Abstract
Nucleophosmin (NPM1) is an abundant nucleolar protein that aids in the maturation of pre-ribosomal particles and participates in oncogenic stress responses through its interaction with the Alternative Reading Frame tumor suppressor (p14ARF). NPM1 mediates multiple mechanisms of phase separation which contribute to the liquid-like properties of nucleoli. However, the effects of phase separation on the structure and dynamics of NPM1 are poorly understood. Here we show that NPM1 undergoes phase separation with p14ARF in vitro, forming condensates that immobilize both proteins. We probed the structure and dynamics of NPM1 within the condensed phase using solid-state NMR spectroscopy. Our results demonstrate that within the condensed phase, the NPM1 oligomerization domain forms an immobile scaffold, while the central intrinsically disordered region and the C-terminal nucleic acid binding domain exhibit relative mobility.
Keywords: NPM1, p14ARF, MAS, ssNMR, phase separation, biomolecular condensates
Graphical Abstract
1. Introduction
It is now recognized that large portions of the cell interior are compartmentalized through the formation of non-membrane bound organelles termed biomolecular condensates [1]. Studies of these structures in vitro and in live cells has demonstrated that many biomolecules perform their biological roles within phase separated structures exhibiting properties ranging from viscous liquids to viscoelastic materials and elastic solids [2]. Furthermore, aberrant phase transitions have been implicated in human neurodegenerative diseases and cancer [3–7]. A key goal is to understand how the structural and dynamic properties of biomolecules enable the collective behavior that underlies phase separation and the formation of biomolecular condensates.
Many proteins that drive the assembly of biomolecular condensates in cells are intrinsically disordered (IDPs) or contain intrinsically disordered regions (IDRs). Solution state NMR spectroscopy, which has been extensively applied in studies of IDPs and IDRs, has also been applied in studies of IDPs and IDRs within condensed phases [8]. However, these studies have been mostly limited to liquid-like phases, as proteins within viscoelastic condensates generally experience rapid transverse relaxation due to extensive cross-linking interactions and slowed molecular tumbling, thus contributing to extensive resonance broadening. By contrast, magic angle spinning (MAS) solid state NMR (ssNMR) effectively eliminates contributions to resonance broadening, including chemical shift anisotropy (CSA), allowing them to be selectively reintroduced by specific pulse schemes in order to alternately probe immobile or dynamic regions of molecules [9]. Accordingly, the last decade has seen a rapid expansion in the application of ssNMR techniques to the study of biomolecular condensates [8].
Here, we focus on condensates formed through interactions between Nucleophosmin (NPM1) and the Alternative Reading Frame (ARF) tumor suppressor (p14ARF in humans and p19ARF in mice). NPM1 is an abundant nucleolar protein comprised of an N-terminal oligomerization domain (OD), a small, helical C-terminal nucleic acid binding domain (CTD) and a central, polyampholytic IDR. These structural properties contribute to NPM1’s ability to undergo phase separation in vitro and are important for cellular function. NPM1 contributes to the liquid-like properties of nucleoli, aids in the maturation and export of pre-ribosomal particles and, through its interactions with ARF, mediates oncogenic stress responses [10, 11]. During interphase, ARF is expressed at low levels and is localized to the granular component of the nucleolus through interactions with NPM1 [12]. NPM1 sequesters ARF in high molecular weight complexes [13], which protect it from degradation [13–15], and are required for the maintenance of ribosome biogenesis homeostasis. ARF deletion results in a NPM1-dependent surge in ribosome biogenesis, protein synthesis and increased nucleolar size [16], three characteristic phenotypes of cancer cells [17]. ARF protein levels rise in response to oncogene activation [18]. And increased ARF expression leads to the nucleolar sequestration of MDM2, a negative regulator of p53, and NPM1, resulting in cell cycle arrest through p53-dependent and - independent mechanisms, respectively [19]. Cellular stress can also modulate the fluid features of nucleoli and induce transitions to what may be solid-like structures. For example, a diverse array of stressors can induce the formation of so-called nucleolar detention centers, which immobilize essential proteins including MDM2, in amyloid bodies [20]. When overexpressed in nucleoli, EGFP-tagged p14ARF exhibits dynamics consistent with inclusion within a viscoelastic medium, e.g. the incomplete recovery of fluorescence after photo bleaching and a slow half-time for recovery relative to other nucleolar proteins [21]. Together, these studies support a mechanism wherein ARF acts as a modulator of the liquid-like properties of the nucleolar matrix through its interaction with NPM1, thus exerting control over the cell through alteration of ribosome biogenesis and activation of p53 during oncogenic stress events.
Here we show that NPM1 and p14ARF undergo phase separation in vitro. We used confocal fluorescence microscopy to characterize the material properties of the condensates and ssNMR to probe the structure and dynamics of NPM1 within the condensed phase matrix. Our results show that NPM1 and p14ARF experience limited diffusion within condensates. And within the condensed phase, the OD of NPM1 forms an immobilized scaffold, while the central IDR and CTD remain relatively mobile.
2. Materials & Methods
2.1. Protein Expression and Purification
A synthetic gene encoding p14ARF was cloned into a modified pET28 expression vector containing an N-terminal polyhistidine tag and tobacco etch virus (TEV) protease cleavage site. p14ARF was expressed in batch cultures of E. coli BL21 cells grown at 37 °C in LB medium supplemented with 30 μg/ml of Kanamycin. At OD600nm = 0.8, 0.5 mM Isopropyl β-D-1 thiogalactopyranoside (IPTG) was added, cells were incubated at 37 °C for an additional 3 hours and harvested by centrifugation at 3,800 rpm at 4 °C. Cells were resuspended in 50 mM Tris pH 8.0, 500 mM NaCl, 5 mM beta-mercaptoethanol, and SIGMAFAST protease inhibitor cocktail tablets (Sigma), and disrupted by sonication. The lysate was cleared by centrifugation at 30,000 rpm at 4 °C and Urea was added to a final concentration of 6 M. The cell pellet, which contained additional p14ARF protein within inclusion bodies, was resuspended in 6 M GuHCl, 0.1% Triton, 5 mM beta-mercaptoethanol and subjected to mechanical disruption followed by additional sonication. This fraction was cleared by centrifugation at 30,000 rpm at 4 °C and the supernatant was removed and combined with the initial lysate. p14ARF was purified by Ni2+-affinity chromatography on an ÄKTA FPLC (GE) using a linear gradient of 50 mM Tris pH 8.0, 500 mM NaCl, 5 mM beta-mercaptoethanol and 500 mM Imidazole. The Ni-column eluent was further purified by HPLC using an H2O/CH3CN/0.1% trifluoroacetic acid solvent system. Fractions containing pure p14ARF were lyophilized and stored at 4 °C.
The cloning, expression, and purification of NPM1 was performed as described [22, 23] with the following modifications. For isotopic labeling of NPM1, cells were grown in MOPS-based minimal media containing [U13C6]-D-glucose and 15NH4Cl (Cambridge Isotope Laboratories) [24]. Following purification, lyophilized NPM1 protein was refolded by resuspension in 6 M guanidinium HCl with 2 mM dithiothreitol (DTT), and dialysis against 10 mM sodium phosphate pH 7.0, 150 mM NaCl, and 2 mM DTT. Proper folding into the pentameric structure was confirmed by circular dichroism spectroscopy and dynamic light scattering [23].
2.2. Confocal Microscopy
p14ARF was fluorescently labeled with a Succinimidyl Ester derivative of Alexa Fluor 405 (Thermo Fisher) using amide chemistry as specified by the manufacturer [25]. Conjugation of Alexa Fluor 488 (Thermo Fisher) to NPM1 was performed as previously described [26]. To prepare NPM1-p14ARF droplets for microscopy analysis, p14ARF was resuspended from a lyophilized powder using DMSO and added directly to NPM1 in solution at room temperature (~23 °C), such that the total protein concentrations were 20 μM NPM1 and 20 μM p14ARF. The final buffer contained 10 mM Na-P pH 7.0, 150 mM NaCl, 2mM DTT, 1.67% DMSO. Low concentrations of DMSO had no effect on the structure of NPM1 as confirmed by solution state NMR (Fig. S1). To prevent unwanted photo-physical artifacts associated with Fӧrster resonance energy transfer (FRET), droplet suspensions were prepared in parallel, with only one labeled species at <10% of the total protein concentration. Following phase separation, droplet suspensions were incubated for 1 hour at room temperature and transferred to 16-well CultureWell chambered slides (Grace BioLabs, Bend, OR, USA) pre-coated with PlusOne Repel Silane ES (GE Healthcare, Pittsburgh, PA, USA) and Pluronic F-127 (Sigma- Aldrich, St. Louis, MO, USA).
To determine the concentration of NPM1 and p14ARF inside droplets and whether prolonged ultracentrifugation alters the condensed phase molar ratio, the following experiment was performed. Suspensions of droplets were prepared and split into two aliquots. One was ultracentrifuged at 100,000 rpm (436,000 xg) at 4 °C for 22 hours, while the second was stored at 4 °C. Calibration curves were prepared from images of uncongugated Alexa Fluor 405 and Alexa Fluor 488 dye solutions. Serial dilutions were performed for each dye. And for each dilution, 3D images were collected with a 0.23 μm step size along the Z-axis. Fluorescence intensities were averaged over the entire field of view and over all Z planes and plotted as a function of the known dye concentration. 3D images of NPM1-p14ARF droplets were collected over multiple imaging areas (>5) using the same instrument parameters. And for each Z plane, images were segmented into light phase and condensed phase regions of interest (ROI) based on fluorescence intensity using the analyze particles function in image J [27]. The output was then manually checked to ensure that only ROIs from in focus droplets were used for further analysis. The condensed phase protein concentrations were then determined using the standard curves and the mean fluorescence intensities within selected ROIs, which were corrected for dilution and for the effects of viscosity on dye quantum yields [22]. The correction factors used for Alexa Fluor 405 and Alexa Fluor 488 were 1.62 and 0.73, respectively. The condensed phase concentrations were calculated as the average and standard deviation of all analyzed ROIs. The values reported represent the average and propagation of error from 4 replicates.
For fluorescence recovery after photo-bleaching (FRAP) experiments, circular areas (0.78 μm2) within droplets were photobleached to ~ 50% of the initial fluorescence intensities and monitored every ~500 ms over the course of 200 s. Images were processed using Slidebook 6.0 (Intelligent Imaging Innovations, Gottingen, Germany). ROI intensities were normalized and corrected for global photobleaching during image acquisition [28]. For NPM1 and p14ARF, recovery curves were calculated from the average and standard deviation of n=12 droplets collected over three imaging areas.
To determine the thermal stability of NPM1-p14ARF droplets, a suspension of droplets containing Alexa Fluor 488 labeled NPM1 was sandwiched between two glass slides and imaged using confocal fluorescence microscopy at temperatures ranging from 4–50 °C. The sample temperature was controlled using a PE100 inverted peltier stage for Zeiss microscopes (Linkam Scientific Instruments) and monitored with an external digital thermometer (Omega), which was attached directly to the microscope slide. For each temperature, the fluorescence intensities were extracted from all droplets within 5 imaging areas using Image J and averaged.
To determine the effects of prolonged ultracentrifugation on the morphology and porosity of the NPM1-p14ARF condensed phase a suspension of droplets was subjected to 18 hours of ultracentrifugation at 100,000 rpm (436,000 xg) at 4 °C and the permeability of the sedimented condensed phase to fluorescently tagged polymers was assessed. Following ultracentrifugation, 10 μl of droplets were transferred to a microscope slide and 0.5 μl of either mPEG-Rhodamine, 10K (Creative PEGWorks) or Ficol-Fluorescein, 70K (Creative PEGWorks) was added such that the final polymer concentration was 6 μM. Following 1 hour incubation at room temperature, the droplets were imaged by confocal fluorescence and differential interference contrast microscopy.
All microscopy was performed on a 3i Marianas spinning disk confocal microscope (Intelligent Imaging Innovations Inc., Denver, CO, USA). For condensed phase protein concentration and FRAP measurements a 100X oil immersion objective (N.A. 1.4) was used. For temperature stability measurements and droplet permeability assays 20X (N.A. 0.75) and 10X (N.A. 0.45) air objectives were used, respectively.
2.3. Solution-State NMR Experiments
Solution state NMR experiments were performed on a Bruker AVANCE NEO spectrometer operating at a proton Larmor frequency of 800.13 MHz. This spectrometer was equipped with a 5 mm triple-resonance 1H/13C/15N TCI cryo-probe. Spectra were processed in Topspin 4.0 and analyzed in Sparky. Additional experimental details are provided in the supplemental figure legends.
2.4. Solid-State NMR Experiments
For ssNMR analysis, a 50 ml suspension of [U13C,U15N]-NPM1-p14ARF droplets was prepared as described in section 2.2 with the exception that the final buffer contained 10 mM Na-P pH 7.0, 150 mM NaCl, 2 mM TCEP, 1.67% DMSO-d6, and 0.02% NaN3. The droplets were pelleted by ultracentrifugation at 30,000 rpm for 18 hours at 4 °C. Most of the light phase was decanted, and the condensed phase was transferred to a 1.7 ml Eppendorf tube by resuspending the condensed phase with a pipettor several times followed by centrifugal sedimentation at 21,000 rpm on a benchtop centrifuge at 4 °C. This resulted in 100 μl of condensed phase, ~85 μl (~140 nmoles of [U13C,U15N]-NPM1) of which was transferred into a 3.2 mm ssNMR rotor with a 90 μl capacity by centrifugation using a custom rotor packing device.
All ssNMR spectra were collected on a Bruker AVANCE NEO spectrometer operating at a proton Larmor frequency of 600.13 MHz. This spectrometer was equipped with a 3.2 mm triple-resonance 1H/13C/15N CPMAS probe with cryogenically cooled NMR coils and preamplifiers (BioSolids CryoProbe™). The 1H/13C/15N CPMAS cryoprobe provides a >3-fold enhancement of 13C and 15N sensitivity (per unit volume) as compared to a conventional room temperature probe (data not shown). All experiments were collected at 11.5 kHz MAS. Spectra were processed in Topspin 4.0 and mNova (mestrelab) and analyzed in Sparky. Chemical shifts were calibrated using adamantane as an external reference by setting the up-field resonance to 38.5 ppm.
For the 13C-13C-DARR experiment, carbon and proton 90-degree RF fields were set to 60 and 71 kHz respectively for hard pulses, cross polarization and heteronuclear decoupling. During cross polarization (CP) the carbon RF field was constant, while the proton RF field was linearly ramped over an amplitude of 30 %. The proton contact RF field was optimized to match the carbon RF field at the first spinning Hartmann-Hahn condition. The CP contact time was optimized to 1250 μs. Acquisition of the 13C signal was done under Swept-frequency two-pulse phase modulation (SW-TPPM). 2104 and 1024 increments were collected for the direct and indirect dimension, respectively, over a spectral window of 301 and 228 ppm, respectively. The recycle delay was set to 3.5 s and the total experimental time was 20 hours and 30 minutes.
For the NCA and 2D NCACX experiments, proton to nitrogen cross polarization was achieved with at a nitrogen RF field of 41 kHz. The proton contact field was optimized at the first Hartmann-Hahn matching condition by applying a tangential modulation of the proton RF field. The CP contact time was optimized at 700 μs. During the N-Ca polarization transfer, the nitrogen RF field was set to 4 KHz, while the corresponding 13C field was optimized around a value of 16 kHz using a tangential modulation of the RF field. The optimal contact time during the N-Ca transfer was found at 3500 μs. Hard pulses for 13C and 15N were set to 64 and 41 kHz, respectively, while the proton hard pulse and decoupling RF field were set to 80 kHz. In the 2D NCACX experiment, a 50 ms DARR mixing scheme was applied at proton RF field matching the spinning speed. 4 (NCA) or 16 (2D NCACX) scans per increment were acquired for a total experimental time of 15 min (NCA) or 1 hour (2D NCACX) recording 2024 × 64 increments over a spectral window of 301 × 47 ppm. Nitrogen and Ca offsets were set to 122.5 and 60 ppm, respectively.
Similar acquisition parameters were used for the NCO, 3D NCOCX, and 3D NCACX experiments with the following exceptions. For the NCO, that the 15N and CO RF fields were set to ~18.5 kHz and ~30 kHz respectively for an optimized contact time of 2 ms. The 15N and CO offsets were set to 124 and 173 ppm respectively. For the 3D NCOCX, The DARR mixing was done under 11.5 kHz of proton irradiation for a total mixing period of 50 ms. 16 scans per increment were acquired for a total experimental time of 1 day and 17 hours recording 1818 × 92 × 32 increments over a spectral window of 301 × 38 × 47 ppm. And for the 3D NCACX, the DARR mixing was done under 11.5 kHz of proton irradiation for a total mixing period of 20 ms. 32 scans per increment were acquired for a total experimental time of 15 hours, recording 1362 × 60 × 16 increments over a spectral window of 301 × 40 × 47 ppm.
For the 2D INEPT-through-bond-correlation-spectroscopy (TOBSY) experiment,1H-13C magnetization transfer via INEPT was used to excite mobile sites, after which 6 ms of TOBSY 13C-13C mixing was applied using the P916 condition [29] with a 37 kHz 13C radio frequency field. To remove the effect of residual 1H–13C interactions during indirect and direct 13C detection, WALTZ-64 decoupling was employed. This spectrum was acquired with 512 complex points in the indirect dimension and 1362 points in the direct dimension, with dwell times of 60 ms and 12 ms, respectively, using a spectral window of 185 × 301 ppm, respectively. 32 scans were acquired for each increment, with a repetition delay of 1.5 s. Acquisition parameters are summarized in Table S1 and additional experimental details are provided in the figure legends.
3. Results and Discussion
3.1. Characterization of NPM1-p14ARF condensates using fluorescence microscopy
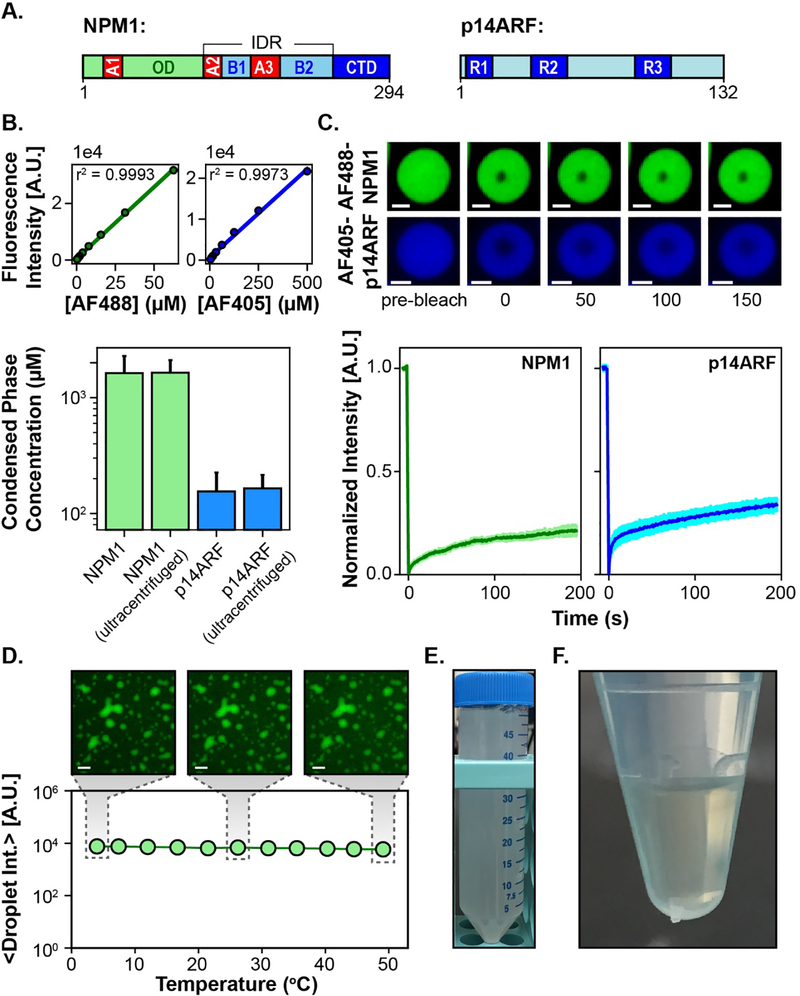
We have previously shown that NPM1 can undergo liquid-liquid phase separation with proteins containing Arg-rich motifs [30]. ARF, a highly basic protein (pI ~12) that possesses several Arg-motifs (Fig. 1A), can also undergo phase separation with NPM1. Using confocal fluorescence microscopy, we determined that NPM1 and p14ARF are enriched to concentrations of 1632 +/− 655 μM and 155 +/− 70 μM within the condensed phase, respectively (Fig. 1B). This would suggest an ~2:1 ratio of NPM1 pentamers to p14ARF molecules, consistent with our previous studies of NPM1 phase separation with the nucleolar protein Surf6 [26]. However, unlike NPM1 and Surf6, which form a viscous liquid phase composed of slowly diffusing molecules, FRAP analyses revealed that within NPM1-p14ARF condensates, most NPM1 and p14ARF molecules are immobilized on the seconds timescale (Fig. 1C). These diffusive dynamics are suggestive of viscoelastic material properties.
Fig. 1.
(A) Schematic representations of NPM1 and p14ARF, where the NPM1 acidic and basic tracts are denoted as A1–3 and B1–2, respectively, and p14ARF arginine motifs are denoted as R1–3. (B, top) Standard curves for unconjugated Alexa Fluor 488 and Alexa Fluor 405. Error bars representing the standard deviation are smaller than the markers. (B, bottom) Concentrations of NPM1 and p14ARF in non-centrifuged droplets and droplets that were ultracentrifuged at 100,000 rpm (436,000 xg). Error bars represent the propagation of error from 4 replicates (C, top) FRAP of Alexa Fluor 488 labeled NPM1 and Alexa Fluor 405 labeled p14ARF within individual droplets, where the scale bar = 2 μm. (C, bottom) FRAP recovery curves for Alexa Fluor 488 labeled NPM1 and Alexa Fluor 405 labeled p14ARF, where the error bars represent the standard deviation of n=12 droplets each. (D) Average fluorescence intensity of Alexa Fluor 488 labeled NPM1 within the NPM1-p14ARF condensed phase plotted as a function of temperature. Error bars representing the standard deviation are smaller than the markers. Representative images are shown above, where the scale bar = 20 μm. (E) Image of a 50 ml suspension of [U13C, U15N]-NPM1-p14ARF droplets collected immediately after phase separation. (F) The [U13C, U15N]-NPM1-p14ARF condensed phase after centrifugal sedimentation.
In principle, the equilibria of two phase systems are intrinsically temperature sensitive [31]. For example, fluctuations around the critical point may dissolve droplets and fluctuations about sol-gel lines may result in the irreversible aggregation of phase separated proteins. The MAS rotor in our experimental setup can be maintained at 4 °C to >40 °C in the NMR spectrometer. However, samples can experience a 10–15 °C temperature increase as they are accelerated to 13,000 rpm or higher. Therefore, we tested the temperature-sensitivity of NPM1-p14ARF condensates. Droplets were imaged at temperatures ranging from 4–50 °C. No change in droplet morphology or fluorescence intensity of Alexa Fluor labeled NPM1 was observed, indicating that NPM1-p14ARF condensates are stable over this temperature range (Fig. 1D).
We also determined whether ultracentrifugal force alters the pore size of the condensed phase or the molar ratio of condensed NPM1 and p14ARF. Droplets were subjected to prolonged ultracentrifugation at 100,000 rpm (436,000 xg) and their permeability to fluorescently tagged probe molecules of increasing hydrodynamic radii was assessed (Fig. S2). No substantial change in droplet permeability was observed. And the ultracentrifuged condensed phase exhibited no morphological differences as determined by differential interference contrast microscopy (e.g., the presence of large aggregates or fibrils within the condensed phase). Furthermore, ultracentrifugation did not significantly change the concentration of NPM1 and p14ARF within droplets, which was 1644 +/− 463 and 165 +/− 51 μM, respectively (Fig. 1B).This suggests that ultracentrifugal force does not irreversibly disturb the “meshwork” of protein-protein interactions that constitute the condensed phase.
3.2. CP-MAS 2D & 3D correlation spectroscopy
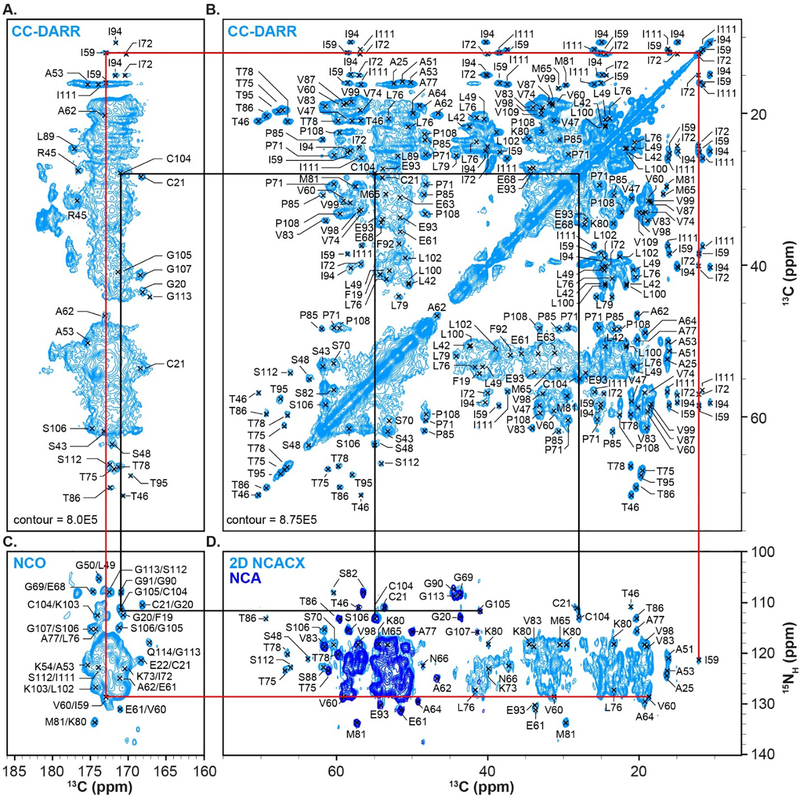
To produce a sample of isolated NPM1-p14ARF condensed phase for ssNMR analysis, a 50 ml suspension of [U13C, U15N]-NPM1-p14ARF droplets was prepared and subsequently pelleted by centrifugation (Fig. 1E, F). We began our ssNMR study by probing the rigid regions of NPM1 using cross-polarization. Initially, 2D 13C-13C correlation spectra were acquired using dipolar-assisted rotational resonance (DARR) [32], which allowed us to assess the spectral quality of CP spectra and provided several amino acid specific assignments for well resolved spin systems (Fig. 2A, B). This was particularly useful for identifying resonances for residues containing methyl groups, which were generally sharp and well resolved. These included 2 of 8 Met, 4 of 10 Ile, and 5 of 10 Thr spin systems. We also collected NCA, NCO and 2D NCACX correlation spectra (Fig. 2C,D) [33, 34], which together with the CC-DARR spectrum, provided several unambiguous sequential assignments, including C104-G105 & I59-V60. However, establishing resonance assignments utilizing 2D correlation spectra was hampered by resonance overlap, particularly in the carbonyl region of the CC-DARR spectrum.
Fig. 2.
(A, B) The 13C-13C-DARR experiment collected with 50 ms of DARR mixing time, Tset = 4.8 °C. The spectrum displayed is the sum of two experiments collected with 4 and 16 scans. (C) The NCO experiment, collected with 4 scans, Tset = 4.8 °C. (D) Overlay of the NCA experiment collected with 4 scans, Tset = 4.8 °C (dark blue) and the 2D NCACX correlation experiment (light blue) collected with 16 scans and 50 ms of DARR mixing time and Tset = 4.8 °C. Sequential assignments for C104-G105 & I59-V60 are indicated by the black and red lines, respectively.
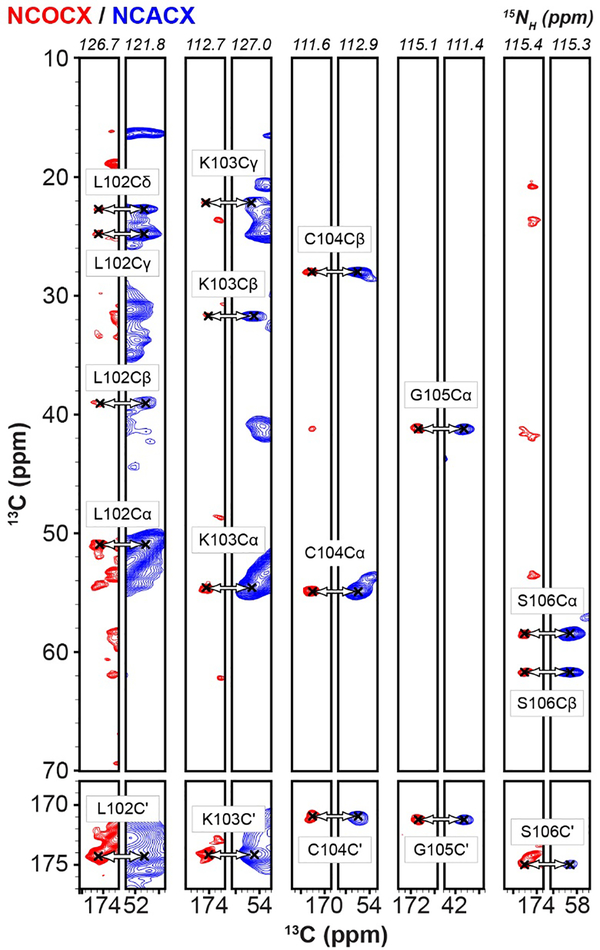
Site-specific assignments of backbone and sidechain resonances were obtained using 3D NCACX and 3D NCOCX spectra [33, 34]. Representative strips from these experiments are shown in Fig. 3. In total, resonances for 73 of the 294 residues in NPM1 were assigned (Table S2). These corresponded exclusively to residues within the N-terminal OD, suggesting that this domain forms an immobile scaffold within the NPM1-p14ARF condensed phase matrix.
Fig. 3.
Representative strip plots of the 3D NCOCX spectrum, collected with 16 scans and 50 ms of DARR mixing, set T = 5 °C (red) and the 3D NCACX spectrum, collected with 32 scans and 20 ms of DARR mixing, Tset = 4.8 °C (blue).
3.3. Scalar coupling based 2D correlation spectroscopy
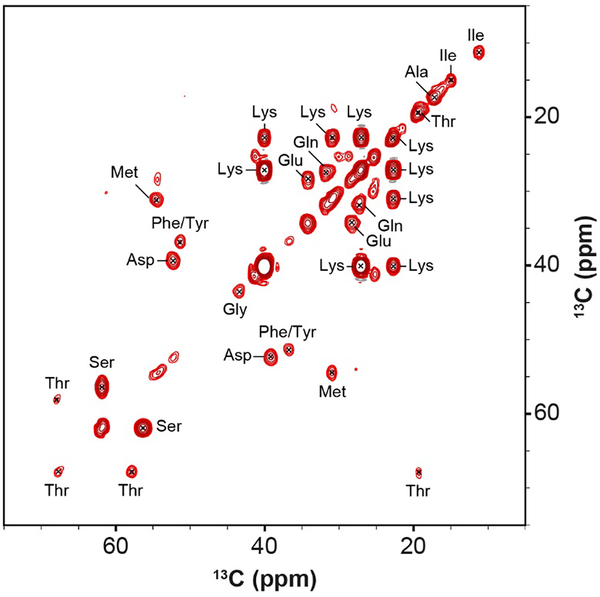
To probe mobile regions of NPM1 within the condensed phase, we performed the 13C-13C INEPT-TOBSY experiment [29, 35] (Fig. 4). We detected weak resonances for Met, Gly, Ile and Thr and strong resonances for Ala, Asp, Glu, Lys, and Ser spin systems. While site-specific assignment of these peaks was not possible, these latter amino acid types are enriched in flexible regions within the OD and the central IDR (Table S3). Spin systems for specific amino acids found only in the CTD (Table S3) were absent, suggesting that within the NPM1-p14ARF condensed phase, the NPM1 CTD experiences line broadening due to transient interactions with other polypeptide chains within the condensate, or due to motion that is occurring on an intermediate timescale, which is undetectable to the scalar coupling and cross polarization based methods utilized here.
Fig. 4.
The aliphatic region of the INEPT-TOBSY experiment collected with 32 scans, 6 ms of P916 TOBSY mixing, 13C RF = 37 kHz, and Tset = −4 °C.
3.4. Secondary Structure Analysis
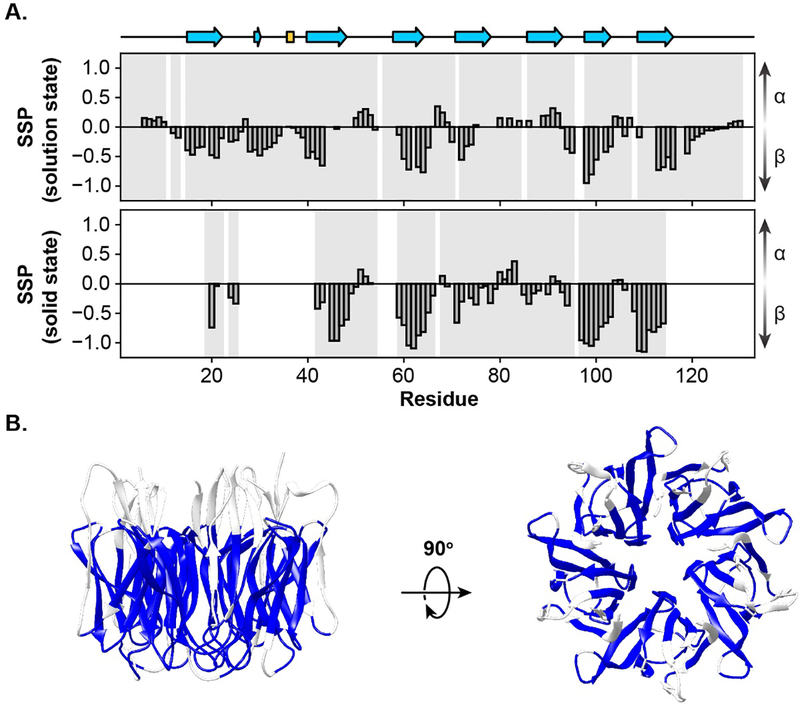
To determine whether the structure of NPM1 is altered upon phase separation with p14ARF, we analyzed secondary structure using the secondary structure propensity (SSP) algorithm. SSP combines different chemical shifts into a single residue-specific secondary structure propensity score, where values of 1 and −1 indicate fully formed α-helical and β-sheet structures, respectively [36] (Fig. 5A). SSP scores were calculated using the Cα and Cβ chemical shifts obtained from CP-MAS ssNMR experiments, revealing several regions of β-sheet structure. These were in excellent agreement with the SSP scores for NPM1 1–130 aa (N130) calculated using solution state NMR chemical shift values and also agreed well with the structure of the OD determined by X-ray crystallography [23]. Secondary chemical shifts (ΔδCα-ΔδCβ) calculated using the random coil chemical shifts from the RefDB [37], also showed good agreement with values obtained from solution state NMR (Fig. S3). Thus, phase separation with p14ARF does not dramatically alter the secondary structure of the NPM1 OD. To highlight these regions, the residues detected in CP-MAS experiments are mapped to the X-ray crystal structure of the NPM1 OD in Fig. 5B.
Fig. 5.
(A, top) The secondary structure calculated from the X-ray crystal structure of the NPM1 OD (PDB: 4N8M) using DSSP and the SSP scores for N130 calculated from solution state Cα and Cβ chemical shifts. (A, bottom) The SSP scores for NPM1 within the NPM1-p14ARF condensed phase calculated from solid state Cα and Cβ chemical shifts. The completeness of assignment is highlighted in light grey. (B) The X-ray crystal structure of the NPM1 OD, where residues that were assigned using CP-MAS ssNMR are highlighted in blue.
4. Conclusion
Here we provide the first observations of NPM1 phase separation with the p14ARF tumor suppressor. Upon mixing equimolar amounts of the two proteins, the system rapidly de-mixes to form a condensed phase containing a roughly 2:1 stoichiometry of NPM1 pentamers to p14ARF molecules. ssNMR analysis demonstrates that within this condensed phase, NPM1 forms a rigid scaffold of immobilized pentamers, while the IDR and CTD experience relative mobility. These results stand in contrast to other recent ssNMR studies of viscoelastic condensed phases, wherein cross-linking interactions induce disorder to order transitions [38, 39], and highlight the dynamic nature of NPM1’s IDR and CTD. These domains act as key mediators of the protein-protein and protein-RNA interactions that drive NPM1 phase separation in vitro and contribute to NPM1 function in the cell [22, 30]. We and others have proposed that NPM1’s nucleolar function is modulated through dynamic rearrangements of its interaction networks [26]. ARF exerts control over ribosome biogenesis in part by reducing NPM1’s molecular mobility in the nucleolus [19]. Our results suggest that within nucleoli, higher order NPM1-ARF assemblies would give rise to a viscoelastic nucleolar matrix. Such a state may enable the sequestration of both components, while the retention of molecular dynamics within the NPM1 IDR and CTD may provide a means for sequestered NPM1-ARF complexes to dynamically respond to changes in the surrounding nucleolar milieu. These results also highlight the utility of ssNMR for probing the structure and dynamics of proteins within condensates. The continued application of these techniques will be helpful for elucidating the molecular determinants of phase separation and the formation of biomolecular condensates.
Supplementary Material
Highlights.
NPM1 and p14ARF undergo heterotypic phase separation
ssNMR analysis was used to probe NPM1 within the condensed phase
NPM1 oligomerization domains form a rigid scaffold amidst mobile C-terminal domains
Acknowledgements
We acknowledge Cheon-Gil Park and Nazia Ahmed for assistance with cloning and protein preparation, respectively. Confocal fluorescence microscopy images were collected at the Cell & Tissue Imaging Center at SJCRH, supported by SJCRH and the NIH (NCI P30 CA021765). We thank Victoria Frohlich, Sharon King, Aaron Pitre, and Jennifer Peters for technical help with microscopy. Solution state NMR spectra were collected at the St. Jude NMR Center. This work was supported by the US NIH (R01 GM115634, R35 GM131891; to R.W. K.); US National Cancer Institute Cancer Center Support Grant P30CA21765 (to SJCRH); and ALSAC (to SJCRH). E.G. was supported by an NIH Research Supplement to Promote Diversity in Health-related Research (R01 GM115634-02S1).
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Special Note
We dedicate this article to Professor James Prestegard on the occasion of his 75th birthday and for the inspiration Jim provided to Dr. Kriwacki while he was a user of Yale’s NMR facility in the 1980s and a graduate student in Chemistry at Yale University in the 1990s. Jim’s inquisitive spirit, keen insights, and patience as a mentor inspired and shaped many generations of NMR scientists, first at Yale and later at the University of Georgia. Thanks for your mentorship, Jim, and best wishes for the future.
Richard Kriwacki.
Data Availability
2D and 3D NMR data is available in Sparky format on the Kriwacki lab website at https://www.stjuderesearch.org/site/lab/kriwacki/.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Banani SF, et al. , Biomolecular condensates: organizers of cellular biochemistry. Nature Reviews Molecular Cell Biology, 2017. 18: p. 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weber SC, Sequence-encoded material properties dictate the structure and function of nuclear bodies. Current Opinion in Cell Biology, 2017. 46: p. 62–71. [DOI] [PubMed] [Google Scholar]
- 3.Ambadipudi S, et al. , Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein Tau. Nature Communications, 2017. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain A and Vale RD, RNA phase transitions in repeat expansion disorders. Nature, 2017. 546: p. 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White MR, et al. , C9orf72 Poly(PR) Dipeptide Repeats Disturb Biomolecular Phase Separation and Disrupt Nucleolar Function. Molecular Cell, 2019. 74: p. 713–728.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong S, et al. , Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science (New York, N.Y.), 2018. 361: p. eaar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel A, et al. , A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell, 2015. 162: p. 1066–1077. [DOI] [PubMed] [Google Scholar]
- 8.Mitrea DM, et al. , Methods for Physical Characterization of Phase Separated Bodies and Membrane-Less Organelles. Journal of molecular biology, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matlahov I and van der Wel PCA, Hidden motions and motion-induced invisibility: Dynamics-based spectral editing in solid-state NMR. Methods, 2018. 148: p. 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feric M, et al. , Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell, 2016. 165: p. 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maggi LB, et al. , ARF tumor suppression in the nucleolus, in Biochimica et Biophysica Acta - Molecular Basis of Disease. 2014. p. 831–839. [DOI] [PubMed] [Google Scholar]
- 12.Lindström MS, et al. , Immunolocalization of Human p14ARF to the Granular Component of the Interphase Nucleolus. Experimental Cell Research, 2000. 256: p. 400–410. [DOI] [PubMed] [Google Scholar]
- 13.Bertwistle D, Sugimoto M, and Sherr CJ, Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Molecular and cellular biology, 2004. 24: p. 985–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuo M-L, et al. , N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes & development, 2004. 18: p. 1862–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korgaonkar C, et al. , Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Molecular and cellular biology, 2005. 25: p. 1258–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apicelli AJ, et al. , A non-tumor suppressor role for basal p19ARF in maintaining nucleolar structure and function. Molecular and cellular biology, 2008. 28: p. 1068–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derenzini M, Montanaro L, and Treré D, What the nucleolus says to a tumour pathologist, in Histopathology. 2009, John Wiley & Sons, Ltd; (10.1111). p. 753–762. [DOI] [PubMed] [Google Scholar]
- 18.Sherr CJ, Divorcing ARF and p53: an unsettled case. Nature Reviews Cancer, 2006. 6: p. 663–673. [DOI] [PubMed] [Google Scholar]
- 19.Brady SN, et al. , ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Molecular and cellular biology, 2004. 24: p. 9327–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Audas, Timothy E, Jacob Mathieu D. and Lee S, Immobilization of Proteins in the Nucleolus by Ribosomal Intergenic Spacer Noncoding RNA. Molecular Cell, 2012. 45: p. 147–157. [DOI] [PubMed] [Google Scholar]
- 21.Kinor N and Shav-Tal Y, The dynamics of the alternatively spliced NOL7 gene products and role in nucleolar architecture. Nucleus, 2011. 2: p. 229–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitrea DM, et al. , Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nature Communications, 2018. 9: p. 842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitrea DM, et al. , Structural polymorphism in the N-terminal oligomerization domain of NPM1. Proceedings of the National Academy of Sciences, 2014. 111: p. 4466–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neidhardt FC, Bloch PL, and Smith DF, Culture Medium for Enterobacteria. Journal of Bacteriology, 1974. 119(3): p. 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thermofisher, Amine-Reactive Probes | 2 Labeling Proteins. Molecular Probes By Life Technologies, 1996. 1996: p. 1–11.
- 26.Ferrolino MC, et al. , Compositional adaptability in NPM1-SURF6 scaffolding networks enabled by dynamic switching of phase separation mechanisms. Nature Communications, 2018. 9: p. 5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindelin J, et al. , Fiji: an open-source platform for biological-image analysis. Nature Methods, 2012. 9: p. 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Day CA, et al. , Analysis of protein and lipid dynamics using confocal fluorescence recovery after photobleaching (FRAP). Current Protocols in Cytometry, 2012: p. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardy EH, Verel R, and Meier BH, Fast MAS Total Through-Bond Correlation Spectroscopy. Journal of Magnetic Resonance, 2001. 148: p. 459–464. [DOI] [PubMed] [Google Scholar]
- 30.Mitrea DM, et al. , Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife, 2016. 5: p. e0126420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berry J, Brangwynne CP, and Haataja M, Physical principles of intracellular organization via active and passive phase transitions, in Reports on Progress in Physics. 2018. [DOI] [PubMed] [Google Scholar]
- 32.Takegoshi K, Nakamura S, and Terao T, 13C–1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chemical Physics Letters, 2001. 344: p. 631–637. [Google Scholar]
- 33.Baldus M, et al. , Cross polarization in the tilted frame: Assignment and spectral simplification in heteronuclear spin systems. Molecular Physics, 1998. 95: p. 1197–1207. [Google Scholar]
- 34.Hong M and Griffin RG, Resonance assignments for solid peptides by dipolar-mediated 13C/15N correlation solid-state NMR [7], in Journal of the American Chemical Society. 1998. p. 7113–7114. [Google Scholar]
- 35.Baldus M and Meier BH, Total Correlation Spectroscopy in the Solid State. The Use of Scalar Couplings to Determine the Through-Bond Connectivity. Journal of Magnetic Resonance, Series A, 1996. 121: p. 65–69. [Google Scholar]
- 36.Marsh JA, et al. , Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Science, 2006. 15: p. 2795–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Neal S, and Wishart DS. RefDB: A database of uniformly referenced protein chemical shifts. in Journal of Biomolecular NMR. 2003. [DOI] [PubMed] [Google Scholar]
- 38.Ader C, et al. , Amyloid-like interactions within nucleoporin FG hydrogels. Proceedings of the National Academy of Sciences, 2010. 107: p. 6281–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostylev MA, et al. , Liquid and Hydrogel Phases of PrPC Linked to Conformation Shifts and Triggered by Alzheimer’s Amyloid-β Oligomers. Molecular Cell, 2018. 0: p. 426–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.