Abstract
Derangement in bone mineral density (BMD) caused by glucocorticoid is well-known. The present study aimed to find key biological pathways associated with low BMD after glucocorticoid treatment in asthmatics using gene expression profiles of peripheral blood cells. We utilized immortalized B cells (IBCs) from 32 childhood asthmatics after multiple oral glucocorticoid bursts and peripheral blood mononuclear cells (PBMCs) from 17 adult asthmatics after a long-term use of oral glucocorticoid. We searched co-expressed gene modules significantly related with the BMD Z score in childhood asthmatics and tested if these gene modules were preserved and significantly associated with the BMD Z score in adult asthmatics as well. We identified a gene module composed of 199 genes significantly associated with low BMD in both childhood and adult asthmatics. The structure of this module was preserved across gene expression profiles. We found that the cellular metabolic pathway was significantly enriched in this module. Among 18 hub genes in this module, we postulated that 2 genes, CREBBP and EP300, contributed to low BMD following a literature review. A novel biologic pathway identified in this study highlighted a gene module and several genes as playing possible roles in the pathogenesis of glucocorticoid- induced derangement in BMD.
Subject terms: Genetics, Asthma
Introduction
Derangement in bone mineral density (BMD) caused by glucocorticoids is well-known. Glucocorticoid treatment is the most common cause of secondary osteoporosis1 and can induce a dosage-dependent reduction in bone mineral accretion in childhood asthmatics2. We previously reported that polymorphisms located in CRHR1, TBCD and RAPGEF5 were associated with low BMD in childhood asthmatics after a long-term treatment of glucocorticoids3–5. However, so far, there has been no comprehensive study to elucidate biologic mechanisms underlying glucocorticoid-induced derangement in BMD in asthmatics who are frequently treated by glucocorticoid.
A network based analysis is an emerging and promising approach and provides new insights into BMD genetics by grouping genes into modules which are enriched for particular biological processes6,7. The present study aimed to find key biological pathways associated with low BMD after glucocorticoid treatment in patients with asthma using weighted gene co-expression network analysis (WGCNA).
For this purpose, we used gene expression profiles of immortalized B cells (IBCs) from childhood asthmatics after multiple oral glucocorticoid bursts and those of peripheral blood mononuclear cells (PBMCs) from adult asthmatics after a long-term use of oral glucocorticoids. Peripheral blood is an easily accessible tissue, although it is not directly involved in bone biology. However, it was suggested that molecular profiling of peripheral blood might reflect physiological and pathological events occurring in different tissues of the body8. Moreover, recent reports showed that PBMCs could be used to assess glucocorticoid sensitivity or efficacy of osteoporosis treatment9,10. Taken together, we may use gene expression profiles of peripheral blood cells for the purpose of searching what biologic pathways are related to glucocorticoid-induced derangement in BMD.
Results
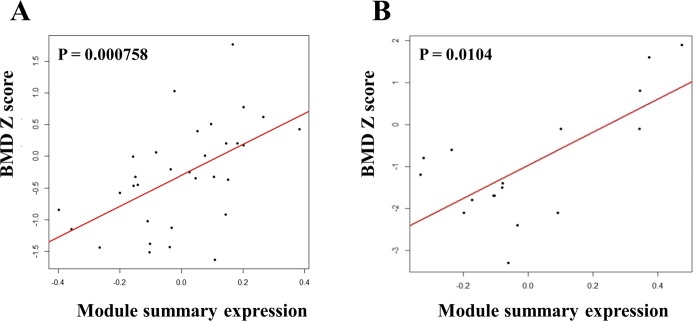
A total of 32 childhood asthmatics whose IBCs and BMD profiles were available and 17 adult asthmatics enrolled (Table 1). Applying WGCNA to the 5,000 genes expressed in Sham-treated IBCs, we could identify 10 modules with various sizes ranging from 125 genes in the purple module to 1,372 genes in the turquoise module (Fig. 1). To emphasize the impact of strong correlations over weak ones in the network construction, we chose an empirical soft threshold of 6, representing a strong model fit for scale-free topology (R2 > 0.8, Figure S1). A total of 63 genes could not be assigned to a module as a membership gene, and were grouped into the grey module which was not considered for further analysis. Eigengene values of two modules (blue and magenta modules) showed significant associations with the BMD Z score in multivariate linear regression analysis. Eigengene value of the blue module consisted of 794 genes was negatively associated with the BMD Z score (P = 0.00294), whereas that of the magenta module containing 199 genes showed a positive association (P = 0.000758) (Fig. 2 and Figure S2).
Table 1.
Characteristics of subjects enrolled.
| Childhood asthmatics† (n = 32) | Adult asthmatics‡ (n = 17) | |
|---|---|---|
| Gender, male (%) | 23 (71.9) | 6 (35.3) |
| Age, year | 8.63 (1.93) | 60.11 (11.71) |
| Smoking, yes (%) | Not applicable | 3 (17.65) |
| Body mass index, kg/m2 | 17.71 (3.21) | 23.72 (5.18) |
| Vitamin D, ng/mL | 31.81 (9.97) | 14.71 (7.72) |
| Tanner stage (%) | ||
| I/II/III/IV/V | 24 (75.0)/6 (18.8)/1(3.1)/1 (3.1) | Not applicable |
| Baseline BMD Z-score | −0.10 (2.23) | Not available |
| Ethnicity (%) | ||
| Non-Hispanic white | 32 (100) | 0 |
| Asian | 0 | 17 (100) |
| Cumulative dose of PD, mg | 1188.40 (608.05) | 3130.53 (1552.09) |
| Final BMD Z-score | −0.29 (0.81) | −0.97 (1.37) |
Data indicate mean (Standard deviation); BMD, Bone mineral density; PD, Prednisone; †Characteristics measured at enrollment of the CAMP study except cumulative dose of prednisone and the final BMD Z-score; ‡Characteristics measured at enrollment of the present study.
Figure 1.
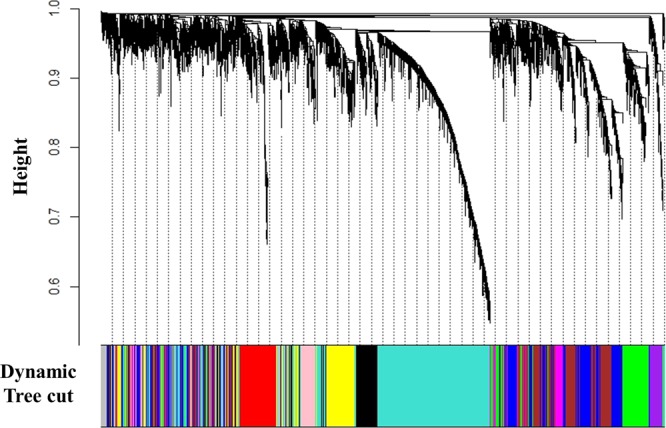
Co-expression modules identified in gene expression profiles of Sham-treated immortalized B cells from childhood asthmatics. Top panel shows a dendrogram of 5,000 genes. Bottom panel shows colors corresponding to the cluster membership labels for each gene.
Figure 2.
Associations between eigengene values of the bone mineral density Z score-associated common module and the bone mineral density Z score. (A) Childhood asthmatics. (B) Adult asthmatics. Both P values were adjusted by covariates.
To assess the reproducibility and generalizability, we tested if our results were replicated in other gene expression profiles from adult asthmatics (Sham-treated PBMCs). Replication analysis was done in two ways: preservation of module (the consistency of network module structure across gene expression profiles) and association with the target phenotype (association between corresponding eigengene values and the BMD Z score). Among 10 modules identified in the discovery cohort, three modules (turquoise, magenta and purple modules) were significantly preserved in the replication cohort (Fig. 3). Module preservation statistics and P values are presented in Table S1. Multivariate linear regression analysis showed that eigengene value of the magenta module was significantly associated with the BMD Z score in the replication cohort (P = 0.0104) (Fig. 2). Based on these findings, we defined the magenta module identified in gene expression profiles from childhood asthmatics as the BMD Z score-associated common module. Among 199 membership genes in the BMD Z score-associated common module, 18 genes (ARMC5, ATP2A2, CCNK, CREBBP, EP300, EP400, GTF3C1, IPO13, MTF1, NOL8, NUP188, PCF11, RFX5, SDAD1, SETD1A, SLC25A22, UBAP2L, WDR59) were taken as hub genes due to their high connectivity and association with the BMD Z score (Figure S3 and Table S2).
Figure 3.
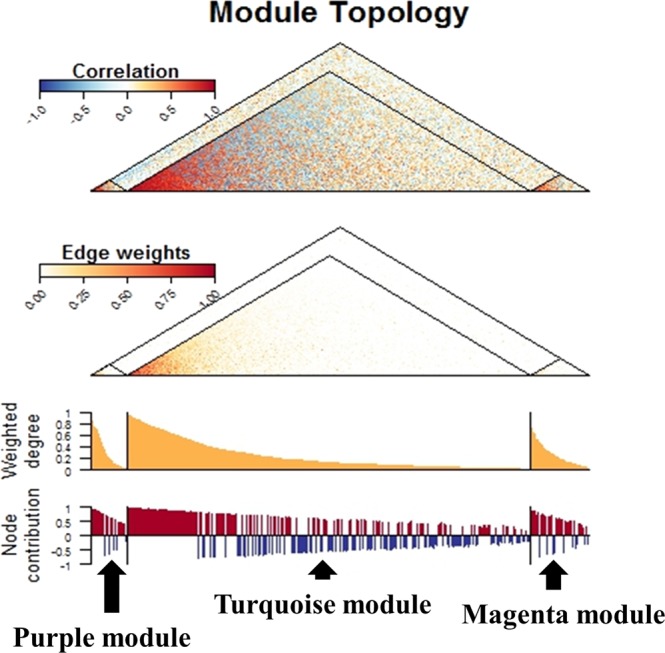
Preservation of modules identified in gene expression profiles of Sham-treated immortalized B cells from childhood asthmatics in gene expression profiles of Sham-treated peripheral mononuclear cell from adult asthmatics. The first (top) panel shows a heatmap of pair-wise correlations among the genes comprising the turquoise, magenta, and purple modules. The second panel shows a heatmap of the edge weights (connections) among the genes comprising the three modules. The third panel shows the distribution of scaled weight degrees (relative connectedness) among the genes comprising the three modules. The fourth panel shows the distribution of node contributions (correlation to module eigengene) among the genes comprising the three modules. Genes are ordered from left to right based on their weighted degree in the discovery cohort, so as to highlight the consistency of the network properties in the replication cohort.
To evaluate in vitro perturbation of modules identified in gene expressions of Sham-treated IBCs by Dex, we measured changes of the module structure between Sham- and Dex-treated IBCs. Among the 10 modules, only the magenta module identified in Sham-treated IBCs was not preserved in Dex-treated IBCs (a maximal permutation P = 0.096, Table S3). In addition, a co-expression network of the 18 hub genes showed distinct differences between them (Fig. 4).
Figure 4.
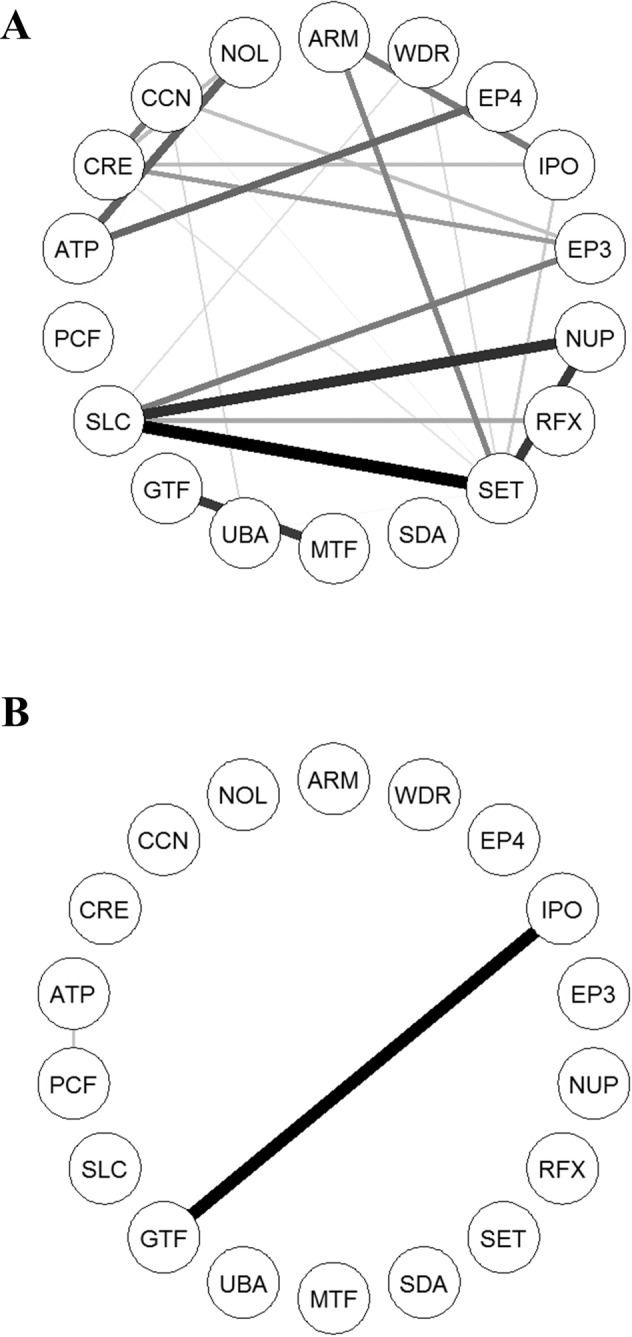
Co-expression network of hub genes of the bone mineral density Z score-associated common module. (A) Sham-treated IBCs from childhood asthmatics. (B) Dex-treated IBCs from childhood asthmatics. For clarity, only the edges corresponding to the Pearson correlation coefficient >0.8 were shown. The edge width is proportional to the Pearson correlation coefficient between two nodes. The network was visualized using qgraph R package. ARM, ARMC5; ATP, ATP2A2; CCN, CCNK; CRE, CREBBP; EP3, EP300; EP4, EP400; GTF, GTF3C1; IPO, IPO13; MTF, MTF1; NOL, NOL8; NUP, NUP188; PCF, PCF11; RFX, RFX5; SDA, SDAD1; SET, SETD1A; SLC, SLC25A22; UBA, UBAP2L; WDR, WDR59.
To assign biological meaning to interpretability of the BMD Z score-associated common module, we performed pathway enrichment analyses in GO biological process categories with its membership genes. To increase the specificity in the enrichment results, we set a more stringent threshold of overlap (intersection of query and test sets ≥ 3) and excluded consideration of inferred electronic annotations (i.e., annotations created by in silico curation methods of potentially lower quality). The most significantly enriched pathway was the cellular metabolic pathway (GO:0044237, FDR adjusted P value = 2.75E-7). This pathway consists of 10,751 genes and 150 genes out of 199 genes belong to the BMD Z score-associated common module overlapped. Enriched pathways identified are shown in Table 2. As we could not find datasets to replicate our findings, we performed a pathway analysis using the top-200 single nucleotide polymorphisms (SNPs) (Table S4) that were significantly associated with the bone mineral accretion in childhood asthmatics treated by oral prednisone bursts in our previous genome-wide association study4. We performed a pathway analysis using genes mapped to 200 SNPs and identify the cellular macromolecular metabolic process (GO:0044260) as the most significant candidate causal pathway (FDR P value = 0.02).
Table 2.
Biological pathways enriched in the bone mineral density Z score-associated common module.
| ID | Name† | Depth‡ | P value§ |
|---|---|---|---|
| GO:1903901 | Negative regulation of viral life cycle | 1 | 0.0418 |
| GO:0045071 | Negative regulation of viral genome replication | 1 | 0.014 |
| GO:0006475 | Internal protein amino acid acetylation | 1 | 0.0448 |
| GO:0018394 | Peptidyl-lysine acetylation | 1 | 0.0453 |
| GO:0018393 | Internal peptidyl-lysine acetylation | 2 | 0.0394 |
| GO:0018076 | N-terminal peptidyl-lysine acetylation | 2 | 0.0287 |
| GO:0016570 | Histone modification | 1 | 0.0291 |
| GO:0016573 | Histone acetylation | 2 | 0.0352 |
| GO:0010646 | Regulation of cell communication | 1 | 0.0485 |
| GO:0051169 | Nuclear transport | 1 | 0.041 |
| GO:0006913 | Nucleocytoplasmic transport | 2 | 0.0374 |
| GO:0002886 | Regulation of myeloid leukocyte mediated immunity | 1 | 0.0321 |
| GO:0002279 | Mast cell activation involved in immune response | 1 | 0.0453 |
| GO:0051640 | Organelle localization | 1 | 0.0335 |
| GO:0043300 | Regulation of leukocyte degranulation | 1 | 0.0336 |
| GO:0051656 | Establishment of organelle localization | 1 | 0.0052 |
| GO:0033003 | Regulation of mast cell activation | 1 | 0.0283 |
| GO:0033006 | Regulation of mast cell activation involved in immune response | 2 | 0.0153 |
| GO:0043303 | Mast cell degranulation | 1 | 0.0453 |
| GO:0043304 | Regulation of mast cell degranulation | 2 | 0.0153 |
| GO:0050657 | Nucleic acid transport | 1 | 0.0335 |
| GO:0051236 | Establishment of RNA localization | 1 | 0.0354 |
| GO:0050658 | RNA transport | 2 | 0.0335 |
| GO:0006405 | RNA export from nucleus | 3 | 0.0287 |
| GO:0071166 | Ribonucleoprotein complex localization | 1 | 0.0219 |
| GO:0071426 | Ribonucleoprotein complex export from nucleus | 2 | 0.0205 |
| GO:0008150 | Biological process | 1 | 0.00234 |
| GO:0071840 | Cellular component organization or biogenesis | 2 | 0.0279 |
| GO:0044085 | Cellular component biogenesis | 3 | 0.00104 |
| GO:0008152 | Metabolic process | 2 | 0.00000288 |
| GO:0044238 | Primary metabolic process | 3 | 0.000013 |
| GO:0071704 | Organic substance metabolic process | 3 | 0.0000188 |
| GO:0009058 | Biosynthetic process | 3 | 0.000516 |
| GO:0006807 | Nitrogen compound metabolic process | 3 | 0.0000273 |
| GO:0009987 | Cellular process | 2 | 0.000425 |
| GO:0044237 | Cellular metabolic process | 3 | 0.000000275 |
| GO:0043933 | Macromolecular complex subunit organization | 1 | 0.0278 |
| GO:0071822 | Protein complex subunit organization | 2 | 0.019 |
| GO:0065003 | Macromolecular complex assembly | 1 | 0.0289 |
| GO:0006461 | Protein complex assembly | 2 | 0.0278 |
| GO:0010629 | Negative regulation of gene expression | 1 | 0.0138 |
| GO:0000122 | Negative regulation of transcription from RNA polymerase II promoter | 1 | 0.0272 |
| GO:0002520 | Immune system development | 1 | 0.0138 |
| GO:0048534 | Hematopoietic or lymphoid organ development | 2 | 0.0167 |
| GO:0030097 | Hemopoiesis | 3 | 0.0225 |
| GO:0006325 | Chromatin organization | 1 | 0.034 |
| GO:0031648 | Protein destabilization | 1 | 0.0356 |
| GO:0033554 | Cellular response to stress | 1 | 0.00328 |
| GO:0036294 | Cellular response to decreased oxygen levels | 1 | 0.0448 |
| GO:0071456 | Cellular response to hypoxia | 2 | 0.038 |
| GO:0043618 | Regulation of transcription from RNA polymerase II promoter in response to stress | 1 | 0.0428 |
| GO:0061418 | Regulation of transcription from RNA polymerase II promoter in response to hypoxia | 2 | 0.0466 |
| GO:0051649 | Establishment of localization in cell | 1 | 0.0302 |
†Gene ontology biological pathway, ‡Only depth 1~3 were presented, §Benjamini-Hocherg FDR P value.
Discussion
The purpose of this study was to find key biological pathways associated with glucocorticoid-induced BMD derangement including a reduction in bone mineral accretion in childhood asthmatics and a reduction in BMD in adult asthmatics after long-term treatment of glucocorticoid. Using gene expression profiles on blood cells, we could identify a key gene module composed of 199 genes whose eigengene value showed a significant association with the BMD Z score in both childhood and adult asthmatics and whose connectivity was preserved as well.
Membership genes in this module were significantly overrepresented in the cellular metabolic pathway (GO:0044237). A previous report exploring differential gene expression profiles on PBMC by the graph clustering approach and GO term enrichment analysis showed 9 gene clusters associated with osteoporosis11. Nine clusters included various GO pathways, such as, response to virus (GO:0009615), immune response (GO:0006955), response to hypoxia (GO:0001666), and response to extracellular stimulus (GO:0009991) which were similar with GO pathways identified in this study. The cellular metabolic pathway representing the chemical reactions and pathways by which individual cells transform chemical substances never been recognized in previous studies focused on osteoporosis. To get a further insight, we performed a pathway analysis using the genes mapped to top-200 SNPs that were significantly associated with the bone mineral accretion in childhood asthmatics in our previous genome-wide association study4. We identified the cellular macromolecular metabolic process (GO:0044260) as the most important candidate causal pathway. This pathway is a sub-pathway of the cellular metabolic pathway (GO:0044237)12. These SNPs could not be directly linked to gene expression/modules identified in the present study, as genomics data were generated from different populations. However, we thought that this result would be an additional evidence suggesting a possible role of the cellular metabolic pathway in osteoporosis pathogenesis. Cellular metabolism is an important part of the bone biology and homeostasis13,14, but, so far, no direct evidence connecting this pathway to glucocorticoid-induced BMD derangement has been issued, although the cellular amine metabolic pathway (GO:0033240), a sub-pathway of the cellular metabolic pathway, was reported to be enriched in gene expressions of PBMC from patients with osteoporosis compared to normal control15. Further studies are needed to confirm our observations.
It was not easy to give biological meanings to our findings at the pathway level, as identified pathways were diverse and contained many genes (for example, the cellular metabolic pathway included 9416 genes12). For this reason, we selected highly connected intramodular18 hub genes. By reviewing the literatures, we selected two genes of putative interest: EP300 encoding E1A binding protein p300 and CREBBP coding CREB binding protein (CBP). CBP and p300 are highly related transcriptional co‐activators possessing histone acetyltransferase activity and are known to participate in the activities of hundreds of different transcription factors involved in a variety of cell functions16. Up-regulation of miR-132-3p could inhibit osteoblast differentiation in part by decreasing p300 expression17, and CBP could mediate osteoblast-adipocyte lineage plasticity by interaction with Maf18,19. In addition, both CBP and p300 were known to be important co-factors mediating transforming growth factor-beta and bone morphogenic protein signaling which have fundamental roles in bone homeostasis20, and to modulate parathyroid hormone regulation of osteoblast21. Interestingly, it has been also reported that these two proteins are involved in steroid receptor signaling22. Steroid receptors require coactivators for efficient activation of target gene expression and these coactivators might contribute to tissue specific responses to steroid hormone. Likewise, CBP and p300 might function additively or antagonistically to each other depending on their relative concentrations and type of target tissue, to influence the sensitivity of tissues to glucocorticoids23. In addition, it was reported that CREBBP mutations resulted in impaired expression of GC receptor responsive gene and impaired histone acetylation and transcriptional regulation24,25. Given that CBP and p300 are involved not only in bone homeostasis but also in glucocorticoid receptor signaling, a gene module identified in this study which contained CREBBP and EP300 may play an important role in glucocorticoid-induced derangement in BMD.
Among other genes belong to the hub genes, ARMC5 encoding armadillo repeat containing 5 and IPO13 encoding importin 13 are of interest. ARMC5 inactivation affects steroid production and cell survival in vitro and adrenal hyperplasia associated with severe Cushing syndrome26. As well known, Cushing’s syndrome results in osteoporosis, which is analogue to glucocorticoid-induced osteoporosis27. IPO13 is a steroid-inducible gene and the glucocorticoid receptor is a cargo substrate for importin 1328. IPO13 is thus possibly associated with glucocorticoid signaling, although its role in glucocorticoid-induced osteoporosis is unknown.
The strength of our study was that we searched biological mechanisms underlying glucocorticoid-induced derangement in BMD with a network based methodology for the first time. A recent review highlighted an importance of network-based approach in genomic studies for osteoporosis29. Considering that the modularity of networks is inherent to cell biology and thus molecular interactions organized into functional unit are more relevant to explain biological phenomena30, the present study might provide a comprehensive and holistic understanding of glucocorticoid-induced derangement in BMD. To generate biological networks, we utilized WGCNA in the present study. WGCNA, unlike the other biological networks, is a way to organize data in an unbiased manner and provides a gene module summary which can be related with clinical phenotypes29. In a systems genetics aspect, if a Bayesian structure learning algorithm is to be applied to WGNCA, the direction of the flow of molecular information would be elucidated, which enables us figure out the causal relationship. Recently, such an attempt identified novel pathways in patients with the late-onset Alzheimer’s disease pathology and coronary artery disease31,32. However, to our knowledge, Bayesian structure learning algorithm in WGNCA has not yet been applied in the field of bone mineral density and steroid response. Another important thing is that we can analyze co-expressions across a set of perturbations with WGCNA. In the present study, we found that a structure of the BMD Z score-associated common module was disrupted by in vitro perturbation using gene expression profiles of Dex-treated IBCs. Eigengene values of this module showed significant positive associations with the BMD Z score in both childhood and adult asthmatic (Fig. 2). We assume that a disruption of this protective gene module by glucocorticoid may result in BMD reduction in asthmatics treated by glucocorticoid for a long time, which provides a functional relevance of our observations. In addition, we confirmed that a module significantly associated with the BMD Z score preserved in different conditions (bone mineral accretion in childhood asthmatics and osteoporosis in adult asthmatics), which may increase the generalizability of observations in this study.
There are some limitations in our study. First, we utilized gene expression profiles of blood cells instead of the cells directly involved in bone biology. We could find a publicly available gene expression dataset of Sham- and Dex-treated osteoblast (GSE21727)33. This dataset was generated from cultured trabecular bone cells obtained from ~100 unrelated Caucasian donors treated by Sham and Dex (100 nM, 2 hour later). Using this dataset, we could find that the magenta module (the BMD Z score-associated common module) identified in this study was preserved in gene expressions of Sham-treated osteoblasts (a maximal permutation P = 0.0000599; Table S5 and Figure S4). Similar to Dex-treated IBCs, this module was not preserved in gene expressions of Dex-treated osteoblasts (a maximal permutation P = 0.253; Data were not shown). These findings suggested that a gene modules of this study might be important across different tissue types (blood cells and osteoblast), although GSE21727 was a small dataset (one subject with three replicates). Evidently, previous reports showed that tissue-specific genes can be expressed in non-tissue-specific manner34,35, and peripheral blood cells expressed approximately over 80% of the genes encoded by the human genome36. In addition, it was demonstrated that osteoclasts could be generated from PBMC population37,38 and PBMC could be used to assess efficacy of osteoporosis treatment10. Taken together, peripheral blood cells may be a surrogate for bone tissue and recapitulate bone cell biology in part, although further studies will be needed. Secondly, a small number of participants, different ethnicity and age (Non-Hispanic white childhood asthmatics vs. Asian elderly asthmatics), and different ways of exposure to glucocorticoid (short-term burst vs. chronic use) were considered before generalizing our findings. Previously, age-dependent alterations in osteoblast and osteoclast activity were reported39,40. In general, short-term glucocorticoid use was associated with mild adverse effects, whereas long-term use might be associated with more serious sequel including osteoporosis41. For this reason, we provided information on the hub genes and enriched biologic pathways of genes belong to the blue module, a childhood asthmatics-specific gene module, in the supplementary information (Figure S5, Table S6, and Table S7). Currently, there are no publicly available datasets to validate our findings. However, a replication analysis should be done when datasets become available. In addition, an integrative analysis using other potentially relevant -omics data such as, miRNAs, lncRNAs, and methylation profiles is warranted in the future.
In conclusion, the potentially significant genes and pathways identified in our analysis may help further elucidate of the molecular mechanisms and provide novel insights regarding glucocorticoid-induced derangement in BMD in asthmatics.
Methods
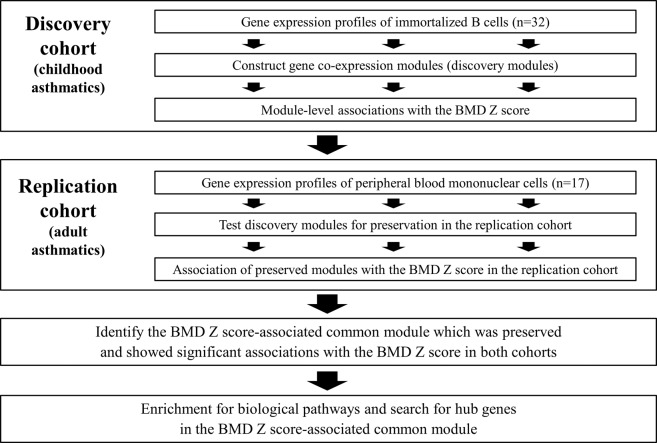
All experimental protocols were approved by the Institutional Review Board of the corresponding institutions. This study was approved by the Institutional Review Board of the Brigham and Women’s Hospital (2002-P-00331/41) and the Seoul National University Hospital (H-1508-095-695). Informed consents were obtained from all study participants and a parent and/or legal guardian, if subjects were under 18. And all methods were carried out in accordance with relevant guidelines and regulations. The overall study design is presented in Fig. 5.
Figure 5.
Overall study design.
Study populations and bone mineral density measurements
The discovery cohort consisted of Non-Hispanic white children from the Childhood asthma management program (CAMP) trial whose IBCs were available42. In asthma exacerbations, short courses of oral prednisone were prescribed per protocol to them. Each burst consisted of 2 mg/kg per day (up to 60 mg) of prednisone for 2 days followed by 1 mg/kg per day (up to 30 mg) for 2 days. Subsequent maintenance of prednisone was allowed when the improvement was not sufficient. BMD measurements of the lumbar spine (L1-L4) were performed annualy during the study period by means of dual-energy x-ray absorptiometry [Hologic (Hologic Inc., Waltham, MA, US) or Lunar (GE Healthcare, Madison, WI, US)]. The BMD Z score that represents a difference between the measured bone density in a subject and the average bone density in age- and sex-matched controls was calculated by using the CAMP internal ref. 2. In the present study, the final BMD Z score measured after mean follow-up duration of 4.3 years was used as a target phenotype.
The replication cohort was drawn from adult asthmatics who were treated at Seoul National University Hospital, Seoul, Korea. All of them received more than 15 mg of prednisone per day continuously for at least one year before enrollment for the control of their symptoms. Doses above 10 mg of prednisone per day result in significant bone loss43. At the enrollment, BMD was measured at the lumbar spine (L1-L4) by dual-energy X-ray absorptiometry [Lunar (GE healthcare, Madison, WI, US)]. Like childhood asthmatics, the BMD Z score was used as a target phenotype.
Gene expression arrays
As previously described44 IBCs derived from childhood asthmatics from the CAMP trial were cultured in RPMI 1640 medium (Sham-treated IBCs) and treated with dexamethasone (10−6 M) for 6 hours (Dex-treated IBCs). Dex-treated IBCs were used to measure in vitro perturbation of gene expressions by Dex. PBMCs from adult asthmatics were cultured in RPMI 1640 medium for 6 hours (Sham-treated PBMCs). Gene expression levels were measured using the Illumina HumanRef8 v2 BeadChip (Illumina, San Diego, CA, US) for childhood asthmatics and the Affymetrix GeneChip Human Gene 2.0 ST (Affymetrix, Santa Clara, CA, US) for adult asthmatics. We removed probes with bad chromosome annotation and probes in X or Y chromosome. We then did variance stabilizing transformation and quantile normalization respectively to reduce the effects of technical noises and to make the distribution of expression level for each array closer to normal distribution.
Statistical analysis
Analysis was performed with R version 3.4.3 (www.r-project.org). We performed WGCNA on each gene expression profile with the R package “WGCNA”45. To allow comparability of data from different microarray platforms, we first calculated the mean expression value of each gene in two expression profiles (Sham-treated IBCs and Sham-treated PBMCs) after collapsing probes by genes. We then compared the mean expression level of genes of them and selected top-5000 genes in common with the highest correlation. Using 5,000 genes we first characterized unsigned correlation networks and their relationships with each other and found gene modules in gene expression profiles from childhood asthmatics. A module was defined as a group of highly interconnected genes45. We computed eigengene values of modules identified and performed multivariate linear regression analysis adjusted by baseline age, gender, BMD Z score, vitamin D level, Tanner stage, and body mass index Z score. Eigengene values can be effective and biologically meaningful tools for studying the relationships between modules of a gene co-expression network and phenotypes46. The BMD Z score-associated module was defined when its eigengene value was significantly associated with the BMD Z score, a target phenotype. We next tested if the BMD Z score-associated modules identified from childhood asthmatics and were preserved in gene expression profiles from adult asthmatics using the R package “NetRep”47. NetRep can quantify the preservation of gene co-expression modules across different datasets and produce unbiased p values based on a permutation approach to score module preservation without assuming data are normally distributed47. The BMD Z score-associated common module denoted a module which was identified in gene expression profiles from childhood asthmatics and significantly preserved in gene expression profiles from adult asthmatics. We then calculated an eigengene value (the first principle component) of the BMD Z score-associated common module in adult asthmatics and performed multivariate linear regression analysis adjusted by age, gender, vitamin D level, body mass index, and smoking status.
We then performed Gene ontology (GO) pathway overrepresentation analyses of the BMD Z score-associated common modules using “g:Profiler” (database version: r1730_e88_eg35)48. g:Profiler (https://biit.cs.ut.ee/gprofiler/) provides an adjusted P value calculated in a manner that accounts for the hierarchical relationships among the tested gene sets. Finally, we searched the key driving hub genes in the BMD Z score-associated common modules, as an occurrence of certain disease is less likely associated with the abnormality of the whole genes involved in the biological pathway. Hub genes were defined by module connectivity, measured by absolute value of the Pearson’s correlation (cor.geneModuleMembership > 0.8) and clinical trait relationship, measured by absolute value of the Pearson’s correlation (cor.geneTraitSignificance >0.2)45,49. For the pathway analysis of SNPs, we selected the top-200 SNPs (Table S) from our previous report4 and put those SNPs into ICSNPathway (http://icsnpathway.psych.ac.cn/). The ICSNPathway enables us to identify candidate causal SNPs and pathways from genome-wide association study by one analytical framework50. Parameters used were as follow: specify SNP, P value < 1.0E-5; linkage disequilibrium cutoff, r2 = 0.8; Rule of mapping SNPs to genes, within gene; Population, CEU; Pathway gene set size, minimum = 5 and maximum = 100; FDR cutoff, 0.05.
Supplementary information
Acknowledgements
This work was supported by the National Institutes of Health, US (R01 NR013391, R01 HL127332, and U01 HL065899) to KGT, by the Seoul National University Hospital [0420153010 (2015–1079)] to PHW, and by the Kangdong Sacred Heart Hospital Fund (2015-10) to LSY.
Author contributions
Heung-Woo Park, Suh-Young Lee, and Kelan G. Tantisira: Conception and design of the study, data generation, analysis, interpretation, and preparation of the manuscript. Ha-Kyeong Won, Byung-Keun Kim, Sae-Hoon Kim, Yoon-Seok Chang, and H. William Kelly: Data generation, and critical revision of the manuscript. Sang-Heon Cho: Data generation, analysis, interpretation, and critical revision of the manuscript.
Data availability
Due to the regulation of the Ethics Review Board of our institute, we could not deposit our data in publicly available repositories. However, immediately following publication, individual participant data that underlie the results reported in this article will be able to be shared after de-identification with researchers who will provide a methodologically sound proposal. Proposals should be directed to guinea71@snu.ac.kr.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56656-9.
References
- 1.Weinstein RS. Glucocorticoid-induced bone disease. N Engl J Med. 2011;365:62–70. doi: 10.1056/NEJMcp1012926. [DOI] [PubMed] [Google Scholar]
- 2.Kelly HW, et al. Effect of long-term corticosteroid use on bone mineral density in children: a prospective longitudinal assessment in the childhood Asthma Management Program (CAMP) study. Pediatrics. 2008;122:e53–61. doi: 10.1542/peds.2007-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones TS, et al. CRHR1 polymorphisms predict bone density in survivors of acute lymphoblastic leukemia. J Clin Oncol. 2008;26:3031–3037. doi: 10.1200/JCO.2007.14.6399. [DOI] [PubMed] [Google Scholar]
- 4.Park HW, et al. Genetic risk factors for decreased bone mineral accretion in children with asthma receiving multiple oral corticosteroid bursts. J Allergy Clin Immunol. 2015;136:1240–1246. doi: 10.1016/j.jaci.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park HW, et al. A genetic factor associated with low final bone mineral density in children after a long-term glucocorticoids treatment. Pharmacogenomics J. 2017;17:180–185. doi: 10.1038/tpj.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farber CR. Identification of a gene module associated with BMD through the integration of network analysis and genome-wide association data. J Bone Miner Res. 2010;25:2359–2367. doi: 10.1002/jbmr.138. [DOI] [PubMed] [Google Scholar]
- 7.Chen YC, et al. Integrative analysis of genomics and transcriptome data to identify potential functional genes of BMDs in females. J Bone Miner Res. 2016;31:1041–1049. doi: 10.1002/jbmr.2781. [DOI] [PubMed] [Google Scholar]
- 8.Mohr S, Liew CC. The peripheral-blood transcriptome: new insights into disease and risk assessment. Trends Mol Med. 2007;13:422–432. doi: 10.1016/j.molmed.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Chapin WJ, et al. Peripheral blood IRF1 expression as a marker for glucocorticoid sensitivity. Pharmacogenet. Genomics. 2015;25:126–133. doi: 10.1097/FPC.0000000000000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stuss M, et al. Assessment of OPG/RANK/RANKL gene expression levels in peripheral blood mononuclear cells (PBMC) after treatment with strontium ranelate and ibandronate in patients with postmenopausal osteoporosis. J Clin Endocrinol Metab. 2013;98:E1007–1011. doi: 10.1210/jc.2012-3885. [DOI] [PubMed] [Google Scholar]
- 11.Xiao H, Shan L, Zhu H, Xue F. Detection of significant pathways in osteoporosis based on graph clustering. Mol Med Rep. 2012;6:1325–1332. doi: 10.3892/mmr.2012.1082. [DOI] [PubMed] [Google Scholar]
- 12.Gene Ontology Browser, GO:0044237. Available at: http://www.informatics.jax.org/vocab/gene_ontology/GO:0044237. Accessed November 14, 2019.
- 13.Florencio-Silva R, Sasso GR, Sasso-Cerri E, Simões MJ, Cerri PS. Biology of Bone Tissue: Structure, Function, and Factors That Influence Bone Cells. Biomed Res Int. 2015;2015:421746. doi: 10.1155/2015/421746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marks SC, Jr, Popoff SN. Bone Cell Biology: The Regulation of Development, Structure, and Function in the Skeleton. Am J Anat. 1988;183:1–44. doi: 10.1002/aja.1001830102. [DOI] [PubMed] [Google Scholar]
- 15.Li JJ, Wang BQ, Fei Q, Yang Y, Li D. Identification of candidate genes in osteoporosis by integrated microarray analysis. Bone Joint Res. 2016;5:594–601. doi: 10.3892/mmr.2012.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 17.Hu Z, et al. miRNA-132-3p inhibits osteoblast differentiation by targeting Ep300 in simulated microgravity. Sci Rep. 2015;5:18655. doi: 10.1038/srep18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berendsen AD, Olsen BR. Osteoblast-adipocyte lineage plasticity in tissue development, maintenance and pathology. Cell Mol Life Sci. 2014;71:493–497. doi: 10.1007/s00018-013-1440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishikawa K, et al. Maf promotes osteoblast differentiation in mice by mediating the age-related switch in mesenchymal cell differentiation. J Clin Invest. 2010;120:3455–3465. doi: 10.1172/JCI42528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu M, Chen G, Li YP. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang D, Guo J, Divieti P, Shioda T, Bringhurst FR. CBP/p300-interacting protein CITED1 modulates parathyroid hormone regulation of osteoblastic differentiation. Endocrinology. 2008;149:1728–1735. doi: 10.1210/en.2007-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutti D, Kaul A, Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol Cell Biol. 2000;20:2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kino T, Nordeen SK, Chrousos GP. Conditional modulation of glucocorticoid receptor activities by CREB-binding protein (CBP) and p300. J. Steroid Biochem. Mol. Biol. 1999;70:15–25. doi: 10.1016/S0960-0760(99)00100-4. [DOI] [PubMed] [Google Scholar]
- 24.Mullighan CG, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu NZ, Cidlowski JA. Glucocorticoid receptor isoforms generate transcription specificity. Trends Cell Biol. 2006;16:301–307. doi: 10.1016/j.tcb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Assié G, et al. ARMC5 mutations in macronodular adrenal hyperplasia with Cushing’s syndrome. N Engl J Med. 2013;369:2105–2014. doi: 10.1056/NEJMoa1304603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaltsas G, Makras P. Skeletal diseases in Cushing’s syndrome: osteoporosis versus arthropathy. Neuroendocrinology. 2010;92(Suppl 1):60–64. doi: 10.1159/000314298. [DOI] [PubMed] [Google Scholar]
- 28.Tao T, Lan J, Lukacs GL, Haché RJ, Kaplan F. Importin 13 regulates nuclear import of the glucocorticoid receptor in airway epithelial cells. Am J Respir Cell Mol Biol. 2006;35:668–680. doi: 10.1165/rcmb.2006-0073OC. [DOI] [PubMed] [Google Scholar]
- 29.Al-Barghouthi BM, Farber CR. Dissecting the genetics of osteoporosis using systems approaches. Trends Genet. 2019;35:55–67. doi: 10.1016/j.tig.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 31.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mäkinen VP, et al. Integrative genomics reveals novel molecular pathways and gene networks for coronary artery disease. PLoS Genet. 2014;10:e1004502. doi: 10.1371/journal.pgen.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gene Expression Omnibus, GSE21727. Available at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21727. Accessed November 14, 2019.
- 34.helly J, Kaplan JC, Maire P, Gautron S, Kahn A. Transcription of the dystrophin gene in human muscle and non-muscle tissue. Nature. 1988;333:858–860. doi: 10.1038/333858a0. [DOI] [PubMed] [Google Scholar]
- 35.Kimoto Y. A single human cell expresses all messenger ribonucleic acids: the arrow of time in a cell. Mol Gen Genet. 1998;258:233–239. doi: 10.1007/s004380050727. [DOI] [PubMed] [Google Scholar]
- 36.Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Komano Y, Nanki T, Hayashida K, Taniguchi K, Miyasaka N. Identification of a human peripheral blood monocyte subset that differentiates into osteoclasts. Arthritis Res Ther. 2006;8:R152. doi: 10.1186/ar2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, Jiang H, Zhang H, Hou J. Generating osteoclasts from human peripheral blood mononuclear cells. Int J Pathol Clin Med. 2008;28:10–13. doi: 10.1007/978-1-61779-415-5_11. [DOI] [Google Scholar]
- 39.Becerikli M, et al. Age-dependent alterations in osteoblast and osteoclast activity in human cancellous bone. J Cell Mol Med. 2017;21:2773–2781. doi: 10.1111/jcmm.13192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai P, et al. Loss of Rictor with aging in osteoblasts promotes age-related bone loss. Cell Death Dis. 2016;13:e2408. doi: 10.1038/cddis.2016.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buchman AL. Side effects of corticosteroid therapy. J Clin Gastroenterol. 2001;33:289–294. doi: 10.1097/00004836-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 42.Childhood Asthma Management Program Research Group. Long-term effects of budesonide or nedocromil in children with asthma. The Childhood Asthma Management Program Research Group. N. Engl. J. Med.343, 1054–1063, 10.1056/NEJM200010123431501 (2000). [DOI] [PubMed]
- 43.Sambrook PN. How to prevent steroid induced osteoporosis. Ann Rheum Dis. 2005;64:176–178. doi: 10.1136/ard.2003.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu W, et al. Pharmacogenomics: novel loci identification via integrating gene differential analysis and eQTL analysis. Hum Mol Genet. 2014;23:5017–5024. doi: 10.1093/hmg/ddu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ritchie SC, et al. A scalable permutation approach reveals replication and preservation patterns of network modules in large datasets. Cell Syst. 2016;3:71–82. doi: 10.1016/j.cels.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 48.Reimand Jüri, Arak Tambet, Adler Priit, Kolberg Liis, Reisberg Sulev, Peterson Hedi, Vilo Jaak. g:Profiler—a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Research. 2016;44(W1):W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, Article17, 10.2202/1544-6115.1128 (2015). [DOI] [PubMed]
- 50.Zhang K., Chang S., Cui S., Guo L., Zhang L., Wang J. ICSNPathway: identify candidate causal SNPs and pathways from genome-wide association study by one analytical framework. Nucleic Acids Research. 2011;39(suppl):W437–W443. doi: 10.1093/nar/gkr391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Due to the regulation of the Ethics Review Board of our institute, we could not deposit our data in publicly available repositories. However, immediately following publication, individual participant data that underlie the results reported in this article will be able to be shared after de-identification with researchers who will provide a methodologically sound proposal. Proposals should be directed to guinea71@snu.ac.kr.