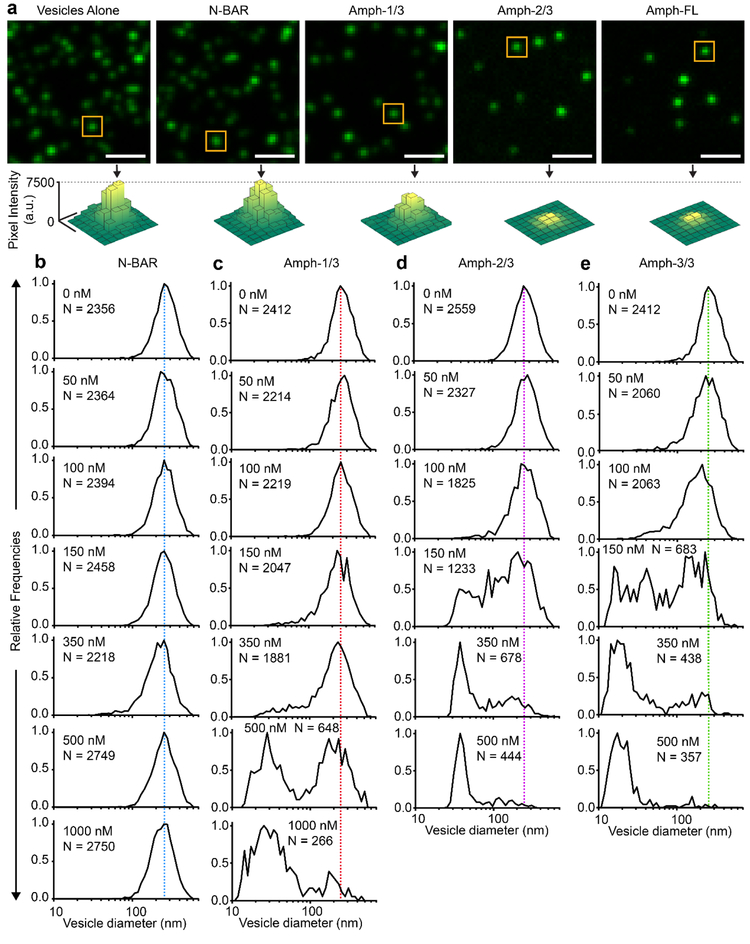
To examine the impact of the disordered domain length on membrane remodeling by Amphiphysin1, we utilized an assay similar to the one described in Figure 1b. Here instead of incubating tethered-vesicles with protein-containing solutions, vesicles and proteins were incubated together at 37°C for 30 minutes prior to tethering. Vesicles were incubated with protein at a lipid concentration of 10 μM. After tethering, vesicles were visualized using fluorescence microscopy, Figure 2a. Vesicles were fluorescently labeled using Oregon Green DHPE (Oregon Green 488 1,2,-dihexadecanoyl-sn-glycero-3-phosphoethanolamine). However, Amph-FL and its truncation mutants were not fluorescently labeled. Notably, as shown in Figure 2a, the tethering of vesicles became more sparse as the length of the disordered domain increased. We attribute this trend to the increased radius of the IDP domains. As they become larger, they begin to physically obstruct interactions between the biotinylated lipid head groups and NeutrAvidin on the passivated cover slip. When vesicles were incubated with 150 nM N-BAR, we observed no significant change in average fluorescence intensity for vesicles. This result suggests that the vesicle diameter also remained unchanged, indicating that N-BAR did not drive substantial membrane vesiculation. However, in the presence of 150 nM Amph-1/3, Amph-2/3, or Amph-FL, average fluorescence intensities decreased, which corresponded to decreases in the average vesicle diameter. More prominent decreases were observed for Amph-2/3 and Amph-FL, which contain longer disordered domains. These results suggest that membrane vesiculation became more substantial as the length of the disordered domain increased.
Figure 2: As the length of the disordered domain increases, membrane vesiculation becomes more potent.
(a) Representative fluorescent images of vesicles tethered in the absence of protein and after incubation with 150 nM protein. Fluorescence intensity was adjusted separately in each image to achieve maximum contrast. Images with constant fluorescence intensity scales are provided in Supplemental Figure S3. Yellow boxes highlight representative vesicles with fluorescence intensities near the population median. Surface plots represent the distribution of pixel intensity values within the yellow boxes. (b-e) Histograms representing vesicle diameter distributions before and after exposure to (b) N-BAR, (c) Amph-1/3, (d) Amph-2/3, and (e) Amph-FL at various concentrations. N corresponds to the number of vesicles processed to generate each histogram. Scale bars in a represent 2 μm.
To quantify the ability of these proteins to induce membrane vesiculation, we monitored the distribution of vesicle diameter as a function of protein concentration for Amph-FL and its truncation mutants, Figures 2b–e. Vesicle diameters were estimated by comparing the mean Oregon Green DHPE intensity for tethered vesicles to the mean diameter obtained from dynamic light scattering, as outlined in the methods section. When the concentration of N-BAR was increased from 0 to 1000 nM, vesicle diameter distributions remained constant, centered around 240 nm, Figure 2b. Amph-1/3 displayed a greater capacity to induce membrane vesiculation than N-BAR. As Amph-1/3 concentration was increased from 0 to 1000 nM, the 240 nm diameter population disappeared and a sub-100 nm diameter population emerged, Figure 2c. Amph-2/3 displayed an even greater capacity to induce membrane vesiculation - as protein concentration was increased from 0 to 500 nM, a population centered at 40 nm diameter emerged as the 240 nm diameter population disappeared, Figure 2d. Amph-FL displayed the greatest capacity to induce membrane vesiculation – as protein concentration was increased from 0 to 500 nM, a population centered at 20 nm diameter emerged as the 240 nm diameter population disappeared, Figure 2e. In Figures 2b–e, the average diameter of vesicle populations decreased as the length of the IDP domain was increased, indicating that increased IDP domain length successively enhanced membrane vesiculation.
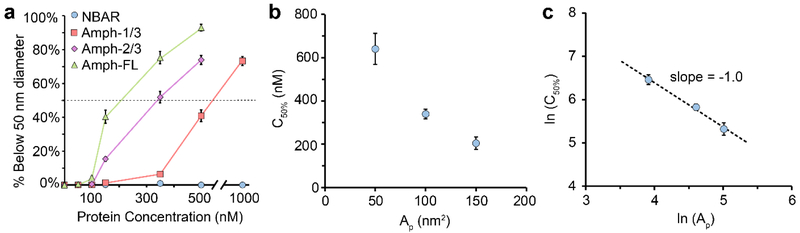
The results in Figure 2 for N-BAR and Amph-FL are in line with those in our recent publication.28 However, the results for Amph-1/3 and Amph-2/3 represent the first observation of an intermediate degree of membrane vesiculation upon truncation of the disordered domain. This finding strongly suggests that the length of the disordered domain is a critical modulator of the vesiculation process. These results are summarized in Figure 3a, where the percentage of each vesicle population that was below 50 nm in diameter is plotted as a function protein concentration for each protein. For vesicles exposed to the N-BAR domain, 0% had a diameter of 50 nm or less, even when protein concentration was increased to 1 μM. This result clearly indicates that the N-BAR domain alone is incapable of driving vesiculation. In contrast, for Amph-1/3, Amph-2/3, and Amph-FL, vesicle diameters were reduced as protein concentration increased such that 50% of the populations fell below 50 nm diameter. The protein concentration associated with this transition will be referred to as C50%. Based on the plots in Figure 3a, we estimated the C50% for Amph-1/3, Amph-2/3, and Amph-FL to be roughly 640 nM, 340 nM, and 210 nM, respectively.
Figure 3: Disordered domain projected area and vesiculation concentration vary inversely.
(a) Percentage of the vesicle populations in Figures 2b–e with diameters below 50 nm as a function of protein concentration. (b) Experimentally observed C50% as a function of Ap. (c) Log-log plot of C50% and Ap. The dashed line represents a line of best fit. Error bars in a represent the standard error of the mean for 10–18 fields of view. Each field of view had dimensions 23 × 23 μm2. Error bars in b represent the propagated errors from the linear interpolations performed in a, and error bars in c represent the propagated errors from b.
The trend of decreasing C50% with increasing length of the disordered domain can be attributed to the propensity of N-BAR domains to concentrate disordered domains, generating local pressure in the plane of the membrane.28 N-BAR domains alone are known to assemble into scaffolds that can generate and stabilize membrane tubules. However, the presence of the disordered domain in proximity to the N-BAR scaffolds is likely to result in steric interactions among the disordered domains. The resulting build-up of steric pressure can be alleviated by membrane deformation20, 21, ultimately resulting in membrane vesiculation.28 However, the relationship between vesiculation and the length of the disordered domain has not been previously considered.
Previous studies of membrane deformation by protein crowding have suggested that membrane vesiculation will occur when proteins reach a critical fractional coverage of the membrane surface, about 0.2–0.4. At this coverage, the pressure arising from molecular collisions becomes non-negligible according to crowded particle theory.41 Fractional coverage can be defined as the product of the projected area of the protein on the membrane surface, Ap, and the number of bound proteins per membrane area, Cmem. Our previous studies have shown that the affinities of N-BAR and Amph-FL for the membrane surface are approximately equivalent.28 Therefore, when fractional coverage of the membrane surface is well below one, Cmem should be approximately proportional to concentration in solution for each protein. If we assume that membrane vesiculation takes place at approximately the same coverage for each protein, then the product of Ap and Cmem during vesiculation should be approximately constant. Therefore Ap should be inversely proportional to Cmem during the vesiculation process. Assuming that Cmem and the concentration of the protein in solution are proportional to one another, the concentration in solution during vesiculation, C50% should be inversely proportional to Ap, Equation 3. This inverse relationship was observed in Figure 3b.
| (3) |
When C50% and Ap were plotted on a log-log plot and fit a linear regression, a slope of −1.0 ± 0.2 was obtained, Figure 3c. This result suggests that the inverse scaling argument provided a reasonable approximation to our data. This result also suggests and that steric congestion of the membrane surface by the disordered domains can, to the first order, explain the observed vesiculation of the membrane as the length of the disordered domains increases.
While these data are useful for quantifying membrane vesiculation, they do not reveal the precise nature of the membrane remodeling process. Notably, the N-BAR domain alone is expected to drive the formation of membrane tubules.10–12 However this phenomenon cannot be observed directly with the tethered-vesicle assay, owing to the limitation imposed by optical diffraction. Moreover, it is possible that the proteins with intermediate disordered domain lengths may generate a mixture of tubules and vesicles, an effect which also cannot be observed with the tethered-vesicle assay. Therefore, to clearly differentiate between tubule formation and vesiculation, we utilized transmission electron microscopy to directly visualize membrane deformation in the presence of Amph-FL and its truncation mutants.
Impact of disordered domain length on formation of membrane tubules.
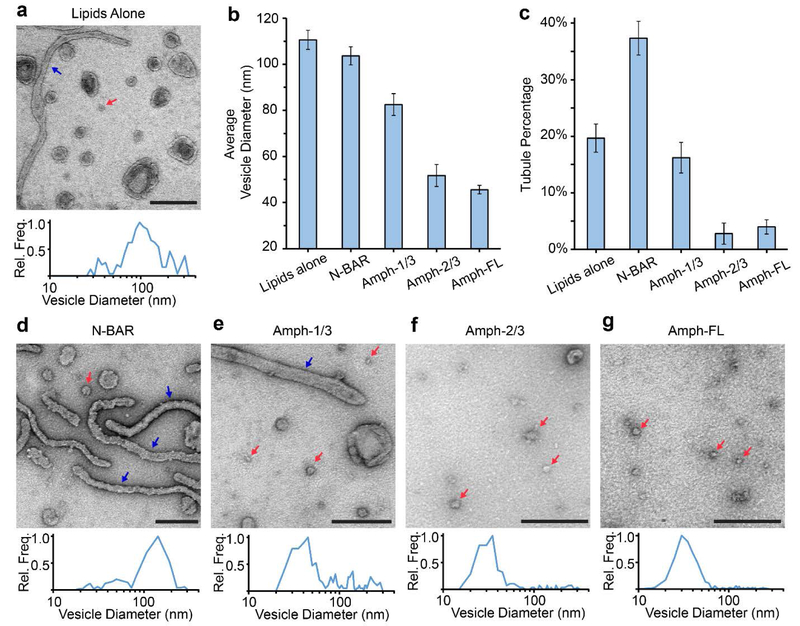
We used negative stain transmission electron microscopy (TEM) to directly visualize membrane morphology after remodeling. With non-staining EM methods, such as cryo-EM, it can be more difficult to resolve small vesicles due to the low intrinsic contrast of lipid bilayers. Vesicles and proteins were incubated at 37°C for 30 minutes prior to staining. In the absence of protein, vesicles appeared as rounded objects with an average diameter of 111 ± 4 nm (S.E., N=238), Figures 4a and 4b. Membrane tubules were present as well, Figure 4a, blue arrow. Tubules accounted for approximately 20% of the structures measured, Figure 4c. When vesicles were incubated with 5 μM N-BAR at a lipid concentration of 100 μM, tubules became more prominent, as expected based on previous findings.,9, 42 accounting for nearly 40% of the observed structures, Figures 4c and 4d. The average diameter of these tubules was 49 ± 1 nm (S.E., N=131). The vesicles that remained had an average diameter of 104 ± 4 nm (S.E., N=219), Figure 4b. The size distributions of vesicles in the presence and absence of N-BAR were similar, which is consistent with the result obtained from the fluorescence-based assay in Figures 2a–b. When vesicles were exposed to 5 μM Amph-1/3, small vesicular structures became more prominent, Figure 4e, and had an average diameter of 83 ± 5 nm (S.E., N=174), Figure 4b. Approximately 15% of the observable population of structures was comprised of tubules, Figure 4c. Small vesicular structures were also prominent in samples incubated with Amph-2/3 and Amph-FL, Figures 4f and 4g, respectively. Vesicles exposed to 5 μM Amph-2/3 had an average diameter of 52 ± 5 nm (S.E., N=131), while vesicles exposed to 5 μM Amph-FL had an average diameter of 46 ± 2 nm (S.E., N=402). When vesicles were exposed to either Amph-2/3 or Amph-FL, tubules made up less than 5% of the population of lipid structures.
Figure 4: The disordered domain of Amphiphysin1 progressively decreased formation of lipid tubules and increased membrane vesiculation.
(a) Representative TEM image of vesicles in the absence of protein and a corresponding histogram of the measured vesicle diameter distribution. (b) Average vesicle diameters in the absence of protein and after incubation with 5 μM protein. (c) Percentage of tubules present in the absence of protein and after incubation with 5 μM protein. (d-g) Representative TEM images of vesicles and tubules after exposure to 5 μM (d) N-BAR, (e) Amph-1/3, (f) Amph-2/3, or (g) Amph-FL. Corresponding histograms of measured vesicle diameter distributions are also provided in d-g. Red arrows highlight representative vesicles with diameters that are ≤ 50 nm in diameter. Blue arrows highlight tubules. All scale bars represent 250 nm. Histograms in a, d-g contained 131–402 total vesicles. Error bars in b and c represent the standard error of the mean. In b, N = 131–402 vesicles and in c, N = 10–20 fields of view per condition. Each field of view had dimensions 1932×1932 nm2.
Though the results from Figure 4 are qualitatively consistent with the results from Figure 2, the vesicle diameter distributions reported using TEM, Figure 4b, differ from the distributions reported using fluorescence microscopy, Figure 2b–e. We attribute this disparity to limitations of the TEM-based assay. In particular, as the diameter of the vesicles decreases, they become more difficult to resolve using TEM. Vesicles that were not clearly resolved were excluded from our analysis, likely resulting in an overestimate of the average diameter of the smallest vesicles. Additionally, some of the larger vesicles may be destroyed during the sample preparation process, likely resulting in an underestimate of the average diameter of the largest vesicles. Despite these issues, data from the TEM images and the fluorescence images are in qualitative agreement concerning the shift in the vesicle size distribution as a function of the length of the disordered c-terminal domain.
The results of the electron microscopy experiments show that membrane vesiculation increases with increasing length of the Amphiphysin1’s disordered domain, resulting in a smaller average vesicle diameter. This result is in qualitative agreement with the results obtained from the fluorescence-based assay of vesicle diameter in Figures 2a and 2c–e. Notably, the trend reverses for formation of lipid tubules, which were observed less frequently as the length of the disordered domain increased, Figure 4c. This result suggests that N-BAR-mediated lipid tubules were gradually destabilized as the length of the disordered domain increased, resulting in membrane vesiculation.
CONCLUSIONS
Curvature sensing and curvature generation are related yet distinct biophysical phenomena. In the regime of sparsely adsorbed proteins, curvature sensing is at its strongest.43 However, as the coverage of the membrane surface by proteins increases, the proteins begin to generate curvature.20, 21 The current paradigm is that structured protein domains are responsible for these phenomena. However, our recent work has demonstrated that the large conformational entropy of intrinsically disordered domains enables them to make substantial contributions to curvature sensing and remodeling, despite their lack of stable structure.14, 15, 28 Building on this work, here we probe the mechanistic relationship between structured and disordered remodeling proteins. Specifically, we quantified Amphiphysin1’s ability to sense and drive membrane curvature as its disordered C-terminal domain was progressively shortened.
When the length of an intrinsically disordered protein is increased, the number of unique conformations that it can occupy increases, resulting in an increase in conformation entropy. The results of our curvature sensing studies revealed that the free energy change associated with binding of a disordered protein to a more highly curved membrane surface increased linearly with increasing length. This finding, which can be predicted by modeling the disordered protein as a freely-joined chain,14 provides a useful first approximation of the potential of disordered proteins to sense curvature.
As the length of an intrinsically disordered protein increases, its radius of gyration will also generally increase.15 Therefore, the concentration of membrane-bound proteins required to crowd the membrane surface decreases with increasing length of the intrinsically disordered domain. As the fraction of the membrane surface covered by proteins increases, steric pressure among disordered domains becomes non-negligible, eventually driving membrane vesiculation as shown previously.16, 28 The results of the present study revealed that the concentration of protein in solution required to drive substantial membrane vesiculation was inversely proportional to the disordered domain’s projected area. This scaling is consistent with a steric mechanism of membrane curvature generation.
In nature, the lengths of intrinsically disordered domains in proteins that contain BAR domains can vary substantially.44, 45 A recent study reported domains for which the number of disordered residues, Nres, varied from 50 to 400,24 similar to the range of domain lengths considered in the present study of Amphiphysin1. Here we conclude that disordered domains significantly contribute to sensing and driving membrane curvature when their footprint on the membrane surface exceeds that of the BAR domain. By passing this threshold, the disordered domain has the opportunity to interact with the membrane surface and with neighboring proteins. These interactions give rise to entropic constraints that underlie the ability of disordered domains to participate in curvature sensing and membrane remodeling.14, 28
Our findings provide a rationale for exploring other proteins in the BAR domain family, such as the F-BAR-containing proteins PSTPIP244 and FCHo124, or the I-BAR-containing proteins IRSp5346 and MIM/ABBA47. The curvature sensing properties of several other proteins that contain both BAR domains and disordered domains have previously been examined. These include PICK148 and Endophilin A1.8 In these studies, the disordered domains were isolated and shown to fold into amphipathic helices in the presence of lipids. These transitions provide a rationale for the ability of these domains to sense membrane curvature. These results are complementary to our own recent findings on the ability of disordered domains to sense and drive membrane curvature by entopic and electrostatic mechanisms.14–17 In a broader context, our work highlights the significance of studying the synergistic relationship between structured and disordered protein domains. Disordered domains are ubiquitous among the protein machinery responsible for membrane trafficking and are often coupled to structured domains within the same protein molecule.24 It has become increasingly apparent that the disordered domains cannot be neglected in biophysical studies of protein-lipid interactions. Moreover, disordered domains, which make up approximately a third of the proteome,49 represent an extensive class of proteins, suggesting that additional sensors and drivers of membrane curvature remain to be discovered. Therefore, an important goal for future work is to more fully understand the biophysical roles of disordered domains in membrane remodeling.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the National Institutes of Health through R01GM120549 to Stachowiak, F32GM128316 to Zeno, and F31GM121013 to Snead. The authors would like to thank Prof. Tobias Baumgart for providing plasmids that code for full-length Amphiphysin1. These plasmids were used to generate all of the truncation mutants in this work.
Footnotes
CONFLICTS OF INTEREST
There are no conflicts to declare.
REFERENCES
- 1.McMahon HT and Gallop JL, Nature, 2005, 438, 590–596. [DOI] [PubMed] [Google Scholar]
- 2.Conner SD and Schmid SL, Nature, 2003, 422, 37. [DOI] [PubMed] [Google Scholar]
- 3.Hurley JH, Boura E, Carlson L-A and Różycki B, Cell, 2010, 143, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mierzwa B and Gerlich DW, Developmental cell, 2014, 31, 525–538. [DOI] [PubMed] [Google Scholar]
- 5.Helfrich W, Zeitschrift für Naturforschung C, 1973, 28, 693–703. [DOI] [PubMed] [Google Scholar]
- 6.Stachowiak JC, Brodsky FM and Miller EA, Nature cell biology, 2013, 15, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerberg J and Kozlov MM, Nature Reviews Molecular Cell Biology, 2006, 7, 9. [DOI] [PubMed] [Google Scholar]
- 8.Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger P-Y, Kunding AH, Castillo J, Gether U, Hedegård P and Stamou D, Nature chemical biology, 2009, 5, 835–841. [DOI] [PubMed] [Google Scholar]
- 9.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR and McMahon HT, Science, 2004, 303, 495–499. [DOI] [PubMed] [Google Scholar]
- 10.Frost A, Unger VM and De Camilli P, Cell, 2009, 137, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mim C and Unger VM, Trends in Biochemical Sciences, 2012, 37, 526–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simunovic M, Voth GA, Callan-Jones A and Bassereau P, Trends in cell biology, 2015, 25, 780–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adam J, Basnet N and Mizuno N, Scientific reports, 2015, 5, 15452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeno WF, Baul U, Snead WT, DeGroot AC, Wang L, Lafer EM, Thirumalai D and Stachowiak JC, Nature communications, 2018, 9, 4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeno WF, Thatte AS, Wang L, Snead WT, Lafer EM and Stachowiak JC, Journal of the American Chemical Society, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snead WT, Hayden CC, Gadok AK, Zhao C, Lafer EM, Rangamani P and Stachowiak JC, Proceedings of the National Academy of Sciences, 2017, 114, E3258–E3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM and Stachowiak JC, Nature communications, 2015, 6, 7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das RK and Pappu RV, Proceedings of the National Academy of Sciences, 2013, 110, 13392–13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann H, Soranno A, Borgia A, Gast K, Nettels D and Schuler B, Proceedings of the National Academy of Sciences, 2012, 109, 16155–16160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stachowiak JC, Hayden CC and Sasaki DY, Proceedings of the National Academy of Sciences, 2010, 107, 7781–7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA and Hayden CC, Nature cell biology, 2012, 14, 944. [DOI] [PubMed] [Google Scholar]
- 22.Kalthoff C, Alves J, Urbanke C, Knorr R and Ungewickell EJ, Journal of Biological Chemistry, 2002, 277, 8209–8216. [DOI] [PubMed] [Google Scholar]
- 23.Dyson HJ and Wright PE, Nature Reviews Molecular Cell Biology, 2005, 6, 197–208. [DOI] [PubMed] [Google Scholar]
- 24.Pietrosemoli N, Pancsa R and Tompa P, PLoS Computational Biology, 2013, 9, e1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dafforn TR and Smith CJ, EMBO Reports, 2004, 5, 1046–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhatia VK, Madsen KL, Bolinger PY, Kunding A, Hedegård P, Gether U and Stamou D, The EMBO journal, 2009, 28, 3303–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owen DJ, Collins BM and Evans PR, Annual review of cell and developmental biology, 2004, 20, 153–191. [DOI] [PubMed] [Google Scholar]
- 28.Snead WT, Zeno WF, Kago G, Perkins RW, Richter JB, Zhao C, Lafer EM and Stachowiak JC, Journal of Cell Biology, 2019, 218, 664–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campelo F and Malhotra V, Annual Review of Biochemistry, 2012, 81, 407–427. [DOI] [PubMed] [Google Scholar]
- 30.Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen J-S, McLeod I, McDonald WH, Gygi SP, Yates III JR and Gould KL, Molecular Cell, 2010, 39, 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dephoure N, Gould KL, Gygi SP and Kellogg DR, Molecular Biology of the Cell, 2013, 24, 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Momin N, Lee S, Gadok A, Busch D, Bachand G, Hayden C, Stachowiak J and Sasaki D, Soft Matter, 2015, 11, 3241–3250. [DOI] [PubMed] [Google Scholar]
- 33.Aguet F, Antonescu CN, Mettlen M, Schmid SL and Danuser G, Developmental cell, 2013, 26, 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamou D, Duschl C, Delamarche E and Vogel H, Angewandte Chemie International Edition, 2003, 42, 5580–5583. [DOI] [PubMed] [Google Scholar]
- 35.Kunding AH, Mortensen MW, Christensen SM and Stamou D, Biophysical Journal, 2008, 95, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campelo F, McMahon H and Kozlov M, Biophysical Journal, 2008, 95, 2325–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antonny B, Annual Review of Biochemistry, 2011, 80, 101–123. [DOI] [PubMed] [Google Scholar]
- 38.Nowicki W, Nowicka G and Narkiewicz-Michałek J, Polymer, 2009, 50, 2161–2171. [Google Scholar]
- 39.Phillips R, Kondev J, Theriot J and Garcia H, Physical biology of the cell, Garland Science, 2012. [Google Scholar]
- 40.Krishnan TS, Das SL and Kumar PS, Soft Matter, 2019, 15, 2071–2080. [DOI] [PubMed] [Google Scholar]
- 41.Mansoori G, Carnahan NF, Starling K and Leland T Jr, The Journal of Chemical Physics, 1971, 54, 1523–1525. [Google Scholar]
- 42.Gallop JL, Jao CC, Kent HM, Butler PJG, Evans PR, Langen R and McMahon HT, The EMBO journal, 2006, 25, 2898–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorre B, Callan-Jones A, Manzi J, Goud B, Prost J, Bassereau P and Roux A, Proceedings of the National Academy of Sciences, 2012, 109, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roberts-Galbraith RH and Gould KL, Cell Cycle, 2010, 9, 4091–4097. [DOI] [PubMed] [Google Scholar]
- 45.Salzer U, Kostan J and Djinović-Carugo K, Cellular and Molecular Life Sciences, 2017, 74, 2413–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heung M-Y, Visegrady B, Fütterer K and Machesky LM, European Journal of Cell Biology, 2008, 87, 699–708. [DOI] [PubMed] [Google Scholar]
- 47.Lee SH, Kerff F, Chereau D, Ferron F, Klug A and Dominguez R, Structure, 2007, 15, 145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herlo R, Lund VK, Lycas MD, Jansen AM, Khelashvili G, Andersen RC, Bhatia V, Pedersen TS, Albornoz PB and Johner N, Cell reports, 2018, 23, 2056–2069. [DOI] [PubMed] [Google Scholar]
- 49.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF and Jones DT, Journal of Molecular Biology, 2004, 337, 635–645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.