Abstract
Modern crop breeding is based on the use of genetically and phenotypically diverse plant material and, consequently, a proper understanding of population structure and genetic diversity is essential for the effective development of breeding programs. An example is avocado, a woody perennial fruit crop native to Mesoamerica with an increasing popularity worldwide. Despite its commercial success, there are important gaps in the molecular tools available to support on-going avocado breeding programs. In order to fill this gap, in this study, an avocado ‘Hass’ draft assembly was developed and used as reference to study 71 avocado accessions which represent the three traditionally recognized avocado horticultural races or subspecies (Mexican, Guatemalan and West Indian). An average of 5.72 M reads per individual and a total of 7,108 single nucleotide polymorphism (SNP) markers were produced for the 71 accessions analyzed. These molecular markers were used in a study of genetic diversity and population structure. The results broadly separate the accessions studied according to their botanical race in four main groups: Mexican, Guatemalan, West Indian and an additional group of Guatemalan × Mexican hybrids. The high number of SNP markers developed in this study will be a useful genomic resource for the avocado community.
Subject terms: Plant breeding, Plant genetics
Introduction
Avocado (Persea americana Mill.) is a subtropical evergreen tree native to Mesoamerica. Avocado belongs to the Lauraceae, a family in the order Laurales that, together with the orders Canellales, Piperales and Magnoliales, is included in the Magnoliid clade of early-divergent angiosperms1. This pantropical family has about 50 genera and 2500 to 3000 species. Besides avocado, only a few species in the family have economic importance and these include mainly spices [bay laurel (Laurus nobilis L.) and cinnamon (Cinnamomum verum J.Presl)], camphor (C. camphora (L.) J.Presl) and timber trees (Nectandra spp., Ocotea spp. and Phoebe spp.).
Traditionally, avocado genotypes have been classified in three horticultural races or subspecies mainly related to ecological preferences and botanical characteristics2. The Mexican and Guatemalan subspecies are adapted to highland areas in Central America (cold climates), being the Guatemalan race more susceptible to low temperatures. The West Indian subspecies is adapted to low-land areas in the same region (tropical climates).
Avocado market demand has increased exponentially in recent years and in 2017 avocado world production was close to 6 million tons. Most of the production is concentrated in a few countries (Mexico, Dominican Republic, Peru, Indonesia, Colombia, Brazil), Mexico being the largest producer with 34% of the total world production (more than 2 million tons)3. However, in spite of the increasing importance of this crop, there are important bottlenecks for efficient breeding and development of new avocado cultivars, due to the absence or poor availability of molecular resources and phenotypic data and to the limited genetic pool in breeding programs worldwide. Developing new high quality avocado cultivars is an urgent need in this crop since approximately 90% of the avocado production worldwide depends on a single cultivar, ‘Hass’, that originated as a chance seedling in California ninety years ago4.
Different types of genetic markers have been utilized in avocado for genotype fingerprinting, paternity analyses, diversity and phylogenetic studies, linkage map construction and screening for traits of interest. Initial works included minisatellites5, Variable Number of Tandem Repeats (VNTRs)6, Random Amplified Polymorphic DNA (RAPDs)7 and Restriction Fragment Length Polymorphism (RFLPs)8,9. More recently, Single Sequence Repeats (SSRs), which are codominant and highly polymorphic facilitating the study of intraspecific relations and diversity, have been specifically developed in avocado and used for fingerprinting and diversity analyses10–19. However, in spite of the inherent advantages of SSR markers, their frequency of distribution is not uniform over the genome and their use in association analyses is problematic20. Moreover, it is difficult to compare SSRs from different populations or systems, and the analyses are laborious and costly compared to new sequencing technologies (NGS)21. Indeed, Single Nucleotide Polymorphism (SNP) markers are becoming the marker of choice in crop genetic studies with different aims: linkage mapping, analysis of quantitative trait loci (QTL), association studies, marker-assisted selection (MAS) or genomic selection (GS)22. The advantages of SNPs include the large number of markers that can be generated at a reduced cost, the fact that they are the most frequent source of variation in eukaryotic genomes, their bi-allelic nature that offers accuracy in variant calling, their high reproducibility or their reduced cost that makes them accessible to most laboratories23–25. Those advantages are specially relevant in woody perennial crops since their application would significantly reduce time and cost of breeding programs.
Up to now, NGS applied to avocado research has been reduced to transcriptome analyses26,27 and the development of SNPs to characterize genetic diversity28–30. In addition, very recently, a first avocado nuclear genome sequence has been published31. In order to provide additional high quality SNPs for the avocado research community, in this work a collection of 71 avocado accessions representing the three classical botanical races were genotyped and characterized using newly developed SNP markers. Those markers were mapped to a draft genome of the most important avocado cultivar worldwide, ‘Hass’, in order to increase the quality of the markers developed.
Results
Development of an avocado draft genome for mapping the raw reads
A draft genome of the avocado ‘Hass’ variety was developed to assist with read mapping and SNP calling. The sequencing of ‘Hass’ DNA produced 487.54 million raw Illumina reads (73.13 Gb) and 487.21 million processed reads (72.15 Gb). The estimated haploid genome size for ‘Hass’ ranged from 1.33 Gb (17-mer) to 1.63 Gb (73-mer) with an estimated genomic heterozygosity ranging from 1.05% (73-mer) to 1.41% (17-mer). The stats are summarized in Table 1. The assembly size represents 77% of the estimated genome size (1.33 Gb). The total number of sequences indicates highly fragmented assemblies in which the average sequence size (0.54 Kb) and the L50 (0.68 Kb) are below the average plant gene length (e.g. 2.01 Kb for Arabidopsis thaliana) and, consequently, no gene structural annotation could be performed32.
Table 1.
Summary of the Persea americana Mill. cv ‘Hass’ draft genome assembly.
| Assembly Statistics | Contigs | Scaffolds |
|---|---|---|
| Total assembly size (Gb) | 1.03 | 1.01 |
| Total assembled sequences | 2,096,006 | 1,852,224 |
| Longest sequence length (Kb) | 57.80 | 160.08 |
| Average sequence length (Kb) | 0.49 | 0.54 |
| N50 index (sequences) | 475,145 | 377,224 |
| L50 length (Kb) | 0.56 | 0.68 |
GBS sequencing, mapping and variant calling
GBS (Genotyping-By-Sequencing) libraries for 71 avocado accessions (Table 2) were constructed and sequenced by Illumina HiSeq 2500 (1 × 100) and Illumina HISeq 4000 (2 × 150). The sequencing produced 405.93 million raw Illumina reads. After processing (see Methods), 345.37 million reads were obtained with differences among accessions in the number of reads (Supplementary Fig. S1 ). A higher number of processed reads is often associated to a higher number of mapped reads to each of the GBS locations. These reads of the individual genotypes were mapped onto the reference genome to retain only mapped reads to a unique localization in the genome. Such uniquely mapped reads represented approximately 80% of the total. Finally, 1,070,902 variants were detected. Of those, 945,064 were SNPs, 22,321 were InDels, 69,500 were MNPs (multi-nucleotide polymorphisms) and 6,604 were complex (as combination of the previous types).
Table 2.
List of the 71 Avocado accessions studied with SNPs in this work.
| Accesions | SampleID | Code | Germplasm collection | Previous race assignment | Race assignment predicted from the results of this work |
|---|---|---|---|---|---|
| 0028(Ardith) | 2835 | 1 | South Africa | GxM85 | GxM |
| A0.25 | A02554 | 2 | South Africa | Unknown | GxM |
| A0.68 | A06852 | 3 | South Africa | Unknown | GxM |
| 87.17.1 | 871728 | 4 | South Africa | Unknown | GxM |
| 1.14.2 | 114218 | 5 | South Africa | Unknown | GxWI |
| Alcaraz | ALCA74 | 6 | Spain | Unknown | GxM |
| Bacon | BACO39 | 7 | South Africa | GxM12, M11,41 or G40 | GxM |
| Bernecker | BERN18 | 8 | USA | WI86 | WI |
| Beta | BETA19 | 9 | USA | GxWI87 | GxWI |
| A0.57 | A05720 | 10 | South Africa | GxM12 | GxM |
| Butler | BUTL16 | 11 | USA | WI85 | WI |
| C.A. Bueno | CABU95 | 12 | Spain | Unknown | M |
| Catalina | CATA11 | 13 | USA | WI85 | WI |
| Choquette | CHOQ9 | 14 | USA | GxWI85 | GxWI |
| Cilfam | CILF46 | 15 | South Africa | Unknown | GxM |
| Colin V-33 | COLI31 | 16 | South Africa | GxM85 | GxM |
| Collinred B | COLL1 | 17 | USA | GxWI85 | GxWI |
| Collinson | COLL36 | 18 | USA | GxWI85 | GxWI |
| Dusa | DUSA33 | 19 | Spain | GxM12 | GxM |
| Edranol | EDRA63 | 20 | South Africa | Hybrid4 or G4 | GxM |
| Fuchsia | FUCH17 | 21 | USA | WI85 | GxMxWI |
| Fuerte | FUER16 | 22 | South Africa | GxM12 or M40 | GxM |
| G-6 | G692 | 23 | Spain | M12 | MxWI |
| Gem | GEM77 | 24 | Spain | GxM12 or G41 | GxM |
| Gottfried | GOTT04 | 25 | South Africa | M88 | MxWI |
| Grace | GRAC26 | 26 | South Africa | Unknown | GxM |
| Gwen | GWEN40 | 27 | South Africa | GxM85 or G40 | GxM |
| H287 | H28757 | 28 | South Africa | Unknown | GxM |
| Hansie | HANS05 | 29 | South Africa | Unknown | M |
| Hass | HASS38 | 30 | Spain | GxM11,31 or G12 | GxM |
| Hass | HASS55 | 31 | South Africa | GxM11,31 or G12 | GxM |
| Iriet | IRIE34 | 32 | Spain | GxM11 | GxM |
| A0.67 | A06729 | 33 | South Africa | Unknown | GxM |
| Lamb Hass | LAHA24 | 34 | South Africa | GxM11,12 | GxM |
| La Piscina | LAPI93 | 35 | Spain | Unknown | M |
| Largo | LARG24 | 36 | USA | WI85 | GxWI |
| Linda | LIND50 | 37 | South Africa | G85 | G |
| Lisa | LISA23 | 38 | USA | MxWI85 | GxMxWI |
| Lyon | LYON25 | 39 | South Africa | Hybrid41 or G85 | GxM |
| Maluma | MALU85 | 40 | Spain | GxM4 | GxM |
| Melendez 2 | MELE12 | 41 | USA | GxWI85 | GxWI |
| Mike | MIKE30 | 42 | South Africa | Unknown | G |
| Monroe | MONR10 | 43 | USA | MxWI85 or GxWI85 | GxWI |
| Mrs Tooley | MRTO08 | 44 | South Africa | Unknown | GxMxWI |
| Murrieta Green | MUGR27 | 45 | South Africa | G41 | G |
| Nabal | NABA21 | 46 | South Africa | G85 | G |
| Negra de la Cruz | NECR31 | 47 | South Africa | M89 | GxM |
| Nimlioh | NIML09 | 48 | South Africa | G85 | G |
| Nn10 | NN1068 | 49 | South Africa | G41 | GxM |
| NN63 | NN6310 | 50 | South Africa | G41 | GxM |
| Pinkerton | PINK45 | 51 | South Africa | GxM12 or G40 | GxM |
| Pollock | POLL6 | 52 | USA | WI85 | WI |
| Reed | REED89 | 53 | Spain | G41 | GxM |
| Regal | REGA11 | 54 | South Africa | Unknown | GxM |
| Rincon | RINC12 | 55 | South Africa | Unknown | GxM |
| RR-86 | RR8691 | 56 | Spain | Unknown | GxMxWI |
| Rustenburg Round | RURO36 | 57 | South Africa | Unknown | GxMxWI |
| Russell | RUSS22 | 58 | USA | WI85 | WI |
| Ryan | RYAN13 | 59 | South Africa | GxM85 | GxM |
| Semil 43 | SEMI14 | 60 | USA | GxWI86 | GxWI |
| Shepard | SHEP42 | 61 | South Africa | G41 | GxM |
| Teague | TEAG60 | 62 | South Africa | M41,85 | GxM |
| Telez | TELE66 | 63 | South Africa | Unknown | MxWI |
| Thomas | THOM90 | 64 | South Africa | M12 | MxWI |
| Toro Canyon | TOCA96 | 65 | South Africa | M12 or GxM16 | GxM |
| Trapp | TRAP2 | 66 | USA | WI85 | WI |
| TX531 | TX5344 | 67 | South Africa | Hybrid41 or G85 | GxM |
| Vero Beach n° 1 | VERO4 | 68 | USA | MxWI85 | MxWI |
| Waldin | WALD28 | 69 | USA | WI85 | WI |
| Wester | WEST5 | 70 | USA | WI85 | WI |
| Yon | YON3 | 71 | USA | GxWI85 | GxWI |
The race codes stand for: G = Guatemalan; M = Mexican; WI = West Indian. Interracial hybrids are indicated with a cross.
SNP development
After filtering (see Methods), 7,108 SNPs with no missing data, of which 19.45% were private (Supplementary Table S1), were detected for the 71 accessions (Table 2). The SNPs were categorized according to nucleotide substitutions: 61.04% were transitions [C/T (2195) or A/G (2144)] and 38.96% transversions [A/C (778), C/G (646), A/T (666), G/T (679)]. The transition/transversion ratio was 1.57, similar to the results reported in other species33–35. The mean of observed heterozygosity was 0.16 whereas the mean of expected heterozygosity was 0.17 and the average frequency of minor alleles was 0.11, although, for the samples studied, the population was not in Hardy-Weinberg equilibrium. This last result was expected taking into account that the material studied does not represent a randomly obtained population.
Diversity and population structure using filtered SNPs
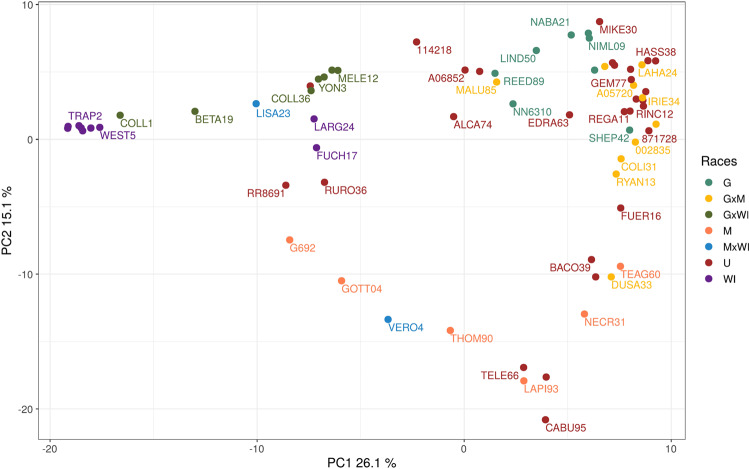
Distinct relationships among accessions were obtained with different analyses of the filtered SNPs. A first approximation to study genetic structure was obtained using principal component analysis (PCA) for the complete set of biallelic SNPs (Fig. 1). The first two components explained more than 40% of the variation (26.1% and 15.1%). Three differentiated groups that correspond with the three different horticultural races were observed. As expected, interracial hybrid accessions could be observed between the three main groups.
Figure 1.
Principal component analysis (PCA) of 71 avocado accessions with 7108 SNPs using the R software version 3.5.1 with the package ggplot2 version 374. Each genotype is represented with its sampleID (Table 2). The colors explain the race of the accessions according to the literature: turquoise green: G, yellow: GxM, dark green: GxWI, orange: M, red: U, orange: M, blue: MxWI, and purple: WI. (G: Guatemalan, M: Mexican, WI: West Indian and U: Unknown).
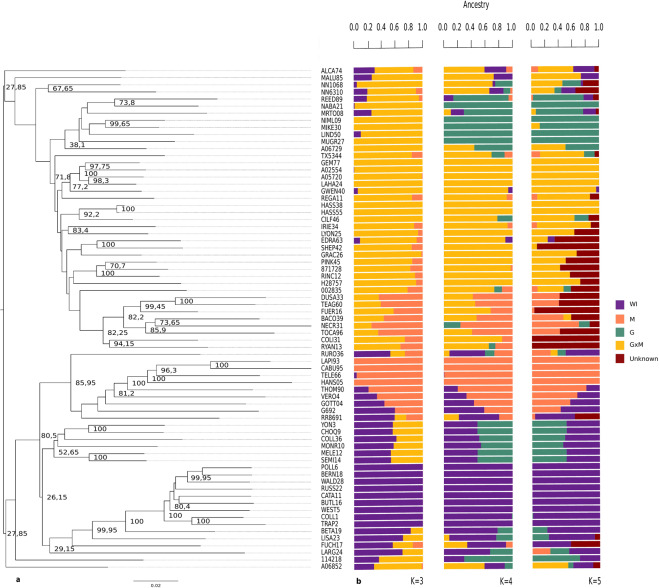
Prevosti’s distance36 was used to evaluate the genetic structure as a second approximation. This distance determines the fraction of different sites between samples. It was plotted as a dendrogram based on Neighbor Joining (NJ) showing the relationships between genotypes (Fig. 2a). Two main clusters weakly supported by bootstrap values (27.8) were revealed in the dendrogram. One of the clusters was composed of a big strongly supported subgroup (71.8) which included mainly Guatemalan x Mexican (GxM) hybrid genotypes (‘Pinkerton’, ‘Lyon’, ‘Iriet’, ‘Gem’, ‘Hass’, ‘Lamb Hass’, among others), a few genotypes categorized as Mexican (‘Teague’, ‘Negra de la Cruz’), as well as genotypes considered as Guatemalan (‘Shepard’), and a genotype of unknown race (‘TX531’). Another subgroup (bootstrap value of 38.1) included mainly accessions considered as Guatemalan (‘Reed’, ‘Nabal’, ‘Nimlioh’, ‘Linda’, ‘Murrieta Green’) and it was close to genotypes of unknown race (‘A0.67’, ‘Mike’,‘Mrs Tooley’). Moreover, the other two genotypes that are reported as Guatemalan (‘NN10’, ‘NN63’) form a strongly supported cluster (67.6), whereas ‘Maluma’ and ‘Alcaraz’ appear isolated of these subgroups.
Figure 2.
(a) Dendrogram based on Neighbour Joining (NJ) plotted using Figtree78 showing genetic relationships among 71 avocado accessions. Node labels represent bootstrap values (only values cited in the manuscript and values >70% are shown) out of 2000 bootstrap replicates. (b) Barplots describing the population stratification of the most probable number of clusters K = 4, followed by K = 3 and K = 5 were estimated with the ADMIXTURE software37. At K = 4, the avocado races were shown with different colors: orange: M; green: G; yellow: GxM hybrids; purple: WI; maroon: unknown. (G: Guatemalan, M: Mexican, WI: West Indian).
The second cluster was formed by two genotypes of unknown origin (‘A0.68’ and ‘1.14.2’) and a strongly supported group (bootstrap value of 80.5) composed of two subgroups. One of them (well supported with a bootstrap value of 85.9), contained genotypes considered as Mexican (‘G-6’, ‘Thomas’, ‘Gottfried’), a MxWI hybrid (‘Vero Beach No. 1’), as well as genotypes of unknown race (‘RR-86’, ‘Telez’, ‘Rustenburg Round’, ‘C.A. Bueno’ and ‘Hansie’). The other subgroup was weakly supported (bootstrap value of 26.1) and was composed of two subgroups. One of them (29.1 bootstrap value), contained mostly West Indian genotypes (‘Pollock’, ‘Bernecker’, ‘Waldin’, ‘Russel’, ‘Catalina’, ‘Butler’, ‘Wester’, ‘Trapp’, ‘Fuchsia’,‘Largo’), together with some Guatemalan × West Indian (GxWI) (‘Beta’, ‘Collinred B’) or Mexican x West Indian (MxWI) (‘Lisa’) hybrids. The other subgroup was also weakly supported (52.6), and was represented by GxWI hybrids (‘Yon’, ‘Choquette’, ‘Collinson’, ‘Melendez 2’ and ‘Semil 43’) and a MxWI hybrid (‘Monroe’).
An admixture analysis using the ADMIXTURE software37 was performed after the PCA analysis. The most favorable number of clusters was 4, followed by 3 and 5 although the differences among the number of populations were small with a cross-validation error between 0.28 and 0.29. At K = 4, the division between genotypes reported as Mexican, West Indian and Guatemalan was evident. Furthermore, a separated cluster was formed with the GxM hybrid genotypes (Fig. 2b). In order to have a broader view of the genetic structure of the populations, the STRUCTURE software38 and STRUCTURE HARVESTER39 were also implemented. In agreement with the ADMIXTURE results, K = 4 was revealed as the most probable number of clusters (Supplementary Figs. S2 and S3b) but, in this case, accessions considered as Guatemalan and as GxM hybrids were not clearly differentiated.
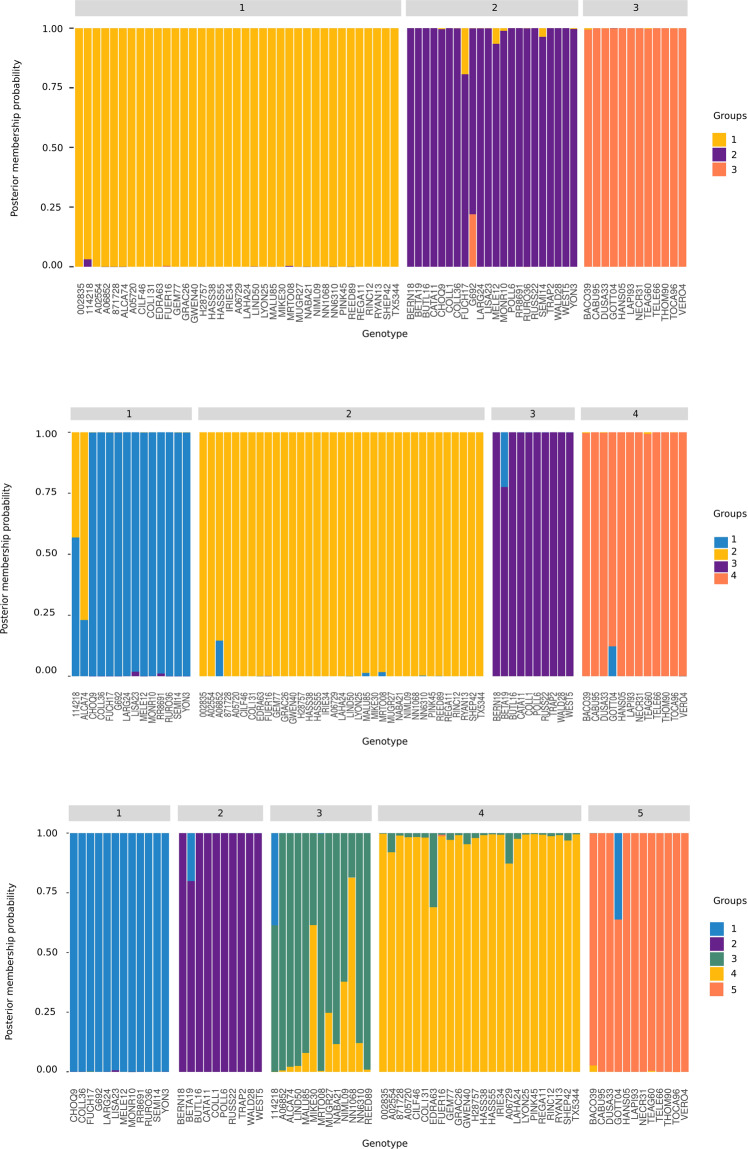
In order to describe the diversity between pre-defined groups, Discriminant Analysis of Principal Components (DAPC) was performed to obtain the number of clusters. These results were consistent with the cross-validation errors (ADMIXTURE) and Evanno algorithm (STRUCTURE) regarding the number of clusters (K). K = 4 was again revealed as the most likely scenario, closely followed by K = 3 and K = 5 (Fig. 3) (Supplementary Table S2). At K = 3, accessions were divided in agreement with the other methods (ADMIXTURE and STRUCTURE). One group included mainly Guatemalan race accessions and GxM hybrids. A second group consisted of West Indian race accessions, GxWI hybrids and MxWI hybrids. The third group included Mexican race genotypes, GxM hybrids and MxWI hybrids (Supplementary Table S2). For K = 4, the West Indian race accessions were divided into two groups, one which included mainly pure West Indian genotypes and another one which included mainly GxWI hybrid genotypes. For K = 5, Guatemalan genotypes and GxM hybrid genotypes were split into two different groups (Supplementary Table S2).
Figure 3.
Discriminant analysis of principal components (DAPC) to infer group structure for the number of groups K = 3–5 (obtained with the function find.clusters.) (Table S3) and produced using the R software version 3.5.1. Each genotype is a bin on the x-axis, and the assigned probability of population membership is shown as a stacked bar chart. Each population is shown in different color. Overall for K = 3, group 1: GxM, group 2: WI, group 3: M; for K = 4, group 1: GxWI and MxWI, group 2: GxM, group 3: WI, group 4: M; for K = 5, group 1: GxWI and MxWI, group 2: WI, group 3: G, group 4: GxM, group 5: M.
In order to validate the pre-defined clusters shown above, the fixation index (Fst value) was calculated for every pair of populations using the pre-defined groups (K = 3–5) by DAPC (Supplementary Table S2). In all cases, a contrast between populations was shown and supported the previous analysis. For K = 4, the lowest value was 0.18 between groups two (mostly genotypes considered as GxM hybrids, and some cultivars considered Guatemalan) and one (mostly cultivars considered as GxWI hybrids). The highest value was 0.61 between groups three (mostly cultivars considered as West-Indian) and two (mostly cultivars considered as GxM hybrids) (Table 3).
Table 3.
Fst genetic differentiation of 71 avocado accessions grouped by K = 4.
| Group1 [GxWI] | Group2 [G] + [GxM] | Group3 [WI] | Group4 [M] | |
|---|---|---|---|---|
| Group1 (GxWI) | 0 | 0.18 | 0.39 | 0.23 |
| Group2 | 0.18 | 0 | 0.61 | 0.33 |
| Group3 (WI) | 0.39 | 0.61 | 0 | 0.48 |
| Group4 (M) | 0.23 | 0.33 | 0.48 | 0 |
The most represented race per group is shown inside the parentheses.
Nucleotide diversity was also studied for each cluster using different indexes (Pi and Watterson’s Theta) (Table 4). For K = 4, Pi ranged from 270.14 to 515.27, and Watterson’s Theta ranged from 304.74 to 471.15. A higher diversity was obtained in the cluster with mainly Mexican genotypes, followed by the cluster with mainly West Indian and Guatemalan genotypes, whereas a lower diversity was shown in the group with mainly GxM hybrids.
Table 4.
Nucleotide diversity statistics according to population structure (K = 3, K = 4, and K = 5) performed by DAPC.
| Groups | Number of accessions | Pi | Watterson’s Theta | |
|---|---|---|---|---|
| K = 3 | 1 (GxM) | 37 | 273.65 | 307.58 |
| 2 (WI) | 22 | 543.69 | 521.76 | |
| 3 (M) | 12 | 515.27 | 471.15 | |
| K = 4 | 1 (GxWI) | 14 | 419.23 | 467.9 |
| 2 (GxM) | 35 | 270.14 | 304.74 | |
| 3 (WI) | 10 | 417.75 | 434.08 | |
| 4 (M) | 12 | 515.27 | 471.15 | |
| K = 5 | 1 (GxWI) | 12 | 420.06 | 458.96 |
| 2 (WI) | 10 | 417.75 | 434.08 | |
| 3 (G) | 13 | 293.23 | 303.88 | |
| 4 (GxM) | 24 | 234.76 | 264.03 | |
| 5 (M) | 12 | 515.27 | 471.15 |
The accessions belonging to each group are specified in the Supplementary Table S3.The most represented race per group is shown inside the parentheses.
The genetic diversity per group established by DAPC and minor allele frequencies were also analyzed. The highest observed heterozygosity (0.20) was shown in the cluster with mainly Mexican race cultivars and, in the case of minor allele frequencies, the highest values (0.11) were observed in the same group (Table 5).
Table 5.
Proportion of observed heterozygosity (Ho) and average minor allele frequency for K = 3, K = 4, and K = 5.
| Groups | Number of accessions | Proportion observed heterozygosity (Ho) | Average Minor allele frequency | |
|---|---|---|---|---|
| K = 3 | 1(GxM) | 37 | 0.14 | 0.08 |
| 2(WI) | 22 | 0.15 | 0.10 | |
| 3(M) | 12 | 0.20 | 0.11 | |
| K = 4 | 1(GxWI) | 14 | 0.19 | 0.11 |
| 2(GxM) | 35 | 0.14 | 0.08 | |
| 3(WI) | 10 | 0.10 | 0.07 | |
| 4(M) | 12 | 0.2 | 0.11 | |
| K = 5 | 1(GxWI) | 12 | 0.19 | 0.11 |
| 2(WI) | 10 | 0.10 | 0.07 | |
| 3(G) | 13 | 0.14 | 0.10 | |
| 4(GxM) | 24 | 0.14 | 0.10 | |
| 5(M) | 12 | 0.20 | 0.11 |
The most represented race per group is shown inside the parenthesis.
Assignment of genotypes of unknown or confusing pedigree to established groups
Based on the above analyses, the assignment of some genotypes of unknown or confusing pedigree to racial groups could be established. Among known genotypes with ambiguous racial assignments, examples include ‘Bacon’, ‘Edranol’, ‘Fuerte’, ‘Gem’, ‘Gwen’, ‘Hass’, ‘Lyon’, ‘Pinkerton’, ‘Toro Canyon’ and ‘TX531’ which have been considered by different authors as pure Mexican40, Guatemalan4,12,41 or GxM hybrids4,11,12 (Table 2). The ADMIXTURE results obtained in this work indicate that all are indeed GxM hybrids, although in ‘Edranol’ a West Indian component was also found. Some samples whose pedigree was unknown (‘A0.25’, ‘A0.68’, ‘87.17.1’, ‘1.14.2’ and ‘Alcaraz’) seem to be GxM hybrids although some probably are three-race hybrids with a low proportion of West Indian heritage. Other accessions (‘Mike’ and ‘Mrs Tooley’) seem to be pure Guatemalan whereas others (‘Hansie’ and ‘C.A. Bueno’) appear as pure Mexican.
Discussion
Although numerous crop breeding programs are benefiting from new molecular genotyping approaches, these advances are slower in most woody perennial species and especially in tropical and subtropical fruit crops since, in most cases, no previous significant genomic information is available. Regarding avocado, in spite of the different ongoing breeding programs and different types of molecular markers that have been developed and used in the last two decades5,8,10,14–19,28–31,40,42,43, there is still a need to generate additional markers that can be used at a large scale, especially to link molecular markers to most of the traits of agronomic interest, that are controlled by multiple genes. Thereby, the use of new approaches such as high throughput sequencing can fill this gap in order to speed up avocado breeding as has occurred in other crops.
A draft ‘Hass’ avocado genome for diversity analyses
In this study an avocado (cv. ‘Hass’) fragmented genome with small contigs was developed. This fragmentation presents several limitations for genomic studies, such as the impossibility to perform a gene structure annotation, and, consequently, its use for gene discovery. Nevertheless, this draft genome allowed aligning the reads from a reduced-representation approach, and obtaining a high number of molecular markers. Since the use of non-reference variant calling approaches such as Stacks44, TASSEL-UNEAK45 and GBS-SNP-CROP46 can increase the possibilities of variant miscalls46–48 the approach followed in this work using a fragmented genome draft is appropriate to reduce this problem. Previous studies have developed some SNP markers in avocado28–31,43 but, to our knowledge, this is the first time that an avocado draft genome has been used to facilitate SNP calling from a reduced-representation sequencing. Current work is underway to generate a reference genome of avocado starting from the draft ‘Hass’ genome developed in this work.
Diversity analyses and population structure
A total of 7,108 Single-Nucleotide Polymorphism (SNPs) were detected for the 71 accessions studied using a ‘Hass’ draft genome to align the reads. These molecular markers showed a higher proportion of transition substitutions (61.10%) over transversions (38.89%). This is commonly known as ‘transitions bias’ and it is explained by the fact that transitions are more conservative on proteins and has been reported in previous studies with different crops including avocado28,49–51. Probably due to the lack of sterility barriers between the avocado horticultural races, a low percentage (19.45%) of private SNPs was observed.
The average observed heterozygosity (0.16) was lower than the results reported in other studies based on simple sequence repeat (SSR) markers15–17 and with different accessions than those analyzed in this work. These differences have been obtained in other studies50,52 and were expected considering the nature of SSRs49,53. A lower level of observed heterozygosity was also reported compared to other woody perennial crops such as peach, litchi or olive54–56. These differences could be due to the kind of accessions considered. Thus, avocado market worldwide is currently dominated by a single cultivar, ‘Hass’, whereas in other fruit crops, as peach and olive, a wide range of cultivars is grown around the world. ‘Hass’ or ‘Hass’ descendants, such as ‘Gwen’, are part of the pedigree of different varieties in the GxM group (the most representative in this study) and this biased selection could result in a decrease of heterozygosity.
In this work, different analyses utilizing SNP markers (PCA, Neighbour-Joining, ADMIXTURE, STRUCTURE, and DAPC) were performed. These show a clear separation between horticultural races, although with exceptions in some STRUCTURE and DAPC results, in which a clear distinction between genotypes considered as Guatemalan and GxM hybrids was not obtained for K = 4 in contrast to ADMIXTURE with which a separation between those two groups was found. This difficulty in separating both groups was expected since Guatemalan genes predominate in current avocado germplasm57. Moreover, as there are not sterility barriers among the botanical races, admixture between different races may have occurred during avocado evolutionary history and domestication processes2. In any case, overall, the clustering inferred with DAPC resulted in lower admixture among accessions than that inferred with either STRUCTURE or ADMIXTURE. Similar results of genetic admixture underestimation with DAPC have been shown in other studies and could be due to overestimation of posterior membership probability by DAPC58,59. Interestingly at K = 5 a new subgroup is obtained with ADMIXTURE (Fig. 2b) in the GxM group. This new group could represent accessions with a higher Mexican component.
The group with mainly Mexican race accessions shows the highest genetic diversity and the highest proportion of private SNPs (46.42%) (Supplementary Table S3) together with a high observed heterozygosity. Similar results were also obtained in other studies11,12,16. Regarding the genetic diversity results, it should be noted that the group with mainly Guatemalan accessions and the group with mainly Mexican accessions show a higher genetic diversity than the GxM hybrid group, despite their lower sample size. The results obtained also show a clear separation of West Indian accessions from the two other horticultural races as has been reported in previous studies9,16,18,40 using a lower number of molecular markers. This is expected taking into account that the Mexican and Guatemalan races have a common ecological niche, in the tropical highlands, whereas the West Indian race is adapted to lowlands in Central America2.
Assignment of genotypes of unknown pedigree to established groups
In avocado the main criteria to assign genotypes to the three specific botanical races have been based on morphological traits and, since most of the accessions are developed from chance seedlings, their pedigree is unknown. The approach followed in this work allowed the assignment of some unknown or unclear genotypes to established groups. In agreement with previous works40, admixture among the three botanical races are shown for some cultivars, although GxM genotypes involve most of the accessions studied. These hybrids represent the most important avocado cultivars grown worldwide.
In this study, the development of a high number of SNPs after mapping the raw read to a draft avocado (cv. ‘Hass’) genome has allowed the genotyping and efficient discrimination of avocado accessions revealing a clear grouping based on racial origin. The SNP markers developed are a public resource that will be useful for future studies of avocado germplasm management and characterization, Genetic Selection (GS), Marker Assisted Selection (MAS), Genome Wide Association Studies (GWAS) or Quantitative Trait Loci (QTL) analyses and, consequently, helping to significantly reduce breeding costs in this crop. However, this progress will need additional studies to increase the number of available markers in order to have an optimum number of markers in the different avocado breeding populations.
Methods
Plant material
Seventy one avocado (Persea americana Mill.) accessions were selected and young leaves were collected in the field. The accessions analyzed combine genotypes from the different avocado races obtained from breeding programs (such as ‘Gem’, ‘Gwen’, ‘Iriet’ or ‘Lamb Hass’), commercial varieties (‘Bacon’, ‘Choquette’, ‘Edranol’, ‘Fuerte’, ‘Hass’ or ‘Reed’), rootstocks (‘Dusa’, ‘Thomas’ or ‘Toro Canyon’) and local Spanish accessions with interest as possible source of new rootstocks (‘La Piscina’ or ‘C.A. Bueno’). Those accessions are maintained in three different germplasm collections: IHSM La Mayora (IM; Algarrobo Costa, Spain), Westfalia Fruit (WF; Tzaneen, South Africa) and the US National Avocado Germplasm Repository (UA; Miami, FL, US) (Table 2). Two different samples of ‘Hass’ from two different germplasm collections were included in the analyses as control of the results obtained.
DNA extraction, library preparation, sequencing and processing the raw reads
DNA from leaves of each accession was isolated using a Qiagen DNeasy Plant Mini Kit following the manufacturer’s guidelines. The DNA purity and concentration were determined using NanoDrop spectrophotometer and Qubit 2.0 Fluorometer. The optimization of a library enzyme was performed on a ‘Hass’ genomic DNA sample digested with PstI, EcoT221, and ApeKI restriction enzymes. The DNA fragment distribution was assessed with Agilent 2100 Bioanalyzer System. Libraries were prepared using Sonah et al.60 protocol digesting 100 ng genomic DNA of each variety with ApeKI. The resulting libraries were sequenced with the Illumina HiSeq 2500 platform (1 × 100) at the Duke Center for Genomics and Computational Biology and the Illumina HiSeq 4000 platform (2 × 150) at the Novogene Corporation.
The raw reads were demultiplexed using GBSx package61. Then reads were processed to remove possible adapter sequences, discard reads shorter than 50 bases and filter low-quality regions by using Fastq-mcf software version 1.04.80762 (-l 50 and -q 30).
A draft avocado (cv.‘Hass’) genome assembly
In order to map the reads to a draft avocado genome, the ‘Hass’ genotype was sequenced (2 × 150) with a depth of 100X using the Illumina platform. The genome size and heterozygosity were estimated using the Kmer distribution approach described in Liu et al. 201363. In brief, Kmer distributions for 19, 25, 31, 37, 43, 55, 61, 67, 73 and 85-mers were calculated with Jellyfish and then loaded in the GenomeScope web portal64. Two different assemblers were used to assemble the Illumina reads, Minia65 and SOAPdenovo266. Although both of them use algorithms for de novo short read assemblies, Minia requires lower computational resources that SOAPdenovo2 and filters false positives65. Kmer sizes ranging from 17 to 115-mers (steps of 8) were used with both assemblers. The assembled contigs stats were compared across the different conditions and assemblers and the assembly produced by Minia65 with a Kmer of 115 was selected as the one that produced the most contiguous assembly as reported in other studies65. Contigs were scaffolded using SSPACE v3.067.
Mapping, SNP discovery and filtering
The generated reads were mapped with BWA version 0.7.10-r78968 with default parameters. Unmapped reads were removed using Samtools version 1.3.169 and BAM files were produced with the retained reads. All BAM files were merged by Bamaddrg (https://github.com/ekg/bamaddrg), and Samtools package version 1.3.169 was used to sort and index BAM files. FreeBayes version 0.9.2070 was run to detect variants and remove SNPs with mapping quality lower <20 and read depth <5. The raw SNPs obtained were further filtered using the VCFtools package version 0.1.12.71 removing no biallelic SNPs, missing data and SNPs within 1000 bp distance. Before and after filtering, a summary statistic was generated using Vcf-stats version 0.1.1271. Finally, only SNP variants were retained and their diversity was analyzed using Adegenet package version 2.1.172 and Hardy-Weinberg equilibrium was tested using pegas package version 0.1073.
Analysis of the genetic structure of diverse avocado accessions
In order to show the usefulness of the SNPs generated, the genetic relationships, genetic structure and group divergence of 71 avocado accessions were thoroughly analyzed using different methods such as PCA, NJ distance tree, DAPC and Bayesian clustering as well as genetic properties of these populations through parameter such as Fst, Pi and Watterson’s theta.
PCA was performed using Adegenet package version 2.1.172 and was plotted using ggplot2 packages version 374 in RStudio version 1.1.45375 and R version 3.5.1.
Prevosti’s distance ( where is the number of loci considered, Pajk the frequency of the allele arrangement k in the locus j in the population a, and Pajk the corresponding value in the population b36) matrix and Neighbor-joining (NJ) tree were generated via the Poppr package version 2.8.276,77 with 2000 bootstrap replicates using the SNP data set. The figures were plotted with FigTree version 1.4.478.
The population structure was studied with three different approaches (ADMIXTURE, STRUCTURE and DAPC). The three programs basically assign each of the accessions to one or more ancestral populations or clusters. They differ in how the data are processed and the algorithm used. Thus, maximum likelihood estimation of individual ancestries was analyzed with ADMIXTURE version 1.337 that was run iterating K from 1 to 20. This analysis is based on the same statistical model as STRUCTURE although it performs a maximum likelihood estimation of individuals instead of a Bayesian approach and, consequently, allows a faster cluster estimation from a large SNP dataset. Furthermore, in order to choose the optimum number of populations (K), a cross-validation approach was used for all the Single Nucleotide Polymorphism (SNPs). Each chosen value of K was plotted using RStudio version 1.1.45375 and R version 3.5.1. The STRUCTURE program was run five times per each number of populations (K). Each run was implemented with a burn-in period of 20000 steps followed by 200000 Monte Carlo Markov chain replicates79–81 Evanno et al.82 method was used to determine the most probable number of K with the software STRUCTURE HARVESTER39. Subsequently, since STRUCTURE-like approaches assume that markers are not linked and that populations are panmictic38, Discriminant Analysis of Principal Components (DAPC) was also applied in order to identify and describe well-defined clusters of genetically related genotypes using the R package Adegenet version 2.1.172. To perform this analysis, data were transformed using PCA. The find.clusters function was used to identify the number of clusters. The Bayesian Information Criterion (BIC) was calculated to associate with the correct number of subgroups, and a cross-validation function (XvalDapc) was used to corroborate the best number of PCA retained. Before this analysis, the files were read using read.vcf and converted into Genind and Genlight class with VcfR2genind and VcfR2genlight.
Finally, the Fixation index (Fst) which allows differentiating populations with ranges between 0 (no differentiation) and 1 (complete differentiation)83 was also obtained with the R package PopGenome version 2.6.184 to analyze group distinction. Moreover, Nucleotide diversity statistics Pi and Watterson’s theta were estimated considering the grouping produced by DAPC, K = 3, K = 4, and K = 5 and were also determined with the same package.
Supplementary information
Acknowledgements
This work was supported by Ministerio de Economía y Competitividad- European Regional Development Fund. (AGL2016-77267-R). AT was supported by an FPI fellowship from Ministerio de Economía y Competitividad (BES-2014-068832). We thank T. Hasing for help in library preparation and Y. Verdún for technical assistance. The authors acknowledge Advanced Research Computing at Virginia Tech for providing computational resources and technical support that have contributed to the results reported within this paper. The authors also thank Therese Bruwer and Zelda van Rooyen (Westfalia Fruit, South Africa) for providing some of the leaf material used in this study.
Author contributions
J.I.H., A.B., A.T. and A.J.M. conceived the experimental design. A.T. participated in the sample collection and DNA extraction. A.T. and A.S. prepared the libraries. A.T. and A.B. analyzed the data. All the authors discussed the results and contributed to the preparation of the final manuscript.
Data availability
The ‘Hass’ draft genome raw reads have been deposited at NCBI under the BioProject PRJNA564097. The GBS dataset is deposited under PRJNA564105. Most of the analyses have been carried out using R software 3.5.1. All scripts have been deposited at https://github.com/IHSMFruitCrops/Hass-genotyping.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56526-4.
References
- 1.Chase MW, et al. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181(1):1–20. doi: 10.1111/boj.12385. [DOI] [Google Scholar]
- 2.Schaffer, B., Wolstenholme, B. N. & Wiley, A. W. Introduction in The Avocado: Botany, Production, and Uses. (eds. Schaffer, B., Wolstenholme, B. N & Whiley, A. W.) 1–9 (CABI, Wallingford, UK, 2013).
- 3.FAO. Statistics Division of Food and Agriculture Organization of the United Nations (FAOSTAT) http://www.fao.org/faostat/es/#data/QC (Accessed September 13th 2019).
- 4.Crane, J. H. et al. Cultivars and rootstocks in The Avocado: Botany, Production, and Uses (eds. Schaffer, B., Wolstenholme, B. N & Whiley, A. W.) 1–9 (CABI, Wallingford, UK, 2013).
- 5.Lavi U, Hillel J, Vainstein A. Application of DNA fingerprints for identification and genetic analysis of avocado. J. Am. Soc. Hort. Sci. 1991;116:1078–1081. doi: 10.21273/JASHS.116.6.1078. [DOI] [Google Scholar]
- 6.Mhameed S, et al. Level of heterozygosity and mode of inheritance of variable number of tandem repeat loci in avocado. J. Am. Soc. Hort. Sci. 1996;121:778–782. doi: 10.21273/JASHS.121.5.768. [DOI] [Google Scholar]
- 7.Fiedler J, Bufler G, Bangerth F. Genetic relationships of avocado (Persea americana Mill.) using RAPD markers. Euphytica. 1998;101:249–255. doi: 10.1023/A:1018321928400. [DOI] [Google Scholar]
- 8.Furnier GR, Cummings MP, Clegg MT. Evolution of the avocados as revealed by DNA restriction site variation. J. Hered. 1990;81:183–188. doi: 10.1093/oxfordjournals.jhered.a110963. [DOI] [Google Scholar]
- 9.Davis J, Henderson D, Kobayashi M, Clegg MT, Clegg MT. Genealogical relationships among cultivated avocado as revealed through RFLP analysis. J. Hered. 1998;89:319–323. doi: 10.1093/jhered/89.4.319. [DOI] [Google Scholar]
- 10.Sharon D, et al. An integrated genetic linkage map of avocado. Theor. Appl. Genet. 1997;95:911–921. doi: 10.1007/s001220050642. [DOI] [Google Scholar]
- 11.Schnell RJ, et al. Evaluation of avocado germplasm using microsatellite markers. J. Am. Soc. Hort. Sci. 2003;128:881–889. doi: 10.21273/JASHS.128.6.0881. [DOI] [Google Scholar]
- 12.Ashworth VETM, Clegg MT. Microsatellite markers in avocado (Persea americana Mill.): genealogical relationships among cultivated avocado genotypes. J. Hered. 2003;94:407–415. doi: 10.1093/jhered/esg076. [DOI] [PubMed] [Google Scholar]
- 13.Ashworth VETM, Kobayashi MC, De La Cruz M, Clegg MT. Microsatellite markers in avocado (Persea americana Mill.): development of dinucleotide and trinucleotide markers. Sci. Hortic. 2004;101:255–267. doi: 10.1016/j.scienta.2003.11.008. [DOI] [Google Scholar]
- 14.Borrone WJ, Schnell RJ, Viola HA, Ploetz RC. Seventy microsatellite markers from Persea americana Miller (avocado) expressed sequences tags. Mol. Ecol. Notes. 2007;7:439–444. doi: 10.1111/j.1471-8286.2006.01611.x. [DOI] [Google Scholar]
- 15.Alcaraz ML, Hormaza JI. Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Hereditas. 2007;144:244–253. doi: 10.1111/j.2007.0018-0661.02019x. [DOI] [PubMed] [Google Scholar]
- 16.Gross-German E, Viruel MA. Molecular characterization of avocado germplasm with a new set of SSR and EST-SSR markers: genetic diversity, population structure, and identification of race-specific markers in a group of cultivated genotypes. Tree Genet. Genomes. 2013;9:539–555. doi: 10.1007/s11295-012-0577-5. [DOI] [Google Scholar]
- 17.Guzmán LF, et al. Genetic structure and selection of a core collection for long term conservation of avocado in Mexico. Front. Plant. Sci. 2017;8:243. doi: 10.3389/fpls.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boza JE, et al. Genetic differentiation, races and interracial admixture in avocado (Persea americana Mill.), and Persea spp. evaluated using SSR markers. Genet. Resour. Crop. Ev. 2018;65:1195–1215. doi: 10.1007/s10722-018-0608-7. [DOI] [Google Scholar]
- 19.Ge Y, et al. Transcriptome sequencing of different avocado ecotypes: de novo transcriptome assembly, annotation, identification and validation of EST-SSR Markers. Forests. 2019;10:411. doi: 10.3390/f10050411. [DOI] [Google Scholar]
- 20.Ching A, et al. SNP frequency, haplotype structure and linkage disequilibrium in elite maize inbred lines. BMC Genetics. 2002;3:19. doi: 10.1186/1471-2156-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasheed A, et al. Crop breeding chips and genotyping plataforms: progress, challenge, and perspectives. Mol. Plant. 2017;10:1047–1064. doi: 10.1016/j.molp.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Scheben A, Batley J, Edwards D. Genotyping-by-sequencing approaches to characterize crop genomes: choosing the right tool for the right application. Plant Biotecnol. J. 2017;15:149–161. doi: 10.1111/pbi.12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studer, B. & Kölliker, R. SNP Genotyping Technologies. In Diagnostics in Plant Breeding (eds. Lübberstedt, T. & Varshney, R. K.) (Springer Science + Business Media Dordrecht, 2013).
- 24.Chagné D, et al. Development of a set of SNP markers present in expressed genes of the apple. Genomics. 2008;92:353–358. doi: 10.1016/j.ygeno.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Tan HW, Fang W. Developing single nucleotide polymorphism (SNP) markers from transcriptome sequences for identification of longan (Dimocarpus longan) germplasm. Hortic. Res. 2015;2:14065. doi: 10.1038/hortres.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibarra-Laclette E, et al. Deep sequencing of the Mexican avocado transcriptome, an ancient angiosperm with a high content of fatty acids. BMC Genomics. 2015;16:599. doi: 10.1186/s12864-015-1775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vergara-Pulgar C, et al. De novo assembly of Persea americana cv. “Hass“ transcriptome during fruit development. BCM Genomics. 2019;20:108. doi: 10.1186/s12864-019-5486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn DN, et al. Application of genomic tools to avocado (Persea americana) breeding: SNP discovery for genotyping and germplasm characterization. Sci. Hortic. 2019;246:1–11. doi: 10.1016/j.scienta.2018.10.011. [DOI] [Google Scholar]
- 29.Ge Y, et al. Genome-wide assessment of avocado germplasm determined from Specific Length Amplified Fragment sequencing and transcriptomes: population structure, genetic diversity, identification, and application of race-specific markers. Genes. 2019;10:215. doi: 10.3390/genes10030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinstein M, et al. Genetic diversity of avocado (Persea americana Mill.) germplasm using pooled sequencing. BMC Genomics. 2019;20:379. doi: 10.1186/s12864-019-5672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rendón-Anaya M, et al. The avocado genome informs deep angiosperm phylogeny, highlights introgressive hybridization, and reveals pathogen-influenced gene space adaptation. PNAS. 2019;116:17081–17089. doi: 10.1073/pnas.1822129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wortman JR, et al. Annotation of the Arabidopsis genome. Plant Physiol. 2003;132:461–468. doi: 10.1104/pp.103.022251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soorni A, Fatahi R, Salami SA, Haak DC, Bombarely A. Assessment of genetic diversity and population structure in Iranian cannabis germplasm. Sci Rep. 2017;7:15668. doi: 10.1038/s41598-017-15816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shearman JR, et al. SNP identification from RNA sequencing and linkage map construction of rubber tree for anchoring the draft genome. PLoS. One. 2015;10:e0121961. doi: 10.1371/journal.pone.0121961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pootakham W, et al. Genome-wide SNP discovery and identification of QTL associated with agronomic traits in oil palm using genotyping-by-sequencing (GBS) Genomics. 2015;105:288–295. doi: 10.1016/j.ygeno.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 36.Prevosti A, Ocaña J, Alonso G. Distance between populations of Drosophila subobscura based on chromosome arrangement frequencies. Theor. Appl. Genet. 1975;45:231–241. doi: 10.1007/BF00831894. [DOI] [PubMed] [Google Scholar]
- 37.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 40.Chen H, Morrell PL, Ashworth VETM, Clegg MT. Tracing the geographic origins of major avocado cultivars. J. Hered. 2009;100:56–65. doi: 10.1093/jhered/esn068. [DOI] [PubMed] [Google Scholar]
- 41.Variety Database of the Univ. of California at Riverside, http://ucavo.ucr.edu/ (Accessed September 13th 2019) (2019).
- 42.Lavi U, Cregan PB, Hillel J. Application of DNA markers for identification and breeding of fruit trees. Plant Breed. Rev. 1994;12:195–226. [Google Scholar]
- 43.Chen H, Morrell PL, de la Cruz M. Nucleotide diversity and linkage disequilibrium in wild avocado (Persea americana Mill.) J Hered. 2008;99:382–389. doi: 10.1093/jhered/esn016. [DOI] [PubMed] [Google Scholar]
- 44.Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: Building and genotyping loci de novo from short-read sequences. G3-Genes Genom. Genet. 2011;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu F, et al. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS. Genet. 2013;9:e1003215. doi: 10.1371/journal.pgen.1003215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melo ATO, Bartaula R, Hale L. GBS-SNP-CROP: a reference-optional pipeline for SNP discovery and plant germplasm characterization using variable length, paired-end genotyping-by-sequencing data. BMC Bioinformatics. 2016;17:29. doi: 10.1186/s12859-016-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leggett RM, MacLean D. Reference-free SNP detection: dealing with the data deluge. BMC Genomics. 2014;15:S10. doi: 10.1186/1471-2164-15-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berthouly-Salazar C, et al. Genotyping-by-Sequencing SNP identification for crops without a reference genome: using transcriptome based mapping as an alternative strategy. Front. Plant. Sci. 2016;7:777. doi: 10.3389/fpls.2016.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taranto F, D´Agostino N, Greco B, Cardi T, Tripoli P. Genome-wide SNP discovery and population structure analysis in pepper (Capsicum annum) using genotyping by sequencing. BMC Genomics. 2016;17:943. doi: 10.1186/s12864-016-3297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pootakham W, et al. Construction of high-density integrated genetic linkage map of rubber tree (Hevea brasiliensis) using genotyping-by-sequencing (GBS) Genomics. 2015;6:367. doi: 10.3389/fpls.2015.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kujur A, et al. Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea. Front. Plant. Sci. 2015;6:162. doi: 10.3389/fpls.2015.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Micheletti D, et al. Whole-Genome Analysis of diversity and SNP-major gene association in peach germplasm. Plant. Genome. 2015;5:92–102. doi: 10.1371/journal.pone.0136803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helyar SJ, et al. Application of SNPs for population genetics of nonmodel organisms: new opportunities and challenges. Mol. Ecol. Resour. 2011;1:123–36. doi: 10.1111/j.1755-0998.2010.02943.x. [DOI] [PubMed] [Google Scholar]
- 54.Aranzana MJ, Illa E, Howad W, Arús P. A first insight into peach [Prunus persica (L.) Batsch] SNP variability. Tree Genet. Genomes. 2012;8:1359–1369. doi: 10.1007/s11295-012-0523-6. [DOI] [Google Scholar]
- 55.Biton I, et al. Development of a large set of SNP markers for assessing phylogenetic relationships between the olive cultivars composing the Israel olive germplasm collection. Mol. Breed. 2015;35:107. doi: 10.1007/s11032-015-0304-7. [DOI] [Google Scholar]
- 56.Liu W, et al. Identifying litchi (Litchi chinensis Sonn.) cultivars and their genetic relationships using single nucleotide polymorphism (SNP) markers. PLoS. One. 2015;10:e0135390. doi: 10.1371/journal.pone.0135390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chanderbali, A. S., Soltis, D. E.,Soltis, P. S. & Wolstenholme, B. N. Taxonomy and botany in The Avocado: Botany, Production, and Uses. (eds. Schaffer, B., Wolstenholme, B. N & Whiley, A. W.) 32–50 (CABI, Wallingford, UK, 2013).
- 58.Söderquist P, et al. Admixture between released and wild game birds: a changing genetic landscape in European mallards (Anas platyrhynchos) Eur. J. Wildl. Res. 2017;63:98. doi: 10.1007/s10344-017-1156-8. [DOI] [Google Scholar]
- 59.Frosch C, et al. The genetic legacy of multiple beaver reintroductions in Central Europe. PLoS. One. 2014;9:e97619. doi: 10.1371/journal.pone.0097619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonah H, et al. An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS. One. 2013;8:e54603. doi: 10.1371/journal.pone.0054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herten K, Hestand MS, Vermeesch JR, Van Houdt JKJ. GBSX: a toolkit for experimental design and demultiplexing genotyping by sequencing experiments. BMC Bioinformatics. 2015;16:73. doi: 10.1186/s12859-015-0514-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aronesty E. Comparison of sequencing utility programs. Open Bioinforma. J. 2013;7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
- 63.Liu, B. et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. Preprint at, https://arxiv.org/abs/1308.2012 (2013).
- 64.Vurture GW, et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics. 2017;33:2202–2204. doi: 10.1093/bioinformatics/btx153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chikhi R, Rizk G. Space-efficient and exact de Bruijn graph representation based on a Bloom filter. Algorithm. Mol. Biol. 2013;8:22. doi: 10.1186/1748-7188-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo RB, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217x-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–9. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 68.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transformation. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garrison E. & Marth G. Haplotype-based variant detection from short-read sequencing. Preprint at, http://arxiv.org/abs/1207.3907 (2012).
- 71.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 73.Paradis E. Pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 74.Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag New York, 2009).
- 75.R core Team. R: a language and environment for statistical computing. R foundation for statistical computing, Vienna; https://www.R-project.org (Accessed September 13th 2019) (2018).
- 76.Kamvar ZN, Tabina JF, Grünwald NJ. Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ Prepr. 2014;2:e281. doi: 10.7717/peerj.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kamvar ZN, Brooks JC, Grünwald NJ. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015;6:208. doi: 10.3389/fgene.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rambaut, A. FigTree version 1.4.4, http://tree.bio.ed.ac.uk/software/figtree/ (Accessed September 13th 2019).
- 79.Larrañaga N, et al. A Mesoamerican origin of cherimoya (Annona cherimola Mill.): Implications for conservation of plant genetic resources. Mol. Ecol. 2017;26:4116–4130. doi: 10.1111/mec.14157. [DOI] [PubMed] [Google Scholar]
- 80.Martin C, Herrero M, Hormaza JI. Molecular characterization of apricot germplasm from an old stone collection. PLoS. One. 2011;6:e23979. doi: 10.1371/journal.pone.0023979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pritchard, J. K., Wen, X. & Falush, D. Documentation for structure software: version 2.3. Preprint at, http://burfordreiskind.com/wp-content/uploads/Structure_Manual_doc.pdf (Accessed September 13th 2019) (2010).
- 82.Evanno G, Regnaut S, GOUDET J. Detecting the number of clusters of individuals using the software: STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 83.Hahn, M. W. Population structure in Molecular Population Genetics. (eds Sinauer Associates) 81–83 (Oxford University Press. U.S.A., 2018).
- 84.Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014;31:1929–36. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hofshi, R. Avocado database, http://www.avocadosource.com/AvocadoVarieties/QueryDB.asp (Accessed September 13th 2019).
- 86.U.S. National Plant Germplasm System, https://npgsweb.ars-grin.gov/gringlobal/search.aspx? (Accessed September 13th 2019).
- 87.Avocado information database, https://www.myavocadotrees.com/beta-avocado.html (Accessed September 13th 2019).
- 88.Wolfe, H. S., Toy, L. R. & Stahl, A. L. Avocado production in Florida. Fl. Agr. Ext. Serv. Bull. 141 (1949).
- 89.Ben-Ya’cov, A., Zilberstaine, M., Goren, M. & Tomer, E. The Israeli avocado germplasm bank: where and why the items had been collected. In Proc. V World Avocado Congress. Spain. October 19–24 (2003).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ‘Hass’ draft genome raw reads have been deposited at NCBI under the BioProject PRJNA564097. The GBS dataset is deposited under PRJNA564105. Most of the analyses have been carried out using R software 3.5.1. All scripts have been deposited at https://github.com/IHSMFruitCrops/Hass-genotyping.