Abstract
Quantitative real-time PCR (qPCR) is commonly used for deciphering gene functions. For effective qPCR analyses, suitable reference genes are needed for normalization. The objective of this study is to identify the appropriate reference gene(s) for qPCR analyses of the leaves and roots of ramie (Boehmeria nivea L.), an important natural fiber crop. To accomplish this goal, we investigated the expression patterns of eight common plant qPCR reference genes in ramie leaves and roots under five abiotic stresses, five hormonal treatments, and one biotic stress. The relative expression stabilities of the eight genes were evaluated using four common but different approaches: geNorm, NormFinder, BestKeeper, and RefFinder. Across the 11 tested conditions, ACT1 was the most stably expressed among the eight genes while GAPDH displayed the biggest variation. Overall, while variations in the suggested reference genes were found for different tissue x treatment combinations, our analyses revealed that together, genes ACT1, CYP2, and UBQ can provide robust references for gene expression studies of ramie leaves under most conditions, while genes EF-1α, TUB, and ACT1 can be used for similar studies of ramie roots. Our results should help future functional studies of the genes in ramie genome across tissues and environmental conditions.
Subject terms: Gene expression analysis, Plant molecular biology
Introduction
RNA abundance analysis is an important approach for studying gene functions. Frequently used methods for quantifying RNA abundance include Northern blotting1, ribonuclease protection assay (RPA)2, semi-quantitative reverse transcription polymerase chain reaction (sRT-PCR)3, and quantitative real-time PCR (qPCR)4. Among these methods, qPCR has become an increasingly prevalent method due to its high specificity, high sensitivity, wide dynamic range, and relatively low cost. qPCR experiment is often used to validate gene expression data obtained from microarray hybridization or from transcriptomics based on next-generation sequencing platforms. However, the usefulness and interpretation of qPCR results depend heavily on a number of factors, including the quality and quantity of extracted RNA samples, the efficiency of the reverse transcription reaction, random errors in experiments, and the reference gene selected for normalization and comparison5–7. Among these factors, the expression stability of the reference gene is crucial for minimizing systematic biases and for accurately normalizing target gene expression using qPCR. Therefore, identifying suitable and reliable internal reference genes represents a critical step in qPCR analyses.
Many studies have been conducted to identify reference genes for qPCR in different organisms, targeting different tissues, developmental stages, and environmental conditions. For plants, a number of genes have been frequently used as reference genes for qPCR, including the following house-keeping genes: 18S rRNA, ACT (β or γ actin), TUB (α or β tubulin), EF-1α (elongation factor 1α), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and UBQ (poly-ubiquitin)8–12. In addition, several new genes were recently identified as stably expressed in different plants and plant tissues under selected conditions. These new candidate reference genes include F-box (a F-box family protein), SAND (a SAND family protein), PP2A (protein phosphatase 2 A), PEPKR1 (phosphoenolpyruvate carboxylase-related kinase 1), TIP41 (Tap42-interacting protein of 41 kDa), eIF-4α (eukaryotic translation initiation factor 4α), CRKs (CDPK-related kinases), ABC (ATP-binding cassette transporter), CYP2 (cyclophilin2), ELF1 (eukaryotic elongation factor 1, α or β), IDE (insulin-degrading enzyme), CBP20 (carotenoid-binding protein 20), UBC (ubiquitin-conjugating enzyme), and SamDC (S-adenosylmethionine decarboxylase gene)8–11.
Ramie (Boehmeria nivea L.) or “China grass” is an important natural fiber crop mainly grown in China, India, and Southeast Asian and other Pacific Rim countries12. Aside from its long stem fiber, other components of the plant are also of significant economic value, including a high level of crude protein contents in leaves and shoots that make them an ideal source of feed for beef cattle and geese13. However, various biotic and abiotic stresses, such as root-lesion nematodes14, fungal diseases15–18, viral infections19, ramie moth20, drought, flooding21, heavy metal contamination22, and nutrient deficiency23 often limit the productivity of ramie plants. At present, the detailed mechanisms underlying the ramie plants’ responses to these biotic and abiotic stresses are largely unknown. Using a variety of molecular tools, a few recent studies identified gene expression differences associated with ramie plants’ responses to drought, nutrient deficiency, cadmium (Cd) contamination, and infections by the fungal pathogen Colletotrichum gloeosporioides, the nematode Pratylenchus coffeae, and the ramie moth Cocytodes coerulea Guenée12,24–33. However, in their confirmations through qPCR, these studies used different reference genes and/or analyzed different plant tissues and/or stress conditions. As a result, it is difficult to compare observations from different studies.
The objectives of this study are to analyze the expression stability of a set of candidate reference genes in ramie plants under a variety of conditions and to identify the most suitable reference gene(s) for future qPCR analyses of gene expressions in this plant. Based on results from previous studies of ramie24–27,30–33 and in other plants8–11,34, the following eight genes showed relatively stable expressions under different conditions: 18S rRNA, ACT1, GAPDH, α-TUB, EF1α, UBQ, F-box, and CYP2. We thus selected these genes for evaluation as potential reference genes for future studies of gene expression in ramie using qPCR. Specifically, the expression stability of these genes in the roots and leaves of ramie plants under different abiotic stresses (high temperature, low temperature, drought, Cd contamination, and high-salt stress), hormonal stimuli (salicylic acid [SA], Benzothiadiazole [BTH], methyl jasmonate [MeJA], ethephon [ETH], and gibberellin [GA3]), and biotic stress (infection by the parasitic oomycete Pythium vexans) were analyzed using four commonly used analytical programs (geNorm, NormFinder, BestKeeper, and RefFinder). Our analyses identified specific reference gene(s) for each given condition for future gene expression studies of ramie leaves and roots.
Materials and Methods
Plant sample preparation and treatment
An elite ramie cultivar, Zhongzhu No. 2, was used in this study. The seedlings were prepared via the stem cutting propagation method to obtain genetically identical seedlings of similar age and size25. The ramie seedlings were grown at 25 ± 1 °C with a 12 h photoperiod, 75 ± 1% relative humidity (RH), under light intensity of 5000 lux, in a greenhouse within the Institute of Bast Fiber Crops, Chinese Academy of Agricultural Science (IBFC, CAAS). When the seedlings were approximately 20 cm in height, they were used in the following treatments. For drought, salt, and heavy metal stress treatments, ramie plants were subjected to 200 mL of 20% PEG 8000, 200 mM of NaCl, and 400 µM CdCl2 for 4 hours, respectively. For cold and heat shock treatments, plants were transferred to a light incubator at temperatures of 4 °C and 40 °C for 4 h, respectively. For hormone treatments, ramie plants were subjected to MeJA (100 µM), SA (100 µM), BTH (100 µM), ETH (100 ppm), and GA3 (1 mM) respectively for 4 h, either by foliar spraying or by root soaking8,34. For the biotic stress treatment, ramie roots were inoculated with P. vexans isolate HF1 and incubated for three days using the method described previously16. All of the treatments were each performed in three biological replicates. Untreated plants grown for the same time intervals were collected as controls. The harvested plant materials (roots, leaves) from each of the treatments were washed with MINIQ-filtered water, immediately frozen in liquid nitrogen, and stored at −80 °C until use.
Candidate reference genes selection and primer design
Eight candidate reference genes (18S rRNA, ACT1, GAPDH, α-TUB, EF1α, UBQ, F-box, and CYP2) were selected for this study based on two sets of criteria. The first was the relative stability of their expressions under different stresses, including drought, Cd, infection by root-lesion nematodes, and feeding by ramie moth larvae as revealed by transcriptome analyses using the next generation sequencing platform in ramie (Supplementary Table S1)24–27,30–33. The second criterion was that these eight genes had been used as internal reference genes for qPCR experiments in many other plants8–11,34. The sequences of the eight genes in ramie were obtained from either the NCBI database or the assembled transcriptome sequences described in previous studies24–27,30,31,33. Information about these eight genes is shown in Supplementary Table S2. Gene specific primers (Table 1) were designed using Primer3Plus (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi)35.
Table 1.
Information about the eight candidate reference genes and primer sequences used for quantitative real-time PCR (qPCR) in this study.
| Gene symbol | Gene description | Gene ID | Primer sequences (5′-3′)* | Amplicon length (bp) | Tm (°C) | PCR efficiency (%) | Correlation coefficient (R2) |
|---|---|---|---|---|---|---|---|
| 18S | 18S rRNA | AF206870.1 | tgacggagaattagggttcga/ccgtgtcaggattgggtaatt | 100 | 60 | 99 | 0.9743 |
| ACT | Actin1 | DQ665832.2 | gttgaaccctaaggctaacagag/ggaatccagcacgataccag | 139 | 60 | 100 | 0.9944 |
| GAPDH | glyceraldehyde-3- phosphate dehydrogenase | comp37700_c0 | tggagacaagaaacagcaccct/cggcaattccgccatttaac | 121 | 60 | 100 | 0.9635 |
| EF-1α | Elongation factor 1α | comp36076_c0 | tggccgtccttggaaatacc/ctcctgggcatcgtgacttt | 121 | 60 | 99 | 0.9578 |
| αTUB | α-tubulin | comp24081_c0 | tgcatttcggtccacatcg/catcatcacctccgccaac | 132 | 60 | 100 | 0.9222 |
| CYP2 | Cyclophilin2 | comp26478_c0 | cggcgagtctatctacgga/tgacgatttccattccctcg | 200 | 60 | 100 | 0.9626 |
| UBQ | Ubiquitin | comp20072_c0 | agacgagcataacatttcctgc/gccgtactcttgccgattac | 151 | 60 | 100 | 0.9273 |
| F-box | F-box family protein | comp30228_c0 | atagagggcgtaggctgagg/tccttcgggtggttatgttc | 127 | 60 | 98 | 0.9889 |
*The primer sequences represent forward/reverse primers for each gene.
RNA extraction and cDNA synthesis
For both root and leaf samples from each treatment, the total RNA was extracted using an EASYspin Plus Total RNA Kit (Aidlab, Beijing, China), following the manufacturer’s protocol. The concentration and purity of total RNA was determined using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was examined using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). First-strand cDNAs were synthesized from 1 μg of total RNA from the leaf or root samples, using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany).
qPCR amplification
To analyze expression stability of the eight candidate genes, we first determined the specificity and PCR efficiency for each of the designed primer pairs. To test the specificity of the primers to the target genes, PCR was performed using cDNA as the template. All PCR products were examined by agarose [1.5% (w/v)] gel electrophoresis. For each primer pair, the appearance of a single band of the expected size on the agarose gel was considered consistent with the primers being specific for the target gene. To test for PCR amplification efficiency, we followed the method described previously based on standard dilution curves in qPCR36, using the combined cDNA sample from the roots and leaves of control plants. All qPCR was performed using LightCycler 480 SYBR Green I Master, Roche LightCycler 96-well plates and Roche LightCycler 480 II (Roche Diagnostics, Mannheim, Germany). Each reaction mixture contained 10 μL SYBR Green I Master Mix, 3 μL diluted cDNA, 1 μL of forward primer (10 μM), 1 μL of reverse primer (10 μM), and 5 µL ddH2O in a total volume of 20 μL. The following amplification conditions were applied for all eight genes: 1 cycle at 95 °C for 10 min, 45 cycles at 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 15 s, followed by 1 cycle of 95 °C for 10 s, 65 °C for 60 s and 95 °C for 1 s. RNase-free water was used as a negative control. In our analyses, each biological sample (i.e. a treatment x tissue combination) had three biological replicates, and each biological replicate had three technical replicates.
Gene expression stability and statistical analysis
For each qPCR reaction, we obtained a Ct (threshold-cycle) value. The mean ± standard deviation (SD) of the Ct value for each tissue x treatment combination was calculated and the statistical significance of the Ct value differences between tissues and among treatments was obtained using the Student’s t-test. The Ct values for both types of tissues (i.e. leaves and roots) under the tested conditions were then analyzed by GeNorm37, NormFinder38, BestKeeper39 and RefFinder40 to evaluate and rank the expression stability of the eight candidate reference genes. These four programs were chosen because they use different formulae to calculate and rank expression stabilities and that they have been widely used to rank candidate reference genes for qPCR experiments across all major groups of organisms. Among the four programs, RefFinder is the most comprehensive. It is a web-based program that integrates four computational programs (geNorm, Normfinder, BestKeeper, and the comparative delta-Ct method) to compare and rank the tested candidate reference genes. Specifically, it uses the ranking information from each program, assigns an appropriate weight to each individual gene, and calculates the geometric mean of their weights for the overall final ranking40. Previous studies e.g.34,36 have found that these programs provide complementary information to help researchers in their final selection of reference genes for specific purposes.
Results
Specificity and amplification efficiency of qPCR primers
The specificity of the gene-specific primers was determined by agarose gel electrophoresis of PCR amplicons and melting curve analysis. Agarose gel electrophoresis of PCR products of the eight candidate reference genes all showed a single band at the expected sizes (Fig. 1). In addition, each of the melting curves of these eight genes in ramie samples showed a single peak in qPCR, and the amplification curves of the nine repeats (three biological replicates x three technical replicates) for each gene had excellent repeatability (Supplementary Fig. S1). These results indicated that the primers were specific for their respective target genes. The PCR amplification efficiencies of the eight primer pairs ranged from 0.98 to 1.0 (Table 1), meeting the requirements for qPCR experiments. These results indicated that the primers for all eight candidate reference genes can be used for analyzing their expression patterns through qPCR.
Figure 1.
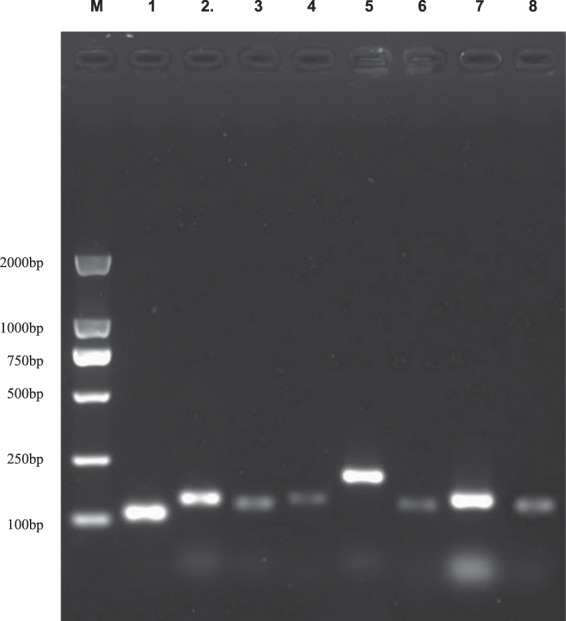
PCR products of eight candidate reference genes. Lane M: AL2000 DNA ladder, lane 1: 18S, lane 2: ACT1, lane 3: GAPDH, lane 4: αTUB, lane 5: CYP, lane 6: F-box, lane 7: UBQ, lane 8: EF-1α.
Expression profiling of candidate reference genes
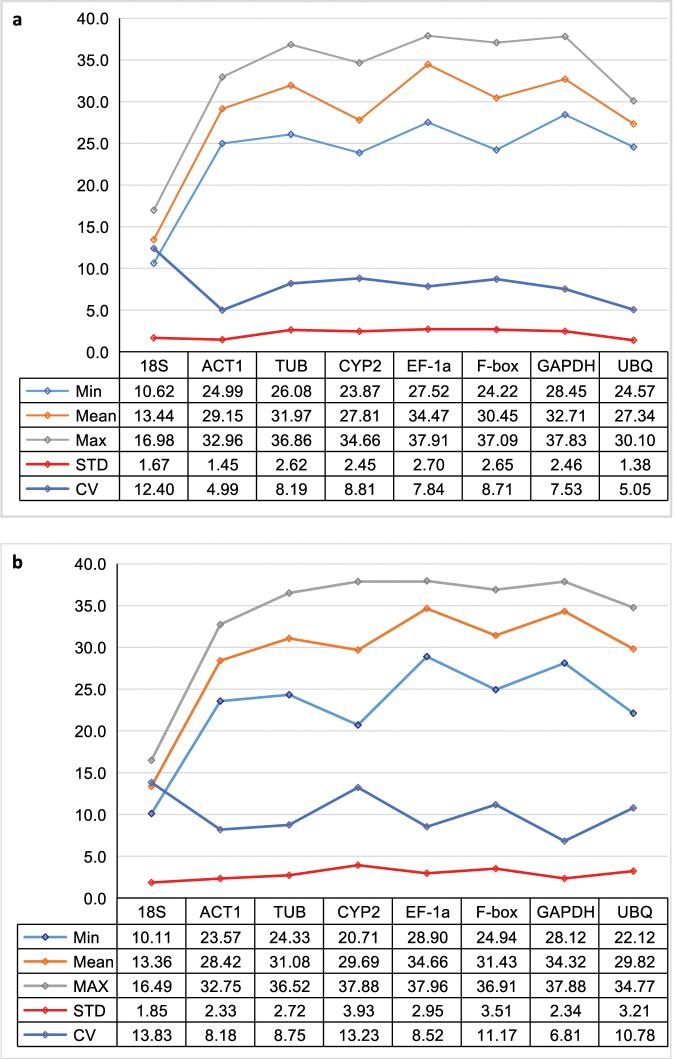
In qPCR, Ct values are used to quantify the expression levels of genes, with a low Ct value indicating a high gene expression level and vice versa. The expression profiles of the eight candidate reference genes in the roots and leaves of ramie in different treatments are summarized in Fig. 2. All eight candidate genes showed variations in expression levels, with different genes showing different ranges. Among the eight genes, the Ct values ranged from 9.6 to 37.9 among the root samples and 10.6 to 37.8 among the leaf samples. The 18S rRNA gene showed the lowest average Ct values [13.44 ± 1.67 (mean ± standard deviation) in leaves and 13.36 ± 1.85 in roots], consistent with its high concentration in individual cells, while the EF-1α showed the highest average Ct value (34.47 ± 2.70 in the leaves and 34.66 ± 2.95 in the roots).
Figure 2.
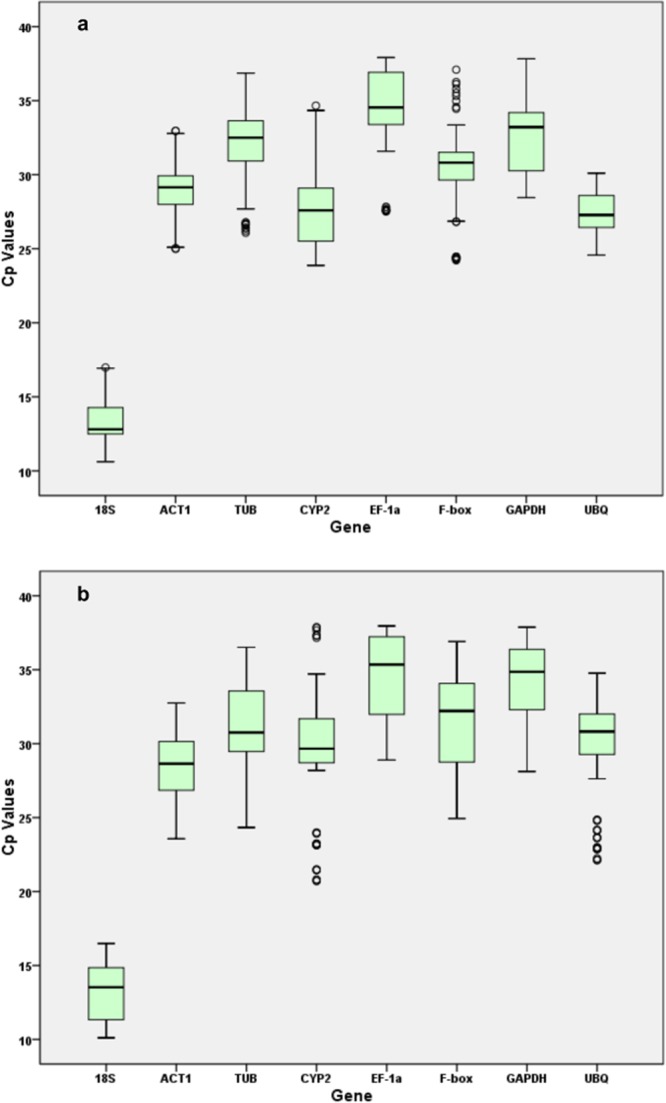
Box-plot graphs of Ct values for all candidate reference genes in leaves (a) and roots (b) of ramie under different conditions. Variations are displayed as medians (lines), 25th to 75th percent (boxes) and ranges (whiskers). The gene abbreviations are the same as in Table 1.
Based on the means and standard deviations of Ct values, the overall coefficient of variation (CV) of the Ct values was calculated for each gene across all tested conditions. Low CV values indicated relatively low variability and high stability across the treatment conditions. The CV values of the eight candidate reference genes among all root samples were overall low and similar, ranging from 6.81% (for GAPDH) to 13.83% (for 18S rRNA); among all leaf samples the range was from 4.99 (for ACT1) to 12.40% (for 18S rRNA), respectively (Fig. 3). On the basis of CV values, the stability ranking of the eight candidate reference genes in root samples across all treatment conditions was: GAPDH > ACT1 > EF-1α > TUB > UBQ > F-box > CYP2 > 18S rRNA, while in the leaf samples, the stability ranking was ACT1 > UBQ > GAPDH > EF-1α > TUB > F-box > CYP2 > 18S rRNA. However, there were wide variations among treatments and groups of treatments in the relative rankings of expression stability among the eight genes. Below we summarize the main rankings by the four different analytical methods.
Figure 3.
Data statistics of Ct values of eight candidate reference genes in leaves (a) and roots (b) of ramie. Total number of Ct values for each reference gene is 108.
Analysis of candidate reference genes by genorm
The stability of potential reference genes was first examined via geNorm software and the expression stability (M) of all eight candidates was calculated. A candidate gene with an M value <1.5 is considered to be a good reference gene for qPCR37. In all leaf samples, ACT1 and UBQ were the most stably expressed genes, with the same M value of 1.13, while EF-1α, with an M value of 2.29, was the least stably expressed gene. Among the ramie samples subjected to abiotic stresses (including low and high temperatures, high salt, heavy metal, and drought), the CYP2 and F-box genes were the most stably expressed in their leaves, both with an M value of 0.62, while the other six candidates showed less stable expression, all with M values over 1.73. In the leaf samples of ramie plants treated with different hormones (SA, BTH, MeJA and GA3), the 18S rRNA and ACT1 genes were the most stably expressed, both with an M value of 0.84. However, in the leaf samples of ramie infected with P. vexans, the F-box and UBQ genes were the most stably expressed, both having an M value of 0.54. For the leaves of ramie plants under other treatments, 18S rRNA, ACT1, UBQ, and CYP2 were the top ranked, showing relatively stable expression levels within individual treatments, while in most cases, the expression level of GAPDH was the least stable (Table 2).
Table 2.
Suitability ranking, based on the geNorm program, of the eight candidate genes as potential reference genes for qPCR experiments involving leaves and roots of ramie plants under different treatments.
| Tissue | Rank | Total | Abiotic | Hormone | P. vexans | SA | BTH | JA | ETH | GA3 | Heat | Cold | NA | Cd | PEG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | 1 |
ACT1 (1.13) |
CYP2 (0.62) |
18S (0.84) |
F-box (0.54) |
EF-1a (0.11) |
18S (0.16) |
ACT1 (0.15) |
UBQ (0.18) |
18S (0.42) |
18S (0.10) |
18S (0.21) |
CYP2 (0.29) |
CYP2 (0.40) |
ACT1 (0.51) |
| 2 |
UBQ (1.13) |
F-box (0.62) |
ACT1 (0.84) |
UBQ (0.54) |
GAPDH (0.11) |
ACT1 (0.16) |
UBQ (0.15) |
TUB (0.18) |
F-box (0.42) |
CYP2 (0.10) |
TUB (0.21) |
UBQ (0.29) |
F-box (0.40) |
GAPDH (0.51) |
|
| 3 |
GAPDH (1.71) |
ACT1 (1.73) |
UBQ (0.99) |
EF-1a (0.71) |
CYP2 (0.14) |
F-box (0.18) |
F-box (0.28) |
CYP2 (0.21) |
ACT1 (0.51) |
UBQ (0.48) |
EF-1a (0.32) |
F-box (0.38) |
EF-1a (0.50) |
UBQ (0.64) |
|
| 4 |
TUB (1.91) |
UBQ (1.83) |
CYP2 (1.17) |
18S (0.83) |
TUB (0.16) |
EF-1a (0.26) |
18S (0.37) |
F-box (0.26) |
UBQ (0.52) |
F-box (0.58) |
CYP2 (0.41) |
GAPDH (0.49) |
ACT1 (0.63) |
TUB (0.69) |
|
| 5 |
CYP2 (2.01) |
GAPDH (2.04) |
GAPDH (1.32) |
GAPDH (0.89) |
F-box (0.17) |
TUB (0.32) |
GAPDH (0.45) |
GAPDH (0.38) |
EF-1a (0.56) |
ACT1 (0.70) |
F-box (0.43) |
EF-1a (0.56) |
TUB (0.76) |
EF-1a (0.77) |
|
| 6 |
F-Box (2.03) |
TUB (2.2) |
TUB (1.45) |
TUB (0.95) |
ACT1 (0.25) |
CYP2 (0.45) |
CYP2 (0.59) |
18S (0.45) |
TUB (0.60) |
TUB (0.77) |
GAPDH (0.47) |
ACT1 (0.62) |
UBQ (0.82) |
18S (0.89) |
|
| 7 |
18S (2.19) |
EF-1a (2.41) |
F-box (1.69) |
ACT1 (1.03) |
18S (0.36) |
UBQ (0.58) |
TUB (0.75) |
EF-1a (0.55) |
CYP2 (0.68) |
EF-1a (0.9) |
ACT1 (0.57) |
18S (0.73) |
18S (0.96) |
CYP2 (1.02) |
|
| 8 |
EF-1a (2.29) |
18S (2.57) |
EF-1a (1.79) |
CYP2 (1.44) |
UBQ (0.52) |
GAPDH (0.71) |
EF-1a (0.86) |
ACT1 (0.76) |
GAPDH (0.76) |
GAPDH (1.07) |
UBQ (0.63) |
TUB (0.83) |
GAPDH (1.26) |
F-box (1.10) |
|
| Root | 1 |
ACT1 (1.11) |
F-box (1.08) |
18S (1.18) |
ACT1 (0.17) |
TUB (0.19) |
ACT1 (0.33) |
EF-1a (0.3) |
EF-1a (0.43) |
GAPDH (0.37) |
EF-1a (0.24) |
F-box (0.14) |
EF-1a (0.38) |
ACT1 (0.35) |
F-box (0.37) |
| 2 |
TUB (1.11) |
UBQ (1.08) |
UBQ (1.18) |
TUB (0.17) |
GAPDH (0.19) |
CYP2 (0.33) |
TUB (0.3) |
CYP2 (0.43) |
CYP2 (0.37) |
TUB (0.24) |
18S (0.14) |
GAPDH (0.38) |
CYP2 (0.35) |
ACT1 (0.37) |
|
| 3 |
F-box (1.65) |
ACT1 (1.54) |
ACT1 (1.31) |
F-box (0.24) |
F-box (0.21) |
TUB (0.39) |
18S (0.35) |
18S (0.51) |
UBQ (0.42) |
ACT1 (0.43) |
ACT1 (0.23) |
ACT1 (0.5) |
EF-1a (0.43) |
GAPDH (0.42) |
|
| 4 |
EF-1a (1.71) |
EF-1a (1.57) |
CYP2 (1.38) |
EF-1a (0.39) |
EF-1a (0.26) |
18S (0.42) |
UBQ (0.43) |
GAPDH (0.58) |
18S (0.58) |
UBQ (0.49) |
UBQ (0.38) |
18S (0.68) |
18S (0.54) |
TUB (0.50) |
|
| 5 |
GAPDH (1.78) |
TUB (1.69) |
TUB (1.43) |
CYP2 (0.57) |
CYP2 (0.27) |
UBQ (0.51) |
GAPDH (0.47) |
TUB (0.66) |
F-box (0.83) |
18S (0.56) |
TUB (0.46) |
TUB (0.85) |
UBQ (0.66) |
18S (0.71) |
|
| 6 |
18S (1.9) |
GAPDH (1.79) |
GAPDH (1.5) |
UBQ (0.63) |
ACT1 (0.33) |
F-box (0.61) |
F-box (0.58) |
ACT1 (0.71) |
EF-1a (0.94) |
F-box (0.64) |
CYP2 (0.5) |
F-box (0.97) |
GAPDH (0.79) |
EF-1a (0.77) |
|
| 7 |
UBQ (1.98) |
18S (1.91) |
F-box (1.76) |
GAPDH (0.72) |
UBQ (0.41) |
EF-1a (0.74) |
ACT1 (0.67) |
F-box (0.74) |
ACT1 (1.03) |
CYP2 (0.74) |
EF-1a (0.54) |
CYP2 (1.05) |
F-box (0.95) |
UBQ (0.80) |
|
| 8 |
CYP2 (2.08) |
CYP2 (2.19) |
EF-1a (1.9) |
18S (0.81) |
18S (0.49) |
GAPDH (0.86) |
CYP2 (0.89) |
UBQ (0.79) |
TUB (1.12) |
GAPDH (1.01) |
GAPDH (0.88) |
UBQ (1.11) |
TUB (1.14) |
CYP2 (1.48) |
For all samples of ramie roots, ACT1 and TUB (both with an M value of 1.11) were the most stably expressed based on geNorm, while CYP2 was the least stably expressed. In the root samples of ramie under abiotic stresses, F-box and UBQ were the most stably expressed genes, with the same M value of 1.08, while CYP2 displayed the least stable expression with an M value of 2.19. For the ramie root samples under different hormonal treatments, the 18S rRNA and ACT1 were the most stably expressed genes, with the same M value of 0.84. In root samples infected with P. vexans, all eight candidate genes showed M values below 1.5; the ranking of the 8 genes based on M values was ACT1/TUB (0.17) < F-box (0.240) < EF-1α (0.39) < CYP2 (0.57) < UBQ (0.63) < GAPDH (0.72) < 18S rRNA (0.81). For the roots of ramie plants under other single treatments, EF-1α and CYP2 were overall the most stably expressed, while in most cases GAPDH was the least stably expressed (Table 2).
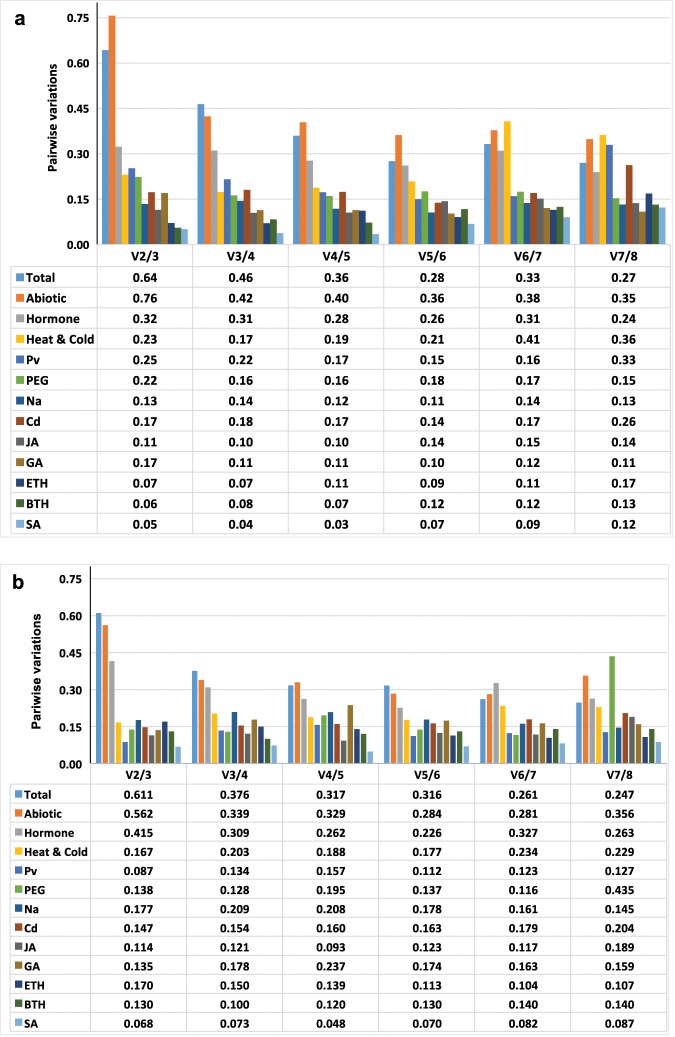
To determine the optimal number of reference genes for gene expression studies, we performed a stepwise calculation of the pairwise variation (Vn/Vn + 1) between sequential normalization factors using geNorm. In this analysis, a Vn/Vn + 1 < 0.15 indicates that introducing an additional reference gene for normalization is not necessary37. Among the eight genes, the most stable reference gene sets varied among samples and treatments (Fig. 4). For the leaf samples of ramie plants subjected to SA, BTH, ETH, GA, and cold or high salt conditions, the V2/V3 values were all lower than 0.15, indicating that two reference genes were sufficient for normalization. For leaf samples treated with PEG, infected with P. vexans, under abiotic stress, or subjected to a hormonal treatment, no Vn/Vn+1 value was <0.15, i.e. no optimal reference gene number was suggested by the program geNorm (Fig. 4a). A variable pattern was also noted for the root samples. For example, in the root samples of ramie plants subjected to SA, BTH, GA3, or JA treatments or under Cd, PEG, or cold stresses, the V2/V3 values were all lower than 0.15, indicating that two reference genes should be used for normalization. In contrast, for the root samples of ramie plants subjected to high temperature stress or stimulated by ETH, three and four reference genes were recommended for qPCR analyses of gene expressions, respectively (Fig. 4b).
Figure 4.
Determination of the optimal reference gene number by the geNorm program based on calculations using the pairwise variation (Vn/Vn + 1) approach. The optimal number of reference genes (n) is reached when Vn/Vn + 1 < 0.15. (a) leaves; (b) roots.
Analysis of reference genes by NormFinder
Analyses using the model-based approach NormFinder revealed the stability value of each gene under various treatments. Overall, the most stable reference gene for all the leaf and root samples was ACT1, with stability values of 0.658 and 0.382, respectively. Individually, ACT1 was the most stably expressed gene in both the leaf and root samples under abiotic stresses. However, under hormonal treatments, CYP2 (0.419) and TUB (0.407) were the most stably expressed genes for leaves and roots, respectively. In the leaf and root samples of ramie plants infected by P. vexans or subjected to cold or heat stress, F-box and ACT1 were the most stably expressed genes, respectively. Based on NormFinder, GAPDH showed among the least stable expression under most conditions, both in the root and leaf samples (Table 3). Among individual treatments, the most suitable reference genes identified by NormFinder were similar to those selected by geNorm except for the GA3-treated plants for both the leaf and root samples. Thus, the reference genes recommended by NormFinder were highly consistent with those obtained by geNorm analysis.
Table 3.
Suitability ranking, based on the NormFinder program, of the eight candidate genes as potential reference genes for qPCR experiments.
| Tissue | Rank | Total | Abiotic | Hormone | P.vexans | SA | BTH | JA | ETH | GA3 | Heat | Cold | NA | Cd | PEG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | 1 |
ACT1 (0.66) |
ACT1 (0.52) |
CYP2 (0.42) |
F-box (0.13) |
GAPDH (0.04) |
ACT1 (0.05) |
18S (0.13) |
UBQ (0.05) |
ACT1 (0.12) |
18S (0.04) |
CYP2 (0.07) |
CYP2 (0.07) |
TUB (0.24) |
EF-1a (0.18) |
| 2 |
UBQ (0.93) |
UBQ (0.59) |
GAPDH (0.43) |
UBQ (0.19) |
CYP2 (0.05) |
F-box (0.10) |
ACT1 (0.31) |
TUB (0.06) |
UBQ (0.18) |
CYP2 (0.04) |
F-box (0.17) |
UBQ (0.10) |
F-box (0.42) |
TUB (0.24) |
|
| 3 |
GAPDH (1.04) |
F-box (1.15) |
TUB (0.55) |
EF-1a (0.39) |
ACT1 (0.11) |
18S (0.17) |
UBQ (0.35) |
CYP2 (0.21) |
EF-1a (0.24) |
UBQ (0.37) |
18S (0.22) |
F-box (0.31) |
ACT1 (0.45) |
UBQ (0.40) |
|
| 4 |
F-box (1.12) |
GAPDH (1.18) |
ACT1 (0.78) |
18S (0.46) |
EF-1a (0.13) |
EF-1a (0.19) |
GAPDH (0.37) |
GAPDH (0.25) |
TUB (0.31) |
F-box (0.51) |
TUB (0.25) |
EF-1a (0.44) |
CYP2 (0.45) |
GAPDH (0.53) |
|
| 5 |
CYP2 (1.15) |
CYP2 (1.39) |
18S (1.10) |
TUB (0.54) |
TUB (0.18) |
TUB (0.22) |
CYP2 (0.39) |
F-box (0.27) |
F-box (0.37) |
TUB (0.56) |
EF-1a (0.30) |
ACT1 (0.45) |
UBQ (0.55) |
ACT1 (0.60) |
|
| 6 |
TUB (1.19) |
EF-1a (1.49) |
UBQ (1.17) |
GAPDH (0.63) |
F-box (0.24) |
CYP2 (0.42) |
F-box (0.43) |
18S (0.27) |
18S (0.44) |
EF-1a (0.61) |
ACT1 (0.46) |
GAPDH (0.49) |
18S (0.59) |
CYP2 (0.71) |
|
| 7 |
EF-1a (1.43) |
TUB (1.76) |
F-box (1.27) |
ACT1 (0.94) |
18S (0.44) |
UBQ (0.63) |
TUB (0.68) |
EF-1a (0.64) |
GAPDH (0.59) |
ACT1 (0.74) |
GAPDH (0.46) |
18S (0.53) |
EF-1a (0.66) |
18S (0.74) |
|
| 8 |
18S (1.46) |
18S (1.87) |
EF-1a (1.28) |
CYP2 (1.82) |
UBQ (0.67) |
GAPDH (0.72) |
EF-1a (0.74) |
ACT1 (0.93) |
CYP2 (0.61) |
GAPDH (1.05) |
UBQ (0.52) |
TUB (0.72) |
GAPDH (1.43) |
F-box (0.76) |
|
| Root | 1 |
ACT1 (0.38) |
ACT1 (0.22) |
TUB (0.41) |
ACT1 (0.07) |
TUB (0.13) |
CYP2 (0.03) |
18S (0.11) |
EF-1a (0.17) |
18S (0.18) |
TUB (0.08) |
EF-1a (0.02) |
ACT1 (0.24) |
ACT1 (0.12) |
ACT1 (0.13) |
| 2 |
TUB (0.75) |
EF-1a (0.65) |
CYP2 (0.52) |
TUB (0.11) |
GAPDH (0.13) |
18S (0.13) |
EF-1a (0.13) |
CYP2 (0.21) |
GAPDH (0.43) |
ACT1 (0.14) |
F-box (0.03) |
F-box (0.46) |
CYP2 (0.2) |
F-box (0.13) |
|
| 3 |
GAPDH (0.94) |
F-box (0.88) |
ACT1 (0.55) |
F-box (0.13) |
F-box (0.15) |
ACT1 (0.27) |
TUB (0.27) |
18S (0.33) |
F-box (0.48) |
EF-1a (0.15) |
ACT1 (0.07) |
EF-1a (0.54) |
EF-1a (0.26) |
GAPDH (0.15) |
|
| 4 |
UBQ (1.08) |
UBQ (0.93) |
GAPDH (0.66) |
EF-1a (0.39) |
ACT1 (0.17) |
F-box (0.34) |
UBQ (0.34) |
GAPDH (0.40) |
EF-1a (0.51) |
UBQ (0.20) |
UBQ (0.18) |
GAPDH (0.59) |
UBQ (0.41) |
TUB (0.18) |
|
| 5 |
F-box (1.09) |
TUB (0.99) |
UBQ (1.18) |
CYP2 (0.45) |
CYP2 (0.24) |
TUB (0.40) |
GAPDH (0.35) |
TUB (0.41) |
ACT1 (0.63) |
18S (0.36) |
18S (0.23) |
TUB (0.59) |
18S (0.52) |
18S (0.68) |
|
| 6 |
EF-1a (1.14) |
GAPDH (1.09) |
18S (1.20) |
UBQ (0.51) |
EF-1a (0.27) |
UBQ (0.55) |
ACT1 (0.50) |
ACT1 (0.42) |
CYP2 (0.71) |
F-box (0.53) |
TUB (0.47) |
CYP2 (0.62) |
GAPDH (0.57) |
EF-1a (0.71) |
|
| 7 |
CYP2 (1.32) |
18S (1.52) |
F-box (1.33) |
GAPDH (0.59) |
UBQ (0.33) |
EF-1a (0.63) |
F-box (0.53) |
F-box (0.55) |
UBQ (0.73) |
CYP2 (0.79) |
CYP2 (0.53) |
18S (0.65) |
F-box (0.95) |
UBQ (0.79) |
|
| 8 |
18S (1.32) |
CYP2 (1.94) |
EF-1a (1.42) |
18S (0.69) |
18S (0.48) |
GAPDH (0.77) |
CYP2 (1.04) |
UBQ (0.58) |
TUB (0.86) |
GAPDH (1.25) |
GAPDH (1.31) |
UBQ (0.78) |
TUB (1.12) |
CYP2 (2.41) |
Analysis of reference genes by BestKeeper
The excel-based BestKeeper algorithm was also used to evaluate the expression stability of the eight candidate reference genes. This approach initially uses data on the standard deviation (SD) and co-efficient of variation (CV) of the average Ct values for the specific treatments. The lower the SD and CV values, the more stable its expression is among the treatments. In this analysis, genes with SD > 1 were considered as undesirable reference genes. The genes with SD values less than 1 were then analyzed to derive a BestKeeper Index. When all the treatments were considered together, the Ct SD values were all greater than 1 for both the leaf and root samples. Thus, our results based on this criterium alone suggested that no single gene could be used as the reference gene across all the treatments. However, there were big variations among the genes for subsets of the samples and tested conditions, with some showing greater promise as candidate reference genes than others. For example, in ramie leaves stimulated by hormonal treatments and under abiotic stresses, both 18S rRNA and UBQ showed low SD values and relatively stable expression levels. For root samples, none of the eight genes showed stable expression in groups of ramie plants subjected to hormonal treatments, under abiotic stresses, or in the total samples. However, among the single treatments, genes 18S rRNA, EF-1α, F-box, and TUB showed relatively stable expression (SD < 1) in most cases. In contrast, genes CYP2, UBQ, and GAPDH displayed relatively unstable expression under many conditions, for both the leaf and root samples (Table 4). Taken together, results from BestKeeper analyses suggest that different treatments will require separate considerations for reference genes in qPCR experiments (Table 4).
Table 4.
Ranking the expression stabilities of eight candidate reference genes in ramie leaves and roots based on the Bestkeeper program.
| Tissue | Rank | Total | Abiotic | Hormone | P. vexans | SA | BTH | JA | ETH | GA3 | Heat | Cold | NA | Cd | PEG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | 1 |
ACT1 1.09 ± 3.75 |
UBQ 0.92 ± 3.34 |
18S18S 0.44 ± 3.55 |
F-box 0.36 ± 1.12 |
TUB 0.02 ± 0.08 |
F-box 0.19 ± 0.63 |
ACT1 0.10 ± 0.35 |
CYP2 0.37 ± 1.47 |
18S 0.33 ± 2.55 |
CYP2 0.25 ± 0.94 |
ACT1 0.12 ± 0.44 |
18S 0.15 ± 1.15 |
CYP2 0.31 ± 1.10 |
EF-1a 0.12 ± 0.31 |
| 2 |
UBQ 1.12 ± 4.10 |
ACT1 1.16 ± 3.89 |
ACT1 0.82 ± 2.87 |
UBQ 0.48 ± 1.64 |
F-box 0.05 ± 0.21 |
EF-1a 0.22 ± 0.64 |
GAPDH 0.12 ± 0.37 |
UBQ 0.37 ± 1.49 |
F-box 0.39 ± 1.24 |
TUB 0.37 ± 1.08 |
TUB 0.19 ± 0.62 |
TUB 0.31 ± 0.95 |
Box 0.34 ± 1.09 |
TUB 0.39 ± 1.10 |
|
| 3 |
18S 1.23 ± 9.22 |
18S 1.30 ± 8.91 |
UBQ 1.08 ± 4.00 |
18S 0.50 ± 3.77 |
EF-1a 0.09 ± 0.31 |
18S 0.24 ± 1.91 |
UBQ 0.16 ± 0.57 |
ACT1 0.38 ± 1.40 |
UBQ 0.43 ± 1.61 |
EF-1a 0.56 ± 1.52 |
EF-1a 0.24 ± 0.70 |
UBQ 0.42 ± 1.53 |
GAPDH 0.41 ± 1.28 |
F-box 0.62 ± 1.73 |
|
| 4 |
CYP2 1.76 ± 6.32 |
EF-1a 1.52 ± 4.20 |
CYP2 1.34 ± 5.02 |
TUB 0.52 ± 1.58 |
GAPDH 0.15 ± 0.49 |
ACT1 0.25 ± 0.84 |
F-box 0.19 ± 0.62 |
TUB 0.41 ± 1.34 |
CYP2 0.58 ± 2.10 |
UBQ 0.74 ± 2.69 |
CYP2 0.36 ± 1.27 |
CYP2 0.66 ± 2.24 |
ACT1 0.48 ± 1.61 |
CYP2 0.71 ± 2.13 |
|
| 5 |
F-box 1.83 ± 6.00 |
F-box 2.11 ± 6.79 |
TUB 1.54 ± 4.95 |
GAPDH 0.57 ± 1.71 |
CYP2 0.15 ± 0.59 |
TUB 0.30 ± 0.91 |
18S 0.21 ± 1.69 |
GAPDH 0.49 ± 1.58 |
GAPDH 0.60 ± 1.78 |
18S 0.78 ± 4.76 |
18S 0.50 ± 4.34 |
F-box 0.83 ± 2.67 |
EF-1a 0.6 ± 1.76 |
18S 0.72 ± 5.13 |
|
| 6 |
GAPDH 1.93 ± 5.93 |
GAPDH 2.25 ± 6.76 |
GAPDH 1.55 ± 4.77 |
EF-1a 0.67 ± 1.85 |
ACT1 0.37 ± 1.32 |
GAPDH 0.58 ± 1.72 |
CYP2 0.74 ± 2.64 |
F-box 0.51 ± 1.69 |
ACT1 0.70 ± 2.42 |
F-box 0.80 ± 2.70 |
GAPDH 0.55 ± 1.89 |
EF-1a 0.87 ± 2.52 |
18S 0.67 ± 4.83 |
UBQ 0.81 ± 2.84 |
|
| 7 |
EF-1a 2.01 ± 5.83 |
CYP2 2.30 ± 8.08 |
F-box 2.08 ± 7.05 |
ACT1 0.77 ± 2.52 |
18S 0.65 ± 4.96 |
CYP2 0.80 ± 2.86 |
TUB 0.89 ± 2.74 |
EF-1a 0.58 ± 1.76 |
EF-1a 0.83 ± 2.45 |
ACT1 0.92 ± 3.08 |
UBQ 0.60 ± 2.33 |
GAPDH 0.97 ± 2.96 |
TUB 0.94 ± 2.67 |
GAPDH 0.85 ± 2.34 |
|
| 8 |
TUB 2.01 ± 6.30 |
TUB 2.53 ± 7.74 |
EF-1a 2.12 ± 6.49 |
CYP2 0.94 ± 3.26 |
UBQ 0.89 ± 3.23 |
UBQ 1.04 ± 3.79 |
EF-1a 0.94 ± 2.64 |
18S 0.66 ± 5.68 |
TUB 0.86 ± 2.65 |
GAPDH 0.94 ± 2.65 |
F-box 0.76 ± 2.53 |
ACT1 1.03 ± 3.62 |
UBQ 1.14 ± 4.11 |
ACT1 0.93 ± 2.95 |
|
| Root | 1 |
18S 1.60 ± 11.96 |
18S 1.62 ± 12.28 |
UBQ 1.22 ± 3.96 |
F-box 0.30 ± 0.93 |
TUB 0.04 ± 0.14 |
F-box 0.20 ± 0.63 |
TUB 0.23 ± 0.67 |
EF-1a 0.01 ± 0.03 |
18S 0.20 ± 1.32 |
EF-1a 0.22 ± 0.73 |
18S 0.03 ± 0.28 |
EF-1a 0.13 ± 0.35 |
18S 0.11 ± 0.86 |
CYP2 0.08 ± 0.21 |
| 2 |
ACT1 1.88 ± 6.64 |
GAPDH 2.48 ± 7.30 |
CYP2 1.36 ± 4.45 |
EF-1a 0.31 ± 0.86 |
F-box 0.05 ± 0.20 |
GAPDH 0.30 ± 0.82 |
EF-1a 0.26 ± 0.71 |
CYP2 0.24 ± 0.81 |
F-box 0.34 ± 1.00 |
TUB 0.26 ± 0.89 |
F-box 0.09 ± 0.35 |
ACT1 0.25 ± 0.80 |
F-box 0.22 ± 0.65 |
EF-1a 0.13 ± 0.34 |
|
| 3 |
GAPDH 1.96 ± 5.71 |
ACT1 2.82 ± 9.97 |
18S 1.41 ± 10.07 |
TUB 0.33 ± 1.10 |
GAPDH 0.09 ± 0.27 |
18S 0.32 ± 2.22 |
UBQ 0.28 ± 0.88 |
18S 0.32 ± 3.03 |
EF-1a 0.42 ± 1.12 |
18S 0.51 ± 4.45 |
ACT1 0.25 ± 1.03 |
GAPDH 0.40 ± 1.09 |
EF-1a 0.35 ± 1.00 |
UBQ 0.20 ± 0.61 |
|
| 4 |
TUB 2.21 ± 7.10 |
EF-1a 3.12 ± 9.14 |
ACT1 1.53 ± 5.34 |
ACT1 0.38 ± 1.34 |
CYP2 0.18 ± 0.62 |
EF-1a 0.38 ± 1.09 |
18S 0.45 ± 3.01 |
GAPDH 0.33 ± 0.97 |
GAPDH 0.50 ± 1.35 |
ACT1 0.51 ± 2.00 |
EF-1a 0.43 ± 1.41 |
18S 0.60 ± 3.74 |
ACT1 0.53 ± 1.80 |
18S 0.27 ± 1.89 |
|
| 5 |
EF-1a 2.39 ± 6.91 |
TUB 3.20 ± 10.32 |
GAPDH 1.54 ± 4.40 |
UBQ 0.44 ± 1.46 |
EF-1a 0.19 ± 0.64 |
CYP2 0.51 ± 1.69 |
GAPDH 0.50 ± 1.41 |
ACT1 0.50 ± 1.91 |
CYP2 0.73 ± 2.31 |
UBQ 0.68 ± 2.82 |
UBQ 0.50 ± 2.16 |
TUB 0.83 ± 2.37 |
CYP2 0.72 ± 2.25 |
TUB 0.66 ± 2.01 |
|
| 6 |
UBQ 2.43 ± 8.14 |
F-box 4.17 ± 13.39 |
TUB 1.65 ± 5.23 |
GAPDH 0.47 ± 1.34 |
ACT1 0.35 ± 1.27 |
TUB 0.65 ± 2.09 |
ACT1 0.59 ± 1.91 |
TUB 0.53 ± 1.76 |
ACT1 0.74 ± 2.51 |
F-box 0.96 ± 3.57 |
TUB 0.64 ± 2.51 |
F-box 0.90 ± 2.62 |
GAPDH 0.99 ± 2.76 |
ACT1 0.78 ± 2.55 |
|
| 7 |
F-box 2.81 ± 8.95 |
UBQ 4.29 ± 14.90 |
EF-1a 2.20 ± 6.34 |
CYP2 0.61 ± 2.04 |
UBQ 0.38 ± 1.22 |
ACT1 0.70 ± 2.41 |
F-box 0.69 ± 1.98 |
UBQ 0.57 ± 2.00 |
UBQ 0.76 ± 2.45 |
GAPDH 1.04 ± 3.32 |
CYP2 0.75 ± 3.33 |
CYP2 1.16 ± 3.52 |
UBQ 1.03 ± 3.20 |
GAPDH 0.89 ± 2.48 |
|
| 8 |
CYP2 3.00 ± 10.02 |
CYP2 5.58 ± 18.83 |
F-box 2.36 ± 7.51 |
18S 0.96 ± 7.64 |
18S 0.61 ± 4.24 |
UBQ 0.97 ± 3.13 |
CYP2 1.58 ± 4.90 |
F-box 0.62 ± 2.00 |
TUB 0.88 ± 2.64 |
CYP2 1.17 ± 5.16 |
GAPDH 1.58 ± 5.16 |
UBQ 1.26 ± 3.91 |
TUB 1.84 ± 5.65 |
F-box 0.95 ± 2.63 |
Note: Data after gene symbols are Std ± CV%.
Analysis of reference genes by RefFinder
As described briefly above, RefFinder is a comprehensive web-based tool for reference gene identification that integrates geNorm, NormFinder, Delta Ct and BestKeeper approaches40. In this study, RefFinder was employed to evaluate the eight candidate reference genes in leaf and root samples under different conditions. The result showed that ACT1 was the most stably expressed gene when all the leaf and root samples were considered together. Individually, it was also the most stable one in the ramie leaf and root samples under various abiotic stresses. However, under other conditions, different reference genes were recommended by RefFinder. For example, in ramie treated with different hormones, CYP2 and TUB showed the most stable expression in leaf and root samples, respectively. In the leaves and roots of ramie plants infected with P. vexans, UBQ and TUB were the top ranked reference genes respectively. In most other single treatments, ACT1, CYP2, and UBQ displayed relatively stable expressions in leaves, while EF-1α, TUB, and ACT1 showed relatively stable expression in roots. GAPDH displayed the least stable expression in most cases for both the leaf and root samples (Table 5).
Table 5.
Ranking the expression stabilities of eight candidate reference genes in ramie leaves and roots based on the RefFinder program.
| Tissue | Rank | Total | Abiotic | Hormone | P.vexans | SA | BTH | JA | ETH | GA3 | Heat | Cold | NA | Cd | PEG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leaf | 1 |
ACT1 (1.00) |
ACT1 (1.57) |
CYP2 (3.50) |
UBQ (1.19) |
GAPDH (1.5) |
ACT1 (1.41) |
ACT1 (1.41) |
UBQ (1.19) |
ACT1 (1.97) |
18S (1.19) |
TUB (1.86) |
CYP2 (1.41) |
box (1.41) |
EF-1a (1.50) |
| 2 |
UBQ (1.68) |
UBQ (2.00) |
18S (6.73) |
F-box (1.41) |
EF-1a (2.45) |
box (1.86) |
18S (1.86) |
TUB (1.57) |
UBQ (2.38) |
CYP2 (1.41) |
CYP2 (2.11) |
UBQ (1.86) |
CYP2 (1.68) |
TUB (2.38) |
|
| 3 |
GAPDH (3.83) |
F-box (2.59) |
ACT1 (1.57) |
18S (3.46) |
CYP2 (2.63) |
18S (2.06) |
UBQ (2.06) |
CYP2 (2.71) |
18S (2.45) |
UBQ (3.41) |
18S (2.63) |
F-box (3.41) |
TUB (2.94) |
GAPDH (3.25) |
|
| 4 |
F-box (4.43) |
CYP2 (3.50) |
GAPDH (4.86) |
EF-1a (3.83) |
TUB (2.99) |
EF-1a (3.72) |
F-box (4.36) |
F-box (4.23) |
F-box (2.94) |
F-box (4.43) |
F-box (3.31) |
18S (4.30) |
ACT1 (3.46) |
UBQ (3.57) |
|
| 5 |
CYP2 (5.00) |
GAPDH (4.86) |
TUB (6.96) |
TUB (4.95) |
F-box (4.16) |
TUB (5.00) |
GAPDH (4.47) |
GAPDH (4.73) |
EF-1a (4.05) |
TUB (4.95) |
EF-1a (3.87) |
EF-1a (4.86) |
EF-1a (4.53) |
ACT1 (3.76) |
|
| 6 |
TUB (5.63) |
EF-1a (5.24) |
UBQ (2.00) |
GAPDH (5.48) |
ACT1 (5.05) |
CYP2 (6.24) |
CYP2 (5.73) |
18S (6.24) |
TUB (5.09) |
EF-1a (5.24) |
GAPDH (4.74) |
GAPDH (5.42) |
UBQ (5.73) |
CYP2 (5.42) |
|
| 7 |
18S (5.86) |
18S (6.73) |
F-box (2.59) |
ACT1 (7.00) |
18S (7.00) |
UBQ (7.24) |
TUB (7.00) |
EF-1a (6.74) |
GAPDH (6.51) |
ACT1 (6.44) |
ACT1 (6.74) |
TUB (5.66) |
18S (6.74) |
F-box (5.86) |
|
| 8 |
EF-1a (7.20) |
TUB (6.96) |
EF-1a (2.54) |
CYP2 (8.00) |
UBQ (8.00) |
GAPDH (7.44) |
EF-1a (8.00) |
ACT1 (8.00) |
CYP2 (7.48) |
GAPDH (8.00) |
UBQ (8.00) |
ACT1 (5.89) |
GAPDH (8.00) |
18S (6.88) |
|
| Root | 1 |
ACT1 (1.19) |
ACT1 (1.73) |
TUB (2.34) |
TUB (1.41) |
TUB (1.57) |
CYP2 (1.41) |
EF-1a (1.19) |
EF-1a (1.19) |
18S (1.41) |
TUB (1.19) |
F-box (1.68) |
EF-1a (1.57) |
ACT1 (1.32) |
ACT1 (1.78) |
| 2 |
TUB (2.00) |
F-box (2.71) |
UBQ (2.45) |
ACT1 (1.57) |
F-box (1.73) |
18S (2.38) |
TUB (2.06) |
CYP2 (1.41) |
GAPDH (2.00) |
EF-1a (1.86) |
18S (2.11) |
ACT1 (1.57) |
CYP2 (2.00) |
F-box (1.93) |
|
| 3 |
GAPDH (3.41) |
EF-1a (2.83) |
18S (2.66) |
F-box (2.28) |
GAPDH (1.86) |
ACT1 (2.71) |
18S (2.21) |
18S (3.00) |
F-box (3.08) |
ACT1 (2.63) |
ACT1 (2.28) |
GAPDH (2.63) |
EF-1a (2.71) |
TUB (3.72) |
|
| 4 |
18S (4.14) |
UBQ (3.25) |
CYP2 (2.91) |
EF-1a (4.23) |
CYP2 (4.73) |
F-box (3.31) |
UBQ (3.72) |
GAPDH (4.23) |
CYP2 (3.83) |
UBQ (3.94) |
EF-1a (3.44) |
F-box (3.83) |
18S (3.16) |
EF-1a (3.83) |
|
| 5 |
F-box (4.68) |
18S (4.30) |
ACT1 (3.08) |
CYP2 (5.23) |
EF-1a (4.90) |
TUB (4.16) |
GAPDH (5.00) |
ACT1 (5.18) |
EF-1a (4.12) |
18S (4.40) |
UBQ (3.94) |
TUB (5.23) |
UBQ (4.68) |
GAPDH (3.83) |
|
| 6 |
UBQ (5.14) |
GAPDH (4.56) |
GAPDH (4.43) |
UBQ (5.42) |
ACT1 (5.18) |
EF-1a (5.66) |
F-box (6.24) |
TUB (5.48) |
ACT1 (5.44) |
F-box (6.00) |
TUB (5.73) |
18S (5.29) |
GAPDH (6.24) |
18S (3.98) |
|
| 7 |
EF-1a (5.42) |
TUB (5.00) |
F-box (7.24) |
GAPDH (7.00) |
UBQ (7.00) |
UBQ (6.16) |
ACT1 (6.74) |
F-box (7.24) |
UBQ (5.66) |
CYP2 (7.24) |
CYP2 (6.74) |
CYP2 (6.19) |
F-box (6.44) |
UBQ (5.66) |
|
| 8 |
CYP2 (7.74) |
CYP2 (8.00) |
EF-1a (7.74) |
18S (8.00) |
18S (8.00) |
GAPDH (7.74) |
CYP2 (8.00) |
UBQ (7.74) |
TUB (8.00) |
GAPDH (7.74) |
GAPDH (8.00) |
UBQ (8.00) |
TUB (8.00) |
CYP2 (8.00) |
Using RefFinder, we also analyzed and ranked the suitable reference genes across root and leaf samples. As shown in Supplementary Table S3, ACT1, F-box, and 18S rRNA were ranked as the most suitable reference genes under most conditions (hormone stimuli, hot, cold, high salt, or heavy metal stress, and P. vexans infection), while CYP2 and UBQ were ranked the least suitable for most of the treatments.
Discussion
Ramie is an excellent source of long, natural fiber. It has attracted increasing attention from farmers, consumers and researchers in many parts of the globe. In China, many types of research are being conducted on ramie, including germplasm collection and assessment, domestication, and breeding for high yield, long fiber, and stress tolerance24,25,41–44. One of the emerging topics of research is to identify the key genes and metabolic pathways involved in ramie’s growth and its response to environmental stresses45–50. Gene expression profiling represents an important approach for understanding the roles of various genes during these processes. qPCR is a reliable and sensitive technique for measuring gene expression levels. However, accurate interpretations of qPCR results depend on the stability of reference genes used for data normalization. So far, several genes have been used as reference genes for normalizing gene expression data in ramie for a few selected conditions. However, the appropriateness of these genes as references has not been critically evaluated30–32. Indeed, previous studies in other organisms suggested that different tissues, different developmental stages and different environmental conditions may require different reference genes in order to accurately interpret the expressions of specific genes in qPCR experiments8–11. In this study, we screened eight candidate genes in ramie for their potential use as reference genes. These eight genes were chosen based on the ramie transcriptome data that showed limited variations in their relative abundance when ramie plants were subjected to several selected treatments, such as drought, Cd, ramie moth and root lesion nematode infestations. In addition, all eight genes have been used as reference genes for qPCR analyses of gene expression data in other plants, with several (e.g. ACT1, TUB) being commonly used across the Eukaryotic Domain.
Generally, an ideal reference gene is one that is stably expressed in different tissues of different cultivars under a wide range of environmental conditions. Based on the combined rankings of four programs, among the eight genes, on an individual gene basis, ACT1 showed overall the most stable expression in leaf and root samples. Thus, if only a single gene were to be used as an internal reference, ACT1 would be the most suitable reference gene for both leaf and root samples under a variety of conditions. Indeed, actin family genes are well-known reference genes across the Eukaryote Domain. However, our analyses indicated that for certain conditions/tissues, another gene or a combination of genes was more suitable as an internal reference in order to provide accurate normalization in qPCR experiments. For example, in ramie plants stimulated by hormones, TUB showed the highest stability and should be selected as the reference gene for analyzing gene expressions in the roots. Similar findings were reported for other plants, with genes displaying variable stability under different conditions. For example, when tomato leaves were subjected to treatments of nitrogen (N) starvation, low temperature, and suboptimal light during growth, among their eight screened candidate genes [ACT1, TUB, EF1, GAPDH, phosphoglycerate kinase (PGK), ribosomal protein L2 (RPL2), ubiquitin (UBI), and a catalytic subunit of protein phosphatase 2 A (PP2Acs)], GAPDH and PGK ranked at the top during light stress but poorly during N starvation and cold stress51. In contrast, EF1 ranked the best during N starvation and cold stress but poorly during light stress. Four genes ACT1, UBI, RPL2, and PP2Acs all appeared to be relatively stably expressed when all stress conditions were considered. Similar to what we found, no gene in tomato was identified that exhibited such a constant level of expression as to outperform all other candidates under all individual experimental conditions51. Indeed, similar results were also found for carrot leaves and roots8,52. In one study, ACT1 and TUB were determined to be the most suitable reference genes for carrot leaves under different abiotic stresses and hormone stimuli8. In another, among the nine screened candidates, ACT1 showed overall the most stable expression in carrot roots and leaves while GAPDH was the least stable52. Their overall analyses suggested ACT1 or different combinations of ACT1, EF-1α, the eukaryotic translation initiation factor 4α (eIF-4α), TUB, or UBQ were needed to normalize gene expression during carrot development52.
For both the leaf and root samples, there was no unanimity among the four analytical programs in ranking the most suitable reference gene(s) across all treatments. However, in most cases, one candidate gene was usually ranked as the most suitable reference gene by two to three algorithms, indicating that they were potentially good reference genes for these treatments. The degree of agreement for specific sets of treatments ranged from unanimity to complete disagreement. For instance, in the roots of ramie treated by SA or ETH, TUB and EF-1a were determined to be the most suitable reference genes, respectively, by all four analytical methods, indicating that these should be the preferred reference genes in these environmental conditions. In contrast, in the leaves of ramie under cold stress, 18S rRNA, CYP2, ACT1, and TUB were ranked as the most suitable reference genes according to geNorm, NormFinder, BestKeeper, and RefFinder, respectively.
While environmental influences on gene expression are well documented, even for reference genes, in recent years, emerging evidence indicates that most genes also show tissue specific expression levels53–59. For example, many genes, even housekeeping genes, such as OsTubA1 in rice55, GhTUB1 in cotton56, α- and β-tubulin genes in Populus57 showed tissue-preferential expression. The ACT1 gene showed different expression levels in the leaflet, flower, and young and mature fruits of blackberry and raspberry58. PmAct1 and PmAct2 showed distinct expression levels in different organs of beach plum59. In our analysis, the most stably expressed genes in the roots and leaves of ramie often differed even under the same growth conditions. Our results are consistent with those reported above and show that different ramie tissues under the same growth condition may require different internal reference genes during qPCR. However, to our knowledge, our study is among the first to demonstrate that different tissue x growth condition combinations may require different internal reference genes. Indeed, the suitable reference genes across the root and leaf samples of ramie plants under particular treatments as suggested by RefFinder were often inconsistent with those recommended separately for the roots or leaves. Taken together, our results indicate that there may not be a set of superior reference genes for all tissues of ramie, and that these suitable reference genes should be determined empirically according to experimental conditions and tissues.
Among the eight candidate genes screened in our study, most have a history of being used as internal reference genes for quantifying gene expressions through a variety of techniques. The relatively new one is F-box. F-box was recently shown to be the most stable reference gene under different experimental conditions in citrus fruits and in the common ivy (Hedera helix)60,61. In our study, we evaluated the F-box gene as a potential internal reference in ramie and found that it was among the most stably expressed in certain tissue x treatment combinations, including leaf samples of ramie plants subjected to P. vexans infection, BTH treatment, or Cd stresses and root samples of ramie plants subjected to SA treatment and cold or drought stresses. In contrast, GAPDH, a commonly used reference gene, performed relatively poorly under several experimental conditions, such as BTH, heat, cold, and salt stress treatments. Similar results have been reported for some other plants. For example, among those screened, GAPDH was the least suitable reference gene in purple false brome (Brachypodium distachyon), switchgrass (Panicum virgatum), qianhu (Peucedanum praeruptorum), and ryegrass (Lolium multiflorum) under abiotic stresses11,45,53,62.
Conclusions and perspectives
In this study, we selected eight candidate genes based on ramie transcriptome data and from the common list of qPCR reference genes used in the plant kingdom to search for suitable reference genes for use in qPCR analysis under different conditions for ramie leaves and roots. Our analyses showed that each of the eight genes showed the highest ranking in at least one tissue x experimental treatment combination as suggested by at least one of the analytical programs. Among these eight genes, three (ACT1, CYP2, and UBQ) displayed relatively stable expression in the leaves under most experimental conditions, while EF-1α, TUB, and ACT1 showed relatively stable expression in ramie roots under most experimental conditions. The most stable reference genes in leaf samples were often different from those in root samples, even under the same experimental conditions. We believe that the most stable reference genes screened in this study will improve the accuracy and standardization of investigations of ramie target gene expression under different stress conditions by qPCR analysis.
Note that the materials used in this study included only the roots and leaves of one ramie variety at the seedling stage. Even though we screened a diversity of biotic and abiotic stress conditions, the recommended reference genes identified here may not be the most suitable for other tissues (e.g. stems and flowers), other cultivars/varieties, and/or other developmental stages. In addition, although the eight genes selected here for analyses represent the commonly used reference genes across the plant kingdom and are among the most stably expressed based on transcriptome data, we cannot exclude the possibility that there might be more suitable reference genes in the ramie genome. Indeed, the variations in gene expressions observed here among the eight genes suggest that appropriate validations of candidate reference genes should be conducted for different tissues, genotypes, and developmental stages before a specific reference gene(s) is chosen for normalization of gene expression patterns.
Supplementary information
Acknowledgements
The study was supported by the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2015-IBFC); the Natural Science Foundation of Hunan Province (2017JJ2298); the Project for Monitoring and Prevention of Crop Pests, Disease, and Mice from the Ministry of Agriculture (S158); and the National Natural Science Foundation of China (81973430). We would like to thank Editage (www.editage.jp) for English language editing of the original version. We thank Heather Yoell of McMaster University for proofreading the revised version.
Author contributions
Y.Y. and Z.Y. conceived and designed the experiments; Y.Y., Q.B., Y.K.C., C.G., L.Z., Z.L., Y.C., J.C., X.S. and L.G. performed the experiments; Y.Y. and G.Z. analyzed the data; Z.Y. contributed reagents and materials; Y.Y. and G.Z. wrote the paper; J.X. revised the manuscript.
Data availability
We confirm that all the data associated with this manuscript are freely available and are presented either within the main manuscript file or in the Supplementary Materials section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yongting Yu, Gang Zhang and Zhun Yan.
Yongting Yu and Zhun Yan are deceased.
Supplementary information
is available for this paper at 10.1038/s41598-019-56640-3.
References
- 1.Srivastava RA. Analysis of RNA by Northern blotting using riboprobes. Methods Mol. Biol. 1998;86:103–112. doi: 10.1385/0-89603-494-1:103. [DOI] [PubMed] [Google Scholar]
- 2.Tymms MJ. Quantitative measurement of mRNA using the RNase protection assay. Methods Mol. Biol. 1995;37:31–46. doi: 10.1385/0-89603-288-4:31. [DOI] [PubMed] [Google Scholar]
- 3.de Toledo SM, Azzam EI, Gasmann MK, Mitchel RE. Use of semiquantitative reverse transcription-polymerase chain reaction to study gene expression in normal human skin fibroblasts following low dose-rate irradiation. Int. J. Radiat. Biol. 1995;67:135–143. doi: 10.1080/09553009514550171. [DOI] [PubMed] [Google Scholar]
- 4.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 5.Bustin SA. Real-time, fluorescence-based quantitative PCR: a snapshot of current procedures and preferences. Expert Rev. Mol. Diagn. 2005;5:493–498. doi: 10.1586/14737159.5.4.493. [DOI] [PubMed] [Google Scholar]
- 6.Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol. Med. 2002;8:257–260. doi: 10.1016/S1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- 7.Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Hematol. 2002;30:503–512. doi: 10.1016/S0301-472X(02)00806-8. [DOI] [PubMed] [Google Scholar]
- 8.Tian C, et al. Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PloS One. 2015;10:e0117569. doi: 10.1371/journal.pone.0117569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang L, et al. Identification of candidate reference genes in perennial ryegrass for quantitative RT-PCR under various abiotic stress conditions. PloS one. 2014;9:e93724. doi: 10.1371/journal.pone.0093724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le DT, et al. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PloS one. 2012;7:e46487. doi: 10.1371/journal.pone.0046487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong, S. Y., Seo, P. J., Yang, M. S., Xiang, F. & Park, C. M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8 (2008). [DOI] [PMC free article] [PubMed]
- 12.Liu TM, Zhu SY, Tang QM, Yu YT, Tang SW. Identification of drought stress-responsive transcription factors in ramie (Boehmeria nivea L. Gaud) BMC Plant Biol. 2013;13:130. doi: 10.1186/1471-2229-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kipriotis E, Xiong HP, Vafeiadakis T, Kiprioti M, Alexopoulou E. Ramie and kenaf as feed crops. Ind. Crop. Prod. 2015;68:126–130. doi: 10.1016/j.indcrop.2014.10.002. [DOI] [Google Scholar]
- 14.Yu YT, et al. J. Northwest A&F Univ. (Nat. Sci. Ed.) 2011. Identification of a nematode isolate from rot root of ramie; pp. 105–109. [Google Scholar]
- 15.Yu YT, et al. First report of black leaf spot caused by Alternaria alternata on ramie in China. J. Phytopath. 2016;164:358–361. doi: 10.1111/jph.12428. [DOI] [Google Scholar]
- 16.Yu YT, et al. First report of brown root rot caused by Pythium vexans on ramie in Hunan, China. Can. J. Plant Path. 2016;38:405–410. doi: 10.1080/07060661.2016.1230150. [DOI] [Google Scholar]
- 17.Yang M, et al. Identification of the pathogen causing leaf spot disease on Boehmeria nivea. Acta Phytopathol. Sin. 2014;44:318–322. [Google Scholar]
- 18.Wang XX, et al. First report of anthracnose caused by Colletotrichum gloeosporioides on ramie in China. Plant Dis. 2010;94:1508–1508. doi: 10.1094/PDIS-08-10-0556. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Zhang XY, Qian YJ. Molecular characterization of Ramie mosaic virus isolates detected in Jiangsu and Zhejiang provinces, China. Acta virologica. 2009;54:225–228. doi: 10.4149/av_2010_03_225. [DOI] [PubMed] [Google Scholar]
- 20.Zeng LB, Xue ZD, Yu YT, Yan ZY, Yang RL. Changes of occurrence and control of Cocytodes caerulea Guenée. Hunan Agr Sci. 2013;10:23–24. [Google Scholar]
- 21.Peng DX. Effect of draught and flooding on the development of ramie plant. China’s Fiber Crop. 1992;4:17–21. [Google Scholar]
- 22.Yang B, et al. Constitutional tolerance to heavy metals of a fiber crop, ramie (Boehmeria nivea), and its potential usage. Environ. Poll. 2010;158:551–558. doi: 10.1016/j.envpol.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 23.Li LY, Cui GX, Zou XM, Liu F. Root growth, fiber yield and quality for difference potassium efficiency ramie genotypes under lower potassium stress. Chin. Agr. Sci. Bull. 2006;22:246–249. [Google Scholar]
- 24.Zeng L, et al. Transcriptome analysis of ramie (Boehmeria nivea L. Gaud.) in response to ramie moth (Cocytodes coerulea Guenee) infestation. BioMed Res. Int. 2016;2016:3702789. doi: 10.1155/2016/3702789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu YT, et al. Identification of ramie genes in response to Pratylenchus coffeae infection challenge by digital gene expression analysis. Int. J. Mol. Sci. 2015;16:21989–22007. doi: 10.3390/ijms160921989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu TM, Zhu SY, Tang QM, Tang SW. Genome-wide transcriptomic profiling of ramie (Boehmeria nivea L. Gaud) in response to cadmium stress. Gene. 2015;558:131–137. doi: 10.1016/j.gene.2014.12.057. [DOI] [PubMed] [Google Scholar]
- 27.Zhu SY, Tang SW, Tang QM, Liu TM. Genome-wide transcriptional changes of ramie (Boehmeria nivea L. Gaud) in response to root-lesion nematode infection. Gene. 2014;552:67–74. doi: 10.1016/j.gene.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Deng G, et al. Comparative proteome analysis of the response of ramie under N, P and K deficiency. Planta. 2014;239:1175–1186. doi: 10.1007/s00425-014-2040-3. [DOI] [PubMed] [Google Scholar]
- 29.Wang XX, et al. Characterization by suppression subtractive hybridization of transcripts that are differentially expressed in leaves of anthracnose-resistant ramie cultivar. Plant Mol. Biol. Rep. 2012;30:547–555. doi: 10.1007/s11105-011-0361-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu TM, Zhu SY, Tang QM, Tang SW. Identification of 32 full-length NAC transcription factors in ramie (Boehmeria nivea L. Gaud) and characterization of the expression pattern of these genes. Mol. Genet. Genomics. 2014;289:675–684. doi: 10.1007/s00438-014-0842-4. [DOI] [PubMed] [Google Scholar]
- 31.Ma XF, et al. Cloning and tissue expression of Actin1 gene in different fiber development phases of ramie [Boehmeria nivea (Linn.) Gaud] Acta Agron. Sin. 2010;36:101–108. doi: 10.3724/SP.J.1006.2010.00101. [DOI] [Google Scholar]
- 32.Tian ZJ, Yi R, Chen JR, Guo QQ, Zhang XW. Cloning and expression of cellulose synthase gene in ramie [Boehmeria nivea (Linn.) Gaud.] Acta Agron. Sin. 2008;34:76–83. [Google Scholar]
- 33.An X, et al. Transcriptome profiling and identification of transcription factors in ramie (Boehmeria nivea L. Gaud) in response to PEG treatment, using illumina paired-end sequencing technology. Int. J. Mol. Sci. 2015;16:3493–3511. doi: 10.3390/ijms16023493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang P, et al. Selection of suitable reference genes for RT-qPCR normalization under abiotic stresses and hormone stimulation in Persimmon (Diospyros kaki Thunb) PloS one. 2016;11:e0160885. doi: 10.1371/journal.pone.0160885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Untergasser A, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Spiegelaere W, et al. Reference Gene Validation for RT-qPCR, a Note on Different Available Software Packages. PLoS One. 2015;10(3):e0122515. doi: 10.1371/journal.pone.0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnology Lett. 2004;26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 40.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012;80:75–84. doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 41.Liu C, et al. Draft genome analysis provides insights into the fiber yield, crude protein biosynthesis, and vegetative growth of domesticated ramie (Boehmeria nivea L. Gaud) DNA Res. 2018;25:173–181. doi: 10.1093/dnares/dsx047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ullah S, et al. Interactive effect of gibberellic acid and NPK fertilizer combinations on ramie yield and bast fibre quality. Sci. Rep. 2017;7:10647. doi: 10.1038/s41598-017-09584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satya P, et al. Start codon targeted (SCoT) polymorphism reveals genetic diversity in wild and domesticated populations of ramie (Boehmeria nivea L. Gaudich.), a premium textile fiber producing species. Meta gene. 2015;3:62–70. doi: 10.1016/j.mgene.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu T, Tang S, Zhu S, Tang Q, Zheng X. Transcriptome comparison reveals the patterns of selection in domesticated and wild ramie (Boehmeria nivea L. Gaud) Plant Mol. Biol. 2014;86:85–92. doi: 10.1007/s11103-014-0214-9. [DOI] [PubMed] [Google Scholar]
- 45.Liu Q, et al. Reference gene selection for quantitative real-Time reverse-transcriptase PCR in annual ryegrass (Lolium multiflorum) subjected to various abiotic stresses. Molecules. 2018;23:172. doi: 10.3390/molecules23010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Yongting, Zhang Gang, Li Zhimin, Cheng Yi, Gao Chunsheng, Zeng Liangbin, Chen Jia, Yan Li, Sun Xiangping, Guo Litao, Yan Zhun. Molecular Cloning, Recombinant Expression and Antifungal Activity of BnCPI, a Cystatin in Ramie (Boehmeria nivea L.) Genes. 2017;8(10):265. doi: 10.3390/genes8100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An X, et al. Senescence is delayed when ramie (Boehmeria nivea L.) is transformed with the isopentyl transferase (ipt) gene under control of the SAG12 promoter. FEBS Open Bio. 2017;7:636–644. doi: 10.1002/2211-5463.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang C, et al. A ramie bZIP transcription factor BnbZIP2 is involved in drought, salt, and heavy metal stress response. DNA Cell Biol. 2016;35:776–786. doi: 10.1089/dna.2016.3251. [DOI] [PubMed] [Google Scholar]
- 49.Zheng JS, et al. Characterization of a glutamine synthetase gene BnGS1-2 from ramie (Boehmeria nivea L. Gaud) and biochemical assays of BnGS1-2-over-expressing transgenic tobacco. Acta Physiol. Plant. 2015;37:1742. doi: 10.1007/s11738-014-1742-2. [DOI] [Google Scholar]
- 50.Liu, T., Zhu, S., Tang, Q. & Tang, S. Identification of a CONSTANS homologous gene with distinct diurnal expression patterns in varied photoperiods in ramie (Boehmeria nivea L. Gaud). Gene560 (2015). [DOI] [PubMed]
- 51.Lovdal T, Lillo C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Analytical Biochem. 2009;387:238–242. doi: 10.1016/j.ab.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 52.Wang GL, et al. Comparison of nine reference genes for real-time quantitative PCR in roots and leaves during five developmental stages in carrot (Daucus carota L.) J. Hortic. Sci. Biotechnol. 2016;91:264–270. doi: 10.1080/14620316.2016.1148372. [DOI] [Google Scholar]
- 53.Zhao Y, et al. Selection of reference genes for gene expression normalization in Peucedanum praeruptorum Dunn under abiotic stresses, hormone treatments and different Tissues. PloS one. 2016;11:e0152356. doi: 10.1371/journal.pone.0152356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin Y, et al. Validation of potential reference genes for qPCR in maize across abiotic stresses, hormone treatments, and tissue types. PloS one. 2014;9:e95445. doi: 10.1371/journal.pone.0095445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeon JS, et al. Tissue-preferential expression of a rice alpha-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol. 2000;123:1005–1014. doi: 10.1104/pp.123.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li XB, Cai L, Cheng NH, Liu JW. Molecular characterization of the cotton GhTUB1 gene that is preferentially expressed in fiber. Plant Physiol. 2002;130:666–674. doi: 10.1104/pp.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oakley RV, Wang YS, Ramakrishna W, Harding SA, Tsai CJ. Differential expansion and expression of alpha- and beta-tubulin gene families in. Populus. Plant Physiol. 2007;145:961–973. doi: 10.1104/pp.107.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang CH, Wang XM, Wang LB, Wu WL, Li WL. cDNA fragment cloning and expression analysis of actin gene in Rubus spp. Biotechnology. 2011;21:10–15. [Google Scholar]
- 59.Zhang CH, et al. cDNA fragment cloning and expression analysis of two actin genes in Beach plum (Prunus maritima Marshall) Biotechnology. 2012;22:27–31. [Google Scholar]
- 60.Mafra V, et al. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PloS one. 2012;7:e31263. doi: 10.1371/journal.pone.0031263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun HP, Li F, Ruan QM, Zhong XH. Identification and validation of reference genes for quantitative real-time PCR studies in Hedera helix L. Plant Physiol. Biochem. 2016;108:286–294. doi: 10.1016/j.plaphy.2016.07.022. [DOI] [PubMed] [Google Scholar]
- 62.Jiang X, et al. Reference gene selection for Real-time quantitative PCR normalization in switchgrass (Panicum virgatum L.) root tissue. J. Agr. Biotechnol. 2014;22:55–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We confirm that all the data associated with this manuscript are freely available and are presented either within the main manuscript file or in the Supplementary Materials section.