Abstract
Objective:
This study aimed to develop a gene-expression signature for identification of lymph node (LN) metastasis in esophageal squamous cell carcinoma (ESCC) patients.
Summary of Background Data:
LN metastasis is recognized as the most important independent risk factor for therapeutic decision-making of ESCC patients.
Methods:
A bioinformatic approach was used to analyze RNA sequencing profiles of ESCC patients, and to develop a gene-expression signature for identifying LN metastasis. The robustness of this panel was assessed in 2 independent patient cohorts (n = 56 and 224).
Results:
We initially prioritized a 16-gene signature out of the total 20,531 mRNAs. The model estimated by these 16 genes discriminated LN status with an area under the curve (AUC) of 0.77 [95% confidence interval (95% CI), 0.68–0.87, 5-fold cross-validation]. Subsequently, a reduced and optimized 5-gene panel was trained in a clinical cohort, which effectively distinguished ESCC patients with LN metastasis (cohort-1: AUC, 0.74; 95% CI, 0.58–0.89; cohort-2, T1–T2: AUC, 0.74; 95% CI, 0.63–0.86), and was significantly superior to preoperative computed tomography (AUC, 0.61; 95% CI, 0.50–0.72). Furthermore, a combination signature comprising of the 5-gene panel together with the lymphatic vessel invasion (LVI) and venous invasion (VI) demonstrated a significantly improved diagnostic performance compared with individual clinical variables, in both cohorts (cohort-1: AUC, 0.87; 95% CI, 0.78–0.96; cohort-2: AUC, 0.76; 95% CI, 0.65–0.88).
Conclusion:
Our novel 5-gene panel is a robust diagnostic tool for LN metastasis, especially in early-T stage ESCC patients, with a promising clinical potential.
Keywords: biomarkers, esophageal squamous cell carcinoma, gene expression, lymph node
Esophageal cancer (EC) is one of the most aggressive malignancies worldwide and is the sixth leading cause of cancer-related mortality with an estimated 400,000 deaths in 2012.1 ECs consist of 2 main histological subtypes, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). Among these, ESCC accounts for 80% of all ECs,2 and is highly prevalent in eastern Asia, as well as eastern and southern Africa.3 Currently, ESCC diagnosis is achieved by endoscopy, combined with esophagography, endoscopic ultrasonography, computed tomography (CT), and 18F-fluorodeoxyglucose positron emission tomography (PET). These modalities are also used to evaluate the clinical stage of ESCC patients.4 However, unfortunately, these procedures are inadequate for evaluating the lymph node (LN) status of ESCC patients.5–7
Because esophagus is anatomically juxtaposed to the cardiopulmonary structures, ESCC LN metastasis results in much poorer prognosis than other cancer types.8 Therefore, LN metastasis is recognized as the most critical independent risk factor for ESCC prognosis.9 Due to the fact that lymphatic flow through the esophagus submucosal layer is abundant and multidirectional,8 ESCC LN metastasis often spreads to nodes relatively far from the primary tumor site. For example, recurrent laryngeal nerve chain nodes, especially those on the right side, can be found involved in some ESCC patients, even though their primary tumors are still confined within the submucosal layer.10 Furthermore, currently, esophagectomy with adequate LN dissection is one of the most effective surgical treatments for patients with advanced ESCC without distant metastasis.11–13 However, radical LN dissection increases postoperative morbidity and mortality, and is particularly unsuitable for elderly patients and those with cardiopulmonary diseases.14,15 Therefore, availability of a robust diagnostic marker for LN metastasis will significantly improve treatment modalities for patients before surgery and may circumvent unnecessary excessive LN dissection, which often results in severe complications.
Although several molecular biomarkers for LN metastasis have been identified, the majority of these markers have limited detection potential.16,17 Herein, we performed a systematic and comprehensive analysis of the molecular characteristics of LN status, and subsequently developed a novel gene-expression signature for the detection of ESCC LN metastasis. The robustness of the signature was initially verified in a publicly available dataset, followed by comprehensive validation in 2 independent clinical cohorts.
METHODS
Public Dataset and Gene-expression Signature Identification
The entire workflow for the discovery of signature genes and the development of a gene panel is shown in supplementary figure S1, http://links.lww.com/SLA/B355. To identify a gene-expression signature for detection of ESCC LN metastasis, we first investigated genome-wide gene-expression profiles of primary tumors, with or without LN metastasis from The Cancer Genome Atlas (TCGA). In total, RNA-sequencing data from 96 patients, of whom 54 were LN metastasis negative (LN−) and 38 LN metastasis positive (LN+), were used for analysis. The pathological T staging numbers for these cases were T1 (LN−, 3; LN+, 5)T2(LN−, 23; LN+, 8), T3 (LN−, 25; LN+, 24), and T4 (LN−, 3; LN+, 1)The. TCGA gene expression data (level 3 RNA-Seq data) for ESCC were downloaded from Firehose Broad GDAC portal (http://gdac.broadinstitute.org/, accessed on December 3, 2016). Scaled estimates in the gene-level RSEM files were first converted to TPM (transcripts per million) by multiplying with 106 and thereafter log2-transformed.
For each of the 20,531 gene transcripts included in the TCGA ESCC RNA-Seq dataset, we performed a Wilcoxon signed-rank test for determining differential gene expression between LN− and LN+ groups, and calculated the log2 fold change and average expression level as additional indicators for prioritizing potential candidates. To evaluate the diagnostic power of the gene panel, we first built a multivariate logistic regression model using selected gene features, and subsequently calculated the area under the curve (AUC) values for each of the receiver operator characteristic (ROC) plots. The robustness of this gene panel was further assessed by a 5-fold cross-validation (repeated 10,000 times). Risk scores were subsequently calculated according to the probability of each patient to be identified as LN+.
In-house Clinical Cohorts
For clinical training and validation of identified gene markers, we enrolled 2 independent patient cohorts totaling 267 cases (supplementary figure S2, http://links.lww.com/SLA/B355). Fifty-six surgically resected ESCC specimens were collected from the National Cancer Center Hospital, Tokyo, Japan (cohort-1) between December 2004 and June 2006. As a second independent validation cohort of early T stage cases, among 224 ESCC cases surgically resected at the Nagoya University Hospital, Nagoya, Japan, between October 2001 and October 2015, 75 consecutively enrolled ESCC patients who were pathologically diagnosed as T1–T2 only were included in this study (cohort-2). Patients were treated with preoperative or postoperative systemic chemotherapy with or without radiotherapy according to the surgeons’ discretion. All procedures associated with the study were approved by the Institutional Review Boards of all participating institutions, and all patients provided written informed consent.
Patient characteristics are summarized in Table 1. The median follow-up duration for all cases after surgery was 32 months (ranging from 0.5 to 127 months). All tumor tissue specimens were histologically confirmed for ESCC diagnosis, and were classified by tumor-node-metastasis (TNM) stages before and after surgery, according to the American Joint Committee on Cancer staging handbook, 7th edition.18
TABLE 1.
Characteristics of Patients and Their Tumors*
| Cohort-1 (n = 56) | Cohort-2 (n = 75) | |||||
|---|---|---|---|---|---|---|
| LN− (n = 13) | LN+ (n = 43) | P‡ | LN− (n = 38) | LN± (n = 37) | P‡ | |
| Age | NA | NA | NA | 67±1.3 | 65±1.5 | 0.31 |
| Sex | > 0.99 | 0.07 | ||||
| Male | 11 | 37 | 30 | 22 | ||
| Female | 2 | 6 | 8 | 15 | ||
| Location | 0.39 | 0.30 | ||||
| Cervical | 0 | 0 | 5 | 1 | ||
| Upper thoracic | 2 | 5 | 3 | 5 | ||
| Middle thoracic | 8 | 19 | 22 | 25 | ||
| Lower thoracic | 3 | 19 | 8 | 6 | ||
| T stage | 0.26 | NA | ||||
| 1–2 | 4 | 7 | 38 | 37 | ||
| 3–4 | 9 | 36 | 0 | 0 | ||
| Tumor size, cm | 6.3±1.0 | 6.5±0.3 | 0.77 | 4.2±0.3 | 3.8±0.4 | 0.44 |
| SCC, ng/mL† | NA | NA | NA | 1.3±0.2 | 1.5 ±0.4 | 0.54 |
| Differentiation | 0.51 | 0.15 | ||||
| Poor | 3 | 15 | 2 | 6 | ||
| Others | 10 | 28 | 36 | 31 | ||
| Examined LN | 56±3.8 | 56±2.5 | 0.95 | 43±3.4 | 45±2.8 | 0.71 |
| LV invasion | 0.05 | 0.04 | ||||
| Positive | 4 | 28 | 17 | 26 | ||
| Negative | 9 | 15 | 20 | 11 | ||
| Venous invasion | 0.004 | 0.07 | ||||
| Positive | 2 | 27 | 7 | 14 | ||
| Negative | 11 | 16 | 30 | 23 | ||
LN− indicates lymph node metastasis negative; LN+, lymph node metastasis positive; LV, lymphatic vessel; NA, not available.
Plus-minus values are means ± standard error of the mean.
Cutoff value is 1.5 ng/mL.
P value is derived from t test, Chi-square test, or Fisher exact test.
RNA Extraction and Quantitative PCR Analysis
Total RNA was extracted from tissue specimens using the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Hilden, Germany), and quantitative PCR (qPCR) assays were performed using SensiFAST SYBR (Bioline, London, UK). The relative expression of target gene was normalized against glyceraldehyde 3-phosphate dehydrogenase using the comparative Ct method.19 The PCR primers used in the current study are shown in supplementary table S1, http://links.lww.com/SLA/B355.
Statistical Analyses
Continuous variables were expressed as mean ± standard error of the mean and were compared using t tests. Categorical variables were compared using the χ2 or Fisher exact tests, as appropriate. In the discovery phase, expression of each gene was analyzed by a Wilcoxon signed-rank test and logistic regression analysis was performed to build a multivariate scoring model. The method of DeLong et al20 was employed to test the statistical significance of the difference between ROC curves. In validation phase, the missing value was imputed using k-nearest neighbors (k = 10) inside each cohort. Overall survival (OS) rates were estimated using the Kaplan-Meier method and compared using a log-rank test. We adhere to the STARD guidelines to report this study.21 All statistical analyses were performed using R (version 3.3.3; http//www.r-project.org/) or Med-Calc version 12.3 (MedCalc Software, Ostend, Belgium). Statistical significance was set at P < 0.05, which was obtained using 2-tailed tests.
RESULTS
Identification of a Gene Expression Signature for Detecting LN Metastasis Using a High-throughput RNA Sequencing-Based Gene Expression Profiling
To develop a gene expression signature for identification of LN metastasis, we first interrogated TCGA dataset with genome-wide RNA sequencing-based gene expression profiling results for 96 ESCC patients. Among all, 92 patients had clinical data available for LN status (54 LN− and 38 LN+ cases), and were selected for the subsequent analysis. Of the total 20,531 annotated genes, 104 were differentially expressed (absolute log2 fold change > 0.5, corresponding P < 0.01, Wilcoxon signed-rank test) (supplementary figure S3, http://links.lww.com/SLA/B355). To reduce the gene signature for a clinically viable and practical assay, we filtered out all genes with low expression levels (average expression level < 3), which resulted in a panel of 26 genes. To eliminate any redundancy and genes with overlapping diagnostic potential, we used statistical approaches that further eliminated 10 genes (absolute pair-wise Pearson correlation coefficients > 0.5), yielding a final 16-gene panel. A heatmap of the identified 16-gene signature and the risk score curve based on a repeated cross-validation are shown in Fig. 1A. The detective power of each of the 16 genes for stratifying LN status of ESCC patients is shown in supplementary table S2, http://links.lww.com/SLA/B355, with AUC values ranging from 0.66 to 0.72 (supplementary figure S4, http://links.lww.com/SLA/B355). Using risk scores based on the detection of multivariate logistic regression with a 5-fold cross-validation (repeated for 10,000 times, and 90% sensitivity as threshold in the training set), we evaluated the performance of the entire 16-gene panel [AUC = 0.77, 95% confidence interval (95% CI) = 0.68–0.87] (Fig. 1B). Compared with other clinical factors such as age (AUC = 0.60, 95% CI = 0.48–0.71), sex (AUC = 0.54, 95% CI = 0.46–0.61), and the anatomical location of the primary tumor (AUC = 0.53, 95% CI = 0.41–0.64), our trained model achieved a superior diagnostic power (all P < 0.01, compared with each factor) (Fig. 1B). Furthermore, stratified analysis confirmed that the diagnostic value of the 16-gene panel emerged as an independent indicator for LN metastasis compared with other clinical factors (supplementary table S3, http://links.lww.com/SLA/B355).
FIGURE 1.
Discovered gene expression signature for lymph node metastasis identification in esophageal squamous cell carcinoma. A, A heatmap of the identified 16-gene signature and the risk score curve generated from the probability in the repeated cross-validation. B, Receiver operating characteristic (ROC) curves comparing the accuracy of the 16-gene signature, age, sex, and tumor location in the TCGA dataset. The ROC curve of the 16-gene panel was based on a 5-fold cross-validation on the TCGA dataset and the average area under the ROC curve (AUC) was 0.77 (95% confidence interval = 0.68–0.87). LN indicates lymph node.
Patient Characteristics of the Clinical Cohorts
A cohort of 131 histologically characterized ESCC cases were enrolled in this study and a total of 6456 LNs were examined (the average number of LNs examined per patient were 49). Among these, 66 cases (50%) were confirmed LN+ following pathological examination. The total number of positive LNs were 359 (5%) and the average number of positive LNs per LN+ patient were 5. The average examined LNs between LN− and LN+ cases did not differ in the both clinical cohorts (Table 1).
Biomarker Panel Training and Validation in Independent Clinical Cohorts
The 16-gene signature identified from the high-throughput data was first measured in the training cohort (cohort-1, 13 LN− and 43 LN+ cases). To prioritize the most robust gene signature, we next performed recursive feature elimination approach based on random forest classification with 5-fold cross-validation on the qPCR data (Fig. 2A). As a result, 5 genes (PLAC8, SLC12A8, CSPG4, TFPI, and TNFSF10) were selected (named “5-gene panel” hereafter). Multivariate logistic regression was subsequently used to build a scoring formula for quantifying the status of LN metastasis: logit (probability) = 4.17+(−0.17) × PLAC8+0.61 × SLC12A8+(−0.39) × CSPG4+0.44 × TFPI+(−0.31) × TNFSF10, and the AUC of the 5-gene panel was 0.74 (95% CI = 0.58–0.89), while the AUC for sex and the tumor location were 0.51 (95% CI = 0.39–0.62) and 0.63 (95% CI 0.46–0.79), respectively (Fig. 2B).
FIGURE 2.
Biomarker panel training and verification with clinical cohorts. A, To identify the most robust gene signature, a recursive feature elimination based on a 5-fold cross-validation of the random forest algorithm was performed on the qPCR data of training cohort [cohort-1, lymph node metastasis negative (LN−), n = 13; lymph node metastasis positive (LN+), n = 43]. With the selected top 5 genes (PLAC8, SLC12A8, CSPG4, TFPI, and TNFSF10), the average accuracy was 0.79. B, A receiver operating characteristic (ROC) curve of the 5-gene panel compared with the ROC curves of sex and tumor location in the cohort-1. C, A ROC curve of the 5-gene panel compared with the ROC curves of age, sex, and tumor location in the cohort-2 (LN−, n = 38; LN+, n = 37).
Next, we assessed the robustness of this 5-gene panel in an independent validation cohort (cohort-2, 38 LN− and 37 LN+ cases). Considering that ESCC without invasion to the adventitia is most likely to be resected completely by surgery and endoscopic therapies can be applied to Tis (carcinoma in situ or high-grade dysplasia) and T1a (tumor invading lamina propria or muscularis mucosae) cases without metastatic LNs, we deliberately evaluated the efficacy of the 5-gene panel in T1–T2 tumors. The AUC of the 5-gene panel in the cohort-2 was 0.74 (95% CI = 0.63–0.86) (Fig. 2C). Compared with other clinical factors such as age (AUC = 0.57, 95% CI = 0.44–0.71), sex (AUC = 0.60, 95% CI = 0.49–0.70), and the tumor location (AUC = 0.59, 95% CI = 0.48–0.70), the 5-gene panel achieved a significantly better detective power (P = 0.02, P = 0.03, and P = 0.02, respectively) (Fig. 2C).
Clinical Significance of the 5-gene Panel for Detecting LN Metastasis in ESCC Patients
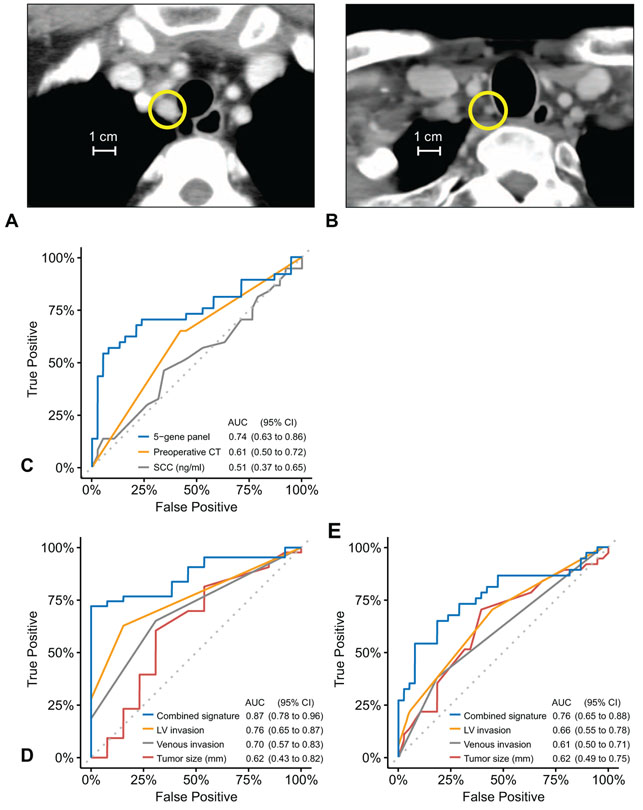
For their clinical management, CT imaging is generally used to diagnose LN metastasis and to determine clinical N stage before therapy in ESCC patients. The criteria such as LN diameter >1 cm in size, ring-enhancement, and heterogeneous enhancement are typically used for CT-based diagnosis of LN metastasis (Fig. 3A). However, in some cases, CT imaging is inadequate for diagnosing LN metastasis, and such patients can only be appropriately diagnosed by postsurgical pathological examination (Fig. 3B). Accordingly, to address this important clinical challenge, we assessed the accuracy of CT imaging and serum SCC antigen as pre-treatment diagnostic methods for detection of LN metastasis, and compared their performance with our 5-gene panel in the cohort-2. Of interest, the diagnostic accuracy of our newly developed 5-gene panel was significantly superior to both CT imaging and SCC (P = 0.08 and P = 0.005, respectively, Table 2, Fig. 3C) for the detection of LN metastasis in ESCC patients.
FIGURE 3.
Comparisons with other clinicopathological valuables. A, A representative computed tomography (CT) image of an esophageal squamous cell carcinoma metastatic lymph node (LN) in the cohort-2. The basis for the diagnosis is a LN diameter greater than 1 cm; the yellow circle indicates the typical metastatic LN. B, A representative CT image of a LN that could not be diagnosed by CT, but only by pathological examination after surgery in the cohort-2. The yellow circle indicates the metastatic LN with a diameter less than 1 cm. C, Receiver operating characteristic (ROC) curves comparing accuracy of the 5-gene panel, preoperative CT imaging based diagnosis and serum SCC in the cohort-2. D, ROC curves comparing the accuracy of the combined signature and each single clinicopathological feature in the cohort-1. E, ROC curves comparing the accuracy of the combined signature and each single clinicopathological feature in the cohort-2. LV indicates lymphatic vessel.
TABLE 2.
Comparisons of 5-gene Panel and Other Pre-treatment Diagnostic Methods
| 5-gene Panel | Preoperative CT | Serum SCC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 95% CI | 95% CI | 95% CI | |||||||
| Best | Low | High | Best | Low | High | Best | Low | High | |
| AUC | 0.74 | 0.63 | 0.86 | 0.61 | 0.50 | 0.72 | 0.51 | 0.37 | 0.65 |
| OR | 15.3 | 3.98 | 58.9 | 2.42 | 0.95 | 6.19 | 1.56 | 0.60 | 4.08 |
| Specificity | 0.92 | 0.74 | 1.00 | 0.57 | 0.43 | 0.73 | 0.68 | 0.11 | 1.00 |
| Sensitivity | 0.57 | 0.43 | 0.81 | 0.65 | 0.51 | 0.81 | 0.43 | 0.03 | 0.97 |
| Accuracy | 0.75 | 0.68 | 0.85 | 0.61 | 0.51 | 0.72 | 0.56 | 0.51 | 0.67 |
| TN | 35 | 28 | 38 | 21 | 16 | 27 | 25 | 4 | 37 |
| TP | 21 | 16 | 30 | 24 | 19 | 30 | 15 | 1 | 34 |
| FN | 16 | 7 | 21 | 13 | 7 | 18 | 20 | 1 | 34 |
| FP | 3 | 0 | 10 | 16 | 10 | 21 | 12 | 0 | 33 |
| NPV | 0.69 | 0.63 | 0.83 | 0.62 | 0.52 | 0.75 | 0.56 | 0.52 | 0.93 |
| PPV | 0.88 | 0.72 | 1.00 | 0.60 | 0.52 | 0.71 | 0.56 | 0.50 | 1.00 |
| 1-specificity | 0.08 | 0 | 0.26 | 0.43 | 0.27 | 0.57 | 0.32 | 0 | 0.89 |
| 1-sensitivity | 0.43 | 0.19 | 0.57 | 0.35 | 0.19 | 0.49 | 0.57 | 0.03 | 0.97 |
| 1-accuracy | 0.25 | 0.15 | 0.32 | 0.39 | 0.28 | 0.49 | 0.44 | 0.33 | 0.49 |
| 1-NPV | 0.31 | 0.18 | 0.37 | 0.38 | 0.25 | 0.48 | 0.44 | 0.07 | 0.48 |
| 1-PPV | 0.13 | 0 | 0.28 | 0.40 | 0.29 | 0.49 | 0.44 | 0 | 0.50 |
AUC indicates area under the curve; CI, confidence interval; CT, computed tomography; FN, false negative; FP, false positive; NPV, negative predictive value; OR, odds ratio; PPV, positive predictive value; TN, true negative; TP, true positive.
The curability of the endoscopic resection for the relatively early ESCC is mainly defined by the presence of the residual lesions. Next, to better define the potential clinical significance of our 5-gene panel for determining its curability without direct pathological LN assessment, we compared the diagnostic ability of our signature by combining it together with lymphatic vessel invasion (LVI) and venous invasion against each of the clinical features associated with LN metastasis. For both clinical validation cohorts, using ROC analysis, this combined signature demonstrated a superior diagnostic power compared with individual clinical features [cohort-1, P = 0.01 (LVI), P = 0.009 (venous invasion), and P = 0.01 (tumor size); cohort-2, P = 0.007 (LVI), P = 0.003 (venous invasion), and P = 0.07 (tumor size), respectively] (Fig. 3D, E, supplementary table S4 and S5, http://links.lww.com/SLA/B355).
Prognostic Value of the Gene Expression Panel in Clinical Cohorts
The presence of LN metastasis was significantly associated with OS in the cohort-1 [hazard ratio (HR) = 4.41, 95% CI 1.3–14.6, P = 0.008] (Fig. 4A). When the patients were categorized into high and low-risk groups with the risk scores derived from the 5-gene panel, using 0.25 as a cutoff threshold that can achieve 90% sensitivity in the cohort-1, the high-risk group of ESCC patients had a significant worse OS (HR = 6.47, 95% CI 0.9–47.6, P = 0.04) (Fig. 4B). The univariate cox regression analysis also revealed that this risk score from the 5-gene panel was significantly associated with OS in the cohort-1 (P = 0.006). However, in the cohort-2, while LN+ group tended to have worse OS (HR = 2.25, 95% CI = 0.9–5.6, P = 0.07) (Fig. 4C), the high-risk group estimated by the 5-gene panel had a slightly better OS, but it did not show statistical significance (HR = 0.5, 95% CI = 0.2–1.2, P = 0.10) (Fig. 4D) (univariate cox regression analysis, P = 0.25). The lack of significant difference in OS in cohort-2 could be attributed by virtue of the achieved curability following esophagectomy with adequate LN dissection for this relatively early T stage ESCCs.
FIGURE 4.
Survival analyses of clinical cohorts. A, A comparison of overall survival (OS) between lymph node metastasis negative (LN−) and positive (LN+) cases in the cohort-1. B, A comparison of OS between high and low-risk group estimated by the 5-gene panel in the cohort-1. C, A comparison of OS between LN− and LN+ cases in the cohort-2. D, A comparison of OS between high and low-risk group estimated by the 5-gene panel in the cohort-2.
DISCUSSION
In this article, we performed a systematic and comprehensive RNA sequencing-based gene expression profiling analysis of ESCC patients, with and without LN metastasis to establish a gene expression panel for identification of LN metastasis. We subsequently evaluated the robustness of our marker panel in a publicly available high-throughput dataset, followed by independent validation in 2 different patient cohorts. We demonstrated that a 5-gene panel was robust in both validation cohorts and was particularly effective in identifying patients with LN metastasis with early-stage T1–T2 ESCCs, which are often the Achilles heel for this malignancy. Furthermore, we demonstrated that our 5-gene panel was superior to currently used clinicopathological features, CT imaging, and SCC tumor antigen levels in diagnosing LN metastasis. Furthermore, a combined signature encompassing our 5-gene panel together with lymphatic vessel and venous invasion was significantly superior to the pre-treatment diagnostic modalities and clinicopathological features associated with LN status in ESCC patients.
From a clinical standpoint, the LN status is vitally important for therapeutic decision-making, especially in relatively early T stage ESCC patients. Without LN metastasis, Tis and T1a ESCC patients can be successfully treated with endoscopic mucosal or submucosal resection. However, if the tumor is most likely to reach LNs, esophagectomy with LN dissection is often required to obtain better curability. Even for cases that apparently do not indicate treatment for endoscopic resection, a more precise information of LN status can significantly contribute to decision making for preoperative chemo(radio)therapy, as recommended by the National Comprehensive Cancer Network Guidelines.22 Currently, lymphadenectomy following pathologic diagnosis is the best method for evaluating LN status of ESCC patients.23 According to the American Joint Committee on Cancer and the Union for International Cancer Control, N category for ESCC is classified based on the number of metastatic LNs.18 It is obvious that the number of identified metastatic LNs improve with increase in the total number of surgically dissected LNs. Thus, inadequate number of dissected LNs could potentially lead to stage migration and subsequent underestimation of the disease severity.24 However, sufficient dissection of LN by esophagectomy with radical lymphadenectomy is technically challenging. Furthermore, patients with ESCC have a higher chance of cardiopulmonary dysfunction, considering that a large proportion of ESCC patients typically have high alcohol and tobacco consumption.25,26 Therefore, availability of robust biomarkers that facilitate categorization of patients with ESCC before surgery based on the LN status will permit individualization of the surgical treatments and improve overall mortality and morbidity of ESCC patients by avoiding excessive lymphadenectomy. In this study, we initially elucidated 16 genes as candidates from TCGA dataset (20,531 genes) using bioinformatics approach. We thereafter trained our gene panel in a clinical cohort and validated in another independent cohort. Although the accuracy of individual genes was relatively modest, the combination of multiple genes revealed remarkable diagnostic accuracy. Collectively, we demonstrated that the potential of a multigene panel for identifying LN metastasis in ESCC is realistic, and can be used in the clinic in not so distance future.
The recent advancements in whole genome and transcriptome sequencing have resulted in molecular characterization of most cancer types.27,28 To the best of our knowledge, this is the first study to develop a multigene panel for determination of ESCC LN status. Although our results indicate that our gene signature can distinguish ESCC LN status preoperatively, there were some inherent limitations to this study. First, not all of surgically resected specimens underwent preoperative systemic chemotherapy. For localized advanced squamous cell carcinoma of the thoracic esophagus, preoperative systemic chemo(radio)therapy with cisplatin and 5-fluorouracil is regarded as the standard treatment.29–31 However, in the present study, not all patients underwent chemotherapy before the surgery and may have influenced the effectiveness of the gene-signature. Second, we were unable to evaluate the 5-gene panel with T1–T2 ESCC in cohort-1 because of the limited number of the cases available. Finally, considering that it is a common practice to endoscopically collect biopsy samples before the surgery, it would have been ideal to validate our gene-signature in endoscopically resected specimens.
In conclusion, we have developed a novel gene expression signature from primary tumor tissues for the identification of LN metastasis. Our panel may have clinical usefulness in stratifying patients with or without ESCC LN metastasis.
Supplementary Material
ACKNOWLEDGMENT
We thank Shusuke Toden for editing the manuscript. We thank Raju Kandimalla, Daisuke Izumi, Takatoshi Matsuyama, and Tsuyoshi Ozawa for discussing the analysis. We thank Jinsei Miyoshi for helping to perform the experiments. We thank Yukiko Niwa and Yoko Nishikawa for collecting clinical samples and information.
This work was supported by the CA72851, CA181572, CA184792, CA187956, and CA202797 grants from the National Cancer Institute, National Institute of Health; RP140784 from the Cancer Prevention Research Institute of Texas; grants from the Sammons Cancer Center and Baylor Foundation as well as funds from the Baylor Scott & White Research Institute, Dallas, TX, awarded to Ajay Goel. This work was also supported by a Start-up Grant for New Faculty (7200455), VPRT grant (9610337) at the City University of Hong Kong, a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (Project No. CityU 21101115); and a grant from The Science Technology and Innovation Committee of Shenzhen Municipality (JCYJ20170307091256048) awarded to Xin Wang.
Footnotes
The authors report no conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s Web site (www.annalsofsurgery.com).
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Rustgi AK, El-Serag HB. Esophageal carcinoma. N Engl J Med. 2014;371:2499–2509. [DOI] [PubMed] [Google Scholar]
- 3.Pickens A, Orringer MB. Geographical distribution and racial disparity in esophageal cancer. Ann Thorac Surg. 2003;76:S1367–S1369. [DOI] [PubMed] [Google Scholar]
- 4.Hsu WH, Hsu PK, Wang SJ, et al. Positron emission tomography-computed tomography in predicting locoregional invasion in esophageal squamous cell carcinoma. Ann Thorac Surg. 2009;87:1564–1568. [DOI] [PubMed] [Google Scholar]
- 5.Ji X, Cai J, Chen Y, et al. Lymphatic spreading and lymphadenectomy for esophageal carcinoma. World J Gastrointest Surg. 2016;8:90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eloubeidi MA, Desmond R, Arguedas MR, et al. Prognostic factors for the survival of patients with esophageal carcinoma in the US: the importance of tumor length and lymph node status. Cancer. 2002;95:1434–1443. [DOI] [PubMed] [Google Scholar]
- 7.Shimizu S, Hosokawa M, Itoh K, et al. Can hybrid FDG-PET/CT detect subclinical lymph node metastasis of esophageal cancer appropriately and contribute to radiation treatment planning? A comparison of image-based and pathological findings. Int J Clin Oncol. 2009;14:421–425. [DOI] [PubMed] [Google Scholar]
- 8.Hosch SB, Stoecklein NH, Pichlmeier U, et al. Esophageal cancer: the mode of lymphatic tumor cell spread and its prognostic significance. J Clin Oncol. 2001;19:1970–1975. [DOI] [PubMed] [Google Scholar]
- 9.Mariette C, Piessen G, Briez N, et al. The number of metastatic lymph nodes and the ratio between metastatic and examined lymph nodes are independent prognostic factors in esophageal cancer regardless of neoadjuvant chemoradiation or lymphadenectomy extent. Ann Surg. 2008;247:365–371. [DOI] [PubMed] [Google Scholar]
- 10.Rice TW. Superficial oesophageal carcinoma: is there a need for three-field lymphadenectomy? Lancet. 1999;354:792–794. [DOI] [PubMed] [Google Scholar]
- 11.Nishihira T, Hirayama K, Mori S. A prospective randomized trial of extended cervical and superior mediastinal lymphadenectomy for carcinoma of the thoracic esophagus. Am J Surg. 1998;175:47–51. [DOI] [PubMed] [Google Scholar]
- 12.Lerut T, Deleyn P, Coosemans W, et al. Surgical strategies in esophageal-carcinoma with emphasis on radical lymphadenectomy. Ann Surg. 1992;216:583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peyre CG, Hagen JA, DeMeester SR, et al. The number of lymph nodes removed predicts survival in esophageal cancer: an international study on the impact of extent of surgical resection. Ann Surg. 2008;248:549–554. [DOI] [PubMed] [Google Scholar]
- 14.Ye T, Sun Y, Zhang Y, et al. Three-field or two-field resection for thoracic esophageal cancer: a meta-analysis. Ann Thorac Surg. 2013;96:1933–1941. [DOI] [PubMed] [Google Scholar]
- 15.Akiyama H, Tsurumaru M, Udagawa H, et al. Radical lymph node dissection for cancer of the thoracic esophagus. Ann Surg. 1994;220:364–372. discussion 372–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato K, Hida Y, Miyamoto M, et al. Overexpression of caveolin-1 in esophageal squamous cell carcinoma correlates with lymph node metastasis and pathologic stage. Cancer. 2002;94:929–933. [PubMed] [Google Scholar]
- 17.Tan HZ, Wu ZY, Wu JY, et al. Single nucleotide polymorphism rs13042395 in the SLC52A3 gene as a biomarker for regional lymph node metastasis and relapse-free survival of esophageal squamous cell carcinoma patients. BMC Cancer. 2016;16:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edge SB, Byrd DR, Compton CC, et al. AJCC Cancer Staging Manual. 7th ed New York, NY: Springer; 2010. [Google Scholar]
- 19.Hur K, Toiyama Y, Schetter AJ, et al. Identification of a metastasis-specific microRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015;107:pii: dju492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 21.Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Comprehensive Cancer Network. Esophageal and Esophagogastric Junction Cancers: NCCN Guidelines. Available at: https://www.nccn.org/professionals/physician_gls/pdf/esophageal.pdf. Accessed March 1, 2017. [Google Scholar]
- 23.Rice TW, Ishwaran H, Hofstetter WL, et al. Esophageal cancer: associations with (pN+) lymph node metastases. Ann Surg. 2017;265:122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandarana M, Jiwnani S, Karimundackal G, et al. Lymphadenectomy in esophageal cancer: the real issues. Ann Thorac Surg. 2014;98:389–390. [DOI] [PubMed] [Google Scholar]
- 25.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. [DOI] [PubMed] [Google Scholar]
- 26.Ohashi S, Miyamoto S, Kikuchi O, et al. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149:1700–1715. [DOI] [PubMed] [Google Scholar]
- 27.Chin L, Meyerson M, Aldape K, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ando N, Kato H, Igaki H, et al. A randomized trial comparing postoperative adjuvant chemotherapy with cisplatin and 5-fluorouracil versus preoperative chemotherapy for localized advanced squamous cell carcinoma of the thoracic esophagus (JCOG9907). Ann Surg Oncol. 2012;19:68–74. [DOI] [PubMed] [Google Scholar]
- 30.Urschel JD, Vasan H. A meta-analysis of randomized controlled trials that compared neoadjuvant chemoradiation and surgery to surgery alone for resectable esophageal cancer. Am J Surg. 2003;185:538–543. [DOI] [PubMed] [Google Scholar]
- 31.Allum WH, Stenning SP, Bancewicz J, et al. Long-term results of a randomized trial of surgery with or without preoperative chemotherapy in esophageal cancer. J Clin Oncol. 2009;27:5062–5067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.