Abstract
The pretreatment of lignocellulosic substrates with cattle rumen fluid was successfully developed to increase methane production. In the present study, a 16S rRNA gene-targeted amplicon sequencing approach using the MiSeq platform was applied to elucidate the effects of the rumen fluid treatment on the microbial community structure in laboratory-scale batch methane fermenters. Methane production in fermenters fed rumen fluid-treated rapeseed (2,077.3 mL CH4 reactor−1 for a 6-h treatment) was markedly higher than that in fermenters fed untreated rapeseed (1,325.8 mL CH4 reactor−1). Microbial community profiling showed that the relative abundance of known lignocellulose-degrading bacteria corresponded to lignocellulose-degrading enzymatic activities. Some dominant indigenous cellulolytic and hemicellulolytic bacteria in seed sludge (e.g., Cellulosilyticum lentocellum and Ruminococcus flavefaciens) and rumen fluid (e.g., Butyrivibrio fibrisolvens and Prevotella ruminicola) became undetectable or markedly decreased in abundance in the fermenters fed rumen fluid-treated rapeseed, whereas some bacteria derived from seed sludge (e.g., Ruminofilibacter xylanolyticum) and rumen fluid (e.g., R. albus) remained detectable until the completion of methane production. Thus, several lignocellulose-degrading bacteria associated with rumen fluid proliferated in the fermenters, and may play an important role in the degradation of lignocellulosic compounds in the fermenter.
Keywords: lignocellulose, rumen, methane fermentation, MiSeq, bioaugmentation
Lignocellulosic biomass, composed of cellulose, hemicellulose, and lignin, is the most abundant natural polymeric carbon source in the world (17) and, thus, is an attractive candidate for use in methane production. However, due to the resistance of lignocellulose to fermentation, methane yield is generally low (36). Three types of biological pretreatment are currently being investigated in an attempt to increase methane production efficiency using lignocellulosic biomass: a pretreatment with enzymes, pure cultures, or mixed cultures (40). An enzymatic pretreatment is convenient because lignocellulolytic enzymes may be directly added to the reactor. However, due to the complications associated with lignocellulose, currently available enzymes alone do not effectively degrade lignocellulose; therefore, enzymes are always used with other pretreatment methods, such as an alkali pretreatment (40). A pretreatment using pure cultures, such as white-rot fungi, is a powerful method because these fungi produce many types of lignocellulolytic enzymes that favorably enhance biogas and bioethanol production (1, 3, 4, 43). However, this method is prolonged (several weeks) and requires sterilization. In contrast, the mixed-culture pretreatment (e.g., with rumen fluid, the liquid fraction of digestate from anaerobic digesters) results in a process that is rapid (several d) and does not require sterilization. We previously investigated the preservation of cattle rumen fluid for application to methane production (38) (Nakai, Y. et al. 2017, Patent No. US 9574213 B2) and its potential for the pretreatment of lignocellulosic substrates, and the findings obtained revealed that the rumen fluid treatment improved the degradation rates of cellulose, hemicellulose, and lignin and achieved 1.5–3.2-fold greater methane production than non-treatment systems (5, 7, 37). However, the (i) fate of the added rumen microbial community during methane production and (ii) the impact of the addition of such a different microbial community on the indigenous bacteria of methane seed sludge currently remain unclear. If the added rumen microbial community survives and continues to contribute to methane fermenter efficiency, it may be possible to utilize rumen microbes for the bioaugmentation of methane fermenters (i.e., one-phase biogas digesters). However, if added rumen microbes rapidly die out within the methane fermenter, they are applicable during only the first (hydrolytic) phase in two-phase (hydrolytic phase+methanogenic phase) biogas digesters.
The present study focused on lignocellulose-degrading bacteria and methanogenic archaea that contribute to the methanogenesis of lignocellulose and investigated the impact of adding rumen fluid containing an endogenous microbial community on the indigenous bacteria of methane seed sludge as well as the fate of the added rumen microbial community via MiSeq next-generation sequencing during methane fermentation. Previous studies (13, 24, 26, 31, 34, 41) reported the addition of lignocellulolytic pure cultures to methane fermenters; however, only a few (33, 44) have described the addition of entire rumen microbial communities or investigated their community structure. Furthermore, these studies were snapshots (i.e., microbial community structures at a specific time) of methane fermenters fed rumen fluid. The present study monitored time-dependent changes in the microbial community structure and investigated the fate of rumen-derived lignocellulose-degrading bacteria and indigenous lignocellulose-degrading bacteria of methane seed sludge during methane fermentation.
Materials and Methods
Feedstock
The stems and leaves of rapeseed (Brassica napus) were air-dried and milled to produce lignocellulosic biomass that was used as the substrate for methane production after the pretreatment with rumen fluid.
Rumen fluid treatment
Rumen contents were orally harvested from cattle, filtered through a mesh strainer (1×1 mm pores) to remove coarse solids, and stored at 37°C. Prepared rapeseed (9.0 g dry weight) was pretreated with 300 mL of rumen fluid at 37°C on a rotary shaker at 170 rpm for 6 or 24 h. The rumen fluid treatment was performed in duplicate. The care and use of animals in the present study were approved by the Institutional Animal Care and Use Committee of Tohoku University.
Methane production
Methane seed sludge for the initiation of the experiment was prepared using anaerobically digested sludge collected from the biogas plant at Tohoku University (Osaki, Japan). Some substrates (6) were then added to the sludge, which was used as the sludge for the start-up of the experiment. The experiment commenced when the sludge stopped biological activity, as assessed by the cessation of biogas production. Batch-type methane production was performed in 1,000-mL reactors (600-mL working volume: 200 mL of pretreated and untreated rapeseed was added to 400 mL of seed sludge) and incubated at 35°C for 32 d. Deionized water was used instead of rumen fluid in the control. The methane production process and 16S rRNA sequencing were performed in singlicate.
Analysis
The biomass chemical composition was analyzed by the Agricultural Product Chemical Research Laboratory in the Tokachi Federation of Agricultural Cooperatives using the detergent fiber method (39) and estimation equations of the National Research Council (NRC) (28). Total solids (TS) and volatile solids (VS) were assessed using standard methods (2). Biogas (CH4, CO2, and H2) concentrations were evaluated using a gas chromatograph (GC-8A; Shimadzu, Kyoto, Japan), volatile fatty acid (VFA) concentrations were measured by a high-performance liquid chromatograph (JASCO, Tokyo, Japan), and chemical oxygen demand (COD) was assessed using a colorimetric method based on Cr, as previously described (5). Lignocellulose (lignin, cellulose, xylan)-degrading enzyme activity was measured as previously described (7). DNA extraction was performed using a Power Soil DNA kit (MO BIO, Carlsbad, CA, USA), 16S rRNA gene MiSeq sequencing was performed using the FwOvAd_341F (forward) and ReOvAd_785R (reverse) primer set, and an analysis by QIIME software with a 97% threshold for the OTU definition was conducted as previously described (7). Taxonomic identification was performed at the phylum and genus levels. The representative (most abundant) sequence of each genus was investigated by the basic local alignment search tool (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm the closest species. All sequence data were deposited in the DRA of the DDBJ database under DRA008816.
Results and Discussion
Feedstock and rumen fluid
The chemical composition of rapeseed was as follows: 39.4% non-fibrous carbohydrate, 21.7% cellulose, 3.3% hemicellulose, 3.2% lignin, and other components (such as protein and ash). The TS and VS compositions of rapeseed were 91.9 and 74.2%, respectively. The TS and VS of rumen fluid after filtration were 1.6 and 0.8%, respectively.
Methane production from lignocellulose pretreated with rumen fluid
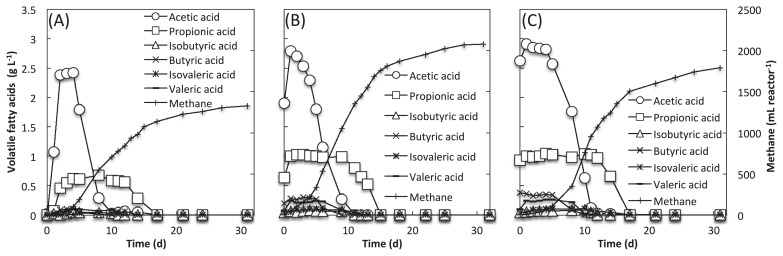
Rapeseed was soaked with rumen fluid for 6 or 24 h prior to the initiation of methane production. Rapeseed was solubilized, resulting in the production of various VFAs, such as acetate and propionate, as metabolites of plant cell wall components (e.g., cellulose, hemicellulose, and lignin) (Table S1). Batch-type methane production was performed using pretreated and untreated (control) rapeseed. VFAs underwent conversion to methane over time (Fig. 1). More methane was produced from pretreated rapeseed (i.e., 2,077.3 mL CH4 reactor−1 from 6-h pretreated rapeseed; 1,790.4 mL CH4 reactor−1 from 24-h pretreated rapeseed) than from untreated control rapeseed (i.e., 1,325.8 mL CH4 reactor−1), which is consistent with previous findings (5).
Fig. 1.
Time course of VFA and methane production in methane fermenters fed untreated rapeseed (A) and rapeseed pretreated with rumen fluid for 6 h (B) and 24 h (C).
Sampling depth, coverage, and taxonomic composition of the microbial community during the methane production process
After quality filtering, paired-end read assembly, quality control, and chimera removal, 2,663,094 V3–V4 16S rRNA sequence reads (average 126,814; minimum 46,723; maximum 197,504) were obtained from raw MiSeq data from 21 samples. The average sequence read length after primer subtraction was 450.9 bp. With a subsample of 46,723 reads per sample, rarefaction curves (Fig. S1) demonstrated that most of the authors’ sampling efforts provided sufficient species coverage to accurately describe the bacterial composition of each sample.
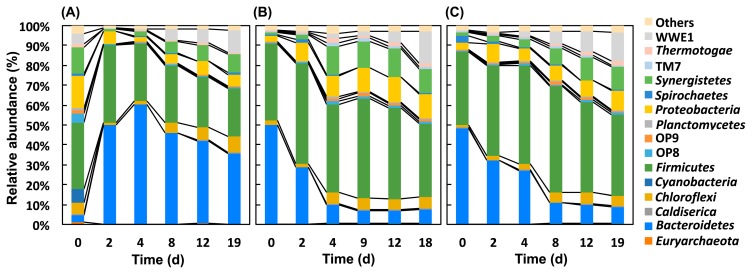
Fifty-one phyla were identified within the microbial community structure during the methane production process. The abundance of 36 of these phyla was <1.0%, and these were included in the “Others” category (Fig. 2). The relative abundance of Synergistetes and WWE1 increased over time. Aminobacterium was the main member of the phylum Synergistetes in the methane fermenter fed untreated rapeseed, whereas Thermovirgaceae, vadinCA02, HA73, and Aminobacterium were the main members in the fermenter fed with rumen fluid-pretreated rapeseed (Fig. S2). The most abundant sequences of Aminobacterium, Thermovirgaceae, and HA73 were associated with known amino acid-degrading bacteria (i.e., members of Aminobacterium [8], Thermovirga [16], and Aminivibrio [19]) (Table S2). The most abundant sequence of vadinCA02 was associated with unknown bacteria. W22 and W5 were detected in the phylum WWE1, and the most abundant sequences were associated with unknown bacteria (Fig. S3 and Table S3). The phylum Bacteroidetes exhibited the most significant changes. Taxa with relative abundance >1.0% that belong to Bacteroidetes are shown in Fig. S4. In the untreated control, Bacteroides, Porphyromonadaceae, and Prevotella were the main members of Bacteroidetes, and their relative abundance increased after the addition of rapeseed. The most abundant gene sequences of these taxa were associated with known plant cell component (i.e., xylan, cellobiose, starch, and protein)-degrading bacteria (i.e., members of Bacteroides [29], Proteiniphilum [14], and Prevotella [25]) (Table S4), and, thus, the increases observed in these bacteria were reasonable. In rumen fluid-pretreated rapeseed, the phylum Bacteroidetes was the most abundant on day 0 because many Bacteroidetes migrated from the pretreatment reactor to the methane fermenter (Fig. 2). Prevotella—originating from pretreatment fluid (Fig. S4)—was detected as the main member of Bacteroidetes on day 0; however, the relative abundance of this genus decreased over time. Similarly, the total number of observed OTUs in fermenters with mixed pretreatment fluids decreased with time to the same level as in the original seed sludge (Fig. S5). Collectively, these results suggest that many rumen-derived microbes may not survive in the methane fermenter.
Fig. 2.
Changes in the phylum-level taxonomic composition of methane fermenters fed untreated rapeseed (A) and rapeseed pretreated with rumen fluid for 6 h (B) and 24 h (C). Fifty-two phyla were identified. The abundance of 37 of these was <1%, and, thus, these phyla were included in the “Others” category.
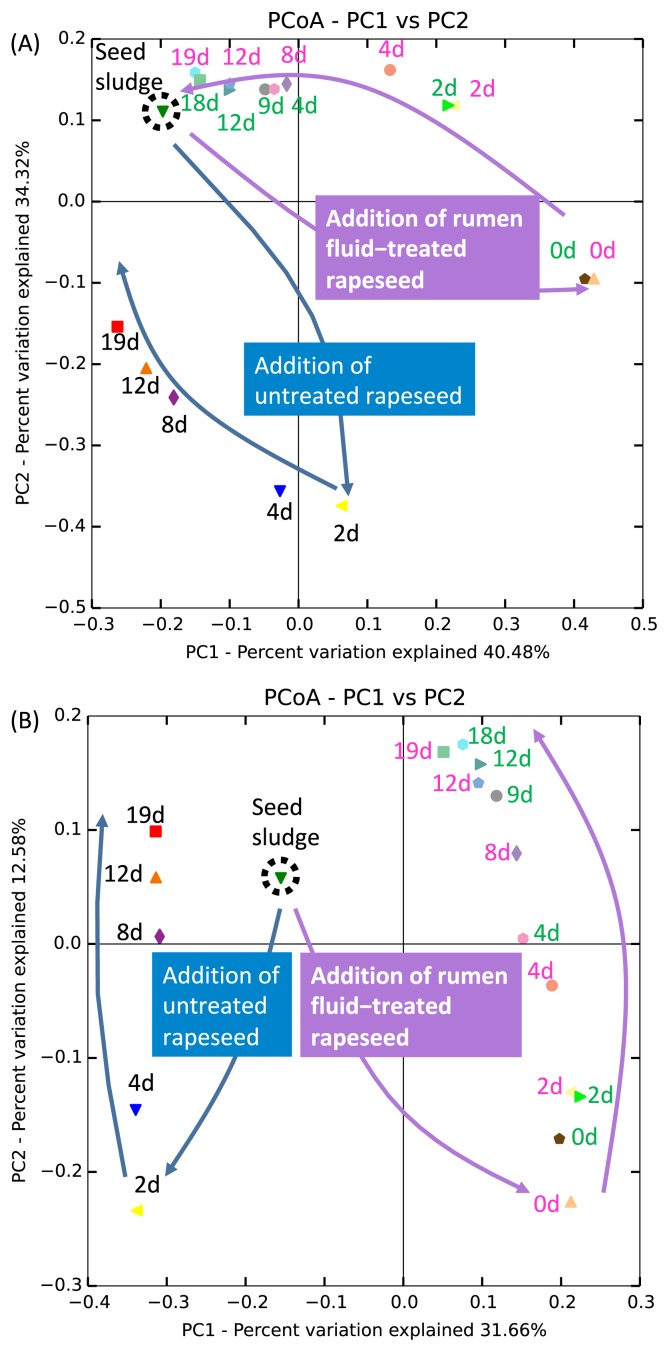
To elucidate community structure alterations caused by the addition of pretreatment fluid, a weighted principal coordinate analysis (PCoA) (i.e., consideration of the relative abundance of each OTU) was performed (Fig. 3A). The results obtained indicated that the structure of the microbial community in the methane fermenter on day 0—immediately after mixing with pretreatment fluid—markedly differed from that of the seed sludge. The microbial community structure of the fermenter contents mixed with pretreatment fluid then roughly returned to that of seed sludge. On the other hand, an unweighted PCoA (Fig. 3B) was performed to elucidate differences in community members (i.e., not considering the relative abundance of each OTU), and the results obtained indicated that the composition in the fermenter mixed with pretreatment fluid did not return to that in the seed sludge. Alpha diversity indices (Shannon, ACE, and Chao1) also indicated that species richness increased due to the addition of pretreatment fluids (Table S5). Thus, these results indicated that although the addition of rumen fluid affected both the relative abundance and composition of the microbial communities of methane seed sludge, its addition had a stronger impact on the compositions of these communities.
Fig. 3.
Principal coordinate analysis (PCoA) of weighted (A) and unweighted (B) UniFrac distances of microbial 16S rRNA sequences (sourced from methane fermentation samples) from the V3–V4 region. d, days. Colors of time points: black=control (i.e., methane fermenter fed untreated rapeseed), green=methane fermenter fed rapeseed pretreated with rumen fluid for 6 h, and pink=methane fermenter fed rapeseed pretreated with rumen fluid for 24 h.
Lignocellulose-degrading bacteria indigenous to methane seed sludge
To estimate the composition of lignocellulose-degrading bacteria originating from methane seed sludge, the species most closely related to the most abundant sequence of each assigned taxon for methane fermentation without the pretreatment (i.e., control) was identified by a homology search of the GenBank database using BLAST. Nine OTUs that are phylogenetically related to known cellulolytic, xylanolytic, and aromatic-degrading bacteria were detected in the methane fermenter without the pretreatment (Table 1). Changes in the relative abundance of each bacterium and its lignocellulose-degrading enzyme activity are shown in Fig. S6. Regarding the cellulose-degrading population, changes in the relative abundance of the two OTUs 14124 and 342612, associated with known cellulolytic bacteria (i.e., members of Cellulosilyticum [12] and Ruminococcus [21]), mirrored the changes in cellulase (CMCase, Avicelase, and β-glucosidase) activities. Regarding the xylan-degrading population, changes in the relative abundance of OTU 572010, associated with known xylanolytic bacteria (i.e., members of Bacteroides [29]), mirrored the changes in cell-bound xylanase activities, whereas changes in the 4 other OTUs (544105, 768947, 14124, and 342612) associated with known xylanolytic bacteria (i.e., members of Ruminofilibacter [30], Prevotella [25], Cellulosilyticum [12], and Ruminococcus [21]) mirrored changes in free xylanase activities. Regarding the aromatic-degrading population, changes in the relative abundance of OTU 625556 mirrored those in manganese peroxidase (MnP) activity. This OTU was related to the aromatic-degrading bacteria Syntrophorhabdus (35); however, sequence homology was low (94%); therefore, OTU 625556 may be a new bacterium related to lignin degradation. The other 3 OTUs (659146, 840291, and 4403651) associated with known aromatic-degrading bacteria (i.e., members of Sedimentibacter [42], Acinetobacter [27], and Pseudomonas [11]) (Fig. S7) did not mirror the dynamics of any lignin-degrading enzyme activities. Changes in the relative abundance of most lignocellulose-degrading bacteria detected during the present study did not contradict those observed in the activities of the relevant enzymes. Therefore, these bacteria may play a direct role in lignocellulose degradation during methane fermentation.
Table 1.
OTUs related to lignocellulose-degrading bacteria in methane seed sludge.
| OTU ID | Relative species of 16S rRNA gene | Substrate | Accession no. | Identity |
|---|---|---|---|---|
| 14124 | Cellulosilyticum lentocellum DSM 5427 | Cellulose, Xylan | NR_074536 | 441/442 (99%) |
| 342612 | Ruminococcus flavefaciens FD-1 | Cellulose, Xylan | AM920691 | 433/440 (98%) |
| 572010 | Bacteroides graminisolvens XDT-1 | Xylan | NR_041642 | 459/460 (99%) |
| 544105 | Ruminofilibacter xylanolyticum S1 | Xylan | DQ141183 | 458/460 (99%) |
| 768947 | Prevotella ruminicola CG41 | Xylan | AB849451 | 459/460 (99%) |
| 659146 | Sedimentibacter hydroxybenzoicus JW/Z-1 | Aromatics | NR_029146 | 421/442 (95%) |
| 625556 | Syntrophorhabdus aromaticivorans UI | Aromatics | NR_041306 | 440/469 (94%) |
| 840291 | Acinetobacter indicus strain A648 | Aromatics | NR_117784 | 465/466 (99%) |
| 4403651 | Pseudomonas balearica VITPS19 | Aromatics | MF164145 | 464/465 (99%) |
OTUs marked with gray color were not detected in rumen fluid but were endemic to methane seed sludge.
Impact of the addition of pretreatment fluid on bacteria indigenous to methane seed sludge
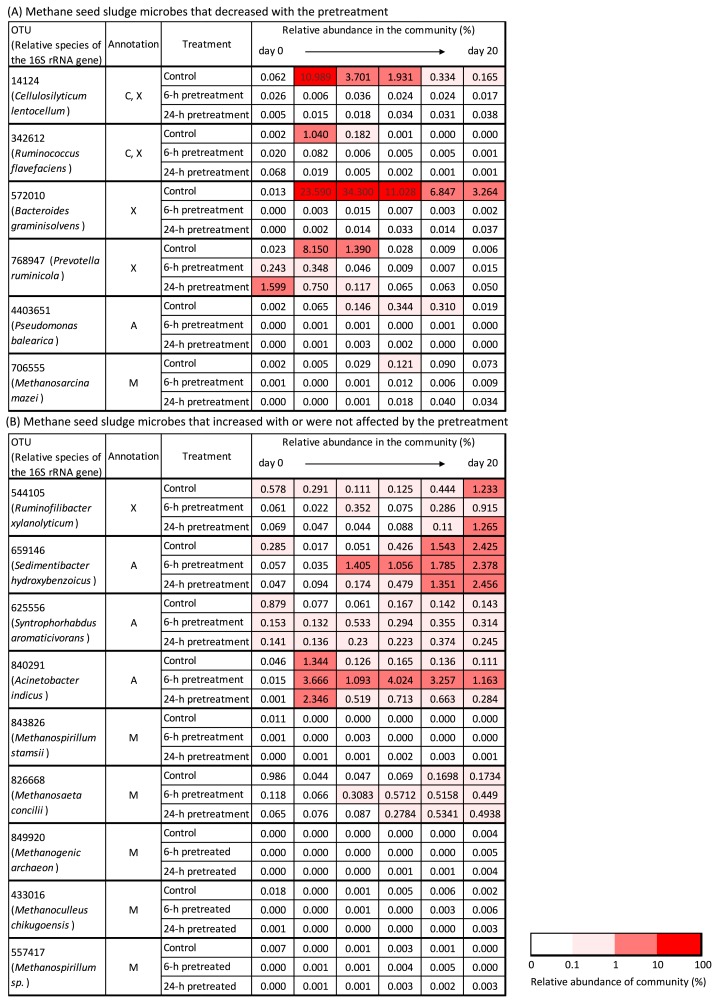
The pretreatment fluid (i.e., rapeseed stem dissolved in rumen fluid) was added to the methane fermenter, and the impact of the addition of this fluid on the above-mentioned indigenous lignocellulose-degrading bacteria of methane seed sludge (Table 1) was investigated. The results obtained for methanogenic archaea (Table S6) were added to these results, and a summary of the results combined are shown in Fig. 4. Six out of 15 OTUs (14124, 342612, 572010, 768947, 4403651, and 706555) no longer increased in relative abundance with the addition of the pretreatment fluid; however, these OTUs increased in the methane fermenter fed untreated rapeseed (i.e., control). OTU14124, 342612, 572010, and 768947 were associated with known cellulolytic and xylanolytic bacteria (i.e., members of Cellulosilyticum [12], Ruminococcus [21], Bacteroides [29], and Prevotella [25]) and decreased by one to four orders of magnitude with the addition of the pretreatment fluid. The pretreatment fluid contained cellulose and hemicellulose (Table S1) as carbon and energy sources, and pH values (Fig. S8) during the methane fermentation period were within the physiological range of these bacteria, which may have grown in the reactor; however, their abundance did not increase. On the other hand, OTU 544105, associated with known xylanolytic bacteria (i.e., members of Ruminofilibacter [30]), increased in abundance, similar to that in the control. Regarding OTUs associated with known aromatic-degrading bacteria, OTU4403651 (i.e., members of Pseudomonas [11]) became undetectable with the addition of the pretreatment fluid at the end, whereas the other 3 OTUs (i.e., members of Sedimentibacter [42], Syntrophohabdus [35], and Acinetobacter [27]) in this category remained detectable. Regarding OTUs associated with known methanogenic archaea, the relative abundance of OTU706555 (i.e., a member of Methanosarcina [10]) decreased by one order of magnitude with the addition of the pretreatment fluid, whereas the other 5 OTUs (i.e., members of Methanospirillum, Methanosaeta, and Methanoculleus) (10) were not notably affected by the addition of the pretreatment fluid. In summary, the addition of the pretreatment fluid had a large impact on the fate of the indigenous cellulolytic and xylanolytic bacteria of methane seed sludge.
Fig. 4.
Impact of the addition of rapeseed pretreated with rumen fluid on the relative abundance of indigenous lignocellulose-degrading bacteria and methanogenic archaea in methane seed sludge. Annotation: C, cellulose-degrading bacteria; X, xylan-degrading bacteria; A, aromatic-degrading bacteria; M, methanogenic archaea.
Fate of added rumen microbes
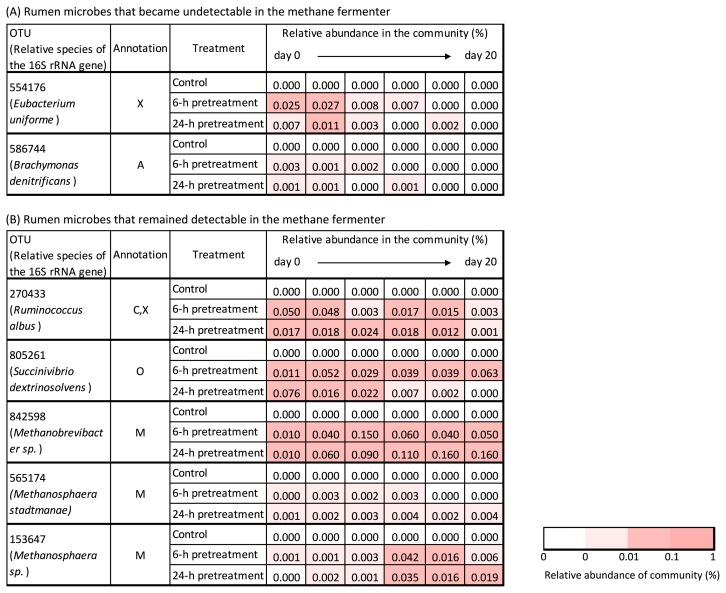
The OTUs associated with known lignocellulose-degrading bacteria and methanogenic archaea detected in the rumen fluid treatment are shown in Table S7. The OTUs endemic to rumen fluid (i.e., not detected in methane seed sludge) are shown in Table S7, and changes in the relative abundance of the added rumen microbes during the methane production process are shown in Fig. 5. Two OTUs (554176, 586744) gradually became undetectable, whereas 5 OTUs (270433, 805261, 842598, 565174, and 153647) remained detectable until the completion of methane production. OTU 270433 was associated with a known cellulolytic rumen bacterium (i.e., Ruminococcus albus [25]). Sequences other than the above-mentioned most abundant sequences were also investigated among OTUs assigned as Ruminococcus, and 4 OTUs were sequences associated with R. albus and remained detectable until the completion of methane production (Fig. S9). Although the present study confirmed the existence of a 16S rRNA gene associated with R. albus, if R. albus is confirmed to actually degrade cellulose in methane fermenters, bioaugmentation using R. albus will be an effective means for methane production from lignocellulose. OTU 805261 was associated with a known cello-oligosaccharolytic rumen bacterium (i.e., a member of Succinivibrio [11]). OTU842598, 565174, and 153647 were associated with known H2-utilizing methanogenic archaea (i.e., members of Methanobrevibacter or Methanosphaera). These methanogenic archaea obtain energy through the reduction of methanol or CO2 to CH4 using H2 as a substrate (10). To achieve the successful degradation of aromatic compounds derived from lignin, the presence of H2-utilizing methanogens (which lower the H2 partial pressure in methane fermenters) is generally required (22, 35). Furthermore, one key reason for the collapse of methane fermentation is propionate accumulation (6, 18). To facilitate propionate degradation, propionate-oxidizing bacteria must thermodynamically coexist with H2-utilizing methanogens (20). The relative abundance of the two OTUs 591709 and 1129757, associated with known propionate-oxidizing bacteria (i.e., members of Syntrophobacter [9, 15]), slightly increased with the addition of the pretreatment fluid (Fig. S10 and Table S8). Therefore, the increase observed in the growth of the above-mentioned H2-utilizing methanogens derived from the pretreatment fluid may promote the continuation of robust methane fermentation.
Fig. 5.
Changes in the relative abundance of lignocellulose-degrading bacteria and methanogenic archaea endemic to rumen fluid during the methane production process. Annotation: C, cellulose-degrading bacteria; X, xylan-degrading bacteria; A, aromatic-degrading bacteria; O, cello-oligosaccharide-degrading bacteria; M, methanogenic archaea.
Some of the OTUs (571178, 99264, 326292, and 589852) in Table S7 associated with known cellulolytic or xylanolytic rumen bacteria (i.e., members of Butyrivibrio [25], Prevotella [25, 33], and Pseudobutyrivibrio [23]), which were also detected in the control, are shown in Fig. S11. These OTUs migrated from the rumen pretreatment reactor to the methane fermenter and were dominant on day 0 in the pretreated methane fermenter. However, their abundance gradually decreased, resulting in the same levels as these bacteria detected in the control at the end of the experiment. Each bacterium of the complex microbial community may have a suitable existence ratio depending on their role.
The present study demonstrated that rumen-derived bacteria associated with known cellulolytic and hemicellulolytic bacteria (i.e., R. albus) remained detectable until the completion of the methane production process. Conversely, indigenous bacteria associated with known cellulolytic and hemicellulolytic bacteria (e.g., Cellulosilyticum lentocellum, Bacteroides graminisolvens, and Prevotella ruminicola) in methane seed sludge markedly decreased after the addition of the pretreatment fluid. In addition, a rumen-derived archaeon associated with known H2-utilizing methanogenic archaea (i.e., Methanobrevibacter sp. and Methanosphaera sp.) also remained detectable until the completion of the methane production process after the addition of pretreatment fluid. Thus, the methane fermenter in the present study became a unique fermenter composed of pretreatment fluid-derived cellulolytic, hemicellulolytic, and methanogenic rumen microbes and indigenous species in methane seed sludge. Differences in the species composition of the methane fermenter suggested by PCoA may be caused by these rumen microbes that survived. If these pretreatment fluid-derived rumen microbes persist in the methane fermenter, they are expected to enhance lignocellulose-based methane productivity. Future studies are needed to elucidate whether these microbes directly contribute to the enhanced efficiency of methane fermentation by confirming the presence of enzyme mRNA (e.g., that of cellulase or hemicellulase) in pretreatment fluid-derived rumen microbes within the methane fermenter.
Conclusion
The rumen fluid pretreatment enhanced methane productivity. The microbial community structure analysis demonstrated that dominant indigenous cellulolytic and hemicellulolytic bacteria in methane seed sludge no longer increased or markedly decreased in relative abundance after the addition of the pretreatment fluid. Conversely, certain rumen-derived cellulolytic, hemicellulolytic, and H2-utilizing methanogenic microbes introduced into the fermenter via the pretreatment fluid inoculation remained detectable until the completion of methane production. Thus, these results showed that the methane fermenter in the present study is a unique fermenter composed of pretreatment fluid-derived rumen microbes and indigenous species in methane seed sludge and also that this microbial community structure may contribute to methane productivity.
SUPPLEMENTARY MATERIAL
Acknowledgements
This study was supported in part by the Japan Society for the Promotion of Science (Grant-in-Aid for JSPS Research Fellows Grant Number 14J08197, Grant-in-Aid for Scientific Research (A) Grant Number 17H01512, and Early-Career Scientists Grant Number 18K18219) and the Next-generation Energies for Tohoku Recovery (NET) Project. The authors declare that they have no competing interests. The authors wish to thank Mr. Kazuya Sato and Ms. Rie Yamamoto, the Field Science Center, Tohoku University, for their technical support during the collection of rumen contents. The authors also thank Assistant Professor Michiaki Omura, Ms. Yukie Ogushi, and Ms. Takako Sasaki for their help with the collection of rapeseed.
References
- 1.Amirta R., Tanabe T., Watanabe T., Honda Y., Kuwahara M., Watanabe T. Methane fermentation of Japanese cedar wood pretreated with a white rot fungus, Ceriporiopsis subvermispora. J Biotechnol. 2006;123:71–77. doi: 10.1016/j.jbiotec.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg A.E., Clesceri L.S., Eaton A.D., editors. APHA (American Public Health Association) Standard methods for the examination of water and wastewater. APHA, AWWA, and WEF; Washington, DC: 1989. pp. 2–79. [Google Scholar]
- 3.Baba Y., Tanabe T., Watanabe T., Honda Y., Watanabe T. Enzymatic saccharification of Japanese cedar wood by combined pretreatments using white rot fungus and organosolvolysis with lactic acid. J Jpn Soc Mater Cycles Waste Manage. 2010;21:219–225. (in Japanese with an English abstract) [Google Scholar]
- 4.Baba Y., Tanabe T., Shirai N., Watanabe T., Honda Y., Watanabe T. Pretreatment of Japanese cedar wood by white rot fungi and ethanolysis for bioethanol production. Biomass Bioenergy. 2011;35:320–324. [Google Scholar]
- 5.Baba Y., Tada C., Fukuda Y., Nakai Y. Improvement of methane production from waste paper by pretreatment with rumen fluid. Bioresour Technol. 2013;128:94–99. doi: 10.1016/j.biortech.2012.09.077. [DOI] [PubMed] [Google Scholar]
- 6.Baba Y., Tada C., Watanabe R., Fukuda Y., Chida N., Nakai Y. Anaerobic digestion of crude glycerol from biodiesel manufacturing using a large-scale pilot plant: Methane production and application of digested sludge as fertilizer. Bioresour Technol. 2013;140:342–348. doi: 10.1016/j.biortech.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Baba Y., Matsuki Y., Mori Y., Suyama Y., Tada C., Fukuda Y., Saito M., Nakai Y. Pretreatment of lignocellulosic biomass by cattle rumen fluid for methane production: Bacterial flora and enzyme activity analysis. J Biosci Bioeng. 2017;123:489–496. doi: 10.1016/j.jbiosc.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Baena S., Fardeau M.L., Labat M., Ollivier B., Thomas P., Garcia J.L., Patel B.K.C. Aminobacterium colombiense gen. nov. sp. nov., an amino acid-degrading anaerobe isolated from anaerobic sludge. Anaerobe. 1998;4:241–250. doi: 10.1006/anae.1998.0170. [DOI] [PubMed] [Google Scholar]
- 9.Boone D.R., Bryant M.P. Propionate-degrading bacterium, Syntrophobacter wolinii sp. nov. gen. nov., from methanogenic ecosystems. Appl Environ Microbiol. 1980;40:626–632. doi: 10.1128/aem.40.3.626-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boone D.R., Garrity G.M. Bergey’s Manual of Systematic Bacteriology: The Archaea and the Deeply Branching and Phototrophic Bacteria. Springer; New York: 2001. [Google Scholar]
- 11.Brenner D.J., Krieg N.R., Staley J.T., Garrity G.M. Bergey’s Manual of Systematic Bacteriology: The Proteobacteria Part B the Gammaproteobacteria. 2nd ed. Springer; New York: 2005. [Google Scholar]
- 12.Cai S., Dong X. Cellulosilyticum ruminicola gen. nov., sp. nov., isolated from the rumen of yak, and reclassification of Clostridium lentocellum as Cellulosilyticum lentocellum comb. nov. Int J Syst Evol Microbiol. 2010;60:845–849. doi: 10.1099/ijs.0.014712-0. [DOI] [PubMed] [Google Scholar]
- 13.Čater M., Fanedl L., Malovrh S., Logar R.M. Biogas production from brewery spent grain enhanced by bioaugmentation with hydrolytic anaerobic bacteria. Bioresour Technol. 2015;186:261–269. doi: 10.1016/j.biortech.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 14.Chen S., Dong X. Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int J Syst Evol Microbiol. 2005;55:2257–2261. doi: 10.1099/ijs.0.63807-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen S., Liu X., Dong X. Syntrophobacter sulfatireducens sp. nov., a novel syntrophic, propionate-oxidizing bacterium isolated from UASB reactors. Int J Syst Evol Microbiol. 2005;55:1319–1324. doi: 10.1099/ijs.0.63565-0. [DOI] [PubMed] [Google Scholar]
- 16.Dahle H., Birkeland N.K. Thermovirga lienii gen. nov., sp. nov., a novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int J Syst Evol Microbiol. 2006;56:1539–1545. doi: 10.1099/ijs.0.63894-0. [DOI] [PubMed] [Google Scholar]
- 17.de Jong E., Gosselink R.J.A. Chapter 17—Lignocellulose-based chemical products. In: Gupta V.K., Tuohy M.G., Kubicek C.P., Saddler J., Xu F., editors. Bioenergy Research: Advances and Applications. Elsevier; Amsterdam: 2014. pp. 277–313. [Google Scholar]
- 18.Harper S.R., Pohland F.G. Recent developments in Hydrogen management during anaerobic biological wastewater treatment. Biotechnol Bioeng. 1986;128:585–602. doi: 10.1002/bit.260280416. [DOI] [PubMed] [Google Scholar]
- 19.Honda T., Fujita T., Tonouchi A. Aminivibrio pyruvatiphilus gen. nov., sp. nov., an anaerobic, amino-acid-degrading bacterium from soil of a Japanese rice field. Int J Syst Evol Microbiol. 2013;63:3679–3686. doi: 10.1099/ijs.0.052225-0. [DOI] [PubMed] [Google Scholar]
- 20.Imai H., Sekiguchi Y., Kamagata Y., Ohashi A., Harada H. Cultivation and in situ detection of a thermophilic bacterium capable of oxidizing propionate in syntrophic association with hydrogenotrophic methanogens in a thermophilic methanogenic granular sludge. Appl Environ Microbiol. 2000;66:3608–3615. doi: 10.1128/aem.66.8.3608-3615.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jindou S., Brulc J.M., Levy-Assaraf M., et al. Cellulosome gene cluster analysis for gauging the diversity of the ruminal cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol Lett. 2008;285:188–194. doi: 10.1111/j.1574-6968.2008.01234.x. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser J.P., Hanselmann K.W. Fermentative metabolism of substituted monoaromatic compounds by a bacterial community from anaerobic sediments. Arch Microbiol. 1982;133:185–194. [Google Scholar]
- 23.Kopecný J., Zorec M., Mrázek J., Kobayashi Y., Marinsek-Logar R. Butyrivibrio hungatei sp. nov. and Pseudobutyrivibrio xylanivorans sp. nov., butyrate-producing bacteria from the rumen. Int J Syst Evol Microbiol. 2003;53:201–209. doi: 10.1099/ijs.0.02345-0. [DOI] [PubMed] [Google Scholar]
- 24.Kovacs K.L., Acs N., Kovacs E., Wirth R., Rakhely G., Strang O., Herbel Z., Bagi Z. Improvement of biogas production by bioaugmentation. BioMed Res Int. 2013 doi: 10.1155/2013/482653. 482653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krause D.O., Denman S.E., Mackie R.I., Morrison M., Rae A.L., Attwood G.T., McSweeney C.S. Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol Rev. 2003;27:663–693. doi: 10.1016/S0168-6445(03)00072-X. [DOI] [PubMed] [Google Scholar]
- 26.Lü F., Ji J., Shao L., He P. Bacterial bioaugmentation for improving methane and hydrogen production from microalgae. Biotechnol Biofuels. 2013;6:92. doi: 10.1186/1754-6834-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malhotra J., Anand S., Jindal S., Rajagopal R., Lal R. Acinetobacter indicus sp. nov., isolated from a hexachlorocyclohexane dump site. Int J Syst Evol Microbiol. 2012;62:2883–2890. doi: 10.1099/ijs.0.037721-0. [DOI] [PubMed] [Google Scholar]
- 28.National Research Council (NRC) Nutrient Requirements of Dairy Cattle. 7th revised ed. National Academy of Sciences; Washington, DC: 2001. [Google Scholar]
- 29.Nishiyama T., Ueki A., Kaku N., Watanabe K., Ueki K. Bacteroides graminisolvens sp. nov., a xylanolytic anaerobe isolated from a methanogenic reactor treating cattle waste. Int J Syst Evol Microbiol. 2009;59:1901–1907. doi: 10.1099/ijs.0.008268-0. [DOI] [PubMed] [Google Scholar]
- 30.Nissilä M.E., Li Y.C., Wu S.Y., Lin C.Y., Puhakka J.A. Hydrogenic and methanogenic fermentation of birch and conifer pulps. Appl Energy. 2012;100:58–65. [Google Scholar]
- 31.Nkemka V.N., Gilroyed B., Yanke J., Gruninger R., Vedres D., McAllister T., Hao X. Bioaugmentation with an anaerobic fungus in a two-stage process for biohydrogen and biogas production using corn silage and cattail. Bioresour Technol. 2015;185:79–88. doi: 10.1016/j.biortech.2015.02.100. [DOI] [PubMed] [Google Scholar]
- 32.Nyonyo T., Shinkai T., Mitsumori M. Improved culturability of cellulolytic rumen bacteria and phylogenetic diversity of culturable cellulolytic and xylanolytic bacteria newly isolated from the bovine rumen. FEMS Microbiol Ecol. 2014;88:528–537. doi: 10.1111/1574-6941.12318. [DOI] [PubMed] [Google Scholar]
- 33.Ozbayram E.G., Akyol Ç., Ince B., Karakoç C., Ince O. Rumen bacteria at work: bioaugmentation strategies to enhance biogas production from cow manure. J Appl Microbiol. 2018;124:491–502. doi: 10.1111/jam.13668. [DOI] [PubMed] [Google Scholar]
- 34.Peng X., Borner R.A., Nges I.A., Liu J. Impact of bioaugmentation on biochemical methane potential for wheat straw with addition of Clostridium cellulolyticum. Bioresour Technol. 2014;152:567–571. doi: 10.1016/j.biortech.2013.11.067. [DOI] [PubMed] [Google Scholar]
- 35.Qiu Y.L., Hanada S., Ohashi A., Harada H., Kamagata Y., Sekiguchi Y. Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl Environ Microbiol. 2008;74:2051–2058. doi: 10.1128/AEM.02378-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawatdeenarunat C., Surendra K.C., Takara D., Oechsner H., Khanal S.K. Anaerobic digestion of lignocellulosic biomass: Challenges and opportunities. Bioresour Technol. 2015;178:178–186. doi: 10.1016/j.biortech.2014.09.103. [DOI] [PubMed] [Google Scholar]
- 37.Takizawa S., Baba Y., Tada C., Fukuda Y., Nakai Y. Pretreatment with rumen fluid improves methane production in the anaerobic digestion of paper sludge. Waste Manage (Oxford, U K) 2018;78:379–384. doi: 10.1016/j.wasman.2018.05.046. [DOI] [PubMed] [Google Scholar]
- 38.Takizawa S., Baba Y., Tada C., Fukuda Y., Nakai Y. Preservation of rumen fluid for the pretreatment of waste paper to improve methane production. Waste Manage (Oxford, U K) 2019;87:672–678. doi: 10.1016/j.wasman.2019.02.043. [DOI] [PubMed] [Google Scholar]
- 39.Van Soest P.J., Wine R.H. Determination of lignin and cellulose in acid detergent fiber with permanganate. J-Assoc Off Anal Chem. 1968;51:780–785. [Google Scholar]
- 40.Wei S. The application of biotechnology on the enhancing of biogas production from lignocellulosic waste. Appl Microbiol Biotechnol. 2016;100:9821–9836. doi: 10.1007/s00253-016-7926-5. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., Guo R.B., Qiu Y.L., Qiao J.T., Yuan X.Z., Shi X.S., Wang C.S. Bioaugmentation with an acetate-type fermentation bacterium Acetobacteroides hydrogenigenes improves methane production from corn straw. Bioresour Technol. 2015;179:306–313. doi: 10.1016/j.biortech.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X., Mandelco L., Wiegel J. Clostridium hydroxybenzoicum sp. nov., an amino acid-utilizing, hydroxybenzoate-decarboxylating bacterium isolated from methanogenic freshwater pond sediment. Int J Syst Evol Microbiol. 1994;44:214–222. doi: 10.1099/00207713-44-2-214. [DOI] [PubMed] [Google Scholar]
- 43.Zheng Y., Zhao J., Xu F., Li Y. Pretreatment of lignocellulosic biomass for enhanced biogas production. Prog Energy Combust Sci. 2014;42:35–53. [Google Scholar]
- 44.Zou Y., Xu X., Li L., Yang F., Zhang S. Enhancing methane production from U. lactuca using combined anaerobically digested sludge (ADS) and rumen fluid pre-treatment and the effect on the solubilization of microbial community structures. Bioresour Technol. 2018;254:83–90. doi: 10.1016/j.biortech.2017.12.054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.