Abstract
Nonalcoholic fatty liver disease (NAFLD) is becoming a major etiological risk factor for hepatocellular carcinoma (HCC) in the United States and other Western countries. In this study, we investigated the role of gene-specific promoter cytosine DNA methylation and gene expression alterations in the development of NAFLD-associated HCC in mice using (1) a diet-induced animal model of NAFLD, (2) a Stelic Animal Model of nonalcoholic steatohepatitis-derived HCC, and (3) a choline- and folate-deficient (CFD) diet (CFD model). We found that the development of NAFLD and its progression to HCC was characterized by down-regulation of glycine N-methyltransferase (Gnmt) and this was mediated by progressive Gnmt promoter cytosine DNA hypermethylation. Using a panel of genetically diverse inbred mice, we observed that Gnmt down-regulation was an early event in the pathogenesis of NAFLD and correlated with the extent of the NAFLD-like liver injury. Reduced GNMT expression was also found in human HCC tissue and liver cancer cell lines. In in vitro experiments, we demonstrated that one of the consequences of GNMT inhibition was an increase in genome methylation facilitated by an elevated level of S-adenosyl-L-methionine. Overall, our findings suggest that reduced Gnmt expression caused by promoter hypermethylation is one of the key molecular events in the development of NAFLD-derived HCC and that assessing Gnmt methylation level may be useful for disease stratification.
Keywords: epigenetics, Gnmt, HCC, NASH, NAFLD
Hepatocellular carcinoma (HCC), the major histological type of primary liver cancer, is the second most lethal cancer worldwide (Bertuccio et al., 2017; Llovet et al., 2016). In the United States, the incidence of HCC has greatly increased over the past 2 decades (Altekruse et al., 2009; Simard et al., 2012). The etiological factors associated with HCC are well-known and include chronic hepatitis B (HBV) and C (HCV) viral infections, chemical exposure, excessive alcohol consumption (Bertuccio et al., 2017), and, recently, nonalcoholic fatty liver disease (NAFLD). The incidence of HCC varies geographically, due to differences in etiology, and is dynamic, due to changes in the contribution of disease-specific etiological risk factors (Choo et al., 2016; Kim et al., 2018). For instance, as a result of advances in immunization and a new generation of antiviral drugs against HBV and HCV, there is a great possibility that these etiological risk factors for HCC will be eliminated in the United States and Western countries (Ioannou et al., 2018; Yoo et al., 2018). On the other hand, it is widely believed that NAFLD will become a predominant liver disease and that nonalcoholic steatohepatitis (NASH), an advanced form of NAFLD, will become a major cause of HCC worldwide (Younes and Bugianesi, 2018). Indeed, Kim et al. (2018) recently reported a 68% increase in HCC associated with NASH in adult patients from 1998–2009 to 2010–2015.
The molecular pathogenesis of HCC is a complex multistage process characterized by the accumulation of genetic and epigenetic alterations in several major cancer-hallmark pathways (Pogribny and Rusyn, 2014; Shibata et al., 2018; The Cancer Genome Atlas Research Network, 2017; Zucman-Rossi et al., 2015). Extensive genomic studies over the past decade have identified the main genetic drivers that facilitate liver cancer initiation and progression (Shibata et al., 2018; Zucman-Rossi et al., 2015). In contrast, the role of epigenetic and transcriptomic drivers of HCC has yet to be uncovered because most functional genomics studies provide only a static snapshot of these abnormalities in full-fledged HCC rather than their dynamic changes during the progression of hepatocarcinogenesis.
Investigating the molecular basis and pathways of the liver carcinogenic process in humans, in general, and NAFLD-associated hepatocarcinogenesis, in particular, is complex and expensive in terms of recruitment and characterization of study cohorts. Additionally, the development of NAFLD-related HCC in humans is difficult to study because it includes a wide range of conditions that are problematical to differentiate noninvasively. Therefore, it is challenging to establish the causality and molecular underpinnings of the disease in studies with humans. In contrast, animal models that mirror human NAFLD pathology overcome many of the shortcomings of the human-only approach and are exceedingly useful for studying the disease pathogenesis, including elucidation of the role of specific genes in the pathogenesis of NAFLD and its progression to HCC (Santhekadur et al., 2018). Among the animal models used to study NAFLD, dietary-based mouse models, using either high-calorie or methyl-donor group deficient diets, are the most widely employed (Jahn et al., 2019; Santhekadur et al., 2018).
Based on these considerations, in this study, we investigated the role of gene-specific promoter cytosine DNA methylation and gene expression alterations in the development of NAFLD-associated HCC in mice using a diet-induced animal model of NAFLD (DIAMOND), which recapitulates the key physiological, metabolic, histologic, transcriptomic, and cell-signaling changes seen in humans with progressive NASH (Asgharpour et al., 2016), and a Stelic Animal Model (STAM) of NASH-derived HCC, which is the first mouse model to depict the sequential evolution of clinical and pathomorphological features of the development of HCC in diabetes-associated NASH patients (Fujii et al., 2013). Additionally, to investigate the effect of genetic background-associated differences in NAFLD pathogenesis, we fed a panel of genetically diverse inbred mice a choline- and folate-deficient (CFD) diet (CFD model), which is an ideal model for the studying the subgroup of NASH patients with histologically advanced NASH (Machado et al., 2015).
MATERIALS AND METHODS
Animals, experimental design, and mouse models of NASH hepatocarcinogenesis
This study used liver tissue samples from male mice subjected to DIAMOND, STAM, and CFD dietary models of NAFLD. In the DIAMOND-associated liver carcinogenesis model, stable male isogenic mice obtained by crossing C57BL/6J and 129S1/SvImJ mice, were fed, ad libitum, a high-fat diet, high carbohydrate diet (42% kcal from fat, 0.1% cholesterol, and a high fructose-glucose solution; TD.88137; Envigo, Madison, Wisconsin) for 24, 36, 44, or 52 weeks. Control mice were fed a standard chow diet (Harlan TD.7012) with normal water. The livers were excised and snap-frozen in liquid nitrogen. The results of clinical chemistry, histological, and transcriptomic analyses are described in detail in Asgharpour et al. (2016) and Cazanave et al. (2017).
In the STAM liver carcinogenesis model, 2-day-old male C57BL/6J mice were injected with streptozotocin (200 μg/mouse). Starting from 4 weeks of age, the mice were continuously fed a high-fat diet (HFD-32; Clea, Tokyo, Japan) throughout the study. Liver samples from male STAM mice at steatotic (6 weeks), NASH-fibrotic (12 weeks), and full-fledged HCC (20 weeks) stages of liver carcinogenesis and from age-matched control C57BL/6J mice were purchased from the Stelic Institute & Co. (Tokyo, Japan).
In the CFD model, male A/J, C57BL/6J, 129S1/SvImJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ mice were used. These strains were selected because they provide an excellent representation of the broad genetic diversity and their genomes have been fully sequenced (Yang et al., 2011). Mice (6 weeks of age) were fed a diet low in methionine (0.17% w/w) and devoid of choline and folic acid (Diet no. 519541, CFD, iron-supplemented, and L-amino acid-defined diet; Dyets, Bethlehem, Pennsylvania) for 12 weeks. Mice in the control group received the same diet supplemented with 0.4% methionine, 0.3% choline bitartrate, and 2 mg/kg folic acid. Diets were stored at 4°C before use and given ad libitum with replacement twice a week. Mice from each strain were euthanized by exsanguination following deep isoflurane anesthesia 12 weeks after diet initiation. The livers were excised, and a slice of the median lobe was fixed in neutral buffered formalin for 48 h for histopathological examination. The remaining liver was snap-frozen immediately in liquid nitrogen and stored at −80°C for subsequent analyses. The results of histological and transcriptomic analyses are described in Tryndyak et al. (2012).
All experiments involving animals were approved by the Animal Care and Use Committee of Virginia Commonwealth University, the Stelic Institute & Co, or the Animal Care and Use Committee of the National Center for Toxicological Research.
Cell lines and cell culture
Immortalized HepG2 and SK-Hep-1 human liver cancer cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, Virginia). The cells were maintained in culture according to the manufacturer’s recommendations.
DNA methylation analyses
Genomic DNA was isolated with DNeasy Blood and Tissue kits (Qiagen, Valencia, California). Methylated DNA immunoprecipitation (MeDIP) was performed with MethylMiner Methylated DNA Enrichment kits (Invitrogen, Carlsbad, California). The methylation status of CpG islands located within the promoters of the selected genes was determined by MeDIP-quantitative polymerase chain reaction (PCR) (MeDIP-qPCR). Reduced representation bisulfite sequencing (RRBS) analysis was used to determine the whole-genome DNA methylation profile by identifying differentially methylated regions (DMRs) in the livers of PWK/PhJ mice fed the CFD diet and in HepG2 and SK-Hep-1 human cells as detailed in Tryndyak et al. (2017).
Quantitative reverse-transcription PCR
Total RNA was extracted from liver tissue samples, and HepG2 and SK-Hep-1 cells using miRNeasy Mini kits (Qiagen). Total RNA (2 μg) was reverse transcribed using random primers and High Capacity cDNA Reverse Transcription kits (Life Technologies, Grand Island, New York). Gene expression was determined by quantitative reverse-transcription PCR (qRT-PCR) using the TaqMan gene expression assays (Life Technologies). The relative amount of each mRNA transcript was determined using the 2−ΔΔCt method (Schmittgen and Livak, 2008).
Western blot analysis
Whole liver tissue lysates or cell lysates containing equal quantities of proteins were separated by 7%–15% SDS-PAGE and transferred to PVDF membranes. The levels of glycine N-methyltransferase (GNMT), DNA methyltransferase 1 (DNMT1), DNA methyltransferase 3A (DNMT3A), DNA methyltransferase 3B (DNMT3B), and ubiquitin-like, containing PHD and RING finger domains 1 (UHRF1) proteins were determined by Western blot analysis as described previously (Tryndyak et al., 2012). AntiDNMT1 (catalog number 5032), anti- DNMT3A (catalog number 3598), antiDNMT3B (catalog number 2161), and antiUHRF1 (catalog number 12387) antibodies were obtained from Cell Signaling Technology (Danvers, Massachusetts), and antiGNMT (catalog number PA5-50333) antibody was obtained from ThermoFisher Scientific (Waltham, Massachusetts). IRDye 800CW-labeled antirabbit or IRDye 680RD-labeled antimouse antibody (LI-COR Biosciences, Lincoln, Nebraska) was used for visualization. Fluorescence was measured using the Odyssey CLx Infrared Imager (LI-COR Biosciences). The images were quantified using ImageStudio 4.0 Software (LI-COR Biosciences). To control for equal loading, the relative amount of the protein of interest was normalized by staining of the membranes with REVERT Total Protein Stain (LI-COR Biosciences; Supplementary Figure 3).
Transfection of HepG2 cells with siRNA
HepG2 cells, seeded in the antibiotic-free medium in 100 mm dishes and grown for 24 h, were transfected in 3 independent biological replicates using 40 nM ON-TARGETplus human GNMT siRNA (Dharmacon, Lafayette, Colorado) and Lipofectamine 3000 (Life Technologies). Seventy-two hours after transfection, adherent cells were harvested by mild trypsinization and their viability was monitored using a Trypan Blue exclusion assay (Sigma-Aldrich; St Louis, Missouri). The cells were reseeded, and the transfection was repeated. Seventy-two hours after the second transfection, adherent cells were harvested by mild trypsinization, washed in phosphate-buffered saline, and immediately frozen at −80°C for subsequent analyses.
Determination of methionine, S-adenosyl-L-methionine, S-adenosyl-L-homocysteine, free homocysteine, and cysteine
The levels of methionine, S-adenosyl-L-methionine (SAM), S-adenosyl-L-homocysteine (SAH), free homocysteine, and free cysteine were measured in the HepG2 cells by high-performance liquid chromatography coupled with coulometric electrochemical detection as previously described (Melnyk et al., 1999, 2000).
Retrieval of data from the online database
GNMT gene expression (level 3) data in human HCC were obtained from The Cancer Genome Atlas (TCGA) database (http://cancergenome.nih.gov).
Statistics
Results are presented as mean ± SD. Statistical analyses were performed using SigmaPlot 13.0 software (Systat Software, Inc, San Jose, Californina). Benjamini-Hochberg adjusted p values (Benjamini and Hochberg, 1995) were calculated and an adjusted p value cutoff ≤ .05 and a fold change threshold > 2.0 were used to generate a list of differentially expressed genes in the livers of mice submitted to DIAMOND model. The differences in gene-specific promoter methylation in the livers between age-matched mice in experimental and control groups were analyzed by an unpaired 2-tailed Student’s t test. The differences between GNMT protein levels in the livers of DIAMOND mice during NAFLD-associated liver carcinogenesis, and Gnmt expression and Gnmt gene-specific methylation in the livers of A/J, C57BL/6J, 129S1/SvImJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ were analyzed by 2-way analysis of variance (ANOVA), with pairwise comparisons being made by the Student-Newman-Keuls method. The GNMT expression data in human HCC were natural log transformed before conducting the analyses to maintain a more equal variance or normal data distribution and were analyzed by the nonparametric Wilcoxon signed-rank test. A difference in data values with a p ≤ .05 was considered significant. Pearson product-moment correlation coefficient was used to determine correlations. A Chi-Square test was applied to evaluate the difference in proportion of DMRs.
RESULTS
Gene-Specific Methylation of Epigenetically Regulated Down-Regulated Genes in HCC in DIAMOND and STAM Mice
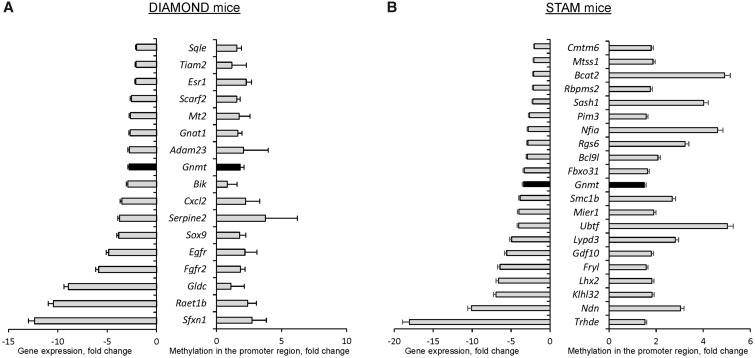
Previously, Cazanave et al. (2017) and de Conti et al. (2017) demonstrated that the development of NAFLD-derived HCC in DIAMOND and STAM mice is characterized by substantial and unique stage-dependent transcriptomic alterations, with a greater number of down-regulated genes being found in HCC. In light of these observations and on the basis of previous reports on the role of a promoter CpG island methylator phenotype in the development of HCC (Shen et al., 2002; Wang et al., 2018), we investigated the status of cytosine DNA methylation in genes that contain promoter CpG islands and are down-regulated in HCC. First, we analyzed differentially expressed genes in NAFLD-derived HCC in DIAMOND mice (52 weeks) (Asgharpour et al., 2016; GSE67679) and found that 50 of down-regulated genes contained CpG islands in their promoter regions. Then, by using MeDIP-qPCR, we analyzed the promoter methylation status of these genes and found 17 hypermethylated genes (Figure 1A). Next, by using a combined analysis of MeDIP-qPCR and gene expression data (de Conti et al., 2017; GSE83596), we identified 21 hypermethylated genes in NAFLD-derived HCC in STAM mice (20 weeks) (Figure 1B). It should be noted that the Gnmt gene was the only gene down-regulated and concurrently hypermethylated in HCC in both STAM and DIAMOND mice (Figure 1).
Figure 1.
Expression and promoter methylation in down-regulated genes containing promoter CpG islands in NAFLD-derived HCC in DIAMOND (52 weeks) (A) and STAM (20 weeks) (B) mice. Lists of hypermethylated and down-regulated genes in NAFLD-derived HCC in (A) DIAMOND (n = 4 controls and n = 5 HCC) and (B) STAM mice (n = 4 controls and n = 4 HCC) were generated based on Benjamini-Hochberg adjusted p values, with a cutoff of .05 and a fold change threshold of 2.0. The gene expression is presented as fold change relative to age-matched control mice. Promoter methylation of differentially expressed genes is presented as an average fold change of individual genes. Only inversely differentially expressed and methylated genes are shown.
Promoter Gene-Specific Methylation of the Genes During NAFLD-Associated Hepatocarcinogenesis
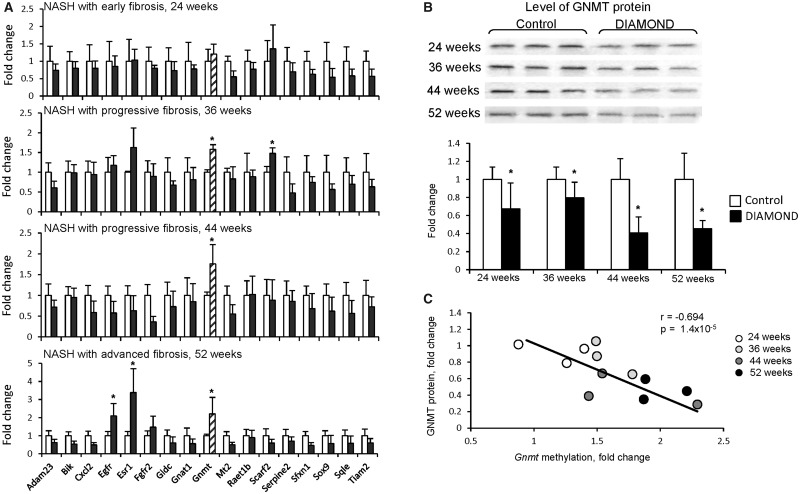
To investigate further the role of the genes found to be hypermethylated in full-fledged HCC, we analyzed their promoter methylation status at different stages of the NAFLD-associated hepatocarcinogenic process in DIAMOND mice: NASH with early fibrosis (24 weeks), NASH with progressive fibrosis (36 and 44 weeks), and NASH with advanced fibrosis (52 weeks). Figure 2A shows that 3 (epidermal growth factor receptor, estrogen receptor 1, and Gnmt) of the 17 genes that were inhibited and concurrently methylated in HCC were also hypermethylated in NASH with advanced fibrosis. This finding supports the hypothesis of simultaneous accumulation of CpG island methylator phenotype alterations in the disease process leading to HCC (Shen et al., 2002; Wang et al., 2018).
Figure 2.
Gnmt gene-specific methylation and expression during NAFLD-associated liver carcinogenesis in DIAMOND mice. A, Gene-specific methylation during NAFLD-associated liver carcinogenesis in the livers of DIAMOND mice (n = 4 controls and n = 5 HCC). The results are presented as an average fold change in methylation of each gene in the livers of DIAMOND mice relative to that in age-matched control mice, which were assigned a value 1. Values are mean ± SD. Asterisks (*) denote a statistically significant (p < .05) difference from the age-matched control mice as determined by a 2-tailed Student’s t test. B, The protein level of GNMT in the livers of DIAMOND mice during NAFLD-associated liver carcinogenesis. The results are presented as an average fold change in GNMT protein levels in the livers of DIAMOND mice relative to that in the age-matched control mice, which were assigned a value 1. Values are mean ± SD (n = 3). Asterisks (*) denote a statistically significant (p < .05) difference from the age-matched control mice as determined by 2-way ANOVA, with pairwise comparisons being made by the Student-Newman-Keuls method. C, Correlation plot of GNMT protein expression and gene promoter methylation in the livers of DIAMOND mice during NAFLD-associated liver carcinogenesis.
Further analysis of gene promoter methylation under the condition of NASH with early fibrosis and NASH with progressive fibrosis showed that only Gnmt exhibited progressive changes towards hypermethylation starting at 36 weeks (Figure 2A). The levels of the Gnmt gene promoter methylation at 36, 44, and 52 weeks were 1.6-, 1.8-, and 2.2-times greater, respectively, than age-matched control mice. In contrast to changes in Gnmt promoter-specific methylation, only sporadic changes in promoter methylation were found in the other genes in these early stages of hepatocarcinogenesis (Figure 2A). A similar stage-dependent Gnmt promoter hypermethylation was also observed during NAFLD-associated liver carcinogenesis in STAM mice (Supplementary Figure 1).
Figure 2B shows that changes in the Gnmt promoter methylation during the development of NAFLD-derived HCC were accompanied by a simultaneous reduction in the level of GNMT protein (Figure 2B) that was inversely correlated (r = −0.694, p = 1.4 × 10−5) with the extent of CpG island promoter methylation (Figure 2C).
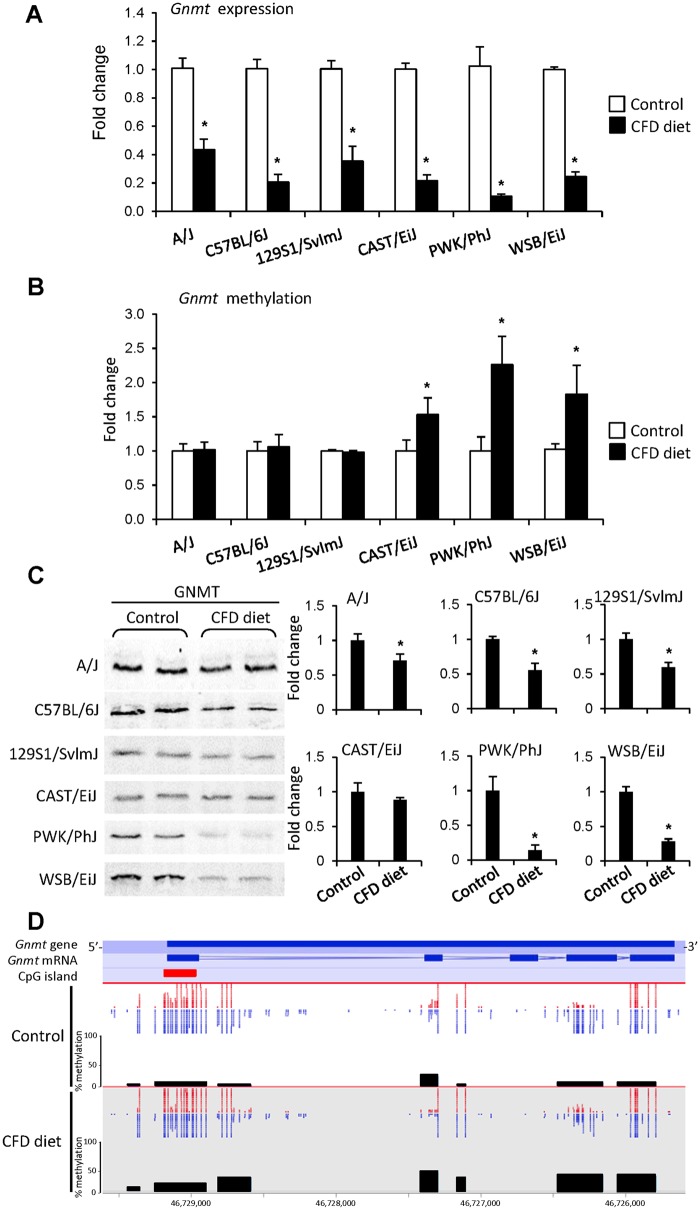
The Effect of a CFD Diet on the Gnmt Promoter Methylation and Gene Expression
Previously, we demonstrated that feeding a panel of genetically diverse inbred mice a CFD diet for 12 weeks resulted in strain-dependent NAFLD-like liver injury with the following order of severity: A/J ≈ C57BL/6J < 129S1/SvImJ ≈ CAST/EiJ < PWK/PhJ < WSB/EiJ (Tryndyak et al., 2012). Herein, we investigated whether or not alterations in Gnmt gene expression and promoter methylation are concordant with the severity of liver injury. Figure 3A shows that feeding the CFD diet caused a substantial inhibition of hepatic Gnmt expression in all strains, while the Gnmt promoter cytosine DNA methylation was increased only in CAST/EiJ and, especially, WSB/EiJ, and PWK/PhJ mice (Figure 3B), 2 mouse strains characterized by the development of advanced NASH-like liver injury (Tryndyak et al., 2012). Changes in Gnmt expression and methylation were accompanied by a marked decrease in the level of GNMT protein (Figure 3C). Hypermethylation of the Gnmt promoter in the livers of PWK/PhJ, the strain that exhibited the most prominent Gnmt down-regulation, was independently confirmed by the RRBS DNA methylation analysis (Figure 3D). It should be noted that PWK/PhJ mice exhibited the highest level of Gnmt expression in the normal livers (Supplementary Figure 3).
Figure 3.
Gnmt expression in the livers of mice fed a CFD diet for 12 weeks. (A) Gnmt expression and (B) gene-specific methylation in the livers of male A/J, C57BL/6J, 129S1/SvImJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ mice fed a CFD diet. The results are presented as an average fold change in the livers of CFD diet-fed mice relative to that in the age-matched control mice, which were assigned a value 1. Values are mean ± SD (n = 5 per strain/diet). Asterisks (*) denote a statistically significant (p < .05) difference from the age-matched control mice as determined by 2-way ANOVA, with pairwise comparisons being made by the Student-Newman-Keuls method. C, The protein level of GNMT in the livers of A/J, C57BL/6J, 129S1/SvImJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ mice. Representative images are shown. The results are presented as an average fold change in GNMT protein levels in the livers of mice fed a CFD diet relative to that in the age-matched control mice, which were assigned a value 1. Values are mean ± SD. Asterisks (*) denotes a statistically significant (p < .05) difference from the age-matched control mice as determined by a 2-tailed Student’s t test. D, RRBS analysis of the Gnmt gene methylation in the livers of control and CFD-fed PWK/PhJ mice. Red dots indicate methylated CpG sites; blue dots indicate unmethylated CpG sites. Y scale shows % methylation of DMRs in the Gnmt gene.
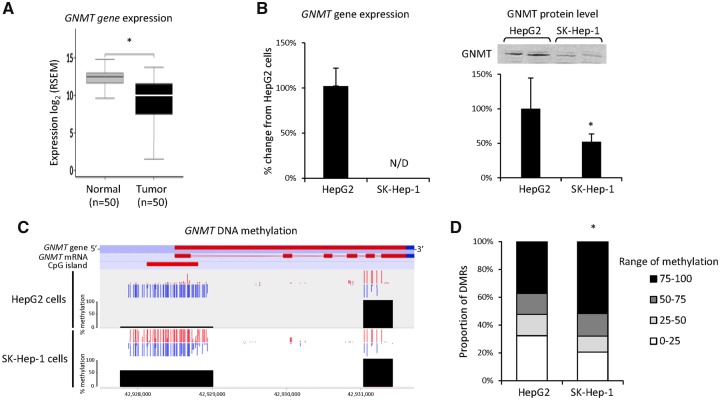
GNMT Methylation and Expression in Human HCC and Liver Cancer Cell Lines
Using the TCGA Research Network database (http://cancergenome.nih.gov; The Cancer Genome Atlas Research Network, 2017), we analyzed the status of GNMT expression and methylation in human HCC. First, we analyzed GNMT gene expression in paired HCC tissue samples and nontumor liver tissue samples obtained from the same HCC patients and showed a lower level of the GNMT transcripts in HCC (Figure 4A). Next, we analyzed GNMT expression and methylation in HepG2 and SK-Hep-1 cells, cancer cell lines derived from different types of human liver tumors. Figure 4B shows that SK-Hep-1 cancer cells exhibited markedly lower GNMT expression compared with HepG2 cells, as evidenced by mRNA and protein levels. RRBS analysis of GNMT gene methylation in these cell lines showed that the CpG island located in the 5’ flanking region of the GNMT gene was predominately unmethylated in HepG2 cells, but was heavily methylated in SK-Hep-1 cells (Figure 4C).
Figure 4.
GNMT expression and cytosine DNA methylation in human HCC and liver cancer cell lines. A, Expression of the GNMT gene in paired HCC and nontumor liver tissue samples obtained from the same HCC patients. Asterisks (*) denotes a statistically significant (p < .05) difference between groups as determined by a Wilcoxon signed-rank test. B, GNMT mRNA and GNMT protein levels in HepG2 (n = 3) and SK-Hep-1 (n = 3) cancer cell lines. N/D = not detected. Representative images are shown. Asterisks (*) denote a statistically significant (p < .05) difference as determined by a 2-tailed Student’s t test. C, RRBS analysis of the GNMT gene methylation in HepG2 and SK-Hep-1 cancer cells. Red dots indicate methylated CpG sites; blue dots indicate unmethylated CpG sites. Y scale shows % methylation of DMRs in the GNMT gene. DMR residing at the 3’-end of the GNMT gene was highly methylated in both cell lines. In contrast, most CpG sites located in CpG island upstream from the gene transcription start site were unmethylated in HepG2 cells. D, Comparison of genome methylation in HepG2 and SK-Hep-1 cells. All DMRs were grouped into 4 quartiles based on the level of DNA methylation. The percentage of DMRs was calculated for each quartile. Asterisks (*) denote a statistically significant (p < .001) difference as determined by a Chi-Square test.
It has been suggested that the loss of GNMT expression in cancer cells may be associated with a DNA methylator phenotype (Huidobro et al., 2013). Therefore, using RRBS DNA methylation analysis, we investigated the status of genome methylation in HepG2 and SK-Hep-1 cells. Figure 4D shows that SK-Hep-1 cells exhibited a more prominent genome methylator phenotype than HepG2 cells. This was evidenced by a higher number of methylated DMRs, especially highly methylated DMRs (75%–100% methylation range), 52% in SK-Hep-1 cells versus 37% in HepG2 cells, and by a smaller number of low methylated DMRs (0%–25% methylation range), 21% in SK-Hep-1 cells versus 32% in HepG2 cells.
Effect of the GNMT Gene Knock-Down on Genomic DNA Methylation
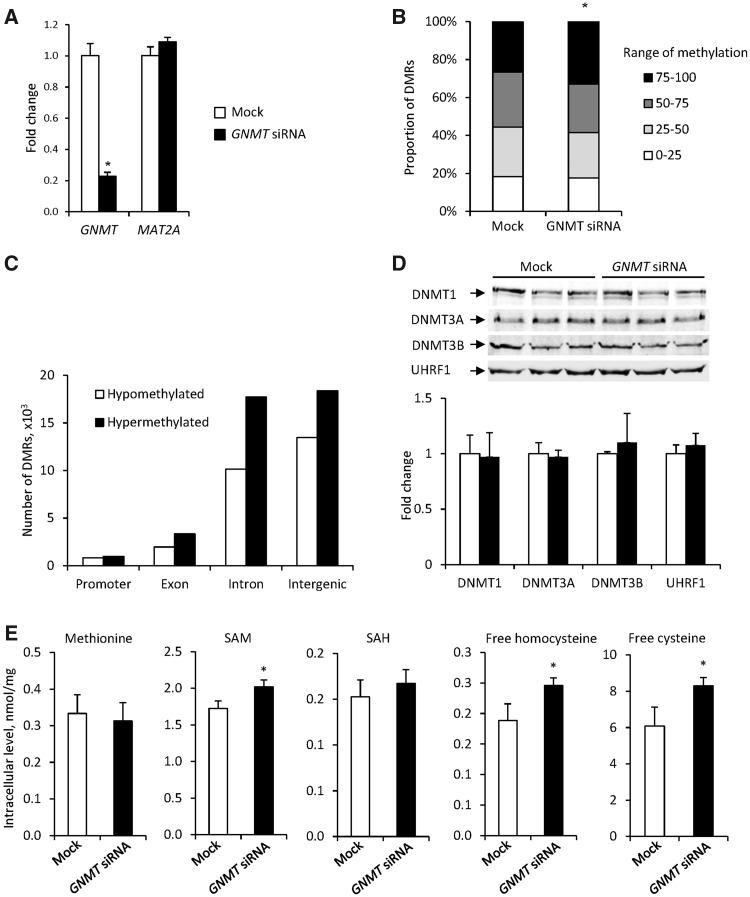
To test further whether or not direct inhibition of GNMT may promote the DNA methylator phenotype, we knocked down the GNMT gene in HepG2 cells, in which GNMT was highly expressed and unmethylated. Figure 5A shows that transfection of HepG2 cells with human GNMT siRNA resulted in a 77% reduction in GNMT expression as compared with that in mock-transfected cells. In contrast, the expression of a nontarget MAT2A gene was not affected in transfected cells. Silencing of GNMT increased the proportion of heavy methylated (75%–100% methylation range) DMRs from 26% to 33% (Figure 5B), with hypermethylation increasing in the exonic, intronic, and intergenic regions of the genome (Figure 5C).
Figure 5.
Effect of GNMT siRNA on genome methylation, expression of DNA methylation-related proteins, and 1-carbon metabolites in HepG2 cells. A, Expression of the GNMT and MAT2A genes in GNMT siRNA-transfected HepG2 cells. The results are presented as an average fold change in GNMT and MAT2A expression in siRNA-transfected HepG2 cells relative to that in mock-transfected cells, which were assigned a value 1. Values are mean ± SD (n = 3). Asterisks (*) denotes a statistically significant (p < .05) difference from the mock-transfected HepG2 cells as determined by a 2-tailed Student’s t test. B, Genome methylation and (C) genome distribution of DMRs in GNMT siRNA- and mock-transfected HepG2 cells. Asterisks (*) denote a statistically significant (p < .001) difference as determined by a Chi-Square test. D, Levels of DNMT1, DNMT3A, DNMT3B, and UHRF proteins in siRNA- and mock-transfected HepG2 cells. E, Levels of methionine, SAM, SAH, free homocysteine, and free cysteine in GNAT siRNA- and mock-transfected HepG2 cells. The results are presented as mean ± SD (n = 3). Asterisks (*) denotes a statistically significant (p < .05) difference from the mock-transfected HepG2 cells as determined by a 2-tailed Student’s t test.
To investigate the mechanisms that may lead to increase of DNA methylation associated with GNMT inhibition, we evaluated the levels of DNMT1, DNMT3A, DNMT3B, and UHRF proteins and 1-carbon metabolites in HepG2 cells transfected with GNMT siRNA. Figure 5D shows that the levels of DNMT1, DNMT3A, DNMT3B, and UHRF proteins were not affected by GNMT siRNA. In contrast, siRNA-mediated GNMT inhibition resulted in a significant increase of the SAM, homocysteine, and cysteine (Figure 5E).
DISCUSSION
The pathogenesis of HCC, one of the few cancer types with a steadily increasing incidence and mortality worldwide, is complex and involves diverse alterations of molecular pathways driven by genetic and epigenetic alterations. Recent progress in the analysis of HCC genomes identified 3 core genetic drivers, TP53, TERT, and WNT signaling, associated with the development of HCC (Shibata et al., 2018; Zucman-Rossi et al., 2015). In contrast, despite well-documented gene-specific DNA methylation aberrations in HCC, epigenetic drivers of liver carcinogenesis are less well defined.
In this study, we investigated the role of alterations in the DNA methylation of gene promoters in the development of HCC associated with NAFLD, the incidence of which has greatly increased in recent years. Using DIAMOND and STAM mice, mouse models of NAFLD-associated liver carcinogenesis that resemble the disease development in humans (Asgharpour et al., 2016; Fujii et al., 2013), we identified several cancer-related genes that were epigenetically down-regulated in NAFLD-derived full-fledged HCC. Among these genes, only the Gnmt gene was down-regulated and hypermethylated in both NAFLD-associated HCC models.
GNMT, a folate-binding protein that catalyzes the transfer of a methyl group from SAM to glycine to form sarcosine and SAH, is the most abundant SAM-dependent hepatic methyltransferase in the liver (Simile et al., 2018; Yeo and Wagner, 1994). GNMT also possesses several important tumor-suppressing features, including the regulation of 1-carbon, lipid, and glucose metabolism, the detoxification of carcinogens, and the regulation of cell proliferation and apoptosis, and is involved in the cellular response against DNA damage (DebRoy et al., 2013; Hughey et al., 2018; Liao et al., 2012; Martínez-Chantar et al., 2008; Simile et al., 2018; Wang et al., 2014; Yen et al., 2013). This suggests that down-regulation of GNMT may substantially compromise any or all of these functions and lead to tumor development. Indeed, results from several studies using Gnmt-null mice have revealed a critical role of Gnmt in the development of HCC (Liao et al., 2012; Martínez-Chantar et al., 2008). This finding was confirmed in clinical studies showing decreased GNMT expression in human HCC (Avila et al., 2000; Chen et al., 1998; Huidobro et al., 2013); however, there is a lack of conclusive information on the mechanism of down-regulation of this gene during liver carcinogenesis.
Several mechanisms have been suggested to be involved in the regulation of GNMT expression. Lee et al. (2009) demonstrated the presence of binding sites for several transcription factors, including the nuclear factor-Y, CCAAT/enhancer-binding protein (C/EBP) β, and xenobiotic response elements, in the GNMT promoter. Previously, we reported a marked decrease of C/EBP β protein in NASH livers from WSB/EiJ and PWK/PhJ mice fed a CFD diet (Pogribny et al., 2013). Two recent reports demonstrating that GNMT is a direct target of miR-224 and miR-873-5p revealed an involvement of microRNAs in GNMT regulation (Fernández-Ramos et al., 2018; Hung et al., 2018). Furthermore, Hung et al. (2018) showed that the level of miR-224 was inversely correlated with GNMT expression in HCC tissue and liver cancer cell lines, and Fernández-Ramos et al. (2018) reported an inverse correlation between the expression of hepatic miR-873-5p and GNMT in cirrhotic and cholestatic patients and animal models. A potential role of epigenetic regulation, specifically, DNA methylation, in GNMT inhibition in human HCC was provided by Huidobro et al. (2013), who demonstrated hypermethylation of the GNMT gene in 20% of primary HCC samples.
In light of this, the results of our study showing (1) a concordant down-regulation of the Gnmt gene and its promoter hypermethylation in full-fledged HCC and (2) their progressive alterations during the NAFLD-associated hepatocarcinogenic process are of great importance. Specifically, our study presents several important new findings on the role of Gnmt in molecular pathogenesis of NAFLD and its progression to HCC. First, using several relevant mouse models of NAFLD we demonstrate the dynamics of Gnmt alterations in NAFLD-related liver carcinogenesis. We show an early appearance of Gnmt down-regulation in NAFLD, the expression of which steadily decreased as the disease advanced. This corresponds to the similar finding by Tsuchida et al. (2018; GSE99010) that indicated an inhibition of Gnmt in diet- and chemical-induced NASH in mice. Second, we demonstrate that progression of NAFLD from simple steatosis to the advanced stages of the disease, eg, NASH, fibrosis, and HCC, is accompanied by progressive promoter hypermethylation of the Gnmt gene. This is an important finding suggesting that evaluation of Gnmt methylation may be useful for monitoring disease stratification. Finally, our study highlights the value of Gnmt as a gene susceptibility biomarker to determine an individual's predisposition to NAFLD and severity of the disease. This suggestion is in good agreement with the report by Tseng et al. (2003) demonstrating that individuals with genetic variations in GNMT causing a lower GNMT expression were overrepresented among HCC patients, and evidence that patients with inborn GNMT deficiency develop spontaneous liver diseases (Augoustides-Savvopoulou et al., 2003; Mudd et al., 2001).
One of the pathological consequences of down-regulation of GNMT is an alteration in cellular 1-carbon metabolism. In this study, we show that inhibition of the GNMT gene results in an increased level of SAM, which is accompanied by hypermethylation of the genome. This finding suggests that inhibition of GNMT expression during liver carcinogenesis may contribute to the establishment of a CpG island methylator phenotype and the inhibition of the critical cancer-related genes favoring the development of HCC. This study, as well as other studies, used only male mouse models of NASH-associated liver carcinogenesis only (Asgharpour et al., 2016; Tsuchida et al., 2018). Future studies are needed to determine if similar alterations exist in female mice.
Overall, we demonstrate that early down-regulation of the Gnmt gene caused by gene promoter hypermethylation is one of the key molecular events in the pathogenesis of NAFLD-derived HCC. The findings of this study indicate that GNMT mRNA, protein, or promoter methylation levels may be early diagnostic biomarkers with great potential to be used as minimally invasive disease-specific markers.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was partly supported by appointment of B.B.M. and L.J. to the Postgraduate Research Program at the NCTR administered by the Oak Ridge Institute for Science and Education (ORISE).
Supplementary Material
The views expressed in this article do not necessarily represent those of the U.S. Food and Drug Administration.
REFERENCES
- Altekruse S. F., McGlynn K. A., Reichman M. E. (2009). Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J. Clin. Oncol. 27, 1485–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgharpour A., Cazanave S. C., Pacana T., Seneshaw M., Vincent R., Banini B. A., Kumar D. P., Daita K., Min H.-K., Mirshahi F., et al. (2016). A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J. Hepatol. 65, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augoustides-Savvopoulou P., Luka Z., Karyda S., Stabler S. P., Allen R. H., Patsiaoura K., Wagner C., Mudd S. H. (2003). Glycine N-methyltransferase deficiency: a new patient with a novel mutation. J. Inherit. Metab. Dis. 26, 745–759. [DOI] [PubMed] [Google Scholar]
- Avila M. A., Berasain C., Torres L., Martín-Duce A., Corrales F. J., Yang H., Prieto J., Lu S. C., Caballería J., Rodés J., et al. (2000). Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J. Hepatol. 33, 907–914. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300. [Google Scholar]
- Bertuccio P., Turati F., Carioli G., Rodriguez T., La Vecchia C., Malvezzi M., Negri E. (2017). Global trends and predictions in hepatocellular carcinoma mortality. J. Hepatol. 67, 302–309. [DOI] [PubMed] [Google Scholar]
- Cazanave S., Podtelezhnikov A., Jensen K., Seneshaw M., Kumar D. P., Min H.-K., Santhekadur P. K., Banini B., Mauro A. G., Oseini A. M., et al. (2017). The transcriptomic signature of disease development and progression of nonalcoholic fatty liver disease. Sci. Rep. 7, 17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-M., Shiu J.-Y. A., Tzeng S. J., Shih L.-S., Chen Y.-J., Lui W.-Y., Chen P.-H. (1998). Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma. Int. J. Cancer 75, 787–793. [DOI] [PubMed] [Google Scholar]
- Choo S. P., Tan W. L., Goh B. K. P., Tai W. M., Zhu A. X. (2016). Comparison of hepatocellular carcinoma in Eastern versus Western populations. Cancer 122, 3430–3446. [DOI] [PubMed] [Google Scholar]
- DebRoy S., Kramarenko I. I., Ghose S., Oleinik N. V., Krupenko S. A., Krupenko N. I. (2013). A novel tumor suppressor function of glycine N-methyltransferase is independent of its catalytic activity but requires nuclear localization. PLoS One 8, e70062.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Conti A., Dreval K., Tryndyak V., Orisakwe O. E., Ross S. A., Beland F. A., Pogribny I. P. (2017). Inhibition of the cell death pathway in nonalcoholic steatohepatitis (NASH)-related hepatocarcinogenesis is associated with histone H4 lysine 16 deacetylation. Mol. Cancer Res. 15, 1163–1172. [DOI] [PubMed] [Google Scholar]
- Fernández-Ramos D., Fernández-Tussy P., Lopitz-Otsoa F., Gutiérrez-de-Juan V., Navasa N., Barbier-Torres L., Zubiete-Franco I., Simón J., Fernández A. F., Arbelaiz A., et al. (2018). MiR-873-5p acts as an epigenetic regulator in early stages of liver fibrosis and cirrhosis. Cell Death Dis. 9, 958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M., Shibazaki Y., Wakamatsu K., Honda Y., Kawauchi Y., Suzuki K., Arumugam S., Watanabe K., Ichida T., Asakura H., et al. (2013). A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med. Mol. Morphol. 46, 141–152. [DOI] [PubMed] [Google Scholar]
- Hughey C. C., Trefts E., Bracy D. P., James F. D., Donahue E. P., Wasserman D. H. (2018). Glycine N-methyltransferase deletion in mice diverts carbon flux from gluconeogenesis to pathways the utilize excess methionine cycle intermediates. J. Biol. Chem. 293, 11944–11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huidobro C., Toraño E. G., Fernández A. F., Urdinguio R. G., Rodríguez R. M., Ferrero C., Martínez-Camblor P., Boix L., Bruix J., García-Rodríguez J. L., et al. (2013). A DNA methylation signature associated with the epigenetic repression of glycine N-methyltransferase in human hepatocellular carcinoma. J. Mol. Med. (Berl) 91, 939–950. [DOI] [PubMed] [Google Scholar]
- Hung J.-H., Li C.-H., Yeh C.-H., Huang P.-C., Fang C.-C., Chen Y.-F., Lee K.-J., Chou C.-H., Cheng H.-Y., Huang H.-D., et al. (2018). MicroRNA-224 down-regulates glycine N-methyltransferase gene expression in hepatocellular carcinoma. Sci. Rep. 8, 12284.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou G. N., Green P. K., Berry K. (2018). HCV eradication induced by direct-acting antiviral agents reduces the risk of hepatocellular carcinoma. J. Hepatol. 68, 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn D., Kircher S., Hermanns H. M., Geier A. (2019). Animal models of NAFLD from hepatologist’s point of view. Biochim. Biophys. Acta Mol. Basis Dis. 1865, 943–953. [DOI] [PubMed] [Google Scholar]
- Kim N. G., Nguyen P. P., Dang H., Kumari R., Garcia G., Esquivel C. O., Nguyen M. H. (2018). Temporal trends in disease presentation and survival of patients with hepatocellular carcinoma: a real-world experience from 1998 to 2015. Cancer 124, 2588–2598. [DOI] [PubMed] [Google Scholar]
- Lee C.-M., Shih Y.-P., Wu C.-H., Chen Y.-M. A. (2009). Characterization of the 5’ regulatory region of the human glycine N-methyltransferase gene. Gene 443, 151–157. [DOI] [PubMed] [Google Scholar]
- Liao Y.-J., Chen T.-L., Lee T.-S., Wang H.-A., Wang C.-K., Liao L.-Y., Liu R.-S., Huang S.-F., Chen Y.-M. A. (2012). Glycine N-methyltransferase deficiency affects Niemann-Pick type C2 protein stability and regulates hepatic cholesterol homeostasis. Mol. Med. 18, 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llovet J. M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G. (2016). Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2, 16018.. [DOI] [PubMed] [Google Scholar]
- Machado M. V., Michelotti G. A., Xie G., de Almeida T. P., Boursier J., Bohnic B., Guy C. D., Diehl A. M. (2015). Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One 10, e0127991.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Chantar M. L., Vázquez-Chantada M., Ariz U., Martínez N., Varela M., Luka Z., Capdevila A., Rodríguez J., Aransay A. M., Matthiesen R., et al. (2008). Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology 47, 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnyk S., Pogribna M., Pogribny I., Hine R. J., James S. J. (1999). A new HPLC method for the simultaneous determination of oxidized and reduced plasma aminothiols using coulometric electrochemical detection. J. Nutr. Biochem. 10, 490–497. [DOI] [PubMed] [Google Scholar]
- Melnyk S., Pogribna M., Pogribny I. P., Yi P., James S. J. (2000). Measurement of plasma and intracellular S-adenosylmethionine and S-adenosylhomocysteine utilizing coulometric electrochemical detection: alterations with plasma homocysteine and pyridoxal 5'-phosphate concentrations. Clin. Chem. 46, 265–272. [PubMed] [Google Scholar]
- Mudd S. H., Cerone R., Schiaffino M. C., Fantasia A. R., Minniti G., Caruso U., Lorini R., Watkins D., Matiaszuk N., Rosenblatt D. S., et al. (2001). Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninaemia. J. Inherit. Metab. Dis. 24, 448–464. [DOI] [PubMed] [Google Scholar]
- Pogribny I. P., Kutanzi K., Melnyk S., de Conti A., Tryndyak V., Montgomery B., Pogribna M., Muskhelishvili L., Latendresse J. R., James S. J., et al. (2013). Strain-dependent dysregulation of one-carbon metabolism in mice is associated with choline- and folate-deficient diet-induced liver injury. FASEB J. 27, 2233–2243. [DOI] [PubMed] [Google Scholar]
- Pogribny I. P., Rusyn I. (2014). Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 342, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhekadur P. K., Kumar D. P., Sanyal A. J. (2018). Preclinical models of non-alcoholic fatty liver disease cure. J. Hepatol. 68, 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1118. [DOI] [PubMed] [Google Scholar]
- Shen L., Ahuja N., Shen Y., Habib N. A., Toyota M., Rashid A., Issa J.-P. J. (2002). DNA methylation and environmental exposures in human hepatocellular carcinoma. J. Natl. Cancer Inst. 94, 755–761. [DOI] [PubMed] [Google Scholar]
- Shibata T., Arai Y., Totoki Y. (2018). Molecular genomic landscapes of hepatobiliary cancer. Cancer Sci. 109, 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard E. P., Ward E. M., Siegel R., Jemal A. (2012). Cancers with increasing incidence trends in the United States: 1999 through 2008. CA Cancer J. Clin. 62, 118–128. [DOI] [PubMed] [Google Scholar]
- Simile M. M., Latte G., Feo C. F., Feo F., Calvisi D. F., Pascale R. M. (2018). Alterations of methionine metabolism in hepatocarcinogenesis: the emergent role of glycine N-methyltransferase in liver injury. Ann. Gastroenterol. 31, 552–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. (2017). Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 169,1327–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryndyak V., Latendresse J. R., Montgomery B., Ross S. A., Beland F. A., Rusyn I., Pogribny I. P. (2012). Plasma microRNAs are sensitive indicators of inter-strain differences in the severity of liver injury induced in mice by a choline- and folate-deficient diet. Toxicol. Appl. Pharmacol. 262, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryndyak V., de Conti A., Doerge D. R., Olson G. R., Beland F. A., Pogribny I. P. (2017). Furan-induced transcriptomic and gene-specific DNA methylation changes in the livers of Fischer 344 rats in a 2-year carcinogenicity study. Arch. Toxicol. 91, 1233–1243. [DOI] [PubMed] [Google Scholar]
- Tseng T.-L., Shih Y.-P., Huang Y.-C., Wang C.-K., Chen P.-H., Chang J.-G., Yeh K.-T., Chen Y.-M., Buetow K. H. (2003). Genotypic and phenotypic characterization of putative tumor susceptibility gene, GNMT, in liver cancer. Cancer Res, 63, 647–654. [PubMed] [Google Scholar]
- Tsuchida T., Lee Y. A., Fujiwara N., Ybanez M., Allen B., Martins S., Fiel M. I., Goossens N., Chou H.-I., Hoshida Y., et al. (2018). A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J. Hepatol. 69, 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-C., Lin W.-L., Lin Y.-J., Tang F.-Y., Chen Y.-M. A., Chiang E.-P. I. (2014). a novel role of the tumor suppressor GNMT in cellular defense against DNA damage. Int. J. Cancer 134, 799–810. [DOI] [PubMed] [Google Scholar]
- Wang Q., Wang G., Liu C., He X. (2018). Prognostic value of CpG island methylator phenotype among hepatocellular carcinoma patients: A systematic review and meta-analysis. Int. J. Surg. 54, 92–99. [DOI] [PubMed] [Google Scholar]
- Yang H., Wang J. R., Didion J. P., Buus R. J., Bell T. A., Welsh C. E., Bonhomme F., Yu A. H.-T., Nachman M. W., Pialek J., et al. (2011). Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 43, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C.-H., Lin Y.-T., Chen H.-L., Chen S.-Y., Chen Y.-M. A. (2013). The multi-functional roles of GNMT in toxicology and cancer. Toxicol. Appl. Pharmacol. 266, 67–75. [DOI] [PubMed] [Google Scholar]
- Yeo E.-J., Wagner C. (1994). Tissue distribution of glycine N-methyltransferase, a major folate-binding protein of liver. Proc. Natl. Acad. Sci. U.S.A. 91, 210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J., Hann H.-W., Coben R., Conn M., DiMarino A. J. (2018). Update treatment for HBV infection and persistent risk for hepatocellular carcinoma: prospect for an HBV cure. Diseases 6, 27.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes R., Bugianesi E. (2018). Should we undertake surveillance for HCC in patients with NAFLD? J. Hepatol. 68, 326–334. [DOI] [PubMed] [Google Scholar]
- Zucman-Rossi J., Villanueva A., Nault J.-C., Llovet J. M. (2015). Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 149, 1226–1239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.