Abstract
Plants are master regulators of rhizosphere ecology, secreting a complex mixture of compounds into the soil, collectively termed plant root exudate. Root exudate composition is highly dynamic and functional, mediating economically important interactions between plants and a wide range of soil organisms. Currently we know very little about the molecular basis of root exudate composition, which is a key hurdle to functional exploitation of root exudates for crop improvement. Root expressed transporters modulate exudate composition and could be manipulated to develop beneficial plant root exudate traits. Using Virus Induced Gene silencing (VIGS), we demonstrate that knockdown of two root-expressed ABC transporter genes in tomato cv. Moneymaker, ABC-C6 and ABC-G33, alters the composition of semi-volatile compounds in collected root exudates. Root exudate chemotaxis assays demonstrate that knockdown of each transporter gene triggers the repulsion of economically relevant Meloidogyne and Globodera spp. plant parasitic nematodes, which are attracted to control treatment root exudates. Knockdown of ABC-C6 inhibits egg hatching of Meloidogyne and Globodera spp., relative to controls. Knockdown of ABC-G33 has no impact on egg hatching of Meloidogyne spp. but has a substantial inhibitory impact on egg hatching of G. pallida. ABC-C6 knockdown has no impact on the attraction of the plant pathogen Agrobacterium tumefaciens, or the plant growth promoting Bacillus subtilis, relative to controls. Silencing ABC-G33 induces a statistically significant reduction in attraction of B. subtilis, with no impact on attraction of A. tumefaciens. By inoculating selected differentially exuded compounds into control root exudates, we demonstrate that hexadecaonic acid and pentadecane are biologically relevant parasite repellents. ABC-C6 represents a promising target for breeding or biotechnology intervention strategies as gene knockdown leads to the repulsion of economically important plant parasites and retains attraction of the beneficial rhizobacterium B. subtilis. This study exposes the link between ABC transporters, root exudate composition, and ex planta interactions with agriculturally and economically relevant rhizosphere organisms, paving the way for new approaches to rhizosphere engineering and crop protection.
Subject terms: Plant signalling, Animal behaviour
Introduction
Plants secrete a complex mixture of water soluble and volatile organic compounds (VOCs) into the soil, collectively termed plant root exudate. Root exudates can enhance the recruitment of beneficial microbes1–5, mobilise nutrients6, sequester toxic compounds in the soil7, and communicate with other plants, and animals8,9. Root exudate composition is dynamic and can be modulated as a factor of development10,11, environment12, physiological state5, and displays marked diversity between species13–16 and cultivars17–21. It is estimated that between 5% and 21% of all photosynthetically-assimilated carbon is released as root exudate22,23.
The process of crop domestication has focused on a small number of desirable traits, relating to plant stature, yield and disease resistance24 often at the expense of other traits. There is evidence that the domestication process has exerted a significant and unintended impact on root exudate composition, and rhizosphere microbe interactions25,26. A general lack of understanding and mechanistic insight represents a major impediment to the exploitation of root exudates for crop improvement. However, it is clear that root exudate composition is an adaptive trait, which can be manipulated. For example, much progress has been made in understanding the interaction between root exudates, and parasitic Striga spp. over recent years. This insight has underpinned efforts to alter exudate strigolactone content, a known germination stimulant and attractant of these economically important and globally distributed parasitic plants17,27–30. Comparatively, less progress has been made in understanding the analogous interaction between root exudate and plant parasitic nematodes (PPNs), in part due to the increased complexity of nematode biology. Nonetheless, recent years have seen renewed interest in this field of research18,31–35.
It is estimated that PPNs reduce crop yields by 12.3%, equating to an estimated $US 80–157 billion in losses each year36–38. PPNs respond transcriptionally, physiologically and behaviourally to plant root exudates, using exudates to trigger egg hatching, and to facilitate host-finding31,35,39–43. Understanding the molecular, chemical and physiological mechanisms underpinning both root exudation and PPN interactions could facilitate the development of aggressive new ex planta control strategies for sustainable intensification of global agriculture, through breeding, rhizosphere engineering and/or biotechnology44–47. The identification of key parasite attractants and repellents could also facilitate the development of new push-pull strategies.
The rhizosphere microbiome has a major influence on crop health48,49, and phenotype50,51. Microbial chemotaxis to plant root exudates is an important factor in the competition for chemical resources in the rhizosphere, and colonisation of plant roots3,52. As such, alteration of root exudate composition could impact on a wide range of interactions. Exploitation of root exudates for improved crop health offers intriguing potential, but requires a detailed study of the link between crop genotype and highly complex, multi-species interactions.
Considerable interest has developed around the manipulation of membrane transporters for crop improvement53,54, and ABC transporters have been implicated directly in modifying root exudate composition5,55. ABC transporters represent one of the single largest gene families in plants, which regulate the sequestration and mobilisation of a vast array of chemistry linked to diverse metabolic, physiological and morphological functions56–60. The ABC transporter gene complement of tomato is numbered at 154, with a considerable proportion expressed in root tissue61. Here we have employed an improved Virus Induced Gene Silencing (VIGS) method to reveal a functional link between two ABC transporter genes, root exudate composition, and rhizosphere interactions with economically important microbes and parasites.
Results
Knockdown of tomato ABC transporter genes by VIGS
VIGS was used to co-silence two genes of interest (ABC-C6 or ABC-G33), alongside a visual reporter gene, Phytoene DeSaturase (PDS). Knockdown of PDS triggers a mild leaf bleaching phenotype due to the accumulation of phytoene, a white carotenoid pigment62. Co-silencing was necessary to identify responsive plants for exudate collection and downstream bioassays; not all plants will trigger a viable RNAi response to VIGS challenge. We observed that plant growth was reduced by over 75% relative to control treatments when using the traditional blunt syringe inoculation method. We therefore sought to develop a less damaging approach to inoculation of A. tumefaciens (containing the VIGS plasmids). We discovered that A. tumefaciens cultures could efficiently invade leaf cells when applied topically to tomato seedling cotyledons with Silwett L-77, which is frequently used to aid A. tumefaciens invasion during floral dip transformation protocols63. Following topical application of A. tumefaciens (containing the VIGS plasmids), bleaching phenotypes typically began to develop after 11 days and peaked at around 21 days post inoculation. Bleaching phenotypes following blunt syringe application began to emerge around 15 days post inoculation, and similarly peaked around 21 days post inoculation. Furthermore, the frequency of plants demonstrating the mild photobleaching phenotype associated with PDS knockdown was between 80% and 100% following topical application. The leaf infiltration method resulted in around 75% of plants demonstrating the co-silenced bleaching phenotype.
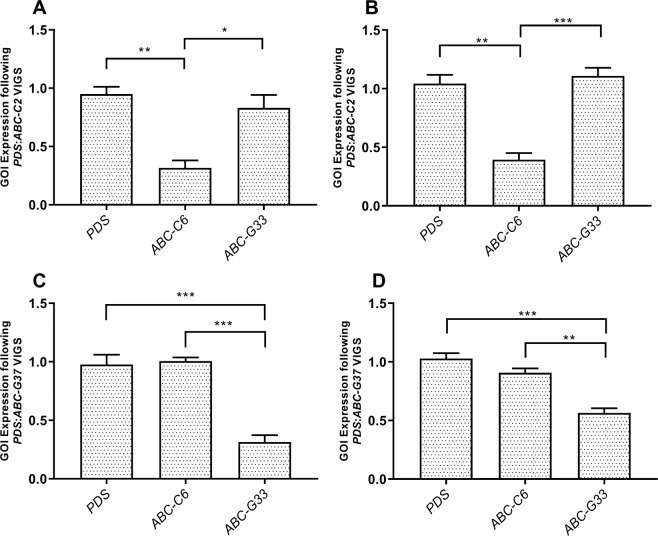
Both inoculation methods triggered robust and specific gene knockdown by week three post inoculation (Fig. 1), with some gene-specific variation around the relative persistence of gene knockdown in topical application treatment groups, relative to syringe inoculation treatment groups. Plants were sampled three weeks post inoculation for further experiments. Transcript abundance of ABC-C6 and ABC-G33 was reduced by 63.29% ± 11.64% and 66.30% ± 8.68% using blunt syringe inoculation. Topical application resulted in transcript knockdown of 64.92% ± 9.44% and 46.40% ± 5.90% for ABC-C6 and ABC-G33 respectively. ABC-C6 transcript levels recovered to control treatment levels by week nine post inoculation when syringe inoculation was employed. In contrast, ABC-G33 transcript abundance remained significantly reduced at week nine following topical inoculation. Due to the improved performance of topical inoculation in terms of knockdown persistence and plant growth rate, we adopted this method for all subsequent experiments. Only plants that displayed co-silenced bleaching phenotypes were taken for further analysis across all experiments.
Figure 1.
VIGS triggers target-specific knockdown of root-expressed ABC transporter genes in tomato. (A) The mean ratio of ABC-C6 abundance relative to the endogenous control gene, elongation factor 1 subunit alpha (EF-α), following blunt syringe inoculation of A. tumefaciens and pTRV plasmids. (B) The mean ratio of ABC-C6 abundance relative to the endogenous control gene following topical application of A. tumefaciens and pTRV plasmids. (C) The mean ratio of ABC-G33 abundance relative to the endogenous control gene following blunt syringe inoculation of A. tumefaciens and pTRV plasmids. (D) The mean ratio of ABC-G33 abundance relative to the endogenous control gene following topical application of A. tumefaciens and pTRV plasmids. Data represent three biological replicates sampled on day 21 post-inoculation, with each replicate consisting of three plants each; error bars represent SEM. One-way ANOVA and Tukey’s HSD tests were used to assess statistical significance between groups (*P < 0.05, **P < 0.01, ***P < 0.001).
Knockdown of ABC transporter genes modulate exudate composition and parasite behaviour
Root exudate was collected from VIGS treatment groups at three weeks post inoculation. Behavioural responses of two root knot nematodes, Meloiodogyne incognita and Meloidogyne javanica, alongside the potato cyst nematode, Globodera pallida were assayed across experimental exudates.
Hatching
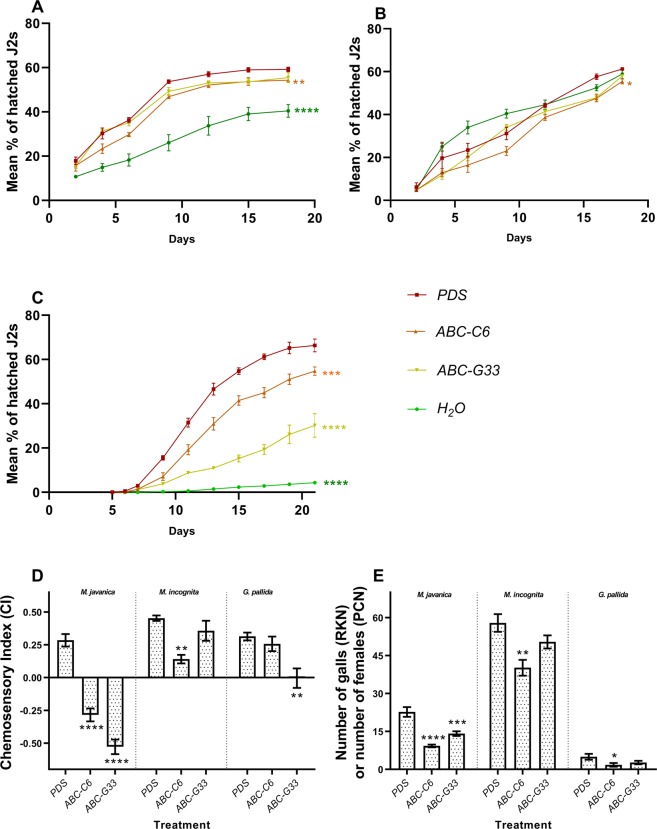
The hatching response of each species was measured as the percentage of emerging infective second stage juveniles (J2s) over time, for which the area under the curve was calculated for comparison of experimental groups (Fig. 2A–C). Area under the curve allows a more robust assessment of hatching phenotypes over time, as it is proportional to both the rate of hatching (gradient) and also the final hatch percentage. Knockdown of ABC-C6 and ABC-G33 triggered a reduction in G. pallida hatching (measured as area under the cumulative percentage hatch [AUCPH]) following knockdown of ABC-C6 and ABC-G33 by 174.40 ± 22.04 (arbitrary units) and 420.10 ± 23.29, respectively (relative to PDS control). Egg hatching was also reduced for M. incognita following knockdown of ABC-C6, by 33.80 ± 7.31. Knockdown of ABC-G33 caused hatching reduction of 16.90 ± 7.11. Knockdown of ABC-C6 also triggered a reduction in hatching of M. javanica of 112.40 ± 33.31, whereas knockdown of ABC-G33 led to a reduction of 59.70 ± 30.83.
Figure 2.
ABC transporter gene knockdown modulates PPN hatching, attraction and invasion. Hatching of J2s in response to experimental treatment (root exudates or water) as a factor of time, for (A) M. incognita; (B) M. javanica; and (C) G. pallida. Error bars represent SEM. (D) Chemosensory challenge of Meloidogyne and Globodera spp. A positive chemosensory index (CI) equates to attraction towards tested exudates, and a negative CI infers repulsion from the exudates. Data represent five biological replicates for each nematode species in each treatment group. (E) The number of galls formed (Meloidogyne spp.) or developing females found on the root surface (G. pallida) six weeks post inoculation on VIGS plants (n = 10 plants per species). Asterisks indicate statistical significance relative to controls following one-way ANOVA and Tukey’s HSD tests: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Attraction
Knockdown of ABC-C6 modified the chemosensory responses of M. incognita and M. javanica to collected exudates (Fig. 2D). Experimentally manipulated exudates were less attractive to M. incognita J2s, whereas M. javanica J2s were repelled by the same exudates. Following knockdown of ABC-C6, chemosensory indices (CI) were reduced by 0.31 ± 0.04 and 0.57 ± 0.07 for M. incognita and M. javanica respectively, relative to PDS control exudates. Following knockdown of ABC-G33, M. javanica was consistently repelled by these exudates with a CI score reduction of 0.81 ± 0.07, whereas M. incognita retained attraction. The CI score for M. incognita to ABC-G33 exudates was reduced by just 0.10 ± 0.08. G. pallida J2s displayed reduced attraction to exudates from ABC-G33 knockdown plants, by 0.32 ± 0.08, whereas for ABC-C6 exudates, the CI was reduced by only 0.06 ± 0.06.
Plant infection
Knockdown of ABC transporter genes also reduced the number of galls produced following infection of VIGS plants with Meloidogyne spp. (Fig. 2E); knockdown of ABC-C6 and ABC-G33 caused a reduction of 17.70 ± 4.68 galls and 7.50 ± 4.34 galls respectively, following infection with M. incognita. The number of galls produced following infection by M. javanica was reduced by 13.43 ± 2.02 and 8.57 ± 2.14 when ABC-C6 and ABC-G33 knockdown plants were challenged, respectively. The number of G. pallida females was reduced by 3.20 ± 1.38 following knockdown of ABC-C6, whereas knockdown of ABC-G33 caused a decrease of 2.30 ± 1.36 cysts per plant. G. pallida retained attraction to exudates collected following ABC-C6 knockdown, however cyst counts were significantly reduced following infection of VIGS plants.
Metabolomic characterisation of exudates following ABC transporter knockdown
Collected exudates were assessed by coupled gas chromatography-mass spectrometry (GC-MS) to identify changes in exudate composition. Several compounds were quantitatively altered in exudates collected following ABC gene knockdown, relative to PDS knockdown controls (Fig. 3). Knockdown of ABC-C6 resulted in reduced abundance of 2-methyloctacosane, and increased abundance of nonadecane and tetradecanoic acid, relative to control treatment (PDS knockdown). Contrastingly, knockdown of ABC-G33 triggered elevated abundance of eicosane, 9-O-pivaloyl-N-acetylcolchinol, heptadecane and octadecanoic acid. Exudate compounds were quantified across experimental groups, and fatty acid composition was confirmed by derivatization using their methyl esters (Supplementary File S1).
Figure 3.
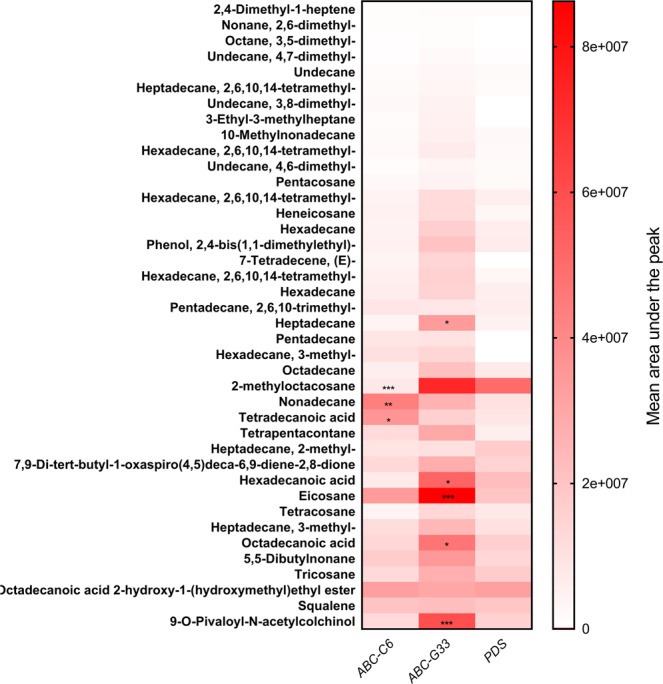
Heatmap showing differences in the relative abundance of identified compounds across experimental exudates. The mean composition of 10 biological replicates (three plants per replicate) is plotted for each experimental group post-VIGS, and has been assessed by two-way ANOVA, and Tukey’s multiple comparison test. Statistical significance is indicated relative to the PDS knockdown control, *P < 0.05, **P < 0.01, ***P < 0.001.
Parasite behavioural responses to selected differentially exuded compounds
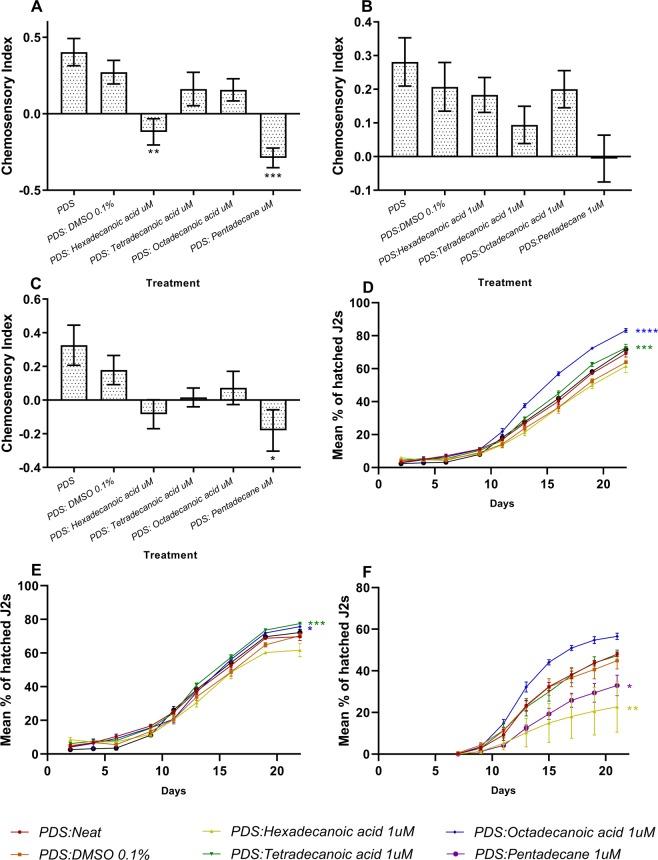
PPN species were assayed for responsiveness to selected commercially available compounds that were differentially exuded following knockdown of either ABC transporter gene. Tetradecanoic acid (elevated following ABC-C6 knockdown), hexadecanoic acid, octadecanoic acid (both elevated following ABC-G33 knockdown), and pentadecane (elevated following knockdown of both ABC-C6 and ABC-G33 relative to controls) were solubilised in 100% dimethyl sulfoxide (DMSO) (1 mM stocks). Each solubilised compound was independently inoculated into control exudates (PDS knockdown) to a final experimental concentration of 1 µM 0.10% DMSO. Inoculated control exudates were then used for egg hatching and chemotaxis assays (Fig. 4).
Figure 4.
PPN responses to selected exudate compounds. Chemotaxis responses to selected compounds inoculated into control plant root exudate (PDS knockdown treatment) for: (A) M. incognita; (B) M. javanica; (C) G. pallida. Data represent the mean of five biological replicates of root exudate, for which ten replicate assays are performed for each nematode species. Asterisks indicate statistical significance in chemosensory index relative to the PDS:DMSO control following one-way ANOVA and Tukey’s HSD tests. Hatch responses following inoculation of control exudate with selected compounds for: (D) M. incognita; (E) M. javanica; (F) G. pallida. The area under the curve of percentage hatch (AUCPH) was estimated by trapezoidal integration and compared by one-way ANOVA and Dunnett’s multiple comparisons test and asterisks indicate statistical significance in hatching relative to the PDS:DMSO control. **P < 0.01; ***P < 0.001; ****P < 0.0001, error bars represent SEM.
Egg hatching of Meloidogyne spp. was enhanced when tetradecanoic acid or octadecanoic acid were inoculated into PDS knockdown exudates. The AUCPH for M. incognita increased by 95 ± 16.82% days and 208.10 ± 17.22% days, respectively (Fig. 4A). For M. javanica the AUCPH increased by 101.70 ± 19.52 and 71.20 ± 21.76, respectively (Fig. 4B). However, the addition of hexadecanoic acid and pentadecane significantly inhibited egg hatching in G. pallida by 178.70 ± 65.22% days and 124.20 ± 31.80% days, respectively (Fig. 4C).
The addition of 1 µM pentadecane to control root exudates reduced the CI of M. incognita by 0.56 ± 0.10. Likewise, the addition of 1 µM hexadecanoic acid reduced the CI of M. incognita by 0.39 ± 0.12. 1 µM tetradecanoic acid, or octadecanoic acid had no statistically significant impact on M. incognita attraction to control root exudates. For M. javanica, a 0.21 ± 0.10 decrease in CI was observed upon addition of 1 µM pentadecane; no statistically significant differences were observed following addition of the other compounds. The CI of G. pallida to root exudates was reduced by 0.36 ± 0.15 with the addition of 1 µM pentadecane.
Knockdown of ABC transporter genes selectively modulates microbial chemotaxis
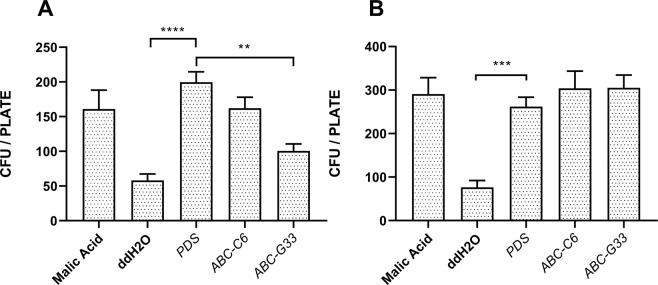
The attraction of B. subtilis and A. tumefaciens to root exudates was assessed following ABC gene knockdown. Both species were significantly more attracted to the positive control, 1 mM malic acid, relative to the negative ddH2O control. Similarly, root exudates collected from the PDS knockdown plants were significantly more attractive to both B. subtilis and A. tumefaciens, than ddH2O (Fig. 5). The attraction of B. subtilis to exudates collected following knockdown of ABC-C6 was statistically unchanged relative to the control treatment. However, knockdown of ABC-G33 triggered a reduced attraction, and a mean reduction of 99 colony forming units (CFUs) relative to exudates from PDS knockdown plants. Knockdown of ABC-C6 and ABC-G33 had no impact on the attraction of A. tumefaciens to collected exudates.
Figure 5.
ABC transporter gene knockdown modulates the attraction of B. subtilis, but not A. tumefaciens. (A) Chemosensory response of B. subtilis (168) to root exudates collected following gene knockdown. (B) Chemosensory response of A. tumefaciens (AGL-1) to root exudates collected following gene knockdown. Data represent ten biological replicates for each species. **P < 0.01; ***P < 0.001; ****P < 0.0001, error bars represent SEM.
Discussion
The predominantly sessile life-style of a plant necessitates substantial molecular and biochemical plasticity to coordinate responses to environmental conditions, and to interact contextually with floral and faunal communities. It is clear that plant genotype influences exudate composition, and organismal interactions17–21,25,26. However, our understanding of the basic biology underpinning these interactions is limited.
ABC transporters modulate root exudate composition, and rhizosphere microbe interactions5,48,55. In this study, we used VIGS to investigate the role of two ABC transporter genes, ABC-C6 and ABC-G33, in modulating tomato root exudate composition, and interactions with three important PPN species. Specifically, we demonstrate that knockdown of ABC-C6 and ABC-G33 transporter genes quantitatively alters tomato root exudate composition and inhibits PPN hatching and attraction behaviours to varying degrees. We assessed the involvement of individual compounds in mediating interactions with the PPN species by inoculating selected, differentially exuded compounds into control exudates. By assessing hatching and chemotaxis responses to these experimentally manipulated exudates, we demonstrate that hexadecanoic acid and pentadecane are inhibitors of G. pallida host-finding and hatching; pentadecane is an inhibitor of Meloidogyne spp. host-finding. Tetradecanoic acid and octadecanoic acid had no impact on host-finding of either species but did enhance egg hatching rates of both Meloidogyne spp. These results suggest that hydrocarbons and fatty acids in root exudate both mediate PPN host finding. However, our findings do not exclude the possibility that other classes of compounds may play a role in J2 host finding.
PPNs are highly damaging parasites with a tremendous impact on agriculture globally. Meloidogyne and Globodera spp. employ an extremely sophisticated and adaptive repertoire of effector molecules to subvert the plant host64. The complexity and diversity of these organisms mean that crop Resistance (R) genes are either unavailable, or are insufficiently durable, to protect crops over the long term65. Our reliance on synthetic chemical pesticides for their management can also have negative consequences to the environment and human health. Novel approaches, which are safe and effective, are required to control these parasites.
Preventing the act of PPN host-finding and invasion represents an attractive intervention strategy for crop protection, and one that could be developed through manipulation of crop root exudate composition. This strategy has been effectively demonstrated against parasitic plants within the Striga genus17,27–30, and could prevent secondary crop infection events. Our data demonstrate that crop plant gene expression can be modulated to alter exudate composition, as well as hatching and attraction of several important PPN species.
The GC-MS dataset reveals several compounds that are elevated in root exudates following ABC gene knockdown. When considering this relative to the behavioural response of PPN species to the exudates, we would hypothesise that elevated compounds function as repellents. Our behavioural data corroborate this for hexadecanoic acid, which is elevated following knockdown of ABC-G33, and pentadecane, which is elevated in exudates following knockdown of both ABC-C6 and ABC-G33. Whilst our analysis of individual compounds has not been exhaustive, these observations do provide some confidence in our ability to predict exudate compound function using these approaches. It is clear however, that several of the compounds we selected for subsequent behavioural characterisation have no influence on PPN behaviour in these assays, and that complex interactions are likely involved. It should also be noted that root exudates will contain many additional compounds, which cannot be identified by GC-MS alone.
pH is known to influence the speciation of organic acids. On the basis of known pKa values, hexadecanoic acid, octadecanoic acid and tetradecanoic acid would be found mostly in the form of conjugate base (control PDS exudates were pH 6.94, and experimental exudates with inoculated compounds ranged between pH 6.60–7.12). Whilst we can’t conclusively determine if the changes in microbe or parasite attraction result from pH related changes on exudate speciation, the fact that different behavioural outcomes are observed in response to exudates that were inoculated with different organic acids (with similar pKa values), indicates that this is unlikely to be the only influencing factor. Certainly, more research is required to determine the relative impact of exudate compositional change, relative to pH impact on speciation. Our experimental design has assessed individual exudate compounds in the context of collected exudates, rather than isolated compounds, which provides additional assurance that speciation is appropriate for the exudate context, and our results are therefore biologically relevant.
It is increasingly apparent that rhizosphere microbes are vital for plant health, and it has been shown that microbial chemotaxis is an important factor in the early colonisation of plant roots, and plant protection3. We assessed the impact that root exudate compositional changes had on the positive chemotaxis of beneficial (B. subtilis66,67) and pathogenic (A. tumefaciens) rhizosphere microbes to the experimental exudates. Our results indicate that A. tumefaciens was attracted to root exudates irrespective of the compositional changes following knockdown of either ABC-C6 or ABC-G33. However, B. subtilis was significantly less attracted to root exudates following knockdown of ABC-G33. These data indicate that approaches to rhizosphere engineering, through the manipulation of root exudate composition, will need to assess a wide range of relevant organismal interactions. In this study, we used the domesticated B. subtilis strain 168 as an experimental model for B. subtilis chemotaxis. B. subtilis 168 does not form biofilms, unlike the undomesticated parental strain B. subtilis NCIB 361068,69. We found that B. subtilis NCIB 3610 would form biofilms within our chemotaxis assay timeframe, making it difficult to enumerate individual cells that were attracted between experimental treatments. However, there may be additional implications for plant-microbe interactions, particularly in terms of biofilm and vir gene induction.
Our study relies upon a new approach to VIGS inoculation, which uses topical application of A. tumefaciens to seedling leaves, in a suspension containing the non-ionic surfactant and wetting agent, Silwet-L77. Unlike blunt syringe inoculation, this approach does not trigger growth stunting effects in the treated plant and promotes longer lasting gene knockdown. In the context of this study, using a transient gene silencing approach, such as VIGS has several conceptual benefits relative to other approaches that constitutively inhibit target gene expression. Transient knockdown can limit downstream secondary impacts in plants relative to constitutively inhibited genes by CRISPR-Cas9 or transgenic RNAi approaches, which could lead to false positive phenotypes. This phenomenon could develop as a function of biochemical knock-on effects that manifest phenotypically, but do not relate exclusively to target gene function, as previously suggested55. Conceptually, we should have higher confidence in phenotypes recorded at earlier time points following loss of function analyses, which VIGS can facilitate. VIGS may also minimise genetic compensation, which occurs through transcriptional rescue of aberrant phenotypes, by counter-balanced expression of related genes. This can generate false negative phenotypes and is especially prominent in large gene families70. Rossi et al. indicate that genetic compensation can be avoided or reduced using transient knockdown70. VIGS typically results in a lower level of target gene knockdown than does transgenic dsRNA production and RNAi in planta71, which may also reduce genetic compensation processes. VIGS is extremely cost-effective relative to other functional genomics approaches, in terms of both reagents and personnel time. It also represents the highest throughput reverse genetics tool available across a number of crop plants, and can be used to rapidly probe parasite interactions72. VIGS can also be used to silence multiple genes simultaneously73.
Collectively, the data generated in this study support efforts to manipulate crop plant genes and promote beneficial rhizosphere interactions. This could occur through breeding, or biotechnology, and could support the sustainable intensification of global agriculture, through the rational and targeted exploitation of crop metabolic potential.
Materials and Methods
Virus induced gene silencing
Solanum lycopersicum (cv. Moneymaker) seedlings were sterilised by rinsing in 1% sodium hypochlorite (from a diluted commercial bleach) for no more than 1 minute. Seeds were then rinsed in sterile ddH2O three times for no more than 2 minutes per wash. ddH2O was removed and seeds were sown on 0.5x MS agar plates (half strength Murashige and Skoog (MS) basal salt mixture, 2 mM Morpholinoethanesulfonic acid (MES), 1.5% agar (w/v), pH 5.7). Seeds were stratified at 4 °C for 48 h in darkness before transfer to 23 °C with 16 h of white light (140–160 μE.m−1.s−1)/8 h. Seedlings were transferred to John Innes number 2 compost upon cotyledon emergence.
The tobacco rattle virus (TRV) VIGS vector, pTRV2, was modified to contain a 200 bp fragment of the tomato PDS gene (Solyc03g123760; pTRV2-PDS). We used a co-silencing system as previously described73,74 by generating pTRV2-PDS-ABC-C6 and pTRV2-PDS-ABC-G33 plasmids containing contiguous 200 bp fragments of the PDS gene sequence, followed by 200 bp of the gene of interest (either ABC-C6 [Solyc08g006880] or ABC-G33 [Solyc01g101070]). Agrobacterium tumefaciens strain GV3101 was used for VIGS throughout. A. tumefaciens cultures were transformed by electroporation, and stored as single use glycerol stocks. Briefly, 30 ng plasmid was added to 50 µl thawed electro-competent A. tumefaciens cells on ice, and then gently mixed by pipette. A single pulse of 2.2 kV was delivered to bacteria in a pre-chilled 1 mm gap cuvette. Cells were suspended in 1 ml LB broth, and incubated at 28 °C for two hours at 200 rpm, before plating 50 µl on LB agar plates with 50 µg/ml kanamycin and 50 µg/ml gentamycin. Colonies were screened for successful transformation by colony PCR using universal pTRV backbone primers (see Table 1). Plants were inoculated with pTRV1/pTRV2 on the third day after transfer, between 2–4 pm. From this point, plants were covered with a foil-lined propagator (to maintain humidity) for approximately 18 h at 18 °C; the lower temperature is necessary to promote VIGS efficacy.
Table 1.
qRT-PCR primer sequences.
| Gene | Tomato Gene ID | Primer Sequences (5′–3′) |
|---|---|---|
| EF-α | Solyc06g005060 | F: TACTGGTGGTTTTGAAGCTG |
| R: AACTTCCTTCACGATTTCATCATA | ||
| PDS | Solyc03g123760 | F: GAAGGCGCTGTCTTATCAGG |
| R: GCTTGCTTCCGACAACTTCT | ||
| ABC-C6 | Solyc08g006880 | F: ACACCCTGGTTATATCTGTTTC |
| R: AAAGACCCAGCAAGTAGTTATAG | ||
| ABC-G33 | Solyc01g101070 | F: GCAATGAGGCCAATGTTAAG |
| R: TTGAAGGTTGTCATGTTCAATG | ||
| pTRV1 |
F: GAGGGGAAACAAGCGGTACA R: TACCTCGTTCCCAAACAGCC |
|
| pTRV2 |
F: ACTCACGGGCTAACAGTGCT R: GACGTATCGGACCTCCACTC |
Preparation of A. tumefaciens cultures for tomato inoculation
For each construct, a single use glycerol stock was thawed and inoculated into 5 ml of LB broth containing 50 µg/ml kanamycin and 50 µg/ml gentamycin. pTRV1 was divided into two cultures (2 × 5 ml). Cultures were then incubated in darkness for 24–48 h at 28 °C, with orbital agitation at 180 rpm, to an OD600 of between 0.75–1. A. tumefaciens cultures were then diluted to a total volume of 50 ml containing 50 µg/ml kanamycin and 50 µg/ml gentamycin, 10 mM MES, and 20 µM acetosyringone. Cultures were then incubated for 24 h at 28 °C, with orbital agitation at 180 rpm, after which they were normalised to an OD600 of 1 in infiltration buffer (200 μM acetosyringone, 10 mM MES, 10 mM MgCl2, pH 5.7). Cultures were covered with foil and incubated for 3 h at room temperature. Immediately before topical application, a pTRV1 culture was mixed in a 1:1 v/v ratio with pTRV2, pTRV2-PDS, pTRV2-PDS-ABC-C6, or pTRV2-PDS-ABC-G33, as appropriate. Silwet-L77 was then added to a final concentration of 0.02%. No Silwet-L77 was added for the blunt syringe leaf infiltration method.
Leaf infiltration and topical application methods
Prior to inoculation, the plants were watered to approximately 0.7 Field Capacity (FC). Leaf infiltration by blunt syringe was conducted as previously described, with a total volume of 0.1 ml injected per plant75,76. For topical application, 0.02% Silwett was added to the cultures immediately prior to application. An autoclaved paintbrush (size 10) was used to apply the culture across both abaxial and adaxial surfaces of the cotyledons, as well as the hypocotyl. Five strokes were administered to each seedling with a freshly inoculated paintbrush. Inoculated plants were maintained at 18 °C, and covered with a foil-lined propagator for approximately 18 h. The propagator lid was then removed, and routine maintenance resumed at 18 °C with a 16 h light, 8 h dark cycle, as before.
Plant phenotype analysis
On the day of A. tumefaciens inoculation, and each subsequent day for six days, photographs were taken of five randomly selected plants in each treatment group to track growth of cotyledons. During this period cotyledon growth was found to be linear, after which growth begins to plateau and true leaves emerge. Thus the rate of cotyledon growth can be expressed as a function of y = mx + c. The rate of growth of each leaf was used in one-way ANOVA and Tukey’s HSD post-hoc tests to compare treatments. Plants were checked daily for leaf bleaching phenotypes, indicative of PDS co-silencing. The number of plants with unambiguous bleaching phenotypes were recorded daily and expressed as a percentage of the total number of plants inoculated.
qRT-PCR analysis of gene transcript knockdown
Plants with bleaching phenotypes were removed from their pots and washed thoroughly under running water to remove soil. Three plants comprised one biological replicate in each treatment group at the times indicated. The plant tissue was wrapped in tin foil and flash frozen. Tissue was then crushed with a pestle and mortar into a fine powder. Frozen tissue was transferred to a 1.5 ml plastic microcentrifuge tube and total RNA was extracted using the Simply RNA Purification kit and Maxwell 16 extraction robot, following the manufacturer’s instructions (Promega). 10 µg total RNA was treated with Turbo DNase according to the manufacturer’s instructions (Ambion). 1 µg of purified RNA was subsequently reverse transcribed into cDNA using the High Capacity RNA-to-cDNA Kit as per manufacturer’s instructions (Applied Biosystems). A reverse transcription reagent (RTr) control, and a reverse transcription minus the reverse transcripase (RT-) control for a randomly selected sample per batch, were included.
cDNA, and controls, were diluted 1/4 using nuclease free water. 2.5 µl template was used for each qRT-PCR reaction in a total of 12 µl with 666 nM of each primer and 1x SensiFAST SYBR No-ROX mix as per manufacturer’s instructions (BIOLINE). Technical PCR reactions for each sample were performed in triplicate for each target using a Rotorgene Q thermal cycler with the following regime: [95 °C × 10 min, 45 × (95 °C × 20 s, 60 °C × 20 s, 72 °C × 25 s) 72 °C × 10 min]. PCR efficiencies of each amplicon, and the corresponding cT value, were calculated using the Rotorgene Q software. Relative quantification of each target amplicon was obtained by an augmented comparative Ct method77, relative to the reference gene EF-α (Solyc06g005060). Ratio-changes in transcript abundance were calculated relative to pTRV2-PDS treated plants. Data were analysed by one-way ANOVA and Tukey’s HSD post hoc test. Oligonucleotide sequences are listed in Table 1.
Exudate collection
Plants were removed from their pots as described above, and cleared of soil by rinsing under running water. Five plants comprised one biological replicate. Plants were bunched together and their roots placed inside a 50 ml screw-top centrifuge tube (Corning), containing 10 ml ddH20. Plants were maintained under a standard 16 h light, 8 h dark regime at 18 °C for the duration of exudate collection. After 24 h, at around 1 pm, plants were removed from the tubes and the exudate was passed through a 0.22 µM filter to remove root border cells and residual soil. Soil control samples (approximately 1 g soil in 10 ml 0.22 µM filtered ddH20) were processed in the same way. For chemosensory and hatching experiments, exudate was stored in centrifuge tubes at 4 °C in darkness until use. For GC-MS metabolomics, exudates were stored at −80 °C immediately after filtering.
Non-targeted exudate metabolite profiling by GC-MS
Root exudates were freeze dried in batches and stored at −80 °C until all samples had been processed (n = 40; 10 biological replicates for each treatment group). Samples were extracted with GC-grade dichloromethane (1 mL) (Sigma–Aldrich, St. Louis, MO, USA), vortexed for 10 s, sonicated for 5 min, and centrifuged at 14,000 rpm for 5 min. The organic phase was dried over anhydrous Na2SO4, concentrated to 50 µL under a gentle stream of N2 and then analysed (1.0 µL) by GC-MS on a 7890 A gas chromatograph linked to a 5975 C mass selective detector (Agilent Technologies, Inc., Santa Clara, CA, USA). The GC was fitted with a HP5 MS low bleed capillary column (30 m × 0.25 mm i.d., 0.25 µm) (J&W, Folsom, CA, USA). Helium at a flow rate of 1.25 ml min-1 served as the carrier gas. The oven temperature was programmed from 35 to 285 °C with the initial temperature maintained for 5 min then 10 °C min-1 to 280 °C, held at this temperature for 20.4 min. The mass selective detector was maintained at ion source temperature of 230 °C and a quadrupole temperature of 180 °C. Electron impact (EI) mass spectra were obtained at the acceleration energy of 70 eV. Fragment ions were analyzed over 40–550 m/z mass range in the full scan mode. The filament delay time was set at 3.3 min. A HP Z220 SFF intel xeon workstation equipped with ChemStation B.02.02. acquisition software was used. The mass spectrum was generated for each peak using Chemstation integrator set as follows: initial threshold = 5, initial peak width = 0.1, initial area reject = 1 and shoulder detection = on. The compounds were identified by comparison of mass spectrometric data and retention times with those of authentic standards and reference spectra published by library–MS databases: National Institute of Standards and Technology (NIST) 05, 08, and 11. Exudate compounds were quantified, and fatty acids detected in the organic phase were confirmed by derivatization using their methyl esters and analysed by GC/MS.
Nematode culture and maintenance
Meloidogyne nematode species were maintained on S. lycopersicum (cv. Moneymaker). Four-week old plants were infected with 1,000 J2s. Eggs were extracted eight weeks post infection by first gently rinsing the root tissue free of soil and placing the tissue in a 500 ml Duran bottle. Tissue was then rinsed in 50% bleach for no more than one minute, and then in water, pouring the liquid through nested 180 µM, 150 µM, and 38 µM sieves arranged from largest pore size to smallest on the bottom. For maximum recovery, plant material was then crushed by hand and rinsed in water through the sieves. Eggs collected in the bottom sieve were pelleted and re-suspended in saturated sucrose. 1 ml ddH20 was added to the solution, and eggs were collected by flotation in the water layer by centrifugation at 2000 rpm for 2 minutes. Eggs were collected and placed in a fine mesh filter hatchery (pore size 38 µm) in 5 ml spring water (autoclaved and sterilised by 0.22 µm filter). 50 µL antibiotic/antimycotic solution (Sigma) was added and eggs were incubated at 23 °C. Freshly hatched J2s were collected every second or third day in hydrophobically lined microcentrifuge tubes for use in chemosensory and infection assays within 48 h of hatch.
Globodera pallida (pathotype PA2/3) cysts were reared on field grown potatoes (cv. Desiree). Two 10 week-old tomato (cv. Moneymaker) plants were grown under greenhouse conditions. 1 L ddH20 was poured into the soil and root stock of each pot, and collected after passing through the pot. Additional ddH20 was poured through the pots until the total collection volume reached 1 L. The collected exudate solution was passed through a 0.22 µM filter and stored in glass jars at 4 °C until use. For chemosensory assays and infection assays, G. pallida cysts were hatched in 1:1 (v/v) tomato exudate diluted in ddH20 and collected at two to three-day intervals in hydrophobically lined microcentrifuge tubes.
Soil infection assays
Tomato plants exhibiting the co-silenced leaf bleaching phenotype were selected at around 21 days post inoculation, and then challenged with 250 J2s. All treatments were blinded from this stage. Six weeks after J2 inoculation, galls and cysts could be identified. These were counted for each experimentally infected plant.
PPN chemosensory assays
Chemosensory assays were conducted as before35,45 by making a solid agar base (3 ml of 1.5% (w/v) agar) in a 60 mm Petri dish. 3 ml of smooth 0.5% (w/v) agar slurry was then poured on top of the solid base. Slurry was prepared by continuous mixing of liquid 0.5% agar until fully cooled. Approximately 150 J2s were then added to the centre of the arena under a microscope. Plates were covered and left in darkness at room temperature (20 °C) overnight on a vibration free bench (for approx. 16 h). The numbers of nematodes observed in the different regions of the arena were then counted under a light microscope. The Chemosensory Index (CI) of each assay was calculated using the formula CI = (A − B)/A + B, where A is the number of worms counted in the root exudate zone of the assay plate, and B is the number of worms counted in the water control zone of the assay plate35,45.
PPN egg hatching assays
Tomato root exudate (collected as described above) was diluted in a 1:1 ratio with ddH2O. 500 µl of diluted exudate was dispensed into wells of a 24-well culture plate. To each exudate sample, between 15 and 20 G. pallida cysts were added. Four biological replicates were prepared for each treatment group. The spaces between wells were half filled with ddH20 and the plates wrapped in parafilm to reduce evaporation and changes of volume throughout the experiment. Plates were incubated at 17 °C for 21 days. Each well was checked daily for signs of nematode emergence. Following the first day of emergence, J2s were counted every 48 h. After approximately 3 weeks, the remaining unhatched eggs/J2s were counted in order to obtain a cumulative percentage hatch rate for each exudate sample. The number of emerging J2s at each time point was converted to a cumulative percentage, which was plotted against time as previously described78. 2000 freshly extracted M. incognita or M. javanica eggs were incubated in 500 µl of experimental plant root exudates. The ratio of unhatched to hatched J2s was then recorded every second or third day. These ratio values were converted into percentages and plotted relative to time, as previously described. A cumulative percentage hatch rate was defined for each sample. For each batch of eggs, triplicate counts of unhatched and hatched J2s were made. Four experimental replicates were used to calculate means. The area under the curve from the cumulative percentage hatch (AUC) was estimated by trapezoidal integration, as described by Campbell & Madden (1990).
Microbe chemotaxis assays
Microbial chemotaxis assays were conducted broadly as in Allard-Massicotte et al.3. B. subtilis (168) and A. tumefaciens (AGL-1) were inoculated onto LB agar plates and spread to single colonies. One day-old colonies were inoculated into 3 ml LB broth and orbitally rotated at 180 rpm, overnight (28 °C for A. tumefaciens and 37 °C for B. subtilis). Cultures were pelleted at 8,000 rpm for 10 min, and the cells were washed in 1.5 ml chemotaxis buffer (10 mM Potassium Phosphate Buffer, pH 7.0), 0.1 mM EDTA, 0.05% glycerol, 5 mM Sodium-d, l-Lactate, 0.14 mM CaCl2, 0.3 mM (NH4)2SO4). Cells were collected by centrifugation and subsequently re-suspended in fresh chemotaxis buffer to an OD600 of 0.002. 200 μl of cell suspension was added to each well of a 96-well plate; ten replicates for each experimental group.
1 μl microcapillary tubes (Sigma-Alrdich) were filled with either: (i) experimental root exudates (ABC-C6, ABC-G33, or PDS), (ii) positive (1 mM malic acid), or negative (ddH2O) controls. Loaded microcapillary tubes were placed into the cell suspension wells of the 96-well plate for 1 h, and maintained at 23 °C. During this time, planktonic B. subtilis or A. tumefaciens cells could migrate towards, and into, the microcapillary tube. Following the 1 h assay timecourse, the capillary tubes were removed. Excess cell suspension was removed from the outside of each capillary tube by rinsing briefly with ddH2O. The 1 µl content of each capillary tube was ejected into 99 µl of chemotaxis buffer by positive pressure. 20 µl of each solution was spread onto a 1.5% LB agar plate. LB plates were sealed with parafilm, and incubated at 28 °C for A. tumefaciens, or 37 °C for B. subtilis for 48 h. Colony forming units were counted for each replicate plate.
Supplementary information
Acknowledgements
VIGS plasmids pTRV1 and pTRV2 were kindly provided by Prof. Ian Graham, University of York. A. tumefaciens AGL-1 was provided by Dr Vladimir Nekrasov, Rothamsted Research. A. tumefaciens strain GV3101 was provided by Dr Penny Hirsch, Rothamsted Research.
Author contributions
D.E.C., S.D., R.W., X.C., B.T. and M.S. conducted the experimental work. J.J.D., D.E.C., S.D. and R.W. wrote the manuscript and prepared figures. The study was conceived by J.J.D., D.E.C. and D.E.C., S.D., R.W., X.C., B.T., A.G.M. and J.J.D. were involved in the interpretation of data. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Deborah Elizabeth Cox and Steven Dyer.
Supplementary information
is available for this paper at 10.1038/s41598-019-56493-w.
References
- 1.Kim B, Song GC, Ryu CM. Root exudation by aphid leaf infestation recruits root-associated Paenibacillus spp. to lead plant insect susceptibility. J.Microbiol Biotechnol. 2016;26(3):549–57. doi: 10.4014/jmb.1511.11058. [DOI] [PubMed] [Google Scholar]
- 2.Li B, et al. Root exudates drive interspecific facilitation by enhancing nodulation and N2 fixation. PNAS. 2016;113(23):6496–501. doi: 10.1073/pnas.1523580113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allard-Massicotte R, et al. Bacillus subtilis early colonization of Arabidopsis thaliana roots involves multiple chemotaxis receptors. MBio. 2016;7(6):e01664–16. doi: 10.1128/mBio.01664-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan J, et al. Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6. Sci Rep. 2015;5:13438. doi: 10.1038/srep13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badri DV, et al. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 2009;151(4):2006–2007. doi: 10.1104/pp.109.147462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waters BM, Amundsen K, Graef G. Gene expression profiling of iron deficiency chlorosis sensitive and tolerant soybean indicates key roles in phenylpropanoids under alkalinity stress. Front Plant Sci. 2018;9:10. doi: 10.3389/fpls.2018.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Andrade LRM, Ikeda M, Do Amaral LIV, Ishizuka J. Organic acid metabolism and root excretion of malate in wheat cultivars under aluminium stress. Plant Physiol Biochem. 2011;49(1):55–60. doi: 10.1016/j.plaphy.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Hiltpold I, Baroni M, Toepfer S, Kuhlmann U, Turlings TCJ. Selection of entomopathogenic nematodes for enhanced responsiveness to a volatile root signal helps to control a major root pest. J.Exp Biol. 2010;213:2417–2423. doi: 10.1242/jeb.041301. [DOI] [PubMed] [Google Scholar]
- 9.Bertin C, Yang XH, Weston LA. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil. 2003;256:67–83. doi: 10.1023/A:1026290508166. [DOI] [Google Scholar]
- 10.Chaparro Jacqueline M., Badri Dayakar V., Bakker Matthew G., Sugiyama Akifumi, Manter Daniel K., Vivanco Jorge M. Root Exudation of Phytochemicals in Arabidopsis Follows Specific Patterns That Are Developmentally Programmed and Correlate with Soil Microbial Functions. PLoS ONE. 2013;8(2):e55731. doi: 10.1371/journal.pone.0055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrne, J. T., Maher, N. J. & Jones, P. W. Comparative responses of Globodera rostochiensis and G. pallida to hatching chemicals. J.Nematol. 33(4), 195–202 not available (2001). [PMC free article] [PubMed]
- 12.Giles CD, et al. Response-based selection of barley cultivars and legume species for complementarity: Root morphology and exudation in relation to nutrient source. Plant Sci. 2017;255:12–28. doi: 10.1016/j.plantsci.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Zwetsloot MJ, Kessler A, Bauerle TL. Phenolic root exudate and tissue compounds vary widely among temperate forest tree species and have contrasting effects on soil microbial respiration. New Phytol. 2018;218(2):530–541. doi: 10.1111/nph.15041. [DOI] [PubMed] [Google Scholar]
- 14.Bowsher AW, Ali R, Harding SA, Tsai CJ, Donovan LA. Evolutionary divergences in root exudate composition among ecologically-contrasting Helianthus species. PLoS One. 2016;11(1):e1048280. doi: 10.1371/journal.pone.0148280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cieslinksi G, Van Rees CJ, Szmigielska AM, Huang PM. Low molecular weight organic acids released from roots of durum wheat and flax into sterile nutrient solutions. J.Plant Nutr. 1997;20:753–764. doi: 10.1080/01904169709365291. [DOI] [Google Scholar]
- 16.Fletcher JS, Hegde RS. Release of phenols by perennial plant roots and their potential importance in bioremediation. Chemosphere. 1995;31(4):3009–3016. doi: 10.1016/0045-6535(95)00161-Z. [DOI] [Google Scholar]
- 17.Mohemed Nasreldin, Charnikhova Tatsiana, Fradin Emilie F, Rienstra Juriaan, Babiker Abdelgabar G T, Bouwmeester Harro J. Genetic variation in Sorghum bicolor strigolactones and their role in resistance against Striga hermonthica. Journal of Experimental Botany. 2018;69(9):2415–2430. doi: 10.1093/jxb/ery041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kihika R, et al. Parasitic nematode Meloidogyne incognita interactions with different Capsicum annum cultivars reveal the chemical constituents modulating root herbivory. Sci Rep. 2017;7:2903. doi: 10.1038/s41598-017-02379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monchgesang S, et al. Natural variation of root exudates in Arabidopsis thaliana-linking metabolomic and genomic data. Sci Rep. 2017;6:29033. doi: 10.1038/srep29033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monchgesang S, et al. Plant to plant variability in root metabolite profiles of 19 Arabidopsis thaliana accessions is substance-class-dependent. Int J Mol Sci. 2016;17(9):E1565. doi: 10.3390/ijms17091565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Micallef SA, Shiaris MP, Colon-Carmona A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J.Exp Bot. 2009;60:1729–1742. doi: 10.1093/jxb/erp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones DL, Nguyen C, Finlay RD. Carbon flow in the rhizosphere: carbon trading at the soil-root interface. Plant Soil. 2009;321:5–33. doi: 10.1007/s11104-009-9925-0. [DOI] [Google Scholar]
- 23.Marschner, H. Mineral nutrition of higher plants (2nd Edition.) ISBN: 9780124735439 (Academic Press, 1995).
- 24.Meyer RS, Duval AE, Jensen HR. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 2012;196(1):29–48. doi: 10.1111/j.1469-8137.2012.04253.x. [DOI] [PubMed] [Google Scholar]
- 25.Iannucci A, Fragasso M, Beleggia R, Nigro F, Papa R. Evolution of the crop rhizosphere: impact of domestication on root exudates in tetraploid wheat (Triticum turgium L.) Front Plant Sci. 2017;8:2124. doi: 10.3389/fpls.2017.02124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulgarelli D, et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17(3):392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, et al. Structural analysis of HTL and D14 proteins reveals the basis for ligand selectivity in Striga. Nat Commun. 2018;9(1):3947. doi: 10.1038/s41467-018-06452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gobena D, et al. Mutation in sorghum LOW GERMINATION STIMULANT 1 alters strigolactones and causes Striga resistance. PNAS. 2017;114(17):4471–4476. doi: 10.1073/pnas.1618965114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohemed N, et al. Evaluation of field resistance to Striga hermonthica (Del.) Benth. In Sorghum bicolor (L.) Moench. The relationship with strigolactones. Pest Manag Sci. 2016;71(11):2082–2090. doi: 10.1002/ps.4426. [DOI] [PubMed] [Google Scholar]
- 30.Jamil M, Charnikhova T, Houshyani B, van Ast A, Bouwmeester HJ. Genetic variation in strigolactone production and tillering in rice and its effect on Striga hermonthica infection. Planta. 2012;235(3):473–484. doi: 10.1007/s00425-011-1520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Čepulytė R, Danquah WB, Bruening G, Williamson VM. Potent Attractant for Root-Knot Nematodes in Exudates from Seedling Root Tips of Two Host Species. Sci Rep. 2018;8:10847. doi: 10.1038/s41598-018-29165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoysted GA, Bell CA, Lilley CJ, Urwin PE. Aphid Colonization Affects Potato Root Exudate Composition and the Hatching of a Soil Borne Pathogen. Front Plant Sci. 2018;9:1278. doi: 10.3389/fpls.2018.01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murungi LK, et al. Identification of key root volatiles signaling preference of tomato over spinach by the root knot nematode Meloidogyne incognita. J.Agric Food Chem. 2018;66:7328–7336. doi: 10.1021/acs.jafc.8b03257. [DOI] [PubMed] [Google Scholar]
- 34.Kirwa HK, Murungi LK, Beck JJ, Torto B. Elicitation of Differential Responses in the Root-Knot Nematode Meloidogyne incognita to Tomato Root Exudate Cytokinin, Flavonoids, and Alkaloids. J.Agric Food Chem. 2018;66(43):11291–11300. doi: 10.1021/acs.jafc.8b05101. [DOI] [PubMed] [Google Scholar]
- 35.Warnock ND, et al. Exogenous RNA interference exposes contrasting roles for sugar exudation in host-finding by plant pathogens. Int J Parasitol. 2016;46(8):473–7. doi: 10.1016/j.ijpara.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Coyne DL, et al. Plant-Parasitic Nematodes and Food Security in Sub-Saharan Africa. Annu Rev Phytopathol. 2018;56:381–403. doi: 10.1146/annurev-phyto-080417-045833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones JT, et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol. 2013;14(9):946–961. doi: 10.1111/mpp.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicol J. M., Turner S. J., Coyne D. L., Nijs L. den, Hockland S., Maafi Z. Tahna. Genomics and Molecular Genetics of Plant-Nematode Interactions. Dordrecht: Springer Netherlands; 2011. Current Nematode Threats to World Agriculture; pp. 21–43. [Google Scholar]
- 39.Palomares-Rius JE, et al. Gene expression changes in diapause or quiescent potato cyst nematode, Globodera pallida, eggs after hydration or exposure to tomato root diffusate. PeerJ. 2016;4:e1654. doi: 10.7717/peerj.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang G, et al. Effects of tomato root exudates on. Meloidogyne incognita PLoS ONE. 2016;11(4):e0154675. doi: 10.1371/journal.pone.0154675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zasada, I. A., Peetz, A., Wade, N., Navarre, R. A. & Ingham, R. E. Host status of different potato (Solanum tuberosum) varieties and hatching in root exudates of Globodera ellingtonae. J.Nematol. 45(3), 195–201 not available. PMID: 241157804 (2016). [PMC free article] [PubMed]
- 42.Duarte A, Maleita C, Abrantes I, Curtis R. Tomato root exudates induce transcriptional changes of Meloidogyne hispanica genes. Phytopathol Mediterr. 2015;54(1):104–108. doi: 10.14601/Phytopathol_Mediterr-14595. [DOI] [Google Scholar]
- 43.Teillet A. Transcriptional changes of the root-knot nematode Meloidogyne incognita in Response to Arabidopsis thaliana root signals. PLoS ONE. 2013;8(4):e61259. doi: 10.1371/journal.pone.0061259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahkami AH, White RA, Handakumbura PP, Jansson C. Rhizosphere engineering: enhancing sustainable plant ecosystem productivity. Rhizosphere. 2017;3(2):233–243. doi: 10.1016/j.rhisph.2017.04.012. [DOI] [Google Scholar]
- 45.Warnock ND, et al. Nematode neuropeptides as transgenic nematicides. PLoS Pathog. 2017;13(2):e1006237. doi: 10.1371/journal.ppat.1006237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dessaux Y, Grandclement C, Faure D. Enginerring the rhizosphere. Trends in Plant Sci. 2016;21(3):266–278. doi: 10.1016/j.tplants.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Jones Peter, Devine Ken. Effects of hatching factors on potato cyst nematode hatch and in-egg mortality in soil and in vitro. Nematology. 2001;3(1):65–74. doi: 10.1163/156854101300106900. [DOI] [Google Scholar]
- 48.Sasse J, Martinoia E, Northen T. Feed your friends: do plant exudates shape the root microbiome? Trends in Plant Sci. 2018;23(1):25–41. doi: 10.1016/j.tplants.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends in Plant Sci. 2012;17(8):478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Hubbard CJ, et al. The effect of rhizosphere microbes outweighs host plant genetics in reducing insect herbivory. Mol Ecol. 2018;28(7):1801–1811. doi: 10.1111/mec.14989. [DOI] [PubMed] [Google Scholar]
- 51.Lu T, et al. Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome. 2018;6(1):231. doi: 10.1186/s40168-018-0615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Weert S, et al. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol Plant Microbe Interact. 2002;15(11):1173–80. doi: 10.1094/MPMI.2002.15.11.1173. [DOI] [PubMed] [Google Scholar]
- 53.Lane TS, et al. Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology. BMC Biotechnol. 2016;16(1):47. doi: 10.1186/s12896-016-0277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schroeder JI, et al. Using membrane transporters to improve crops for sustainable food production. Nature. 2013;497:60–66. doi: 10.1038/nature11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Badri DV, et al. Altered profile of secondary metabolites in the root exudates of Arabidopsis ATP-Binding Cassette (ABC) transporter mutants. Plant Physiol. 2008;146(2):762–771. doi: 10.1104/pp.107.109587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adebesin F, et al. Emission of volatile organic compounds from petunia flowers is facilitated by an ABC transporter. Science. 2017;356(6345):1386–1388. doi: 10.1126/science.aan0826. [DOI] [PubMed] [Google Scholar]
- 57.Hwang J-U, et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle. Mol Plant. 2016;9(3):338–355. doi: 10.1016/j.molp.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 58.Martinoia E, Meyer S, De Angeli A, Nagy R. Vacuolar transporters in their physiological context. Annu Rev Plant Biol. 2012;63:183–213. doi: 10.1146/annurev-arplant-042811-105608. [DOI] [PubMed] [Google Scholar]
- 59.Yazaki K. ABC transporters involved in the transport of plant secondary metabolites. FEBS Lett. 2006;580(4):1183–91. doi: 10.1016/j.febslet.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 60.Liu G, Sanches-Fernandez R, Li Z-S, Rea PA. Enhanced multispecificity of Arabidopsis vacuolar multidrug resistance-associated protein-type ATP-binding cassette transporter, AtMRP2. J Biol Chem. 2001;276:8648–8656. doi: 10.1074/jbc.M009690200. [DOI] [PubMed] [Google Scholar]
- 61.Ofori PA, et al. Genome-wide analysis of ATP binding cassette (ABC) transporters in tomato. PLoS ONE. 2018;13(7):e0200854. doi: 10.1371/journal.pone.0200854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Winzer T, et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- 63.Clough Steven J., Bent Andrew F. Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. The Plant Journal. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 64.Mitchum MG, et al. Nematode effector proteins: an emerging paradigm of parasitism. New Phytol. 2013;199:879–894. doi: 10.1111/nph.12323. [DOI] [PubMed] [Google Scholar]
- 65.Davies Laura J., Elling Axel A. Resistance genes against plant-parasitic nematodes: a durable control strategy? Nematology. 2015;17(3):249–263. doi: 10.1163/15685411-00002877. [DOI] [Google Scholar]
- 66.Adam M, Heuer H, Hallmann J. Bacterial antagonists of fungal pathogens also control root-knot nematodes by induced systemic resistance of tomato plants. PLoS ONE. 2014;9(2):e90402. doi: 10.1371/journal.pone.0090402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu M, et al. Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World J. Microbiol. Biotechnol. 2014;30:835–845. doi: 10.1007/s11274-013-1486-y. [DOI] [PubMed] [Google Scholar]
- 68.Kesel S, et al. Direct comparison of physical properties of Bacillus subtilis NCIB 3610 and B-1 biofilms. Appl Environ Microbiol. 2016;82:2424–2432. doi: 10.1128/AEM.03957-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeigler DR, et al. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190(21):6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rossi A, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 71.Albert Markus, Belastegui-Macadam Xana, Kaldenhoff Ralf. An attack of the plant parasiteCuscuta reflexainduces the expression ofattAGP, an attachment protein of the host tomato. The Plant Journal. 2006;48(4):548–556. doi: 10.1111/j.1365-313X.2006.02897.x. [DOI] [PubMed] [Google Scholar]
- 72.Dubreuil G, et al. Tobacco rattle virus mediates gene silencing in a plant parasitic root-knot nematode. J Exp Bot. 2009;60(14):4041–4050. doi: 10.1093/jxb/erp237. [DOI] [PubMed] [Google Scholar]
- 73.Orzaez Diego, Medina Aurora, Torre Sara, Fernández-Moreno Josefina Patricia, Rambla José Luis, Fernández-del-Carmen Asun, Butelli Eugenio, Martin Cathie, Granell Antonio. A Visual Reporter System for Virus-Induced Gene Silencing in Tomato Fruit Based on Anthocyanin Accumulation. Plant Physiology. 2009;150(3):1122–1134. doi: 10.1104/pp.109.139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stratmann JW, Hind SR. Gene silencing goes viral and uncovers the private life of plants. Entomol Exp Appl. 2011;140(2):91–102. doi: 10.1111/j.1570-7458.2011.01147.x. [DOI] [Google Scholar]
- 75.Liu Y, Schiff M, Dinesh‐Kumar SP. Virus‐induced gene silencing in tomato. The Plant Journal. 2002;31(6):777–786. doi: 10.1046/j.1365-313X.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 76.Senthil-Kumar M, Mysore KS. Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc. 2014;9(7):1549–62. doi: 10.1038/nprot.2014.092. [DOI] [PubMed] [Google Scholar]
- 77.Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29(9), e45 not available. PMID: 11328886 (2001). [DOI] [PMC free article] [PubMed]
- 78.Campbell, C. L. & Madden, L. V. Introduction to Plant Disease Epidemiology. ISBN: 0471832367 (John Wiley & Sons,1990).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.