Abstract
MicroRNAs (miRNAs) have been found to play an important role in breast cancer, functioning either as potential oncogenes or tumor suppressor genes, but their role in the prognosis of patients remains unclear. The aim of the present review study is to highlight recent preclinical and clinical studies performed on both circulating and tissue-specific miRNAs and their potential role as prognostic markers in breast cancer. We systematically searched the PubMed database to explore the prognostic value of miRNAs in breast cancer. After performing the literature search and review, 117 eligible studies were identified. We found that 110 aberrantly expressed miRNAs have been associated with prognosis in breast cancer. In conclusion, the collective data presented in this review indicate that miRNAs could serve as novel prognostic tools in breast cancer, while the clinical application of these findings has yet to be verified.
Keywords: breast cancer, microRNAs, prognosis, biomarkers
INTRODUCTION
Breast carcinoma is the leading cause of cancer death in women worldwide [1]. According to the GLOBOCAN 2018 worldwide estimates of cancer incidence and mortality, in 2018, about 2,088,849 new cases were diagnosed and approximately 626,679 women were predicted to die from the disease [2]. These data support the need to develop more efficient strategies for preventive intervention, evaluation of therapy, and prediction of prognosis [3].
Undoubtedly, TNM staging is of great prognostic value; however, considering all the limitations of the currently available prognostic strategies, it is overall recognized that new affordable more accurate methods indicative of molecular characteristics of tumors are needed to achieve personalized treatment [4]. Still, it remains difficult to achieve these goals, because of the absence of refined (sensitive and specific) biomarkers for disease monitoring and for addressing breast cancer on an individual basis.
MicroRNAs are a small class of endogenous, evolutionarily conserved, single-stranded noncoding RNAs, with a length of approximately 19–24 nucleotides [5]. Interaction between miRNAs and mRNAs, within the 3′untranslated region of the target genes, leads to the degradation or inhibition of mRNA translation [6]. In the past few years, miRNAs have attracted considerable attention in the cancer research field, due to their regulatory actions in multiple levels [7, 8]. Depending on the target gene that they regulate, miRNAs can either serve as “tumor suppressor miRs” by repressing oncogenes or as “onco-miRs” by targeting tumor suppressor genes. However, a number of miRNAs play both tumor suppressor and onco-miR roles depending on the cellular context and tumor type [9].
Particularly in breast cancer, microRNAs (miRNAs or miRs) have been proposed as promising biomarkers because they can be readily detected in tumor biopsies (non-circulating miRNAs) and can also be identified in blood, plasma, serum, and saliva (circulating miRNAs) [10]. Furthermore, circulating miRNAs are bound to lipoproteins such as HDL, are associated with Argonaute 2 (Ago2) protein, or are packaged into exosome-like microparticles, micro-vesicles, and apoptotic bodies [11]. Therefore, they are protected from endogenous RNAase activity, and hence they are reliable.
Several lines of evidence have proven that in breast cancer, the expression levels of miRNAs are altered due to key mechanisms, such as epigenetic control, transcription factors, or the effect of mutated proteins [10]. According to previous publications [12], miRNAs are considered as tumor suppressive or protective when they are down-regulated in cancer compared to their normal counterpart, or else, they are termed oncogenic miRNAs or onco-miRs. In this context, miRNAs are increasingly recognized as promising biomarkers, given the fact that they are easy to isolate, and they maintain their structural stability under different conditions of sample processing and isolation. A prognostic biomarker should indicate a patient’s outcome, for example disease recurrence or disease progression, independent of the treatment regimen that was followed, and they are highly desirable for personalized or precise patient treatment [13].
The aim of the present review is to highlight recent preclinical and clinical studies performed on both circulating and tissue-specific miRNAs and therefore to identify their potential role as prognostic markers in breast cancer. We will particularly focus on the potential role of miRNAs in breast cancer prognosis, and on how miRNAs have the potential to answer actual clinical needs, such as identification of biomarkers for prognosis, in order to achieve the goal of individualized breast cancer treatment.
RESULTS
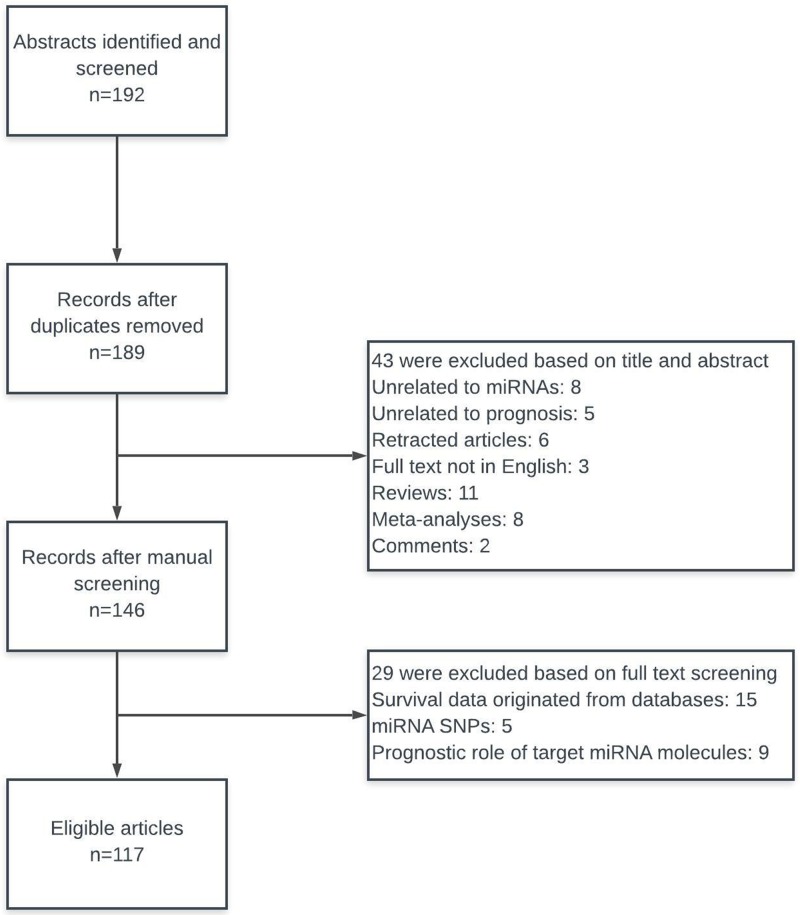
The search strategy retrieved 192 articles. Of these articles, 42 were irrelevant, 11 were reviews, eight (8) were meta-analyses, six (6) were retracted articles, three (3) were not in English, three (3) were duplicates, two (2) were comments and 117 were eligible. The aforementioned steps concerning the selection of studies are illustrated in detail in Figure 1. Therefore, a total of 117 articles were eligible for this systematic review and the prognostic role of 110 miRNA molecules is described (Table 1). Furthermore, we retrieved five studies, in which authors have identified six distinct microRNA signatures with prognostic value in breast cancer (Table 2).
Figure 1. Flow diagram of the study selection process.
Table 1. List of prognostic microRNAs in breast cancer.
| Prognostic microRNA | Breast cancer type | Detection method | Prognostic value | Role | Biological sample | References |
|---|---|---|---|---|---|---|
| let-7 | BC not
classified |
qRT-PCR | potential prognostic
biomarker as altered levels of miR-let-7 are associated with metastases risk |
tumor
suppressor |
serum | [56] |
| let-7-3p | TNBC | NGS, qRT-
PCR |
independent prognostic
factor for OS, DFS |
onco-miR | FFPE | [57] |
| let-7b | luminal subtype | qRT-PCR,
LNA-ISH, TMAs |
independent prognostic
factor for OS associated with luminal tumors |
tumor
suppressor |
FFPE | [58] |
| let-7c/miR-
99a/miR-125b cluster |
estrogen-
dependent BC cell line |
Nanostring,
qRT-PCR, luciferase assay |
potential prognostic factor
for OS in the luminal A subtype |
tumor
suppressor |
cell lines | [59] |
| miR-1 | ER-positive,
stage IV BC |
PCR,
microarray, ISH, IHC |
independent worse
prognostic factor of DFS and BC-specific survival associated with stage, lymph node metastasis, distant metastasis, histological grade, ER status, PR status and Ki-67 |
onco-miR | FFPE | [60] |
| miR-7 | BC not
classified |
qRT-PCR | potential prognostic factor
for OS, DFS predictive of an adverse response to tamoxifen therapy |
onco-miR | fresh frozen
tissue, cell lines |
[61] |
| miR-9 | TNBC,
BC not classified |
qRT-PCR | prognostic factor of DFS
and DMFS, OS |
onco-miR | FFPE, fresh
frozen tissue, cell lines |
[62, 63] |
| miR-10b | BC not
classified, TNBC |
qRT-PCR | independent prognostic
factor for DFS associated with distant metastasis, occurrence in TNBC, associated with genico- obstetric history |
onco-miR | FFPE, fresh
frozen tissue, cell lines |
[17, 40, 41, 64] |
| miR-15a | TNBC | qRT-PCR | prognostic factor for OS,
DFS |
tumor
suppressor |
fresh frozen
tissue |
[65] |
| miR-16 | triple
possitive BC |
qRT-PCR,
Western blot, luciferase report assay, MTS assay |
potentially tumor
suppressive effect on cancer progression of ER positive breast cancers, impairment of cell proliferation |
tumor
suppressor |
FFPE | [45] |
| miR-19a | newly
diagnosed IBC stage III, IBC stage IV, non-IBC stage II-IV and HER2+ BC |
qRT-PCR | potential prognostic factor
for OS, DFS in patients with metastatic HER2(+) IBC. |
tumor
suppressor |
serum, cell
lines |
[66] |
| miR-19b | BC not
classified |
qRT-PCR | prognostic factor for OS
associated with distant metastasis and TNM stage |
onco-miR | fresh frozen
tissue, cell lines |
[67] |
| miR-20b-5p | BC not
classified |
microRNA
arrays |
potential prognostic factor
for DFS, correlated with the presence of breast tumor interstitial fluid |
onco-miR | FFPE,
interstitial breast tumor fluids, serum |
[68] |
| miR-21 | stage II/III
BC, HER2 positive, TNBC |
qRT-PCR,
microarray, luciferase report assay |
independent prognostic
factor of OS, DFS, prognostic biomarker for resistance to trastuzumab, to predict lymph node metastases occurrence in TNBC, to predict high grade in non TNBC possible, prognostic factor in daughter of patients, associated with genico- obstetric history |
onco-miR | FFPE, serum,
fresh frozen tissue, cell lines |
[16-27] |
| miR-22 | BC not
classified |
qRT-PCR,
ISH, luciferase report assay |
potential prognostic factor
for OS, DFS, associated with EMT/metastasis |
both | FFPE, cell lines | [69, 70] |
| miR-24-2*. | BC cell
lines |
qRT-PCR | associated with tumor
suppressive activity through the suppression of cellular survival |
tumor
suppressor |
cell lines, fresh
frozen mouse tissue |
[71] |
| mir-24-3p | BC not
classified (stage I-III) |
Nanostring
technology |
potential prognostic
biomarker of occult metastasis |
onco-miR | plasma | [72] |
| miR-27a | BC not
classified |
ISH, IHC | independent prognostic
factor for OS, DFS |
onco-miR | FFPE | [73] |
| miR-27b-3p | TNBC | qRT-PCR | independent prognostic
factor for OS, DMF survival |
onco-miR | FFPE | [74] |
| miR-29a | BC not
classified |
qRT-PCR,
microarray |
asocciated with
poor response and chemotherapy resistance |
onco-miR | FFPE, cell lines | [75] |
| miR-29b | lobular
and ductal subtypes |
qRT-PCR | prognostic factor for OS,
DFS |
tumor
suppressor |
fresh frozen
tissue |
[76, 77] |
| miR-30a | TNBC | NGS,
qRT-PCR, microarray, luciferase assay |
independent prognostic
factor for OS, DFS |
tumor
suppressor |
FFPE, cell lines | [57, 78] |
| miR-30a-3p | TNBC | qRT-PCR | prognostic factor for OS,
RFS |
tumor
suppressor |
FFPE | [57] |
| miR-30a-5p | TNBC | NGS | prognostic factor for OS,
RFS |
tumor
suppressor |
FFPE | [57] |
| miR-30c-5p | TNBC | qRT-PCR | prognostic factor for
RFS |
tumor
suppressor |
FFPE | [57] |
| miR-30e* | ESR1-/
ERBB2- tumors |
microarray,
ISH |
prognostic factor for DFS | tumor
suppressor |
fresh frozen
tissue |
[79] |
| miR-34a | BC not
classified TNBC |
qRT-PCR,
TMAs |
prognostic factor for OS,
associated with response and chemotherapy resistance |
both | FFPE, plasma,
cell lines |
[75, 80, 81] |
| miR-34b | TNBC | qRT-PCR | prognostic factor for OS,
DFS |
onco-miR | FFPE | [82] |
| miR-34c | TNBC | qRT-PCR | independent risk factor
for OS |
tumor
suppressor |
Plasma | [81] |
| miR-93-5p | BC not
classified |
microRNA
arrays |
potential prognostic factor
for DFS, correlated with the presence of breast tumor interstitial fluid |
onco-miR | FFPE,
interstitial breast tumor fluids, serum |
[68] |
| miR-95-3p | TNBC | qRT-PCR | prognostic factor for OS,
RFS in patients treated with anthracycline-based chemotherapy |
onco-miR | FFPE | [57] |
| miR-96 | BC cell
lines |
qRT-PCR | potential prognostic factor
for OS associated with EMT and regulation of growth factors involved in G1/S-phase transition |
onco-miR | cell lines | [44] |
| miR-99a | BC not
classified |
qRT-PCR | potential prognostic factor
for OS, independent risk factor for breast cancer |
tumor
suppressor |
serum | [83] |
| miR-122 | BC not
classified (stage II-III) |
qRT-PCR,
NGS |
potential prognostic
factor for disease relapse, predictor of metastasis |
onco-miR | serum | [84] |
| miR-124 | BC not
classified |
qRT-PCR | prognostic factor for
OS associated with advanced TNM stage, lymph node metastasis and poorer pathological differentiation, associated with age at diagnosis (>50 years old) |
tumor
suppressor |
FFPE, fresh
frozen tissue |
[85, 86] |
| miR-125a-5p | BC not
classified |
microarray,
qRT-PCR, luciferase assay, ISH, IHC |
potential prognostic factor
for OS, progression-free survival (PRS) |
tumor
suppressor |
serum, cell
lines |
[87] |
| miR-125b | HER2
positive BC, stage II/III |
qRT-PCR, ISH | prognostic factor for
OS, DFS, associated with aromatase inhibitor esistant breast cancers |
onco-miR | FFPE, serum,
cell lines |
[26, 88, 89] |
| miR-126-5p | BC not
classified |
microRNA
arrays |
potential prognostic factor
for DFS |
onco-miR | FFPE,
interstitial breast tumor fluids, serum |
[68] |
| miR-127 | BC not
classified |
qRT-PCR | prognostic factor of OS | tumor
suppressor |
fresh frozen
tissue, cell lines |
[90] |
| miR-128-3p | TNBC | qRT-PCR | prognostic factor for RFS | tumor
suppressor |
FFPE | [57] |
| miR-129-5p | BC not
classified |
qRT-PCR,
luciferase report assay |
potential prognostic factor
for OS, DFS, associated with EMT |
tumor
suppressor |
FFPE, fresh
frozen tissue, cell lines |
[91] |
| miR-133a | BC not
classified |
qRT-PCR,
TMA, ISH, Luciferase assay |
potential prognostic factor
for DFS associated with migration and invasion |
tumor
suppressor |
FFPE, fresh
frozen tissue, cell lines |
[92] |
| miR-140 | BC not
classified |
qRT-PCR,
microarray |
asocciated with
poor response and chemotherapy resistance |
onco-miR | FFPE, cell lines | [75] |
| miR-141 | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, PFS associated with circulating tumor cells status |
onco-miR | plasma | [33, 34] |
| miR-143 | Triple
possitive BC |
qRT-PCR,
Western blot, luciferase report assay, MTS assay |
potentially tumor
suppressive effect on cancer progression of ER positive breast cancers, impairment of cell proliferation |
tumor
suppressor |
FFPE | [45] |
| miR-144 | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, PFS |
tumor
suppressor |
plasma | [34] |
| miR-145 | BC not
classified |
qRT-PCR | potential prognostic factor
for DFS, OS (3-year survival rate) |
tumor
suppressor |
fresh frozen
tissue |
[93, 94] |
| miR-146a | BRCA1-
deficient TNBC tumors |
qRT-PCR | potential prognostic factor
for OS |
tumor
suppressor |
FFPE, cell lines | [95] |
| miR-148a | TNBC | qRT-PCR,
microarray |
potential prognostic factor
for OS associated with metastasis |
tumor
suppressor |
Cell lines,
mouse models |
[96] |
| miR-155 | TNBC,
BC not classified |
qRT-PCR,
microarray, luciferase report assay |
prognostic factor of
DMFS, associated with lymph node metastasis |
both | FFPE, fresh
frozen tissue, cell lines |
[62, 97] |
| miR-182 | BC not
classified, TNBC |
qRT-PCR | potential prognostic factor
to predict lymph node metastases occurrence in TNBC, associated with genico-obstetric history, related with hormonal receptors |
onco-miR | FFPE, serum | [17, 98] |
| miR-183/182/96
cluster |
BC not
classified |
qRT-PCR, ISH | potential prognostic factor
for OS, DFS |
onco-miR | breast tissues
not classified, cell lines |
[99] |
| miR-187 | BC not
classified |
TMA, ISH | independent prognostic
factor FOR breast cancer– specific survival (BCSS) |
onco-miR | FFPE, cell lines | [100] |
| miR-193b | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, PFS |
onco-miR | plasma | [34] |
| miR-195-5p | BC not
classified |
microRNA
arrays |
potential prognostic factor
for DFS |
onco-miR | FFPE,
interstitial breast tumor fluids, serum |
[68] |
| miR-199a-5p | TNBC | NGS | prognostic factor for OS | tumor
suppressor |
FFPE | [57] |
| miR-199b-5p | BC not
classified (-II stage) |
qRT-PCR,
assays in vitro |
potential prognostic factor
for OS associated with TNM stage and lymph node metastasis |
tumor
suppressor |
fresh frozen
tissue and cell lines |
[101] |
| miR-200a | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, PFS, associated with circulating tumor cells status, potential to detect the onset of metastasis |
onco-miR | plasma | [33, 34] |
| miR-200b | BC not
classified |
qRT-PCR,
microRNA arrays, ISH, TMA, luciferase report assay |
potential prognostic factor
for OS (independent), PFS associated with advanced clinical stage, metastasis, cell proliferation, apoptosis, cell cycle distribution and circulating tumor cells status, potential to detect the onset of metastasis |
both | FFPE, plasma,
cell lines |
[33, 34, 46, 47] |
| miR-200c | BC not
classified |
qRT-PCR,
microRNA arrays |
prognostic factor of OS,
DFS, potential to detect the onset of metastasis, associated with circulating tumor cells status |
onco-miR | fresh frozen
tissue, plasma |
[33, 34, 102] |
| miR-200c/141
cluster |
BC not
classified, TNBC |
qRT-PCR,
CAT reporter assay, siRNA transfection, Western blot |
poor prognostic factor
in TNBC, promoting metastasis |
onco-miR | FFPE, cell
lines, xenograft animal model |
[103] |
| miR-203 | BC not
classified, ER positive BC |
microRNA
arrays, qRT- PCR, Western blot, luciferase report assay, MTS assay |
potential prognostic factor
for OS, PFS associated with EMT and circulating tumor cells status |
both | FFPE, plasma,
cell lines |
[33, 34, 44, 45] |
| miR-203-5p | TNBC | NGS | prognostic factor for OS | onco-miR | FFPE | [57] |
| miR-203a | ductal
in situ, invasive ductal and lobular carcinoma |
qRT-PCR | potential prognostic
marker associated with increased stage in invasive lobular carcinomas |
tumor
suppressor |
FFPE | [104] |
| miR-204 | BC not
classified |
qRT-PCR | potential prognostic factor
for OS, DFS, correlated with chemotherapeutic resistance |
tumor
suppressor |
FFPE | [105] |
| miR-205 | BC not
classified |
qRT-PCR,
LNA-ISH, TMAs, IHC |
potential prognostic
factor for OS associated with tumours of ductal morphology, for OS and DFS in early breast cancer |
tumor
suppressor |
FFPE | [21, 58] |
| miR-206 | BC not
classified |
qRT-PCR,
luciferase report assay |
potential prognostic factor
for OS |
both | fresh frozen
tissue, cell lines |
[94, 106, 107, |
| miR-210 | early first
primary BC, TNBC |
qRT-PCR,
microarray |
independent prognostic
factor for OS, DFS, associated with poor clinical outcome in ER- positive, tamoxifen-treated BC patients, involved in cell proliferation, migration and invasion, Potential to detect the onset of metastasis prior to clinical diagnosis, associated with circulating tumor cells status |
onco-miR | FFPE, fresh
frozen tissue, plasma, cell lines (Breast cancer and tumor-educated macrophages) |
[33-39] |
| miR-210-3p | BC cell
lines |
qRT-PCR | potential prognostic factor
for OS associated with EMT and regulation of growth factors involved in G1- to S-phase transition |
onco-miR | cell lines | [44] |
| miR-215 | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, PFS, Potential to detect the onset of metastasis prior to clinical diagnosis |
tumor
suppressor |
plasma | [34] |
| miR-218 | BC not
classified |
qRT-PCR | prognostic factor for OS
associated with lymph node metastases, higher grades, |
tumor
suppressor |
fresh frozen
tissue |
[108] |
| miR-221 | BC not
classified |
qRT-PCR | prognostic factor for DFS,
OS, RFS |
onco-miR | FFPE, fresh
frozen tissue, cell lines |
[41, 63, 109] |
| miR-221-3p | TNBC | qRT-PCR | prognostic factor for DFS | tumour
suppressor |
FFPE, cell lines | [110] |
| miR-222 | BC not
classified |
qRT-PCR,
TMA |
potential prognostic
factor related to lymph node metastasis, down- regulation of the estrogen receptor, EMT, tumor progression, poor response and chemotherapy resistance |
onco-miR | FFPE, fresh
frozen tissue, cell lines |
[75, 109] |
| miR-222-3p | BC not
classified |
qRT-PCR,
microarray |
independent prognostic
factor for DFS postoperatively |
onco-miR | serum | [111] |
| miR-301a | BC not
classified, TNBC |
qRT-PCR,
microarray, ISH |
prognostic factor for DFS,
OS |
onco-miR | FFPE | [112, 113] |
| miR-320a | BC not
classified |
chromogenic
ISH |
potential prognostic factor
for OS for invasive breast cancer |
tumor
suppressor |
FFPE | [114] |
| miR-324-5p | TNBC | NGS | prognostic factor for OS | onco-miR | FFPE | [57] |
| miR-329 | BC not
classified |
qRT-PCR | independent prognostic
factor for OS |
tumor-
suppressor |
serum, fresh
frozen tissue, cell lines |
[115] |
| miR-330-3p | BC not
classified |
qRT-PCR | potential prognostic factor
for OS |
onco-miR | fresh frozen
tissue |
[116] |
| miR-339-5p | BC not
classified |
qRT-PCR,
TMA, ISH |
independent prognostic
factor for OS, DFS |
tumor
suppressor |
FFPE, cell lines | [117] |
| miR-361-5p | BC not
classified, TNBC |
TMAs, ISH | prognostic factor for DFS | tumor
suppressor |
FFPE | [118] |
| miR-365 | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS |
miR-365,
onco-miR |
plasma | [34] |
| miR-370 | BC not
classified |
qRT-PCR,
TMA |
potential prognostic factor
for DFS |
onco-miR | FFPE | [119] |
| miR-374a | BC not
classified, IDC stage II |
qRT-PCR,
TMAs, Luciferase assay, MTT assays, IHC |
potential prognostic factor
for DFS, contributes to tumorigenicity and progression |
onco-miR | FFPE, fresh
frozen tissue, cell lines, xenograft mouse models |
[120, 121] |
| miR-375 | BC not
classified, stage II-III locally advanced and IBC patients |
qRT-PCR,
microRNA arrays, NGS |
potential prognostic factor
for OS, PFS associated with circulating tumor cells status, related to hormonal receptors |
both | serum, plasma | [33, 34, 84, 98] |
| miR-409-3p | BC not
classified |
qRT-PCR | independent prognostic
factor for OS associated with advanced TNM stage, lymph node metastasis, and poorer pathological differentiation |
tumor
suppressor |
fresh frozen
tissue |
[112] |
| miR-423 | BC not
classified |
qRT-PCR,
microarray |
asocciated with
poor response and chemotherapy resistance |
onco-miR | FFPE, cell lines | [75] |
| miR-429 | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, PFS |
miR-429
onco-miR |
plasma | [34] |
| miR-451 | BC cell
lines |
qRT-PCR | potential factor associated
with cell survival and endocrine resistance |
tumor
suppressor |
cell lines | [123] |
| miR-454 | BC not
classified (stage I-III) |
TMA, ISH | potential prognostic
factor for OS (especially in TNBC) and DFS, associated with response to anthracycline |
onco-miR | FFPE | [124] |
| miR-454-3p | BC not
classified |
microRNA
arrays |
potential prognostic factor
for DFS |
onco-miR | FFPE,
interstitial breast tumor fluids, serum |
[68] |
| miR-486-5p | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, Potential to detect the onset of metastasis prior to clinical diagnosis |
tumor
suppressor |
plasma | [34] |
| miR-493 | TNBC | TMAs, ISH | prognostic factor for DFS | tumour
suppressor |
FFPE | [125] |
| miR-494 | node-
negative BC |
ISH | 8.5-fold risk of breast
cancer death (association trend-not clinical significance) |
tumour
suppressor |
fresh frozen
tissue |
[126] |
| miR-497 | BC not
classified, TNBC |
qRT-PCR,
luciferase assay |
potential prognostic factor
for OS |
tumor
suppressor |
fresh frozen
tissue, cell lines, orthotopic mouse models |
[127, 128] |
| miR-548c-5p | TNBC | qRT-PCR, ISH | independent prognostic
factor for OS, DFS |
onco-miR | FFPE | [39] |
| miR-574 | BC not
classified |
qRT-PCR,
microarray |
asocciated with
poor response and chemotherapy resistance |
onco-miR | FFPE, cell lines | [75] |
| miR-574-3p | BC not
classified |
qRT-PCR,
NGS |
potential prognostic factor
for OS, DFS |
tumor
suppressor |
FFPE | [129] |
| miR-588 | BC not
classified |
qRT-PCR | prognostic factor of OS | tumour
suppressor |
fresh frozen
tissue, cell lines |
[130] |
| miR-590-3p | BC cell
lines |
qRT-PCR,
luciferase report assay |
associated with breast
cancer cells viability, growth and apoptosis |
tumor
suppressor |
cell lines | [131] |
| miR-597 | BC not
classified |
qRT-PCR | prognostic factor of OS | tumor
suppressor |
fresh tissue | [132] |
| miR-601 | BC not
classified |
qRT-PCR | prognostic factor for
DFS associated with cell proliferation and metastasis |
tumor
suppressor |
FFPE, cell lines | [133] |
| miR-638 | BC not
classified, BRCA1- deficient TNBC tumors |
qRT-PCR | independent prognostic
factor for OS associated with lymph node metastasis and TNM stage |
tumor
suppressor |
FFPE, fresh
frozen, cell lines |
[95, 134] |
| miR-644a | BC cell
lines |
qRT-PCR,
luciferase report assay |
associated with tumor
progression and distant metastasis-free survival |
tumor
suppressor |
cell lines | [135] |
| miR-660-5p | BC not
classified |
qRT-PCR,
NGS |
potential prognostic factor
for OS, DFS |
onco-miR | FFPE | [129] |
| miR-711 | BC not
classified |
qRT-PCR | independent prognostic
factor for OS, DFS, associated with breast cancer cells’ proliferation, colony formation, invasion |
onco-miR | FFPE, cell lines | [136] |
| miR-744 | BC not
classified |
qRT-PCR,
microarray |
associated with
poor response and chemotherapy resistance |
onco-miR | FFPE, cell lines | [75] |
| miR-801 | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, PFS associated with circulating tumor cells status |
onco-miR | plasma | [33, 34] |
| miR-874 | BC not
classified |
qRT-PCR | prognostic factor for OS | tumour
suppressor |
fresh frozen
tissue, cell lines |
[137] |
| miR-940 | IDC, TNBC | qRT-PCR | prognostic factor for OS | tumor suppressor | serum | [138] |
| miR-1179 | BC not
classified |
RT-PCR | independent prognostic
factor for OS |
tumor
suppressor |
breast tissue
not classified, cell lines |
[139] |
| miR-1247-5p | BC not
classified |
qRT-PCR | independent prognostic
indicator for DFS, OS |
tumor
suppressor |
FFPE, fresh
frozen tissue, cell lines |
[140, 141] |
| miR-1260 | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS |
onco-miR | plasma | [34] |
| miR-1274a | BC not
classified |
microRNA
arrays, qRT- PCR |
potential prognostic factor
for OS, PFS |
onco-miR | plasma | [34] |
| miR-1274b | BC not
classified |
microRNA
arrays |
potential prognostic factor
for DFS |
onco-miR | FFPE,
interstitial breast tumor fluids, serum |
[68] |
| miR-1825 | BC not
classified |
microRNA
arrays |
potential prognostic factor
for DFS |
onco-miR | FFPE,
interstitial breast tumor fluids, serum |
[68] |
| miR-3178 | BC not
classified |
qRT-PCR,
microarray |
associated with
poor response and chemotherapy resistance |
onco-miR | FFPE, cell lines | [75] |
| miR-4653-3p | HR+ BC
(stage I~III) |
qRT-PCR | potential prognostic
biomarker for DFS for patients treated with adjuvant tamoxifen |
tumor
suppressor |
FFPE | [142] |
| miR-6780b | BC not
classified |
qRT-PCR,
microarray |
associated with
poor response and chemotherapy resistance |
onco-miR | FFPE, cell lines | [75] |
Abbreviations: quantitative reverse transcriptase real-time polymerase chain reaction (qRT-PCR), In situ hybridization (ISH), locked nucleic acid probe in situ hybridization (LNA-ISH), Immunohistochemistry (IHC), epithelial-mesenchymal transition (EMT), formalin-fixed paraffin embedded (FFPE), Next Generation Sequencing (NGS), overall survival (OS), relapse free survival (RFS), disease free survival (DFS), progress free survival (PFS), breast cancer (BC), triple negative breast cancer (TNBC), Inflammatory breast cancer (IBC), Invasive Ductal Carcinoma (IDC), estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor type 2 (HER2).
Table 2. List of prognostic microRNA signatures in breast cancer.
| miRNA signature | Breast cancer type | Detection method | Prognostic value | Role | Biological sample | References |
|---|---|---|---|---|---|---|
| miR-183-5p,
miR-194-5p, miR-1285-5p signature |
BC not
classified |
microarrays,
qRT-PCR |
potential
prognostic factor for OS in young breast cancer patients (age <35 years) |
miR-183-5p onco-miR
miR-194-5p onco-miR miR-1285-5p tumor suppressor |
FFPE | [48] |
| miR-21, miR-
30c, miR-181a, miR-181c, miR-125b, miR-7, miR- 200a, miR- 135b, miR-22 and miR-200c signature |
HR
positive, HER2 negative |
qRT-PCR | potential
prognostic factor for DRFS |
10-miRNA-based
classifier as a prognostic model |
FFPE | [49] |
| miR-155, miR-
493, miR-30e and miR-27a signature |
TNBC | qRT-PCR, IHC | potential
prognostic factor for OS associated with taxanes resistance |
miR-155 tumor
suppressor miR-493 tumor suppressor miR-30e onco-miR miR-27a onco-miR |
FFPE | [50] |
| miR-16, 155,
125b, 374a signature |
TNBC | qRT-PCR | potential
prognostic factor for OS |
miR-16 tumor
suppressor miR-155 tumor suppressor miR-125b onco-miR miR-374a tumor suppressor |
FFPE | [51] |
| miR-16, 125b,
374a, 374b, 421, 655, 497 signature |
TNBC | qRT-PCR | potential
prognostic factor for DDFS |
miR-16 tumor
suppressor miR-125b onco-miR miR-374a tumor suppressor miR-374b tumor suppressor miR-421 onco-miR miR-655 onco-miR miR-497 tumor suppressor |
FFPE | [51] |
| miR-191-5p,
miR-214-3p, miR-451a, and miR-489 signature |
BC not
classified |
qRT-PCR,
microarray |
independent
prognostic factor for OS, DFS |
miR-191-5p onco-miR
miR-214-3p tumor suppressor miR-451a tumor suppressor miR-489 tumor suppressor |
FFPE, cell lines | [52] |
Abbreviations: breast cancer (BC), quantitative reverse transcriptase real-time polymerase chain reaction (qRT-PCR), formalin-fixed paraffin embedded (FFPE), overall survival (OS), distant disease-free survival (DDFS), distant recurrence free survival (DRFS).
According to our results, presented in Table 1, the majority of publications have not taken into account the distinct breast cancer subtypes during the development of their research protocol, since in 60.8% of studies breast cancer samples were not classified. The remaining 25.8% focused on Triple Negative Breast Cancer (TNBC) samples or involved Luminal A (5.0%), Luminal B (1.7%) and HER2-positive (1.7%) breast cancer samples. Of note, 5.0% of the selected studies accessed the prognostic value of miRNAs through experiments performed on breast cancer cell lines. Different detection methods, as well as different sample types were used for the detection of the prognostic miRNA expression levels (i.e., paraffin-fixed, formalin-fixed, freshly frozen tumors, plasma or serum). Concerning the detection methods, quantitative reverse transcriptase real-time polymerase chain reaction (qRT-PCR) was used in 35,8% of the eligible studies, while in 21,7% of the studies qRT-PCR was performed along with Microarray analysis. Additionally, next generation Sequencing technologies (9,2%), in situ hybridization techniques (9,2%), luciferase report assays (6,7%) or a combination of various techniques (10,8%) were employed.
DISCUSSION
We conducted a comprehensive systematic literature review to unfold the utility of miRNA biomarkers that can be evaluated for predicting prognosis in breast cancer patients. We have identified 117 studies that investigate the potential correlation between miRNA profile expression in breast cancer tissue and in the circulation and their possible use as prognostic factors. Interestingly, most of the miRNAs found to be associated with prognosis in breast cancer, were assessed in only a single study. Six miRs (miR-10b, miR-200b, miR-21, miR-203, miR-375, and miR-210) were evaluated in at least four studies and the discussion will be mainly focused on these molecules, based on an effort to merely provide some important information on the most commonly researched molecules in accordance with our systematic literature review.
MiR-21 is one of the most extensively studied cancer-related miRNAs and its aberrant expression and deregulation may play a pivotal role in the majority of cancers [14]. miR-21 may serve as a key regulator of oncogenic processes, including tumor growth, migration, and invasion [15], through targeting the pro-apoptotic phosphatase and tensin homolog (PTEN) and promoting tumor cell proliferation [16]. According to our initial search results, we retrieved 12 studies [16-27] and four meta-analyses [28-31] focusing on the prognostic value of miR-21, which collectively provide robust evidence that miR-21 up-regulation is associated with poor outcomes in cancer patients.
Mir-210 has multiple functions in cancer cells and is involved in angiogenesis, cell cycle regulation, DNA damage repair, mitochondrial metabolism, and immune response [32]. According to our search results, including seven studies [33-39], high expression of miR-210 has been significantly associated with poor survival in patients with breast cancer. Notably, single miR-210 assay has been proposed as an independent prognostic factor in this disease.
Concerning miR-10b, it has been presented as a potential biomarker that could play a predictive role in lymph node metastases occurrence across TNBC and in the incidence of high-grade tumors in non-TNBC cases [17]. Elevated expression of miR-10b in breast tumor tissue samples has been associated with adverse outcome, which is further supported from data derived from in vitro studies [40]. Finally, a survival analysis of 230 breast tissue samples has shown that high levels of miR-10b result to a short relapse free survival (RFS) of breast cancer, acting as an independent prognostic factor of RFS [41]. Our results, emphasize the oncogenic role of miR-10b and indicate that its high expression may be correlated with poor survival in breast cancer, while a recent metanalysis further strengthens our findings [30].
MiR-200 family members function as regulators of the epithelial to mesenchymal transition (EMT), which is one of the initial steps in tumor metastasis [42]. Specifically, miR-200b and miR-203 have both been characterized as tumor suppressors in multiple breast tumor types [43]. However, there seems to be an inconsistency in the existing literature, since we retrieved two studies that have found that higher expression of circulating miR-200b and miR-203 are associated with worse outcome [33, 34], further substantiated by a study on breast cancer cell lines [44]. However, other studies on breast cancer tissue samples and cell lines presented inverse results [45-47]. These discrepancies exhibit the diverse regulatory roles of miR-200 family members, depending on the cellular context and type of biological sample (blood VS tissue), and highlight the potential prognostic impact of these EMT regulating miRNA molecules in breast cancer.
Furthermore, our search retrieved five studies that have found six miRNA signatures to be useful for predicting the outcome of breast cancer [48-52]. Coordinated regulation of multiple miRNAs of potential prognostic value, has helped researchers identify panels of prognostic microRNAs for breast cancer. The discovery of microRNA expression signatures shows considerable promise for determining the prognosis of individuals with breast cancer. Similar miRNA signatures have been identified in a variety of other cancers, including acute myeloid leukemia, chronic lymphocytic leukemia, colon cancer, pancreatic cancer, and non-small cell lung cancer [53]. These reports highlight that this class of RNA molecules is showing substantial potential to be used as prognostic biomarkers for cancer.
Among the limitations of this effort, it should be stressed that this process was essentially driven by the search algorithm, which focused mainly on titles of the published literature, in an effort to provide more relevant results. Furthermore, clear heterogeneity was observed in our results, due to differences in patient characteristics (ethnicity, age, tumor stage, grade and subtype) and the use of different isolation and detection methods, cut-off values for miRNA expression levels, sample preparation methods and sample types (i.e., paraffin-fixed, formalin-fixed, freshly frozen tumors, plasma or serum).
Based on the results of this systematic review, we consider that miRNA detection may address the need for independent, easily accessible, prognostic molecular markers for breast cancer management in clinical practice, by assessing the impact of aberrant miRNA expression on patients’ survival. Our work sums up all the available data on prognostic miRNAs and can also act as a valuable reference point for future studies. Furthermore, while prognostic studies can assist in answering important questions concerning specific patient outcomes, their vigorous and careful design is a necessary condition for ensuring the reliability of results [54]. It should be stressed out that the thorough validation of prognostic factors is a necessary and unavoidable process in order to maximize certainty in predicting future breast cancer patients’ outcomes. Therefore, extensive validation studies focusing on particular miRNAs or miRNA signatures should be performed to relate baseline clinical and experimental variables to outcome. Eventually, all the reviewed molecular studies may help in bringing prognostic miRNAs closer to the clinical practice.
MATERIALS AND METHODS
Methods of search strategy and study eligibility
This systematic review was conducted in accordance with the PRISMA guidelines [55] and in line with the a priori protocol agreed on and signed by EZ and FZ. Eligible studies were sought in PubMed without any restriction of publication language; the end-of-search date was January 28, 2019. The following search algorithm was used: breast[ti] AND (carcinoma OR carcinomas OR cancer OR cancers OR neoplasm OR neoplasms) AND (microRNA[ti] OR miR[ti] OR miRNA[ti] OR microRNAs[ti] OR miRs[ti] OR miRNAs[ti]) AND (prognosis[ti] OR prognostic[ti] OR survival[ti] OR outcome[ti] OR mortality[ti]). Eligible articles included studies examining the prognostic role of microRNAs in breast cancer. Only prospective and retrospective studies as well as case reports were considered eligible. In instances where multiple (overlapping) publications stemming from the same study were identified, the larger size study and the one with longer follow-up were included, unless the reported outcomes were mutually exclusive. Authors working independently and blindly to each other in pairs (E.Z., F.Z.) performed the selection of eligible studies; in case of disagreement, consensus with the whole team was reached.
Data extraction
The extraction of data comprised general information, including the name of the miRNA molecule, the breast cancer type in which its expression was determined, method of detection, the sample type that was used, its prognostic value in breast cancer, its function in cancer (onco-miR or tumor suppressor-miR) and the author-year of publication. Data were independently extracted and analyzed by a pair of reviewers (E.Z. and F.Z.), with one reviewer being blinded to the other; if needed, the final decision was reached by a team consensus.
Eligible literature met the following criteria: (1) measured miR expression levels in tumor or blood samples or human cell lines and (2) only articles in English. Publications were excluded if they had one or more of the following criteria: (1) studies referring to the prognostic role of single nucleotide polymorphisms (SNPs) in miRNA genes affecting their function; (2) studies that refer to the prognostic role of target miRNA molecules (molecules regulated by miRs); (3) studies based solely on a bioinformatics approach or a computational algorithm, with survival data originated from databases without subsequent biological validation and (4) review papers, meta-analyses, comments, letters or duplicate publications.
Footnotes
CONFLICTS OF INTEREST
The authors declare that they have no potential conflicts of interest.
REFERENCES
- 1. World Health Organization. Media Centre-Cancer (Fact Sheet). https://www.who.int/mediacentre/factsheets/fs297/en/. 2017. Accessed Feb 1 2017.
- 2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018; 68:394–424. 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3. Hinestrosa MC, Dickersin K, Klein P, Mayer M, Noss K, Slamon D, Sledge G, Visco FM. Shaping the future of biomarker research in breast cancer to ensure clinical relevance. Nat Rev Cancer. 2007; 7:309–15. 10.1038/nrc2113. [DOI] [PubMed] [Google Scholar]
- 4. Nalejska E, Mączyńska E, Lewandowska MA. Prognostic and predictive biomarkers: tools in personalized oncology. Mol Diagn Ther. 2014; 18:273–84. 10.1007/s40291-013-0077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J, Zhang KY, Liu SM, Sen S. Tumor-associated circulating microRNAs as biomarkers of cancer. Molecules. 2014; 19:1912–38. 10.3390/molecules19021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoruker EE, Aydoğan F, Gezer U, Saip P, Dalay N. Analysis of circulating microRNAs during adjuvant chemotherapy in patients with luminal A breast cancer. Mol Clin Oncol. 2015; 3:954–58. 10.3892/mco.2015.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McGuire A, Brown JA, Kerin MJ. Metastatic breast cancer: the potential of miRNA for diagnosis and treatment monitoring. Cancer Metastasis Rev. 2015; 34:145–55. 10.1007/s10555-015-9551-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goh JN, Loo SY, Datta A, Siveen KS, Yap WN, Cai W, Shin EM, Wang C, Kim JE, Chan M, Dharmarajan AM, Lee AS, Lobie PE, et al. microRNAs in breast cancer: regulatory roles governing the hallmarks of cancer. Biol Rev Camb Philos Soc. 2016; 91:409–28. 10.1111/brv.12176. [DOI] [PubMed] [Google Scholar]
- 9. Muluhngwi P, Klinge CM. Identification of miRNAs as biomarkers for acquired endocrine resistance in breast cancer. Mol Cell Endocrinol. 2017; 456:76–86. 10.1016/j.mce.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 10. Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics. 2015; 5:1122–43. 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011; 108:5003–08. 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y, Zhang Y, Li Q, Li J, Ma X, Xing J, Rong S, Wu Z, Tian Y, Li J, Jia L. MiRNAs Predict the Prognosis of Patients with Triple Negative Breast Cancer: A Meta-Analysis. PLoS One. 2017; 12:e0170088. 10.1371/journal.pone.0170088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li G, Hu J, Hu G. Biomarker Studies in Early Detection and Prognosis of Breast Cancer. Adv Exp Med Biol. 2017; 1026:27–39. 10.1007/978-981-10-6020-5_2. [DOI] [PubMed] [Google Scholar]
- 14. Pfeffer SR, Yang CH, Pfeffer LM. The Role of miR-21 in Cancer. Drug Dev Res. 2015; 76:270–77. 10.1002/ddr.21257. [DOI] [PubMed] [Google Scholar]
- 15. Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009; 37:918–25. 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- 16. Dong G, Liang X, Wang D, Gao H, Wang L, Wang L, Liu J, Du Z. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. 2014; 31:57. 10.1007/s12032-014-0057-x. [DOI] [PubMed] [Google Scholar]
- 17. Medimegh I, Omrane I, Privat M, Uhrhummer N, Ayari H, Belaiba F, Benayed F, Benromdhan K, Mader S, Bignon IJ, Elgaaied AB. MicroRNAs expression in triple negative vs non triple negative breast cancer in Tunisia: interaction with clinical outcome. PLoS One. 2014; 9:e111877. 10.1371/journal.pone.0111877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008; 14:2348–60. 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Mu L, Yu H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat. 2009; 117:131–40. 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 20. Lee JA, Lee HY, Lee ES, Kim I, Bae JW. Prognostic Implications of MicroRNA-21 Overexpression in Invasive Ductal Carcinomas of the Breast. J Breast Cancer. 2011; 14:269–75. 10.4048/jbc.2011.14.4.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Markou A, Yousef GM, Stathopoulos E, Georgoulias V, Lianidou E. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem. 2014; 60:197–205. 10.1373/clinchem.2013.210542. [DOI] [PubMed] [Google Scholar]
- 22. Toraih EA, Mohammed EA, Farrag S, Ramsis N, Hosny S. Pilot Study of Serum MicroRNA-21 as a Diagnostic and Prognostic Biomarker in Egyptian Breast Cancer Patients. Mol Diagn Ther. 2015; 19:179–90. 10.1007/s40291-015-0143-6. [DOI] [PubMed] [Google Scholar]
- 23. Usmani A, Shoro AA, Memon Z, Hussain M, Rehman R. Diagnostic, prognostic and predictive value of MicroRNA-21 in breast cancer patients, their daughters and healthy individuals. Am J Cancer Res. 2015; 5:2484–90. [PMC free article] [PubMed] [Google Scholar]
- 24. Yan LX, Liu YH, Xiang JW, Wu QN, Xu LB, Luo XL, Zhu XL, Liu C, Xu FP, Luo DL, Mei P, Xu J, Zhang KP, Chen J. PIK3R1 targeting by miR-21 suppresses tumor cell migration and invasion by reducing PI3K/AKT signaling and reversing EMT, and predicts clinical outcome of breast cancer. Int J Oncol. 2016; 48:471–84. 10.3892/ijo.2015.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yadav P, Mirza M, Nandi K, Jain SK, Kaza RC, Khurana N, Ray PC, Saxena A. Serum microRNA-21 expression as a prognostic and therapeutic biomarker for breast cancer patients. Tumour Biol. 2016; 37:15275–82. 10.1007/s13277-016-5361-y. [DOI] [PubMed] [Google Scholar]
- 26. Liu B, Su F, Chen M, Li Y, Qi X, Xiao J, Li X, Liu X, Liang W, Zhang Y, Zhang J. Serum miR-21 and miR-125b as markers predicting neoadjuvant chemotherapy response and prognosis in stage II/III breast cancer. Hum Pathol. 2017; 64:44–52. 10.1016/j.humpath.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 27. Badr M, Said H, Louka ML, Elghazaly HA, Gaballah A, Atef Abd El Mageed M. MicroRNA-21 as a predictor and prognostic factor for trastuzumab therapy in human epidermal growth factor receptor 2-positive metastatic breast cancer. J Cell Biochem. 2019; 120:3459–66. 10.1002/jcb.27620. [DOI] [PubMed] [Google Scholar]
- 28. Pan F, Mao H, Deng L, Li G, Geng P. Prognostic and clinicopathological significance of microRNA-21 overexpression in breast cancer: a meta-analysis. Int J Clin Exp Pathol. 2014; 7:5622–33. [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y, Zhang Y, Pan C, Ma F, Zhang S. Prediction of poor prognosis in breast cancer patients based on microRNA-21 expression: a meta-analysis. PLoS One. 2015; 10:e0118647. 10.1371/journal.pone.0118647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang Y, Zhou X, Ji J, Chen L, Cao J, Luo J, Zhang S. High expression levels of miR-21 and miR-210 predict unfavorable survival in breast cancer: a systemic review and meta-analysis. Int J Biol Markers. 2015; 30:e347–58. 10.5301/jbm.5000160. [DOI] [PubMed] [Google Scholar]
- 31. Jinling W, Sijing S, Jie Z, Guinian W. Prognostic value of circulating microRNA-21 for breast cancer: a systematic review and meta-analysis. Artif Cells Nanomed Biotechnol. 2017; 45:1–6. 10.1080/21691401.2016.1216856. [DOI] [PubMed] [Google Scholar]
- 32. Qin Q, Furong W, Baosheng L. Multiple functions of hypoxia-regulated miR-210 in cancer. J Exp Clin Cancer Res. 2014; 33:50. 10.1186/1756-9966-33-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Madhavan D, Zucknick M, Wallwiener M, Cuk K, Modugno C, Scharpff M, Schott S, Heil J, Turchinovich A, Yang R, Benner A, Riethdorf S, Trumpp A, et al. Circulating miRNAs as surrogate markers for circulating tumor cells and prognostic markers in metastatic breast cancer. Clin Cancer Res. 2012; 18:5972–82. 10.1158/1078-0432.CCR-12-1407. [DOI] [PubMed] [Google Scholar]
- 34. Madhavan D, Peng C, Wallwiener M, Zucknick M, Nees J, Schott S, Rudolph A, Riethdorf S, Trumpp A, Pantel K, Sohn C, Chang-Claude J, Schneeweiss A, Burwinkel B. Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis. 2016; 37:461–70. 10.1093/carcin/bgw008. [DOI] [PubMed] [Google Scholar]
- 35. Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008; 14:1340–48. 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 36. Rothé F, Ignatiadis M, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, Badran B, Fayyad-Kazan H, Desmedt C, Harris AL, Piccart M, Sotiriou C. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011; 6:e20980. 10.1371/journal.pone.0020980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Toyama T, Kondo N, Endo Y, Sugiura H, Yoshimoto N, Iwasa M, Takahashi S, Fujii Y, Yamashita H. High expression of microRNA-210 is an independent factor indicating a poor prognosis in Japanese triple-negative breast cancer patients. Jpn J Clin Oncol. 2012; 42:256–63. 10.1093/jjco/hys001. [DOI] [PubMed] [Google Scholar]
- 38. Bleckmann A, Leha A, Artmann S, Menck K, Salinas-Riester G, Binder C, Pukrop T, Beissbarth T, Klemm F. Integrated miRNA and mRNA profiling of tumor-educated macrophages identifies prognostic subgroups in estrogen receptor-positive breast cancer. Mol Oncol. 2015; 9:155–66. 10.1016/j.molonc.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boukerroucha M, Josse C, ElGuendi S, Boujemla B, Frères P, Marée R, Wenric S, Segers K, Collignon J, Jerusalem G, Bours V. Evaluation of BRCA1-related molecular features and microRNAs as prognostic factors for triple negative breast cancers. BMC Cancer. 2015; 15:755. 10.1186/s12885-015-1740-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chang CH, Fan TC, Yu JC, Liao GS, Lin YC, Shih AC, Li WH, Yu AL. The prognostic significance of RUNX2 and miR-10a/10b and their inter-relationship in breast cancer. J Transl Med. 2014; 12:257. 10.1186/s12967-014-0257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eissa S, Matboli M, Shehata HH, Essawy NO. MicroRNA-10b and minichromosome maintenance complex component 5 gene as prognostic biomarkers in breast cancer. Tumour Biol. 2015; 36:4487–94. 10.1007/s13277-015-3090-2. [DOI] [PubMed] [Google Scholar]
- 42. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008; 10:593–601. 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 43. Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol Ther. 2010; 10:219–22. 10.4161/cbt.10.3.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fisher JN, Terao M, Fratelli M, Kurosaki M, Paroni G, Zanetti A, Gianni M, Bolis M, Lupi M, Tsykin A, Goodall GJ, Garattini E. MicroRNA networks regulated by all-trans retinoic acid and Lapatinib control the growth, survival and motility of breast cancer cells. Oncotarget. 2015; 6:13176–200. 10.18632/oncotarget.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu X, Zhang X, Dhakal IB, Beggs M, Kadlubar S, Luo D. Induction of cell proliferation and survival genes by estradiol-repressed microRNAs in breast cancer cells. BMC Cancer. 2012; 12:29. 10.1186/1471-2407-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ye F, Tang H, Liu Q, Xie X, Wu M, Liu X, Chen B, Xie X. miR-200b as a prognostic factor in breast cancer targets multiple members of RAB family. J Transl Med. 2014; 12:17. 10.1186/1479-5876-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yao Y, Hu J, Shen Z, Yao R, Liu S, Li Y, Cong H, Wang X, Qiu W, Yue L. MiR-200b expression in breast cancer: a prognostic marker and act on cell proliferation and apoptosis by targeting Sp1. J Cell Mol Med. 2015; 19:760–69. 10.1111/jcmm.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hironaka-Mitsuhashi A, Matsuzaki J, Takahashi RU, Yoshida M, Nezu Y, Yamamoto Y, Shiino S, Kinoshita T, Ushijima T, Hiraoka N, Shimizu C, Tamura K, Ochiya T. A tissue microRNA signature that predicts the prognosis of breast cancer in young women. PLoS One. 2017; 12:e0187638. 10.1371/journal.pone.0187638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gong C, Tan W, Chen K, You N, Zhu S, Liang G, Xie X, Li Q, Zeng Y, Ouyang N, Li Z, Zeng M, Zhuang S, et al. Prognostic Value of a BCSC-associated MicroRNA Signature in Hormone Receptor-Positive HER2-Negative Breast Cancer. EBioMedicine. 2016; 11:199–209. 10.1016/j.ebiom.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gasparini P, Cascione L, Fassan M, Lovat F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL, Huebner K. microRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014; 5:1174–84. 10.18632/oncotarget.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cascione L, Gasparini P, Lovat F, Carasi S, Pulvirenti A, Ferro A, Alder H, He G, Vecchione A, Croce CM, Shapiro CL, Huebner K. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One. 2013; 8:e55910. 10.1371/journal.pone.0055910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen X, Wang YW, Zhu WJ, Li Y, Liu L, Yin G, Gao P. A 4-microRNA signature predicts lymph node metastasis and prognosis in breast cancer. Hum Pathol. 2018; 76:122–32. 10.1016/j.humpath.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 53. Grady WM, Tewari M. The next thing in prognostic molecular markers: microRNA signatures of cancer. Gut. 2010; 59:706–08. 10.1136/gut.2009.200022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Halabi S, Owzar K. The importance of identifying and validating prognostic factors in oncology. Semin Oncol. 2010; 37:e9–18. 10.1053/j.seminoncol.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. 2009; 6:e1000100. 10.1371/journal.pmed.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Elghoroury EA, ElDine HG, Kamel SA, Abdelrahman AH, Mohammed A, Kamel MM, Ibrahim MH. Evaluation of miRNA-21 and miRNA Let-7 as Prognostic Markers in Patients With Breast Cancer. Clin Breast Cancer. 2018; 18:e721–26. 10.1016/j.clbc.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 57. Turashvili G, Lightbody ED, Tyryshkin K, SenGupta SK, Elliott BE, Madarnas Y, Ghaffari A, Day A, Nicol CJ. Novel prognostic and predictive microRNA targets for triple-negative breast cancer. FASEB J. 2018; fj201800120R. 10.1096/fj.201800120R. [DOI] [PubMed] [Google Scholar]
- 58. Quesne JL, Jones J, Warren J, Dawson SJ, Ali HR, Bardwell H, Blows F, Pharoah P, Caldas C. Biological and prognostic associations of miR-205 and let-7b in breast cancer revealed by in situ hybridization analysis of micro-RNA expression in arrays of archival tumour tissue. J Pathol. 2012; 227:306–14. 10.1002/path.3983. [DOI] [PubMed] [Google Scholar]
- 59. Bailey ST, Westerling T, Brown M. Loss of estrogen-regulated microRNA expression increases HER2 signaling and is prognostic of poor outcome in luminal breast cancer. Cancer Res. 2015; 75:436–45. 10.1158/0008-5472.CAN-14-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Minemura H, Takagi K, Miki Y, Shibahara Y, Nakagawa S, Ebata A, Watanabe M, Ishida T, Sasano H, Suzuki T. Abnormal expression of miR-1 in breast carcinoma as a potent prognostic factor. Cancer Sci. 2015; 106:1642–50. 10.1111/cas.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Uhr K, Sieuwerts AM, de Weerd V, Smid M, Hammerl D, Foekens JA, Martens JW. Association of microRNA-7 and its binding partner CDR1-AS with the prognosis and prediction of 1st-line tamoxifen therapy in breast cancer. Sci Rep. 2018; 8:9657. 10.1038/s41598-018-27987-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jang MH, Kim HJ, Gwak JM, Chung YR, Park SY. Prognostic value of microRNA-9 and microRNA-155 expression in triple-negative breast cancer. Hum Pathol. 2017; 68:69–78. 10.1016/j.humpath.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 63. Cheng CW, Yu JC, Hsieh YH, Liao WL, Shieh JC, Yao CC, Lee HJ, Chen PM, Wu PE, Shen CY. Increased Cellular Levels of MicroRNA-9 and MicroRNA-221 Correlate with Cancer Stemness and Predict Poor Outcome in Human Breast Cancer. Cell Physiol Biochem. 2018; 48:2205–18. 10.1159/000492561. [DOI] [PubMed] [Google Scholar]
- 64. Parrella P, Barbano R, Pasculli B, Fontana A, Copetti M, Valori VM, Poeta ML, Perrone G, Righi D, Castelvetere M, Coco M, Balsamo T, Morritti M, et al. Evaluation of microRNA-10b prognostic significance in a prospective cohort of breast cancer patients. Mol Cancer. 2014; 13:142. 10.1186/1476-4598-13-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shinden Y, Akiyoshi S, Ueo H, Nambara S, Saito T, Komatsu H, Ueda M, Hirata H, Sakimura S, Uchi R, Takano Y, Iguchi T, Eguchi H, et al. Diminished expression of MiR-15a is an independent prognostic marker for breast cancer cases. Anticancer Res. 2015; 35:123–27. [PubMed] [Google Scholar]
- 66. Anfossi S, Giordano A, Gao H, Cohen EN, Tin S, Wu Q, Garza RJ, Debeb BG, Alvarez RH, Valero V, Hortobagyi GN, Calin GA, Ueno NT, et al. High serum miR-19a levels are associated with inflammatory breast cancer and are predictive of favorable clinical outcome in patients with metastatic HER2+ inflammatory breast cancer. PLoS One. 2014; 9:e83113. 10.1371/journal.pone.0083113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li C, Zhang J, Ma Z, Zhang F, Yu W. miR-19b serves as a prognostic biomarker of breast cancer and promotes tumor progression through PI3K/AKT signaling pathway. Onco Targets Ther. 2018; 11:4087–95. 10.2147/OTT.S171043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Halvorsen AR, Helland Å, Gromov P, Wielenga VT, Talman MM, Brunner N, Sandhu V, Børresen-Dale AL, Gromova I, Haakensen VD. Profiling of microRNAs in tumor interstitial fluid of breast tumors - a novel resource to identify biomarkers for prognostic classification and detection of cancer. Mol Oncol. 2017; 11:220–34. 10.1002/1878-0261.12025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pandey AK, Zhang Y, Zhang S, Li Y, Tucker-Kellogg G, Yang H, Jha S. TIP60-miR-22 axis as a prognostic marker of breast cancer progression. Oncotarget. 2015; 6:41290–306. 10.18632/oncotarget.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen B, Tang H, Liu X, Liu P, Yang L, Xie X, Ye F, Song C, Xie X, Wei W. miR-22 as a prognostic factor targets glucose transporter protein type 1 in breast cancer. Cancer Lett. 2015; 356:410–17. 10.1016/j.canlet.2014.09.028. [DOI] [PubMed] [Google Scholar]
- 71. Martin EC, Elliott S, Rhodes LV, Antoon JW, Fewell C, Zhu Y, Driver JL, Jodari-Karimi M, Taylor CW, Flemington EK, Beckman BS, Collins-Burow BM, Burow ME. Preferential star strand biogenesis of pre-miR-24-2 targets PKC-alpha and suppresses cell survival in MCF-7 breast cancer cells. Mol Carcinog. 2014; 53:38–48. 10.1002/mc.21946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Khodadadi-Jamayran A, Akgol-Oksuz B, Afanasyeva Y, Heguy A, Thompson M, Ray K, Giro-Perafita A, Sánchez I, Wu X, Tripathy D, Zeleniuch-Jacquotte A, Tsirigos A, Esteva FJ. Prognostic role of elevated mir-24-3p in breast cancer and its association with the metastatic process. Oncotarget. 2018; 9:12868–78. 10.18632/oncotarget.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tang W, Zhu J, Su S, Wu W, Liu Q, Su F, Yu F. MiR-27 as a prognostic marker for breast cancer progression and patient survival. PLoS One. 2012; 7:e51702. 10.1371/journal.pone.0051702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shen S, Sun Q, Liang Z, Cui X, Ren X, Chen H, Zhang X, Zhou Y. A prognostic model of triple-negative breast cancer based on miR-27b-3p and node status. PLoS One. 2014; 9:e100664. 10.1371/journal.pone.0100664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen X, Lu P, Wang DD, Yang SJ, Wu Y, Shen HY, Zhong SL, Zhao JH, Tang JH. The role of miRNAs in drug resistance and prognosis of breast cancer formalin-fixed paraffin-embedded tissues. Gene. 2016; 595:221–26. 10.1016/j.gene.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 76. Shinden Y, Iguchi T, Akiyoshi S, Ueo H, Ueda M, Hirata H, Sakimura S, Uchi R, Takano Y, Eguchi H, Sugimachi K, Kijima Y, Natsugoe S, Mimori K. miR-29b is an indicator of prognosis in breast cancer patients. Mol Clin Oncol. 2015; 3:919–23. 10.3892/mco.2015.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Papachristopoulou G, Papadopoulos EI, Nonni A, Rassidakis GZ, Scorilas A. Expression Analysis of miR-29b in Malignant and Benign Breast Tumors: A Promising Prognostic Biomarker for Invasive Ductal Carcinoma With a Possible Histotype-Related Expression Status. Clin Breast Cancer. 2018; 18:305–312.e3. 10.1016/j.clbc.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 78. Cheng CW, Wang HW, Chang CW, Chu HW, Chen CY, Yu JC, Chao JI, Liu HF, Ding SL, Shen CY. MicroRNA-30a inhibits cell migration and invasion by downregulating vimentin expression and is a potential prognostic marker in breast cancer. Breast Cancer Res Treat. 2012; 134:1081–93. 10.1007/s10549-012-2034-4. [DOI] [PubMed] [Google Scholar]
- 79. D’Aiuto F, Callari M, Dugo M, Merlino G, Musella V, Miodini P, Paolini B, Cappelletti V, Daidone MG. miR-30e* is an independent subtype-specific prognostic marker in breast cancer. Br J Cancer. 2015; 113:290–98. 10.1038/bjc.2015.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Peurala H, Greco D, Heikkinen T, Kaur S, Bartkova J, Jamshidi M, Aittomäki K, Heikkilä P, Bartek J, Blomqvist C, Bützow R, Nevanlinna H. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer. PLoS One. 2011; 6:e26122. 10.1371/journal.pone.0026122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zeng Z, Chen X, Zhu D, Luo Z, Yang M. Low Expression of Circulating MicroRNA-34c is Associated with Poor Prognosis in Triple-Negative Breast Cancer. Yonsei Med J. 2017; 58:697–702. 10.3349/ymj.2017.58.4.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Svoboda M, Sana J, Redova M, Navratil J, Palacova M, Fabian P, Slaby O, Vyzula R. MiR-34b is associated with clinical outcome in triple-negative breast cancer patients. Diagn Pathol. 2012; 7:31. 10.1186/1746-1596-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Li J, Song ZJ, Wang YY, Yin Y, Liu Y, Nan X. Low levels of serum miR-99a is a predictor of poor prognosis in breast cancer. Genet Mol Res. 2016; 15. 10.4238/gmr.15038338. [DOI] [PubMed] [Google Scholar]
- 84. Wu X, Somlo G, Yu Y, Palomares MR, Li AX, Zhou W, Chow A, Yen Y, Rossi JJ, Gao H, Wang J, Yuan YC, Frankel P, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 2012; 10:42. 10.1186/1479-5876-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dong LL, Chen LM, Wang WM, Zhang LM. Decreased expression of microRNA-124 is an independent unfavorable prognostic factor for patients with breast cancer. Diagn Pathol. 2015; 10:45. 10.1186/s13000-015-0257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Oltra SS, Peña-Chilet M, Vidal-Tomas V, Flower K, Martinez MT, Alonso E, Burgues O, Lluch A, Flanagan JM, Ribas G. Methylation deregulation of miRNA promoters identifies miR124-2 as a survival biomarker in Breast Cancer in very young women. Sci Rep. 2018; 8:14373. 10.1038/s41598-018-32393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hsieh TH, Hsu CY, Tsai CF, Long CY, Chai CY, Hou MF, Lee JN, Wu DC, Wang SC, Tsai EM. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget. 2015; 6:494–509. 10.18632/oncotarget.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Luo Y, Wang X, Niu W, Wang H, Wen Q, Fan S, Zhao R, Li Z, Xiong W, Peng S, Zeng Z, Li X, Li G, et al. Elevated microRNA-125b levels predict a worse prognosis in HER2-positive breast cancer patients. Oncol Lett. 2017; 13:867–74. 10.3892/ol.2016.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Vilquin P, Donini CF, Villedieu M, Grisard E, Corbo L, Bachelot T, Vendrell JA, Cohen PA. MicroRNA-125b upregulation confers aromatase inhibitor resistance and is a novel marker of poor prognosis in breast cancer. Breast Cancer Res. 2015; 17:13. 10.1186/s13058-015-0515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang S, Li H, Wang J, Wang D, Yao A, Li Q. Prognostic and biological significance of microRNA-127 expression in human breast cancer. Dis Markers. 2014; 2014:401986. 10.1155/2014/401986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yu Y, Zhao Y, Sun XH, Ge J, Zhang B, Wang X, Cao XC. Down-regulation of miR-129-5p via the Twist1-Snail feedback loop stimulates the epithelial-mesenchymal transition and is associated with poor prognosis in breast cancer. Oncotarget. 2015; 6:34423–36. 10.18632/oncotarget.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wu ZS, Wang CQ, Xiang R, Liu X, Ye S, Yang XQ, Zhang GH, Xu XC, Zhu T, Wu Q. Loss of miR-133a expression associated with poor survival of breast cancer and restoration of miR-133a expression inhibited breast cancer cell growth and invasion. BMC Cancer. 2012; 12:51. 10.1186/1471-2407-12-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Liu HT, Xu YT, Li HY, Zhao J, Zhai HY, Chen Y. Loss of microRNA-145 expression is involved in the development and prognosis of breast cancer complicated by type 2 diabetes mellitus. Int J Biol Markers. 2016; 31:e368–74. 10.5301/jbm.5000220. [DOI] [PubMed] [Google Scholar]
- 94. Quan Y, Huang X, Quan X. Expression of miRNA-206 and miRNA-145 in breast cancer and correlation with prognosis. Oncol Lett. 2018; 16:6638–42. 10.3892/ol.2018.9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zavala V, Pérez-Moreno E, Tapia T, Camus M, Carvallo P. miR-146a and miR-638 in BRCA1-deficient triple negative breast cancer tumors, as potential biomarkers for improved overall survival. Cancer Biomark. 2016; 16:99–107. 10.3233/CBM-150545. [DOI] [PubMed] [Google Scholar]
- 96. Xu X, Zhang Y, Jasper J, Lykken E, Alexander PB, Markowitz GJ, McDonnell DP, Li QJ, Wang XF. MiR-148a functions to suppress metastasis and serves as a prognostic indicator in triple-negative breast cancer. Oncotarget. 2016; 7:20381–94. 10.18632/oncotarget.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, Cheng JQ. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014; 33:679–89. 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ali OS, Shabayek MI, Seleem MM, Abdelaziz HG, Makhlouf DO. MicroRNAs 182 and 375 Sera Expression as Prognostic Biochemical Markers in Breast Cancer. Clin Breast Cancer. 2018; 18:e1373–79. 10.1016/j.clbc.2018.07.020. [DOI] [PubMed] [Google Scholar]
- 99. Song C, Zhang L, Wang J, Huang Z, Li X, Wu M, Li S, Tang H, Xie X. High expression of microRNA-183/182/96 cluster as a prognostic biomarker for breast cancer. Sci Rep. 2016; 6:24502. 10.1038/srep24502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mulrane L, Madden SF, Brennan DJ, Gremel G, McGee SF, McNally S, Martin F, Crown JP, Jirström K, Higgins DG, Gallagher WM, O’Connor DP. miR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin Cancer Res. 2012; 18:6702–13. 10.1158/1078-0432.CCR-12-1420. [DOI] [PubMed] [Google Scholar]
- 101. Fang C, Wang FB, Li Y, Zeng XT. Down-regulation of miR-199b-5p is correlated with poor prognosis for breast cancer patients. Biomed Pharmacother. 2016; 84:1189–93. 10.1016/j.biopha.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 102. Tuomarila M, Luostari K, Soini Y, Kataja V, Kosma VM, Mannermaa A. Overexpression of microRNA-200c predicts poor outcome in patients with PR-negative breast cancer. PLoS One. 2014; 9:e109508. 10.1371/journal.pone.0109508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Jin T, Suk Kim H, Ki Choi S, Hye Hwang E, Woo J, Suk Ryu H, Kim K, Moon A, Kyung Moon W. microRNA-200c/141 upregulates SerpinB2 to promote breast cancer cell metastasis and reduce patient survival. Oncotarget. 2017; 8:32769–82. 10.18632/oncotarget.15680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Gomes BC, Martins M, Lopes P, Morujão I, Oliveira M, Araújo A, Rueff J, Rodrigues AS. Prognostic value of microRNA-203a expression in breast cancer. Oncol Rep. 2016; 36:1748–56. 10.3892/or.2016.4913. [DOI] [PubMed] [Google Scholar]
- 105. Li W, Jin X, Zhang Q, Zhang G, Deng X, Ma L. Decreased expression of miR-204 is associated with poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 2014; 7:3287–92. [PMC free article] [PubMed] [Google Scholar]
- 106. Li Y, Hong F, Yu Z. Decreased expression of microRNA-206 in breast cancer and its association with disease characteristics and patient survival. J Int Med Res. 2013; 41:596–602. 10.1177/0300060513485856. [DOI] [PubMed] [Google Scholar]
- 107. Hesari Z, Nourbakhsh M, Hosseinkhani S, Abdolvahabi Z, Alipour M, Tavakoli-Yaraki M, Ghorbanhosseini SS, Yousefi Z, Jafarzadeh M, Yarahmadi S. Down-regulation of NAMPT expression by mir-206 reduces cell survival of breast cancer cells. Gene. 2018; 673:149–58. 10.1016/j.gene.2018.06.021. [DOI] [PubMed] [Google Scholar]
- 108. Ahmadinejad F, Mowla SJ, Honardoost MA, Arjenaki MG, Moazeni-Bistgani M, Kheiri S, Teimori H. Lower expression of miR-218 in human breast cancer is associated with lymph node metastases, higher grades, and poorer prognosis. Tumour Biol. 2017; 39:1010428317698362. 10.1177/1010428317698362. [DOI] [PubMed] [Google Scholar]
- 109. Falkenberg N, Anastasov N, Rappl K, Braselmann H, Auer G, Walch A, Huber M, Höfig I, Schmitt M, Höfler H, Atkinson MJ, Aubele M. MiR-221/-222 differentiate prognostic groups in advanced breast cancers and influence cell invasion. Br J Cancer. 2013; 109:2714–23. 10.1038/bjc.2013.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Deng L, Lei Q, Wang Y, Wang Z, Xie G, Zhong X, Wang Y, Chen N, Qiu Y, Pu T, Bu H, Zheng H. Downregulation of miR-221-3p and upregulation of its target gene PARP1 are prognostic biomarkers for triple negative breast cancer patients and associated with poor prognosis. Oncotarget. 2017; 8:108712–25. 10.18632/oncotarget.21561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wang Y, Yin W, Lin Y, Yin K, Zhou L, Du Y, Yan T, Lu J. Downregulated circulating microRNAs after surgery: potential noninvasive biomarkers for diagnosis and prognosis of early breast cancer. Cell Death Discov. 2018; 4:21. 10.1038/s41420-018-0089-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yu H, Li H, Qian H, Jiao X, Zhu X, Jiang X, Dai G, Huang J. Upregulation of miR-301a correlates with poor prognosis in triple-negative breast cancer. Med Oncol. 2014; 31:283. 10.1007/s12032-014-0283-2. [DOI] [PubMed] [Google Scholar]
- 113. Zheng JZ, Huang YN, Yao L, Liu YR, Liu S, Hu X, Liu ZB, Shao ZM. Elevated miR-301a expression indicates a poor prognosis for breast cancer patients. Sci Rep. 2018; 8:2225. 10.1038/s41598-018-20680-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yang H, Yu J, Wang L, Ding D, Zhang L, Chu C, Chen Q, Xu Z, Zou Q, Liu X. miR-320a is an independent prognostic biomarker for invasive breast cancer. Oncol Lett. 2014; 8:1043–50. 10.3892/ol.2014.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Li P, Dong J, Zhou X, Sun W, Huang H, Chen T, Ye B, Zheng Z, Lu M. Expression patterns of microRNA-329 and its clinical performance in diagnosis and prognosis of breast cancer. Onco Targets Ther. 2017; 10:5711–18. 10.2147/OTT.S147974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wang H, Chen SH, Kong P, Zhang LY, Zhang LL, Zhang NQ, Gu H. Increased expression of miR-330-3p: a novel independent indicator of poor prognosis in human breast cancer. Eur Rev Med Pharmacol Sci. 2018; 22:1726–30. 10.26355/eurrev_201803_14587. [DOI] [PubMed] [Google Scholar]
- 117. Wu ZS, Wu Q, Wang CQ, Wang XN, Wang Y, Zhao JJ, Mao SS, Zhang GH, Zhang N, Xu XC. MiR-339-5p inhibits breast cancer cell migration and invasion in vitro and may be a potential biomarker for breast cancer prognosis. BMC Cancer. 2010; 10:542. 10.1186/1471-2407-10-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Cao ZG, Huang YN, Yao L, Liu YR, Hu X, Hou YF, Shao ZM. Positive expression of miR-361-5p indicates better prognosis for breast cancer patients. J Thorac Dis. 2016; 8:1772–79. 10.21037/jtd.2016.06.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sim J, Ahn H, Abdul R, Kim H, Yi KJ, Chung YM, Chung MS, Paik SS, Song YS, Jang K. High MicroRNA-370 Expression Correlates with Tumor Progression and Poor Prognosis in Breast Cancer. J Breast Cancer. 2015; 18:323–28. 10.4048/jbc.2015.18.4.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Li JY, Zhang Y, Zhang WH, Jia S, Kang Y, Tian R. Effects of differential distribution of microvessel density, possibly regulated by miR-374a, on breast cancer prognosis. Asian Pac J Cancer Prev. 2013; 14:1715–20. 10.7314/APJCP.2013.14.3.1715. [DOI] [PubMed] [Google Scholar]
- 121. Zhang J, He Y, Yu Y, Chen X, Cui G, Wang W, Zhang X, Luo Y, Li J, Ren F, Ren Z, Sun R. Upregulation of miR-374a promotes tumor metastasis and progression by downregulating LACTB and predicts unfavorable prognosis in breast cancer. Cancer Med. 2018; 7:3351–62. 10.1002/cam4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cao GH, Sun XL, Wu F, Chen WF, Li JQ, Hu WC. Low expression of miR-409-3p is a prognostic marker for breast cancer. Eur Rev Med Pharmacol Sci. 2016; 20:3825–29. [PubMed] [Google Scholar]
- 123. Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene. 2012; 31:39–47. 10.1038/onc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Cao ZG, Li JJ, Yao L, Huang YN, Liu YR, Hu X, Song CG, Shao ZM. High expression of microRNA-454 is associated with poor prognosis in triple-negative breast cancer. Oncotarget. 2016; 7:64900–09. 10.18632/oncotarget.11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Yao L, Liu Y, Cao Z, Li J, Huang Y, Hu X, Shao Z. MicroRNA-493 is a prognostic factor in triple-negative breast cancer. Cancer Sci. 2018; 109:2294–301. 10.1111/cas.13644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Gurvits N, Autere TA, Repo H, Nykänen M, Kuopio T, Kronqvist P, Talvinen K. Proliferation-associated miRNAs-494, -205, -21 and -126 detected by in situ hybridization: expression and prognostic potential in breast carcinoma patients. J Cancer Res Clin Oncol. 2018; 144:657–66. 10.1007/s00432-018-2586-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Liu J, Zhou Y, Shi Z, Hu Y, Meng T, Zhang X, Zhang S, Zhang J. microRNA-497 Modulates Breast Cancer Cell Proliferation, Invasion, and Survival by Targeting SMAD7. DNA Cell Biol. 2016; 35:521–29. 10.1089/dna.2016.3282. [DOI] [PubMed] [Google Scholar]
- 128. Zhong H, Yang J, Zhang B, Wang X, Pei L, Zhang L, Lin Z, Wang Y, Wang C. LncRNA GACAT3 predicts poor prognosis and promotes cell proliferation in breast cancer through regulation of miR-497/CCND2. Cancer Biomark. 2018; 22:787–97. 10.3233/CBM-181354. [DOI] [PubMed] [Google Scholar]
- 129. Krishnan P, Ghosh S, Wang B, Li D, Narasimhan A, Berendt R, Graham K, Mackey JR, Kovalchuk O, Damaraju S. Next generation sequencing profiling identifies miR-574-3p and miR-660-5p as potential novel prognostic markers for breast cancer. BMC Genomics. 2015; 16:735. 10.1186/s12864-015-1899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Yu M, Zhang X, Li H, Zhang P, Dong W. MicroRNA-588 is downregulated and may have prognostic and functional roles in human breast cancer. Med Sci Monit. 2017; 23:5690–96. 10.12659/MSM.905126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Abdolvahabi Z, Nourbakhsh M, Hosseinkhani S, Hesari Z, Alipour M, Jafarzadeh M, Ghorbanhosseini SS, Seiri P, Yousefi Z, Yarahmadi S, Golpour P. MicroRNA-590-3P suppresses cell survival and triggers breast cancer cell apoptosis via targeting sirtuin-1 and deacetylation of p53. J Cell Biochem. 2019; 120:9356–68. 10.1002/jcb.28211. [DOI] [PubMed] [Google Scholar]
- 132. Zhang XY, Liu DJ, Yuan RB, Zhang DH, Li SR, Zhang SH, Zhang LY. Low expression of miR-597 is correlated with tumor stage and poor outcome in breast cancer. Eur Rev Med Pharmacol Sci. 2018; 22:456–60. 10.26355/eurrev_201801_14195. [DOI] [PubMed] [Google Scholar]
- 133. Hu JY, Yi W, Wei X, Zhang MY, Xu R, Zeng LS, Huang ZJ, Chen JS. miR-601 is a prognostic marker and suppresses cell growth and invasion by targeting PTP4A1 in breast cancer. Biomed Pharmacother. 2016; 79:247–53. 10.1016/j.biopha.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 134. Li M, Wang J, Liu H. Downregulation of miR-638 promotes progression of breast cancer and is associated with prognosis of breast cancer patients. Onco Targets Ther. 2018; 11:6871–77. 10.2147/OTT.S182034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Raza U, Saatci Ö, Uhlmann S, Ansari SA, Eyüpoğlu E, Yurdusev E, Mutlu M, Ersan PG, Altundağ MK, Zhang JD, Doğan HT, Güler G. Şahin Ö. The miR-644a/CTBP1/p53 axis suppresses drug resistance by simultaneous inhibition of cell survival and epithelial-mesenchymal transition in breast cancer. Oncotarget. 2016; 7:49859–77. 10.18632/oncotarget.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hu JY, Yi W, Zhang MY, Xu R, Zeng LS, Long XR, Zhou XM, Zheng XS, Kang Y, Wang HY. MicroRNA-711 is a prognostic factor for poor overall survival and has an oncogenic role in breast cancer. Oncol Lett. 2016; 11:2155–63. 10.3892/ol.2016.4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zhang L, Yan DL, Yang F, Wang DD, Chen X, Wu JZ, Tang JH, Xia WJ. DNA methylation mediated silencing of microRNA-874 is a promising diagnosis and prognostic marker in breast cancer. Oncotarget. 2017; 8:45496–505. 10.18632/oncotarget.17569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Liu W, Xu Y, Guan H, Meng H. Clinical potential of miR-940 as a diagnostic and prognostic biomarker in breast cancer patients. Cancer Biomark. 2018; 22:487–93. 10.3233/CBM-171124. [DOI] [PubMed] [Google Scholar]
- 139. Li WJ, Xie XX, Bai J, Wang C, Zhao L, Jiang DQ. Increased expression of miR-1179 inhibits breast cancer cell metastasis by modulating Notch signaling pathway and correlates with favorable prognosis. Eur Rev Med Pharmacol Sci. 2018; 22:8374–82. 10.26355/eurrev_201812_16535. [DOI] [PubMed] [Google Scholar]
- 140. Zeng B, Li Y, Feng Y, Lu M, Yuan H, Yi Z, Wu Y, Xiang T, Li H, Ren G. Downregulated miR-1247-5p associates with poor prognosis and facilitates tumor cell growth via DVL1/Wnt/β-catenin signaling in breast cancer. Biochem Biophys Res Commun. 2018; 505:302–08. 10.1016/j.bbrc.2018.09.103. [DOI] [PubMed] [Google Scholar]
- 141. Zhang P, Fan C, Du J, Mo X, Zhao Q. Association of miR-1247-5p expression with clinicopathological parameters and prognosis in breast cancer. Int J Exp Pathol. 2018; 99:199–205. 10.1111/iep.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Zhong X, Xie G, Zhang Z, Wang Z, Wang Y, Wang Y, Qiu Y, Li L, Bu H, Li J, Zheng H. MiR-4653-3p and its target gene FRS2 are prognostic biomarkers for hormone receptor positive breast cancer patients receiving tamoxifen as adjuvant endocrine therapy. Oncotarget. 2016; 7:61166–82. 10.18632/oncotarget.11278. [DOI] [PMC free article] [PubMed] [Google Scholar]