Summary
T cell stimulation is metabolically demanding. To exit quiescence, T cells rely on environmental nutrients, including glucose and the amino acids glutamine, leucine, serine and arginine. The expression of transporters for these nutrients is tightly regulated and required for T cell activation. In contrast to these amino acids, which are essential or require multi-step biosynthesis, alanine can be made from pyruvate by a single transamination. Here we show that extracellular alanine is nevertheless required for efficient exit from quiescence during naïve T cell activation and memory T cell restimulation. Alanine deprivation leads to metabolic and functional impairments. Mechanistically, this vulnerability reflects low expression of alanine aminotransferase the enzyme required for interconverting pyruvate and alanine, while activated T cells instead induce alanine transporters. Stable isotope tracing reveals that alanine is not catabolized but instead supports protein synthesis. Thus, T cells depend on exogenous alanine for protein synthesis and normal activation.
Introduction
In health, naive and memory T cells are retained in a quiescent state. In response to pathogen exposure, these cells quickly exit quiescence and enter a cycle of rapid cell growth and robust proliferation to generate large numbers of effector cells to fight off threats and restore homeostasis. The transition from quiescence to activation is driven by metabolic adaptation of these cells to meet their changing needs. With activation, glycolysis is highly induced, and glucose-derived carbon fuels multiple synthetic pathways, including the pentose phosphate and serine biosynthetic pathways, both of which contribute to de novo generation of nucleotides. In parallel, glucose-derived pyruvate feeds into the tricarboxylic acid cycle (TCA cycle), where it supplies a carbon source for lipid synthesis and electrons for ATP synthesis via the generation of citrate (Berod et al., 2014) and the electron transport chain, respectively.
To adequately fuel these metabolic pathways, which often share common resources, activated T cells rely on extracellular pools of precursor nutrients. For example, glutamine-derived carbon feeds into the TCA cycle to compensate for the export of citrate from the mitochondria to the cytosol, where it is used for fat synthesis (Berod et al., 2014). While glutamine is not an essential amino acid and can therefore be synthesized by mammalian cells, successful T cell activation depends on access to extracellular glutamine and its uptake (Carr et al., 2010; Nakaya et al., 2014). Similarly, although serine can be synthesized from the glycolytic intermediate 3-phosphoglycerate, extracellular serine remains the primary carbon source for mitochondrial one carbon metabolism, the main pathway in T cells for supplying one carbon units for purine and thymidine biosynthesis (Ron-Harel et al., 2016). Accordingly, serine deprivation from the diet is sufficient to impair T cell proliferation and effector functions (Ma et al., 2017). These studies emphasize how T cells depend on extracellular nutrients, including non-essential amino acids, to meet their rapidly changing metabolic needs upon activation.
Controlling the uptake to these nutrients is a key mechanism for regulating T cell activation (Geiger et al., 2016; Patsoukis et al., 2015; Ramsay and Cantrell, 2015; Rodriguez et al., 2007; Rollings et al., 2018; Wieman et al., 2007; Wofford et al., 2008). In this study, building on work done nearly 40 years ago describing a role for alanine in supporting T cell activation (Rotter et al., 1979), we identify alanine as an extracellular nutrient critical for T cells to transition from quiescence to activation. Our findings show that the nonessential amino acid alanine is not synthesized by T cells during early activation. Instead, naive T cells rely on extracellular alanine pools for their initial activation, and memory T cells need extracellular alanine for successful restimulation. Alanine deprivation negatively affects cell growth, proliferation and effector functions, and skews activation-induced metabolic reprogramming. Interestingly, in contrast to certain cancer cells that catabolize alanine (Sousa et al., 2016), T cells appear to exclusively use extracellular alanine for protein synthesis.
Results
Extracellular alanine is essential for initial activation of naïve T cells
We discovered T cell reliance on extracellular alanine for activation while investigating why dialyzed serum inhibited the activation of T cells in vitro. To identify extracellular nutrients that are required for T cell activation, we utilized a well-established system for ex vivo stimulation. Sorted naïve CD62L+CD25-CD44loCD4+ T cells were cultured with soluble anti-CD3 and antigen presenting cells (APCs) to stimulate the T cell receptor (TCR) and co-stimulatory signals required for proper activation (Figure 1A) in the presence of standard “complete” FBS (cFBS) or dialyzed FBS (dFBS). As expected, T cell stimulation resulted in increased T cell size in cFBS. However, the increase in cell size was blunted in dFBS (Figure 1B). To assess changes in effector function, we quantified secreted cytokine levels in spent culture media after cells were activated and cultured for 72 hr. The concentrations of the proinflammatory cytokines IL17, IFNγ and IL6 were all significantly reduced in the spent media from cells activated in the presence of dFBS (Figure 1C). Moreover, cells cultured in dFBS media proliferated less in the 72 hr following activation (Figure 1D). These findings suggest that at least one factor that is important for T cell stimulation is lost through FBS dialysis. To validate these findings in an orthogonal system, we stimulated sorted naive CD4+ T cells with plate bound anti-CD3 and anti-CD28 (Supplementary Figure 1A). As with APC-driven stimulation, culture in dFBS media impaired cell growth (Supplementary Figure 1B) and proliferation (Supplementary Figure 1C). Notably, alanine deprivation did not have a profound effect on expression of early activation markers, as demonstrated with CD69 (Supplementary Figure 1D). Since this simplified system proved suitable for ex vivo analysis of purified T cell cultures, it was used for all subsequent experiments.
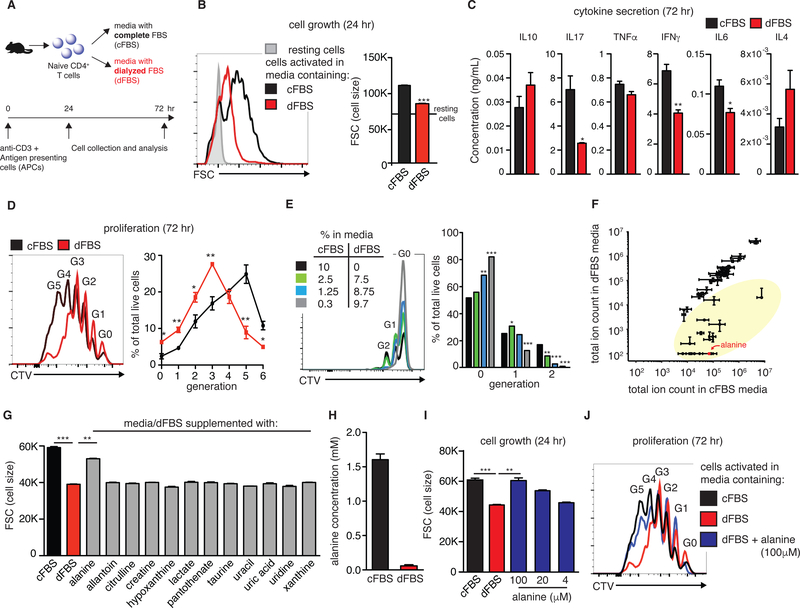
Figure 1: Alanine deprivation delays early T cell activation.
(A) Scheme of experimental design. Purified naïve CD4+ T cells were activated in vitro in growth media containing 10% of either complete or dialyzed FBS (cFBS or dFBS, respectively). (B) A representative flow-cytometry plot and its quantitation, showing activation-induced growth in cell size. (C) Cytokine secretion into growth media was analyzed by flow cytometry at 72 hr post-activation (n=3). (D,E) Proliferation (number of cell divisions detected by dilution of CellTrace) was assessed in cells activated under conditions described in A (D) and in cells activated for 48 hr in media prepared with various fractions of cFBS and dFBS (E) (n=4). (F) Growth media prepared with either 10% cFBS or dFBS was analyzed by LC-MS (using reverse-phase chromatography; n=2). Metabolites highlighted in yellow have reduced concentrations in dFBS media. (G) Rescue cell growth experiments at 24hr post-activation using media supplemented with selected metabolites absent in dFBS media (at final concentrations of 100 μM; n=3). (H) Calculated alanine concentration in cFBS and dFBS as determined by LC-MS using an internal standard ([15N]-alanine, n=3 for three different standard concentrations). (I) Activation-induced growth in cell size in different media conditions (n=3) (J) Proliferation in different media conditions with alanine supplementation (n=3). Results are mean ± SEM. *p<0.05, **p<0.01, ***p<0.001 (Student’s t-test comparing each of the conditions to dFBS media). All results are representatives of two or more independent experiments.
Standard T cell culture media contains 10% serum. Reducing the cFBS portion to 2.5% (offsetting with 7.5% dFBS to maintain total serum concentration) led to no observable effect on post-activation proliferation. However, further dilution of the cFBS fraction caused a substantial delay in proliferation (Figure 1E). Our findings suggest that early T cell activation requires one or more components found in cFBS that are reduced by >75% via dialysis. To identify this unknown component(s), growth media (RPMI) prepared with either 10% cFBS or 10% dFBS was analyzed by liquid chromatography mass spectrometry (LC-MS) in negative ionization mode. Chromatographic separation was achieved via two approaches: reverse phase ion-pairing chromatography (Figure 1F) and HILIC chromatography (Supplementary Figure 1E), with metabolites identified based on exact mass and retention time matched to authenticated standards. A set of 12 metabolites was independently identified by both LC-MS methods as being reduced by >75% in the dFBS media (Supplementary Figure 1F).
To determine which of the identified metabolites is essential for early T cell activation, naïve T cells were activated using media containing cFBS, dFBS or dFBS supplemented with each of the 12 candidate metabolites. Cells were collected at 24 hr post-activation for analysis of cell growth by flow cytometry. Strikingly, only alanine supplementation effectively rescued cell growth in dFBS media (Figure 1G). Although dialysis depletes every amino acid from serum, alanine is the only one of the 20 proteogenic amino acids not present in commercial RPMI media. Thus, serum supplementation supplies all of the alanine present in growth media.
To further investigate the dependency of proper T cell activation on extracellular alanine, naïve T cells were stimulated in media made with cFBS, dFBS, or dFBS + alanine (Supplementary Figure 1G). Results were confirmed across four different lots of dFBS. The concentration of alanine for supplementation was selected based on its normal serum concentration in humans (140 – 580 μM) and measurement of 1.5 mM alanine in pure FBS using an isotopic internal standard (Figure 1H and Supplementary Figure 1H), implying an alanine concentration in medium with 10% cFBS of 150 μM. 100 μM alanine completely rescued activation-induced growth and proliferation (Figures 1I and 1J, respectively).
Alanine is not an important catabolic substrate
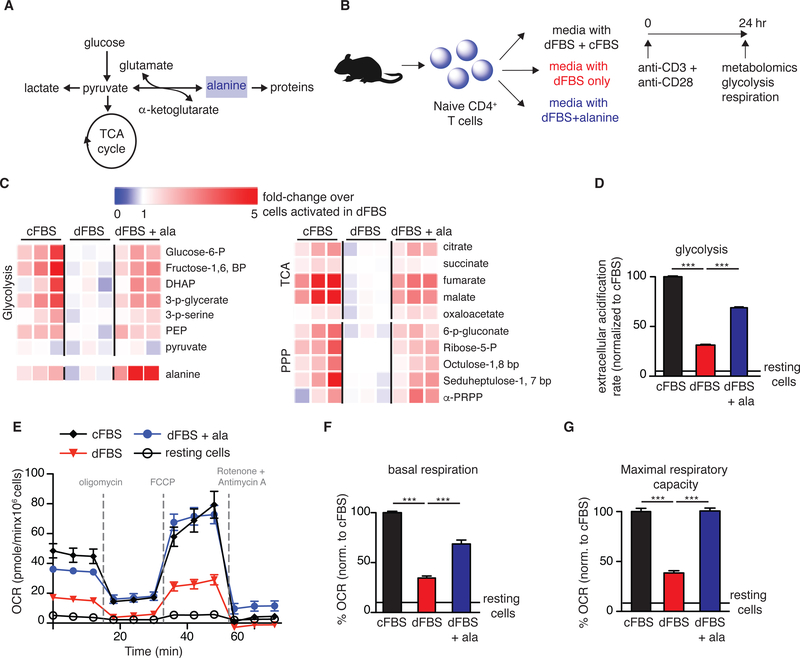
T cell activation causes dramatic metabolic upregulation, including simultaneous increases in aerobic glycolysis and mitochondrial metabolism. Therefore, we hypothesized that alanine could be an essential source of pyruvate for feeding into the TCA cycle during early T cell activation (Figure 2A). To test the effect of alanine deprivation on metabolic reprogramming, naïve T cells were stimulated using the different media conditions and collected at 24 hr post-activation for analysis of metabolic activity (Figure 2B). Cellular alanine, and metabolic intermediates in glycolysis, the pentose phosphate pathway, and the TCA cycle (Figure 2C) were lower in T cells activated in dFBS only media compared to media supplemented with cFBS, and were rescued by alanine supplementation. Consistent with these findings, at 24 hr post-activation, extracellular acidification rate (ECAR; Figure 2D), mitochondrial basal respiration and maximal respiratory capacity (Figure 2E – 2G) were suppressed in T cells stimulated in dFBS media, and rescued by alanine. Notably, the effect of alanine deprivation on metabolic reprogramming was not immediate, as at 9 hr post-activation, which precedes most activation-induced protein synthesis, concentrations of cellular metabolites were similar in T cells activated with and without alanine (Supplementary Figure 2A). Thus, the widespread metabolic reprogramming that arises from T cell activation is impaired in the absence of extracellular alanine.
Figure 2: Alanine deprivation skews T cell metabolism.
(A) Schematic showing metabolic fates of alanine in T cells. (B) Experimental design for evaluating metabolic impact of alanine in T cell activation. Media was supplemented with 10% of either dFBS, a mix of cFBS/dFBS (1:3) or dFBS with 100 μM alanine. (C) Changes in metabolite abundance in activated CD4+ T cells, normalized to levels in cells activated in dFBS. (D-G) Evaluation of T cell metabolic function by the Seahorse Extracellular Flux Analyzer (n=8–10 replicates/condition): (D) Extracellular acidification rate under blockade of mitochondrial respiration (measure of maximal glycolytic capacity), (E) changes in oxygen consumption rate (OCR) in activated T cells, (F) quantification of basal respiration, and (G) oxygen consumption rate following mitochondrial uncoupling (maximal respiratory capacity). Results are mean ± SEM of at least 2 individual experiments. ***p<0.001 (Student’s t-test comparing each of the conditions to dFBS media).
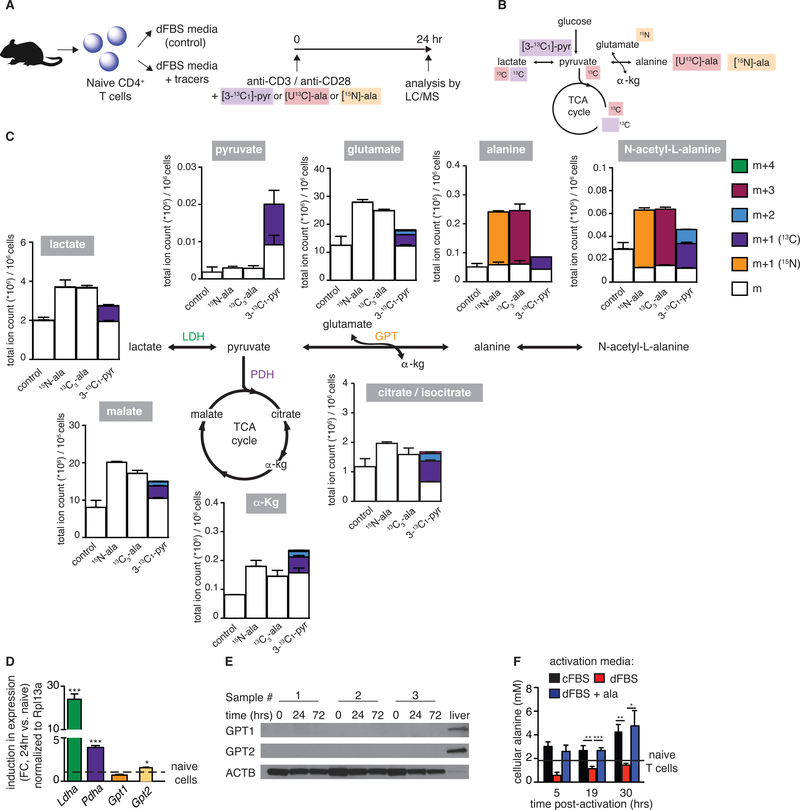
To directly test the catabolic fate of alanine, we cultured T cells in dFBS media with two different isotopically labeled forms of alanine ([U-13C]-alanine or [15N]-alanine; Figure 3A). Transamination of [U-13C]-alanine will give rise to ([U-13C]-pyruvate), which can subsequently be reduced to lactate or oxidized to enter the TCA cycle, in both cases giving rise to 13C-labeled products. Similarly, transamination of [15N]-alanine will give rise to [15N]-glutamate (Figure 3B). Interestingly, no 13C-labeling from alanine was detected in pyruvate or lactate, TCA cycle intermediates, or glutamate. Indeed, the only substantially 13C-labeled aqueous metabolites detected in these tracing experiments were alanine itself and its acetylated derivative (N-acetyl-L-alanine) (Figure 3C and Supplementary Figure 2B). As positive controls, we detected extensive labeling of TCA cycle intermediates and glutamate from both pyruvate (Figure 3C and Supplementary Figure 2B) and glutamine (Supplementary Figure 2C, 2D). Though a significant amount of alanine is labeled from 13C-pyruvate, this likely arises from exaggerated transaminase activity driven by the artificially high concentration of media pyruvate (1 mM), which is not present in the media for any other condition. To rule out the possibility that alanine catabolism is defective selectively in dFBS media, we performed alanine tracing experiments in cFBS media. Again, we found no evidence of alanine catabolism by the recently activated T cells (Supplementary Figure 2E). The lack of pyruvate and TCA intermediate labeling from 13C-alanine (at concentrations that fully rescue activation) implies the absence of meaningful alanine to pyruvate transaminase activity. We therefore concluded that T cells do not substantially catabolize alanine and the decreased levels of central carbon metabolites in cells deprived of alanine were mediated, at least in part, by a reduction in glucose uptake (Supplementary Figure 2F, 2G).
Figure 3: Activated T cells do not catabolize alanine.
(A) Experimental design for tracing the catabolic fate of alanine in activated T cells. Media was supplemented with dFBS and either of the following isotopic tracers: [3-13C1]-pyr (1mM), [U-13C]-ala (100μM) or [15N-ala] (100μM). (B) Schematic of anticipated incorporation of the tracers into relevant metabolic pathways. (C) Graphs summarize the normalized total ion counts of different isotopomers of representative metabolites in T cells activated in the presence of different tracers (A) (n=2). Isotope labeling data have been corrected for 13C natural abundance. LDH: lactate dehydrogenase, PDH: pyruvate dehydrogenase, GPT: glutamate-pyruvate transaminase (also known as alanine transaminase). (D) Expression of LDH (Ldha) and PDH (Pdha) complex subunits and of GPT isoforms (Gpt1, Gpt2) were measured by qPCR. Fold-change in induced expression (activated vs. naive T cells) was normalized to Rpl13a (n=3).(E) Protein quantitation of GPT1 and GPT2 by western blot, comparing naïve and activated T cells. Samples of mouse liver were used as positive control. (F) Cellular alanine concentration determined by LC-MS using an internal standard ([U-13C]-alanine) (n=3). Results are representative of at least 2 independent experiments.
T cells are low in alanine transaminase, the enzyme required for both alanine catabolism and synthesis
Alanine can be synthesized from glucose through pyruvate transamination by the enzyme alanine aminotransferase, also known as glutamate-pyruvate transaminase (GPT; Figure 3C). To examine the extent of alanine production, naïve T cells were activated in standard cFBS media, supplemented with uniformly labeled glucose [U-13C-glc]. At 5 hr post-activation, only 1–2% of cellular alanine was labeled (Supplementary Figure 3A). Thus, newly synthesized alanine is not a major source of alanine during early activation. Interestingly, in an independent experiment, we observed a small amount of alanine accumulation in the media over time (Supplementary Figure 3B). However, it is unclear if this apparent excretion of de novo alanine is deliberate, or is an artifact arising from diffusion of intracellular alanine into the alanine-free (since dFBS lacks alanine) culture media. In addition to alanine synthesis, the major fates of pyruvate are oxidation to acetyl-CoA via pyruvate dehydrogenase (PDH) and reduction to lactate via lactate dehydrogenase (LDH) (Figure 3C). qPCR analysis showed massive upregulation of genes in the PDH and LDH complexes with T cell activation, but a slight suppression or low induction in Gpt1 and Gpt2 mRNA, respectively (Figure 3D). Notably, Gpt2 mRNA was induced later in the activation process, and its levels were significantly higher at 48 and 72 hr post-activation compared to naïve T cells (Supplementary Figure 3C). Our recently published proteomic data showed a greater than 5-fold induction in protein levels of subunits in the PDH and LDH complexes during T cell activation (Ron-Harel et al., 2016), while GPT levels remained undetectable throughout (Supplementary Figure 3D). Western blot analysis further verified that neither GPT1 nor GPT2 were induced during T cell activation (Figure 3E). Consistent with this observation, T cell activation in dFBS resulted in a decrease in cellular alanine pools (Figure 3F). Although GPT protein levels remain low even at 72 hr post-activation, we detected some alanine synthesis from glucose at 24 hr (Supplementary Figure 3A) and saw a slight increase in cellular alanine concentrations over time in cells activated in dFBS (Figure 3F), possibly driven by the increase in cellular pyruvate pool size by 24 hr post-activation (Supplementary Figure 3E). These results suggest that during early activation, low alanine transaminase activity does not meet the cellular needs for alanine, and therefore T cells depend on extracellular alanine pools for proper activation.
Extracellular alanine is used for protein synthesis
In contrast to the small changes in alanine transaminase expression, we measured a large induction of some of the alanine transporters during T cell activation, supporting the observation that T cells depend on extracellular alanine. Alanine is classified as a neutral amino acid. Of the five neutral amino acid transporters, two were highly induced upon T cell activation based on analysis of our T cell proteome dataset: SLC1A5 (also known as ASCT2), and sodium-dependent neutral amino acid transporter 1 (SNAT1) (Supplementary Figure 3F). In T cells, ASCT2 mainly regulates glutamine uptake (Nakaya et al., 2014), whereas the net effect of SNAT1 deletion in human CD4+ T cells is reduced alanine uptake (Matheson et al., 2015). Therefore, we focused our validation on SNAT1 as the major mediator of alanine uptake in CD4+ T cells. SNAT1 expression and protein levels were highly induced with T cell activation (Supplementary Figures 3G, 3H).
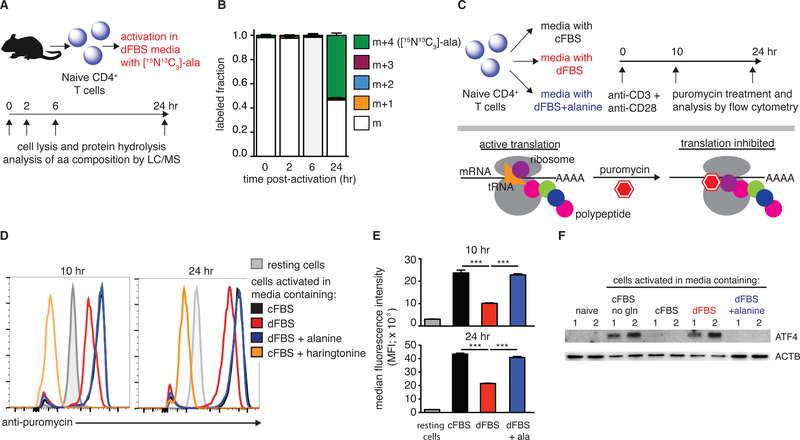
As T cell activation did not substantially engage in alanine catabolism, we hypothesized that T cells take up alanine via SNAT1 to support the large boost in protein synthesis that occurs upon T cell activation (Figure 2A). To validate the incorporation of extracellular alanine into proteins, naïve T cells were activated ex vivo in dFBS media supplemented with [U-15N13C]-alanine. Cells were collected at different time points post-activation, followed by protein hydrolysis and analysis of protein-derived alanine by LC/MS (Figure 4A). Strikingly, the contribution of extracellular alanine to total protein remains low for the first 6 hr, but by 24 hr, over 50% of alanine in proteins was labeled. Furthermore, the labeled alanine was exclusively of the m+4 isotopomer, indicating direct incorporation of extracellular alanine (Figure 4B). Thus, extracellular alanine is taken up by T cells upon activation and is directly used for protein synthesis without metabolic interconversion by transamination.
Figure 4: Alanine deprivation inhibits activation-induced protein synthesis.
(A) Experimental design for measuring incorporation of extracellular alanine into proteins during T cell activation. (B) Alanine labeled fraction in total cell proteome of activated T cells (n=2). (C) Experimental design to quantify active protein translation during T cell activation. Purified naïve CD4+ T cells were activated in different media conditions and were treated with puromycin (binds active translation complexes) at 10 and 24 hr, followed by flow cytometry analysis. (D) Representative flow cytometry plots of >3 experiments that measured active translation in the different conditions. T cells treated with harringtonine (to disrupt translation) serve as negative controls. (E) Quantitation of anti-puromycin median fluorescence intensity (MFI; n=3). (F) Representative western blot images measuring ATF4 levels in naïve T cells, and T cells activated for 3 hr in the indicated media conditions. Cells activated in glutamine-depleted media serve as a positive control. Results are representative of at least 3 independent experiments.
To evaluate whether extracellular alanine is also needed for protein synthesis during the first 6 hr of activation, we grew cells initially in dFBS, before adding back alanine. Statistically significant decreases in cell size were observed for any delay of 3 hr or more (Supplementary Figures 3I, 3J). Thus, T cells require extracellular alanine, possibly for protein synthesis, after only a few hours post-activation.
To provide further evidence for the requirement of alanine for protein synthesis, we quantified the number of actively translating ribosomes, taking advantage of the fact that puromycin selectively binds to actively translating ribosomes and its binding can be quantified using fluorescent anti-puromycin antibody (Arguello et al., 2018; Dadehbeigi and Dickson, 2013; Goodman and Hornberger, 2013) (Figure 4C). Our analysis demonstrated a reduction in active protein translation in T cells stimulated in media containing dFBS compared to cFBS, and a complete rescue with alanine supplementation (Figure 4D, 5E). The specificity of the fluorescent signal was validated by addition of harringtonine, an inhibitor of translation that disrupts ribosome-mRNA complexes. Treatment with harringtonine abolished the signal (Figure 4D).
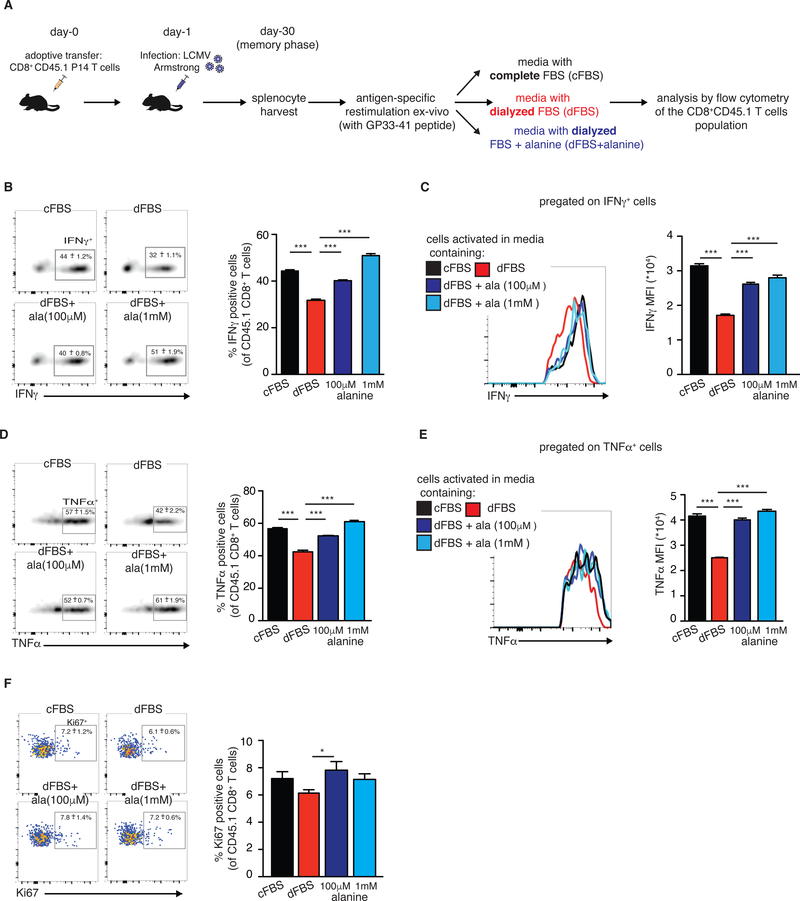
Figure 5: Extracellular alanine is essential for memory T cell restimulation.
(A) Experimental design for assessing the need for extracellular alanine during memory T cell restimulation. (B) Representative flow cytometry plots showing intracellular IFNγ staining following restimulation, and quantification of IFNγ positive cells. (C) Representative flow cytometry histograms and quantification of IFNγ MFI. (D) Representative flow cytometry plots showing intracellular TNFα staining following restimulation and quantification of TNFα positive cells. (E) Representative flow cytometry histograms and quantification of TNFα MFI. (F) Representative flow cytometry plots showing intracellular Ki67 staining following restimulation, and quantification of Ki67 positive cells. Representative plots of two independent experiments, n=5 mice. Results are mean ± SEM. *p<0.05, **p<0.01, ***p<0.001 (Student’s t-test comparing each of the conditions to dFBS media).
Uncharged tRNAs activate GCN2 to inhibit translation (Harding et al., 2000; Pakos-Zebrucka et al., 2016). GCN2 induces expression of ATF4 that upregulates expression of amino acid transporters and metabolic genes (Yang et al., 2018). To test whether this stress response is activated by alanine deprivation, naïve CD4+ T cells were stimulated in media containing either cFBS, dFBS or dFBS + alanine. Glutamine deprived media and naïve cells were used as positive and negative controls, respectively. Cells were collected 3 hr post-activation and analyzed by western blot. ATF4 levels were induced in cells deprived of alanine (dFBS media) and completely reversed by alanine supplementation (Figure 4F). In sum, upon activation, naïve T cells upregulate alanine uptake to fuel protein synthesis.
Alanine depletion inhibits memory T cell restimulation
We further examined the role of alanine in supporting of naïve, activated, and memory CD8+ T cells. To this end, we used a well-established in vivo model of acute lymphocytic choriomeningitis virus (LCMV) infection (Figure 5A). Since our initial observations were made in naïve CD4+ T cells, we first validated that naïve CD8+ T cells were similarly dependent on extracellular alanine for initial activation. Naïve CD8+ T cells were stimulated ex vivo with plate-bound anti-CD3/anti-CD28 in the indicated media conditions (Supplementary Figure 4A). Stimulation-induced cell growth and proliferation were impaired in CD8+ T cells activated in dFBS media compared to cells activated in cFBS media, and rescued by alanine supplementation (Supplementary Figure 4B, C, respectively). CD8+ T cells, like CD4+ T cells, induced SNAT1 levels with activation (Supplementary Figure 4D). Moreover, analysis by puromycin treatment (as described in Figure 4C) indicated reduced active translation in CD8+ T cells deprived of alanine and a complete rescue with alanine supplementation (Supplementary Figure 4E).
Since T cells can eventually proliferate in dFBS (Figures 1D, 1J), we hypothesized that extracellular alanine would not be required for effector T cell function. To assess the need for extracellular alanine during antigen-specific T cell activation, P14 TCR transgenic CD8+ T cells specific for LCMV peptide GP33–41 were transferred into wild type C57BL/6 recipients and infected with LCMV Armstrong, which causes an acute viral infection. Splenocytes were harvested on day 8 post LCMV infection, during the effector phase of the immune response to LCMV, cultured in vitro for 5 hr in dFBS or cFBS media with GP33–41 LCMV peptide (Supplementary Figure 5A), and effector functions assessed by flow cytometry (See Supplementary Figure 5B for gating strategy). No significant differences were seen in production of effector molecules: granzyme B (GzB), IFNγ and TNFα, or proliferation (assessed by staining against Ki67) as a function of alanine availability (Supplementary Figure 5C-F). Thus, once metabolically active, T cells do not depend on extracellular alanine.
Unlike active effector T cells, memory cells are metabolically quiescent. To explore their requirements for extracellular alanine, we analyzed splenocytes from LCMV infected mice at day 30-post infection (memory phase). The splenocytes were incubated ex vivo for 5 hr under the indicated media conditions in the presence of the GP33–41 LCMV peptide (Figure 5A). Splenocytes were then stained and analyzed by flow cytometry to assess antigen-specific T cell responses. Restimulation of memory T cells under conditions of alanine deprivation (dFBS media) caused a significant reduction in production of the proinflammatory cytokines IFNγ and TNFα. The inhibitory effect of alanine deprivation manifested as a reduced percentage of cytokine-expressing T cells (Figures 5B and 5D for IFNγ and TNFα, respectively) and lower cytokine production per cell (Figures 5C and 5E for IFNγ and TNFα, respectively). Production of both cytokines was completely restored with alanine supplementation (Figure 5B-5E). Interestingly, GzB production was not negatively affected by alanine deprivation, and was even slightly increased in dFBS media (Supplementary Figure 5G, 5H), suggesting that GzB might be pre-formed and stored in the cells, prior to restimulation. Proliferation rates, measured by staining against the endogenous marker Ki67, were low among the restimulated memory T cells under all media conditions (Figure 5F), potentially due to the short stimulation (5 hr). Yet, a minor reduction in Ki67+ cells was observed under alanine deprivation (Figure 5F). Thus, extracellular alanine is required for effector cytokine production by memory CD8+ T cells upon restimulation.
Discussion
In this study, we identify alanine as a semi-essential amino acid for T cells during activation. Extracellular alanine is needed early in the activation program (3–6 hr) of both naïve and memory T cells, and failure to meet this requirement leads to impaired metabolic reprogramming, including reduced glucose uptake and decreased levels of metabolites in central carbon metabolism. This demand for extracellular alanine stems mainly from the requirement for alanine monomers for protein production, as alanine taken up during T cell stimulation is not catabolized and is directly incorporated into proteins.
How does nutrient availability control T cell activation? T cell activation induces rapid cell growth, proliferation and secretion of effector molecules. To fuel these biosynthetic and energy demands, T cells evolve to rely on the resources in their environment: activation signals induce expression of glucose and amino acid transporters as the cells require sustained uptake of nutrients (Rollings et al., 2018; Sinclair et al., 2013). Moreover, nutrient-sensitive signaling pathways such as mTOR, O-GlcNAC and cMYC regulate T cell activation and differentiation (Hukelmann et al., 2016; Sinclair et al., 2013; Swamy et al., 2016), thus connecting metabolite availability with function. Reliance on extracellular alanine joins other nutrient dependencies of activated T cells established in recent years, including glucose (Cham and Gajewski, 2005), glutamine (Nakaya et al., 2014), leucine (Ananieva et al., 2016), serine (Ma et al., 2017), and arginine (Geiger et al., 2016; Rodriguez et al., 2007). In all cases, restriction of the specific metabolite inhibited T cell activation and/or effector functions. Recent studies suggest that metabolic restriction helps cancer cells escape immune surveillance. Specifically, low glucose availability within the tumor microenvironment inhibits anti-tumor immunity (Chang et al., 2015).
Alanine is the second most abundant amino acid in the circulation, after glutamine, which makes it very accessible to T cells, especially those in organs with ample blood supply. Interestingly, analysis of changes in abundance of cell surface proteins during HIV infection of human CD4+ T cells identified a specific downregulation of SNAT1 on infected T cells, and subsequent reduction of alanine uptake (Matheson et al., 2015). That study further suggested that alanine is catabolized by T cells, and that downregulation of SNAT1 reduced glutamate pools. Similarly, a study in pancreatic cancer cells showed reliance on extracellular alanine as a source of carbon for the TCA cycle (Sousa et al., 2016). In our studies, we could not detect any evidence of alanine catabolism by T cells.
Our finding that T cells depend on extracellular pools of alanine is surprising, especially since alanine is a non-essential amino acid, for which mammalian cells possess the necessary synthetic machinery. Moreover, activated T cells are highly engaged in glycolysis and glutaminolysis, resulting in significant flux through pyruvate and glutamate, respectively, the immediate substrates for alanine production. Using estimated mammalian cell concentrations of pyruvate and glutamate under standard tissue culture conditions, production of alanine from pyruvate is thermodynamically favorable, with delta G ~ −10 KJ/mol using the Weizmann Equilibrium Calculator (Flamholz et al., 2012; Park et al., 2016). This suggests that appreciable GPT expression during early activation would facilitate alanine production. Therefore, we speculate that GPT expression may be intentionally suppressed during T cell activation.
It is possible that T cells sacrifice the ability to produce alanine early in activation to avoid depleting cellular pyruvate pools, which may be more critical for fueling the mitochondrial biosynthetic program (Buck et al., 2016; Ron-Harel et al., 2016) and/or for accepting electrons from NADH to allow regeneration of NAD+. In this respect, it is important to note that glycolysis induces flux through pyruvate without increasing net pyruvate. Production of pyruvate requires transfer of electrons from NADH into the mitochondrion, and ultimately to molecular oxygen. While an efficient source of ATP, this process can bottleneck due to low oxygen availability (as can occur in ischemic infection sites or tumors) or inefficient machinery for electron transfer from the cytosol to mitochondrion (as may occur early in T cell activation, when mitochondria are relatively scarce). Indeed, increasing evidence identifies oxidized carbon as a limiting reagent in multiple cellular contexts (Birsoy et al., 2015; Gui et al., 2016). By taking in exogenous alanine, T cells may conserve oxidized carbon for more pressing needs, including synthesis of aspartate, the amino acid that is least abundant in the circulation.
In sum, we find that access to extracellular alanine is essential during early T cell activation and memory T cell restimulation to fuel protein synthesis. Serum alanine concentrations are known to fluctuate in mammals by as much as 2–3 fold depending on the fed-state, dietary composition and feeding schedule for the animal (Knipfel et al., 1972; Krebs, 1972). The local concentrations of alanine in different tissue niches have not been thoroughly annotated. Moreover, alanine is depleted in some tumors (Kamphorst et al., 2015). It is possible that control of local alanine levels through the uptake or secretion of alanine by resident cells in the lymphatic tissue, or other tissues where resident memory T cells get re-activated (such as the skin, the lungs or the brain), may impact T cell activation. Similarly, alanine availability in tumors may affect anti-tumor immunity. Taken together, our findings suggest that T cells may be sensitive to changing alanine concentrations during physiological or disease states. Thus, interventions that target alanine uptake may provide a therapeutic strategy for modulating T cell activation or memory function.
LEAD CONTACT AND MATERIALS AVAILABILITY
This study did not generate new reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marcia Haigis (marcia_haigis@hms.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Wild type C57BL/6 mice were purchased from the Jackson Laboratory (Bar Harbor, ME). P14 (Taconic B6.Cg-Tcratm1Mom Tg(TcrLCMV)327Sdz) were backcrossed 10 generations to Jackson C57BL/6J and crossed to the CD45.1 congenic background (B6.SJL-Ptprca Pepcb/BoyJ). 7–10-week-old female mice were used for all experiments. Mice were housed in specific pathogen-free conditions at Harvard Medical School and used in accordance with animal care guidelines from the Harvard Medical School Standing Committee on Animals and the National Institutes of Health.
METHODS DETAILS
Culture and stimulation of naïve CD4+ T cells
Naïve CD4+CD62LhiCD44loCD25- or CD8+CD62LhiCD44loCD25- cells were purified from splenocytes using CD4+ or CD8+ magnetic beads (Miltenyi Biotec) and sorted by flow cytometry (purity >99%) as previously described (Ron-Harel et al., 2016). Naïve T cells were cultured at 37°C and 5% CO2 in complete RPMI media (RPMI, supplemented with 10% FBS, 10 mM HEPES, 10 U/ml penicillin/streptomycin, and 50 μM 2-mercaptoethanol (all from Life Technologies)). When indicated, dialyzed FBS was used in combination or as a replacement of standard FBS, and alanine (Sigma Aldrich), isotopically labeled alanine (Cambridge Isotope Laboratories, Inc.) or isotopically labeled pyruvate (Cambridge Isotope Laboratories, Inc.) was added to achieve the denoted concentrations. For glutamine labeling experiments, glutamine-free RPMI (Gibco) was supplemented with [U-13C,15N]-Glutamine (2 mM, Cambridge Isotope Laboratories, Inc.). Cells were activated in vitro with 4 μg/mL plate-bound anti-CD3 (clone 145–2C11; BioXCell) and anti-CD28 (clone 37.51; BioXCell), or co-cultured with antigen presenting cells (splenocytes that were depleted of T cells using anti-CD4 and anti-CD8 magnetic beads (Miltenyi Biotec.), with 2 μg/mL soluble anti-CD3. 50K T cells were incubated with 250K APCs per well, in a 96 well plate. In some experiments, IL-7 (5 ng/mL; R&D Systems) was added to culture media. In metabolite rescue experiments, media containing 10% dFBS was supplemented with denoted metabolites (achieving a final concentration of 100 μM, which was chosen to exceed the likely concentrations of these metabolites in media prepared with 10% cFBS, based on reported estimates of these metabolites in serum). Identical results were obtained when supplementing dFBS media with 10 μM of the same metabolites, except for alanine, which failed to rescue T cell activation at this concentration.
Metabolite measurements by LC-MS
For analysis of intracellular metabolites by liquid chromatography mass spectrometry, activated CD4+ T cells were collected into Eppendorf tubes, centrifuged at 9,500 g for 20 sec at room temperature, resuspended in 1 mL PBS, and promptly centrifuged again under the same conditions. The supernatant was quickly removed and the pellets were extracted with 50–100 μl extraction buffer (either 80% methanol/20% water or 40% acetonitrile/40% methanol/20% water + 0.5% formic acid). Total time from perturbation of the cells to quenching was ~ 1 min. Samples extracted w/formic acid buffer were incubated 5 min on ice following by pH neutralization with 15% w/v ammonium hydroxide. Samples were stored at −80°C, followed by centrifugation at 4°C (~15,000 g) to remove insoluble cell components. For serum and media metabolite analysis, 10 μL of sample was diluted into 40 μL of methanol cooled on dry ice. The mixture was centrifuged at 17,000 g for 5 min at 4°C, and the resulting supernatants were diluted and analyzed directly by LC-MS. For experiments where absolute metabolite concentrations were obtained, isotopically labeled internal standards were added to the extraction buffer.
Metabolites were analyzed using a quadrupole-orbitrap mass spectrometer (Q Exactive Plus, Thermo Fisher Scientific, Waltham, MA), coupled to hydrophilic interaction chromatography (HILIC) with LC separation on a XBridge BEH Amide column, or a stand-alone orbitrap (Thermo-Fisher Exactive) coupled to reversed-phase ion-pairing chromatography with LC separation on a Waters column. Both mass spectrometers were operating in negative ion mode and were coupled to their respective liquid chromatography methods via electronspray-ionization. Data were analyzed using the MAVEN software suite (Clasquin et al., 2012). Metabolite abundances were normalized to cell count. Unless otherwise indicated, isotopic labeling of metabolites arising from incubation with 13C or 15N labeled nutrients were corrected for natural abundance, as previously described (Su et al., 2017).
Isolation of protein and hydrolysis into amino acid monomers
Cell pellets were mixed in 200 μl of lysis buffer (2% SDS, 150mM NaCl, 50mM Tris (pH 8.5), proteinase inhibitor mix (Roche, South San Francisco, CA, USA), 5mM DTT, and incubated on ice for 10 min, followed by incubation at 60°C for 45 min. Samples were allowed to cool to room temperature and incubated for 45 min with iodoacetamide to a final concentration of 14 mM. Samples were mixed with 3 parts ice-cold methanol, 1 part chloroform and 2.5 parts H2O, and centrifuged at 4000 g for 10 min. Following removal of the top layer, 3 parts of ice-cold methanol were added, followed by centrifugation at 4000 g for 5 min. Following removal of the top layer, the samples were mixed with 3 parts of ice-cold acetone, vortexed, and centrifuged at 4000 g for 5 min. The pellet was then washed one more time in 2 mL ice-cold acetone, and stored at (−80°C) prior to chemical hydrolysis. The resulting protein pellet was resuspended in 6 N HCl/acetic acid (50:50, 100 μL) and was heated at 95°C for one hour, using a slightly modified version of a reported protocol for protein hydrolysis (Tsugita and Scheffler, 1982). The resulting aqueous solution was diluted into 40% acetonitrile/40% methanol/20% water and analyzed by LC-MS.
LCMV infection and ex vivo stimulation of effector and memory T cells
Spleens were isolated from P14 TCR transgenic mice and naïve CD8+ T cells were purified using the naïve CD8+ MACS kit (>95% purity; Miltenyi Biotec). Cells were transferred intravenously (500 cells) to C57BL/6 wild type mice on day 0. The following day (day 1), the mice were intraperitoneally infected with 2×105 PFU LCMV Armstrong, and sacrificed at day 8 or 30 post infection for analysis. Spleens were isolated from LCMV-infected mice, dissociated, and ACK-lysed to remove red blood cells. Two million cells/well were plated in a 96 U bottom plate and incubated at 37°C for 4 hr with Golgi Stop (BD Biosciences) at 0.06% and LCMV GP33–41 peptide at 1 μg/mL. Cells were then harvested and processed for analysis by flow cytometry.
Flow cytometry
In vitro activated cells or cells isolated from LCMV-infected mice were collected, resuspended in staining buffer (PBS containing 1% fetal bovine serum and 2 mM EDTA), and stained directly with labeled antibodies from Biolegend against CD8 (YTS156.7.7), CD45.1 (A20), and CD69 (H1.2F3). For intracellular staining, the FoxP3 fix/perm kit (eBioscience) was used after surface staining, followed by staining against: IFNγ (XMG1.2), GzB (GB11), TNFα (MP6-XT22) and Ki67 (16A8). For assessment of proliferation, cells were loaded with CellTrace violet (Molecular Probes), according to the manufacturer’s protocol. Analysis of cytokine levels in growth media was performed using the Th1/Th2/Th17 cytometric bead array (BD Biosciences), following the manufacturer’s protocol. All flow cytometry was analyzed with an LSRII (BD biosciences) using standard filter sets, and FlowJo software (TreeStar).
Glucose uptake
Naïve CD4+ T cells were activated with anti-CD3/CD28 antibodies in complete RPMI media supplemented with either 10% dialyzed FBS or 10% complete FBS. After 24 hours, cells were washed and starved of glucose by incubation in glucose-free, serum-free RPMI media for 30 minutes before incubation with 50 μg/mL 2-deoxy-2-[(7-nitro-2,1,3-benzoxadiazol-4-yl)amino]-d-glucose (2-NBDG) for 20 min. Uptake was measured by flow cytometric evaluation of signal in the FITC channel. Fluorescence intensity was normalized to cell size, using the forward-side scatter.
Real-time PCR
Total DNA was isolated using DNeasy kit (Qiagen). cDNA was synthesized using the iSCRIPT kit (BioRad). Quantitative PCR analysis was performed with SYBR green, using a LightCycler 480 (Roche). Primers used: Snat1: F: 5’- TGAGACTGCCCAAATGCCTAAGGA-3’, R: 5’- TCTCTGTTGCAGAAAGCTGGTGGA −3’; Pdha: F: 5’- CCACCTCATCACTGCCTATC-3’, R: 5’- CCTTTAGCACAACCTCCTCTT-3’; Ldha: F: 5’-ACAAACTCAAGGGCGAGATG-3, R: 5’- GGAGTTCGCAGTTACACAGTAG-3; Gpt1: 5’- GGAAGGTGCTAACTCTGGATAC-3’, R: 5’- GGCACGGATAACCTCAGTAAA-3’; Gpt2: F: 5’-AACTGTATCCGTGAAGATGTGGC-3’, R: 5’-CAGGTAAATGTTGTCTGGGTCTG-3’;
Protein content analysis by western blot
Cells were lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.5 and 0.5% NP-40) supplemented with protease inhibitor cocktail (Roche). Lysates were mixed with sample buffer (Tris-HCL pH 6.8, 30% glycerol, 10% SDS, 0.012% bromophenol blue) under reducing conditions, separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, and transferred to nitrocellulose membrane. The membranes were incubated for 1 hr in Tris-Buffered Saline (TBS) containing 0.1% Tween-20 and 5% nonfat milk, followed by an overnight incubation with the respective primary antibodies at 4°C. The membranes were then washed and incubated with horseradish peroxidase-conjugated secondary antibody for 1 hr, washed and developed using an enhanced chemiluminescence reagent (Thermo Scientific). Primary antibodies used: rabbit anti-GPT1 (16897–1-AP, Proteintech), rabbit anti-GPT2 (16757–1-AP, Proteintech), rabbit anti-ATF4 (D4B8, #11815, Cell Signaling), rabbit anti SNAT1 (D9L2P, #36057, Cell Signaling), rabbit anti-ACTB (A2066, Sigma-Aldrich), HRP conjugated rabbit anti-ACTB (13E5, Cell Signaling).
Mitochondrial respiration and glycolysis
Oxygen consumption rate and extracellular acidification rate were measured from cells in non-buffered DMEM containing 5 mM glucose, 2 mM L-glutamine, and 1 mM sodium pyruvate, under basal conditions and in response to mitochondrial inhibitors: 1 μM oligomycin, 1 μM FCCP, 100 nM rotenone, and 1 μM antimycin A (All from Sigma) on the XFe96 Extracellular Flux Analyzer (Agilent Technologies).
Analysis of active protein synthesis with puromycin treatment
T cells were activated in vitro for up to 24 hr at the indicated times, puromycin (10 μg/mL) was added to the culture for 10 min at 37° C. Some cells were treated with harringtonine (1 μg/mL) for 15 min at 37° C, prior to addition of puromycin. Cells were then washed with cold PBS, processed for intracellular staining of puromycin (AF604 anti-puromycin, EMD Millipore Corp), and immediately analyzed by flow cytometry.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
All statistical analyses were performed using Prism (Graphpad Software). We used unpaired Student’s t-test when comparing two groups, and one-way analysis of variance (ANOVA) followed by Tukey-Kramer’s post hoc analysis, when comparing 3 groups.
DATA AND CODE AVAILABILITY
The proteomics dataset used in this study is available at Ron-Harel et al., 2016. All used software is available and listed in the Key Resource Table. This study did not generate code or new datasets.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD3e (145–2C11) | BioXCell | BE0001–1 |
| Anti-mouse CD28 (37.51) | BioXCell | BE0015–1 |
| APC/cy7 anti-mouse CD8b (YTS156.7.7) | Biolegend | 126619 |
| PE anti-mouse CD45.1 (H1.2F3) | Biolegend | 110707 |
| PerCP/cy5.5 anti-mouse CD69 (H1.2F3) | Biolegend | 104521 |
| APC anti-mouse IFNγ (XMG1.2) | Biolegend | 505809 |
| FITC anti-mouse GzB (GB11) | Biolegend | 515403 |
| PerCP/cy5.5 anti-mouse TNFα (MP6-XT22) | Biolegend | 506321 |
| PerCP/cy5.5 anti-mouse Ki67 (16A8) | Biolegend | 652423 |
| BV421 donkey anti-rabbit IgG (Poly4064) | Biolegend | 406410 |
| Rabbit anti-GPT1 | Proteintech | 16897–1-AP |
| Rabbit anti-GPT2 | Proteintech | 16757–1-AP |
| Rabbit anti-ATF4 (D4B8) | Proteintech | 11815 |
| Rabbit anti-SNAT1 (D9L2P) | Cell Signaling | 36057 |
| Rabbit anti-ACTB | Sigma Aldrich | A2066 |
| HRP conjugated rabbit anti-ACTB (13EB) | Cell signaling | 5125 |
| AF604 anti-puromycin | EMD Millipore Corp | MABE343-AF647 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| L-Alanine | Sigma Aldrich | A7627–100G |
| 15N labeled alanine | Cambridge Isotope Laboratories, Inc. | NLM-454 |
| 3–13C labeled pyruvate | Cambridge Isotope Laboratories, Inc. | CLM-1575 |
| U-13C-15N glutamine | Cambridge Isotope Laboratories, Inc. | CNLM-1275 |
| RPMI 1640 | Thermo Fisher | 11875–119 |
| RPMI 1640 (no glutamine) | Thermo Fisher | 21870100 |
| FBS | Corning | 45000–734 |
| Dialyzed FBS (1698218, 1743489) | Thermo Fisher | 26400044 |
| Oligomycin A | Sigma Aldrich | 75351 |
| FCCP | Sigma Aldrich | C2920 |
| Rotenone | Sigma Aldrich | R8875 |
| Antimycin A | Sigma Aldrich | A8674 |
| 2-NBDG | Thermo Fisher | N13195 |
| Puromycin | InvivoGen | ant-pr-1 |
| CellTrace violet | Thermo Fisher | C34557 |
| Golgi stop | BD Biosciences | 554724 |
| Complete Mini Protease Inhibitor | Roche | 11873580001 |
| Recombinant IL-7 | R&D systems | 407-ML |
| Critical Commercial Assays | ||
| CD4 (L3T4) Microbeads, mouse | Miltenyi Biotec | 130–117-043 |
| CD8 (Ly-2) Microbeads, mouse | Miltenyi Biotec | 130–117-044 |
| Foxp3/transcription factor staining buffer set | eBioscience | 00–5523-00 |
| Th1/Th2/Th17 cytometric bead array, mouse | BD Biosciences | 560485 |
| iScript cDNA synthesis kit | BioRad | 1708891 |
| DNeasy blood and tissue kit | QIAGEN | 69506 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | The Jackson Laboratory | 000664 |
| B6.Cg-Tcratm1Mom Tg(TcrLCMV)327Sdz/TacMmjax | The Jackson Laboratory | 37394-JAX |
| B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | 002014 |
| Oligonucleotides | ||
| Primer Snat1, F: TGAGACTGCCCAAATGCCTAAGGA, R: TCTCTGTTGCAGAAAGCTGGTGGA | This paper | N/A |
| Primer Pdha, F: CCACCTCATCACTGCCTATC, R: CCTTTAGCACAACCTCCTCTT | This paper | N/A |
| Primer Ldha, F: ACAAACTCAAGGGCGAGATG, R: GGAGTTCGCAGTTACACAGTAG | This paper | N/A |
| Primer Gpt1, F: GGAAGGTGCTAACTCTGGATAC, R: GGCACGGATAACCTCAGTAAA | This paper | N/A |
| Primer Gpt2, F: AACTGTATCCGTGAAGATGTGGC, R: CAGGTAAATGTTGTCTGGGTCTG | This paper | N/A |
| Software and Algorithms | ||
| Prism | GraphPad Sofwate | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | FlowJo | https://www.flowjo.com |
| MAVEN software suite | Clasquin et al., 2012) | https://maven.apache.org |
Supplementary Material
Acknowledgements
The authors thank Matthew Coxe for his help with performing experiments. This study was supported by grants from the Ludwig Center at Harvard Medical School, NIH grant U54-CA225088 (A.H.S. and M.C.H.), NIH grant R01CA213062 (M.C.H.), and by NSF Graduate Research Fellowship Program (G.N.). NRH was supported by The European Molecular Biology Organization long-term postdoctoral fellowship, and the Israeli National Postdoctoral Award Program for Advancing Women In Science.
References:
- Ananieva EA, Powell JD, and Hutson SM (2016). Leucine Metabolism in T Cell Activation: mTOR Signaling and Beyond. Advances in nutrition 7, 798S–805S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello RJ, Reverendo M, Mendes A, Camosseto V, Torres AG, de Pouplana Ribas L., van de Pavert SA, Gatti E, and Pierre P. (2018). SunRiSE - measuring translation elongation at single-cell resolution by means of flow cytometry. Journal of cell science 131. [DOI] [PubMed] [Google Scholar]
- Berod L, Friedrich C, Nandan A, Freitag J, Hagemann S, Harmrolfs K, Sandouk A, Hesse C, Castro CN, Bahre H, et al. (2014). De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nature medicine 20, 1327–1333. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, and Sabatini DM (2015). An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O’Sullivan D, Geltink Klein R.I., Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, et al. (2016). Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 166, 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, Sowell RT, Kaech SM, and Pearce EL (2017). Metabolic Instruction of Immunity. Cell 169, 570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr EL, Kelman A, Wu GS, Gopaul R, Senkevitch E, Aghvanyan A, Turay AM, and Frauwirth KA (2010). Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. Journal of immunology 185, 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cham CM, and Gajewski TF (2005). Glucose availability regulates IFN-gamma production and p70S6 kinase activation in CD8+ effector T cells. Journal of immunology 174, 4670–4677. [DOI] [PubMed] [Google Scholar]
- Chang CH, Qiu J, O’Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. (2015). Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell 162, 1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasquin MF, Melamud E, and Rabinowitz JD (2012). LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinformatics Chapter 14, Unit14 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadehbeigi N, and Dickson AJ (2013). Application of a nonradioactive method of measuring protein synthesis in industrially relevant Chinese hamster ovary cells. Biotechnology progress 29, 1043–1049. [DOI] [PubMed] [Google Scholar]
- Flamholz A, Noor E, Bar-Even A, and Milo R. (2012). eQuilibrator--the biochemical thermodynamics calculator. Nucleic acids research 40, D770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, Kogadeeva M, Picotti P, Meissner F, Mann M, et al. (2016). L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 167, 829–842 e813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, and Hornberger TA (2013). Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exercise and sport sciences reviews 41, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui DY, Sullivan LB, Luengo A, Hosios AM, Bush LN, Gitego N, Davidson SM, Freinkman E, Thomas CJ, and Vander Heiden MG (2016). Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell metabolism 24, 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, and Ron D. (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Molecular cell 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Hukelmann JL, Anderson KE, Sinclair LV, Grzes KM, Murillo AB, Hawkins PT, Stephens LR, Lamond AI, and Cantrell DA (2016). The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nature immunology 17, 104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, et al. (2015). Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer research 75, 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipfel JE, Keith MO, Christensen DA, and Owen BD (1972). Diet and feeding interval effects on serum amino acid concentrations of growing swine. Canadian Journal of Animal Sciences 52, 143–153. [Google Scholar]
- Krebs HA (1972). Some aspects of the regulation of fuel supply in omnivorous animals. Advances in enzyme regulation 10, 397–420. [DOI] [PubMed] [Google Scholar]
- Ma EH, Bantug G, Griss T, Condotta S, Johnson RM, Samborska B, Mainolfi N, Suri V, Guak H, Balmer ML, et al. (2017). Serine Is an Essential Metabolite for Effector T Cell Expansion. Cell metabolism 25, 482. [DOI] [PubMed] [Google Scholar]
- Matheson NJ, Sumner J, Wals K, Rapiteanu R, Weekes MP, Vigan R, Weinelt J, Schindler M, Antrobus R, Costa AS, et al. (2015). Cell Surface Proteomic Map of HIV Infection Reveals Antagonism of Amino Acid Metabolism by Vpu and Nef. Cell host & microbe 18, 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, Blonska M, Lin X, and Sun SC (2014). Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 40, 692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, and Gorman AM (2016). The integrated stress response. EMBO reports 17, 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JO, Rubin SA, Xu YF, Amador-Noguez D, Fan J, Shlomi T, and Rabinowitz JD (2016). Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nature chemical biology 12, 482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsoukis N, Bardhan K, Chatterjee P, Sari D, Liu B, Bell LN, Karoly ED, Freeman GJ, Petkova V, Seth P, et al. (2015). PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nature communications 6, 6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay G, and Cantrell D. (2015). Environmental and metabolic sensors that control T cell biology. Frontiers in immunology 6, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez PC, Quiceno DG, and Ochoa AC (2007). L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 109, 1568–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollings CM, Sinclair LV, Brady HJM, Cantrell DA, and Ross SH (2018). Interleukin-2 shapes the cytotoxic T cell proteome and immune environment-sensing programs. Science signaling 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron-Harel N, Santos D, Ghergurovich JM, Sage PT, Reddy A, Lovitch SB, Dephoure N, Satterstrom FK, Sheffer M, Spinelli JB, et al. (2016). Mitochondrial Biogenesis and Proteome Remodeling Promote One-Carbon Metabolism for T Cell Activation. Cell metabolism 24, 104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter V, Yakir Y, and Trainin N. (1979). Role of L-alanine in the response of human lymphocytes to PHA and Con A. Journal of immunology 123, 1726–1731. [PubMed] [Google Scholar]
- Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, and Cantrell DA (2013). Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nature immunology 14, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, Kremer D, Hwang RF, Witkiewicz AK, Ying H, et al. (2016). Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Lu W, and Rabinowitz JD (2017). Metabolite Spectral Accuracy on Orbitraps. Analytical chemistry 89, 5940–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DM, and Cantrell DA (2016). Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nature immunology 17, 712–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugita A, and Scheffler JJ (1982). A rapid method for acid hydrolysis of protein with a mixture of trifluoroacetic acid and hydrochloric acid. European journal of biochemistry 124, 585–588. [DOI] [PubMed] [Google Scholar]
- Wieman HL, Wofford JA, and Rathmell JC (2007). Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Molecular biology of the cell 18, 1437–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wofford JA, Wieman HL, Jacobs SR, Zhao Y, and Rathmell JC (2008). IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood 111, 2101–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Xia R, Yue C, Zhai W, Du W, Yang Q, Cao H, Chen X, Obando D, Zhu Y, et al. (2018). ATF4 Regulates CD4(+) T Cell Immune Responses through Metabolic Reprogramming. Cell reports 23, 1754–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics dataset used in this study is available at Ron-Harel et al., 2016. All used software is available and listed in the Key Resource Table. This study did not generate code or new datasets.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD3e (145–2C11) | BioXCell | BE0001–1 |
| Anti-mouse CD28 (37.51) | BioXCell | BE0015–1 |
| APC/cy7 anti-mouse CD8b (YTS156.7.7) | Biolegend | 126619 |
| PE anti-mouse CD45.1 (H1.2F3) | Biolegend | 110707 |
| PerCP/cy5.5 anti-mouse CD69 (H1.2F3) | Biolegend | 104521 |
| APC anti-mouse IFNγ (XMG1.2) | Biolegend | 505809 |
| FITC anti-mouse GzB (GB11) | Biolegend | 515403 |
| PerCP/cy5.5 anti-mouse TNFα (MP6-XT22) | Biolegend | 506321 |
| PerCP/cy5.5 anti-mouse Ki67 (16A8) | Biolegend | 652423 |
| BV421 donkey anti-rabbit IgG (Poly4064) | Biolegend | 406410 |
| Rabbit anti-GPT1 | Proteintech | 16897–1-AP |
| Rabbit anti-GPT2 | Proteintech | 16757–1-AP |
| Rabbit anti-ATF4 (D4B8) | Proteintech | 11815 |
| Rabbit anti-SNAT1 (D9L2P) | Cell Signaling | 36057 |
| Rabbit anti-ACTB | Sigma Aldrich | A2066 |
| HRP conjugated rabbit anti-ACTB (13EB) | Cell signaling | 5125 |
| AF604 anti-puromycin | EMD Millipore Corp | MABE343-AF647 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| L-Alanine | Sigma Aldrich | A7627–100G |
| 15N labeled alanine | Cambridge Isotope Laboratories, Inc. | NLM-454 |
| 3–13C labeled pyruvate | Cambridge Isotope Laboratories, Inc. | CLM-1575 |
| U-13C-15N glutamine | Cambridge Isotope Laboratories, Inc. | CNLM-1275 |
| RPMI 1640 | Thermo Fisher | 11875–119 |
| RPMI 1640 (no glutamine) | Thermo Fisher | 21870100 |
| FBS | Corning | 45000–734 |
| Dialyzed FBS (1698218, 1743489) | Thermo Fisher | 26400044 |
| Oligomycin A | Sigma Aldrich | 75351 |
| FCCP | Sigma Aldrich | C2920 |
| Rotenone | Sigma Aldrich | R8875 |
| Antimycin A | Sigma Aldrich | A8674 |
| 2-NBDG | Thermo Fisher | N13195 |
| Puromycin | InvivoGen | ant-pr-1 |
| CellTrace violet | Thermo Fisher | C34557 |
| Golgi stop | BD Biosciences | 554724 |
| Complete Mini Protease Inhibitor | Roche | 11873580001 |
| Recombinant IL-7 | R&D systems | 407-ML |
| Critical Commercial Assays | ||
| CD4 (L3T4) Microbeads, mouse | Miltenyi Biotec | 130–117-043 |
| CD8 (Ly-2) Microbeads, mouse | Miltenyi Biotec | 130–117-044 |
| Foxp3/transcription factor staining buffer set | eBioscience | 00–5523-00 |
| Th1/Th2/Th17 cytometric bead array, mouse | BD Biosciences | 560485 |
| iScript cDNA synthesis kit | BioRad | 1708891 |
| DNeasy blood and tissue kit | QIAGEN | 69506 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | The Jackson Laboratory | 000664 |
| B6.Cg-Tcratm1Mom Tg(TcrLCMV)327Sdz/TacMmjax | The Jackson Laboratory | 37394-JAX |
| B6.SJL-Ptprca Pepcb/BoyJ | The Jackson Laboratory | 002014 |
| Oligonucleotides | ||
| Primer Snat1, F: TGAGACTGCCCAAATGCCTAAGGA, R: TCTCTGTTGCAGAAAGCTGGTGGA | This paper | N/A |
| Primer Pdha, F: CCACCTCATCACTGCCTATC, R: CCTTTAGCACAACCTCCTCTT | This paper | N/A |
| Primer Ldha, F: ACAAACTCAAGGGCGAGATG, R: GGAGTTCGCAGTTACACAGTAG | This paper | N/A |
| Primer Gpt1, F: GGAAGGTGCTAACTCTGGATAC, R: GGCACGGATAACCTCAGTAAA | This paper | N/A |
| Primer Gpt2, F: AACTGTATCCGTGAAGATGTGGC, R: CAGGTAAATGTTGTCTGGGTCTG | This paper | N/A |
| Software and Algorithms | ||
| Prism | GraphPad Sofwate | https://www.graphpad.com/scientific-software/prism/ |
| FlowJo | FlowJo | https://www.flowjo.com |
| MAVEN software suite | Clasquin et al., 2012) | https://maven.apache.org |