Summary
Multi-subunit SMC ATPases control chromosome superstructure and DNA topology, presumably by DNA translocation and loop extrusion. Chromosomal DNA is entrapped within the tripartite SMC/kleisin ring. Juxtaposed SMC heads (‘J heads’) or engaged SMC heads (‘E heads’) split the SMC/kleisin ring into ‘S’ and ‘K’ sub-compartments. Here, we map a DNA binding interface to the S compartment of E heads Smc/ScpAB and show that head/DNA association is essential for efficient DNA translocation and for traversing highly-transcribed genes in Bacillus subtilis. We demonstrate that in J heads Smc/ScpAB chromosomal DNA resides in the K compartment but is absent from the S compartment. Our results imply that the DNA occupancy of the S compartment changes during the ATP hydrolysis cycle. We propose that DNA translocation involves DNA entry into and exit out of the S compartment possibly by DNA transfer between compartments and DNA segment capture.
Keywords: Chromosome condensation, Condensin, SMC, kleisin, Smc/ScpAB, MukB, Rad50, Cohesin, DNA loop extrusion
Introduction
Lengthwise condensation of DNA molecules into rod-shaped chromatids is a prerequisite for the faithful segregation of eukaryotic chromosomes (Belmont, 2006; Kschonsak and Haering, 2015; Nasmyth, 2001). Several lines of evidence suggest that chromosome condensation relies on the formation of a series of radial loops along the chromosome axis (Earnshaw and Laemmli, 1983; Gibcus et al., 2018; Marsden and Laemmli, 1979; Naumova et al., 2013). Condensation is mediated by a SMC complex called condensin (Hirano, 2016; Houlard et al., 2015). Condensin supports ATP-dependent processive enlargement of DNA loops in vitro, apparently by holding onto one DNA double helix and translocating along another, a process referred to as DNA loop extrusion (Ganji et al., 2018). The related cohesin complex organizes chromosomes during interphase, presumably using a similar principle (Merkenschlager and Nora, 2016; Rao et al., 2017).
SMC complexes also promote chromosome segregation in bacteria (Gruber, 2011; Hirano, 2016). MukBEF – a distantly related SMC complex – appears to generate a series of DNA loops covering large parts of the Escherichia coli chromosome (Lioy et al., 2018). The more widely-distributed Smc/ScpAB complex initiates DNA translocation mainly or exclusively from within a defined chromosomal region – surrounding the single replication origin (Gruber and Errington, 2009; Minnen et al., 2016; Sullivan et al., 2009). Rather than promoting lengthwise DNA compaction, DNA translocation by Smc/ScpAB aligns the two chromosome arms flanking the replication origin (Minnen et al., 2016; Tran et al., 2017; Wang et al., 2017; Wang et al., 2015). By doing so, it may help DNA topoisomerase IV to untangle nascent sister chromosomes and enable efficient chromosome segregation during fast growth (Bürmann and Gruber, 2015; Gruber and Errington, 2009; Gruber et al., 2014; Wang et al., 2014).
In multiple bacteria including Bacillus subtilis (Bs), Smc/ScpAB is recruited to the chromosome by ParB protein bound to parS sites, which are located in the replication origin region (Gruber and Errington, 2009; Minnen et al., 2011; Sullivan et al., 2009). Smc/ScpAB then translocates onto parS flanking DNA in a directional and processive manner. Recruitment to parS, release from parS and DNA translocation are intimately linked with the Smc ATPase cycle (Minnen et al., 2016; Wang et al., 2018; Wilhelm et al., 2015).
Smc proteins harbor a ~50 nm antiparallel coiled coil ‘arm’ with a hinge dimerization domain at one end and a head domain at the other end. Two Smc proteins interact via the hinge domain and by co-aligning the Smc arms, thus also bringing the heads in close proximity (juxtaposed heads or J heads) (Figure 1A) (Diebold-Durand et al., 2017; Hirano and Hirano, 2004; Soh et al., 2015). The ScpA subunit (kleisin) connects both Smc heads, while the central region of ScpA associates with a dimer of ScpB proteins (Bürmann et al., 2013; Palecek and Gruber, 2015). The pentameric Smc/ScpAB complex thus folds into a highly-elongated, rod-shaped particle. While the circumference of the tripartite SMC/kleisin ring allows for a large lumen, the alignment of the Smc arms in the absence of ATP binding (J heads) strongly reduces the lumen by closing the S compartment (Figure 1A). J heads Smc/ScpAB represents the predominant conformation of Bs Smc as judged by in vivo cysteine cross-linking, except when ATP hydrolysis is reduced in the E1118Q (‘EQ’) Walker B mutant (Diebold-Durand et al., 2017).
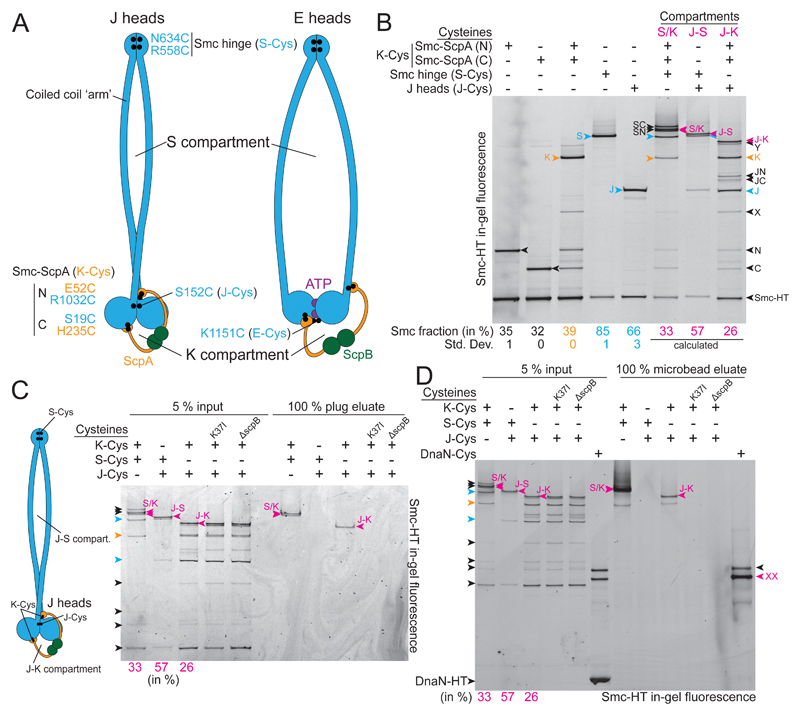
Figure 1. Locating DNA duplexes in J heads Smc/ScpAB.
(A) Schematic view of J heads (left panel) and E heads Smc/ScpAB (right panel). Cysteine residues are indicated as dots in black colors. The carboxy-terminal HaloTag fusion on Smc is omitted for simplicity.
(B) Identification and quantification of cross-linked species of Smc-HaloTag (‘Smc-HT’; labelled with HaloTag-TMR) using cysteine pairs in isolation and combination. Fully cross-linked species of Smc-HT are indicated in magenta colors and denoted as ‘S/K’ for the SMC/kleisin ring, ‘J-S’ for the J-S compartment and ‘J-K’ for the J-K compartment. Species with Smc hinge cross-linked only (‘S’) or J heads cross-linked only (‘J’) are labelled in blue colors, while the species having both Smc/ScpA interfaces cross-linked only (‘K’) is shown in yellow colors. All other species are indicated in black colors: Smc-neck/ScpA-N (‘N’), Smc-cap/ScpA-C, combination of J-Cys and Smc-neck/ScpA-N (‘JN’), combination of J-Cys and Smc-cap/ScpA-C (‘JC’), combination S-Cys and Smc-neck/ScpA-N (‘SN’), combination S-Cys and Smc-cap/ScpA-C (‘SC’). Species ‘X’ and ’Y’ are derived from alternative Smc/ScpAB having ScpA associated with neck and cap of a single Smc protomer. Annotation of X, SN, SC and S/K species with help of published data (Bürmann et al., 2013; Wilhelm et al., 2015). Mean fraction (and standard deviation) of species N, C, K, S and J were quantified from three replicate experiments (Table S2). The fraction of J-S, J-K and S/K species was calculated using efficiencies determined for isolated J-Cys, K-Cys and S-Cys residues as appropriate.
(C) Chromosome entrapment assay in agarose plugs with J-Cys strains. Cells harboring Smc-HT were cross-linked with BMOE using cysteine pairs at the indicated protein-protein interfaces and incubated with HaloTag-OG substrate (‘input’). Intact chromosomes were isolated in agarose plugs. Input material and co-isolated proteins (‘eluate’) were analyzed by SDS-PAGE and in-gel fluorescence detection. K37I, Walker A ATP binding mutant of Smc (‘KI’).
(D) Chromosome entrapment assay in agarose microbeads with J-Cys strains. As in (C) using agarose microbeads and HaloTag-TMR. A mix of cells carrying wt DnaN (80 %), DnaN-HT (10 %) and DnaN(Cys)-HT (10 %) is included as positive and negative control for entrapment. Note: untagged cells are added to reduce the DnaN-HT signal to the levels of Smc-HT. Arrowheads in magenta colors denote circular protein species. ‘XX’ indicates the double cross-linked, circular DnaN-HT species. The single cross-linked, X-shaped DnaN-HT species exhibits slightly slower mobility (labeled in black colors) (Wilhelm et al., 2015).
See also Figure S1 and Table S2.
The active ABC ATPase is formed by two nucleotide binding domains (the Smc heads) and two ATP molecules (Hirano et al., 2001; Hopfner, 2016). Each ATP molecule binds to the Walker box motifs of one head and to the ABC signature motif of the other head, thus gluing the two heads together (engaged heads or E heads) (Lammens et al., 2004). The two Smc arms emerge from E heads at a distance from one another and at an open angle (Figure 1A) (Diebold-Durand et al., 2017; Kamada et al., 2017). Head engagement thus opens the S compartment (at least partially). Disengagement of E heads upon ATP hydrolysis merges S and K compartments into a single lumen enclosed by the SMC/kleisin (‘S/K’) ring.
While recent studies have provided insights into the molecular architecture of SMC/kleisin complexes, their association with DNA is still poorly understood. Whether and how the opening, closure, loading and unloading of DNA compartments is related to DNA translocation is largely unclear. Here, we map locations for physical DNA binding and DNA entrapment in Bs Smc/ScpAB. We characterize a DNA binding site located at the Smc heads in E heads Smc/ScpAB which is accessible from within the S compartment. Residues supporting head/DNA association are critical for Smc function in Bs and appear conserved in highly diverged SMC-like proteins (Liu et al., 2016; Seifert et al., 2016; Woo et al., 2009). Head/DNA association is essential for Smc DNA translocation but has only limited impact on the rate of ATP hydrolysis. In contrast, a previously identified hinge/DNA association – capable of more strongly stimulating the Smc ATPase – is dispensable for Smc function. We observe DNA entrapment in the K compartment but not in the S compartment of J heads Smc/ScpAB. DNA is, however, detected in the S compartment of Smc/ScpAB when ATP hydrolysis is hindered by the EQ mutation. These findings put forward the notion that DNA transactions involving transfer of DNA between S and K compartments may form the basis for DNA translocation and loop extrusion.
Results
Smc/ScpAB entraps chromosomal DNA in the J-K but not the J-S compartment
We have recently developed an assay for the detection of DNA entrapment in Smc/ScpAB by co-isolating cross-linked Smc species with intact chromosomal DNA in agarose plugs (Wilhelm et al., 2015; Wilhelm and Gruber, 2017). Cysteine cross-linking of the three S/K ring interfaces is used to preserve topological Smc/DNA association in the presence of protein-denaturing agents (Gligoris et al., 2014; Wilhelm et al., 2015). Here, we determined the location of chromosomal DNA in Smc/ScpAB by covalently closing the S and the K compartment (Figure 1A). We initially focused on J heads Smc/ScpAB using Smc residue S152C (‘J-Cys’) (Figure 1A) (Diebold-Durand et al., 2017). J-Cys allows formation of covalently closed J-S and J-K compartments when combined with a cysteine pair at the Smc hinge (‘S-Cys’) and cysteine pairs at both SMC/kleisin interfaces (‘K-Cys’), respectively (Figure 1A). We estimated the efficiency of in vivo cross-linking by the bis-maleimide compound BMOE after quenching the cross-linking reaction, preparation of cell lysates and detection of in-gel fluorescence of Smc-HaloTag (‘HT’) species (Figure 1B). In cells lacking other cysteine pairs, the K-Cys cross-linked species was formed in 39 % of Smc-HT proteins, while S-Cys and J-Cys yielded 85 % and 66 % of cross-linked species, respectively. We calculated the fraction of Smc species expected for fully cross-linked S/K rings (33 %), J-S compartments (57 %) or J-K compartments (26 %) (Figure 1B) (Bürmann et al., 2013; Diebold-Durand et al., 2017). Direct estimation of these fractions from gel images yielded roughly comparable numbers (J-S, 43 ±4 % and J-K, 19 ±2 %, respectively) implying normal formation of the S/K ring in J heads Smc/ScpAB (Table S2). We conclude that covalently closed forms of J-S and J-K compartments are readily observed by in-gel fluorescence detection.
All linear and X-shaped cross-linking intermediates were efficiently eliminated during the chromosome co-entrapment assay. However, we robustly detected co-isolation of the J-K compartment and the S/K ring species with chromosomal DNA (Figure 1C). The isolation of the J-K species was abolished in the absence of ScpB and in the Smc(K37I) mutant, which is unable to bind and hydrolyze ATP (Figure 1C). DNA localization in the J-K compartment is thus likely the result of a physiological loading reaction. J-K species formed using a cysteine residue located at positions along the Smc coiled coil instead of J-Cys (for example D280C) were also retained in agarose plugs (Figure S1A). On the contrary, the J-S compartment species (formed with J-Cys or D280C) were reproducibly eliminated from agarose plugs implying that DNA is rarely or never located between the Smc arms when Smc heads are juxtaposed (Figure 1C, S1A). Aiming to detect possible rare events of DNA entrapment, we next optimized the chromosome entrapment assay. The improved version of the assay is based on agarose microbeads (rather than plugs) and facilitates up-scaling and stringent washing (Figure S1B). With the improved assay, we again observed DNA entrapment in the S/K ring and in the J-K compartment but not in the J-S compartment (Figure 1D). Together, these experiments demonstrate that a fraction of chromosomal Smc/ScpAB display juxtaposed heads. Chromosomal DNA is entrapped exclusively in the K compartment of J heads Smc/ScpAB. Close alignment of the Smc arms presumably excludes DNA from the interarm space (Diebold-Durand et al., 2017).
DNA may occupy the S compartment at another stage of the ATP hydrolysis cycle. To address this possibility, we next investigated E heads Smc/ScpAB. Residue K1151C (‘E-Cys’) is a reporter for E heads Smc/ScpAB. E-Cys cross-linking is very inefficient in otherwise wild-type cells, presumably due to transient or rare head engagement (Figure S1C) (Diebold-Durand et al., 2017; Minnen et al., 2016). As a result, we failed to identify cross-linked species corresponding to E-S and E-K compartments in the input material. We were also unable to recover any such species from agarose eluates (Figure S1C).
Altogether, these experiments demonstrate that the K compartment of Smc/ScpAB is used for DNA entrapment and indicate an absence of DNA entrapment in the S compartment (see below for Smc mutants). The parts of the Smc protein that form the barrier of the S compartment, however, have previously been implicated in Smc/DNA association. Such physical association may involve DNA passing through the S compartment or not.
High-affinity hinge/DNA association is dispensable for Smc function
DNA binding has been reported for isolated hinge domains of multiple SMC complexes (Alt et al., 2017; Arumugam et al., 2003; Chiu et al., 2004; Griese and Hopfner, 2011; Griese et al., 2010; Hirano and Hirano, 2006; Soh et al., 2015; Uchiyama et al., 2015). The substitution of three lysine residues (K666, K667 and K668) at the hinge/coiled coil junction for glutamate strongly reduces DNA binding and DNA-stimulated hydrolysis of ATP in purified preparations of Bs Smc (Hirano and Hirano, 2006) (Figure 2A). The physiological importance of the lysine residues, however, has remained elusive so far. Since the triple glutamate mutation (here denoted as ‘3E-hng’) was not tolerated well in recombinant Smc fragments, we also generated the corresponding triple alanine mutant (‘3A-hng’). We purified Smc hinge domains with adjacent coiled coils (‘SmcH-CC100’) to measure hinge/DNA association using 40 bp fluorescein-labelled dsDNA by fluorescence anisotropy (Figure 2A-C, S2A). Robust DNA binding by wild-type SmcH-CC100 was observed, as reported previously (Soh et al., 2015). In contrast, the 3A-hng mutant exhibited strongly reduced DNA binding even at elevated protein concentrations (Figure 2C).
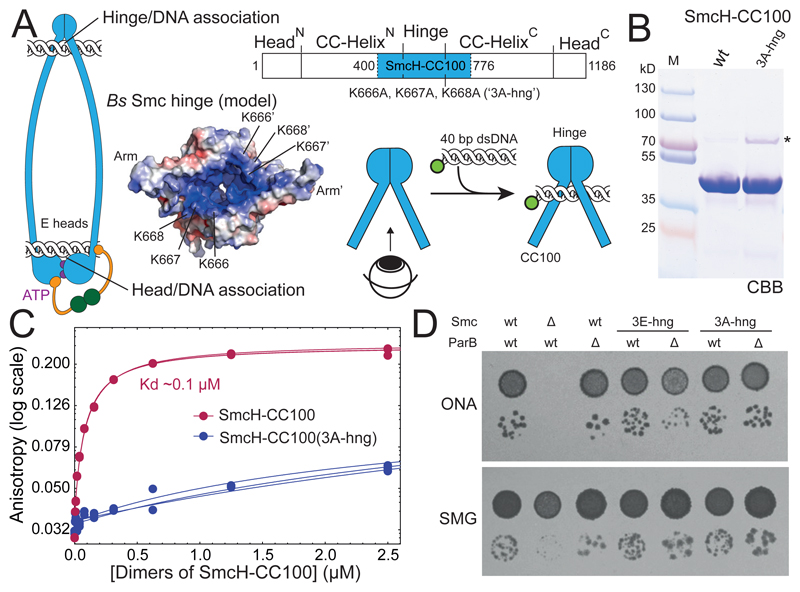
Figure 2. Hinge/DNA association is dispensable.
(A) Schematic representation of E heads Smc/ScpAB with DNA bound to hinge and heads. Schematic view of the SmcH-CC100 construct carrying the K666A, K667A, K668A (‘3A-hng’) mutation. Surface-charge electrostatic potential is shown for the coiled coil-proximal surface of the Bs Smc hinge (modelled from PDB: 1GXL). Scheme for DNA binding to the coiled coil-proximal surface of the hinge.
(B) Coomassie Brilliant Blue (‘CBB’) staining of purified SmcH-CC100 protein. The asterisk denotes an impurity present in the 3A-hng variant. Size of marker proteins (lane M) is given in kilodalton (kD).
(C) DNA binding as measured by fluorescence anisotropy with increasing protein concentration using fluorescein-labelled 40 bp dsDNA at 50 nM concentration. Data points from three experiments are shown as dots. The lines correspond to a non-linear (see STAR*Methods). Note: the substoichiometric amounts of a protein impurity in 3A-hng (see in (B)) may contribute to the observed residual DNA binding but is unlikely to strongly hinder DNA binding of the more abundant SmcH-CC100(3A-hng) protein.
(D) Colony formation by smc(3E-hng) and smc(3A-hng) mutant strains. Dilutions (81-fold and 59,000-fold) of overnight cultures were spotted on nutrient-rich medium (ONA) and nutrient-poor medium (SMG) and grown at 37 °C for 16 and 24 hours, respectively.
See also Figure S2 and Table S3.
Next, we introduced the mutations into the endogenous smc locus by allelic replacement. Cells carrying smc(3A-hng) efficiently formed colonies on nutrient-rich medium (‘ONA’) at 37 °C, demonstrating normal or near-normal Smc function (Figure 2D). To sensitize for partial loss of Smc activity, we deleted the gene encoding for the Smc loader protein ParB (Gruber and Errington, 2009; Wang et al., 2018). Even in the absence of ParB, smc(3A-hng) supported growth on nutrient-rich medium well (Figure 2D). Similar results were obtained with smc(3E-hng), albeit colonies grew slightly slower when parB was deleted (Figure 2D, S2B). The positively-charged surface at the Smc hinge is thus dispensable for Bs Smc function. Moreover, the Smc hinge domain can be replaced altogether by the structurally unrelated Rad50 Zinc hook (Bürmann et al., 2017). These findings imply that Smc/ScpAB must harbor another site (or multiple other sites) for DNA binding to support its putative DNA motor function.
Identification of surface-exposed Smc head residues required for Smc function
Several other DNA binding interfaces have been suggested for SMC/kleisin complexes (Kschonsak et al., 2017; Roy et al., 2015; Zabrady et al., 2016). In case of MukB and the SMC-like protein Rad50, DNA binding has been mapped at the opposite end of the long coiled-coil arms. On the coiled coil-proximal surface of engaged head domains, a DNA binding interface is formed by both heads contributing positively-charged residues for DNA binding (Liu et al., 2016; Seifert et al., 2016). Reverting the charge of such residues in yeast Rad50 protein renders cells sensitive to DNA damaging agents – similar to Rad50 deletion mutants – implying that DNA binding to Rad50 heads is important for the DNA repair function of the Rad50-Mre11 complex (Seifert et al., 2016). In case of MukB, the physiological relevance of the identified DNA binding residues remains to be determined (Woo et al., 2009).
To assess whether such a DNA binding interface may exist in canonical SMC proteins, we selected conserved Smc head residues with positively-charged side chains (Figure 3A). We mutated five lysine and four arginine residues individually to glutamate. All mutant alleles supported wild type-like growth on nutrient rich medium with the exception of smc(R57E) (Figure S3A). The R57 side chain is buried between engaged Smc heads. We thus excluded R57 from further analysis of potential DNA binding residues. We had difficulties deleting the parB gene in five of the eight remaining mutants indicating that the charge-reversal compromises Smc function (Table S4). To elucidate whether these five residues fulfill largely redundant or additive functions, we decided to systematically test combinations of mutations. First, however, we changed from glutamate to alanine substitutions aiming to reduce any undesired impact on protein folding and stability. The five single alanine mutants did not display obvious growth phenotypes (Figure S3B). Likewise, the ten double-alanine mutants grew robustly on nutrient rich medium (Figure S3C). However, when we combined three or more mutations, we identified alleles with strong growth phenotypes (Figure 3B-C). There is no obvious pattern between the degree of growth inhibition and the nature of the underlying mutations. We suspect that all five positively-charged residues contribute to Smc function and that some residue combinations perform better than others. We selected mutant K60A, R120A and K122A (‘3A-hd’) for further characterization. Amongst the triple-alanine mutants, 3A-hd showed a particularly severe growth phenotype. However, 3A-hd was not as severely impaired in growth as the quintuple mutant (‘5A-hd’) (Figure 3C). This may indicate residual DNA binding capability in 3A-hd. However, we were not able to detect differences in the DNA binding of 3A-hd and 5A-hd. All tested triple-, quadruple- and quintuple-alanine proteins were expressed at similar levels as the wild-type protein (Figure 3B-C).
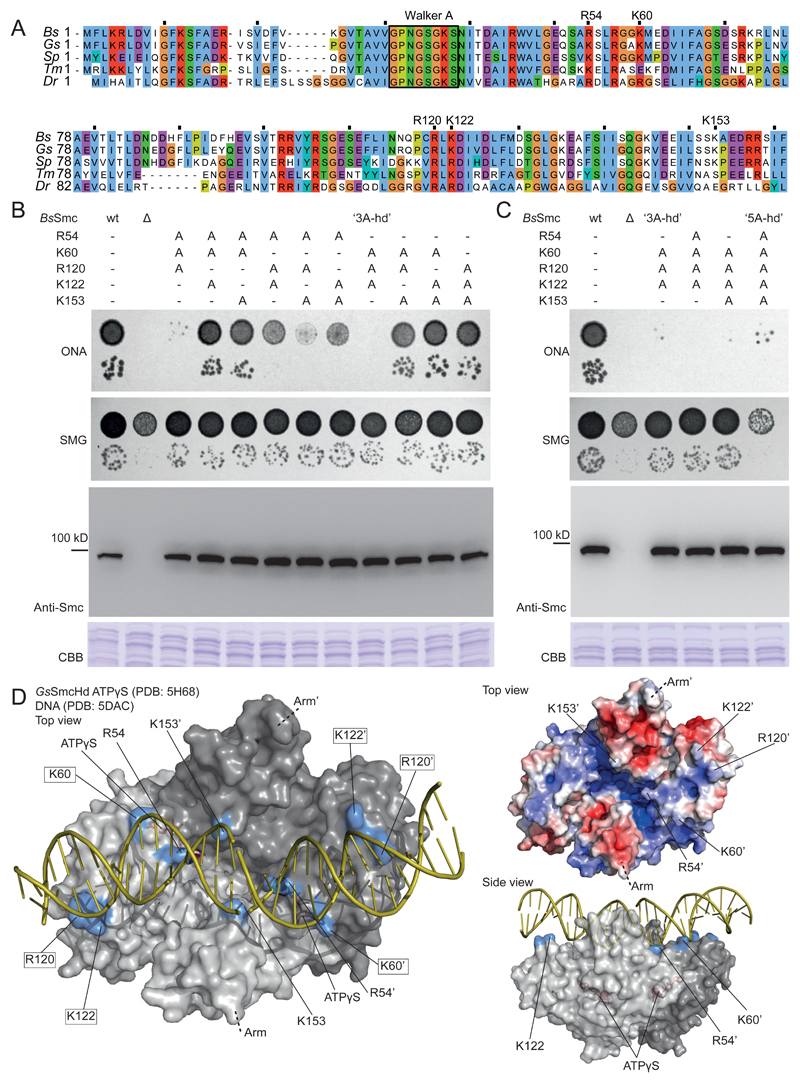
Figure 3. Identification of surface-exposed Smc head residues required for Smc function.
(A) Alignment of N-terminal sequences of five bacterial Smc proteins (Bs, Bacillus subtilis; Gs, Geobacillus stearothermophilus; Sp, Streptococcus pneumoniae; Tm, Thermotoga maritima; Dr, Deinococcus radiodurans). The conserved Walker A box motif is indicated for reference. Residues chosen for detailed analysis are denoted.
(B) Characterization of smc alleles harboring triple alanine mutations. Top panel: Colony formation by dilution (81-fold and 59,000-fold) spotting as in Figure 2D. Bottom panels: Cellular expression levels of Smc variants determined by immunoblotting with polyclonal antibodies raised against Bs Smc. CBB staining of cell extracts on separate gels is shown as control for uniform protein extraction.
(C) Smc alleles harboring selected quadruple mutations and the quintuple alanine mutation. As in (B).
(D) Surface representation of the structure of the Gs SmcHd-ATPγS complex (PDB:5H68) (in gray colors) superimposed onto Rad50Hd-ATPγS-DNA (PDB:5DAC) (only DNA is shown - in yellow colors) (Kamada et al., 2017; Seifert et al., 2016). The side chains of putative DNA binding residues are marked in blue colors. ATPγS is shown in stick representation in red colors. Residues mutated in 3A-hd are marked by boxes (left panel only). Surface-charge electrostatic potential is shown for the Gs SmcHd-ATPγS complex (PDB:5H68) (top right panel).
See also Figure S3
We next aligned the structure of Gs ATPγS-bound Smc heads (PDB: 5H68) with two available Rad50-Mre11-DNA co-crystal structures (PDB: 5DAC and 5DNY) (Kamada et al., 2017; Liu et al., 2016; Seifert et al., 2016). In the superimpositions (RMSD 3.1 and 2.3 Å, respectively) the identified Smc residues are in proximity of the negatively-charged DNA backbone (Figure 3D, S3D) being consistent with them supporting DNA binding. The arrangement of putative DNA binding residues in MukB (K146, R216 and R218 in Haemophilus ducreyi) bears resemblance with the one in Smc/Rad50, despite local structural differences between MukB and Smc/Rad50 heads (Woo et al., 2009).
Head/DNA association
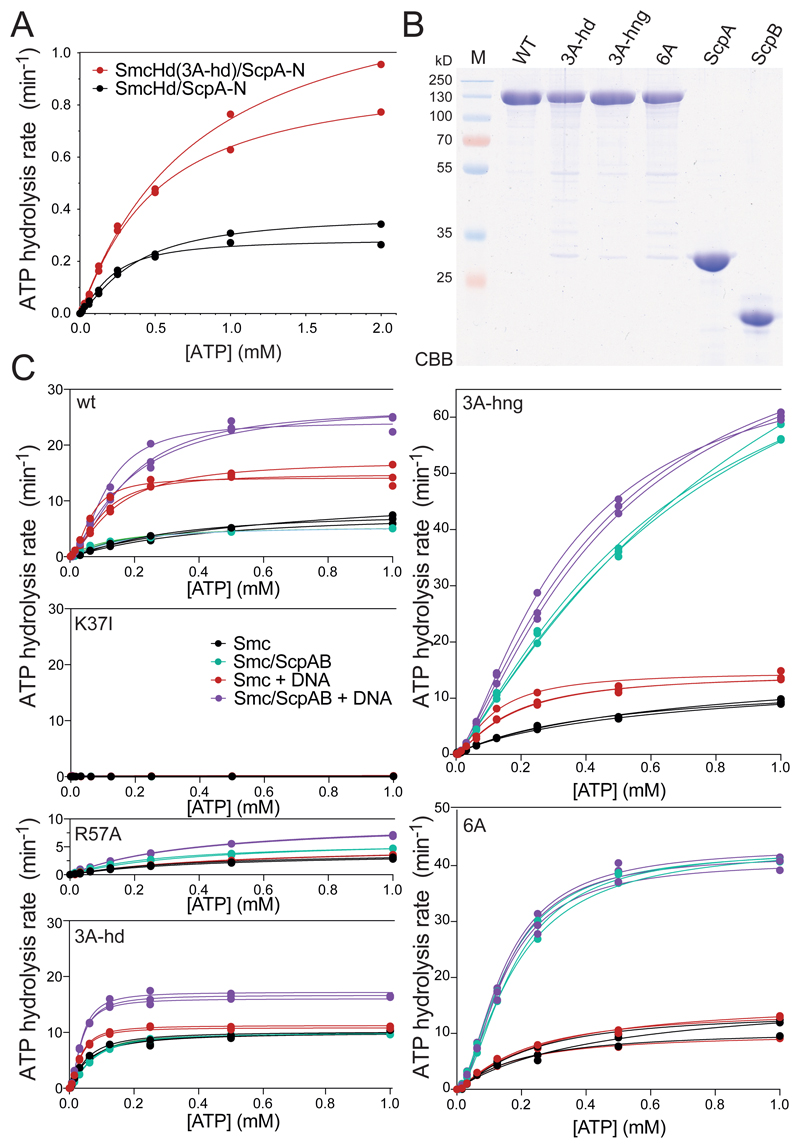
To test whether Smc proteins indeed associate with DNA using these residues, we purified Bs Smc heads with a short coiled-coil fragment (‘SmcHd’) together with a His-tagged N-terminal fragment of ScpA (‘ScpA-N’) (Figure 4A-B). The latter provides a tag for affinity purification and stabilizes recombinant SmcHd preparations (Bürmann et al., 2013). We then measured ATP dependent dimerization of SmcHd/ScpA-N and SmcHd(EQ)/ScpA-N using analytical gel filtration. In absence of ATP, all variants eluted as a single peak at the volume expected for the monomeric complex (Figure 4C, S4). In presence of ATP, an additional faster eluting species was detected for wild-type and 3A-hd variants of SmcHd(EQ)/ScpA-N presumably corresponding to the ATP-induced dimer form. The dimer peak was slightly more pronounced in the 3A-hd mutant. The positively-charged surface in wild-type Smc may thus hinder head engagement somewhat. Protein preparations lacking the EQ mutation did not produce appreciable levels of the dimer form, presumably due to concomitant ATP hydrolysis.
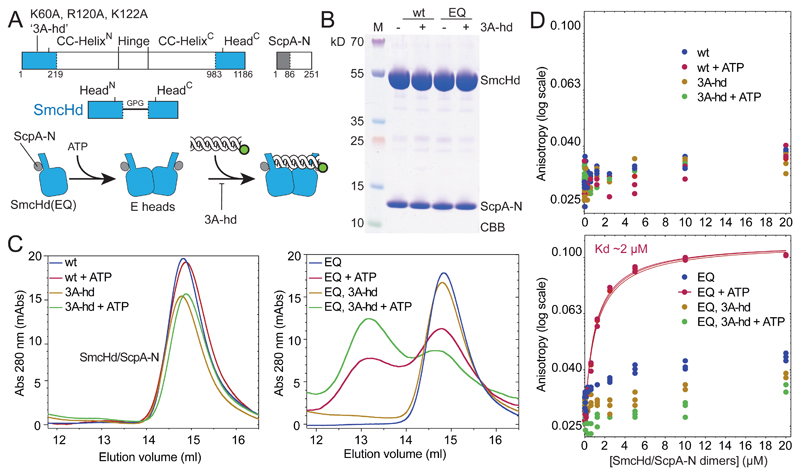
Figure 4. ATP dependent head/DNA association.
(A) Construction of SmcHd/ScpA-N with and without the ‘3A-hd’ mutations (K60A, R120A, K122A). The amino-terminal His6-tag on ScpA-N is omitted from the scheme for simplicity.
(B) CBB staining of purified preparations of SmcHd/ScpA-N constructs with and without EQ and 3A-hd mutations.
(C) Gel filtration profiles of SmcHd/ScpA-N preparations eluted from a Superdex 200 10/300 column. Samples were eluted in buffer supplemented with or without 1 mM ATP. Elution curves for proteins without and with the EQ mutation are shown in the left and right panels, respectively.
(D) DNA binding measurements using fluorescence anisotropy titrations as in Figure 2C. Data points for proteins without and with the EQ mutation are displayed in the top and bottom panels, respectively. Data points and fits for three experiments are shown. The estimated Kd for head/DNA association is calculated under the assumption of complete dimerization of SmcHd(EQ)/ScpA-N. The actual Kd may thus be somewhat lower.
See also Figure S4 and Table S3.
Proteins lacking the EQ mutation displayed poor DNA association with or without ATP as measured by fluorescence anisotropy (Figure 4D, S4B). SmcHd(EQ)/ScpA-N also associated poorly with DNA in the absence of ATP but showed robust DNA binding when ATP was added. The corresponding 3A-hd variant, however, failed to exhibit detectable DNA binding even in the presence of ATP. We conclude that ATP engaged Smc heads support DNA binding via a DNA interface formed at their coiled coil-proximal surface.
Head/DNA association has a minor impact on the rate of ATP hydrolysis
Head/DNA association may play a structural or regulatory role during the ATP hydrolysis cycle. Since the Smc ATPase activity is essential for Bs Smc function (Minnen et al., 2016; Wang et al., 2018), a pronounced defect in ATP hydrolysis could explain the lethality of the 3A-hd mutation. To address this possibility, we measured ATP hydrolysis using an enzyme coupled assay, first using the purified preparations of SmcHd/ScpA-N. The 3A-hd mutant displayed mildly increased – rather than reduced – ATP hydrolysis rate (Figure 5A). This increase is likely explained by the higher efficiency of head engagement (Figure 4C, S4A). [Of note, we observed a quadratic change in the ATP hydrolysis rate (Vmax) with increasing SmcHd concentrations indicating that protein dimerization is the rate-limiting step in ATP hydrolysis by SmcHd/ScpA-N (but not full-length Smc) (Figure S5A).]
Figure 5. Rates of ATP hydrolysis by Smc heads and Smc holo-complexes.
(A) ATP hydrolysis rate of SmcHd/ScpA-N at 10 μM protein concentration and increasing concentrations of ATP as measured by an enzyme-coupled assay. The lines represent a non-linear regression fit of the Hill equation (see STAR*Methods). Data points and fits for two experiments are shown.
(B) CBB staining of purified preparations of untagged, full-length Smc, ScpA and ScpB proteins.
(C) ATP hydrolysis rates of full-length Smc proteins and holo-complexes at 0.15 μM Smc dimer concentration and equivalent concentrations of ScpB dimers and ScpA monomers. As in (A) using protein preparations shown in (B). DNA stimulation was measured by addition of 3 μM 40 bp dsDNA. Data points and fits from three experiments are displayed.
See also Figure S5, and Table S5.
We also measured ATP hydrolysis in a more physiological setting using full-length Smc, ScpAB and DNA (Figure 5B). An ATP binding mutant of Smc (K37I) did not support appreciable levels of ATP hydrolysis. Wild-type Bs Smc dimers showed a significantly higher basal ATPase activity when compared to the SmcHd/ScpA-N sub-complex (Figure 5A-C, S5A). This activity was further stimulated by DNA and even more by DNA and ScpAB. The 3A-hd mutation had only limited impact on the Smc ATP hydrolase rate: While basal activity was slightly increased, and DNA stimulation slightly reduced, the ATP hydrolysis rate in the presence of DNA and ScpAB was roughly comparable to wild-type Smc under the same conditions. Since Smc mutants with much stronger defects in ATP hydrolysis (R57A) (Figure 5C) display rather mild growth phenotypes (Minnen et al., 2016; Wang et al., 2018), the loss of Smc ATPase activity or its gross deregulation is an unlikely cause of lethality. Head/DNA association must have a crucial role apart from regulating ATP hydrolysis.
The Smc(3A-hd) ATPase showed residual DNA stimulation, which presumably arises from hinge/DNA association. To determine whether this is indeed the case, we analyzed Smc(3A-hng) and a sextuple alanine mutant protein combining hinge and head alanine mutations. DNA stimulation of the Smc ATPase was abolished in Smc(6A)/ScpAB (Figure 5C). The Smc ATPase thus becomes insensitive to the presence of DNA when hinge/DNA and head/DNA associations are reduced, strongly implying that there are no other DNA binding sites on Smc/ScpAB capable of regulating the Smc ATPase (in the absence of hinge/DNA and head/DNA association). Of note, Smc(3A-hng) and Smc(6A) proteins showed abnormally high rates of ATP hydrolysis in Smc/ScpAB holo-complexes even in the absence of DNA (discussed below).
Head/DNA association is required for DNA translocation
Aiming to better define the cause of lethality, we next characterized the chromosomal association and distribution of Smc(3A-hd). We were curious whether head/DNA association is critical for the recruitment of Smc to chromosomal loading sites, for the initiation of DNA translocation or for its processivity.
We tested for chromosomal association by the chromosome entrapment assay using cysteine pairs for the covalent closure of the S/K ring. Smc(3A-hd) protein associated normally with ScpA as judged by the cross-linking pattern seen in the input material (Figure 6A). The circular species derived from Smc(3A-hd) proteins, however, was completely depleted from the agarose microbeads, similar to covalent rings obtained for the ATP binding mutant Smc (K37I) (Figure 6A) or for non-functional Mini-Smc proteins with artificially shortened coiled coils (Bürmann et al., 2017; Wilhelm et al., 2015). The Smc(3A-hng) protein displayed DNA entrapment in the S/K ring, albeit with reduced levels when compared to wt Smc. The reduction is likely at least in part due to synthetic lethal effects of 3A-hng, S-Cys and K-Cys mutations (Figure S6A).
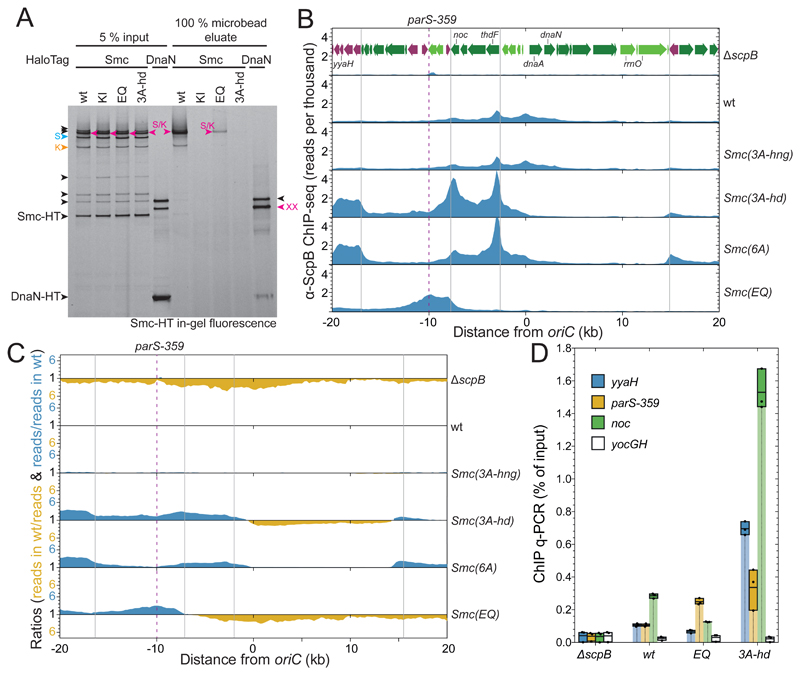
Figure 6. Chromosomal association and distribution of Smc(3A-hd).
(A) Chromosome entrapment assay using agarose microbeads. Smc-HT and DnaN-HT species labelled by HaloTag-TMR were analyzed by SDS-PAGE and in-gel fluorescence detection. As in Figure 1D. All Smc-HT samples harbor K-Cys and S-Cys for cross-linking of the two Smc/ScpA interfaces and the Smc hinge, respectively. The circular S/K species is indicated in magenta colors. All other labeling as in Figure 1B.
(B) ChIP-Seq profiles for the replication origin region represented as normalized reads per thousand total reads. ChIP was performed with a polyclonal antiserum raised against the ScpB protein. Gene organization in the origin region is shown in the ΔscpB panel. Genes in highly-transcribed operons are displayed in green colors. Gray lines indicate the 3’ end of highly transcribed operons. The position of parS-359 is given by a dashed line.
(C) Ratiometric analysis of wild-type and mutant ChIP-Seq profiles shown in (B). For bins with read counts greater than the wild-type sample, the ratio was plotted above the genome coordinate axis (in blue colors). Otherwise the inverse ratio was plotted below the axis (in yellow colors).
(D) α-ScpB ChIP-qPCR. Selected loci in the replication origin region and at the terminus were analyzed by quantitative PCR. Each dot represents a data point from one out of three experiments. The solid boxes span from the lowest to the highest value obtained; the horizontal line corresponds to the mean value calculated from the three biological replicates.
See also Figure S6.
The Smc(3A-hd) protein fails to associate normally with the chromosome, either because it never loads onto the chromosome or because it only entraps small DNA loops which are released during the chromosome entrapment assay. To address these two possibilities, we next performed chromatin immunoprecipitation with α-ScpB antiserum followed by quantitative PCR (ChIP-qPCR) or deep sequencing (ChIP-Seq). The genome-wide distribution of wild-type Smc/ScpAB displays characteristic shallow gradients along the two chromosome arms (Figure S6B-C). The gradient arises from chromosomal loading of Smc/ScpAB at parS sites and subsequent active translocation onto flanking DNA. Smc(EQ) accumulates at parS sites but is depleted from other regions of the chromosome, due to a loading or translocation defect (Minnen et al., 2016) (Figure 6B-C, S6B). We observed that Smc(3A-hng) and Smc(3A-hd) proteins produced very different ChIP-Seq profiles. The Smc(3A-hng) profile was virtually identical to the wild-type profile suggesting that normal hinge/DNA association is not required for efficient loading and translocation. The Smc(3A-hd) profile on the other hand displayed very high enrichment in the replication origin region and virtually complete depletion from chromosome arms (Figure S6B-C).
While Smc(EQ) peaks overlap well with ParB peaks at parS sites (Minnen et al., 2016), Smc(3A-hd) reached maximal enrichment at a distance from parS. The most prominent Smc(3A-hd) peaks were located 8 kb downstream and 2 and 6 kb upstream of the parB gene, which harbors the high-affinity parS-359 site. Enrichment of Smc(3A-hd) directly at parS-359 was comparatively low (Figure 6B-C). These findings suggest that Smc(3A-hd) is efficiently recruited to parS and released onto flanking DNA. Smc(3A-hd) then fails to support efficient DNA translocation, conceivably due to compromised DNA motor activity. Intriguingly, the regions displaying highest enrichment of Smc(3A-hd) were found at the 3’ end of highly-transcribed gene operons in head-on orientation (i.e. operons oriented against the presumed direction of Smc translocation) (Figure 6B). Adjacent co-directional genes (e.g. 10-20 kb upstream of parB), however, displayed very low levels of Smc(3A-hd) enrichment (Figure 6B-C). Thus, Smc(3A-hd)/ScpAB may translocate slowly or diffuse passively on the DNA being pushed backward by head-on transcription complexes and pushed forward by co-directional transcription. In contrast, wild-type Smc/ScpAB efficiently bypasses head-on transcription, albeit residual accumulation at 3’ ends of genes is seen in the ChIP-Seq profiles (Figure 6B).
To ensure that these observations are not artefacts caused by deep sequencing or the lack of ChIP-Seq calibration, we analyzed selected ChIP samples by qPCR. Consistent with the ChIP-Seq profiles, we found that Smc(3A-hd) is more enriched at loci adjacent to parS-359 than at parS-359 itself (Figure 6D, S6D). Enrichment levels of Smc(3A-hd) are substantially higher than Smc(wt) and Smc(EQ) levels as seen by ChIP-Seq. Together, these results imply that Smc/ScpAB complexes can slowly translocate or diffuse away from parS sites without being topologically associated with chromosomal DNA. This suggests that Smc(3A-hd)/ScpAB entraps the base of a DNA loop (‘pseudo-topological’ association) rather than a single DNA duplex (Srinivasan et al., 2018).
A dynamic interplay between hinge/DNA and head/DNA association
We noticed that the Smc(6A) mutant had a slightly more normal chromosomal distribution than Smc(3A-hd) in the ChIP-Seq profiles (Figure 6B-C), consistent with it supporting limited DNA translocation. Moreover, we discovered that Smc(6A) supports substantially better growth on nutrient-rich medium than Smc(3A-hd) (Figure 7A and S7A). Rather than aggravating the phenotype, the hinge alanine mutations partially suppress the defects caused by the head alanine mutations. Smc(3A-hng)/ScpAB complexes exhibited artificially high rates of ATP hydrolysis in the absence of DNA (Figure 5C). The hinge mutation thus appears to bypass the requirement for DNA binding to achieve high ATP turn-over – possibly by emulating the DNA-bound state. The hinge mutation also influences the coiled coil architecture of SmcH-CC100 as measured by A715C cross-linking (Figure 7B). Wild-type SmcH-CC100 gives robust cross-linking of A715C, which is strongly reduced by pre-incubation with DNA (Figure 7B) (Soh et al., 2015). The 3A-hng variant, however, displayed poor cross-linking in absence and presence of DNA, despite forming hinge domain dimers efficiently (Figure S2A). The 3A-hng protein thus preferentially adopts a ‘open-arms’ configuration (Figure 7B). We propose that hinge/DNA association or head/DNA association stabilize an open conformation of Smc/ScpAB and that the 3A-hng mutation stabilizes this conformation as well or even better than association with DNA. Consistent with this notion we find that 3A-hng exhibited higher levels of E heads Smc(EQ)/ScpAB (as measured by in vivo cross-linking of E-Cys) (Figure S7C) (Minnen et al., 2016). The 3A-hng mutation, however, was not able to suppress the lethality of the 5A-hd mutation (Figure S7A).
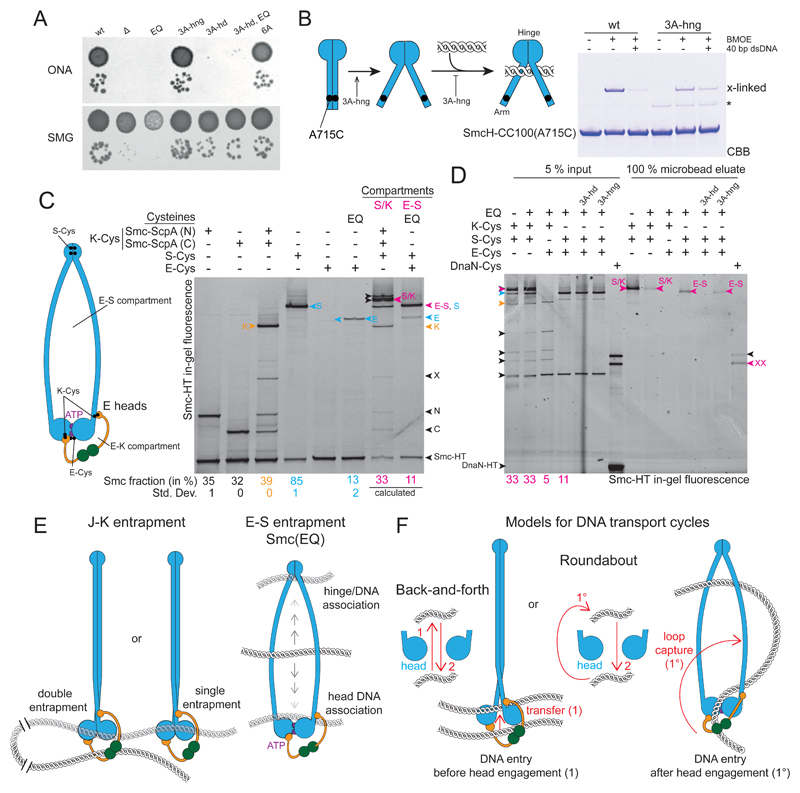
Figure 7. Hinge and head mutations promote S compartment opening and DNA entrapment.
(A) Dilution (81-fold and 59000-fold) spotting of 3A-hng, 3A-hd and 6A mutants. As in Figure 2D. A corresponding ONA image after 12 h incubation is shown in Figure S7A.
(B) Cysteine cross-linking of purified SmcH-CC100(A715C) protein in presence and absence of 10 μM 40 bp dsDNA. The asterisk marks an impurity; see also Figure 2B.
(C) Identification and quantification of cross-linked species of E heads Smc/ScpAB. using cysteine pairs in isolation and combination. As in Figure 1B. Circular species are indicated in magenta colors and denoted as ‘S/K’ for the SMC/kleisin ring and ‘E-S’ for the E-S compartment. Species with E heads cross-linked only (‘E’) are labelled in blue colors. All other labeling as in Figure 1B. Note that species S and E-S co-migrate in the gel. Mean fraction (and standard deviation) of species E in Smc(EQ) was quantified from three replicate experiments. Quantification of all other species as in Figure 1B (Table S2). The fraction of E-S species was calculated under the assumption of normal hinge dimerization in E heads Smc(EQ)/ScpAB (Bürmann et al., 2017).
(D) DNA entrapment in the E-S compartment of Smc(EQ)/ScpAB. Chromosome entrapment assay with agarose microbeads. As in Figure 1D. Arrowheads in magenta color denote circular protein species, S/K for the SMC/kleisin ring and E-S for the E-S compartment.
(E) Summary of main findings.
(F) Models for DNA transport cycles: DNA entry into the S compartment by passage between separated Smc heads is shown on the left-hand side (‘Back-and-forth’). DNA entry into the S compartment during head engagement by loop capture displayed on the right-hand side (‘Roundabout’).
See also Figure S7 and Table S2.
Together, these findings suggest that head/DNA association has essential structural roles during the Smc DNA motor cycle. Defects due to loss of head/DNA association are partially suppressed by the hinge alanine mutations which cause structural alterations in the Smc arms and reduced DNA binding.
DNA entrapment in the E-S compartment
The above results strongly suggest that DNA at least transiently resides in the S compartment to allow for head/DNA association. Since we were unable to evaluate DNA entrapment in the E-S compartment due to inefficient E-Cys cross-linking in wild-type cells, we next made use of the Smc(EQ) mutant to artificially increase levels of E-Cys cross-linking. A small fraction of Smc(EQ) protein is known to entrap DNA in the S/K ring (Wilhelm et al., 2015) (Figure 6A). E-Cys was cross-linked with an efficiency of 13 % under the conditions of the assay (Figure 7C). E-S and E-K compartment species are difficult to discern by in-gel fluorescence detection. However, assuming normal hinge domain dimerization and Smc/ScpA interactions (Bürmann et al., 2017), E-Cys cross-linking is expected to give rise to 5 % and 11 % of E-K and E-S species, respectively, in Smc(EQ)/ScpAB (Figure 7C). The E-S species of Smc(EQ) was retained in agarose beads, whereas the E-K species was not detectable in the eluate. The S/K and E-S species of Smc(EQ) yielded comparable levels in the agarose eluate (Figure 7D), being consistent with the notion that DNA entrapped in the S/K ring is located mostly or exclusively in the E-S compartment. [Of note: While the level of the S/K species is higher in the input material when compared to the E-S species, only a fraction of it is supposedly derived from E heads Smc(EQ)/ScpAB.] The 3A-hd mutation eliminated DNA entrapment in the E-S compartment of Smc(EQ), while 3A-hng had little influence (Figure 7D). These findings demonstrate that DNA duplexes reside in the E-S compartment of Smc/ScpAB at least when the ATP hydrolysis step is hindered by the EQ mutation. Whether DNA entrapment in the E-S compartment of wild-type Smc/ScpAB occurs during chromosomal loading, during every Smc ATPase cycle or not at all remains to be determined.
Discussion
DNA loop extrusion provides a powerful explanation for striking features of long-range chromosome organization in bacteria and in eukaryotes. The wide conservation of SMC genes in bacteria and archaea implies that loop extrusion factors have emerged early during evolution. While SMC proteins have diversified to support a variety of functions, the basic mechanism underlying DNA translocation and loop extrusion has likely remained unaltered. Structural conservation of the SMC/kleisin core supports this notion. Head/DNA association is another essential and potentially widely conserved feature of the tripartite S/K ring, which SMC proteins share with distantly related chromosomal ABC ATPases including the Rad50/Mre11 nuclease and possibly also the recombination protein RecF (Liu et al., 2016; Seifert et al., 2016; Tang et al., 2018; Woo et al., 2009).
Transient DNA occupancy of the interarm space
DNA entrapment is an inherent property of many or all SMC/kleisin complexes (Cuylen et al., 2011; Kanno et al., 2015; Srinivasan et al., 2018; Wilhelm et al., 2015). Our findings, however, challenge the view that SMC proteins harbor long, flexible arms to embrace chromosomal DNA fibers. We provide direct evidence for the residence of chromosomal DNA in the K compartment of J heads Smc/ScpAB and for DNA exclusion from the interarm space (i.e. the S compartment) (Figure 7E). This feature seems conserved in yeast cohesin (Chapard et al., 2018). The long SMC arms may thus serve an entirely different purpose such as enabling efficient DNA translocation by supporting large translocation steps. Consistent with the notion of a mechanical function of SMC arms, we recently found that they tolerate changes in end-to-end distance but require a defined end-to-end orientation (Bürmann et al., 2017). However, we do not think that the S compartment is permanently devoid of DNA. We showed that head/DNA association is critical for chromosome loading and DNA translocation. As the head/DNA interface is positioned at the coiled coil-proximal surface of E heads, head/DNA association in all likelihood involves a DNA double helix occupying the S compartment of Smc/ScpAB (Figure 7E). The observation of DNA entrapment in the E-S compartment of Smc(EQ) complexes provides further support for transient DNA occupancy of the interarm space. Moreover, DNA entrapment between SMC arms may occur in conformations of Smc/ScpAB and yeast cohesin that are not accessible by J-Cys or E-Cys cross-linking and thus not measured in these experiments.
How does DNA enter and exit the S compartment?
We propose that DNA duplexes are loaded into and unloaded from the S compartment during the ATP hydrolysis cycle (Figure 7F). DNA entry into the S compartment may occur after head engagement in E heads Smc/ScpAB when the head/DNA interface is fully formed. In this case, loading of the S compartment cannot be achieved by transfer of a DNA duplex from the K to the S compartment as the passage between the two compartments is blocked by engaged heads. DNA might instead access the S compartment in the form of a small DNA loop (‘Roundabout’ model, Figure 7F). Alternatively, the DNA may enter the S compartment prior to head engagement by passing between disengaged Smc heads (‘Back-and-forth’ model). Such DNA transfer is expected to be impeded by Smc arm alignment and may require a so far undefined driving force. DNA exit from the S compartment is linked to ATP hydrolysis, which promotes head disengagement and dissolution of the head/DNA interface. In this case, the gate between the compartments is open thus facilitating simple transfer of DNA duplexes between the Smc heads into the K compartment. Altogether, the proposed DNA transactions form reaction cycles that move DNA duplexes between or around Smc heads (Figure 7F). The DNA segment capture model involves a roundabout cycle (Diebold-Durand et al., 2017; Marko et al., 2018). Whether and how a back-and-forth cycle may support DNA translocation is unclear.
Physical DNA contacts promoting Smc/ScpAB translocation
Bs Smc/ScpAB harbors a hinge/DNA and a head/DNA interface (Figure 7E) (Hirano and Hirano, 2006; Soh et al., 2015). While the nature of hinge/DNA association has remained elusive, several observations suggest that like head/DNA association also hinge/DNA association involves DNA residing in the S compartment ofE heads Smc/ScpAB (Figure 2A). Small loops of DNA occupying the S compartment may simultaneously engage with the hinge/DNA interface and the head/DNA interface (Marko et al., 2018). DNA loop capture has previously been reported for preparations of SMC complexes and may be a widely shared feature (Kim and Loparo, 2016; Kimura et al., 1999; Kumar et al., 2017; Marko et al., 2018; Sun et al., 2013). However, we cannot rule out the possibility that different DNA duplexes engage with hinge and heads. Our results suggest that head/DNA association is critical, whereas hinge/DNA association is dispensable. A surprising implication is that the coordination of a single strong DNA binding site with the Smc ATPase may suffice for efficient DNA translocation [at least in Smc(3A-hng)]. Such a scenario is difficult to reconcile with any model based on walking along DNA or DNA hand-over. While the findings are largely compatible with the DNA-segment-capture model (Diebold-Durand et al., 2017; Hassler et al., 2018; Marko et al., 2018), a simple version of the model does not provide an explanation for striking phenotypic differences between 3A-hd and 3A-hng mutants. According to the model, the entropic and energetic costs of DNA bending need to be compensated by hinge/DNA and head/DNA association (Marko et al., 2018). To explain the strong phenotypic consequences of 3A-hd, one needs to invoke that head/DNA association has additional functions such as fostering head engagement or opening of the S compartment.
Hinge/head communication
Strong growth phenotypes caused by loss of head/DNA association (in 3A-hd) are partially suppressed by hinge alanine mutations (3A-hng). There are two plausible explanations: (1) Hindering hinge/DNA association alleviates the requirement for head/DNA association. (2) Structural changes caused by the hinge mutations alleviate the need for head/DNA association. We favor the second possibility as multiple observations indicate structural alterations in Smc(3A-hng) proteins (Figure 7B, S7C). If so, then the structural alterations in Smc(3A-hd) may not only circumvent the requirement for head/DNA association but also for hinge/DNA association. Hinge/DNA association may accordingly be essential in wild-type Smc, despite its loss being well tolerated in Smc(3A-hng). In the alternative scenario (#1), hinge and head domains may compete for DNA binding, for example to hand DNA over from one binding site to the other. This may be supported by the folding of the SMC rod at an defined position (‘elbow’) as recently observed in MukB and cohesin (Bürmann et al., 2019). However, such folding has not yet been reported for Smc/ScpAB despite numerous structural studies (Kamada et al., 2013; Kamada et al., 2017; Melby et al., 1998; Soh et al., 2015). Moreover, the observations that the hinge domain can be substituted for a structurally unrelated domain and that the adjacent coiled coil can be shortened or extended are difficult to reconcile with such a putative SMC arm folding and DNA hand-over mechanism (Bürmann et al., 2017).
Altogether, our results provide an account of physical and steric DNA association by Smc/ScpAB. We locate chromosomal DNA in sub-compartments of the large S/K ring and identify head/DNA association as an essential prerequisite for Smc activity.Future models for DNA translocation by Smc/ScpAB (and possibly also other SMC complexes) will need to provide adequate explanations for these findings.
STAR Methods
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-ScpB-His6 rabbit antiserum | Gruber Lab | COD003 |
| Anti-Smc polyclonal rabbit antibody, affinity purified | Gruber Lab | COD008 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Adenosine Triphosphate (ATP) | Sigma-Aldrich | Cat#A6419-10G |
| Benzonase | Sigma-Aldrich | Cat#A1014 |
| Bis(malemido)ethane (BMOE) | Thermo Scientific | Cat#22323 |
| Breathe-Easy | Diversified Biotech | Cat#BEM-1 |
| BsaI | New England Biolabs | Cat#R0559L |
| Certified Low Melt Agarose | Bio-Rad Laboratories | Cat#161-3111 |
| Dynabeads Protein-G | Life Technologies | Cat#10004D |
| Erythromycin | AppliChem | Cat#A2275,0005 |
| GlycolBlue | Ambion | Cat#AM9515 |
| HaloTag-Oregon Green Ligand | Promega | Cat#G2801 |
| HaloTag-TMR Ligand | Promega | Cat#G8251 |
| HisTrap 5 mL | GE Healthcare | Cat#17-5247-01 |
| HiTrap Blue HP | GE Healthcare | Cat#17-0407-01 |
| HiTrap Butyl HP | GE Healthcare | Cat#28-4110-01 |
| HiTrap Q HP | GE Healthcare | Cat#17-1153-01 |
| HiTrap Heparin HP | GE Healthcare | Cat#17-0407-01 |
| HiLoad 16/60 S75 | GE Healthcare | Cat#28-9893-33 |
| HiLoad 16/60 S200 pg | GE Healthcare | Cat#28989335 |
| HiLoad 10/300 S200 GL | GE Healthcare | Cat#17-5175-01 |
| Lysozyme From Chicken Egg White | Sigma-Aldrich | Cat#L6876-5G |
| Lincomycin Hydrochloride Hydrate | Sigma-Aldrich | Cat#31727-250MG |
| Mineral Oil | Sigma-Aldrich | Cat#M3516-1L |
| Nicotinamide Adenine Dinucleotide (NADH) | Santa Cruz | Cat#205762A |
| NuPAGE LDS Sample Buffer (4X) | Life Technologies | Cat#NP0008 |
| Overnight Express Instant TB Medium | Merck | Cat#71491-5 |
| Phosphoenolpyruvic Acid (PEP) | Sigma-Aldrich | Cat#P7002-100MG |
| Phusion HotStart II DNA Polymerase | Thermo Scientific | Cat#F-549L |
| Protease Inhibitor Cocktail | Sigma-Aldrich | Cat#P8849-5ML |
| Pyruvate Kinase/Lactate Dehydrogenase | Sigma-Aldrich | Cat#P0294-5ML |
| Ready-Lyse Lysozyme Solution | Epicenter | Cat#R1802M |
| RNase A | Sigma-Aldrich | Cat#R5125 |
| Sm DNase | MPIB Core Facility | SmDNase |
| Superose 6 pg XK 16/70 | GE Healthcare | Cat#90-1000-42 |
| Tris(2-carboxyethyl) Phosphine Hydrochloride Solution | Sigma-Aldrich | Cat#646547 |
| T4 DNA Ligase | Thermo Scientific | Cat#EL0016 |
| Critical Commercial Assays | ||
| MasterBlock 96 well, 2 mL | Greiner Bio One | Cat#780270 |
| Takyon SYBR 2xMasterMix Blue dTTP No ROX | Eurogentec | Cat#UF-NSMT-B0701 |
| NuPAGE 4-12 % Bis-Tris Gels | Life Technologies | Cat#NP0323BOX |
| Novex 4-12 % Tris-Glycine Mini Gels | Life Technologies | Cat#XP04125BOX |
| NuPAGE 3-8 % Tris-Acetate Gels | Life Technologies | Cat#EA03755BOX |
| QIAquick PCR Purification Kit | QIAGEN | Cat#28106 |
| Costar Spin-X Centrifuge Tube Filter | Corning | Cat#8163 |
| Ovation Ultralow System V2 | NuGEN | Cat#0344 |
| Deposited Data | ||
| Original data | Mendeley Data | DOI:10.17632/xzmvjdhwgt.1 |
| ChIP-Seq Data | NCBI Sequence Read Archive | PRJNA533030 |
| Experimental Models: Organisms/Strains | ||
| E. coli and B. subtilis strains, see Table S6 | Gruber Lab | N/A |
| Oligonucleotides | ||
| yyaH Fw for qPCR CATATTGCCTTTTCCGTTGG | Gruber Lab | N/A |
| yyaH rv for qPCR CATCCTCAATCACGCTTTCA | Gruber Lab | N/A |
| parS-359 Fw for qPCR AAAAAGTGATTGCGGAGCAG | Gruber Lab | N/A |
| parS-359 Rv for qPCR AGAACCGCATCTTTCACAGG | Gruber Lab | N/A |
| Noc Fw for qPCR TCCATAATCGCCTCTTGGAC | Gruber Lab | N/A |
| Noc Rv for qPCR AAGCGCATGCTTATGCTAGG | Gruber Lab | N/A |
| yocGH Fw for qPCR TCCATATCCTCGCTCCTACG | Gruber Lab | N/A |
| yocGH Rv for qPCR ATTCTGCTGATGTGCAATGG | Gruber Lab | N/A |
| 40 bp random sequence DNA A TTAGTTGTTCGTAGTGCTCGTCTGGCTCTGGATTAC CCGC | Gruber Lab | N/A |
| 40 bp random sequence DNA B GCGGGTAATCCAGAGCCAGACGAGCACTACGAACA ACTAA | Gruber Lab | N/A |
| Recombinant DNA | ||
| Plasmid DNA, see Table S7 | Gruber Lab | N/A |
| Software and Algorithms | ||
| Real Time PCR Miner | Zhao and Fernald, 2005 | http://ewindup.info/miner/ |
| Bowtie2 Galaxy tool | Langmead & Salzberg, 2012 | https://usegalaxy.org |
| GraphPad Prism 7.00 | GraphPad Software | www.graphpad.com |
| ImageQuant TL 1D V8.1 | GE Healthcare | https://www.gelifesciences.com/en/us/shop/protein-analysis/molecular-imaging-for-proteins/imaging-software/imagequant-tl-8-1-p-00110 |
| Seq-Monk | Babraham Bioinformatics | https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/ |
Experimental Model
Bacillus subtilis strains and growth
All Bs strains are derived from the 1A700 isolate. Strain usage is listed in Table S1 and all genotypes and strain numbers are given in Table S6. Naturally competent Bs cells were transformed as described in (Bürmann et al., 2013) with longer starvation incubation time (2 hours) for high efficiency transformation as described in (Diebold-Durand et al., 2017). The transformants were selected on SMG-agar plates with appropriate antibiotics and single-colonies were isolated. The strains were confirmed by PCR and sanger sequencing as required. For dilution spot assays, cells were gown in SMG medium at 37 °C to stationary phase and 92 and 95 -fold dilutions were spotted onto ONA (16 h incubation or as indicated) or SMG (24 h incubation) agar plates (Bürmann et al., 2013).
Protein purification
His6-tagged SmcH-CC100
His6-tagged SmcH-CC100 constructs were purified following the method described in (Soh et al., 2015). The proteins were expressed from pET-28 derived vectors in E. coli BL21-Gold (DE3) for 24 hours at 24 °C in 1 L of autoinduction medium (Overnight Express TB,). Extracts were sonicated in binding buffer (50 mM NaPi, pH 7.4 at 4 °C, 300 mM NaCl, 80 mM imidazole, 10 % glycerol, 1 mM DTT) and bound to a 5 mL HisTrap HP column. Columns were washed with 10 column volumes (CV) of binding buffer, followed by 5 CV of 50 mM NaPi, pH 7.4 / 4 °C, 1 M NaCl, 10 % (v/v) glycerol, 1 mM DTT, followed by 5 CV of binding buffer. Proteins were eluted with 500 mM imidazole, pH 7.4 / 4 °C, 300 mM NaCl, 10 % (v/v) glycerol, 1 mM DTT. The elution fractions were further purified on a Hi-Load Superdex 200 16/600 pg equilibrated in 50 mM Tris-HCl, 200 mM NaCl, 1 mM TCEP, pH 7.4. Peak fractions were concentrated in Vivaspin 15 10K MWCO filters and aliquots were flash frozen in liquid N2 and stored at -80 °C.
His6-ScpA-N/SmcHd
The complex was purified as described in (Bürmann et al., 2013). SmcHd constructs containing residues 1-219 and 983-1186 connected by a GPG linker and His6-ScpA-N (residues 1-86) were co-expressed in 1 L of autoinduction medium ZYM-502 for 24 h at 24 °C. Protein extracts were prepared by sonication in binding buffer (50 mM NaPi, pH 7.4 at 4 °C with 300 mM NaCl, 10 % (v/v) glycerol, protease inhibitor cocktail (PIC) and 40 mM imidazole). The extracts were loaded onto a 5 mL HisTrap column, washed with 10 column volumes of binding buffer and eluted in a gradient up to 500 mM imidazole (pH 7.4 at 4 °C) in binding buffer. Protein complexes were further purified by gel filtration on a Hi-Load Superdex 200 16/600 pg equilibrated in 25 mM Tris-HCl pH 7.5, 200 mM NaCl. Finally, the samples were concentrated, flash frozen and stored at -80 °C.
Full-length Bs Smc
Native Smc proteins were purified as described in (Bürmann et al., 2017). Proteins were expressed from pET-22 or pET-28 derived plasmids in E. coli BL21-Gold (DE3) using ZYM-5052 autoinduction medium for 23 h at 24 °C. Cells were resuspended in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 10 % (w/v) sucrose) supplemented with PIC and sonicated. The supernatant after centrifugation was loaded on two HiTrap Blue HP 5 mL columns connected in series and eluted with a linear gradient of lysis buffer containing 1 M NaCl. The main peak elution fractions where diluted in buffer (50 mM Tris-HCl pH 7.5, 1 mM EDTA, 1 mM DTT) to a conductivity equivalent of 50 mM NaCl (≈ 8 mS/cm). The sample was loaded on a HiTrap Heparin HP 5 mL column and was eluted with a linear gradient of buffer up to 2 M NaCl. Except for the 6A mutant, that was loaded onto a HiTrapQ HP column and eluted under the same conditions. The main peak fractions were pooled and concentrated to 5 mL in Vivaspin 15 10K MWCO filters. The samples were further purified by gel filtration on a XK 16/70 Superose 6 PG column in 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT. Main peak fractions where pooled, concentrated and stored at −80 °C. Protein concentration was determined by absorbance using theoretical molecular weight and molar absorption values.
ScpA
Native ScpA was purified using a modified protocol reported in (Fuentes-Perez et al., 2012). The protein was expressed from a pET-28 derived plasmid, transformed into E. coli BL21-Gold (DE3), cultivated in ZYM-5052 autoinduction medium at 16 °C for 28 h. Cells were harvested and resuspended in lysis buffer (50 mM Tris-HCl pH 7.5, 200 mM NaCl, 5 % glycerol) supplemented with PIC and sonicated. The soluble fraction was loaded onto a 5 mL HiTrapQ ion exchange column and eluted with a gradient up to 2 M NaCl. The peak fractions were mixed with 4 M NaCl buffer to reach a final concentration of 3 M NaCl. The sample was injected into a HiTrap Butyl HP column and eluted in a reverse gradient to 50 mM NaCl. Peak fractions were pooled and concentrated to 5 mL in Vivaspin 15 10K MWCO filters and subsequently purified by gel filtration in a Hi Load 16/600 Superdex 75 pg column, equilibrated in 20 mM Tris-HCl pH 7.5, 200 mM NaCl. Protein was concentrated, aliquoted, flash frozen and stored at -80 °C.
ScpB
Native ScpB was purified using a protocol reported in (Fuentes-Perez et al., 2012). E. coli cells harboring a pET-22 derived plasmid with the coding sequence of ScpB were grown in ZYM-5052 autoinduction medium at 24 °C for 23 h. Cells were harvested and resuspended in lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) supplemented with PIC. The material was sonicated, and the soluble phase was diluted to 50 mM NaCl, loaded onto a 5 mL HiTrap Q HP column and eluted with a gradient up to 2 M NaCl. The sample was diluted in 4 M NaCl buffer to a final concentration of 3 M NaCl. The sample was loaded on two 5 mL HiTrap Butyl column connected in series. The protein was eluted with a reverse gradient down to 50 mM NaCl. The peak fractions were concentrated and purified by gel filtration using a Hi Load 16/600 Superdex 200 pg equilibrated in 50 mM Tris-HCl pH 7.5, 100 mM NaCl. The fractions containing the protein were concentrated, aliquoted, flash frozen and stored at -80 °C.
Method details
Fluorescence anisotropy measurements
Fluorescence anisotropy was measured using a 40 bp dsDNA substrate of random sequence (5’-TTAGTTGTTC GTAGTGCTCG TCTGGCTCTG GATTACCCGC) modified at the 3’-end with fluorescein (Soh et al., 2015). The data was acquired using a Synergy Neo Hybrid Multi-Mode Microplate reader (BioTek) with the appropriate filters in black 96-well flat bottom plates at 25 °C. All measurements were done in 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 2 mM MgCl2 with or without 1 mM ATP.
ATPase assay
ATPase activity measurements were done by a pyruvate kinase/lactate dehydrogenase coupled reaction (Bürmann et al., 2017). ADP accumulation was monitored for 1 h by measuring absorbance changes at 340 nm caused by oxidation of NADH in a Synergy Neo Hybrid Multi-Mode Microplate reader. The reaction mixture contained 1 mM NADH, 3 mM Phosphoenol pyruvic acid, 100 U Pyruvate kinase, 20 U Lactate dehydrogenase and the indicated ATP concentrations. The final protein concentration in the assay was 0.15 μM Smc dimers and 10 μM SmcHd/ScpA-N (unless indicated otherwise) in ATPase assay buffer (50 mM HEPES-KOH pH 7.5, 50 mM NaCl, 2 mM MgCl2, 1 mM NADH). Measurements were carried out at 25 °C.
In vitro cysteine cross-linking
Proteins (2.5 μM dimers for SmcH-CC100 and 0.5 μM for full-length Smc) and double-stranded oligonucleotides (40 bp, 10 μM of each oligonucleotide; (5’-TTAGTTGTTC GTAGTGCTCG TCTGGCTCTG GATTACCCGC)) were mixed in 50 mM Tris, 50 mM NaCl, 2 mM MgCl2 and 0.25 mM TCEP (pH 7.5) at room temperature and incubated for 5 min. BMOE was added (0.5 mM final). Reactions were incubated for another 5 min at room temperature and quenched by addition of 2-mercaptoethanol (14 mM) (Soh et al., 2015).
Chromatin Immunoprecipitation
The procedure closely followed the protocol described in (Bürmann et al., 2017). Cultures of 200 mL SMG were inoculated to OD600 = 0.004 and grown to OD600 = 0.02 at 37 °C. Cells were treated with 20 mL of buffer F (50 mM Tris-HCl pH 7.4, 100 mM NaCl, 0.5 mM EGTA pH 8.0, 1 mM EDTA pH 8.0, 10 % (w/v) formaldehyde) and incubated for 30 min at room temperature with sporadic manual shaking. Cells were harvested by filtration and washed in PBS. The cell biomass equivalent to 2 ml at OD600=1 (2 ‘OD units’) was resuspended in 1 mL TSEMS (50 mM Tris pH 7.4, 50 mM NaCl, 10 mM EDTA pH 8.0, 0.5 M sucrose and protease inhibitor cocktail) containing 6 mg/mL lysozyme from chicken egg white. Protoplasts were prepared by incubating at 37 °C for 30 min with shaking. Protoplasts were washed in 2 mL TSEMS, resuspended in TSEMS, split into 3 aliquots and pelleted. Pellets were flash frozen and stored at −80 °C.
Each pellet was resuspended in 1 mL buffer L (50 mM HEPES-KOH pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 1 % (v/v) Triton X-100, 0.1 % (w/v) Na-deoxycholate) containing 0.1 mg/mL RNase A and PIC. The suspension was sonicated in a Covaris E220 water bath sonicator for 5 min at 4 °C, 100 W, 200 cycles, 10 % load and filling level 5. The sonicated material was centrifuged at 4 °C and 20,000 × g and 100 μL were collected as input reference. The immunoprecipitation was carried out by mixing 750 μL of the extract with 50 μL of Dynabeads Protein-G suspension freshly charged with 50 μL α-ScpB antiserum and incubated for 2 h on a wheel at 4 °C. Beads were washed at room temperature in 1 mL each of buffer L, buffer L5 (buffer L containing 500 mM NaCl), buffer W (10 mM Tris-HCl pH 8.0, 250 mM LiCl, 0.5 % (v/v) NP-40, 0.5 % (w/v) Na-Deoxycholate, 1 mM EDTA pH 8.0) and buffer TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0). Beads were resuspended in 520 μL buffer TES (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1 % (w/v) SDS). The reference sample was mixed with 100 μL buffer L, 300 μL buffer TES and 20 μL 10 % SDS. Formaldehyde cross-links were reversed over-night at 65 °C with vigorous shaking.
For phenol/chloroform extraction, samples were cooled to room temperature, vigorously mixed with 500 μL phenol equilibrated with buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA) and centrifuged for 10 min at 20,000 × g. Subsequently, 450 μL of the supernatant was vigorously mixed with 450 μL chloroform and centrifuged for 10 min at 20,000 × g. For DNA precipitation, 400 μL of the supernatant were mixed with 1.2 μL GlycoBlue, 40 μL of 3 M Na-Acetate pH 5.2 and 1 mL ethanol and incubated for 20 min at −20 °C. Samples were centrifuged at 4 °C and 20,000 × g for 10 min, and the precipitate was washed in 500 μL of 70 % ethanol, dissolved in 250 μL buffer PB (QIAGEN) for 15 min at 55 °C, purified with a PCR purification kit, and eluted in 50 μL buffer EB.
For qPCR, samples were diluted in water (1:10 for IP and 1:1000 for input) and duplicate 10 μL reactions (5 μL master mix, 1 μL of 3 μM primer mix, 4 μL sample) were run in a Rotor-Gene Q machine (QIAGEN) using Takyon SYBR MasterMix and the primer pairs listed in the Key Resource Table.
For deep-sequencing, DNA was fragmented to ~200 bp and libraries were prepared using the Ovation Ultralow Library Systems V2 Kit with 15 PCR cycles. Single-read sequencing was performed on a HiSeq 2500 with 125 bp read length.
In vivo cross-linking
Cultures in 200 mL SMG were grown to exponential phase (OD600 = 0.02) at 37 °C (Bürmann et al., 2013). Cells were harvested by filtration, washed in cold PBS + 0.1 % (v/v) glycerol (‘PBSG’), and split into aliquots of a biomass equivalent to 0.85 OD units. Cells were centrifuged 2 min at 10,000 g, resuspended in 200 μL PBSG and cross-linked by adding 0.5 mM BMOE. Cells were incubated with BMOE for 10 min on ice. The reaction was quenched by the addition of 14 mM 2-mercaptoethanol. Cells were pelleted and resuspended in 30 μL of PBSG containing 75 U/mL ReadyLyse Lysozyme, 750 U/mL Sm DNase, 5 μM HaloTag TMR Substrate and protease inhibitor cocktail (‘PIC’). Lysis was performed at 37 °C for 15 min. After lysis, the material was mixed with 10 μL of 4X LDS-PAGE buffer, samples were incubated for 5 min at 95 °C and resolved by SDS-PAGE. Gels were imaged on an Amersham Typhoon scanner with Cy3 DIGE filter setup.
Chromosome Entrapment Assay
Agarose plug assays were done as described in (Wilhelm and Gruber, 2017). The protocol was adopted for agarose microbeads instead of plugs following procedures described in (Wing et al., 1993). The equivalent of 3.75 OD units was resuspended in 121 μL of PBSG, cross-linked in the presence of 1 mM BMOE and quenched with 28 mM 2-mercaptoethanol. Each sample was split into two aliquots, one of 45 μL labelled as input and the other of 90 μL labelled as beads. The bead sample was mixed with 9 μL of DynaBeads suspension (to label the otherwise translucent microbeads) and PIC (1x final). The samples were prepared one at a time: first, the cell suspension was incubated for 10 s at 45 °C and immediately mixed with 100 μL of molten 2 % low-melt agarose prewarmed to 45 °C. 800 μL of mineral oil at 45 °C was added and vigorously mixed for 1-2 minutes at 4 °C. The mixture was kept at room temperature until all samples were prepared. The oil was removed by centrifugation for 1.5 min at 10,000 g at room temperature. The agarose microbeads were rinsed twice in 1 mL of room temperature PBSG. Cell lysis was achieved by addition of ReadyLyse Lysozyme (4 U/μL final), protease inhibitor cocktail, EDTA pH 8 (1 mM final) and HaloTag-TMR substrate (5 μM final) and incubation for 30 min at 37 °C. Separate input samples were lysed in the presence of Sm DNase (5 U/μL final) without EDTA. The input samples where then mixed with 50 μL of 2X LDS loading buffer and stored at -20 °C.
The agarose beads were then washed twice at room temperature in PBSG supplemented with 1 mM EDTA pH 8 and then incubated twice in 1 mL of TES buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA pH 8.0, 1 % (w/v) SDS) at room temperature on a rotating wheel. The final wash was done over-night at 4 °C in TES buffer on a rotating wheel. SDS and EDTA were removed by rinsing the beads three times with 1 mL PBS. Microbeads were resuspended in 100 μL of PBS and incubated with 750 units of Sm DNase 30 min at 37 °C protected from light. To melt the agarose beads, the mixture was incubated at 70 °C for 1 min with vigorous shaking, and centrifuged at 21,000 rpm for 15 min at 4 °C. The supernatant was transferred to a 0.45 μm Costar Spin-X Tube Filter and spun for 1 min at 10,000 × g. The flow-through was diluted to 1 mL final volume with water and proteins were precipitated with 10 % (w/v) trichloroacetic acid (TCA) in presence of 3 μg of BSA (Koontz, 2014). The precipitate was resuspended in 1x LDS Sample Buffer (NuPage) and heated for 5 min at 95 °C. Samples were loaded on a 3 – 8 % Tris-Acetate gel and run for 1.5 h at 35 mA per gel at 4 °C. Gels were scanned on an Amersham Typhoon scanner with Cy2-DIGE (for HT-Oregon-Green) or Cy3-DIGE (for HT-TMR) filter setting.
Quantification and statistical analysis
Analysis of cross-linking efficiencies
The fluorescence intensities of Smc-HT bands were measured using ImageQuant TL 1D V8.1. Lanes were defined manually. Bands were detected automatically and the band intensities corrected for background signal using the Rolling Ball algorithm with a ball radius set to 129. Values from three replicate experiments were exported to Microsoft Excel for calculation of average fractions and standard deviation.
Dissociation constant calculation
Anisotropy values where obtained directly from the BioTek Synergy Neo measurement software, exported and fit using non-linear regression in GraphPad Prism 7 with the following equation:
Where Aobs is the experimentally measured anisotropy, Amin and Amax are the minimal and maximal anisotropy values respectively, [DNA] is the dsDNA concentration fixed at 50 nM in all our measurements, [Smc] is the variable Smc protein concentration and Kd is the dissociation constant. The fit values of triplicates were averaged and calculated the standard deviation. The values are listed in Table S3.
Steady-state kinetics
The absorbance values were exported and fit to a straight-line equation. The slope values were transformed to rate values using the molar absorption coefficient of NADH. The rates were expressed into absolute values by correcting for the protein concentration. Data was fit using non-linear regression to the Hill equation:
Where v is the ATP hydrolysis rate, Vmax is the maximal rate, [ATP] is the variable ATP concentration, h is the Hill coefficient and K0.5 is the semi-saturation concentration.
For the protein dependence experiments (Fig S4A), in the wt protein we fit the rate dependence on protein concentration to the first order rate equation for kcat:
For the SmcHd/N-ScpA constructs we fit the data to the second order rate equation for kcat.
All equations where fit to the experimental data in GraphPad Prism 7. Fit parameters obtained from the average of triplicates or duplicates and standard deviations are summarized in Table S5.
Analysis of qPCR Data
The threshold cycle (CT) was obtained by analyzing the fluorescence raw data in the Real-Time PCR Miner server (http://ewindup.info/miner/) (Zhao and Fernald, 2005). IP/input ratios were calculated as α 2ΔCT, where ΔCT = CT (Input) – CT and α is a constant determined by extraction volumes and sample dilutions. Data are presented as the mean of duplicates.
Analysis of ChIP-Seq data
Deep-sequencing data from chromatin immuno-precipitation samples were mapped to the Bs reference genome NC_000964 (centered on its first coordinate) using Bowtie for Illumina in the Galaxy project website (https://usegalaxy.org/) (Langmead and Salzberg, 2012). Reads were filtered for mapping quality (MAPQ) greater than 10, reduced to bins of 250 bp, and normalized for total read count in SeqMonk. Data are presented in reads per million (rpm).
For ratiometric analysis, the reduced data of each sample was compared to the reduced data of the wild-type sample (Minnen et al., 2016). For each bin, the larger value was divided by the smaller, and the resulting ratio was plotted above the coordinate axis for value (mutant) ≥ value (WT) and below the axis otherwise. Data was exported and graphically represented in GraphPad Prism 7. See also (Bürmann et al., 2017).
Supplementary Material
Acknowledgements
We are grateful to all members of the Gruber lab for critical comments on the manuscript and to Kim Nasmyth and Christophe Chapard for sharing unpublished results. We thank Yolanda Schaerli for advice on the fabrication of agarose microbeads and Anna Anchimiuk for providing genetic tools. Deep sequencing was performed at the Genome Technology Facility (GTF) at UNIL. This work was supported by a European Research Council Consolidator Grant (‘Chrocodyle’ #724482 to S.G.), the Max Planck Society and the University of Lausanne.
Footnotes
Data availability
ChIP-Seq data reported in this manuscript has been deposited at the NCBI Sequence Read Archive with the accession number PRJNA533030. Other original data is available at Mendeley data under the accession doi: 10.17632/xzmvjdhwgt.1
Author contributions
Conceptualization, L.B.R.A., S.G.; Investigation, L.B.R.A., R.V.N.; Data Curation, Methodology, Validation and Formal analysis, R.V.N.; Writing and Visualization, R.V.N., S.G.; Supervision, Project administration and Funding acquisition, S.G.
Declaration of Interests
The authors declare no competing financial interests related to this work.
References
- Alt A, Dang HQ, Wells OS, Polo LM, Smith MA, McGregor GA, Welte T, Lehmann AR, Pearl LH, Murray JM, et al. Specialized interfaces of Smc5/6 control hinge stability and DNA association. Nature communications. 2017;8 doi: 10.1038/ncomms14011. 14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K. ATP hydrolysis is required for cohesin's association with chromosomes. Current biology : CB. 2003;13:1941–1953. doi: 10.1016/j.cub.2003.10.036. [DOI] [PubMed] [Google Scholar]
- Belmont AS. Mitotic chromosome structure and condensation. Current opinion in cell biology. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Bürmann F, Basfeld A, Vazquez Nunez R, Diebold-Durand ML, Wilhelm L, Gruber S. Tuned SMC Arms Drive Chromosomal Loading of Prokaryotic Condensin. Molecular cell. 2017;65:861–872.e869. doi: 10.1016/j.molcel.2017.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürmann F, Gruber S. SMC condensin: promoting cohesion of replicon arms. Nature structural & molecular biology. 2015;22:653–655. doi: 10.1038/nsmb.3082. [DOI] [PubMed] [Google Scholar]
- Bürmann F, Lee BG, Than T, Sinn L, O'Reilly FJ, Yatskevich S, Rappsilber J, Hu B, Nasmyth K, Lowe J. A folded conformation of MukBEF and cohesin. Nature structural & molecular biology. 2019;26:227–236. doi: 10.1038/s41594-019-0196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürmann F, Shin HC, Basquin J, Soh YM, Gimenez-Oya V, Kim YG, Oh BH, Gruber S. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nature structural & molecular biology. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- Chapard C, Jones R, van Oepen T, Scheinost JC, Nasmyth K. The topology of DNA entrapment by cohesin rings. bioRxiv. 2018 doi: 10.1016/j.molcel.2019.05.023. 495762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu A, Revenkova E, Jessberger R. DNA interaction and dimerization of eukaryotic SMC hinge domains. The Journal of biological chemistry. 2004;279:26233–26242. doi: 10.1074/jbc.M402439200. [DOI] [PubMed] [Google Scholar]
- Cuylen S, Metz J, Haering CH. Condensin structures chromosomal DNA through topological links. Nature structural & molecular biology. 2011;18:894–901. doi: 10.1038/nsmb.2087. [DOI] [PubMed] [Google Scholar]
- Diebold-Durand ML, Lee H, Ruiz Avila LB, Noh H, Shin HC, Im H, Bock FP, Bürmann F, Durand A, Basfeld A, et al. Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization. Molecular cell. 2017;67:334–347.e335. doi: 10.1016/j.molcel.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Laemmli UK. Architecture of metaphase chromosomes and chromosome scaffolds. The Journal of cell biology. 1983;96:84–93. doi: 10.1083/jcb.96.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Perez ME, Gwynn EJ, Dillingham MS, Moreno-Herrero F. Using DNA as a fiducial marker to study SMC complex interactions with the atomic force microscope. Biophysical journal. 2012;102:839–848. doi: 10.1016/j.bpj.2012.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C. Real-time imaging of DNA loop extrusion by condensin. Science (New York, N.Y.) 2018;360:102–105. doi: 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Kanemaki M, Xie L, Paulson JR, Earnshaw WC, Mirny LA, et al. Mitotic chromosomes fold by condensin-dependent helical winding of chromatin loop arrays. bioRxiv. 2018 [Google Scholar]
- Gligoris TG, Scheinost JC, Bürmann F, Petela N, Chan KL, Uluocak P, Beckouet F, Gruber S, Nasmyth K, Lowe J. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science (New York, N.Y.) 2014;346:963–967. doi: 10.1126/science.1256917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griese JJ, Hopfner KP. Structure and DNA-binding activity of the Pyrococcus furiosus SMC protein hinge domain. Proteins. 2011;79:558–568. doi: 10.1002/prot.22903. [DOI] [PubMed] [Google Scholar]
- Griese JJ, Witte G, Hopfner KP. Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic acids research. 2010;38:3454–3465. doi: 10.1093/nar/gkq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S. MukBEF on the march: taking over chromosome organization in bacteria? Molecular microbiology. 2011;81:855–859. doi: 10.1111/j.1365-2958.2011.07764.x. [DOI] [PubMed] [Google Scholar]
- Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Gruber S, Veening JW, Bach J, Blettinger M, Bramkamp M, Errington J. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Current biology : CB. 2014;24:293–298. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler M, Shaltiel IA, Haering CH. Towards a Unified Model of SMC Complex Function. Current biology : CB. 2018;28:R1266–r1281. doi: 10.1016/j.cub.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Anderson DE, Erickson HP, Hirano T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. The EMBO journal. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. The EMBO journal. 2004;23:2664–2673. doi: 10.1038/sj.emboj.7600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Molecular cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensin-Based Chromosome Organization from Bacteria to Vertebrates. Cell. 2016;164:847–857. doi: 10.1016/j.cell.2016.01.033. [DOI] [PubMed] [Google Scholar]
- Hopfner KP. Invited review: Architectures and mechanisms of ATP binding cassette proteins. Biopolymers. 2016;105:492–504. doi: 10.1002/bip.22843. [DOI] [PubMed] [Google Scholar]
- Houlard M, Godwin J, Metson J, Lee J, Hirano T, Nasmyth K. Condensin confers the longitudinal rigidity of chromosomes. Nature cell biology. 2015;17:771–781. doi: 10.1038/ncb3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K, Miyata M, Hirano T. Molecular basis of SMC ATPase activation: role of internal structural changes of the regulatory subcomplex ScpAB. Structure (London, England : 1993) 2013;21:581–594. doi: 10.1016/j.str.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Kamada K, Su'etsugu M, Takada H, Miyata M, Hirano T. Overall Shapes of the SMC-ScpAB Complex Are Determined by Balance between Constraint and Relaxation of Its Structural Parts. Structure (London, England : 1993) 2017;25:603–616.e604. doi: 10.1016/j.str.2017.02.008. [DOI] [PubMed] [Google Scholar]
- Kanno T, Berta DG, Sjogren C. The Smc5/6 Complex Is an ATP-Dependent Intermolecular DNA Linker. Cell reports. 2015;12:1471–1482. doi: 10.1016/j.celrep.2015.07.048. [DOI] [PubMed] [Google Scholar]
- Kim H, Loparo JJ. Multistep assembly of DNA condensation clusters by SMC. Nature communications. 2016;7 doi: 10.1038/ncomms10200. 10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Koontz L. TCA precipitation. Methods in enzymology. 2014;541:3–10. doi: 10.1016/B978-0-12-420119-4.00001-X. [DOI] [PubMed] [Google Scholar]
- Kschonsak M, Haering CH. Shaping mitotic chromosomes: From classical concepts to molecular mechanisms. BioEssays : news and reviews in molecular, cellular and developmental biology. 2015;37:755–766. doi: 10.1002/bies.201500020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kschonsak M, Merkel F, Bisht S, Metz J, Rybin V, Hassler M, Haering CH. Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell. 2017;171:588–600.e524. doi: 10.1016/j.cell.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Grosbart M, Nurse P, Bahng S, Wyman CL, Marians KJ. The bacterial condensin MukB compacts DNA by sequestering supercoils and stabilizing topologically isolated loops. The Journal of biological chemistry. 2017;292:16904–16920. doi: 10.1074/jbc.M117.803312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens A, Schele A, Hopfner KP. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Current biology : CB. 2004;14:1778–1782. doi: 10.1016/j.cub.2004.09.044. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioy VS, Cournac A, Marbouty M, Duigou S, Mozziconacci J, Espeli O, Boccard F, Koszul R. Multiscale Structuring of the E. coli Chromosome by Nucleoid-Associated and Condensin Proteins. Cell. 2018;172:771–783.e718. doi: 10.1016/j.cell.2017.12.027. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sung S, Kim Y, Li F, Gwon G, Jo A, Kim AK, Kim T, Song OK, Lee SE, et al. ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex. The EMBO journal. 2016;35:743–758. doi: 10.15252/embj.201592462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF, De Los Rios P, Barducci A, Gruber S. DNA-segment-capture model for loop extrusion by structural maintenance of chromosome (SMC) protein complexes. bioRxiv. 2018 doi: 10.1093/nar/gkz497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- Melby TE, Ciampaglio CN, Briscoe G, Erickson HP. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. The Journal of cell biology. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annual review of genomics and human genetics. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- Minnen A, Attaiech L, Thon M, Gruber S, Veening JW. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Molecular microbiology. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- Minnen A, Bürmann F, Wilhelm L, Anchimiuk A, Diebold-Durand ML, Gruber S. Control of Smc Coiled Coil Architecture by the ATPase Heads Facilitates Targeting to Chromosomal ParB/parS and Release onto Flanking DNA. Cell reports. 2016;14:2003–2016. doi: 10.1016/j.celrep.2016.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annual review of genetics. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Naumova N, Imakaev M, Fudenberg G, Zhan Y, Lajoie BR, Mirny LA, Dekker J. Organization of the mitotic chromosome. Science (New York, N.Y.) 2013;342:948–953. doi: 10.1126/science.1236083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek JJ, Gruber S. Kite Proteins: a Superfamily of SMC/Kleisin Partners Conserved Across Bacteria, Archaea, and Eukaryotes. Structure (London, England : 1993) 2015;23:2183–2190. doi: 10.1016/j.str.2015.10.004. [DOI] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320.e324. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy MA, Dhanaraman T, D'Amours D. The Smc5-Smc6 heterodimer associates with DNA through several independent binding domains. Scientific reports. 2015;5 doi: 10.1038/srep09797. 9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert FU, Lammens K, Stoehr G, Kessler B, Hopfner KP. Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50. The EMBO journal. 2016;35:759–772. doi: 10.15252/embj.201592934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh YM, Bürmann F, Shin HC, Oda T, Jin KS, Toseland CP, Kim C, Lee H, Kim SJ, Kong MS, et al. Molecular basis for SMC rod formation and its dissolution upon DNA binding. Molecular cell. 2015;57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan M, Scheinost JC, Petela NJ, Gligoris TG, Wissler M, Ogushi S, Collier JE, Voulgaris M, Kurze A, Chan KL, et al. The Cohesin Ring Uses Its Hinge to Organize DNA Using Non-topological as well as Topological Mechanisms. Cell. 2018;173:1508–1519.e1518. doi: 10.1016/j.cell.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Nishino T, Marko JF. The SMC1-SMC3 cohesin heterodimer structures DNA through supercoiling-dependent loop formation. Nucleic acids research. 2013;41:6149–6160. doi: 10.1093/nar/gkt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Liu YP, Shan HH, Tian LF, Zhang JZ, Yan XX. ATP-dependent conformational change in ABC-ATPase RecF serves as a switch in DNA repair. Scientific reports. 2018;8 doi: 10.1038/s41598-018-20557-0. 2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NT, Laub MT, Le TBK. SMC Progressively Aligns Chromosomal Arms in Caulobacter crescentus but Is Antagonized by Convergent Transcription. Cell reports. 2017;20:2057–2071. doi: 10.1016/j.celrep.2017.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama S, Kawahara K, Hosokawa Y, Fukakusa S, Oki H, Nakamura S, Kojima Y, Noda M, Takino R, Miyahara Y, et al. Structural Basis for Dimer Formation of Human Condensin Structural Maintenance of Chromosome Proteins and Its Implications for Single-stranded DNA Recognition. The Journal of biological chemistry. 2015;290:29461–29477. doi: 10.1074/jbc.M115.670794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Brandao HB, Le TB, Laub MT, Rudner DZ. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science (New York, N.Y.) 2017;355:524–527. doi: 10.1126/science.aai8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hughes AC, Brandao HB, Walker B, Lierz C, Cochran JC, Oakley MG, Kruse AC, Rudner DZ. In Vivo Evidence for ATPase-Dependent DNA Translocation by the Bacillus subtilis SMC Condensin Complex. Molecular cell. 2018;71:841–847.e845. doi: 10.1016/j.molcel.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Le TB, Lajoie BR, Dekker J, Laub MT, Rudner DZ. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes & development. 2015;29:1661–1675. doi: 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang OW, Riley EP, Rudner DZ. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Current biology : CB. 2014;24:287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm L, Bürmann F, Minnen A, Shin HC, Toseland CP, Oh BH, Gruber S. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife. 2015;4 doi: 10.7554/eLife.06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm L, Gruber S. A Chromosome Co-Entrapment Assay to Study Topological Protein-DNA Interactions. Methods in molecular biology (Clifton, N.J.) 2017;1624:117–126. doi: 10.1007/978-1-4939-7098-8_10. [DOI] [PubMed] [Google Scholar]
- Wing RA, Rastogi VK, Zhang HB, Paterson AH, Tanksley SD. An improved method of plant megabase DNA isolation in agarose microbeads suitable for physical mapping and YAC cloning. The Plant journal : for cell and molecular biology. 1993;4:893–898. doi: 10.1046/j.1365-313x.1993.04050893.x. [DOI] [PubMed] [Google Scholar]
- Woo JS, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- Zabrady K, Adamus M, Vondrova L, Liao C, Skoupilova H, Novakova M, Jurcisinova L, Alt A, Oliver AW, Lehmann AR, et al. Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic acids research. 2016;44:1064–1079. doi: 10.1093/nar/gkv1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.