Abstract
Recent genome-wide association studies (GWAS) have discovered ten genetic risk variants for abdominal aortic aneurysms (AAA). To what extent these genetic variants contribute to the pathology of aneurysms is yet unknown. The present study aims to investigate whether genetic risk variants are associated with three clinical features: diameter of aneurysm sac, type of artery and aneurysm related-symptoms in aortic and peripheral aneurysm patients. Aneurysm tissue of 415 patients included in the Aneurysm-Express biobank was used. A best-fit polygenic risk score (PRS) based on previous GWAS effect estimates was modeled for each clinical phenotype. The best-fit PRS (including 272 variants at PT = 0.01015) showed a significant correlation with aneurysm diameter (R2 = 0.019, p = 0.001). No polygenic association was found with clinical symptoms or artery type. In addition, the ten genome-wide significant risk variants for AAA were tested individually, but no associations were observed with any of the clinical phenotypes. All models were corrected for confounders and data was normalized. In conclusion, a weighted PRS of AAA susceptibility explained 1.9% of the phenotypic variation (p = 0.001) in diameter in aneurysm patients. Given our limited sample size, future biobank collaborations need to confirm a potential causal role of susceptibility variants on aneurysmal disease initiation and progression.
Subject terms: Epidemiology, Genetics research
Introduction
Abdominal aortic aneurysm disease (AAA) is a vascular pathology, affecting in particular elderly Western men1–3. Besides gender and smoking, a positive family history is a known predisposing factor for occurrence of the disease4,5, indicative of a strong heritable component to AAA.
In the past decade, genome-wide association studies (GWAS) have uncovered ten common genetic variants, i.e. single-nucleotide polymorphisms (SNPs), associated to AAA susceptibility6–12. While these GWAS point out part of the genetic underpinnings of AAA, the extent to which these variants influence the clinical presentation, whether these loci are specific for a vascular bed, or how these loci contribute to the pathology of the disease is still largely unknown.
Previous literature has shown many complex traits to be polygenic in origin, comprising small effects of hundreds or even thousands of common variants that in aggregate explain a substantial proportion of trait susceptibility and heritability13–15. For example, the International Schizophrenia Consortium16 summarized weighted genetic effects13 across nominally associated loci at increasingly liberal p-value thresholds into polygenic risk scores (PRS), and correlated PRS to disease susceptibility, demonstrating that a PRS can provide a reliable genetic indicator for clinical outcome.
In the present study, we investigated whether AAA susceptibility variants in aggregate are associated with three clinical phenotypes (maximum diameter of aneurysm sac, aneurysm related symptoms, and type of artery) within the Aneurysm-Express biobank study17. These phenotypes encompass interventions and are therefore clinically relevant. First, we constructed a weighted PRS based on summary level GWAS data for AAA12 using increasingly liberal p-value thresholds and modeled a best fit PRS. Secondly, we tested the genome-wide significant risk SNPs for association with the selected clinical phenotypes in our cohort of aortic and peripheral aneurysm patients with clinically manifested disease. Our results show that higher PRS associates to larger aneurysm diameter, but no association was found with clinical symptoms or type of artery.
Methods
Aneurysm-Express biobank study
The Aneurysm-Express is a biobank study that contains aneurysm sac tissue from patients undergoing open surgical repair of arterial aneurysms. The study design has been published previously17. Ethical approval for this study (TME/C-01.18) was provided by the Medical Research Ethics Committee of University Medical Center Utrecht, Utrecht, The Netherlands on 10 April 2002, and all research was conducted according to the principles of the Declaration of Helsinki (59th amendment, Seoul 2008) and in accordance with the Dutch Medical Research Involving Human Subjects Act (WMO). Patients were operated in two different Dutch hospitals and all participants gave informed consent. For the present study we used clinical information from consecutive patients who were included between 2003 and 2013. The indications to perform open repair were based on at that time current guidelines3. Patients with arterial aneurysms caused by dissection, connective tissue disorders, and mycotic aneurysms or re-operated patients were excluded from this study. Risk factors and demographic data were obtained from clinical records and questionnaires at time of recruitment.
Genotyping, quality control, and imputation
DNA of 503 patients in Aneurysm-Express biobank study was extracted from whole blood EDTA or (when no blood was available) aneurysm tissue samples following standardized in-house validated protocols for the Aneurysm-Express Genomics Study (AAAGS). Samples were sent for genotyping at the Genomic Analysis Center of the Helmholtz Zentrum Münich (Germany) according to OECD standards under study number M00750 using the Illumina HumanCoreExome BeadArray v1.1 (Illumina Inc., www.illumina.com).
Genotype calling was done with the GenomeStudio V2011.1 software and the Genotyping module version 1.9.4 using the original Illumina cluster and manifest files (humancoreexome-12v1-1_a.egt and HumanCoreExome-12- v1-1-C.bpm). The GenCall score cutoff was 0.15 as recommended by Illumina. The average call rate of all samples was 99.55% across 542,585 variants.
Subsequently, community standard quality control (QC) procedures were applied18 to obtain high quality data. Samples with low average genotype calling and sex discrepancies (compared to the clinical data available) based on GenomeStudio metrics were excluded. The data was filtered on 1) individual (sample) call rate > 97%, 2) SNP call rate >96%, 3) average heterozygosity rate ± 2.5 s.d., 4) relatedness (pi-hat >0.20), 5) Hardy-Weinberg Equilibrium (HWE p < 1.0 × 10−6), and 6) population stratification excluding non-Europeans (based on 1000 G phase 3)19. zCall20 was used to call missing exome-variants after QC. After QC and resulted in 478 samples and 541,569 variants (call rate = 99.99%) remained and were used for imputation.
Autosomal missing genotypes were imputed based on phased integrated data from 1000 Genomes (phase 3, version 5) and Genome of the Netherlands v521 using IMPUTE2 (v2.3.0)22 after pre-phasing genotyped data with SHAPEIT2 (v2.644)23.
Polygenic risk score (PRS)
The PRSice software13 was used for creating the best fit PRS per phenotype by comparing scores across a range of different p-value thresholds. In short, PRSice calculates weighted PRS based on the effect estimates reported in the meta-analysis of GWAS12,24. To this end, variants are pruned based on the linkage disequilibrium (r2 < 0.1, clump, range 500 kb)20 as observed in AAAGS for bins of increasingly liberal p-value thresholds (pT, see Supplemental Table 1), and preferentially retaining variants with lower p-values as reported by the GWAS12 (similar to the–clump algorithm in PLINK25). Odds ratios were natural log transformed to betas (β), we only included variants with MAF > 0.05 and imputation quality info-score > 0.8. In order to verify the predictive value of the PRS of the AAA-GWAS, we additionally modeled a PRS of summary statistics of the attention deficit hyperactivity disorder (ADHD) GWAS unrelated to AAA24, and tested this additional polygenic score for association with the selected clinical parameters in our study cohort.
Individual SNP analysis
We selected ten SNPs that were identified in a recent GWAS meta-analysis for AAA12. These SNPs were tested for association with the clinical features separately. For the SNP lookup we used GWASToolKit (https://github.com/swvanderlaan/GWASToolKit, doi: 10.5281/zenodo.997862), which is a collection of scripts to execute SNPTEST v2.5.326 analyses. Given the limited sample size, we calculated the expected power for this analysis27. In case of a risk allele frequency of <20%, and estimated OR of 1.10, the resulting power is ±80% (Supplemental Fig. 1).
Primary endpoints
The primary outcomes or phenotypes were artery type, symptoms, and diameter of the aneurysm sac. Artery type was defined as either aortic, or peripheral (iliac, femoral, popliteal, and carotid). Symptom status was defined as asymptomatic, or any aneurysm related symptoms like thromboembolic events, local pain or swelling, or rupture of the aneurysm sac. Maximum diameter was measured by experienced radiologists at time of inclusion, by using the double oblique plane of computed tomography angiography.
Statistical analysis
Baseline characteristics were compared between patients with upper 20th percentile and lower 80th percentile of the PRS (see Supplemental Table 4). For continuous baseline variables, the Student’s T-test was used for normally distributed variables and Mann-Whitney U test for non-normally distributed variables. Categorical baseline variables were tested by the Chi-squared test for equal distribution among both groups. PRS and individual SNP analyses were performed using linear and logistic regression models where appropriate, adjusted for sex, age, ancestral background using four principal components, smoking status, and diameter of the aneurysm sac or artery type if applicable. Nagelkerke’s r2 was used as a metric of the variance explained by the polygenic model. Statistical analyses were conducted using SPSS v25.0 (IBM Corp. Released 2017. IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp.), PRSice v2.1.4.13, R v3.4.0 (R Core Team (2017), R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org).
Ethical approval and informed consent
All study patients were included from our Aneurysm-Express biobank. The biobank has been approved by the local ethics committee, and all patients gave informed consent.
Results
In the present study a total of 415 aortic and peripheral aneurysm patients from the Aneurysm-Express biobank were included. The average age was 69 ± 8.1 years, the majority (85%) male, and 349 (84%) patients were treated for an AAA (Table 1).
Table 1.
Baseline characteristics of included study patients.
| AAA n = 349 | Iliac n = 13 | Femoral n = 9 | Popliteal n = 35 | Carotid n = 9 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Male | 292 | 84% | 13 | 100% | 8 | 100% | 34 | 97% | 4 | 44% |
|
Age at surgery in years (range) |
70 |
7.1 (48–89) |
65 |
11.0 (48–86) |
67 |
11.2 (49–87) |
66 |
10.0 (45–83) |
62 |
9.7 (46–75) |
| Aneurysm shape | ||||||||||
| saccular | 16 | 5% | 0 | — | 0 | — | 1 | 3% | 3 | 33% |
| fusiform | 332 | 95% | 13 | 100% | 9 | 100% | 34 | 97% | 6 | 67% |
| Reported aneurysm diameter (mm) | 64 |
13.9 (31–118) |
46 |
12.4 (25–70) |
44 |
25.5 (24–100) |
35 |
20.0 (11–105) |
23 |
10.2 (12–38) |
| Symptoms | ||||||||||
| Ruptured | 28 | 8% | 1 | 8% | 0 | — | 2 | 6% | 0 | — |
| Any aneurysm related symptom | 82 | 24% | 4 | 31% | 4 | 44% | 19 | 54% | 6 | 68% |
| Asymptomatic | 237 | 68% | 7 | 54% | 5 | 56% | 14 | 40% | 3 | 33% |
| Hypertension | 275 | 79% | 7 | 54% | 4 | 44% | 22 | 63% | 4 | 44% |
| Diabetes | 58 | 17% | 1 | 8% | 1 | 11% | 4 | 11% | 1 | 11% |
| Statin use | 237 | 68% | 7 | 54% | 3 | 33% | 19 | 54% | 6 | 67% |
| BMI | 25.9 | 4.1 | 26.4 | 5.2 | 27.7 | 4.6 | 27.2 | 3.6 | 23.7 | 2.6 |
| Smoking | 126 | 36% | 6 | 46% | 2 | 22% | 12 | 34% | 0 | — |
Data are given as numbers (percentage) or mean (standard deviation).
Abbreviations: AAA = abdominal aortic aneurysm, mm = millimeter, BMI = body mass index.
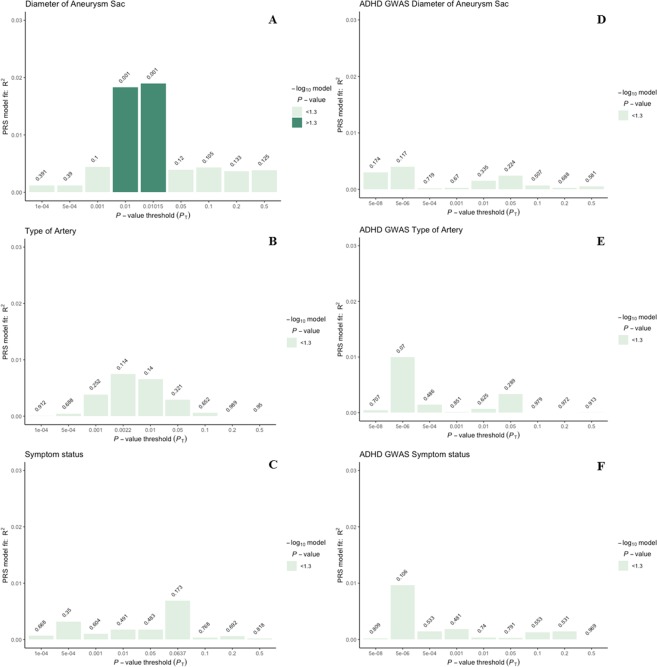
We used summary statistics from the largest meta-GWAS for AAA so far12 to calculate weighted polygenic scores at increasingly liberal p-value thresholds (pT, Supplemental Table 1), and correlated these to artery type, symptoms, and diameter of the aneurysm sac (Fig. 1A–C). A PRS including 272 variants at pT of 0.01015 explained the largest proportion in diameter (R2 = 0.019, p = 0.001, Supplemental Table 2). Diameter per 10.0 millimeter and average PRS is visualized in Fig. 2, showing higher PRS in large aneurysms. Distributions of the AAA-PRS for diameter were comparable between artery types (Kruskal-Wallis test p-value = 0.135, Supplemental Fig. 2). No association was found for artery type and symptom status (Supplemental Table 2). We verified all AAA-PRS findings with another unrelated PRS derived from a GWAS on ADHD24, and correlated these with the selected clinical parameters in our study cohort (Fig. 1D–F). No associations with the ADHD-PRS were observed (Fig. 1D–F, Supplementary Table 3). Clinical characteristics of patients within the upper 20th and lower 80th percentile of the PRS distribution for diameter is shown in Supplemental Table 4.
Figure 1.
PRSice13 generated weighted model for: (A) Diameter of the aneurysm sac, (B) Type of artery, and (C) Symptom status. Models are summarized in Supplementary Tables 2 and 3. (D–F) indicate unrelated PRS derived from ADHD GWAS summary statistics24, showing no association with any of the selected clinical phenotypes in the Aneurysm-Express biobank cohort. Abbreviations: PRS = polygenic risk score, ADHD = attention deficit hyperactivity disorder, GWAS = genome-wide association study.
Figure 2.
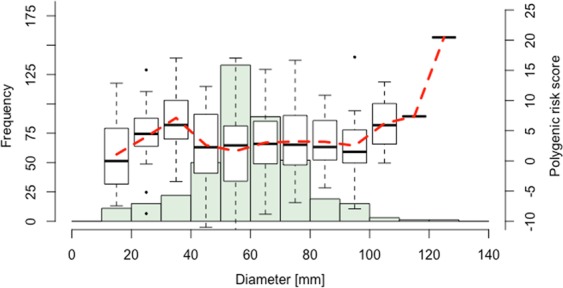
Distribution of maximum diameter of included aneurysms. Boxplots indicate PRS per 10.0 mm diameter, outliers are indicated as dots. Average PRS per diameter is indicated as red dashed line, showing an increase in average PRS from diameters ≥90.0 mm. Abbreviations: PRS = polygenic risk score, mm = millimeter.
Next, we tested ten known AAA SNPs for association with diameter of the aneurysm sac (Table 2), artery type and symptom status (Supplemental Table 5). For diameter, a nominal association was found with rs1466535 (12q13.3, LRP1, p = 0.013). For rs602633 (1p13.3, PSRC1- CELSR2-SORT1), rs1795061 (1q32.3, SMYD2), rs2836411 (21q22.2, ERG) and rs1466535 a concordant effect direction was observed. For six risk variants, the effect direction was discordant with the GWAS results.
Table 2.
Individual SNP analysis of AAA associated SNPs reported by GWAS and the association results for diameter of the aneurysm sac (in millimeters).
| Reported by literature | This study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | BP | Near(est) gene(s) | Alleles | EAF | β a | p | EAF | β | SE | p |
| rs602633 | 1 | 109821511 | PSRC1- CELSR2-SORT1 | T* - G | 0.199 | −0.129 |
6.58 x10−9 |
0.208 | −2.017 | 1.238 | 0.104 |
| rs4129267 | 1 | 154426264 | IL6R | T* - C | 0.370 | −0.132 |
4.76 x10−13 |
0.355 | 1.422 | 1.073 | 0.186 |
| rs1795061 | 1 | 214409280 | SMYD2 | T* - C | 0.337 | 0.123 |
8.80 x10−11 |
0.307 | 0.490 | 1.054 | 0.642 |
| rs10757274 | 9 | 22096055 | CDKN2BAS1/ANRIL | A* - G | 0.462 | −0.216 |
1.54 x10−33 |
0.504 | 0.319 | 1.264 | 0.752 |
| rs10985349 | 9 | 124425243 | DAB2IP | T* - C | 0.195 | 0.158 |
2.40 x10−11 |
0.200 | −0.445 | 1.009 | 0.725 |
| rs1466535 | 12 | 57534470 | LRP1 | G* - A | 0.679 | 0.199b |
9.99 X10−7 |
0.655 | 2.554 | 1.022 | 0.013 |
| rs9316871 | 13 | 22861921 | LINC00540 | A* - G | 0.201 | −0.136 |
4.75 x10−10 |
0.796 | 0.675 | 1.272 | 0.596 |
| rs6511720 | 19 | 11202306 | LDLR | T* - G | 0.096 | −0.218 |
7.90 x10−14 |
0.094 | 2.612 | 1.755 | 0.138 |
| rs3827066 | 20 | 44586023 | PCIF1-ZNF335-MMP9 | T* - C | 0.179 | 0.201 |
2.13 x10−17 |
0.167 | −0.900 | 1.323 | 0.496 |
| rs2836411 | 21 | 39819830 | ERG | T* - C | 0.369 | 0.107 |
5.80 x10−9 |
0.362 | 0.499 | 1.238 | 0.687 |
Abbreviations: SNP = single nucleotide polymorphism, AAA = abdominal aortic aneurysm, Chr = chromosome, BP = base pair, EAF = effect allele frequency, β = beta-coefficient, SE = standard error.
*Effect allele, aβ converted from combined odds ratio’s (discovery and validation phase) of summary statistics of Jones et al.12, bβ converted from discovery phase of summary statistics of Bown et al.7.
Discussion
In this study, we investigated the association of genetic susceptibility variants with clinical phenotypes within a biobank consisting of surgically treated aortic and peripheral aneurysms. Our results demonstrate that polygenic scores of AAA susceptibility are associated with diameter of the aneurysm sac in patients with aneurysms in different vascular beds.
The clinical translation of genetic research has been a topic of interest for the last decades. For rare diseases like Marfan or Ehlers-Danlos syndrome, genetic clinical utility has been proven and applied28, but for more common complex diseases like AAA, the interpretation and translation of large-scale genetic studies is challenging at best29. GWAS-derived association studies were mainly investigated in intracranial and abdominal aneurysm patients30–35. The majority of these studies used a genetic risk score (GRS) based on the index SNPs from GWAS to study additional genetic predicting value to models consisting of demographic characteristics and health parameters. A previous study using a GRS including 4 variants showed that a high GRS was associated with aneurysm growth rate independent of baseline abdominal aortic size36. Recently, a PRS approach, which includes millions of nominally associated SNPs, has proved clinical utility in atherosclerosis, statin therapy, and breast cancer37–39. A polygenic approach can optimize power by using a liberal p-value threshold, enforced by the evidence that additive weak effects of nominally associated common variants explain part of the heritability of, and susceptibility to common complex diseases14,15. Our study uses a polygenic risk approach within a cohort comprising both aortic and peripheral aneurysm patients and shows that a high polygenic risk of AAA is associated to larger diameter of the aneurysm sac, although clinical parameters of patients within the upper 20th and lower 80th percentile of the PRS distribution were not distinctive in post-operative outcome or inflammatory status of the aneurysm (Supplemental Table 4).
After correcting for multiple testing, no effects of individual SNPs were observed, and the majority of the effect directions in our analysis are discordant with the recent GWAS meta-analysis12. This is probably mainly due to limited size and power of both the published GWAS (4,972 cases and 99,858 controls) and our present study (Supplemental Fig. 2). Differences could also be explained by differences in inclusion criteria; the GWAS analysis is performed in patients with AAA ≥ 30 mm, including smaller AAA that have been followed-up and were compared to healthy subjects29. In contrast, our cohort consists solely of symptomatic patients or patients with progressive aneurysmal disease that requires surgical treatment. It is arguably that GWAS identified loci may contribute particularly to initiation of aneurysmal disease, whilst they have discordant effects on disease progression and so other risk variants may have a more prominent role in further deterioration of the vessel wall.
Whether aortic and intracranial aneurysms share genetic susceptibility, was previously investigated in a GWAS of four cohorts32. This study provides evidence that variants at 9p21, 18q11, 15q21, and 2q33 are consistently associated with intracranial, thoracic aneurysms and AAA. Their analysis revealed no additional risk loci associated with joint aneurysms, presumably due to their limited sample size of 3,094 cases and 9,521 controls. The PRS distribution of diameter in the present study was comparable across artery types (Supplemental Fig. 2), this suggests overlapping genetic pathways between peripheral aneurysms and central aneurysms.
The Aneurysm-Express biobank cohort is limited in size when compared to other biobanks, yet unique in its scope, given the availability of arterial aneurysm samples from the open arterial surgery era for analyses. Future studies should focus on biobank collaborations, enrichment, and harmonization to facilitate the in-depth scrutiny and replication of genetic susceptibility on aneurysm and AAA initiation and progression, as well as the exploration of gender differences. Our biobank is surgically driven, and measured diameters may cluster around surgical intervention thresholds (e.g. for AAA ≥ 55 mm) limiting the generalizability, still we observed normally distributed diameters. Although the general pathology of aneurysms in various anatomical locations is similar, the initiating factors might be unique to the site. Artery type was used as composite covariate within the analyses, although individual aneurysm site factors might have confounded our results.
Conclusions
In conclusion, we show that a weighted polygenic score of AAA susceptibility explained 1.9% of the phenotypic variation (p = 0.001) in aneurysm diameter in the Aneurysm-Express biobank study. Future studies should focus on biobank collaborations, enrichment, and harmonization to assess potential impact of AAA susceptibility loci on aneurysmal disease initiation and progression.
Supplementary information
Acknowledgements
This research was financially supported by BBMRI-NL, a Research Infrastructure financed by the Dutch government (NWO 184.021.007). Dr. van der Laan is funded through grants from the Netherlands CardioVascular Research Initiative of the Dutch Heart Foundation (CVON 2011/B019 and CVON 2017-20: Generating the best evidence-based pharmaceutical targets for atherosclerosis [GENIUS I&II]).
Author contributions
C.L., S.L., J.H. and J.S. were responsible for data acquisition, C.L. and S.L. for manuscript writing, C.L., S.L., D.K. and G.B. designed the study protocol. S.L., J.S., G.P. and D.K. supervised the statistical analyses. All authors discussed the results and reviewed the manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available for research partners from the corresponding author.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56230-3.
References
- 1.Singh K, et al. Prevalence of and risk factors for abdominal aortic aneurysms in a population based study: The Tromsø Study. Am J Epidemiol. 2001;154:236–244. doi: 10.1093/aje/154.3.236. [DOI] [PubMed] [Google Scholar]
- 2.Lijftogt N, et al. Failure to Rescue e a Closer Look at Mortality Rates Has No Added Value for Hospital Comparisons but Is Useful for Team Quality Assessment in Abdominal Aortic Aneurysm Surgery in The Netherlands. Eur J Vasc Endovasc Surg. 2018;56:652e661. doi: 10.1016/j.ejvs.2018.06.062. [DOI] [PubMed] [Google Scholar]
- 3.Moll FL, et al. European Society for Vascular Surgery. Management of abdominal aortic aneurysms clinical practice guidelines of the European society for vascular surgery. Eur J Vasc Endovasc Surg. 2011;41:S1–S58. doi: 10.1016/j.ejvs.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Wahlgren CM, et al. Genetic and environmental contributions to abdominal aortic aneurysm development in a twin population. J Vasc Surg. 2010;51:3–7. doi: 10.1016/j.jvs.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 5.Van den Luijtgaarden KM, et al. Risk of abdominal aortic aneurysm (AAA) among male and female relatives of AAA patients. Vascular Medicine. 2017;22:112–118. doi: 10.1177/1358863X16686409. [DOI] [PubMed] [Google Scholar]
- 6.Gretarsdottir S, et al. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nat Genet. 2010;42:692–697. doi: 10.1038/ng.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bown MJ, et al. CARDIoGRAM Consortium; Global BPgen Consortium; DIAGRAM Consortium; VRCNZ Consortium. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am J Hum Genet. 2011;89:619–627. doi: 10.1016/j.ajhg.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradley DT, et al. A variant in LDLR is associated with abdominal aortic aneurysm. Circ Cardiovasc Genet. 2013;6:498–504. doi: 10.1161/CIRCGENETICS.113.000165. [DOI] [PubMed] [Google Scholar]
- 9.Jones GT, et al. A sequence variant associated with sortilin-1 (SORT1) on 1p13.3 is independently associated with abdominal aortic aneurysm. Hum Mol Genet. 2013;22:2941–2947. doi: 10.1093/hmg/ddt141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison SC, et al. Interleukin-6 receptor pathways in abdominal aortic aneurysm. Eur Heart J. 2013;34:3707–3716. doi: 10.1093/eurheartj/ehs354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgadottir A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 12.Jones GT, et al. Meta-Analysis of Genome-Wide Association Studies for Abdominal Aortic Aneurysm Identifies Four New Disease-specific Risk Loci. Circ Res. 2017;120:341–353. doi: 10.1161/CIRCRESAHA.116.308765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Euesden J, et al. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31:1466–8. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torkamani A, et al. The personal and clinical utility of polygenic risk scores. Nature Reviews Genetics. 2018;19:581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 15.Chatterjee N, et al. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 2016;17:392–406. doi: 10.1038/nrg.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia that overlaps with bipolar disorder. Nature. 2009;460(7256):748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurks R, et al. Aneurysm-Express: human abdominal aortic aneurysm wall expression in relation to heterogeneity and vascular events – rationale and design. Eur Surg Res. 2010;45:34–40. doi: 10.1159/000318160. [DOI] [PubMed] [Google Scholar]
- 18.Laurie CC, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genetic Epidemiology. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(10):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldstein JL, et al. zCall: a rare variant caller for array-based genotyping. Bioinformatics. 2012;28(19):2543–2545. doi: 10.1093/bioinformatics/bts479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deelen P, et al. Improved imputation quality of low-frequency and rare variants in European samples using the ‘Genome of The Netherlands’. European Journal of Human Genetics. 2014;22:1321–1326. doi: 10.1038/ejhg.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie B, et al. Fast and accurate genotype imputation in genome‐wide association studies through pre‐phasing. Nature Genetics. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaneau O, et al. A linear complexity phasing method for thousands of genomes. Nature methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 24.Demontis D, et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nature Genetics. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81(9):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchini J, et al. A new multipoint method for genome‐wide association studies by imputation of genotypes. Nature Genetics. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 27.Pulit SL, et al. Associations Claims in the Sequencing Era. Genes. 2014;5:196–213. doi: 10.3390/genes5010196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aday AW, et al. Vascular genetics: presentations, testing and prognostics. Curr Treat Options Cardio Med. 2018;20:103. doi: 10.1007/s11936-018-0703-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wanhainen, A. et al. European Society for Vascular Surgery (ESVS) 2019 Clinical Practice Guidelines on the Management of Abdominal Aorto-iliac Artery Aneurysms, European Journal of Vascular and Endovascular Surgery (2018). [DOI] [PubMed]
- 30.van t Hof FNG, et al. Genetic risk load according to the site of intracranial aneurysms. Neurology. 2014;83:34–39. doi: 10.1212/WNL.0000000000000547. [DOI] [PubMed] [Google Scholar]
- 31.van t Hof FNG, et al. Impact of Inherited Genetic Variants Associated With Lipid Profile, Hypertension, and Coronary Artery Disease on the Risk of Intracranial and Abdominal Aortic Aneurysms. Circ Cardiovasc Genet. 2013;6:264–270. doi: 10.1161/CIRCGENETICS.113.000022. [DOI] [PubMed] [Google Scholar]
- 32.van t Hof FN, et al. Shared Genetic Risk Factors of Intracranial, Abdominal, and Thoracic Aneurysms. J Am Heart Assoc. 2016;14(5):7. doi: 10.1161/JAHA.115.002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiotti, N. et al. Multiple sites of vascular dilation or aneurysmal disease and matrix metalloproteinase genetic variants in patients with abdominal aortic aneurysm. J Vasc Surg, 1–9 (2017). [DOI] [PubMed]
- 34.Adovasio R, et al. Growth Rate of Small Abdominal Aortic Aneurysms and Genetic Polymorphisms of Matrix MetalloProteases-1, −3, and −9. Int J Angiol. 2016;25:93–98.. doi: 10.1055/s-0035-1563603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makrygiannis G, et al. Risk Factor Assessment in a Greek Cohort of Patients With Large Abdominal Aortic Aneurysms. Angiology. 2019;1:35–40. doi: 10.1177/0003319718774474. [DOI] [PubMed] [Google Scholar]
- 36.Ye Z, et al. A multi-locus genetic risk score for abdominal aortic aneurysm. Atherosclerosis. 2016;246:274–279. doi: 10.1016/j.atherosclerosis.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natarajan P, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paquette M, et al. Polygenic risk score predicts prevalence of cardiovascular disease in patients with familial hypercholesterolemia. J. Clin. Lipidol. 2017;11:725–732.e5. doi: 10.1016/j.jacl.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 39.Maas P, et al. Breast cancer risk from modifiable and nonmodifiable risk factors among white women in the United States. JAMA Oncol. 2016;2:1295–1302. doi: 10.1001/jamaoncol.2016.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available for research partners from the corresponding author.