Abstract
The fate of Leishmania infection can be strongly influenced by the host genetic background. In this work, we describe gene expression modulation of the immune system based on dual global transcriptome profiles of bone marrow-derived macrophages (BMDMs) from BALB/c and C57BL/6 mice infected with Leishmania amazonensis. A total of 12,641 host transcripts were identified according to the alignment to the Mus musculus genome. Differentially expressed genes (DEGs) profiling revealed a differential modulation of the basal genetic background between the two hosts independent of L. amazonensis infection. In addition, in response to early L. amazonensis infection, 10 genes were modulated in infected BALB/c vs. non-infected BALB/c macrophages; and 127 genes were modulated in infected C57BL/6 vs. non-infected C57BL/6 macrophages. These modulated genes appeared to be related to the main immune response processes, such as recognition, antigen presentation, costimulation and proliferation. The distinct gene expression was correlated with the susceptibility and resistance to infection of each host. Furthermore, upon comparing the DEGs in BMDMs vs. peritoneal macrophages, we observed no differences in the gene expression patterns of Jun, Fcgr1 and Il1b, suggesting a similar activation trends of transcription factor binding, recognition and phagocytosis, as well as the proinflammatory cytokine production in response to early L. amazonensis infection. Analysis of the DEG profile of the parasite revealed only one DEG among the 8,282 transcripts, indicating that parasite gene expression in early infection does not depend on the host genetic background.
Subject terms: Parasitic infection, Transcriptomics
Introduction
Leishmania is a protozoan parasite and the causative agent of several clinical infections, generically known as leishmaniases. In general, these infections are characterized by cutaneous, mucosal or visceral manifestations1,2. Leishmaniases are considered neglected tropical diseases by the World Health Organization. There is no vaccine available to prevent the disease due to a range of factors, such as diversity among Leishmania species and the interaction of these parasites with the host immune system3–6. Treatment can be complicated since most of the drugs available are expensive and toxic and may require long treatment regimens7,8. Furthermore, resistance to several commonly used drugs has been reported9. In humans, L. amazonensis infection can cause chronic cutaneous lesions, although diffuse cutaneous and visceral manifestations have been reported1,7.
The immune response to Leishmania involves a complex range of cells. Neutrophils and monocytes are first recruited to the site of the insect bite, which leads to the differentiation of macrophages; this differentiation is followed by the recognition and phagocytosis of the parasite, as well as the induction of a range of inflammatory signals. Other phagocytes, such as dendritic cells, also play important roles since they induce the response in other inflammatory tissues. However, macrophages that play a critical roles in the establishment of infection, as they are the main host cells for Leishmania replication inside the phagolysosome10–13. The infection is characterized by Th1 cell-mediated production of interferon gamma (IFN-γ), tumor necrosis factor alpha (TNF-α) and granulocyte macrophage colony-stimulating factor (GM-CSF), which polarizes macrophages to the proinflammatory M1 phenotype and increases nitric oxide synthase 2 (NOS2) and nitric oxide (NO) levels, resulting in parasite control, or by Th2 cell-mediated production of interleukin (IL) 4 (IL4), IL13, IL10, tumor growth factor beta (TGFβ) and macrophage colony-stimulating factor (M-CSF), which polarizes macrophages to an anti-inflammatory M2 phenotype and increases arginase 1 and polyamine production, resulting in parasite replication3,14–17. However, the parasite is able to subvert macrophage killing mechanisms through the modification of host cytokine expression, preventing antigen display by MHC class II molecules and reducing NO production with consequent amastigote differentiation and proliferation11,12.
Leishmania infection in murine models has been extensively characterized and varies according to the parasite species and host genetic background3,18–22. Progressive disease occurs due to impaired cellular immunity, with dysfunction of T cells, macrophages, or both23. Regulation of the host immune response to Leishmania has been well defined in L. major model in which the BALB/c mouse strain is susceptible to infection due to early bursts of IL4 that lead to disease progression. On the other hand, the C57BL/6 mouse strain is resistant to infection due to a dominant Th1-type response leading to infection control13,18–20,24. Experimental murine infections with L. amazonensis have demonstrated distinct susceptibilities compared to those for L. major25,26. L. amazonensis induces severe lesions upon cutaneous inoculation in susceptible BALB/c mice, while the same parasite causes only moderate lesions in resistant C57BL/6 mice21,27. Such variations in infection have been observed as differences in the lesion size, parasite burden, cellular activation and Th1/Th2 ratio between the different infected strains21,25,28.
Furthermore, studies involving knockout mouse strains have revealed interesting data concerning the response of the host to Leishmania infection. Targeted deletion of the Il4 and Il10 genes results in a minimal effects on the development of L. amazonensis29 and L. major infections30, due to reduced IL12 receptor expression, which leads to reduced IL12 responsiveness and, consequently, to impairment of the Th1 response31. In Tlr4- and MyD88- deficient mice, L. amazonensis shows increased in vitro infectivity; in contrast Tlr2-deficient mice exhibited a decreased parasite loads, indicating that this receptor is required for disease progression32.
Based on these findings, we analyzed the modulation of the early immune responses defined by the dual transcriptome profiles of BMDMs from the BALB/c and C57BL/6 mouse strains after infection with L. amazonensis for 4 h. Previous transcriptomic data have revealed novel information about the coordinated response of Leishmania-infected macrophages33–36 and about parasite biology, physiology and gene expression modulation37–42. In this work, we identified a total of 12,641 total mouse transcripts, and analyses of the DEGs profile involved in immune response modulation confirmed the existence of differences between these two hosts that can regulate susceptibility and resistance to L. amazonensis infection. Interestingly, the parasite transcriptome profile showed only one DEG, a noncoding RNA, indicating that the parasite presents no modulation of gene expression in early infection regardless of the host genetic background.
Results
BMDMs from BALB/c mice exhibited a lower infection index than those from C57BL/6 mice at 4 h after infection
BMDMs from the BALB/c and C57BL/6 mouse strains were infected with L. amazonensis (MOI 5:1), and the infection index was analyzed at 4 h after infection. First, no significant differences were observed in the infection rate or the number of intracellular parasites per infected macrophage (Fig. S1A,B). However, the infection index was significantly lower in infected BALB/c than in infected C57BL/6 macrophages (Fig. S1C).
Host transcriptome profiling revealed greater gene expression modulation in BMDMs from C57BL/6 mice than in BALB/c mice in response to L. amazonensis infection
Transcriptomic analyses were performed on five independent biological replicates per analysis of BMDMs from BALB/c and C57BL/6 mice infected or not infected with L. amazonensis for 4 h, using Illumina NovaSeq. 6000 sequencing, which generated millions of reads. The sequencing data are available in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession numbers PRJNA481041 and PRJNA481042 and in the Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRP156183 and SRP156466. The RNA-seq data were aligned to the M. musculus reference genome, and 12,641 transcripts were identified (Table S1).
Analysis of DEGs with a statistical significance threshold of a fold change ≥ 2 and a p-value < 0.05 revealed differential basal backgrounds in non-infected BALB/c vs. non-infected C57BL/6 macrophages; specifically, 313 genes were upregulated, and 254 genes were downregulated. Comparison of BALB/c_La vs. BALB/c macrophages revealed only 20 upregulated genes and 2 downregulated genes. In contrast, comparison of C57BL/6_La vs. C57BL/6 macrophages revealed 358 upregulated genes and 139 downregulated genes, and comparison of BALB/c_La vs. C57BL/6_La macrophages revealed 318 upregulated genes and 434 downregulated genes (Fig. 1).
Figure 1.
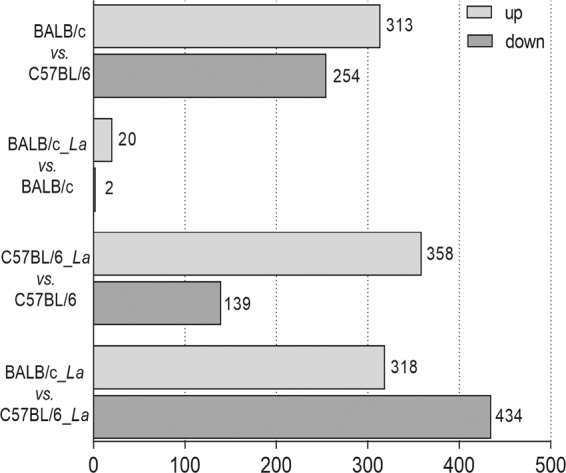
Transcriptome profiles of BMDMs from BALB/c and C57BL/6 mice infected with L. amazonensis. Differential gene expression profiles of BMDMs from BALB/c and C57BL/6 mice infected with L. amazonensis, presented as the numbers of upregulated (light gray) and downregulated (dark gray) transcripts in the following comparisons: non-infected BALB/c vs. non-infected C57BL/6 macrophages; infected BALB/c vs. non-infected BALB/c macrophages; infected C57BL/6 vs. non-infected C57BL/6 macrophages; and infected BALB/c vs. infected C57BL/6 macrophages. The data are from five independent biological replicates, considering a fold change ≥ 2 and a p-value < 0.05. La, L. amazonensis.
In addition, we generated volcano plots comparing the fold changes in expression (log2) with the corresponding adjusted p-values (-log10) (Fig. S2A) and volume plots comparing the fold changes in expression (log2) with the volumes (Fig. S2B). Based on these results, we identified the five most highly modulated transcripts among the comparisons (Table S2). Functional annotation and gene enrichment analyses were performed using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. KEGG enrichment analysis showed the 20 most differentially regulated pathways among the samples (Fig. S2C).
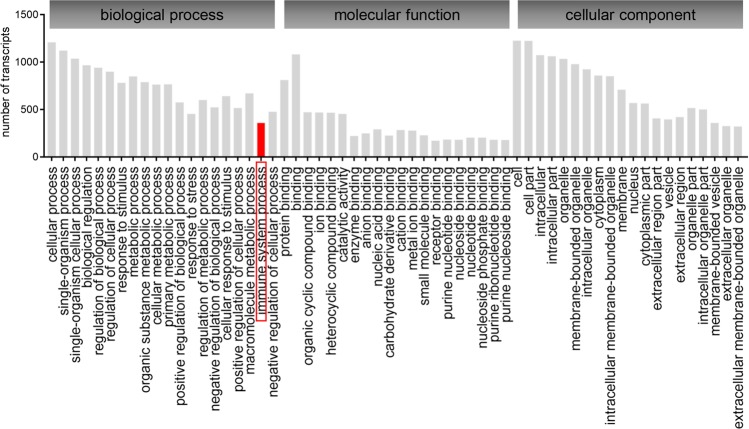
RNA-seq generates a large amount of information that can be analyzed from various perspectives. According to GO enrichment analysis of the DEGs, the most modulated subcategories were associated with biological processes, molecular functions and cellular components (Fig. 2). In this work, we focused on the immune system process term, comprising 361 modulated transcripts (Table S3), to elucidate how the host genetic background differences can define the fate of L. amazonensis infection.
Figure 2.
GO enrichment analysis of DEGs in BMDMs from BALB/c and C57BL/6 mice in response to L. amazonensis infection. The GO enrichment analysis results are presented as numbers of transcripts distributed in three main categories: biological process, molecular function and cellular component. Immune system processes were the focus of this work.
The gene expression modulation patterns revealed higher immune response activation in BMDMs from C57BL/6 mice than in BALB/c mice in response to L. amazonensis infection
Among the 361 modulated transcripts related to immune system processes, 150 of them appeared to differ in expression in non-infected BALB/c vs. C57BL/6 macrophages, indicating the existence of differential basal gene expression in these two host backgrounds, independent of L. amazonensis infection. After L. amazonensis infection, we observed only 10 modulated genes in BALB/c_La vs. BALB/c macrophages; 127 modulated genes in C57BL/6_La vs. C57BL/6 macrophages; and 221 modulated genes in C57BL/6_La vs. BALB/c_La macrophages (Table 1).
Table 1.
Profile of DEGs involved in immune system processes in BMDMs from BALB/c and C57BL/6 in response to L. amazonensis infection.
| comparison | downregulated genes | upregulated genes | p-value | FDR |
|---|---|---|---|---|
| BALB/c vs. C57BL/6 | Adgre1, AF251705, Ang, Apobec3, Asb2, Batf3, Blnk, Bst2, C1qa, C1qb, C1qc, C5ar2, Camp, Ccl5, Ccr2, Cd300a, Cd4, Cd40, Cd79b, Clec1b, Clec2d, Clec4a2, Ctsh, Cxcl10, Cxcl9, Emr1, Erbb2ip, Fcgr4, Fcna, Gbp2, Gbp3, Gbp5, Gbp7, Hfe, Ifit1bl1, Iigp1, Il15, Il18bp, Irf1, Irgm2, Itga4, Itgad, Itgal, Lcn2, Lgals1, Ly86, Marco, Mertk, Mill2, Pde4b, Pnp, Prdm1, S100a8, Samhd1, Skil, Slamf7, Slc11a1, Slc40a1, Smad6, Tfrc, Tgtp1, Tlr8, Tmem176a, Tmem176b, Trim34a, Vcam1, Vegfa, Vsig4, Wwp1 | Ada, Ahcy, Alpk1, Armc6, Batf, Bcl2a1a, Bcl2a1d, Bst1, Ccl2, Ccl24, Ccl3, Ccl4, Ccl7, Ccnb2, Ccr1, Cd109, Cd14, Cd24a, Cd300lf, Cd86, Cdk6, Clec4n, Clec5a, Col3a1, Colec12, Csf1, Ctse, Cx3cr1, Cxcl14, Fam20c, Glo1, Gm8909, Gpr183, H2-Ab1, H2-DMb1, H2-K1, H2-L, H2-Q1, H2-Q2, H2-Q4, H2-Q6, H2-Q8, H2-Q9, H2-T22, H2-T24, Hist1h2bf, Hist1h2bk, Hist1h2bl, Hist2h3c2, Hist1h3a, Hist1h3b, Hist1h3d, Hist1h3g, Hist1h3h, Hist1h3i, Hist1h4a, Hist1h4f, Hist1h4i, Hist4h4, Ifitm1, Ifitm3, Il1rn, Irf7, Kdr, Lat2, Malt1, Mmp14, Myc, Ndrg1, Npy, Oasl1, Pla2g7, Ripk3, Serpine1, Slpi, Spn, Spp1, Stap1, Tnfsf13, Tnfsf8, Top2a | 2.88e−79 | 1.00e−76 |
| BALB/c_La vs. BALB/c | Il1b, Mef2c | Cxcl1, Cxcl2, Cxcl3, Hilpda, Id2, Irg1, Smad6, Tnfrsf26 | 2.43e−8 | 2.24e−6 |
| C57BL/6_La vs. C57BL/6 | Ccr2, Ccr5, Fcgr1, Foxo3, Gcnt1, Gpr183, Hhex, Hist1h2bc, Hist1h2be, Hist1h2bg, Hist1h3e, Hist1h4c, Hist1h4d, Hist1h4h, Hist1h4m, Hist2h3b, Hist2h4, Il16, Lyl1, Mafb, Mapk14, Mef2c, Mertk, Mtus1, Pik3cd, Rassf2, Themis2, Tlr8, Tnfaip8l2, Trim14, Tsc22d3, Zfp36l1, Zfp36l2 | Adora2b, Ampd3, Batf, Bcl2a1a, Bcl2a1d, Bcl3, Birc3, Ccl3, Ccl4, Cd24a, Cd274, Cd40, Cd83, Cd86, Cdkn1a, Cdkn2b, Cebpb, Clec4d, Clec4e, Clec5a, Cxcl1, Cxcl2, Cxcl3, Ednrb, Ezr, Fam20c, Fas, Gadd45g, Gbp5, Gch1, Gpr68, H2-M2, Hcar2, Hilpda, Hmox1, Hsp90aa1, Hyal2, Icam1, Icosl, Id2, Il17ra, Il1rn, Il27, Irak2, Irf1, Irg1, Jag1, Jun, Lcp2, Lilrb4a, Malt1, Mb21d1, Mefv, Mmp14, Nck1, Nfe2l2, Nfkb1, Nfkb2, Nfkbia, Nlrp3, Nod2, Nr1h3, Olr1, Osm, Pde4b, Pmaip1, Ppp4r2, Prdx1, Procr, Ptafr, Rbpj, Rgcc, Ripk2, Rnf19b, Samsn1, Serpine1, Sh2b2, Slamf7, Slc11a2, Smad6, Sod2, Sqstm1,Src, Stx11, Tiparp, Tlr2, Tnf, Tnfaip3, Tnfrsf26, Tnfsf9, Tnip1, Tnip3, Traf3, Trim13 | 1.40e−65 | 2.37e−63 |
| C57BL/6_La vs. BALB/c_La | Adgre1, Ampd3, Ang, Apobec3, Axl, Bcl3, Bcl6, Birc3, Blnk, Bloc1s6, C1qa, C1qb, C1qc, C5ar1, Camp, Casp1, Ccl3, Ccl4, Ccl5, Ccl9, Ccr2, Ccr3, Cd274, Cd38, Cd4, Cd40, Cd79b, Cd83, Cdkn1a, Cebpb, Clec1b, Clec2d, Clec2i, Clec4e, Cnr2, Ctsh, Cxcl1, Cxcl10, Cxcl2, Cxcl3, Cxcl9, Ednrb, Fas, Fcgr4, Fcna, Fyb, Fzd7, Gbp2, Gbp3, Gbp5, Gbp6, Gbp7, Gpr68, H2-Ab1, H2-DMb1, H2-K1, H2-L, H2-M2, Hmox1, Icam1, Icosl, Iigp1, Il10, Il17ra, Il18bp, Il1a, Il1f9, Il27, Irak2, Irak3, Irf1, Irg1, Itgad, Itgal, Jag1, Kdr, Lcn2, Lcp2, Malt1, Mapkapk2, Marco, Mefv, Mertk, Mill2, Nfkb1, Nfkb2, Nfkbia, Nfkbid, Nlrc4, Nlrp3, Nod1, Nr1h3, Olr1, Pde4b, Pmaip1, Pnp, Ppp4r2, Prdm1, Procr, Ptafr, Ptprj, Rab32, Rela, Relb, Rgcc, Ripk2, Rnf19b, S100a8, Sh2b2, Skil, Slamf7, Slc40a1, Smad6, Snx10, Sod2, Sqstm1, Stx11, Tapbpl, Tbk1, Tgtp1, Thbs1, Tlr1, Tlr2, Tmem176a, Tmem176b, Tnf, Tnfaip3, Tnfrsf1b, Tnfsf9, Tnip3, Traf3, Treml4, Trib1, Trim13, Vcam1, Vegfa, Vsig4 | Ada, Ahcy,Aim2, Alpk1, Armc6, Bst1, Ccl2, Ccl24, Ccl7, Ccnb2, Cd109, Cd300lf, Cfb, Clec4n, Csf1, Colec12, Col3a1, Ctse, Cxcl14, Gcnt1, Glo1, Gm8909, Gpr183, H2-Q1, H2-Q2, H2-Q6, H2-Q8, H2-Q9, H2-T22, H2-T24, Hhex, Hist1h2ba, Hist1h2be, Hist1h2bf, Hist1h2bg, Hist1h2bk, Hist1h2bl, Hist1h3a, Hist1h3b, Hist1h3d, Hist1h3e, Hist1h3h, Hist1h3i, Hist1h3g, Hist1h4a, Hist1h4b, Hist1h4d, Hist1h4f, Hist1h4h, Hist1h4i, Hist1h4j, Hist1h4k, Hist1h4m, Hist1h4n, Hist2h3b, Hist2h3c2, Hist2h4, Hist4h4, Ifitm1, Ifitm3, Il16, Il1rn, Irf4, Irf7, Junb, Lgals1, Mmp9, Ndrg1, Npy, Pdgfrb, Pla2g7, Ripk3, Serpine1, Slfn1, Slpi, Spn, Spp1, Tacc3, Tnfsf13, Tnfsf8, Top2a, | 2.02e−125 | 9.61e−123 |
Gene Ontology (GO) enrichment analysis and the profiling of differentially expressed genes (DEGs) involved in the immune system processes in bone marrow-derived macrophages (BMDMs) from BALB/c and C57BL/6 in response to L. amazonensis (La) infection. The analysis was based on p-values and false discovery rates (FDRs).
Furthermore, we categorized the identified molecules according to the main types of immune system processes in response to L. amazonensis infection (Fig. 3). In the BALB/c_La vs. BALB/c comparison, we found that most of the modulated transcripts were immunomodulatory (Il1b, Irg1 and Tnfrsf26) and chemokine signaling molecules (Cxcl1, Cxcl2 and Cxcl3) (Fig. 3 and Table 1). In the C57BL/6_La vs. C57BL/6 comparison, most of the modulated transcripts were immunomodulatory molecules (Clec4d, Clec4e, Clec5a, Il16, Il17ra, Il1rn, Il27, Irak2, Irg1, Lcp2 Mefv, Themis2, Tnf, Tnfaip3, Tnfaip8l2, Tnfrsf26, Tnfsf9, Tnip1 and Tnip3), transcription factors (Batf, Bcl3, Cebpb, Foxo3, Hhex, Id2, Irf1, Jun, Lyl1, Mafb, Nfkb1, Nfkb2, Nfkbia, Tiparp, Trim13, Trim14 and Tsc22d3), adaptor proteins (Malt1, Mef2c, Nck1, Nfe2l2, Nr1h3, Pik3cd, Procr, Ptafr, Rbpj, Rgcc, Sh2b2, Smad6 and Src) and members of recognition pathways (Birc3, Jag1, Lilrb4a, Mapk14, Mb21d1, Nlrp3, Nod2, Ripk2, Tlr2, Tlr8 and Traf3) (Fig. 3 and Table 1).
Figure 3.
Immune response analysis of DEGs in BMDMs from BALB/c and C57BL/6 mice in response to L. amazonensis infection. Pie chart of the modulated molecules involved in the immune response processes grouped into main immune signaling pathways.
The exclusive differential gene expression patterns in BMDMs from C57BL/6 mice appeared to be mostly related to proliferation signaling and transcription factor molecules
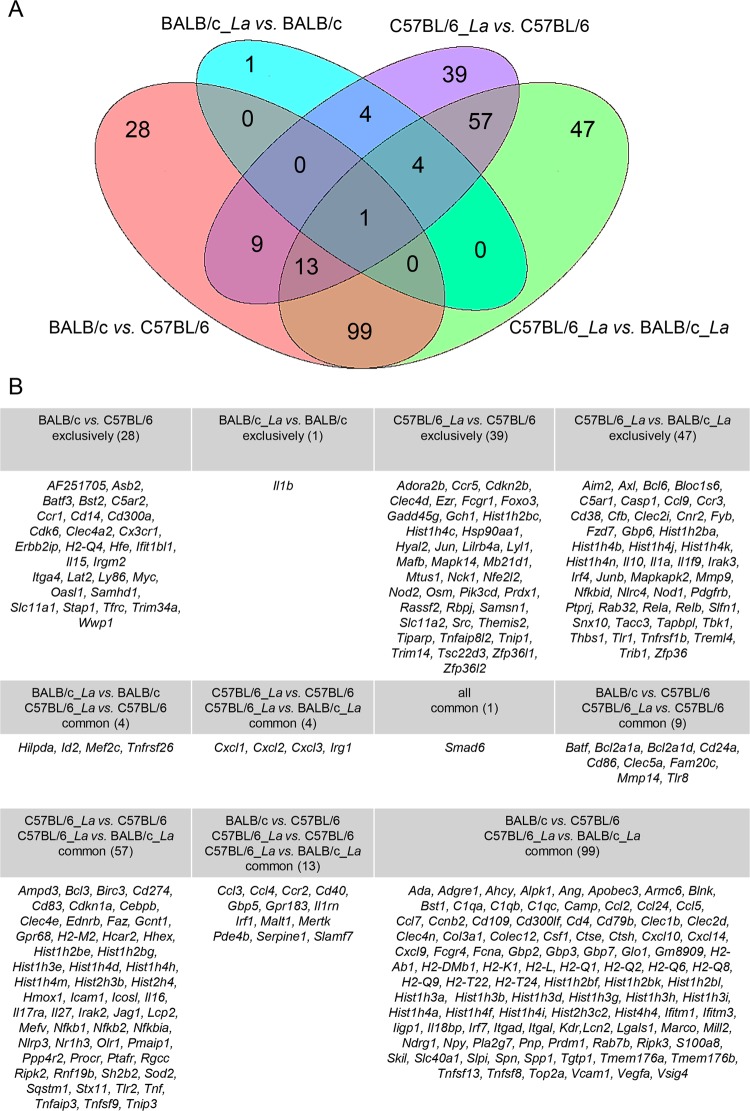
Venn diagram analysis was performed, and based on the results, we grouped the exclusively and commonly modulated genes involved in the immune response. We identified 28 exclusively modulated genes in the comparison of the two host backgrounds (in non-infected macrophages). Additionally, we identified only one exclusively modulated gene in BALB/c_La vs. BALB/c macrophages, 39 exclusively modulated genes in C57BL/6_La vs. C57BL/6 macrophages, and 47 exclusively modulated genes in C57BL/6_La vs. BALB/c_La macrophages. Interestingly, only one gene, Smad6, was common among all comparisons (Fig. 4A).
Figure 4.
Venn diagram analysis of DEGs in BMDMs from BALB/c and C57BL/6 mice in response to L. amazonensis infection. (A) Venn diagram of the 361 DEGs involved in the immune response processes, showing the numbers of exclusively and common genes for each comparison. (B) List of exclusively and common genes according for each comparison in the Venn diagram.
Examination of the immune response modulation associated with L. amazonensis infection and the pattern of exclusively expressed genes in the comparison of BALB/c_La vs. BALB/c macrophages revealed the downregulation of Il1b as unique (Fig. 4B). In contrast, comparison of C57BL/6_La vs. C57BL/6 macrophages revealed a set of 39 modulated genes, among which 22 were upregulated genes and 17 were downregulated. Most of these genes appeared to be involved in the proliferation pathway, such as the downregulated Pik3cd and Mtus1 genes and the upregulated Samsn1, Prdx1, Osm and Hsp90aa1 genes. Another group of genes contained transcription factors, including the downregulated Tsc22d3, Trim14, Mafb and Lyl1 genes and the upregulated Tiparp, Rbpj, Nfe2l2, Jun and Foxo3 genes. We also identified genes involved in recognition and costimulation pathways, as well as genes encoding adaptor molecules: Fcgr1, Mapk14 and Themis2 were downregulated, while Mb21d1/cGas, Nod2, Clec4d, Lilrb4a, Cdkn2b, Nck1, Src and Tnip1 were upregulated. Among apoptosis-related molecules, Rassf2 and Gadd45g appeared downregulated. The immunomodulatory molecules Tnfaip8l2 and Ccr5 were downregulated. The metal transporter Slc11a2 (formerly Nramp2) was upregulated. The histones Hist1h2bc and Hist1h4c, as well as the RNA-binding proteins Zfp36l1 and Zfp36l2, were downregulated. Poorly studied molecules, such as Adora2b, Ezr, Gch1 and Hyal2 were upregulated and were classified as belonging to other pathways (Fig. 5).
Figure 5.
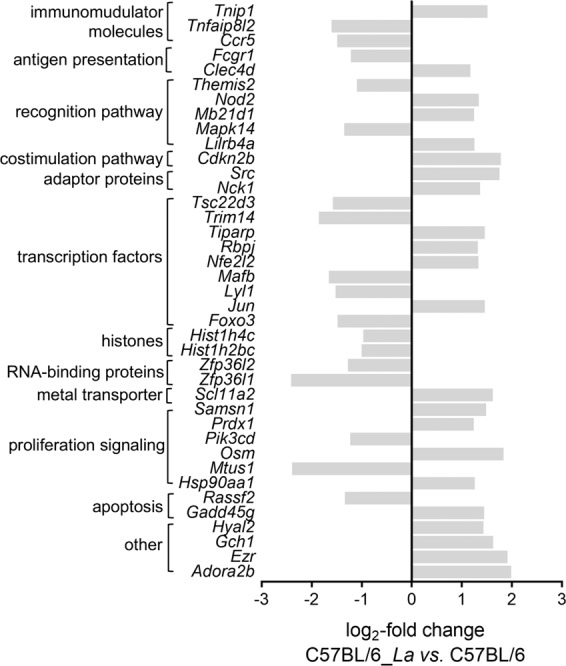
DEGs profile of the exclusively modulated genes involved in the immune response processes in infected C57BL/6 vs. non-infected C57BL/6 BMDMs. The profiles of DEGs are presented as the log2-fold changes in the expression of the 39 exclusively modulated genes involved in the immune response processes in BMDMs from C57BL/6 infected with L. amazonensis vs. non-infected C57BL/6 BMDMs. The genes were classified by their involvement in main immune response signaling pathways or by their identities as regulatory molecules of the immune response pathways. La, L. amazonensis.
RT-qPCR validation assays were performed on some of the most modulated molecules from the RNA-seq data: Il1b, Fcgr1, Ccr5, Smad6, Jun and Mapk14. Comparative analyses showed concordance between the RNA-seq and RT-qPCR data with no statistically significant differences, thus validating the RNA-seq results (Fig. 6).
Figure 6.
RT-qPCR validation of some modulated genes in BALB/c and C57BL/6 BMDMs in response to L. amazonensis infection. Comparative analysis of the relative expression levels of selected genes determined by RNA-seq and validated by RT-qPCR. The bars represent the mean ± SD values of the fold changes in Il1b, Fcgr1, Ccr5, Smad6, Jun and Mapk14 expression determined with five independent biological replicates analyzed in duplicate. The fold changes were calculated through relative quantification using the ΔΔCt method. The data were normalized to Gapdh expression and the relative gene expression was set to 1 for the control (non-infected) samples. Statistical analysis was performed using the t-tests, and no significant differences were observed (p-value < 0.05) between the RT-qPCR and RNA-seq results for the BALB/c_La and C57BL/6_La groups. The bars for Amastin-like (LmxM.33.0960) show the mean after normalization to Gapdh in L. amazonensis infecting BALB/c and L. amazonensis infecting C57BL/6 macrophages. La, L. amazonensis.
Similar to BMDMs, peritoneal macrophages were collected and infected with L. amazonensis, and the gene expression modulation of selected genes was analyzed by RT-qPCR to evaluate whether a similar trend occurred in another macrophage subtype. The infection index appeared significantly lower in C57BL/6_La macrophages than in BALB/c_La macrophages (Fig. S3A), indicating a distinct phenotypic difference between BMDMs and peritoneal macrophages in response to L. amazonensis infection. Comparison of the gene expression in BMDMs and peritoneal macrophage subtypes from BALB/c_La mice revealed lower expression of Smad6 and Mapk14. No modulation of Il1b, Ccr5, Fcgr1 or Jun was observed. On the other hand, we observed lower expression of Smad6, higher expression of Ccr5 and no modulation of Il1b, Mapk14, Fcgr1 and Jun expression in BMDMs compared with peritoneal macrophage subtype from C57BL/6_La (Fig. S3B).
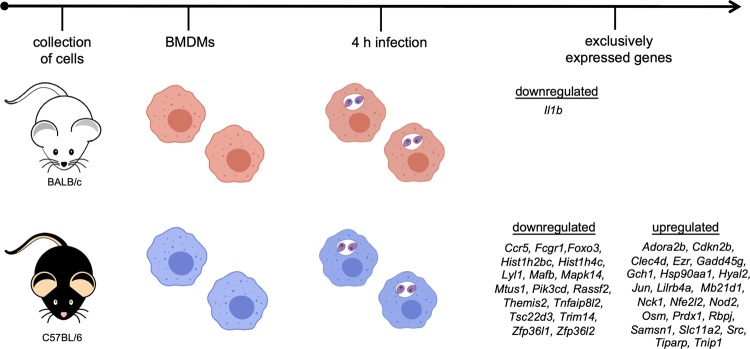
The transcriptomic data presented here corroborate the findings of previous studies on how differential genetic backgrounds from different hosts define susceptibility or resistance to Leishmania infection. The DEGs profiles described in this work represents new knowledge obtained from transcriptome analyses of immune responses between two different host genetic backgrounds. The analyses identified molecular markers that could be linked to susceptibility and resistance to L. amazonensis infection, as illustrated by the schematic representation of the exclusively and DEGs in BMDMs from BALB/c and C57BL/6 mice in response to early L. amazonensis infection (Fig. 7).
Figure 7.
Schematic representation of the exclusive genes and DEGs in BALB/c and C57BL/6 BMDMs in response to L. amazonensis infection. Summary of the data of the exclusive genes and DEGs in BMDMs derived from BALB/c and C657BL/6 mice in response to early L. amazonensis infection.
Parasite transcriptome profiling revealed only one DEG between L. amazonensis infecting BALB/c and L. amazonensis infecting C57BL/6 macrophages
We also analyzed the gene expression of L. amazonensis via alignment to the L. mexicana genome database (Table S1). The sequencing data are available in the NCBI BioProject and SRA databases, as previously described.
After initial assembly, 8,282 parasite transcripts were identified. Analysis of DEGs with significant threshold of a fold change ≥ 2 and a p-value < 0.05, as statistically significant, revealed only one DEG, a noncoding RNA (ncRNA) (LmxM.32.ncRNA:rfamscan:912871–912976), which showed higher expression in infected BALB/c than in infected C57BL/6 macrophages.
Additionally, we performed RT-qPCR validation assays of our RNA-seq data for Amastin-like gene (LmxM.33.0960). Similar to the case for the host comparative analyses, we observed concordance between the RNA-seq and RT-qPCR data (Fig. 6), thus validating the RNA-seq results.
Finally, we observed lower expression of the Amastin-like gene (LmxM.33.0960) in peritoneal macrophages than in BMDMs from BALB/c and C57BL/6 mice (Fig. S3B).
Discussion
The Th1/Th2 paradigm correlating resistance/susceptibility to Leishmania infection has been extensively studied3,14,18–21,23,35,41. Identification of potential biomarkers for leishmaniases can be useful for different approaches, such as diagnosis, prognosis, disease progression monitoring, clinical intervention and host immune response characterization33,34,41,43–45. The host-parasite interaction depends on both host genetic backgrounds33,35,36,41 and the genetic complexity of Leishmania species39,40,46.
L. amazonensis infection elicits different immune responses than those previously described for L. major infection25,26,28,29,31,35. In this work, we present the global transcriptome profiles of BMDMs from BALB/c and C57BL/6 mice non-infected and infected with L. amazonensis, focusing on the modulation of the immune response. In the absence of L. amazonensis infection, we identified significantly different basal gene expression patterns between the two hosts, corroborating with previous findings47. Analysis of the immune response in early L. amazonensis infection revealed 361 modulated genes among the comparisons. Comparison of infected BALB/c to non-infected BALB/c BMDMs revealed low levels of gene expression modulation; this pattern could be related to limited immune response activation, leading to susceptibility of this host to L. amazonensis infection, as previously described21,48. The DEGs involved in immune response modulation comprised mostly immunomodulatory and chemokine signaling molecules, suggesting a link to the inflammation process. In contrast, we observed high levels of gene expression modulation in infected C57BL/6 compared to non-infected C57BL/6 BMDMs. This pattern could be related to increased immune response activation via augmentation of recognition processes and, consequently, activation of signaling cascades, leading to moderate resistance of this host to L. amazonensis infection. Different profiles associated with different host genetic backgrounds have previously been described as being due to different parasite burdens, inflammatory cell populations and cytokine production21,48.
The infection index of BMDMs from BALB/c mice appeared smaller than that of BMDMs from C57BL/6 mice after 4 h of infection. As the infection index represents the number of intracellular parasites multiplied by the percentage of infected macrophages, the biological impact of this difference indicates that at an early stage of infection, C57BL/6 macrophages exhibit greater phagocytosis, which in subsequent times of infections may enable control of parasite replication. Previous studies by our group have demonstrated increased infection index values in BALB/c macrophages after 24 and 48 h of infection; in contrast, the index values of C57BL/6 macrophages appeared to remain stable49,50. However, most gene expression modulation has been described to occur during early Leishmania infection33,38,41,50,51.
The fact that Il1b appeared to be downregulated and was an exclusively modulated gene involved in the immune response in infected BALB/c compared to non-infected BALB/c BMDMs corroborates the important role of this molecule in Leishmania infection. IL1β has previously been identified as an important signaling factor for host resistance to C57BL/6 infection, since this cytokine signals through IL1R and MyD88 to induce NOS2-mediated NO production, which is a major host defense mechanism against Leishmania52. Furthermore, polymorphisms in the Il1b gene are associated with the severity of the disease in patients infected with L. mexicana53. Given these findings, we reinforce the importance of this molecule in Leishmania infection in both hosts52–54.
The 39 exclusively modulated immune response-related genes in infected C57BL/6 compared to non-infected C57BL/6 BMDMs were associated with important signaling pathways, suggesting enhancement of immune response activation resulting in moderated resistance against L. amazonensis infection. The recognition signaling cascade included a large number of modulated molecules, highlighting the importance of the host genetic background in the initial steps of macrophage activation55. Among these molecules, NOD-like receptors play protective roles during Leishmania infection52,56,57. The upregulation of Nod2 in infected C57BL/6 compared to non-infected BMDMs indicates greater macrophage activation in C57BL/6 mice. NOD2 mediates the parasite-induced production of cytokines, such as IL-17 and IFN-γ production, in L. infantum and L. amazonensis infections, whereas NOD1 is not relevant to these infections56,57. Recognition signaling also involves MAPKs, which play important roles against parasitic infections58, driving the switch in macrophage activation from proinflammatory IL12 to anti-inflammatory IL10 cytokines59. Previous studies have demonstrated that signaling during L. amazonensis infection leads to the activation of MAPK1 and MAPK358. MAPK14 has been poorly studied in the context of Leishmania infection, although downregulation of Mapk14 has previously been described to occur in L. braziliensis and L. major infections41,60. Most molecules from the recognition pathway were upregulated, indicating the activation of the downstream steps in recognition signaling cascades.
Immunomodulatory molecules play important roles in macrophage activation and the induction of adaptive immune responses via cytokine production in response to Leishmania infection13,20,61. Among the main cytokines studied, TNF is a multipotent cytokine implicated in a wide range of immune responses occuring in response to many infections62,63. In particular, the TNF-related molecules Tnip1 and Tnfaip8l2 appeared upregulated and downregulated, respectively, indicating signaling cascade activation and repression to maintain immune homeostasis. Leishmania infection can also induce the expression of numerous chemokines26,51,64,65. This event could potentially benefit the parasite due the ability to repress the induction of proinflammatory cytokine expression66. The downregulation of Ccr5 in infected C57BL/6 vs. non-infected C57BL/6 BMDMs could be correlated with the fact that this receptor directs the Th1 immune response and is thus associated with inflammation, cell infiltration and the development of infectious disease67. Previous studies have demonstrated that CCR5 knockout mice exhibit increased resistance to L. major infection68.
Similarly, human macrophage infections with L. amazonensis, L. major and L. panamensis have been shown to elicit immune response modulation of TNF, NF-kB and NOD-like receptor signaling pathways, oxidative stress pathways and proliferation signaling pathways41,69.
The expression of proliferation signals and transcription factor-related molecules was highly modulated according to our data. There are limited descriptions of these molecules; however, they are known to control the expression of many genes required for the effective activation of the immune responses, such as transcriptional activators or repressors, as well as for FOXO transcriptional activity, NF-kB recruitment and Notch signaling70–73.
The release and activation of histones occur in response to stress, leading to Toll-like receptor binding and triggering the activation of multiple signaling pathways74. The downregulation of Hist1h4c and Hist1h2bc could be related to the negative modulation of transcription factors listed above that are involved in macrophage activation.
The metal transporter natural resistance-associated macrophage protein (Nramp) has been associated with resistance to intracellular pathogens due to enhanced NOS2 expression and NO production75,76. Point mutations in Nramp1 promote susceptibility to Leishmania infection by modulating iron acquisition from intracellular compartments76,77, starving pathogens of this essential nutrient and impacting parasite survival and replication78. Although Nramp2 shares a conserved structure and iron transport functions with Nramp1, its role in Leishmania infection has been poorly studied. The upregulation of Slc11a2 (formerly Nramp2) was upregulated in infected C57BL/6 macrophages could be correlated with increased NO production and resistance to L. amazonensis infection.
Apoptosis induced by Leishmania may permit successful infection through modulation of host immunity79. RASSF2 and GADD45G are involved in the regulation of growth and apoptotic processes. Consistent with these findings, we identified downregulation of Rassf2 and the upregulation of Gadd45g as important factors in the modulation of the host immunity in response to L. amazonensis infection.
Molecules not acting in any of the described pathways were classified as “other” due to their limited descriptions in Leishmania infection. Further studies and functional validation could implicate the role of these molecules in host immune modulation in response to L. amazonensis infection, but this study provides only a global transcriptomic view based on the profile of the DEGs involved in immune response modulation in the two different host genetic backgrounds.
Macrophages form a vast and diverse population with considerable plasticity to adapt to different tissues and change in response to environmental variations80–83. The differences between peritoneal macrophages and BMDMs are believed to arise from differential physiological conditions and organ specificity along with the heterogeneity of macrophages83,84. Thus, we compared BMDMs and peritoneal macrophages with regard to some of the modulated genes to reinforce our findings and provide a representation of the in vivo scenario. According to our results, the infection index appeared lower in peritoneal macrophages from C57BL/6 mice than in those from BALB/c mice, indicating a distinct phenotypic difference between the macrophage subtypes in response to early L. amazonensis infection and suggesting that BALB/c mice are more susceptible models than C57BL/6 mice, as previously described21,48. Analyses of gene expression have shown a similar gene expression profiles in the comparison of BMDMs and pre-existing populations, although some differences have also been reported, suggesting that tissue environments dictate the macrophage phenotype required to trigger an effective immune response80,81. In our comparisons we observed nondifferential and differential modulation patterns, indicating that some of the analyzed genes were involved in distinct signaling cascades that lead to a distinct network activity. Smad6 showed a lower gene expression pattern in peritoneal macrophages than in BMDMs in both BALB/c and C57BL/6 mice. Since Smad6 is a regulator of myeloid differentiation85, this expression pattern confirms the differences between the macrophage subtypes. There were no differences in the gene expression patterns of Jun, Fcgr1 and Il1b between macrophage subtypes, suggesting similar trends of activation of transcription factor binding, recognition, phagocytosis and proinflammatory cytokines production. Ccr5 showed high modulation only in peritoneal macrophages from infected C57BL/6 mice, indicating upregulation of this chemokine receptor in this macrophage subtype. Mapk14 showed low modulation only in peritoneal macrophages from infected BALB/c mice, indicating low activation of the cellular response cascade in this macrophage subtype.
Altogether, our findings indicate the need to be cautious in extrapolating findings to in vivo scenarios that may or may not differ from those observed in the present study, especially considering that other immune cells, such as monocytes, neutrophils and lymphocytes, migrates to local cutaneous lesions with Leishmania86–88. Both host and parasite genetic backgrounds also need to be considered in translational approaches to identify biomarkers for the prognosis determination and treatment of the leishmaniases.
Finally, the transcriptome profiling of the parasite revealed only one DEG between L. amazonensis infecting BALB/c macrophages and L. amazonensis infecting C57BL/6 macrophages, a noncoding RNA (LmxM.32.ncRNA:rfamscan:912871–912976). ncRNAs have several functions; for example, they mediate transcription by RNA polymerase II, polyadenylate 3´-ends, regulate transcript expression and are potentially associated with small ribonucleoprotein complexes89. Our observations indicate that during early infection, the parasite exhibits the same gene expression pattern regardless of the host genetic background.
Methods
Animals
Female BALB/c and C57BL/6 mice (6–8 weeks old) were obtained from the Animal Center of the Medical School of the University of São Paulo and were maintained at the Animal Center of the Department of Physiology of the Institute of Bioscience of the University of São Paulo with access to food and water ad libitum.
Leishmania culture
L. amazonensis (MHOM/BR/1973/M2269) was grown at 25 °C in M199 medium (Gibco, Grand Island NY, USA), pH 7.0, supplemented with L-glutamine, 10% heat-inactivated fetal bovine serum, 0.25% hemin, 40 mM NaHCO3, 100 μM adenine, 40 mM HEPES, 100 U/mL penicillin and 100 μg/mL streptomycin, as previously described37–39. The parasites were counted in a Neubauer chamber.
In vitro macrophage infections
BMDMs were obtained from the femurs of BALB/c and C57BL/6 mice through PBS washing, and the cells were collected by centrifugation at 500 x g for 10 min at 4 °C. Lysis of erythrocytes was performed with NH4Cl (145 mM) and Tris-base (200 mM), pH 7.0, followed by incubation on ice for 20 min. After lysis, the cells were washed with cold PBS, centrifuged at 500 x g for 10 min at 4 °C and incubated in RPMI 1640 medium supplemented with penicillin (100 U/mL), streptomycin (100 µg/mL), 2-mercaptoethanol (50 µM), L-glutamine (2 mM), sodium pyruvate (1 mM), 10% fetal bovine serum and 10% L929 conditioned medium as a macrophage stimulating factor source. The cells were differentiated for 7 days at 34 °C in 5% CO2. The BMDMs were used after phenotypic analysis by flow cytometry showed at least 95% F4/80 and CD11b-positive cells, as previously described50. After macrophage differentiation, cellular viability was evaluated with Trypan blue staining (1:1 (v:v)), and the cells were counted in a Neubauer chamber. Approximately 5 × 106 BMDMs from BALB/c and C57BL/6 mice were incubated in sterile 6-well plates (SPL Life Sciences, Korea) overnight at 34 °C in 5% CO2. Non-adherent cells were removed by washing with PBS, and infection was performed with L. amazonensis promastigotes in the stationary growth phase (MOI 5:1). After 4 h of infection, the cultures were washed with PBS; then, RNA was extracted, or the infection index was determinated. Non-infected macrophages maintained in culture under the same conditions were used as the controls. The infections were evaluated by determining the percentage of infected cells after counting 400 panoptic-stained (Laborclin, Parana, Brazil) macrophages. The infection index was determined by multiplying the percentage of infected macrophages by the mean number of intracellular parasites per infected cell90,91. Statistical analyses were performed using Student´s t-test and p-value < 0.05 was considered to indicate a significant difference between infected C57BL/6 macrophages or infected BALB/c macrophages and the corresponding non-infected macrophages.
Peritoneal macrophages were collected from BALB/c and C57BL/6 mice by injection and recovery of 5 mL of RPMI 1640 medium supplemented, as previously described. The cells were recovered by centrifugation at 500 × g for 10 min at 4 °C. Cellular viability was evaluated with Trypan blue staining (1:1 (v:v)), and the cells were counted in a Neubauer chamber. Approximately, 1 × 106 peritoneal macrophages were incubated in sterile 6-well plates (SPL Life Sciences, Korea) overnight at 34 °C in 5% CO2. Non-adherent cells were removed by washing with PBS, and infection was performed with L. amazonensis promastigotes in the stationary growth phase (MOI 5:1). After 4 h of infection, cultures were washed with PBS; then, RNA was extracted or the infection index was determined. Non-infected macrophages maintained in culture under the same conditions were used as the controls. The infections were evaluated as previously described for BMDMs.
Total RNA isolation and library construction
Total RNA was isolated from five independent biological replicates of each infected and non-infected group using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions and as previously described39. The RNA samples were treated with DNase I (1 U per µg of RNA) (Thermo Scientific, Lithuania, EU) at 37 °C for 1 h, and the RNA concentration was determined from the A260/A280 ratio using a NanoDrop ND1000 (Thermo Scientific, USA). In addition, RNA integrity was evaluated using an Agilent 2100 Bioanalyzer and a Pico Agilent RNA 6000 kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions. rRNA depletion was performed using a poly(A) magnetic bead capture protocol and a TrueSeq Stranded Total RNA Sample Prep kit (Illumina) according to the manufacturer´s instructions. Libraries were prepared using a TrueSeq Stranded RNA-seq Library Prep Kit (Illumina), according to the manufacturer’s instructions.
RNA-seq and data analysis
Paired end reads (100 bp) were obtained using an Illumina NovaSeq. 6000 platform at Macrogen Inc. (Seoul, South Korea). Quality control was performed on the sequenced raw reads based on the read quality, total bases, total reads, GC content (%) and basic statistics. The quality of the reads was analyzed using FastQC according to the Phred quality score92. Reads with Phred quality scores lower than 20 were discarded. To reduce bias in the analysis and artifacts, such as low-quality reads and adaptor sequences, Trimmomatic was used93. The trimmed reads were mapped to the reference genome L. mexicana reference genome (MHOMGT2001U1103) with genomic data obtained from TriTrypDB version 36 (www.tritryp.org) and to the M. musculus genome using the TopHat splice-aware aligner94,95. A maximum of two mismatches were allowed. The transcripts were assembled in Cufflinks through read alignment, providing information on the known transcripts. The expression profiles of the assembled transcripts and the abundance estimates for each sample were generated by Cufflinks96. The expression values were calculated as fragments per kilobase of transcript per million mapped reads (FPKM) and are represented as normalized values based on the transcript length and coverage depth97. Gene expression level values were calculated from the transcript counts. DEG analysis was performed for the following comparisons: (1) C57BL/6 vs. BALB/c, (2) BALB/c infected with L. amazonensis vs. BALB/c, (3) C57BL/6 infected with L. amazonensis vs. C57BL/6, and (4) C57BL/6 infected with L. amazonensis vs. BALB/c infected with L. amazonensis. Genes with FPKM values of zero were excluded. Groups under different conditions or with different DEGs were filtered out through statistical hypothesis tests. The false discovery rate (FDR) was controlled by adjusting the p-value using the Benjamini-Hochberg algorithm98. Functional annotation was performed using GO and KEGG analyses. All analyses were performed by Macrogen Inc. (Seoul, South Korea).
RT-qPCR validation
RT-qPCR validation assays were performed using total RNA isolated as previously described above from five biological replicates. Reverse transcription was performed using 2 µg of total RNA as a template, reverse transcriptase and random primers (RevertAid H Minus Reverse Transcriptase Kit, Thermo-Scientific, Canada), according to the manufacturer’s instructions. Equal amounts of cDNA were assessed in total volumes of 25 μL containing Maxima SYBR Green qPCR Master Mix (Thermo Scientific, Lithuania, EU) and primers (200 nM) (Table S4). The mixtures were incubated at 94 °C for 5 min, followed by 40 cycles at 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s. A negative control in the absence of reverse transcriptase was included in the RT-qPCR assays to detect DNA contamination in the RNA samples. The reactions were carried out using a PikoReal Real-time PCR System (Thermo Scientific, Finland). The reactions were performed in duplicate, and analyses were performed using PikoReal Software 2.2 (Thermo Scientific). The fold changes were calculated by relative quantification using the ΔΔCt method99. The data were normalized by Gapdh expression, and the relative gene expression was set to 1 for the control (non-infected) samples. The normalized absolute copy number of the amastin-like gene (LmxM.33.0960) was calculated based on the normalization to a reference, considering the molar mass concentration, according to a standard curve generated from a ten-fold dilution of a quantified PCR product. The normalized Amastin/Gapdh ratio of the absolute number of molecules was used as an expression parameter according to a standard curve generated from a ten-fold serial dilution of a quantified and linearized plasmid containing the target fragment.
Statistical analysis
The experiments were performed with five biological replicates per group and the results are presented as the means ± SDs. DEGs were considered statistically significant considering fold changes ≥ 2, p-value < 0.05 and FDR analysis. RT-qPCR validation assays were performed with five biological replicates, and the results are presented the means ± SDs. Statistical analysis was based on Student´s t-test with p-value < 0.05 indicating statistical significance.
Ethics statement
The experimental protocols for animals were approved by the Animal Care and Use Committee at the Institute of Bioscience of the University of São Paulo (CEUA 233/2015). This study was carried out in strict accordance with the recommended guidelines and the policies for the care and use of laboratory animals of São Paulo State (Lei Estadual 11.977, de 25/08/2005) and the Brazilian government (Lei Federal 11.794, de 08/10/2008).
Supplementary information
Acknowledgements
We would like to thank Juliane Cristina Ribeiro Fernandes and Stephanie Maia Acuña for their comments and suggestions.
Author contributions
Conceived and designed the experiments: J.I.A., S.M.M., L.M.F.W. Performed the experiments: J.I.A., S.M.M., R.A.Z. Analyzed the data: J.I.A., S.M.M., L.M.F.W. Contributed reagents/materials/analysis tools: J.I.A., K.E.M., A.H.N., L.M.F.W. Wrote the draft of the manuscript: J.I.A., S.M.M. and L.M.F.W. Revised the manuscript: J.I.A., S.M.M., R.A.Z., K.E.M., A.H.N., L.M.F.W.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Juliana Ide Aoki, Email: juaoki@usp.br.
Lucile Maria Floeter-Winter, Email: lucile@ib.usp.br.
Supplementary information
is available for this paper at 10.1038/s41598-019-56305-1.
References
- 1.Ashford RW. The leishmaniases as emerging and reemerging zoonoses. Int J Parasitol. 2000;30:1269–1281. doi: 10.1016/S0020-7519(00)00136-3. [DOI] [PubMed] [Google Scholar]
- 2.Burza S, Croft SL, Boelaert M. Leishmaniasis. Lancet. 2018;392:951–970. doi: 10.1016/S0140-6736(18)31204-2. [DOI] [PubMed] [Google Scholar]
- 3.Muxel SM, et al. Arginine and Polyamines Fate in Leishmania Infection. Front Microbiol. 2017;8:2682. doi: 10.3389/fmicb.2017.02682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khamesipour A, Rafati S, Davoudi N, Maboudi F, Modabber F. Leishmaniasis vaccine candidates for development: a global overview. Indian J Med Res. 2006;123:423–438. [PubMed] [Google Scholar]
- 5.Müller, K. E., Solberg, C. T., Aoki, J. I., Floeter-Winter, L. M. & Nerland, A. H. Developing a vaccine for leishmaniasis: how biology shapes policy. Tidsskr Nor Laegeforen137, 10.4045/tidsskr.17.0620 (2018). [DOI] [PubMed]
- 6.Kumar R, Engwerda C. Vaccines to prevent leishmaniasis. Clin Transl Immunology. 2014;3:e13. doi: 10.1038/cti.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366:1561–1577. doi: 10.1016/S0140-6736(05)67629-5. [DOI] [PubMed] [Google Scholar]
- 8.Hotez PJ, Bottazzi ME, Franco-Paredes C, Ault SK, Periago MR. The neglected tropical diseases of Latin America and the Caribbean: a review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl Trop Dis. 2008;2:e300. doi: 10.1371/journal.pntd.0000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croft SL, Sundar S, Fairlamb AH. Drug resistance in leishmaniasis. Clin Microbiol Rev. 2006;19:111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gregory DJ, Olivier M. Subversion of host cell signalling by the protozoan parasite Leishmania. Parasitology. 2005;130(Suppl):S27–35. doi: 10.1017/S0031182005008139. [DOI] [PubMed] [Google Scholar]
- 12.Rossi M, Fasel N. How to master the host immune system? Leishmania parasites have the solutions! Int Immunol. 2018;30:103–111. doi: 10.1093/intimm/dxx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogdan C, Röllinghoff M. The immune response to Leishmania: mechanisms of parasite control and evasion. Int J Parasitol. 1998;28:121–134. doi: 10.1016/S0020-7519(97)00169-0. [DOI] [PubMed] [Google Scholar]
- 14.Mantovani A, et al. The chemokine system in diverse forms of macrophage activation and polarization. Trends in immunology. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 15.Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Frontiers in immunology. 2014;5:614. doi: 10.3389/fimmu.2014.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 18.Von Stebut E, et al. Interleukin 1alpha promotes Th1 differentiation and inhibits disease progression in Leishmania major-susceptible BALB/c mice. J Exp Med. 2003;198:191–199. doi: 10.1084/jem.20030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Stebut E, Udey MC. Requirements for Th1-dependent immunity against infection with Leishmania major. Microbes Infect. 2004;6:1102–1109. doi: 10.1016/j.micinf.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 20.Alexander J, Brombacher F. T helper1/t helper2 cells and resistance/susceptibility to leishmania infection: is this paradigm still relevant? Front Immunol. 2012;3:80. doi: 10.3389/fimmu.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velasquez LG, et al. Distinct courses of infection with Leishmania (L.) amazonensis are observed in BALB/c, BALB/c nude and C57BL/6 mice. Parasitology. 2016;143:692–703. doi: 10.1017/S003118201600024X. [DOI] [PubMed] [Google Scholar]
- 22.Rosas LE, et al. Genetic background influences immune responses and disease outcome of cutaneous L. mexicana infection in mice. Int Immunol. 2005;17:1347–1357. doi: 10.1093/intimm/dxh313. [DOI] [PubMed] [Google Scholar]
- 23.Kong F, et al. Transcriptional Profiling in Experimental Visceral Leishmaniasis Reveals a Broad Splenic Inflammatory Environment that Conditions Macrophages toward a Disease-Promoting Phenotype. PLoS Pathog. 2017;13:e1006165. doi: 10.1371/journal.ppat.1006165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Himmelrich H, Parra-Lopez C, Tacchini-Cottier F, Louis JA, Launois P. The IL-4 rapidly produced in BALB/c mice after infection with Leishmania major down-regulates IL-12 receptor beta 2-chain expression on CD4+ T cells resulting in a state of unresponsiveness to IL-12. J Immunol. 1998;161:6156–6163. [PubMed] [Google Scholar]
- 25.Ji J, Sun J, Qi H, Soong L. Analysis of T helper cell responses during infection with Leishmania amazonensis. Am J Trop Med Hyg. 2002;66:338–345. doi: 10.4269/ajtmh.2002.66.338. [DOI] [PubMed] [Google Scholar]
- 26.Ji J, Sun J, Soong L. Impaired expression of inflammatory cytokines and chemokines at early stages of infection with Leishmania amazonensis. Infect Immun. 2003;71:4278–4288. doi: 10.1128/IAI.71.8.4278-4288.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felizardo TC, Toma LS, Borges NB, Lima GM, Abrahamsohn IA. Leishmania (Leishmania) amazonensis infection and dissemination in mice inoculated with stationary-phase or with purified metacyclic promastigotes. Parasitology. 2007;134:1699–1707. doi: 10.1017/S0031182007003186. [DOI] [PubMed] [Google Scholar]
- 28.Soong L, et al. Role of CD4+ T cells in pathogenesis associated with Leishmania amazonensis infection. J Immunol. 1997;158:5374–5383. [PubMed] [Google Scholar]
- 29.Jones DE, Ackermann MR, Wille U, Hunter CA, Scott P. Early enhanced Th1 response after Leishmania amazonensis infection of C57BL/6 interleukin-10-deficient mice does not lead to resolution of infection. Infect Immun. 2002;70:2151–2158. doi: 10.1128/IAI.70.4.2151-2158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kane MM, Mosser DM. The role of IL-10 in promoting disease progression in leishmaniasis. J Immunol. 2001;166:1141–1147. doi: 10.4049/jimmunol.166.2.1141. [DOI] [PubMed] [Google Scholar]
- 31.Jones DE, Buxbaum LU, Scott P. IL-4-independent inhibition of IL-12 responsiveness during Leishmania amazonensis infection. J Immunol. 2000;165:364–372. doi: 10.4049/jimmunol.165.1.364. [DOI] [PubMed] [Google Scholar]
- 32.Guerra CS, et al. Histopathological analysis of initial cellular response in TLR-2 deficient mice experimentally infected by Leishmania (L.) amazonensis. Int J Exp Pathol. 2010;91:451–459. doi: 10.1111/j.1365-2613.2010.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dillon LA, et al. Simultaneous transcriptional profiling of Leishmania major and its murine macrophage host cell reveals insights into host-pathogen interactions. BMC Genomics. 2015;16:1108. doi: 10.1186/s12864-015-2237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen SM, et al. Meta-transcriptome Profiling of the Human-Leishmania braziliensis Cutaneous Lesion. PLoS Negl Trop Dis. 2016;10:e0004992. doi: 10.1371/journal.pntd.0004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Probst CM, et al. A comparison of two distinct murine macrophage gene expression profiles in response to Leishmania amazonensis infection. BMC Microbiol. 2012;12:22. doi: 10.1186/1471-2180-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ontoria E, et al. Transcriptional Profiling of Immune-Related Genes In. Front Cell Infect Microbiol. 2018;8:197. doi: 10.3389/fcimb.2018.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acuña SM, et al. Arginase expression modulates nitric oxide production in Leishmania (Leishmania) amazonensis. PLoS One. 2017;12:e0187186. doi: 10.1371/journal.pone.0187186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aoki JI, et al. L-arginine availability and arginase activity: Characterization of amino acid permease 3 in Leishmania amazonensis. PLoS Negl Trop Dis. 2017;11:e0006025. doi: 10.1371/journal.pntd.0006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aoki JI, et al. RNA-seq transcriptional profiling of Leishmania amazonensis reveals an arginase-dependent gene expression regulation. PLoS Negl Trop Dis. 2017;11:e0006026. doi: 10.1371/journal.pntd.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rastrojo A, et al. The transcriptome of Leishmania major in the axenic promastigote stage: transcript annotation and relative expression levels by RNA-seq. BMC Genomics. 2013;14:223. doi: 10.1186/1471-2164-14-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernandes, M. C. et al. Dual Transcriptome Profiling of Leishmania-Infected Human Macrophages Reveals Distinct Reprogramming Signatures. MBio7, 10.1128/mBio.00027-16 (2016). [DOI] [PMC free article] [PubMed]
- 42.Fiebig M, Kelly S, Gluenz E. Comparative Life Cycle Transcriptomics Revises Leishmania mexicana Genome Annotation and Links a Chromosome Duplication with Parasitism of Vertebrates. PLoS Pathog. 2015;11:e1005186. doi: 10.1371/journal.ppat.1005186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veras PST, Ramos PIP, de Menezes JPB. In Search of Biomarkers for Pathogenesis and Control of Leishmaniasis by Global Analyses of. Front Cell Infect Microbiol. 2018;8:326. doi: 10.3389/fcimb.2018.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kip AE, et al. Systematic review of biomarkers to monitor therapeutic response in leishmaniasis. Antimicrob Agents Chemother. 2015;59:1–14. doi: 10.1128/AAC.04298-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bahrami F, Harandi AM, Rafati S. Biomarkers of Cutaneous Leishmaniasis. Front Cell Infect Microbiol. 2018;8:222. doi: 10.3389/fcimb.2018.00222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dillon LA, et al. Transcriptomic profiling of gene expression and RNA processing during Leishmania major differentiation. Nucleic Acids Res. 2015;43:6799–6813. doi: 10.1093/nar/gkv656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- 48.Paladi CS, et al. Treatment of Leishmania (Leishmania) amazonensis-infected mice with a combination of a Palladacycle complex and heat-killed Propionibacterium acnes triggers protective cellular immune response. Front Microbiol. 2017;8:333. doi: 10.3389/fmicb.2017.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandes JCR, et al. Melatonin and Leishmania amazonensis infection altered miR-294, miR-30e, and miR-302d i mpacting on Tnf, Mcp-1, and Nos2 e xpression. Front Cell Infect Microbiol. 2019;9:60. doi: 10.3389/fcimb.2019.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muxel SM, Laranjeira-Silva MF, Zampieri RA, Floeter-Winter LM. Leishmania (Leishmania) amazonensis induces macrophage miR-294 and miR-721 expression and modulates infection by targeting NOS2 and L-arginine metabolism. Sci Rep. 2017;7:44141. doi: 10.1038/srep44141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Matte C, Olivier M. Leishmania-induced cellular recruitment during the early inflammatory response: modulation of proinflammatory mediators. J Infect Dis. 2002;185:673–681. doi: 10.1086/339260. [DOI] [PubMed] [Google Scholar]
- 52.Lima-Junior DS, et al. Inflammasome-derived IL-1beta production induces nitric oxide-mediated resistance to Leishmania. Nature medicine. 2013;19:909–915. doi: 10.1038/nm.3221. [DOI] [PubMed] [Google Scholar]
- 53.Fernández-Figueroa EA, et al. Disease severity in patients infected with Leishmania mexicana relates to IL-1β. PLoS Negl Trop Dis. 2012;6:e1533. doi: 10.1371/journal.pntd.0001533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charmoy M, et al. The Nlrp3 inflammasome, IL-1β, and neutrophil recruitment are required for susceptibility to a nonhealing strain of Leishmania major in C57BL/6 mice. Eur J Immunol. 2016;46:897–911. doi: 10.1002/eji.201546015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu D, Uzonna JE. The early interaction of Leishmania with macrophages and dendritic cells and its influence on the host immune response. Front Cell Infect Microbiol. 2012;2:83. doi: 10.3389/fcimb.2012.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nascimento MS, et al. NOD2-RIP2-Mediated Signaling Helps Shape Adaptive Immunity in Visceral Leishmaniasis. The Journal of infectious diseases. 2016;214:1647–1657. doi: 10.1093/infdis/jiw446. [DOI] [PubMed] [Google Scholar]
- 57.Dos Santos JC, et al. The NOD2 receptor is crucial for immune responses towards New World Leishmania species. Sci Rep. 2017;7:15219. doi: 10.1038/s41598-017-15412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z, Mosser DM, Zhang X. Activation of the MAPK, ERK, following Leishmania amazonensis infection of macrophages. J Immunol. 2007;178:1077–1085. doi: 10.4049/jimmunol.178.2.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zambrano-Villa S, Rosales-Borjas D, Carrero JC, Ortiz-Ortiz L. How protozoan parasites evade the immune response. Trends in parasitology. 2002;18:272–278. doi: 10.1016/S1471-4922(02)02289-4. [DOI] [PubMed] [Google Scholar]
- 60.Sousa R, et al. Early Suppression of Macrophage Gene Expression by. Front Microbiol. 2018;9:2464. doi: 10.3389/fmicb.2018.02464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cunningham AC. Parasitic adaptive mechanisms in infection by leishmania. Exp Mol Pathol. 2002;72:132–141. doi: 10.1006/exmp.2002.2418. [DOI] [PubMed] [Google Scholar]
- 62.Murray HW, Jungbluth A, Ritter E, Montelibano C, Marino MW. Visceral leishmaniasis in mice devoid of tumor necrosis factor and response to treatment. Infect Immun. 2000;68:6289–6293. doi: 10.1128/IAI.68.11.6289-6293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roach DR, et al. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J Immunol. 2002;168:4620–4627. doi: 10.4049/jimmunol.168.9.4620. [DOI] [PubMed] [Google Scholar]
- 64.Racoosin EL, Beverley SM. Leishmania major: promastigotes induce expression of a subset of chemokine genes in murine macrophages. Exp Parasitol. 1997;85:283–295. doi: 10.1006/expr.1996.4139. [DOI] [PubMed] [Google Scholar]
- 65.Ritter U, et al. Differential expression of chemokines in patients with localized and diffuse cutaneous American leishmaniasis. J Infect Dis. 1996;173:699–709. doi: 10.1093/infdis/173.3.699. [DOI] [PubMed] [Google Scholar]
- 66.Teixeira MJ, Teixeira CR, Andrade BB, Barral-Netto M, Barral A. Chemokines in host-parasite interactions in leishmaniasis. Trends Parasitol. 2006;22:32–40. doi: 10.1016/j.pt.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 67.Oghumu S, Lezama-Dávila CM, Isaac-Márquez AP, Satoskar AR. Role of chemokines in regulation of immunity against leishmaniasis. Exp Parasitol. 2010;126:389–396. doi: 10.1016/j.exppara.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yurchenko E, et al. CCR5-dependent homing of naturally occurring CD4+ regulatory T cells to sites of Leishmania major infection favors pathogen persistence. J Exp Med. 2006;203:2451–2460. doi: 10.1084/jem.20060956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramírez C, et al. Human macrophage response to L. (Viannia) panamensis: microarray evidence for an early inflammatory response. PLoS Negl Trop Dis. 2012;6:e1866. doi: 10.1371/journal.pntd.0001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, Zhou Y, Graves DT. FOXO transcription factors: their clinical significance and regulation. Biomed Res Int. 2014;2014:925350. doi: 10.1155/2014/925350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta P, Srivastav S, Saha S, Das PK, Ukil A. Leishmania donovani inhibits macrophage apoptosis and pro-inflammatory response through AKT-mediated regulation of β-catenin and FOXO-1. Cell Death Differ. 2016;23:1815–1826. doi: 10.1038/cdd.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tu L, et al. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Auderset F, et al. Redundant Notch1 and Notch2 signaling is necessary for IFNγ secretion by T helper 1 cells during infection with Leishmania major. PLoS Pathog. 2012;8:e1002560. doi: 10.1371/journal.ppat.1002560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen R, Kang R, Fan XG, Tang D. Release and activity of histone in diseases. Cell Death Dis. 2014;5:e1370. doi: 10.1038/cddis.2014.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Canonne-Hergaux F, Gruenheid S, Govoni G, Gros P. The Nramp1 protein and its role in resistance to infection and macrophage function. Proc Assoc Am Physicians. 1999;111:283–289. doi: 10.1046/j.1525-1381.1999.99236.x. [DOI] [PubMed] [Google Scholar]
- 76.Blackwell JM, et al. SLC11A1 (formerly NRAMP1) and disease resistance. Cell Microbiol. 2001;3:773–784. doi: 10.1046/j.1462-5822.2001.00150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mittra B, et al. Iron uptake controls the generation of Leishmania infective forms through regulation of ROS levels. J Exp Med. 2013;210:401–416. doi: 10.1084/jem.20121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huynh C, Sacks DL, Andrews NW. A Leishmania amazonensis ZIP family iron transporter is essential for parasite replication within macrophage phagolysosomes. J Exp Med. 2006;203:2363–2375. doi: 10.1084/jem.20060559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lüder CG, Campos-Salinas J, Gonzalez-Rey E, van Zandbergen G. Impact of protozoan cell death on parasite-host interactions and pathogenesis. Parasit Vectors. 2010;3:116. doi: 10.1186/1756-3305-3-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. 2016;17:34–40. doi: 10.1038/ni.3324. [DOI] [PubMed] [Google Scholar]
- 81.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 82.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang C, et al. Characterization of murine macrophages from bone marrow, spleen and peritoneum. BMC Immunol. 2013;14:6. doi: 10.1186/1471-2172-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tran DD, et al. Transcriptional regulation of immediate-early gene response by THOC5, a member of mRNA export complex, contributes to the M-CSF-induced macrophage differentiation. Cell Death Dis. 2013;4:e879. doi: 10.1038/cddis.2013.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tubo NJ, Jenkins MK. CD4+ T Cells: guardians of the phagosome. Clin Microbiol Rev. 2014;27:200–213. doi: 10.1128/CMR.00097-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Romano A, et al. Divergent roles for Ly6C+CCR2+CX3CR1+ inflammatory monocytes during primary or secondary infection of the skin with the intra-phagosomal pathogen Leishmania major. PLoS Pathog. 2017;13:e1006479. doi: 10.1371/journal.ppat.1006479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ribeiro-Gomes FL, et al. Site-dependent recruitment of inflammatory cells determines the effective dose of Leishmania major. Infect Immun. 2014;82:2713–2727. doi: 10.1128/IAI.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dumas C, Chow C, Müller M, Papadopoulou B. A novel class of developmentally regulated noncoding RNAs in Leishmania. Eukaryot Cell. 2006;5:2033–2046. doi: 10.1128/EC.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aoki JI, Yamashiro-Kanashiro EH, Ramos DCC, Cotrim PC. Efficacy of the tubercidin antileishmania action associated with an inhibitor of the nucleoside transport. Parasitology Research. 2009;104:223–228. doi: 10.1007/s00436-008-1177-z. [DOI] [PubMed] [Google Scholar]
- 91.do Socorro S Rosa MoS, et al. Antileishmanial activity of a linalool-rich essential oil from Croton cajucara. Antimicrob Agents Chemother. 2003;47:1895–1901. doi: 10.1128/AAC.47.6.1895-1901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 98.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 99.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.