Abstract
In cytochrome c oxidase (CytcO) reduction of O2 to water is linked to uptake of eight protons from the negative side of the membrane: four are substrate protons used to form water and four are pumped across the membrane. In bacterial oxidases, the substrate protons are taken up through the K and the D proton pathways, while the pumped protons are transferred through the D pathway. On the basis of studies with CytcO isolated from bovine heart mitochondria, it was suggested that in mitochondrial CytcOs the pumped protons are transferred though a third proton pathway, the H pathway, rather than through the D pathway. Here, we studied these reactions in S. cerevisiae CytcO, which serves as a model of the mammalian counterpart. We analyzed the effect of mutations in the D (Asn99Asp and Ile67Asn) and H pathways (Ser382Ala and Ser458Ala) and investigated the kinetics of electron and proton transfer during the reaction of the reduced CytcO with O2. No effects were observed with the H pathway variants while in the D pathway variants the functional effects were similar to those observed with the R. sphaeroides CytcO. The data indicate that the S. cerevisiae CytcO uses the D pathway for proton uptake and presumably also for proton pumping.
Subject terms: Biophysical chemistry, Oxidoreductases, Bioenergetics
Introduction
Cytochrome c oxidase (CytcO) is a membrane-bound enzyme that catalyzes reduction of O2 to water and uses part of the free energy of this reaction for proton pumping from the negative (n) to the positive (p) side of the membrane. The electron donor for CytcO is cytochrome c (cyt. c), which binds near the electron-entry site, CuA. From CuA electrons are transferred consecutively to heme a, and then to the catalytic site, composed of heme a3 and CuB. For each O2 reduced to water, a total of eight protons are taken up from the n-side of the membrane. Four of these protons are used for reduction of O2 to H2O at the catalytic site (O2 + 4 H+ + 4 e− − > 2 H2O) and the other four protons are taken up from the n side and released to the p side of the membrane (pumped).
In the A-type1,2 bacterial CytcOs, protons are taken up through two pathways named the K pathway, after a conserved Lys residue approximately in the middle of the pathway, and the D pathway, named after a conserved Asp residue at the orifice of the pathway (for review, see3–5) (Fig. 1). The K pathway is used for uptake of ~2 H+ upon reduction of the catalytic site while the D pathway is used for transfer of the remaining 6 protons, i.e. four that are pumped and the remaining two protons used for reduction of O2 to H2O at the catalytic site. In the B-type oxidases all protons are transferred through a single pathway, which approximately overlaps in space with the K pathway in the A-type oxidases3,6,7. Hence, there are alternative pathways for transfer of the pumped protons from the n-side in different oxidases.
Figure 1.
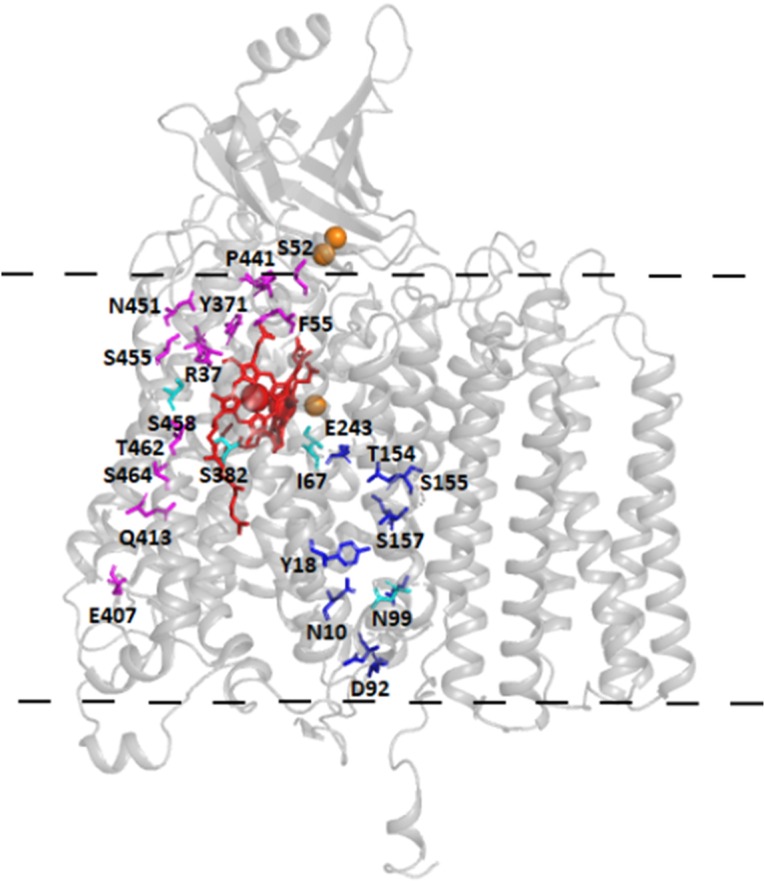
Structural model. The three core subunits of the S. cerevisiae CytcO with the D (blue) and H (magenta) pathway residues marked. Residues that have been substituted in this work are colored in cyan. Copper atoms are colored in orange and hemes in red. The approximate position of the membrane is marked with a dashed line. The structure is based on PDB entry 6HU917.
The mammalian mitochondrial CytcOs belong to the A-class, i.e. they harbor both the K and D proton pathways. However, structural studies have revealed structural changes in another region of the protein, highlighting a putative third functional pathway called the H pathway8 (Fig. 1). The H pathway starts on the n-side, near a His residue (His413, bovine CytcO numbering) and spans the membrane domain close to heme a toward the p-side, via Asp51 and Ser205 (subunit II), and the Tyr440-Ser441 peptide bond, which has been proposed to control unidirectional proton transfer9. It is characterized in its lower part by a water-containing cavity, which was shown to adopt open or closed configuration in different redox and ligand states depending on the interaction of Ser382 with the hydroxyfarnesyl-ethyl group of heme a10. Pumping measurements were performed on mammalian H pathway variants using a chimeric human/bovine mutagenesis system and the authors concluded that the H pathway, and not the D pathway, was the route taken by pumped protons in mammalian CytcOs8,11,12. Equivalent studies in CytcO from R. sphaeroides confirmed that the H pathway is not involved in proton conduction in bacterial CytcOs13. Mammalian and bacterial CytcOs have thus been proposed to operate with different proton-pumping mechanisms. An alternative role has also been suggested for the H pathway as a dielectric well that could modulate the effects of buried charge changes on heme a4.
The bacterial A-type CytcOs are typically composed of the three core subunits I-III, in some cases having an additional fourth subunit that in e.g. R. sphaeroides consists of a single transmembrane helix14. The mammalian CytcOs are bigger with an additional 10–11 accessory subunits15. The S. cerevisiae mitochondrial CytcO is an ideal model of the mammalian counterpart16. It is composed of the three core subunits as well as 9 accessory subunits17–20. An atomic model of S. cerevisiae CytcO determined recently17 confirmed its similarity to mammalian CytcOs, including the core subunits responsible for catalysis and pumping.
In the present study, we prepared structural variants of the S. cerevisiae CytcO in which residues of the proposed D and H pathways were modified. The kinetics of internal electron transfer, were studied in these structural variants. Similar effects were observed with the S. cerevisiae CytcO as with the R. sphaeroides CytcO for the D pathway variants. We did not observe any functional effects of changes in the H pathway of the S. cerevisiae CytcO. Collectively, the data indicate that the mitochondrial S. cerevisiae CytcO displays similar functional characteristics to those seen with bacterial CytcOs.
Results
We investigated two D pathway structural variants (Asn99Asp and Ile67Asn) and two H pathway variants (Ser382Ala and Ser458Ala). These structural variants were chosen from a series of modified yeast strains that displayed apparent respiratory growth defects21. In earlier studies, reduced minus oxidized difference spectra were recorded of samples of each mutant CytcO after affinity chromatography purification. These difference spectra of Asn99Asp and Ser382Ala were identical to that of the wild-type CytcO21 whereas those of Ile67Asn and Ser458Ala both displayed a 2–3 nm shift of their alpha-bands to 601 nm22 and 606 nm, respectively. A comparison of the numbering of these residues in CytcOs from S. cerevisiae, R. sphaeroides and bovine heart mitochondria is shown in Table 1.
Table 1.
Residues in the D and H pathways.
| S. cerevisiae | Bovine | R. sphaeroides |
|---|---|---|
| D pathway | ||
| D92 | D91 | D132 |
| N99 | N98 | N139 |
| I67 | I66 | M107 |
| E243 | E242 | E286 |
| H pathway | ||
| R37 | R38 | R52 |
| S52 | D51 | G92 |
| F55 | Y54 | W95 |
| Y371 | Y371 | Y414 |
| S382 | S382 | S425 |
| E407 | D407 | E450 |
| Q413 | H413 | H456 |
| M428 | Q428 | Q471 |
| I440 | Y440 | Y483 |
| P441 | S441 | I484 |
| N451 | N451 | N494 |
| A454 | S454 | S497 |
| S455 | S455 | S498 |
| S458 | S458 | A501 |
| S464 | A464 | S507 |
| A461 | S461 | S504 |
| A230II | S205II | A261II |
Equivalent residues in the bovine heart and R. sphaeroides CytcOs are listed.
Reaction of the reduced CytcO with O2
To investigate the kinetics of electron transfer in the wild-type and structural variants of the CytcO, the enzyme was fully reduced (by four electrons) and incubated under an atmosphere of carbon monoxide, which binds at heme a3, i.e. the ligand blocks access to O2. The sample was then rapidly (~2 ms) mixed with an O2-saturated solution in a stopped-flow apparatus. Because CO dissociation in the dark is slow (~30 s), the CytcO stays essentially fully reduced over a time scale of milliseconds. About 200 ms after mixing, the ligand was dissociated by means of a short (~10 ns) laser flash, which initiates the reaction of the rescued CytcO with O2 simultaneously in the entire CytcO population. Figure 2 shows absorbance changes associated with reaction of the reduced wild-type CytcO with O2 at 445 nm at pH 7.5 and pH 10. The initial increase in absorbance at t = 0 is associated with CO dissociation. The decrease in absorbance with a time constant of ~40 µs is associated with electron transfer from heme a to the catalytic site forming the peroxy state (PR). This reaction is not associated with any proton uptake from solution and displays a very small pH dependence. In the next step a proton is taken up from solution to form the ferryl state, F with a time constant of ~100 µs at pH 7.5. This reaction is mainly seen at 580 nm where absorbance changes are small and it could not be resolved in this study. At 445 nm the reaction is observed as a small lag before the decrease in absorbance with a time constant of ~1 ms at pH 7.5. In this final step of the reaction the F state decays to form the oxidized CytcO (state O), which is associated with proton uptake from solution and proton pumping.
Figure 2.
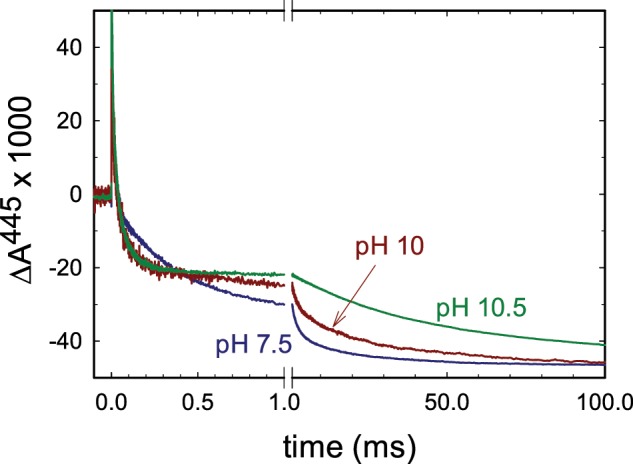
Absorbance changes associated with reaction of the fully reduced CytcO with O2. The sample was illuminated by a laser flash, which results in dissociation of the CO ligand (increase in absorbance at t = 0). The decrease in absorbance is associated with binding of O2 and formation of the PR state with a time constant ~40 µs. The slowest decay in absorbance is associated with the F → O reaction. Representative traces at pH 7.5, 10 and 10.5 are shown. Experimental conditions: mixing ratio of the O2-saturated buffer to enzyme was 5:1. The delay time between mixing and the laser flash was 200 ms. The CytcO concentration was ~1.6 µM after mixing. The composition of the CytcO solution was: 150 mM KCl, 10 mM Bis-tris propane pH 7 and 0.035% DDM. The O2-containing buffer solution was composed of ~1.2 mM O2, 150 mM KCl, 100 mM buffer (MES pH 6, Bis-tris propane pH 7, 7.5, and 9, CHES pH 9.5 and CAPS for pH 10 and 10.5). The traces have been scaled to the same CO-dissociation absorbance change.
Figure 3A shows the pH dependence of the F → O reaction. The rate constant for the wild-type enzyme dropped from ~2·103 s−1 at low pH to ~20 s−1 at pH 10.5. This pH dependence is qualitatively similar to that observed previously with the CytcO from R. sphaeroides or from bovine heart mitochondria23,24. A single kinetic component was observed in the pH range 6–9 and at pH 10.5, but at the intermediate pH values the decay was biphasic with approximately equal amplitudes of the two components. For these points an average of the two rates is plotted in Fig. 3A. With the two H pathway variants, Ser382Ala and Ser458Ala, the rates at pH 7 and pH 10 were similar to those observed with the wild-type CytcO (Table 2).
Figure 3.
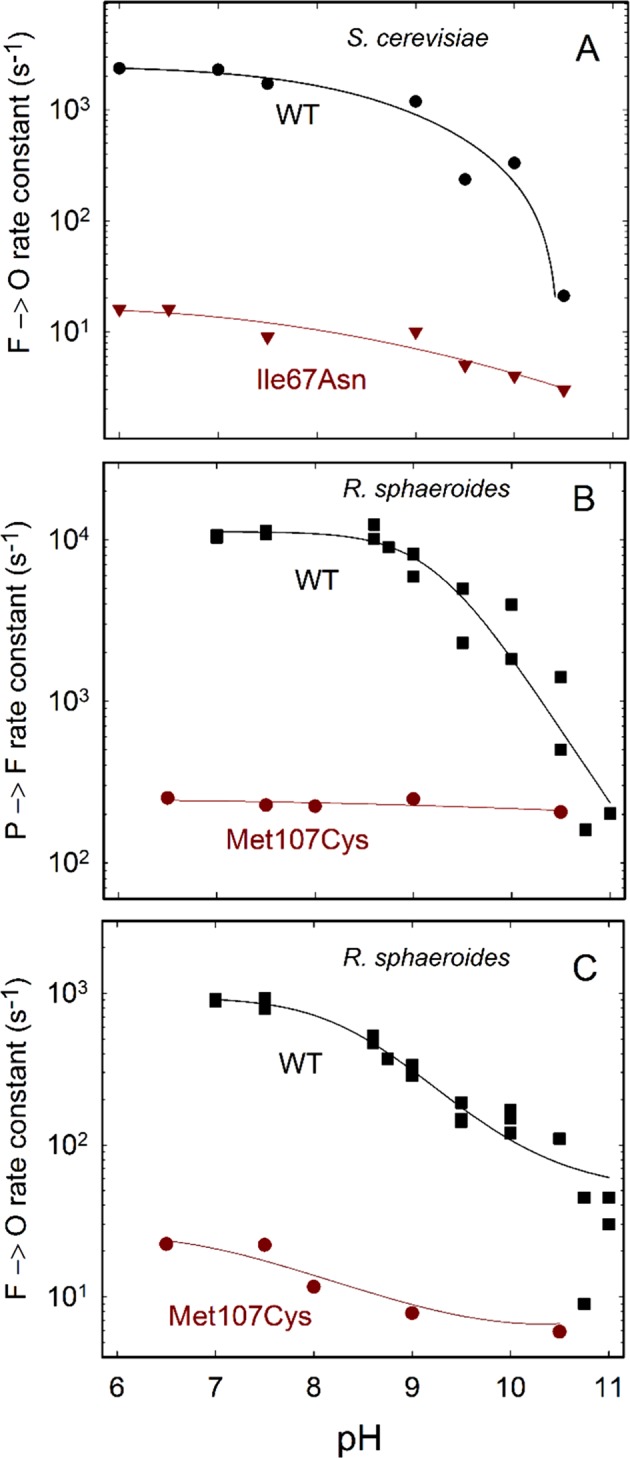
pH dependence of reaction rate constants. (A) The rates were determined by fitting an exponential function to traces obtained at 445 nm, after initiation of the reaction of reduced CytcO with O2. Data with the wild-type and I67N CytcOs. Standard errors ((SD/√n; SD, standard deviation, n, number of measurements) were typically ~10% of the measured value. Experimental conditions were the same as those in Fig. 2. The different buffers used at different pH values are listed in the Fig. 2 legend. (B,C) Data with the R. sphaeroides CytcO. Absorbance changes were measured at 580 nm with the Met107Cys variant and the PR → F (B) and F → O (C) rate constants were determined from a fit of a sum of exponential functions (see e.g.37). Standard errors were typically ~5% of the measured value (SD of ~20 traces measured with 2 samples). The wild-type data are from38. The experiments were carried out in the same way as that in Fig. 2, except that the composition of the CytcO solution was 10 mM Bis-tris propane pH 7.5, 0.05% DDM and 0.1 mM EDTA. The O2-containing buffer solution was composed of ~1.2 mM O2, 100 mM buffer (Bis-tris propane pH 6.5, 7.5, 8 and 9 or CAPS for pH 10.5), 0.05% DDM and 0.1 mM EDTA. The solid line is a guide for the eye.
Table 2.
F → O rate constants. Standard errors are shown.
| Sample | rate constant (s−1) pH 7 | rate constant (s−1) pH 10 |
|---|---|---|
| wild type | 2200 ± 140 (6) | 330 ± 30 (12) |
| N99D | 1200 ± 70 (18) | 780 ± 40 (18) |
| S382A | 1200 ± 60 (8) | 250 ± 150 (8) |
| S458A | 2500 ± 60 (5) | 350 ± 30 (5) |
Number of measurements in parentheses. For the S382A mutant CytcO the error obtained at pH 10 is estimated based on the range of rate constants that could be used to fit an average trace composed of data from eight measurements. In this case, the signal-to-noise ratio did not allow fits of data obtained with the individual samples.
Residue Asn99 is found near Asp92, which is located at the orifice of the D pathway (see Fig. 1). With the Asn99Asp mutant CytcO, we observed F → O rate constants of 1200 ± 100 s−1 and 780 ± 100 s−1 at pH 7 and pH 10 (10 measurements on each of two samples), respectively, i.e. the rate constant was larger with the Asn99Asp than with the wild-type CytcO at pH 10, consistent with earlier observations made with the R. sphaeroides CytcO25.
Residue Ile67 is located at a distance of ~3 Å from Glu243, a key D pathway residue, at the end of the pathway (see Fig. 1). With the Ile67Asn, the F → O rate was about a factor of 100 slower than that seen with the wild-type CytcO, which is consistent with proton uptake through the D pathway in this reactions step. To test the effect of this structural alteration in a bacterial CytcO, we introduced a mutation at the equivalent residue, Met107 in the R. sphaeroides CytcO, which is also located ~3 Å from Glu286 (equivalent of Glu243 in S. cerevisiae). In both S. cerevisiae (Ile67Asn) and R. sphaeroides (Met107Cys) mutation at this site resulted in lower CytcO activity of ~10%22 and ~6%, respectively (see “Materials and Methods”). With the R. sphaeroides mutant CytcO we obtained sufficient amount of CytcO material to be able to perform measurements also at 580 nm where the PR → F reaction step is clearly observed as an increase in absorbance in the time range (Fig. 3B). As seen in the figure, the PR → F reaction rate constant was a factor of ~50 slower than that observed with the wild-type CytcO. Because the absorbance changes associated with the PR → F reaction reflect proton uptake through the D pathway26,27, the data indicate that in the R. sphaeroides CytcO, the amino-acid residue replacement slows proton uptake through this pathway, presumably due to an interaction with Glu286. Note that at high pH, i.e. when proton uptake from solution becomes rate limiting for the PR → F reaction, similar rate constants were observed with the wild-type and mutant CytcOs. Similar to the results obtained with the S. cerevisiae CytcO, the replacement of Met107 by a Cys in R. sphaeroides CytcO resulted in slowing the F → O reaction rate constant by a factor of 50 to ~20 s−1 at pH 7 (Fig. 3C).
Discussion
Results from earlier studies with bacterial CytcOs showed dramatic effects of mutations in the D pathway on the rate of proton transfer and/or the proton-pumping stoichiometry28. In the R. sphaeroides Asn139Asp variant (Asn99Asp in S. cerevisiae) proton pumping was uncoupled from O2 reduction while the activity of the CytcO was slightly increased29. The maximum rate of the F → O reaction at pH < 7 was unaffected by the mutation, but the pH dependence of the reaction rate was altered resulting in an increase in the pKa from 9.4 to ~1125. Mutation of Asn207 to Asp (Asn164Asp in the S. cerevisiae CytcO), near Asn139, also resulted in uncoupling of proton pumping and an altered pH dependence of the F → O reaction25. No effects on proton pumping were observed with the equivalent mutants in the mammalian CytcO, which prompted the authors to conclude that the function of the D pathway is not conserved8,11 and that the H pathway is involved in proton pumping11.
The results from the present study show that the F → O reaction rate at pH 7 was similar with the Asn99Asp CytcO as with the wild-type CytcO (Table 2). As already noted above with the R. sphaeroides CytcO, the characteristic feature of this variant is a shift in the pH dependence of the F → O rate resulting in an elevated pKa value. While this rate constant with the wild-type R. sphaeroides CytcO decreased by a factor of five from ~1000 s−1 to ~200 s−1 upon increasing the pH from 6 to 10, in Asp139Asn variant it decreased from 1200 s−1 to 900 s−1, i.e. by a factor of ~1.325.
With the S. cerevisiae wild-type CytcO, the F → O rate constant decreased from ~2200 s−1 at pH 7 to ~330 s−1 at pH 10, i.e. by a factor of ~7, while with the Asn99Asp CytcO the rate decreased from 1200 s−1 at pH 7 to ~780 s−1 at pH 10, i.e. by a factor of ~1.6 (Table 2). Hence, we observed a similar behaviour with the S. cerevisiae enzyme as with the R. sphaeroides CytcO.
The current study shows that structural alterations close to the D pathway, at the site of residue Ile67 in S. cerevisiae or Met107 in R. sphaeroides CytcO, result in a dramatically slower F → O reaction rate (by factors of ~100 and ~50, respectively). This residue is located at hydrogen-bonding distance from the Glu243 (or Glu286 in the R. sphaeroides CytcO). Results from earlier studies have shown that replacement of Glu286 by other residues results in a dramatic decrease or impaired proton transfer through the D pathway30. Hence, this segment of the D pathway is particularly sensitive to structural alterations. Because with the R. sphaeroides CytcO we observed a ~50-fold slower PR → F reaction rate with the Met107Cys variant (Fig. 3), the effect of the mutation is a dramatically slowed proton transfer through the D pathway. Hence, we conclude that also with the S. cerevisiae CytcO the slowed F → O reaction rate is caused by slowed proton transfer through the D pathway. This interpretation is also consistent with earlier FTIR studies which showed that the low activity of the Ile67Asn variant was linked to perturbations of Glu243 and impairment of its proposed role in proton coupled electron transfer22,31.
The observations discussed above are consistent with data obtained from studies of the Glu243Asp variant32, and, collectively, they indicate that the F → O reaction is associated with proton transfer through the D pathway also in the S. cerevisiae CytcO. We did not observe any effects on the F → O rate constants at pH 7 or pH 10, measured with the H pathway variants (see Table 2). Because proton pumping in CytcO is driven by electron transfer to O2 (and the associated uptake of substrate protons for water formation), if proton pumping would be impaired in the H-pathway mutants, we would expect to observe an effect on the O2-reduction reaction. Because the oxidation kinetics was unperturbed, we conclude that the functionality of the mutants investigated here is unperturbed.
Data from earlier studies showed that the Ser458 to Ala replacement resulted in a decreased respiratory growth21. However, this observation does not imply that proton-pumping would be impaired in the Ser458Ala variant. For example, the mutation could instead yield a fraction CytcO that is not assembled correctly, resulting in slowed overall respiration. In the present study we studied purified CytcO and the observed signals originate from CytcO that bind CO to an intact catalytic site. Results from a study with the R. sphaeroides CytcO in which Ser425, the equivalent of the S. cerevisiae CytcO Ser382 residue, was modified, showed that this residue is not important for proton conduction, but only results in a small increase in the midpoint potential of heme a33.
The data discussed above indicate that the mitochondrial CytcO from S. cerevisiae uses the D pathway for proton transfer from solution to the catalytic site in the F → O reaction step. Even though here we studied electron transfer to O2 at the catalytic site, this electron transfer is linked to proton pumping. Thus, as indicated above, any effect on proton pumping is expected to result in an effect on the intramolecular electron transfer. Mutations of residues in the D pathway displayed qualitatively the same functional characteristics in the S. cerevisiae as with the R. sphaeroides CytcO and the F → O rate was insensitive to changes in the proposed H pathway. Hence, the data suggest that there are no major functional differences between the mitochondrial S. cerevisiae and the bacterial R. sphaeroides CytcOs.
Methods
Preparation of mutants
The construction of the modified yeast strains is described in21.
To prepare the Met107Cys CytcO variant in R. sphaeroides a Quik-Change II site-directed mutagenesis kit (Agilent technologies) was used. The resulting amino-acid replacement was verified by sequencing. The PJS3-SH plasmid was used as a template plasmid for making mutations while pRK415-1 plasmid34 was used for expression35.
Preparation of the S. cerevisiae CytcO
Yeast cells were grown aerobically in YPGal medium at 28 °C and harvested in late log phase as described in21. Mitochondrial membranes were prepared and the S. cerevisiae CytcO was purified as described by Meunier et al.21 with some modifications as outlined here. During solubilization the membranes were diluted to 2 mg/ml in buffer (50 mM KPi, 100 mM KCl and 1.5% n-dodecyl β -D-maltoside (DDM (w/v)) and solubilized for 1 hour. To remove cyt. c the supernatant from the centrifugation step after solubilization was run over a column loaded with a cation-exchanger (Bio-Rex 70, Bio-Rad) equilibrated with 50 mM KPi, 100 mM KCl and 0.035% DDM. After addition of 5 mM imidazole to the flow-through from the ion-exchanger it was loaded on a column filled with 25 ml Ni-resin (NI Sepharose 6 Fast Flow from GE healthcare), pre-equilibrated with (20 mM KPi, 150 mM KCl and 0.035% DDM). The column was washed with four column volumes of wash buffer (20 mM KPi, 150 mM KCl, 10 mM imidazole and 0.035% DDM). This was followed in time by a two-step elution of four column volumes at each imidazole concentration (20 mM KPi, 150 mM KCl, 40 or 100 mM imidazole and 0.035% DDM). The eluted fractions were pooled and concentrated in a 100 kDa cut-off filter (Merck Millipore). Buffer exchange (elution buffer without imidazole) was performed to lower the imidazole content to sub-µM concentrations.
Preparation of the R. sphaeroides CytcO
Expression of the R. sphaeroides CytcO was achieved by growing bacteria aerobically (Sistrom medium) in the dark at 30 °C. After harvesting the cells (at OD550 ≅ 1.5) they were re-suspended (50 mM Tris-buffer, pH 8.0) at 4 °C in the presence of DNase I (0.05 mg/ml final concentration) and passed twice through a continuous-flow cell disruptor (Constant Systems LTD) operating at 170 MPa. The inner membrane fraction was collected by ultracentrifugation (138 000 g for 90 minutes at 4 °C) and 1.5% DDM was added to solubilize the membrane fraction. The histidine-tagged CytcO was purified using Ni2+-NTA affinity chromatography, essentially as described in35,36.
Activity
A Clark-type O2 electrode (Hansatech instruments) was used to measure the oxygen reduction rate of the Met107Cys CytcO. A buffer composed of 50 mM KPi (pH 6.7) and 0.1% DDM was added to the reaction chamber and supplemented with 6 mM ascorbate, 670 µM N,N,N,N-tetramethyl-p-phenylenediamine (TMPD) and 32 µM cyt. c. A CytcO solution (in 100 mM Hepes, pH 7.5 and 0.05% DDM) was added (final concentration of 7 nM) to the reaction chamber and the oxygen-consumption rate during CytcO turnover was monitored (a background oxygen consumption before addition of the CytcO was subtracted). The steady state activity of the Met107Cyt CytcO variant was ~6% of that of the wild-type CytcO, which was ~570 e−/s/CytcO.
Flow-flash measurements
The sample was prepared by exchanging the CytcO buffer to 10 mM (wild-type, Ile67Asn and Met107Cys variants) or 3 mM (Asn99Asp, Ser382Ala and Ser458Ala)) Bis-tris propane pH 7, 150 mM KCl and 0.035%DDM. The CytcO concentration after buffer exchange was 3-10 µM depending on mutant (see figure legends). The sample was transferred to a Thunberg cuvette and air was exchanged for nitrogen. The sample was then reduced with 4 mM ascorbate and 1 µM PMS (electron mediator) (S. cerevisiae CytcO) or 2 mM ascorbate and 1 µM ruthenium hexachloride (electron mediator) (R. sphaeroides CytcO). Nitrogen was then exchanged for CO. The absorbance spectra were recorded in the range 400–700 nm at each step to ensure complete reduction and ligation with CO.
The sample was loaded into one syringe of a modified stopped-flow apparatus (Applied Photophysics). The other syringe was loaded with an oxygen-saturated buffer. The buffer composition for measurements with the S. cerevisiae CytcO: 100 mM (for 1:5 mixing of CytcO:O2 solutions) or 200 mM buffer (for 1:1 mixing) MES for pH 6 and 6.5; Bis-tris propane pH 7, 7.5 and 9; CHES for pH 9.5 and CAPS for pH 10 and 10.5. In addition, the buffer was supplemented with 150 mM KCl and 0.035% DDM. For measurements with R. sphaeroides CytcO: 100 mM Bis-tris propane buffer for pH 6.5, 7.5, 8 and 9; CAPS for pH 10.5 with 0.05% DDM and 0.1 mM EDTA. The CO ligand was dissociated at 0.2 s after mixing, which initiated the reaction. The reaction was monitored by following in time absorbance at specific wavelengths (see figure legends). An amplifier (C11184, Hamamatsu) was used to amplify the signal before recording using a digital oscilloscope.
Acknowledgements
We would like to acknowledge Drs. Pia Ädelroth and Irina Smirnova for valuable advice and discussions. These studies were supported by grants from the Knut and Alice Wallenberg Foundation (KAW), the Swedish Research Council (VR), the Medical Research Council UK (Career Development Award MR/M00936X/1 to A.M.) and the EU COST action CM1306.
Author contributions
M.L.B., J.V., A.M.H. and L.N.Ö. performed experiments. P.B., A.M. and M.L.B. wrote the manuscript. P.B., J.V. and M.L.B. prepared figures. A.M., B.M. and P.B. planned research. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Amandine Maréchal, Email: a.marechal@ucl.ac.uk.
Peter Brzezinski, Email: peterb@dbb.su.se.
References
- 1.Hemp J, Gennis RB. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ. 2008;45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- 2.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochim. Biophys. Acta-Bioenerg. 2001;1505:185–208. doi: 10.1016/S0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 3.Lee HJ, Reimann J, Huang Y, Ädelroth P. Functional proton transfer pathways in the heme-copper oxidase superfamily. Biochimica et Biophysica Acta - Bioenergetics. 2012;1817:537–544. doi: 10.1016/j.bbabio.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 4.Rich PR, Maréchal A. Functions of the hydrophilic channels in protonmotive cytochrome c oxidase. Journal of the Royal Society Interface. 2013;10:183–196. doi: 10.1098/rsif.2013.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzezinski P, Gennis RB. Cytochrome c oxidase: exciting progress and remaining mysteries. J. Bioenerg. Biomembr. 2008;40:521–531. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc. Natl. Acad. Sci. USA. 2009;106:16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Ballmoos C, et al. Dynamics of the KB Proton Pathway in Cytochrome ba3 from Thermus thermophilus. Isr. J. Chem. 2017;57:424–436. doi: 10.1002/ijch.201600136. [DOI] [Google Scholar]
- 8.Tsukihara T, et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc. Natl. Acad. Sci. USA. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamiya K, Boero M, Tateno M, Shiraishi K, Oshiyama A. Possible mechanism of proton transfer through peptide groups in the H-pathway of the bovine cytochrome c oxidase. J. Am. Chem. Soc. 2007;129:9663–9673. doi: 10.1021/ja070464y. [DOI] [PubMed] [Google Scholar]
- 10.Yoshikawa S, Muramoto K, Shinzawa-Itoh K, Mochizuki M. Structural studies on bovine heart cytochrome c oxidase. Biochimica et Biophysica Acta - Bioenergetics. 2012;1817:579–589. doi: 10.1016/j.bbabio.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Shimokata K, et al. The proton pumping pathway of bovine heart cytochrome c oxidase. Proc. Natl. Acad. Sci. USA. 2007;104:4200–4205. doi: 10.1073/pnas.0611627104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aminaka R, et al. Mutagenesis studies on D-pathway function of bovine heart cytochrome c oxidase. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2010;1797:93–94. doi: 10.1016/j.bbabio.2010.04.282. [DOI] [Google Scholar]
- 13.Lee HM, et al. Mutations in the putative H-channel in the cytochrome c oxidase from Rhodobacter sphaeroides show that this channel is not important for proton conduction but reveal modulation of the properties of heme a. Biochemistry. 2000;39:2989–2996. doi: 10.1021/bi9924821. [DOI] [PubMed] [Google Scholar]
- 14.Svensson-Ek M, et al. The X-ray Crystal Structures of Wild-Type and EQ(I-286) Mutant Cytochrome c Oxidases from Rhodobacter sphaeroides. J. Mol. Biol. 2002;321:329–339. doi: 10.1016/S0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 15.Kadenbach B. Regulation of Mammalian 13-Subunit Cytochrome c Oxidase and Binding of other Proteins: Role of NDUFA4. Trends in Endocrinology and Metabolism. 2017;28:761–770. doi: 10.1016/j.tem.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Maréchal A, Meunier B, Lee D, Orengo C, Rich PR. Yeast cytochrome c oxidase: A model system to study mitochondrial forms of the haem-copper oxidase superfamily. Biochimica et Biophysica Acta - Bioenergetics. 2012;1817:620–628. doi: 10.1016/j.bbabio.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartley AM, et al. Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc 1. Nature Structural and Molecular Biology. 2019;26:78–83. doi: 10.1038/s41594-018-0172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strecker V, et al. Supercomplex-associated Cox26 protein binds to cytochrome c oxidase. Biochimica et Biophysica Acta - Molecular Cell Research. 2016;1863:1643–1652. doi: 10.1016/j.bbamcr.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levchenko M, et al. Cox26 is a novel stoichiometric subunit of the yeast cytochrome c oxidase. Biochimica et Biophysica Acta - Molecular Cell Research. 2016;1863:1624–1632. doi: 10.1016/j.bbamcr.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Rathore S, et al. Cryo-EM structure of the yeast respiratory supercomplex. Nature Structural and Molecular Biology. 2019;26:50–57. doi: 10.1038/s41594-018-0169-7. [DOI] [PubMed] [Google Scholar]
- 21.Meunier B, Maréchal A, Rich PR. Construction of histidine-tagged yeast mitochondrial cytochrome c oxidase for facile purification of mutant forms. Biochemical Journal. 2012;444:199–204. doi: 10.1042/BJ20120116. [DOI] [PubMed] [Google Scholar]
- 22.Maréchal A, Meunier B, Rich PR. Assignment of the CO-sensitive carboxyl group in mitochondrial forms of cytochrome c oxidase using yeast mutants. Biochimica et Biophysica Acta - Bioenergetics. 2012;1817:1921–1924. doi: 10.1016/j.bbabio.2012.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Namslauer A, Brzezinski P. Structural elements involved in electron-coupled proton transfer in cytochrome c oxidase. FEBS Lett. 2004;567:103–110. doi: 10.1016/j.febslet.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 24.Oliveberg M, Brzezinski P, Malmström BG. The effect of pH and temperature on the reaction of fully reduced and mixed-valence cytochrome c oxidase with dioxygen. Biochim. Biophys. Acta. 1989;977:322–328. doi: 10.1016/S0005-2728(89)80087-8. [DOI] [PubMed] [Google Scholar]
- 25.Han D, et al. Replacing Asn207 by Aspartate at the Neck of the D Channel in the aa3-Type Cytochrome c Oxidase from Rhodobacter sphaeroides Results in Decoupling the Proton Pump. Biochemistry. 2006;45:14064–14074. doi: 10.1021/bi061465q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Namslauer A, Aagaard A, Katsonouri A, Brzezinski P. Intramolecular proton-transfer reactions in a membrane-bound proton pump: the effect of pH on the peroxy to ferryl transition in cytochrome c oxidase. Biochemsitry. 2003;42:1488–1498. doi: 10.1021/bi026524o. [DOI] [PubMed] [Google Scholar]
- 27.Aagaard A, Brzezinski P. Zinc ions inhibit oxidation of cytochrome c oxidase by oxygen. FEBS Lett. 2001;494:157–160. doi: 10.1016/S0014-5793(01)02299-2. [DOI] [PubMed] [Google Scholar]
- 28.Brzezinski P, Johansson AL. Variable proton-pumping stoichiometry in structural variants of cytochrome c oxidase. Biochimica et Biophysica Acta - Bioenergetics. 2010;1797:710–723. doi: 10.1016/j.bbabio.2010.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Pawate AS, et al. A mutation in subunit I of cytochrome oxidase from Rhodobacter sphaeroides results in an increase in steady-state activity but completely eliminates proton pumping. Biochemistry. 2002;41:13417–13423. doi: 10.1021/bi026582+. [DOI] [PubMed] [Google Scholar]
- 30.Ädelroth P, Svensson EM, Mitchell DM, Gennis RB, Brzezinski P. Glutamate 286 in cytochrome aa3 from Rhodobacter sphaeroides is involved in proton uptake during the reaction of the fully-reduced enzyme with dioxygen. Biochemistry. 1997;36:13824–13829. doi: 10.1021/bi9629079. [DOI] [PubMed] [Google Scholar]
- 31.Maréchal A, Hartley AM, Warelow TP, Meunier B, Rich PR. Comparison of redox and ligand binding behaviour of yeast and bovine cytochrome c oxidases using FTIR spectroscopy. Biochimica et Biophysica Acta - Bioenergetics. 2018;1859:705–711. doi: 10.1016/j.bbabio.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Näsvik Öjemyr L, et al. Reaction of wild-type and Glu243Asp variant yeast cytochrome c oxidase with O2. Biochimica et Biophysica Acta - Bioenergetics. 2014;1837:1012–1018. doi: 10.1016/j.bbabio.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Vilhjálmsdóttir, J., Johansson, A. L. & Brzezinski, P. Structural Changes and Proton Transfer in Cytochrome c Oxidase. Scientific Reports, 5, 10.1038/srep12047 (2015). [DOI] [PMC free article] [PubMed]
- 34.Keen NT, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell DM, Gennis RB. Rapid purification of wildtype and mutant cytochrome c oxidase from Rhodobacter sphaeroides by Ni(2+)-NTA affinity chromatography. FEBS Lett. 1995;368:148–150. doi: 10.1016/0014-5793(95)00626-K. [DOI] [PubMed] [Google Scholar]
- 36.Zhen Y, et al. Overexpression and purification of cytochrome coxidase from Rhodobacter sphaeroides. Protein Expression and Purification. Protein Expr. Purif. 1998;13:326–336. doi: 10.1006/prep.1998.0903. [DOI] [PubMed] [Google Scholar]
- 37.Aagaard A, Namslauer A, Brzezinski P. Inhibition of proton transfer in cytochrome c oxidase by zinc ions: delayed proton uptake during oxygen reduction. Biochim. Biophys. Acta. 2002;1555:133–139. doi: 10.1016/S0005-2728(02)00268-2. [DOI] [PubMed] [Google Scholar]
- 38.Gilderson G, Aagaard A, Brzezinski P. Relocation of an internal proton donor in cytochrome c oxidase results in an altered pK(a) and a non-integer pumping stoichiometry. Biophys. Chem. 2002;98:105–114. doi: 10.1016/S0301-4622(02)00088-1. [DOI] [PubMed] [Google Scholar]


