Abstract
Muscle-specific RING-finger proteins (MuRFs) are E3 ubiquitin ligases that play important roles in protein quality control in skeletal and cardiac muscles. Here we characterized murf gene expression and protein localization in zebrafish embryos. We found that the zebrafish genome contains six murf genes, including murf1a, murf1b, murf2a, murf2b, murf3 and a murf2-like gene that are specifically expressed in skeletal and cardiac muscles of zebrafish embryos. To analyze the subcellular localization, we generated transgenic zebrafish models expressing MurF1a-GFP or MuRF2a-GFP fusion proteins. MuRF1a-GFP and MuRF2a-GFP showed distinct patterns of subcellular localization. MuRF1a-GFP displayed a striated pattern of localization in myofibers, whereas MuRF2a-GFP mainly exhibited a random pattern of punctate distribution. The MuRF1a-GFP signal appeared as small dots aligned along the M-lines of the sarcomeres in skeletal myofibers. To determine whether knockdown of smyd1b or hsp90α1 that increased myosin protein degradation could alter murf gene expression or MuRF protein localization, we knocked down smyd1b or hsp90α1 in wild type, Tg(ef1a:MurF1a-GFP) and Tg(ef1a:MuRF2a-GFP) transgenic zebrafish embryos. Knockdown of smyd1b or hsp90α1 had no effect on murf gene expression. However, the sarcomeric distribution of MuRF1a-GFP was abolished in the knockdown embryos. This was accompanied by an increased random punctate distribution of MuRF1a-GFP in muscle cells of zebrafish embryos. Collectively, these studies demonstrate that MuRFs are specifically expressed in developing muscles of zebrafish embryos. The M-line localization MuRF1a is altered by sarcomere disruption in smyd1b or hsp90α1 knockdown embryos.
Keywords: MuRF, sarcomere organization, transgenic zebrafish
Graphical Abstract
1. Introduction
Skeletal muscle cells contain highly organized sarcomeres, the basic contractile units for muscle contraction. Sarcomeres are assembled together in a process called myofibrillogenesis during muscle cell differentiation and maturation, which involves large numbers of structural and regulatory proteins. Sarcomere assembly requires protein quality control that ensures proper protein folding and removal of misfolded proteins in skeletal and cardiac muscle cells (Kim et al., 2008; Willis et al., 2009a; Portbury et al., 2011; Carlisle et al., 2017; Hnia et al., 2019). When folding is not successful, the misfolded proteins are shuttled to the cellular degradation machinery for destruction by the Ubiquitin/Proteasome System (UPS), which is the primary system for muscle protein degradation (Lecker and Goldberg 2002; Lecker et al., 2004; Attaix et al., 2005; Coux et al., 1996; Solomon and Goldberg, 1996; Taillandier et al., 2004; Sandr, 2013; Sandri et al., 2013). UPS is necessary for maintaining muscle remodeling and homeostasis by balancing the protein syntheses and degradation. Malfunction of protein quality control pathways results in myopathies in skeletal and cardiac muscles (Kitajima et al., 2014; Fanzani et al., 2015; Mearini et al., 2008; Patterson et al., 2007; Willis et al., 2008; Willis et al., 2009a; 2009b; 2010).
E3 ubiquitin ligases play a vital role in determining the selectivity and specificity of the UPS. Three muscle RING finger proteins (MuRFs), namely MuRF1, MuRF2, and MuRF3, have been identified as E3 ligases that are specifically expressed in skeletal and cardiac muscles (Spencer et al., 2000; Centner et al., 2001). MuRF E3 ubiquitin ligases are required for skeletal muscle atrophy (Bodine et al., 2001; 2014; Bonaldo and Sandri, 2013), responsible for ubiquitination and degradation of myofibrillar proteins in skeletal and cardiac muscles, such as myosin-binding protein C, myosin light chain, myosin heavy chain, troponin and titin (Cohen et al., 2009; Kedar et al. 2004; McElhinny et al. 2004; Attaix and Baracos 2010; Mearini et al., 2010). It has been shown that mice lacking MuRF1, MuRF2 or MuRF3 were phenotypically normal under unstressed physiological conditions (Fielitz et al., 2007; Bodine et al., 2001; Lange et al., 2005). However, mice lacking both MuRF1 and MuRF2 develope spontaneous cardiac hypertrophy and show a profound loss of type-II fibers (Witt et al., 2008; Moriscot et al., 2010). In addition, mice deficient for MuRF1 and MuRF3 develop skeletal muscle myopathies with myosin heavy chain accumulation, myofiber fragmentation, and impaired muscle performance (Fielitz et al., 2007). Genetic mutations in MuRF1 and MuRF3 have been implicated in causing protein aggregate myopathies in cardiac and skeletal muscles in human populations (Fletta et al., 2011; Chen et al., 2012; Gumucio and Mendias, 2013; Olive et al., 2015; Jokela et al., 2019).
MuRFs show dynamic patterns of gene expression and protein localization under normal and various myopathy conditions. MuRF1 is up-regulated during skeletal muscle atrophy and drug-induced muscle wasting (Bodine et al., 2001; Aniort et al., 2016), and downregulated during cardiac hypertrophy (Maejima et al., 2014). Mice lacking MuRF1 are resistant to skeletal muscle atrophy (Bodine et al., 2001; Baehr et al., 2011). Subcellular localization analysis reveals that MuRF proteins are not only identified within sarcomeric components such as the Z-band and the M-band but also found in the microtubule, cytoplasm, and nuclei (Spencer, et al, 2000; Centner et al., 2001; McElhinny et al., 2002; 2004; Pizon et al, 2002). Interaction of MuRF1 with titin plays an important role in M-line stability (McElhinny et al., 2002), whereas MuRF2 and MuRF3 appear to be important for microtubule stability and myofibrillogenesis during muscle differentiation (Centner et al. 2001; Chang et al. 2002; McElhinny et al., 2004). During atrophic stress, MuRF1 and MuRF2, and likely MuRF3, translocate in the nucleus and appeare to control muscle gene expression via polyubiquitination and degradation of transcription factors in cardiomyocytes and skeletal muscle (Pizon et al., 2002; Lange et al., 2005; Ochala et al., 2011). Thus, members of the MuRF family may have diverse functions based on their patterns of subcellular localization.
Zebrafish have become a model for studying muscle development and sarcomere assembly (Sanger et al., 2009; Keenan and Currie, 2019). Previous studies have demonstrated that molecular chaperon Hsp90α1 and lysine methyltransferase Smyd1 play vital roles in sarcomere assembly in zebrafish embryos (Tan et al., 2006, Etard et al., 2007; Du et al., 2008; Hawkins et al., 2008; Just et al., 2011; Li et al., 2013; Prill et al., 2016; Cai et al., 2019). hsp90α1 and smyd1 are specifically expressed in developing muscles of zebrafish embryos (Tan et al., 2006; Du et al., 2008; Hawkins et al., 2008; Cai et al., 2019). Loss of hsp90α1 and smyd1 result in defective sarcomere organization and increased myosin protein degradation (Bernick et al., 2012; Li et al., 2013; Cai et al., 2019). It has been suggested that MuRF ubiquitin-proteasome system (UPS) are likely to be involved in myosin protein degradation in the hsp90α1 and smyd1 deficient embryos (Du et al., 2008; Li et al., 2013). However, MuRF expression and protein localization are not well understood in zebrafish. In this study, we analyzed the expression profiles of six murf genes in zebrafish during embryogenesis and characterized their spatial patterns of expression by in situ hybridization. In addition, we generated transgenic zebrafish models expressing MuRF1a-GFP or MuRF2a-GFP fusion proteins and analyzed their subcellular localization in myofibers. We found that murf genes were primarily expressed in skeletal and cardiac muscles of zebrafish embryos. MuRF1a-GFP displayed a sarcomeric M-line localization, which was abolished in smyd1b or hsp90α1 knockdown embryos. This was accompanied by an increased punctuated pattern of MuRF1a-GFP distribution in myofibers of zebrafish embryos. Better understanding the MuRFs expression and subcellular localization may provide an insight into MuRF function in fish muscle development and growth.
2. Material and Methods
Zebrafish lines and maintenance
All zebrafish larvae and adult were maintained in a 28 °C recirculating aquatic system with a photoperiod of 14h light and 10h dark, in the Zebrafish Facility of the Aquaculture Research Center, Institute of Marine and Environmental Technology (Baltimore, MD). Zebrafish embryos over one day were anesthetized in 0.6 mM Tricaine (pH 7.0) before fixation in 4% paraformaldehyde (PFA) to ease pain and facilitate animal handling. This study was performed according to the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All protocols used in this study were approved by the Institutional Animal Care and Use Committee of the University of Maryland Baltimore (Protocol Number: 0419010).
MuRF cDNAs cloning and expression analyses by RT-PCR
A BLAST search was conducted in zebrafish using the human MuRF1 cDNA as a query sequence (NM_032588.3). Six murf genes were identified in the zebrafish genome database. They represent MuRF1 (MuRF1a: NM_001002133.1, MuRF1b: NM_201095.1), MuRF2 (MuRF2a: NM_001002358.1, MuRF2b: NM_001039982.1, and MuRF2-like: NM_001003581.1), and MuRF3 (NM_001045025.2). The full-length sequences encoding all six murfs genes were cloned by RT-PCR from WT zebrafish embryos of 24 hpf. The PCR was carried out using the respective gene-specific MuRF-F1/R1 primers (Table 1). The PCR products were cloned into pGEM-T easy vector and confirmed by DNA sequencing. The gene expression was determined by RT-PCR in zebrafish embryos at 0h, 3h, 6h, 12h, 14h, 19h, 22, 26 hour-post-fertilization (hpf), and 2d, 3d, 4d, and 5 days-post-fertilization (dpf). The PCR was carried out using the respective gene-specific MuRF-F2/R2 primers (Table 1). The PCR products were analyzed on 1.5% agarose gel. The gel images were acquired and photographed by E-Gel Imager (Life Technologies).
Table 1.
Sequences of PCR primers and MOs
| Gene name | Primer sequence |
|---|---|
| MuRF1a | F1:ATGGACATCCAAACGGGTCAAATA R1:TTATTCCTCCTCCTCTTCTTCTTC |
| MuRF1b | R1: TTACTCCTCTTCTTGATCTTCAG F1: ATGGACATTCAGCGAACTGCCTC |
| MuRF2a | F1: ATGAGCGTGACTTTGGATTACTG R1: TTACTTTGGCATATTTAACCAAGA |
| MuRF2b | F1: ATGGACAGCTTGGAGAAGCAACT R1: TTAGGATGATGCGGCGGGTCTGCGT |
| MuRF3 | F1: ATGAACTTCACTTTGGGCTTCAAA R1: TTAGCGCGTCCCTGATTCACCAC |
| MuRF2-like | F1: ATGTCTCTTCCACTGGATATACG R1: CTGACCTGCTGAATGTGTCTTCA |
| MuRF1a | F2: CATCTACAAGCAGCAGTTGGA R2: CTCAGACTTCTGGACCTCATA |
| MuRF1b | F2: AACTTGTCGATTTGAAGTGGT R2: TATTGGACTTTCAGTGGTGCT |
| MuRF2a | F2: TATCTTCCTGCAGAACACGAA R2: TCATCATCCTCATCCTCATCA |
| MuRF2b | F2: AGGAATACAGACACTGGAAGA R2: CATCATCATCATCTCTGATGA |
| MuRF3 | F2: CACAACCAACAGTCTCGATTA R2: CTTACTGAGAAGGTTGGTAAA |
| MuRF2-like | F2: CTTACTGAGAAGGTTGGTAAA R2: TCTGCTTCAGAATCTGGTTCA |
| EF-1α | F: GCATACATCAAGAAGATCGGC R: GCAGCCTTCTGTGCAGACTTTG |
| MuRF1a-EGFP | F: GCGGGATCCATGGACATCCAAACGGGTCAAAT R: GCGGGATCCAATTCCTCCTCCTCTTCTTCTTCCT |
| MuRF2a-EGFP | F: GCGGGATCCATGAGCGTGACTTTGGATTACTG R: GCGGGATCCAGCTTTGGCATATTTAACCAAGAGA |
| EGFP | F: GCACAAGCTGGAGTACAACTACAAC R: GTACAGCTCGTCCATGCCGAGAGT |
| Hsp90α1 ATG-MO | 5’-CGACTTCTCAGGCATCTTGCTGTGT-3’ |
| Smyd1b ATG-MO | 5’-ACTTCCACAAACTCCATTCTGGATC-3’ |
| Standard cont-MO | 5’-CCTCTTACCTCAGTTACAATTTATA 3’ |
The sequences of PCR primers used to isolate respective MuRF genes from zebrafish (Danio rerio) and the MOs used to knock down hsp90α1 or smyd1b expression in zebrafish embryos.
Phylogenetic analysis of murf cDNAs
The phylogenetic tree was constructed using Phylogeny.fr (Dereeper et al., 2008 and 2010) on the website, http://www.phylogeny.fr/version2_cgi/index.cgi. The six zebrafish murf genes could be classified into three classes. Protein multiple alignments were analyzed using Multalin at http://multalin.toulouse.inra.fr/multalin//. The characteristic domains of these MuRFs were analyzed with the Scanprosite tool (http://prosite.expasy.org/scanprosite/).
Construction of Tg(ef1a:MuRF1α-GFP) and Tg(ef1α:MuRF2a-GFP) DNA constructs
Tg(ef1α:MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) DNA constructs were generated by cloning the MuRF1a or MuRF2a coding sequence in-frame upstream of the EGFP coding sequence in the pTol2 vector (Urasaki et al., 2006). The full-length MuRF1a or the MuRF2a coding sequence without the stop codon was generated by PCR using Pfu Turbo DNA polymerase (Agilent) using MuRF 1 a-EGFP-F/R or MuRF2a-EGFP-F/R primers (Table 1). A BamHI site was introduced at both 5’ and 3’ ends of the MuRF1a or MuRF2a coding sequences via the respective PCR primers. The PCR products were digested with BamHI and cloned into the BamHI site of T2A200R150G vector (Urasaki et.al, 2006). The DNA sequences at the MuRF1a-GFP or MuRF2a-GFP junctions were confirmed by sequencing.
Microinjection
DNA constructs of Tg(ef1α-MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) were dissolved in sterile water to a final concentration of 50 or 100 ng/μl. For transient expression analysis, approximately 1–2 nl of DNA construct (50–100 pg) was injected into each embryo at one-or two-cell stage. For the generation of transgenic zebrafish lines, the DNA constructs were mixed with Tol2 transposase mRNA (50 ng/μl). Approximately 1–2 nl of mixed DNA construct/Tol2 transposase mRNA was injected into each embryo at one- or two-cell stage. For microinjection of Morpholino antisense oligos (MOs), the MOs were dissolved in 1× Danieau buffer (Nasevicius and Ekker, 2000) to a final concentration of 0.3–0.5 mM. The MOs were injected into Tg(ef1α-MuRF1a-GFP) or Tg(ef1α:MuRF2a-GFP) transgenic embryos at 1–2 cell stages. The sequences of the smyd1b ATG-MO, Hsp90α1-ATG-MO and standard control MO were listed in Table 1.
Transgenic fish screening
The Tg(ef1α-MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) transgenic zebrafish founders (P1) were screened by examining GFP expression in their F1 embryos at 24 hpf under a fluorescence microscope (Axioplan 2, Zeiss). Adult F1 transgenic fish were identified by PCR using genomic DNA from individual caudal fin. The PCR was carried out using the EGFP-F and EGFP-R primers that specifically amplified the EGFP coding sequence (Table 1).
Whole-mount in situ hybridization
The whole-mount in situ hybridization was performed using digoxigenin-labeled RNA antisense probes as previously described (Du and Dienhart, 2001). The RNA antisense probes were synthesized by in vitro transcription using either T7 or Sp6 RNA polymerase and linearized pGEM-T-zf-murf plasmids as templates (Table 2). The in situ images were acquired using a Leica dissecting microscope MI12 equipped with a cool CCD digital camera (DX8, Olympus).
Table 2.
DNA plasmids for antisense RNA probes
| DNA plasmids | RNA polymerase | Restriction Enzymes |
|---|---|---|
| Murf1a | T7 | NarI |
| Murf1b | SP6 | HindIII |
| Murf2a | T7 | StuI |
| Murf2b | SP6 | NarI |
| Murf3 | SP6 | EcoRV |
| Murf2-like | SP6 | StuI |
The DNA plasmids used to generate the anti-sense probes for in situ hybridization in zebrafish (Danio rerio) embryos. Each plasmid was digested with a specific restriction enzyme and used as template for RNA synthesis using respective T7 or Sp6 RNA polymerase.
Whole-mount nuclear and immunostaining
Immunostaining was carried out on whole-mount zebrafish embryos as previously described (Cai et al., 2019). After fixed in 4% paraformaldehyde in PBS for 1 hour at room temperature, the embryos were washed in PBST for 3×15 min and then digested with 1 mg/mL collagenase (28 hpf for 3.5 min, 30 hpf for 6 min) to increase permeability. Immunostaining was performed with anti-α-actinin (clone EA-53, #A7811, Sigma) and anti-myomesin (mMaC myomesin B4, DSHB) primary antibodies. The secondary antibody was TRITC-conjugated anti-mouse IgG secondary antibody (T7657, Sigma). Hoechst 33258 (Sigma B2883, 1ng/ml in PBST) was used to stain nuclei in fish embryos. After staining, the trunk region was dissected from each embryo and mounted in Vectashield (Vector lab, H-1000) for confocal microscopy (Leica SP8).
3. Results
1. Identification and characterization of murf genes in zebrafish
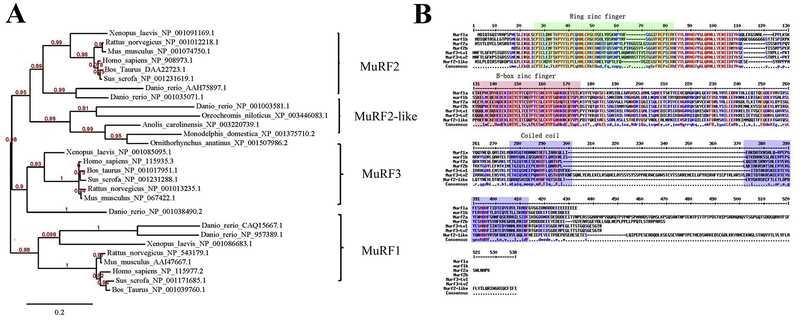
Six murf genes were identified in the zebrafish genome sequence database. The six zebrafish murf genes could be classified into three classes, murf1 (murf1a, murf1b), murf2 (murf2a, murf2b and murf2-like), and murf3 by phylogenetic analysis. The murf1a, murf1b, murf2a, murf2b, murf2-like, and murf3 genes are located on chromosome 6, 9, 2, 24, 20 and 23, respectively. Sequence analysis revealed that the murf1a or murf1b genes had no introns. In contrast, murf2a, 2b, 3, 2-like genes contained 9, 8, 7, 9 exons, respectively. The full-length cDNAs of murf1a, murf1b, murf2a, murf2b, murf3, and murf2-like are predicted to encode MuRF proteins of 345, 348, 443, 366, 359 and 429 aa, respectively. The MuRF proteins show modest sequence identities with their orthologues from other vertebrates. For example, the protein sequence of zebrafish MuRF1a (CAQ15667.1) show 53% and 52% identities with chicken (XP_424369.3) and human MuRF1 orthologues (NP_115977.2), respectively.
All zebrafish MuRF proteins contain three characteristic structural and functional domains, including the ZF_RING_2 domain, ZF_BBOX domain and COS domain (Fig. 1B). The ZF_RING_2 domain, also known as the RING finger domain, contained a Cys3HisCys4 amino acid motif that binds two zinc cations (Borden and Freemont, 1996). The ZF_RING_2 domain contain approximately 40–60 amino acids involved in substrate binding. The B-box-type zinc finger domain is a short protein domain of approximately 40 amino acid residues in length. The COS, known as the C-terminal subgroup one signature domain, is predicted to form two coiled coils (Short and Cox, 2006). The three conserved structural domains make up the tripartite motif, which is essential for MuRF E3 ligase activity.
Figure 1. Phylogenetic analysis of vertebrate MuRFs and protein sequence alignment of MuRFs in zebrafish, Danio rerio..
A. A phylogenetic tree was developed based on MuRF protein sequences from fish to mammals. The MuRF proteins could be divided into three branches. The first branch is made up of the MuRF2 and MuRF2-like subgroup. The other two branches are MuRF1 and MuRF3, respectively.
B. The protein sequence alignment of six MuRFs in zebrafish. MuRF1a, CAQ15667.1; MuRF1b, NP_957389.1; MuRF2a, AAH75897.1; MuRF2b NP_001035071.1; MuRF2-like NP_001003581.1. MuRF3 has two isoforms from alternative splicing named MuRF3_tv1, NP_001038490.2 and MuRF3_tv2 CAI20953.1. The three conserved structural domains are indicated as ring zinc finger domain, B-box zinc finger domain and the coiled coil domain, respectively.
2. Characterization of murf gene expression in zebrafish embryos
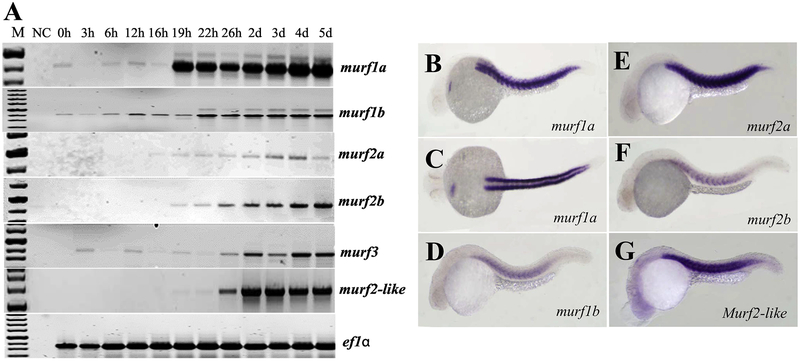
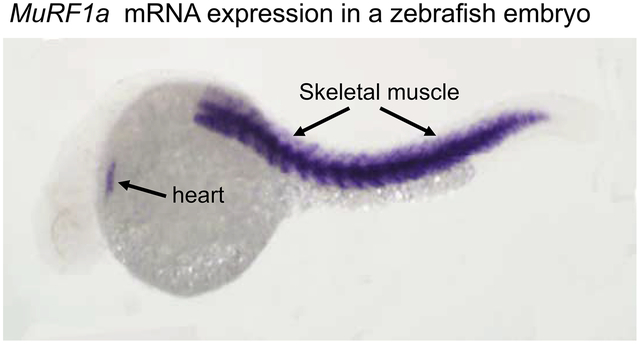
The temporal expression of murf genes in zebrafish embryos was determined by RT-PCR (Fig. 2A). Members of the MuRF family exhibited different patterns of temporal expression, with a strong expression of murf1a starting around the time of myogenesis at 19 hpf. Nevertheless, all murf genes showed increased levels of expression during embryonic development and myogenesis (Fig. 2A). To characterize their spatial pattern of mRNA expression, we performed whole-mount in situ hybridization (Fig. 2B–G). The results showed that five murf genes were expressed in skeletal muscles of zebrafish embryos. In addition, murf1a expression was also detected in the heart primordium of zebrafish embryos at 24 hpf (Fig. 2B, C). Collectively, these data indicate that all five murf genes have an overlapping pattern of expression in skeletal muscles, except murf1a which also showed cardiac muscle expression.
Figure 2. The temporal and spatial patterns of murf gene expression in zebrafish (Danio rerio) embryos.
A. RT-PCR showed the temporal expression of 6 zebrafish murf genes during embryonic development at various stages from newly fertilized eggs to 5 day old larvae. EF1α was included as an internal reference control.
B-G In situ hybridization showed the restricted pattern of expression for each member of the 6 murf genes in zebrafish embryos at 24 hpf. All 6 murf genes were expressed in skeletal muscles of the zebrafish embryos. In addition, murf1a also showed a cardiac muscle expression (Fig. B, C).
3. Characterization of MuRF1a and MuRF2a subcellular localization in muscle fibers of zebrafish embryos
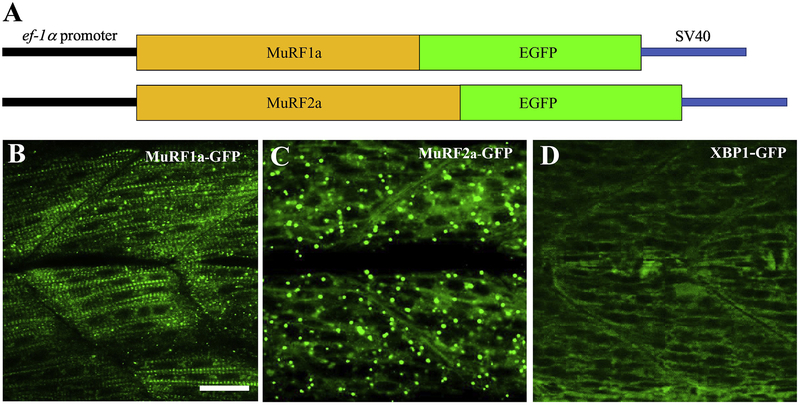
Previous studies have shown that MuRF1 and MuRF2 were localized at the peripheral region of M-lines in chick, mouse and rat cardiac myocytes and skeletal myocytes (Centner et al., 2001; Pizon et al., 2002; McElhinny et al., 2002; 2004). The subcellular localization of MuRF1a and MuRF2a in zebrafish skeletal muscles is not known. It is not clear whether they have similar or distinct patterns of subcellular localization. To analyze the subcellular localization of MuRF1a and MuRF2a in myofibers of zebrafish embryos, we generated Tg(ef1α:MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) transgenic zebrafish models that express the EGFP tagged MuRF1a or MuRF2a fusion protein driven by the elongation factor 1α (EF-1α) gene promoter (Fig. 3A). Three Tg(ef1α:MuRF1a-GFP) and two Tg(ef1α:MuRF2a-GFP) transgenic lines were generated.
Figure 3. Generation of Tg(ef1α:MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) transgenic zebrafish, Danio rerio.
A: Diagram of the Tg(ef1α:MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) transgenes. The full-length coding sequences of zebrafish murf1a or murf2a were cloned in frame upstream of the EGFP coding sequence in the pT2AL200R150G vector. The pT2AL200R150G vector contained an elongation factor-1 alpha (EF-1α) gene promoter. Transgenic zebrafish were generated using these two transgenes.
B-D: Subcellular localization of MuRF1a-GFP (B), MuRF1a-GFP (D), or XBP1-GFP fusion protein in myofibers of the respective transgenic zebrafish embryos at 28 hpf. Scale bar: 30 μm.
The protein localization of MuRF1a-GFP and MuRF2a-GFP was analyzed in the respective transgenic zebrafish embryos. MuRF1a-GFP showed a clear striated pattern of distribution in myofibers of zebrafish embryos at 28 hpf (Fig. 3B). In contrast, MuRF2a-EGFP fusion protein was mainly detected in a random punctate pattern (Fig. 3C). The distinct patterns of MuRF1a and MuRF2a localization was reproducible in multiple Tg(ef1α:MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) transgenic lines. Tg(ef1α:XBP1-GFP) transgenic fish embryos expressing an XBP1-GFP fusion protein showed no such striated or punctate patterns of localization (Fig. 3D) (Li et al., 2015), suggesting that the localization was not caused by the presence of GFP tag.
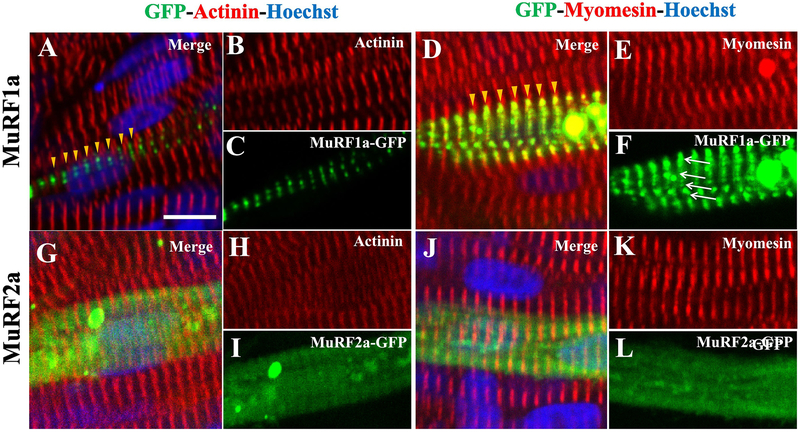
To define the striated distribution of MuRF1a-GFP with respective to the sarcomere structure, we performed immunostaining with anti-α-Actinin and anti-Myomesin antibodies that specifically label the Z-lines and M-lines, respectively. As expected, zebrafish embryos injected with the Tg(ef1α:MuRF 1a-GFP) or the Tg(ef1α:MuRF2a-GFP) transgene showed a mosaic pattern of MuRF1a-GFP and MuRF2a-GFP expression. Immunostaining of the injected embryos showed that MuRF1a-GFP was localized between Z-lines labeled with the anti-α-Actinin antibody (Fig. 4A, C). Immunostaining with anti-Myomesin antibody revealed a co-localization with MuRF1a-GFP at the M-line (Fig. 4D, F). Intriguingly, MuRF1a-GFP appeared as small dots aligned along the M-lines (Fig. 4F). In contrast, Murf2a-GFP displayed a diffused pattern of distribution with no clear sarcomeric localization (Fig. 4G–L). Collectively, these data indicate that MuRF1a-GFP and MuRF2a-GFP have different subcellular localization in myofibers of zebrafish embryos.
Figure 4. Subcellular localization of MuRF1a-GFP and MuRF2a-GFP in myofibers of zebrafish (Danio rerio) embryos.
DNA construct expressing the MuRF1a-GFP or MuRF2a-GFP fusion protein was microinjected into zebrafish embryos. The injected embryos were stained with anti-α-Actinin or anti-Myomesin antibodies that specifically labeled Z-line and M-line, respectively. The nuclei were labeled with Hoechst 33258 (blue).
A-C: Confocal analysis of MuRF1a-GFP and α-Actinin localization in myofibers of zebrafish embryos at 30 hpf. The yellow arrowheads in (A) indicate the striated distribution of MuRF1a-GFP (green) between Z-lines (red). Scale bar: 8 μm.
D-F: Confocal analysis of MuRF1a-GFP and Myomesin localization in myofibers of zebrafish embryos at 30 hpf. The yellow arrowheads in (D) indicate the co-localization of MuRF1a-GFP (green) with M-lines (red). The white arrows in (F) indicate MuRF1a-GFP positive dots aligned along the M-lines (Fig. 4F).
G-L: Confocal analysis of MuRF2a-GFP localization in myofibers stained with anti-α-Actinin (G, H) or anti-Myomesin (J, K) antibodies.
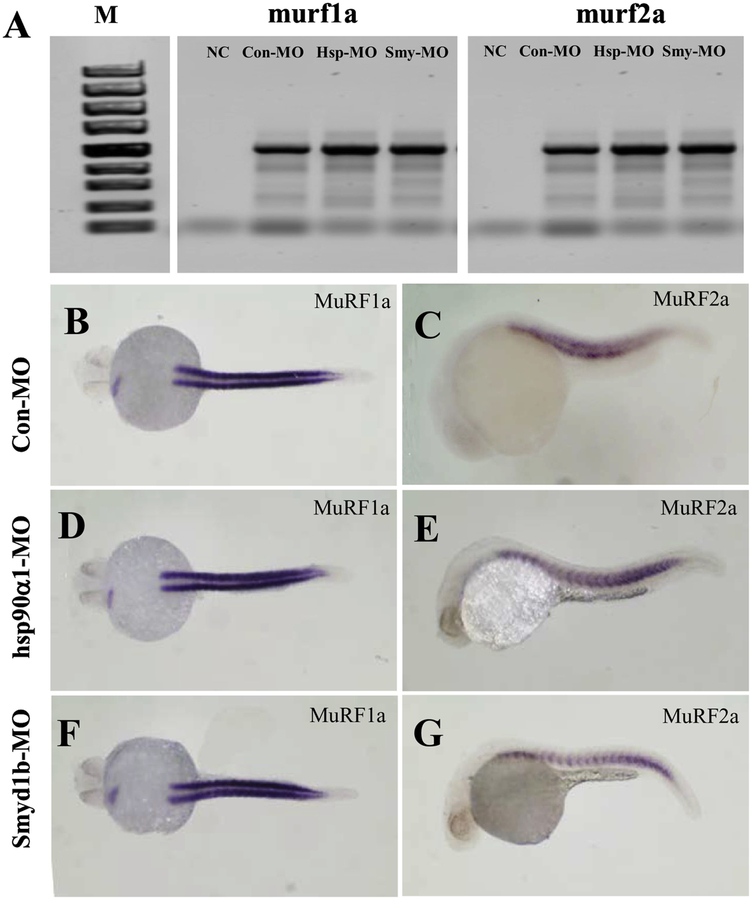
4. Knockdown of hsp90〈1 or smyd1b had no effect on murf gene expression but altered MuRF1a-GFP or MuRF2a-GFP protein localization
It has been reported that loss of hsp90α1 and smyd1 resulted in defective sarcomere organization and increased myosin protein degradation (Bernick et al., 2012. Li et al., 2013). To determine whether knockdown of smyd1b or hsp90α1 altered MuRF expression, we analyzed the murf1a and murf2a mRNA expression in smyd1b or hsp90α1 knockdown embryos by RT-PCR and whole-mount in situ hybridization (Fig. 5). The data showed that knockdown of hsp90〈1 or smyd1b had little or no effect on the mRNA levels of murf1a and murf2a transcripts in zebrafish embryos (Fig. 5).
Figure 5. The effect of hsp90α1 and smyd1b knockdown on murf1a and murf2a gene expression in zebrafish (Danio rerio) embryos.
The hsp90α1 and smyd1b genes were knocked down in zebrafish embryos by gene specific MO injection. murf1a and murf2a gene expression was analyzed by RT-PCR (A) and whole mount in situ hybridization (B-G). A. RT-PCR analysis showed the levels of murf1a and murf2a gene expression in knockdown and control embryos. B-G: Whole mount in situ hybridization showed the murf1a and murf2a transcript signals in control (B and C), hsp90α1 (D and E) or smyd1b (F and G) knockdown embryos at 24 hpf, respectively.
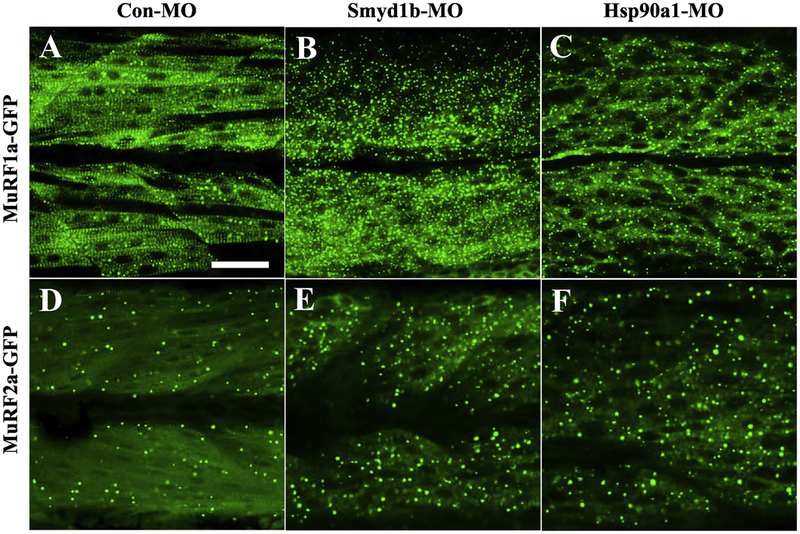
To determine whether knockdown of smyd1b or hsp90〈1 altered MuRF1a-GFP and MuRF2a-GFP protein localization in myofibers of zebrafish embryos, we injected Smyd1b-MO or Hsp90α1-MQ into Tg(ef1α:MuRF1a-GFP) or Tg(Tol2-MuRF2a-GFP) transgenic zebrafish embryos and analyzed their subcellular distribution. The data showed that knockdown of smyd1b or hsp90α1 abolished the M-line localization of MuRF1a-GFP in myofibers of zebrafish embryos (Fig. 6B, C). Intriguingly, this was accompanied by an increased random punctate distribution of MuRF1a-GFP and MuRF2a-GFP fusion proteins in myofibers of the knockdown embryos (Fig. 6B–C, E-F). Collectively, these results revealed that knockdown of hsp90α1 or smyd1b had no effect on murf1a and murf2a mRNA expression, while it dramatically reduced the striated subcellular distribution on MuRF1a-GFP fusion proteins in myofibers.
Figure. 6. The effect of hsp90α1 or smyd1b knockdown on the subcellular localization of MuRF1a-GFP and MuRF2a-GFP in skeletal muscle fibers of zebrafish (Danio rerio) embryos.
hsp90α1 and smyd1b expression was knocked down in Tg(ef1a:MuRF1a-GFP) and Tg(ef1a:MuRF2a-GFP) transgenic fish embryos. The MuRF1a-GFP and MuRF2a-GFP subcellular localization was determined by confocal microscopy in the hsp90α1 and smyd1b knockdown and control embryos.
A-C: MuRF1a-GFP localization in myofibers of zebrafish embryos injected with control-MO (A), Smyd1b-MO (B) or Hsp90α1-M0 (C) at 30 hpf.
D-F: A-C: MuRF2a-GFP localization in myofibers of zebrafish embryos injected with control-MO (D), Smyd1b-MO (E) or Hsp90α1-MQ (F) at 30 hpf.
Scale bar: 50 μm.
4. Discussion
In this study, we characterized the expression and protein localization of MuRFs during muscle development in zebrafish embryos. We showed that the zebrafish genome contains six murf genes that are expressed specifically in skeletal and cardiac muscles of zebrafish embryos. By generating transgenic zebrafish models expressing MurF1a-GFP or MuRF2a-GFP fusion proteins, we were able to study the subcellular localization of MuRF1a and MuRF2a. Our data showed that MuRF1a-GFP or MuRF2a-GFP had distinct patterns of subcellular localization in skeletal muscles. MuRF1a-GFP displayed a sarcomeric pattern of distribution that co-localized with the M-line, whereas MuRF2a mainly appeared in a random punctate pattern. Knockdown of smyd1b or hsp90α1 that induced myosin degradation had no effect on murf gene expression, but dramatically reduced the sarcomeric localization of MuRF1a-GFP and increased the random punctate formation in myofibers of zebrafish embryos. The Tg(ef1α:MurF1a-GFP) and Tg(ef1α:MuRF2a-GFP) transgenic fish provide useful models for studying MuRF action in muscle cell differentiation.
Murf gene structure and expression
Our analyses identified six murf genes in the zebrafish genome, which doubles the number of murf genes (murf1, murf2 and murf3) in mice and human genomes. Our results are in agreement with a previous report (Macqueen et al., 2014), and consistent with the theory of an extra teleost-specific whole-genome duplication in during evolution (Jaillon et al., 2004; Sato and Nishida, 2010). Sequence analysis indicates that murf1a and murf1b are the orthologues of murf1 genes in mammals. However, the gene structure of zebrafish muf1a and murf1b differs significantly from the murf1 genes in mice and humans. The zebrafish muf1a and murf1b genes contain no intron sequence, whereas the mice and human murf1 genes contain 8 introns. Interestingly, all identified teleost murf1 genes are intronless, whereas murf1 genes from spotted gar, a ray-finned fish that split from teleost over 350 million years ago contain the same genomic organization as murf1 in lobe-finned fish and tetrapods (Near et al., 2012). It has been suggested that the intronless gene might be generated by retrotransposition specifically within the teleost lineage, sometime before the common teleost-specific genome duplication occurred 320–350 million years ago (Glasauer and Neuhauss, 2014; Macqueen et al., 2014). Remarkably, the zebrafish murf1a and murf1b show a similar pattern of skeletal and cardiac muscle-specific expression as observed in mice and other vertebrate embryos, suggesting that the murf1 retrogene likely replaced the ancestral murf1 gene at this same locus and thus retaining all the regulatory elements for muscle-specific expression.
In addition to the typical murf1, murf2a and murf3 subfamilies, the zebrafish genome contains a novel murf2-like gene that cannot be found in mammals. Macqueen and colleagues named this novel murf gene as murf4 because it could not be grouped into murf1, murf2 or murf3 subgroups (Macqueen et al., 2014). Our phylogenetic analysis suggest that this novel murf4 gene is more closely related to murf2, thus named it muf2-like gene. The murf2-like/murf4 gene has 9 exons and is expressed in skeletal muscles of zebrafish embryos although its function has yet to be determined.
MuRF-GFP transgenic zebrafish models
Members of the MuRF protein family show dynamic patterns of subcellular localization in skeletal and cardiac muscle cells, ranging from the nucleus to cytoplasm and sarcomeres (Spencer et al., 2000; Centner et al. 2001; Dai and Liew, 2001; McElhinny et al., 2002, 2004). The dynamic localization suggests that MuRFs may have distinct functions with respect to their subcellular localization. By using the unique Tg(ef1α:MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) transgenic zebrafish models generated in this study, we analyzed the MuRF1a-GFP and MuRF2a-GFP protein localization in myofibers of zebrafish embryos. Our data showed that MuRF1a-GFP co-localized primarily with the M-lines in skeletal myofibers. This is consistent with previous findings in chick, mouse and rat cardiac myocytes and skeletal myocytes (McElhinny et al., 2002, 2004).
Tg(ef1α:MuRF1a-GFP) and Tg(ef1α:MuRF2a-GFP) transgenic zebrafish showed normal muscle development and growth. This is in contrast to the transgenic zebrafish model generated using the Tg(myl7:MuRF1a-IRES-GFP) transgene that expressed MuRF1a under the control of a myocardial-specific myl7 promoter derived from the myosin regulatory light chain 7 (Shimizu et al., 2017). Shimizu and colleagues reported that upregulation of MuRF1a expression in cardiac myocytes led to myofibril disarray in zebrafish heart muscles, leading to dilated heart with severe edema in the transgenic zebrafish embryos (Shimizu et al., 2017). The reason for the phenotypic discrepancy is not clear. Given that our transgenic models were generated by different gene constructs, the phenotypic discrepancy could be caused by different levels of ectopic murf1a gene expression in the heart of different transgenic models used in these two studies. Shimizu and colleagues used a strong cardiac muscle-specific myl7 promoter to drive the MuRF1a expression, whereas we used a moderate ubiquitous ef1a promoter to drive MuRF1a expression. Consistent with the idea that overexpression of MuRF1 is detrimental to sarcomere organization, it has been reported that overexpression of MuRF1 disrupted the integrity of titin’s M-line region and the organization of thick filament components in chick cardiac myocytes (McElhinny et al., 2002).
Dynamic MuRF protein localization
We showed in this study that the sarcomeric localization of MuRF1a was abolished in smyd1b or hsp90α1 knockdown zebrafish embryos. Smyd1b or Hsp90α1 are vital regulators of sarcomere assembly. We and others have shown that loss of Smyd1b or Hsp90α1 increased myosin protein degradation leading to sarcomere disruption in skeletal and cardiac muscles of zebrafish embryos (Tan et al., 2006; Du et al., 2008; Hawkins et al., 2008; Just et al., 20121; Li et al., 2013; Prill et al., 2016; Cai et al., 2019). Given that MuRFs directly mediate muscle protein ubiquitination and degradation, we tested whether murf gene expression could be upregulated. Our data showed that knockdown of smyd1b or hsp90α1 had no effect on murf1a and murf2a mRNA expression. However, the subcellular localization of MuRF1a-GFP was significantly altered. It appeared that the sarcomeric localization of MuRF1a-GFP disappeared and replaced with a dramatic increase of random punctate distribution of MuRF1a-GFP in defective myofibers of the smyd1b or hsp90α1 knockdown embryos.
The molecular mechanism underlying the increased MuRF1a-GFP punctate distribution is not clear. The punctate MuRF1a-GFP signals were 1–2 micron in size that was scattered in the skeletal muscle fibers of smyd1b or hsp90α1 knockdown embryos. Similar punctate distribution has been observed in chick and rat cardiac myocytes with GFP-MuRF1 or GFP-MuRF2 overexpression (McElhinny et al., 2002). It has been suggested that these punctate represent aggregates formed via the homo-oligomerization of MuRF proteins (Spencer et al., 2000; Centner et al., 2001). Interestingly, if GFP is only fused with the MuRF RING or tail domain, no aggregates were observed in the cytoplasm or assembled in the M-line region. In contrast, when the COS domain of MuRF1 was fused with GFP, majority of transfected cardiac myocytes were observed aggregated formed in the cytoplasm (McElhinny et al., 2002). The regulation of dynamic localization and the functional significance of forming the punctate aggregates are not clear. A recent study indicates that MuRF1 cellular localization is regulated by SUMO1 mediated post-translational modification (Heras et al., 2019). MuRF1 SUMOylation is essential for its nuclear translocation. It remains to be determined whether a post-translational modification is involved in regulating the sarcomeric distributions of MuRF proteins in myofibers of zebrafish.
Acknowledgments
We thank Amaya Simpson for proofreading the manuscript. This research was supported by grant funding from the National Institute of Health (R01AR072703 to SD). BL was supported by a visiting scholarship from the Shanxi Scholarship Council of China (SSCC). SL was supported by a student fellowship from Chinese Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have no conflicts of interest to disclose.
Declaration of interest: none
References
- Aniort J, Polge C, Claustre A, Combaret L, Béchet D, Attaix D, Heng AE, Taillandier D, 2016. Upregulation of MuRF1 and MAFbx participates to muscle wasting upon gentamicin-induced acute kidney injury. Int. J. Biochem. Cell Biol 79, 505–516. 10.1016/j.biocel.2016.04.006 [DOI] [PubMed] [Google Scholar]
- Attaix D, Baracos VE, 2010. MAFbx/atrogin-1 expression is a poor index of muscle proteolysis. Curr. Opin. Clin. Nutr. Metab. Care 13, 223–224. 10.1097/MCO.0b013e328338b9a6 [DOI] [PubMed] [Google Scholar]
- Attaix D, Ventadour S, Codran A, Béchet D, Taillandier D, Combaret L, 2005. The ubiquitin-proteasome system and skeletal muscle wasting. Essays Biochem 41,173–186. 10.1042/bse0410173 [DOI] [PubMed] [Google Scholar]
- Baehr LM, Furlow JD, Bodine SC, 2011. Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids. J. Physiol 589(19), 4759–4776. 10.1113/jphysiol.2011.212845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernick EP, Zhang PJ, Du S, 2010. Knockdown and overexpression of Unc-45b result in defective myofibril organization in skeletal muscles of zebrafish embryos. BMC Cell Biol 17, 11–70. 10.1186/1471-2121-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ, 2001. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294(5547), 1704–1708. 10.1126/science.1065874 [DOI] [PubMed] [Google Scholar]
- Bodine SC, Baehr LM, 2014. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol Metab 307(6), E469–E484. 10.1152/ajpendo.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaldo P, Sandri M, 2013. Cellular and molecular mechanisms of muscle atrophy. Dis. Model Mech 6(1), 25–39. 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL, Freemont PS, 1996. The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol 6(3), 395–401. [DOI] [PubMed] [Google Scholar]
- Cai M, Han L, Liu L, He F, Chu W, Zhang J, Tian Z, Du SJ, 2019. Defective sarcomere assembly in smyd1a and smyd1b zebrafish mutants. FASEB. J 33(5), 6209–6225. 10.1096/fj.201801578R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle C, Prill K, Pilgrim D, 2017. Chaperones and the Proteasome System: Regulating the Construction and Demolition of Striated Muscle. Int. J. Mol. Sci 19(1), E32 10.3390/ijms19010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, Sorimachi H, Labeit S, 2001. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J. Mol. Biol 306(4), 717–726. 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- Chang W, Webster DR, Salam AA, Gruber D, Prasad A, Eiserich JP, Bulinski JC, 2002. Alteration of the C-terminal amino acid of tubulin specifically inhibits myogenic differentiation. J. Biol. Chem 277, 30690–30698. 10.1074/jbc.M204930200. [DOI] [PubMed] [Google Scholar]
- Chen SN, Czernuszewicz G, Tan Y, Lombardi R, Jin J, Willerson JT, Marian AJ, 2012. Human molecular genetic and functional studies identify TRIM63, encoding Muscle RING Finger Protein 1, as a novel gene for human hypertrophic cardiomyopathy. Circ. Res 111(7), 907–919. 10.1161/CIRCRESAHA.112.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Brault JJ, Gygi SP, Glass DJ, Valenzuela DM, Gartner C, Latres E, Goldberg AL, 2009. During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J. Cell Biol 185(6), 1083–1095. 10.1083/jcb.200901052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coux O, Tanaka K, Goldberg AL, 1996. Strucure and function of the 20S and 26S proteasomes. Annu. Rev. Biochem 65, 801–847. 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- Dai KS, Liew CC, 2001A novel human striated muscle RING zinc finger protein, SMRZ, interacts with SMT3b via its RING domain. J. Biol. Chem 276(26), 23992–23999. 10.1074/jbc.M011208200. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O, 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36, W465–469. 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G, 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol 10, 8 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du SJ, Dienhart M, 2001. Zebrafish tiggy-winkle hedgehog promoter directs notochord and floor plate green fluorescence protein expression in transgenic zebrafish embryos. Dev. Dyn 222(4), 655–666. 10.1002/dvdy.1219. [DOI] [PubMed] [Google Scholar]
- Du SJ, Li H, Bian Y, Zhong Y, 2008. Heat-shock protein 90alphal is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc. Natl. Acad. Sci. USA, 105, 554–559. 10.1073/pnas.0707330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etard C, Behra M, Fischer N, Hutcheson D, Geisler R, Strähle U, 2007. The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90 during myofibrillogenesis. Dev Biol 308, 133–143. [DOI] [PubMed] [Google Scholar]
- Fanzani A, Conraads VM, Penna F, Martinet W, 2012. Molecular and cellular mechanisms of skeletal muscle atrophy: an update. J. Cachexia Sarcopenia Muscle 3(3), 163–179. 10.1007/s13539-012-0074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, Richardson JA, Bassel-Duby R, Olson EN, 2007a. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J. Clin. Invest 117, 2486–2495. 10.1172/JCI32827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foletta VC, White LJ, Larsen AE, Léger B, Russell AP, 2011. The role and regulation of MAFbx/atrogin-1 and MuRF1 in skeletal muscle atrophy. Pflugers Arch 461(3), 325–335. 10.1007/s00424-010-0919-9. [DOI] [PubMed] [Google Scholar]
- Glasauer SM, Neuhauss SC, 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genomics 289(6), 1045–1060. 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- Gumucio JP, Mendias CL, 2013. Atrogin-1, MuRF-1, and sarcopenia. Endocrine 43(1), 12–21. 10.1007/s12020-012-9751-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras G, Namuduri AV, Traini L, Shevchenko G, Falk A, Bergström LS, Jia M, Tian G, Gastaldello S, 2019. Muscle RING-finger protein-1 (MuRF1) functions and cellular localization are regulated by SUMO1 post-translational modification. J Mol Cell Biol 11(5), 356–370. 10.1093/jmcb/mjy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnia K, Clausen T, Moog-Lutz C, 2019. Shaping Striated Muscles with Ubiquitin Proteasome System in Health and Disease. Trends Mol. Med pii: S1471–4914(19)30125–X 10.1016/j.molmed.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Hawkins TA, Haramis AP, Etard C, Prodromou C, Vaughan CK, Ashworth R, Ray S, Behra M, Holder N, Talbot WS, Pearl LH, Strähle U, Wilson SW, 2008. The ATPase-dependent chaperoning activity of Hsp90α regulates thick filament formation and integration during skeletal muscle myofibrillogenesis. Development 135(6), 1147–1156. doi: 10.1242/dev.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biémont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigó R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quétier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Crollius HR, 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431(7011), 946–957. [DOI] [PubMed] [Google Scholar]
- Jokela M, Baumann P, Huovinen S, Penttilä S, Udd B, 2019. Homozygous Nonsense Mutation p.Q274X in TRIM63 (MuRF1) in a Patient with Mild Skeletal Myopathy and Cardiac Hypertrophy. J. Neuromuscul Dis 6(1), 143–146. 10.3233/JND-180350. [DOI] [PubMed] [Google Scholar]
- Just S, Meder B, Berger IM, Etard C, Trano N, Patzel E, Hassel D, Marquart S, Dahme T, Vogel B, Fishman MC, Katus HA, Strähle U, Rottbauer W, 2011. The myosin-interacting protein SMYD1 is essential for sarcomere organization. J Cell Sci 124(Pt 18):3127–36. doi: 10.1242/jcs.084772. [DOI] [PubMed] [Google Scholar]
- Kedar V, McDonough H, Arya R, Li HH, Rockman HA, Patterson C, 2004. Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc. Natl. Acad. Sci. USA 101, 18135–18140. 10.1073/pnas.0404341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan SR, Currie PD, 2019. The Developmental Phases of Zebrafish Myogenesis. J. Dev. Biol 7(2). pii: E12 10.3390/jdb7020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Löwe T, Hoppe T, 2008. Protein quality control gets muscle into shape. Trends Cell. Biol 18(6), 264–272. 10.1016/j.tcb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Kitajima Y, Tashiro Y, Suzuki N, Warita H, Kato M, Tateyama M, Ando R, Izumi R, Yamazaki M, Abe M, Sakimura K, Ito H, Urushitani M, Nagatomi R, Takahashi R, Aoki M, 2014. Proteasome dysfunction induces muscle growth defects and protein aggregation. J. Cell Sci 127(24), 5204–5217. 10.1242/jcs.150961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edstrom L, Ehler E, Udd B, Gautel M, 2005. The kinase domain of titin controls muscle gene expression and protein turnover. Science 308, 1599–1603. 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL, 2002. Slowing muscle atrophy: putting the brakes on protein breakdown. J. Physiol. Lond 545–729. 10.1113/jphysiol.2002.030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, Price SR, Mitch WE, Goldberg AL, 2004. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J 18, 39–51. 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Li H, Zhong Y, Wang Z, Gao J, Xu J, Chu W, Zhang J, Fang S, Du SJ, 2013. Smyd1b is required for skeletal and cardiac muscle function in zebrafish. Mol. Biol. Cell 24(22), 3511–3521. 10.1091/mbc.E13-06-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Z,, Gao LY, Colorni A,, Ucko M,, Fang S, Du SJ, 2015. A transgenic zebrafish model for monitoring xbp1 splicing and endoplasmic reticulum stress in vivo. Mech Dev 137, 33–44. https://doi:10.1016/j.mod.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Macqueen DJ, Fuentes EN, Valdés JA, Molina A, Martin SA, 2014. The vertebrate muscle-specific RING finger protein family includes MuRF4--a novel, conserved E3-ubiquitin ligase. FEBS Lett 588(23):4390–7. https://doi.org10.1016/j.febslet.2014.10.008. [DOI] [PubMed] [Google Scholar]
- Maejima Y, Usui S, Zhai P, Takamura M, Kaneko S, Zablocki D, Yokota M, Isobe M, Sadoshima J, 2014. Muscle-specific RING finger 1 negatively regulates pathological cardiac hypertrophy through downregulation of calcineurin A. Circ. Heart Fail 7(3), 479–490. 10.1161/CIRCHEARTFAILURE.113.000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinny AS, Kakinuma K, Sorimachi H, Labeit S, Gregorio CC, 2002. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. J. Cell Biol 157(1), 125–136. 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinny AS, Perry CN, Witt CC, Labeit S, Gregorio CC, 2004. Muscle-specific RING finger-2 (MURF-2) is important for microtubule, intermediate filament and sarcomeric M-line maintenance in striated muscle development. J. Cell Sci 117, 3175–3188. 10.1242/jcs.01158 [DOI] [PubMed] [Google Scholar]
- Mearini G, Schlossarek S, Willis MS, Carrier L, 2008. The ubiquitin-proteasome system in cardiac dysfunction. Biochim. Biophys. Acta 1782(12), 749–763. 10.1016/j.bbadis.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Mearini G, Gedicke C, Schlossarek S, Witt CC, Krämer E, Cao P, Gomes MD, Lecker SH, Labeit S, Willis MS, Eschenhagen T, Carrier L, 2010. Atrogin-1 and MuRF1 regulate cardiac MyBP-C levels via different mechanisms. Cardiovasc. Res 85(2), 357–366. 10.1093/cvr/cvp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriscot AS, Baptista IL, Bogomolovas J, Witt C, Hirner S, Granzier H, Labeit S, 2010. MuRF1 is a muscle fiber-type II associated factor and together with MuRF2 regulates type-II fiber trophicity and maintenance. J. Struct. Biol 170(2), 344–353. 10.1016/jjsb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC, 2000. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet 26, 216–220. 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, Wainwright PC, Friedman M, Smith WL, 2012. Resolution of ray-finned fish phylogeny and timing of diversification. Proc. Natl. Acad. Sci. USA 109(34), 13698–136703. 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochala J, Gustafson AM, Diez ML, Renaud G, Li M, Aare S, Qaisar R, Banduseela VC, Hedstrom Y, Tang X, Dworkin B, Ford GC, Nair S, Perera S, Gautel M, Larsson L, 2011. Preferential skeletal muscle myosin loss in response to mechanical silencing in a novel rat intensive care unit model: underlying mechanisms. J. Physiol 589, 2007–2026. 10.1113/jphysiol.2010.202044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivé M, Abdul-Hussein S, Oldfors A, González-Costello J, van der Ven PF, Fürst DO, González L, Moreno D, Torrejón-Escribano B, Alió J, Pou A, Ferrer I, Tajsharghi H, 2015. New cardiac and skeletal protein aggregate myopathy associated with combined MuRF1 and MuRF3 mutations. Hum. Mol. Genet 24(13), 3638–3650. 10.1093/hmg/ddv311. [DOI] [PubMed] [Google Scholar]
- Patterson C, Ike C, Willis PW, Stouffer GA, Willis MS, 2007. The bitter end: the ubiquitin-proteasome system and cardiac dysfunction. Circulation 115(11), 1456–1463. 10.1161/CIRCULATIONAHA.106.649863. [DOI] [PubMed] [Google Scholar]
- Pizon V, Iakovenko A, van der Ven PF, Kelly R, Fatu C, Fürst DO, Karsenti E, Gautel M, 2002. Transient association of titin and myosin with microtubules in nascent myofibrils directed by the MURF2 RING-finger protein. J. Cell. Sci 115(23), 4469–4482. 10.1242/jcs.00131. [DOI] [PubMed] [Google Scholar]
- Portbury AL, Willis MS, Patterson C, 2011. Tearin’ up my heart: proteolysis in the cardiac sarcomere. J. Biol. Chem 286(12), 9929–9934. 10.1074/jbc.R110.170571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prill K, Reid PW, Wohlgemuth SL, Pilgrim DB, 2016. Correction: Still Heart Encodes a Structural HMT, Smyd1b, with Chaperone-Like Function during Fast Muscle Sarcomere Assembly. PLoS ONE 11(1): e0148027 10.1371/journal.pone.0148027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, 2013. Protein breakdown in muscle wasting: role of autophagy-lysosome and ubiquitin-proteasome. Int. J. Biochem. Cell Biol 45(10), 2121–2129. 10.1016/j.biocel.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Coletto L, Grumati P, Bonaldo P, 2013. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J. Cell Sci 126(23), 5325–5333. 10.1242/jcs.114041. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Wang J, Holloway B, Du A, Sanger JM, 2009. Myofibrillogenesis in skeletal muscle cells in zebrafish. Cell Motil. Cytoskeleton 66(8), 556–566. 10.1002/cm.20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y & Nishida M, 2010. Teleost fish with specific genome duplication as unique models of vertebrate evolution. Environmental Biology of Fishes 88,169–188. 10.1007/s10641-010-9628-7. [DOI] [Google Scholar]
- Shimizu H, Langenbacher AD, Huang J, Wang K, Otto G, Geisler R, Wang Y, Chen JN, 2017. The Calcineurin-FoxO-MuRF1 signaling pathway regulates myofibril integrity in cardiomyocytes. Elife 19(6), pii: e27955 10.7554/eLife.27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KM, Cox TC, 2006. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem 281(13), 8970–88980. 10.1074/jbc.M512755200. [DOI] [PubMed] [Google Scholar]
- Spencer JA, Eliazer S, Ilaria RL Jr., Richardson JA, Olson EN, 2000. Regulation of microtubule dynamics and myogenic differentiation by MURF, a striated muscle RING-finger protein. J. Cell Biol 150(4), 771–784. 10.1083/jcb.150.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon V, Goldberg AL, 1996. Importance of the ATP-ubiquitinproteasome pathway in the degradation of soluble and myofibrillar proteins in rabbit muscle extracts. J. Biol. Chem 271, 26690–26697. doi: 10.1074/jbc.271.43.26690. 10.1074/jbc.271.43.26690. [DOI] [PubMed] [Google Scholar]
- Taillandier D, Combaret L, Pouch MN, Samuels SE, Béchet D, Attaix D, 2004. The role of ubiquitin-proteasome-dependent proteolysis in the remodelling of skeletal muscle. Proc. Nutr. Soc 63(2), 357–461. 10.1079/PAR2004358. [DOI] [PubMed] [Google Scholar]
- Tan X, Rotllant J, Li H, De Deyne P, Du SJ, 2006. SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc. Natl. Acad. Sci. USA 103, 2713–2718. 10.1073/pnas.0509503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K, 2006. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 174(2), 639–649. 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Schisler JC, Patterson C, 2008. Appetite for destruction: E3 ubiquitin-ligase protection in cardiac disease. Future Cardiol 4(1), 65–75. 10.2217/14796678.4.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Schisler JC, Portbury AL, Patterson C, 2009a. Build it up-Tear it down: protein quality control in the cardiac sarcomere. Cardiovasc. Res 81(3), 439–448. 10.1093/cvr/cvn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Schisler JC, Li L, Rodríguez JE, Hilliard EG, Charles PC, Patterson C, 2009b. Cardiac muscle ring finger-1 increases susceptibility to heart failure in vivo. Circ. Res 105(1), 80–88. 10.1161/CIRCRESAHA.109.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis MS, Zungu M, Patterson C, 2010. Cardiac muscle ring finger-1--friend or foe? Trends Cardiovasc Med 20(1), 12–16. 10.1016/j.tcm.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt CC, Witt SH, Lerche S, Labeit D, Back W, Labeit S, 2008. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO. J 27(2), 350–360. 10.1038/sj.emboj.7601952. [DOI] [PMC free article] [PubMed] [Google Scholar]