Abstract
Objective
This study aims to explore whether miR-195-5p can inhibit proliferation and induce apoptosis of non-small cell lung cancer (NSCLC) cells by targeting CEP55.
Methods
qRT-PCR was used to measure the expression of miR-195-5p in NSCLC cells. MTT assay, colony formation assay, and flow cytometry were used to detect the role of miR-195-5p in NSCLC cells. Western blot was used to measure the protein expression of CEP55, Bax and Bcl-2 in cells. Dual-Luciferase assay was performed to verify the relationship between miR-195-5p and CEP55.
Results
The expression of miR-195-5p was higher in human normal lung cell lines than in NSCLC cells. MiR-195-5p overexpression inhibited cell proliferation, which could block the cell cycle of A549 cell line in the G0/G1 phase. Moreover, overexpression of miR-195-5p increased cell apoptotic rate of A549 cell lines, with the expression of pro-apoptotic protein Bax up-regulated and that of the anti-apoptotic protein Bcl-2 down-regulated. The Dual-Luciferase assay showed that miR-195-5p could specifically target CEP55. Furthermore, CEP55 was down-regulated in NSCLC cells. Overexpression of CEP55 enhanced the proliferation and colony formation ability of A549 cell line. Overexpression of CEP55 can reverse the inhibitory effect of miR-195-5p.
Conclusion
MiR-195-5p inhibits proliferation and induces apoptosis of NSCLC cells by negatively regulating CEP55.
Keywords: non-small cell lung cancer, NSCLC, miR-195-5p, CEP55, cell proliferation, cell apoptosis
Introduction
Lung cancer is one of the malignancies with the highest mortality1 and more than 1 million patients die of lung cancer worldwide every year.2 NSCLC, accounting for 80–85%, is the predominantly common type of lung cancer with an extremely high mortality rate, and the 5-year survival rate of patients is less than 14%.3,4 Moreover, the incidence and mortality rates increase with age. Therefore, finding a more effective method to inhibit or treat lung cancer is a hot research topic.
Centrosome-related protein CEP55 (centrosomal protein, 55 kD) is one of the coiled coil protein family members, and its main functions are to anchor the microtubule and polymerize related proteins, participate in the spindle formation, and then regulate cell proliferation.5 It has been found that CEP55 is expressed in both normal tissues and tumor cells and coupled with centrosomes and intermediates in the cell cycle which plays a role in regulating cell cycle after phosphorylation.6,7 In addition, CEP55 overexpression has been shown to be significantly correlated with tumor staging, invasiveness and metastasis of many malignant tumors,7,8 and can be used as a biomarker for tumor occurrence, progression and prognosis.
MicroRNA is one kind of non-coding single-stranded RNA molecule encoded by endogenous genes with about 22 nucleotides in length in eukaryotes which can regulate the expression of mRNA at the post-transcriptional level. It has been identified as a key regulatory factor of the tumorigenesis in many cancers9,10 and it can inhibit the cell proliferation and metastasis of tumor cells by participating in the regulation of gene expression.11,12 Studies have shown that the expression of miR-195-5p decreased in NSCLC tissues and cell lines. Kaplan–Meier survival analysis shows that the survival time of NSCLC patients with high miR-195-5p expression is significantly longer than that of NSCLC patients with low expression during the 5-year follow-up.13 These results suggest that miR-195-5p can be a potential tumor suppressor of NSCLC.13–15 In this paper, the targeted relationship between miR-195-5p and CEP55 was studied to explore the mechanism of their roles in proliferation and apoptosis of NSCLC, so as to provide a new therapeutic target for clinical treatment of NSCLC.
Materials and Methods
Cell Lines and Cultivation
Human normal lung cell line BEAS-2B and NSCLC cell line H1299 were purchased from Chinese Academy of Sciences, and NSCLC cell line A549 was purchased from key laboratory of department of pathology, Xiamen University. All cells were cultured in RPMI-1640 medium (R8758, Sigma) and maintained in an incubator at 37°C with 5% CO2. When the cells attached with a density of 70–80%, they were digested with 0.25% trypsin (25200072, GIBCO), and then logarithmic growth cells were selected for the experiment.
Vector Construction and Transfection
MiR-195-5p mimics, NC, wt-CEP55 and mut-CEP55 luciferase reporter plasmids, and CEP55 lentiviral expression vector GV358 were provided by Shanghai GenePharma Co., Ltd. The cells were divided into five groups: (1) Control group, cells were only added with transfection agents; (2) Negative control group (NC), cells were transfected unrelated sequences; (3) MiR-195-5p group, cells were transfected with the miR-195-5p mimics; (4) CEP55 group, cells were transfected with CEP55 lentiviral expression vector; (5) MiR-195-5p + CEP55 group.
The cells of each group were seeded into a six-well plate at a density of (2–4) ×105/well. When the cell density reached about 50%, the synthetic fragments and plasmids were transferred into A549 cell lines using LipofectamineTM2000 (11668-027, Beijing Solarbio Science & Technology Co., Ltd.), respectively. And cells were incubated in a 37 °C, 5% CO2 incubator. The medium was changed after 6 h, and cells were collected after transfection for 36–48 h for follow-up experiments.
Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR)
All RNAs were extracted from NSCLC cells and human normal lung cells by Trizol (Thermo Fisher Scientific, Waltham, MA, USA) with its purity and concentration determined. Then, RNA was transcribed into complementary DNA (cDNA) following the instruction of the cDNA kit (Thermo Fisher Scientific, Waltham, MA, USA) and qRT-PCR was performed following the instruction of TB Green® Premix Ex Taq™ II (RR820A, TAKARA). The data were normalized to U6 (for miR-195-5p expression) and GAPDH (for CEP55 mRNA expression) respectively, and calculated by the 2-△△CT method.
MiR-195-5p
Forward Primer: 5′-GGGGTAGCAGCACAGAAAT-3′;
Reverse Primer: 5′- TCCAGTGCGTGTCGTGGA-3′.
U6
Forward Primer: 5′-GCTTCGGCAGCACATATACTAAAAT-3′;
Reverse Primer: 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
CEP55
Forward Primer: 5′-AGTAAGTGGGGATCGAAGCCT-3′;
Reverse Primer: 5′-CTCAAGGACTCGAATTTTCTCCA-3′.
GAPDH
Forward Primer: 5′-GGAGCGAGATCCCTCCAAAAT-3′;
Reverse Primer: 5′-GGCTGTTGTCATACTTCTCATGG-3′.
Western Blot
After 48 h of transfection, the cells were lysed on ice and the total protein was extracted. After measuring the concentration of proteins, protein samples (30 μg) were separated by 10% SDS-PAGE and subsequently transferred onto PVDF membranes. The membrane was blocked with TBST containing 5% skim milk for 1 h. Later, membrane was incubated with primary antibodies including CEP55 rabbit polyclonal antibody (ab170414, 1:5000, abcam, Cambridge, UK), Bax rabbit polyclonal antibody (ab182733, 1:1000, abcam, Cambridge, UK), Bcl-2 rabbit polyclonal antibody (ab32124, 1:1000, abcam, Cambridge, UK) and GAPDH rabbit polyclonal antibody (ab9485, 1:2500, Abcam, Cambridge, UK) overnight at 4°C. TBST was used to wash the membranes for three times with 15 min for each time. Next, the membrane was incubated with secondary antibody of horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG (ab6721, 1:3000, abcam, Cambridge, UK) at room temperature for 1 h. After washing the membranes with TBST, the enhanced chemiluminescence (ECL) kit (P0018FS, Beyotime) was added and ChemiDoc™ Touch Imaging System (12003153, BIO-RAD) was used for exposed photography. The experiment was repeated three times.
Dual-Luciferase Assay
Wt-CEP55 and mut-CEP55 were constructed into luciferase reporter plasmids, and co-transfected into A549 cell line with miR-195-5p mimic and miRNA NC, respectively. After 48 h of cultivation in RPMI-1640 medium containing 10% fetal bovine serum (FBS), the luciferase activities were detected by Dual-Luciferase reporter gene assay kit (RG027, Beyotime) according to manuscript’s information. The experiment was repeated three times.
Cell Proliferation Assay
Cell proliferation was detected by MTT assay (Sigma). The transfected cells were seeded in a 96-well plate at a density of 1×104 cell/well. After cultivation for 24 h, 48 h and 72 h at 37°C, respectively, 20μL MTT (5mg/mL) was added to each well for 4 hr incubation. The supernatant in the well was carefully absorbed. As for suspension cells, the supernatant in the well was absorbed after centrifugation. Then, 150 μL DMSO was added for 15 min to dissolve the crystals. The cell viability was determined by reading OD value at 490 nm using ELISA. All experiments were repeated three times.
Colony Formation Assay
Monolayer cultured NSCLC cells at logarithmic growth phase were digested into single cells using 0.25% trypsin and suspended in RPMI-1640 medium containing 10% FBS. Cells of each group were inoculated with 200 cells per dish in 9 cm culture dishes, respectively, and gently shaken to disperse evenly. The cells were changed with fresh medium every 3 days for 3 weeks. Then, the medium was discarded, and the cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min, transfected with 0.1% crystal violet dye for 10 min and washed with PBS for three times. Finally, the cells were photographed and counted after washing off the dye, and the colony-forming efficiency was calculated (colony forming efficiency = clone number/inoculated cell number ×100%).
Cell Cycle Was Measured by Flow Cytometry
Forty-eight hours after transfection, the cells were digested with 1 mL trypsin for 2–3min, then added 5 mL PBS and blew the bottle wall to make the cells detach into single-cell suspension, which was collected and centrifuged at 1000 rpm for 5 min at 4°C. After discarding the supernatant, 10 mL PBS was added for re-suspension, and cells were fixed with 70% ethanol pre-cooled by ice at 4°C overnights. Next day, the cell suspension was filtered through a 300-mesh sieve, and centrifugated at 1000 rpm for 5 min at 4°C, then the supernatant was discarded. And the cells were fixed with 1 mL of PI solution and stored in darkness at 4°C for 30 min. Flow cytometer (12004279, BIO-RAD) was used to detect cell cycle. The experiment was repeated three times.
Cell Apoptosis Was Detected by Flow Cytometry
Flow cytometry was used to measure apoptosis in accordance with the instructions of FITC-Annexin V apoptosis kit (Hangzhou Haoxin Biotechnology Co., Ltd.). The cells were counted and the concentration of the cells to be detected was adjusted to 5×105–1×106 cells/mL. Then, 1 mL cells were taken and centrifuged at 1000 rpm for 10 min at 4°C, and the supernatant was discarded. Cell suspension was added with 1 mL precooled PBS and the supernatant was discarded after centrifugation at 1000 rpm for 10 min at 4°C. The cells were then resuspended in 200 μL Binding Buffer with10 Μl Annexin V-FITC and 10 μL PI solution at room temperature in darkness for 15 min after which 300 μL Binding Buffer was mixed into the resuspension. Flow cytometry (12004279, BIO-RAD) detected cell apoptosis within 1 h and the experiment was repeated three times.
Statistical Analysis
All the data were processed by SPSS 21.0 and exhibited as mean ± standard deviation. The comparison between two groups was conducted by Student's t-test, and the comparison among multiple groups was conducted by one-way analysis of variance (ANOVA). P<0.05 indicated statistical significance.
Results
Expression of miR-195-5p and CEP55 in Different Cell Lines
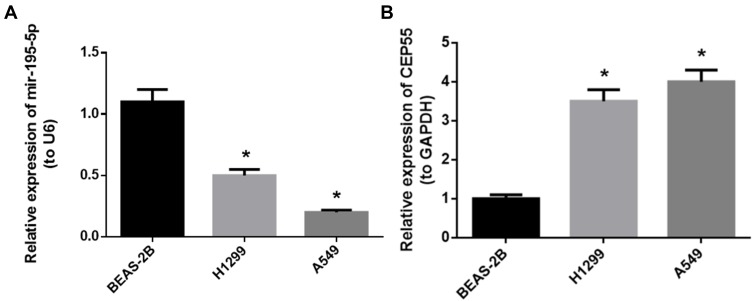
We firstly detected the miR-195-5p and CEP55 expression in human normal lung cell lineBEAS-2B and NSCLC cell lines H1299 and A549 qRT-PCR assay demonstrated that the expression of miR-195-5p was downregulated in H1299 and A549 cell lines compared to BEAS-2B cell line, while the expression of CEP55 mRNA was significantly upregulated in NSCLC cell lines (Figure 1A and B, P<0.05). We selected A549 cell line with a relatively significant difference in expression level for follow-up experiments.
Figure 1.
Expression of miR-195-5p and CEP55 in different cell lines. (A) The expression level of miR-195-5p in each cell line; (B) the expression level of CEP55 mRNA in each cell line.
Note: *p<0.05.
MiR-195-5p Could Inhibit the Proliferation and Induce Apoptosis of NSCLC Cells
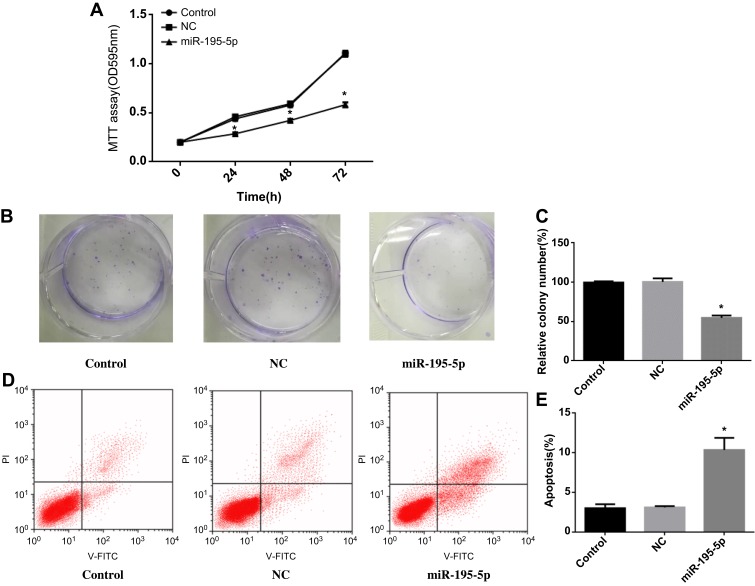
MiR-195-5p was upregulated after treated with miR-195-5p mimics in A549 cell line. MTT assay (Figure 2A) showed that overexpression miR-195-5p inhibited A549 cell proliferation (P<0.05). Meanwhile, the cell colony formation ability of A549 cell line was significantly decreased (Figure 2B and C, P<0.05), indicating that the high expression level of miR-195-5p could inhibit the proliferation of cancer cells. In addition, flow cytometry was used to detect apoptosis in each group, and the results exhibited that overexpression miR-195-5p increased the apoptosis of A549 cell line (Figure 2D and E, P<0.05). These experiments indicated that miR-195-5p overexpression could inhibit the proliferation and promote apoptosis of NSCLC cells.
Figure 2.
MiR-195-5p overexpression can inhibit the proliferation and promote apoptosis of NSCLC cells. (A) MTT assay was used to detect cell proliferation in each group; (B, C) Colony formation assay was used to determine the colony formation ability of each group cells; (D, E) Apoptosis was detected by flow cytometry.
Note: *p<0.05.
CEP55 Overexpression Promoted Proliferation and Slowed Apoptosis of NSCLC Cell Lines
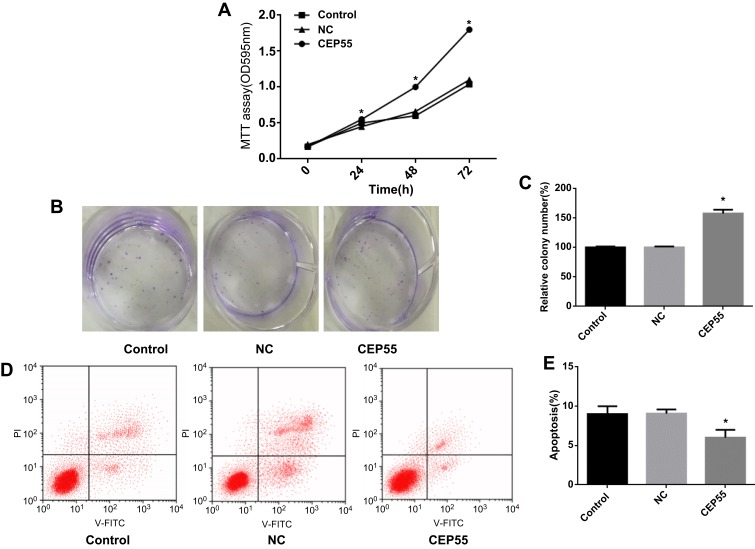
CEP55 was upregulated after treated with lentiviral expression vector GV358 in the A549 cells. MTT method and colony formation assay were used to detect the role of CEP55 in A549 cell line, Compared with NC group, overexpression of CEP55 significantly increased the A549 cell line proliferation (Figure 3A, P<0.05) and colony formation ability (Figure 3B and C, P<0.05). CEP55 overexpression promoted the apoptosis rate in A549 cell line (Figure 3D and E, P<0.05).
Figure 3.
CEP55 overexpression promotes proliferation and slows apoptosis in NSCLC cell lines. (A) MTT assay was used to detect cell proliferation in each group; (B, C) Colony formation assay results of CEP55 group and NC group; (D, E) Apoptosis in CEP55 group and NC group was detected by flow cytometry.
Note: *p<0.05.
MiR-195-5p Could Regulate the Proliferation and Apoptosis of NSCLC by Targeting CEP55
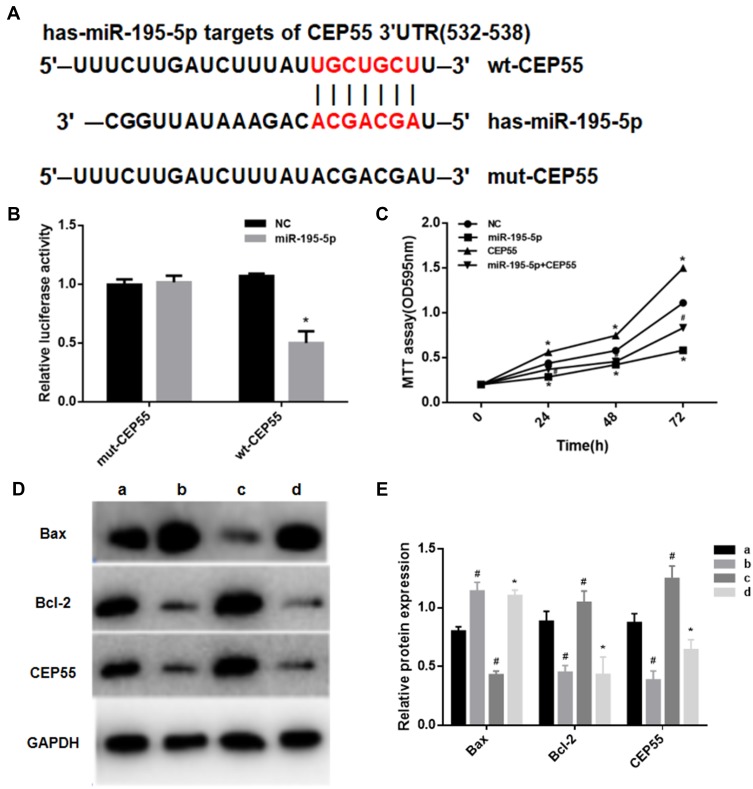
Through the Targetscan.org website, we predicted the region sequences in the 3ʹ-UTR of CEP55 complementary to miR-195-5p, as shown in Figure 4A. Results of dual luciferase assay (Figure 4B) presented that miR-195-5p overexpression decreased the luciferase activity of the wt-CEP55 group (P<0.05), while the luciferase activity of the mut-CEP55 group after transfection with miR-195-5p was not significantly different from that of the mut-cep55 transfected NC group (P>0.05). This suggested that there was a targeted regulatory relationship between miR-195-5p and CEP55.
Figure 4.
MiR-195-5p can induce apoptosis in NSCLC by targeting CEP55. (A) Targeting prediction results of miR-195-5p and CEP55; (B) The results of dual luciferase assay (Note: *represents p<0.05); (C) MTT assay was used to detect cell proliferation (Notes: *represents p<0.05, compared with NC group; # represents p<0.05, compared with CEP55 group); (D, E) Western blot was used to detect the expression levels of CEP55, Bax and Bcl-2 in each group. a Represents NC group; b represents miR-195-5p group; c represents CEP55 group; d represents miR-195-5p+CEP55 group.
Notes: *p<0.05, compared with group c; #p<0.05, compared with group a.
In order to verify that miR-195-5p could directly regulate CEP55 and thus affecting the proliferation and apoptosis of cancer cells, the MTT assays and Western blot assays were performed in this study. When miR-195-5p and CEP55 overexpression vector were co-transfected into A549 cell line, the cell proliferation was significantly decreased compared to the CEP55 group (Figure 4C). Compared with the NC group, the expression of CEP55 and Bcl-2 in the miR-195-5p group were significantly decreased, while that of Bax significantly increased. In the CEP55 group, the expression of Bax decreased and Bcl-2 increased. In the miR-195-5p +CEP55 group, the CEP55 expression level was significantly lower than that of the CEP55 group, and the Bax expression was higher and Bcl-2 expression was lower (Figure 4D and E). These results indicated that miR-195-5p negatively regulated CEP55 expression and induced apoptosis of NSCLC cells.
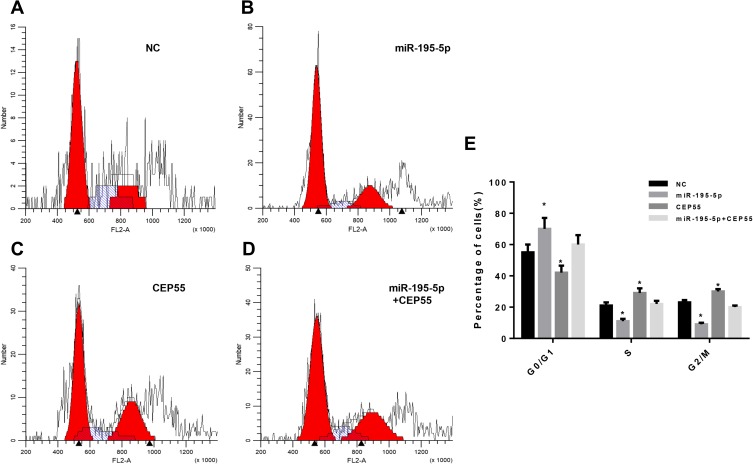
MiR-195-5p Promoted Cell Cycle Arrest of NSCLC by Targeted Inhibiting CEP55 Expression
To investigate the effects of different treatments on the cell cycle of NSCLC, flow cytometry was used to detect the cell cycle of A549 cell line in NC group, miR-195-5p group, CEP55 group and miR-195-5p + CEP55 group after 48 h transfection. The results showed that compared with NC, cells with overexpression miR-195-5p were significantly increased in the G0/G1 phase, while those in S phase and G2/M phase were decreased significantly (Figure 5A, B and E). Compared with the NC group, when CEP55 was overexpressed, the proportion of cells in the G0/G1 phase was decreased, while that in the S phase and G2/M phase was increased significantly, indicating a high proliferation of cancer cells (Figure 5C). In addition, in order to verify whether miR-195-5p could affect the effect of CEP55 on cell cycle, we tested the co-transfection group of miR-195-5p + CEP55. Results showed that compared with the CEP55 group, the number of cells in the G0/G1 phase was increased significantly and that in the S phase and G2/M phase decreased correspondingly (Figure 5D). In summary, our data demonstrated that miR-195-5p overexpression could block lung cancer cells at the G0/G1 phase, while CEP55 overexpression increased the number of cells at S phase and G2/M phase. Meanwhile, the overexpression of miR-195-5p and CEP55 could reduce the effect of the CEP55 overexpression on cell cycle, which may be related to the interaction between the miR-195-5p and CEP55.
Figure 5.
Cell cycle was detected by flow cytometry. (A) Control group; (B) MiR-195-5p group; (C) CEP55 group; (D) MiR-195-5p+CEP55 group; (E) The proportion of each phase of the cell cycle.
Note: *p<0.05, compared with the NC group.
Discussion
Molecular targeted therapy has been widely applied in the treatment of lung cancer. Compared with traditional chemotherapy, molecular targeted therapy is more efficient and less toxic.16 This study mainly discusses the inhibitory effects of miR-195-5p on NSCLC by regulating CEP55, which is of great significance for the discovery of new tumor-related factors and therapeutic targets.
MiR-195-5p is abnormally expressed in various cancers such as breast cancer,17 renal cell carcinoma,18 and colorectal cancer.19 For example, miR-195-5p can inhibit cell proliferation by regulating IRS1 in breast cancer cells,20 and miR-195-5p can weaken cell proliferation by directly targeting cyclin D1 in cervical cancer cells.21 In addition, Liu et al found that the expression of miR-195 was lower in tumor cell tissues through a clinical study of 299 NSCLC samples and negatively correlated with patients’ survival.22 Therefore, we verified the expression of miR-195-5p in NSCLC cell lines and its effect on cell function and found that miR-195-5p showed low expression in H1299 and A549 cell lines. Meanwhile, the overexpression of miR-195-5p significantly inhibited the proliferation of cancer cells and promoted apoptosis, which was basically consistent with previous studies.
CEP55 has been found to play a regulatory role in various cancer cells as an important regulatory protein. For example, the down-regulation of CEP55 can inhibit the proliferation of breast cancer cells.23 CEP55 can promote proliferation and inhibit apoptosis of glioma cells through PI3K/Akt/p21 signal pathways.6 In addition, the expression of CEP55 is inversely correlated with poor prognosis in patients with advanced esophageal squamous cell carcinoma and NSCLC.24,25 So, we also conducted some experiments in NSCLC cells to verify and observe that CEP55 was highly expressed in lung cancer cell lines, and the overexpression of CEP55 promoted proliferation and decreased apoptosis of A549 cell line. Furthermore, we also detected that when CEP55 was overexpressed, the expression level of Bax was down-regulated, while the expression level of Bcl-2 was up-regulated. The results indicated that CEP55 could be used as a target for cancer therapy to affect the proliferation of NSCLC cells, so it may be possible to treat NSCLC by inhibiting CEP55 expression in the future.
MiRNAs often play a stimulating or inhibitory role in cancer cells by regulating downstream target genes. For instance, miR-195-5p can target GDPD5 and increases chemosensitivity and apoptosis in chemoresistant CRC cells.19 MiR-144 inhibits the proliferation, invasion and migration of breast cancer cells by inhibiting CEP55.26 In this study, both miR-195-5p and CEP55 were believed to be related to cancer progression; thus, we speculated that the regulatory function of miR-195-5p on NSCLC cells might also be partly attributed to CEP55. We predicted whether there was a targeted binding region between miR-195-5p and CEP55 through the Targetscan.org website. Both the predicted results and the dual luciferase assay showed that miR-195-5p could directly bind to CEP55 3ʹ-UTR. Western blot and MTT assays also revealed that overexpression of miR-195-5p could inhibit the expression of CEP55 and the effect of CEP55 overexpression on cell proliferation and cell cycle.
This study confirmed that miR-195-5p can negatively regulate CEP55 to inhibit the proliferation and promote the apoptosis of NSCLC cells, so as to play an anticancer role and provide a new target for the treatment of NSCLC. However, the specific molecular mechanism for miR-195-5p/CEP55 axis regulating NSCLC is still unclear which needs to be further explored.
Funding Statement
This study was supported by the Chinese Medicine Science and Technology Program of Zhejiang Province (2018ZA025).
Data Sharing Statement
The data and materials in the current study are available from the corresponding author on reasonable request.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Ethics Approval
The use of the A549 cells was approved by the Institutional Review Boards of the Tongde hospital of Zhejiang.
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138 [DOI] [PubMed] [Google Scholar]
- 2.Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103(3):463–473. doi: 10.1016/j.mcna.2018.12.006 [DOI] [PubMed] [Google Scholar]
- 3.Tonon G, Wong KK, Maulik G, et al. High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci U S A. 2005;102(27):9625–9630. doi: 10.1073/pnas.0504126102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramaniam S, Thakur RK, Yadav VK, Nanda R, Chowdhury S, Agrawal A. Lung cancer biomarkers: state of the art. J Carcinog. 2013;12:3. doi: 10.4103/1477-3163.107958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalimutho M, Sinha D, Jeffery J, et al. CEP55 is a determinant of cell fate during perturbed mitosis in breast cancer. EMBO Mol Med. 2018;10(9). doi: 10.15252/emmm.201708566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li F, Jin D, Tang C, Gao D. CEP55 promotes cell proliferation and inhibits apoptosis via the PI3K/Akt/p21 signaling pathway in human glioma U251 cells. Oncol Lett. 2018;15(4):4789–4796. doi: 10.3892/ol.2018.7934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L, Mei Q, Zhao J, Dai Y, Fu Q. Suppression of CEP55 reduces cell viability and induces apoptosis in human lung cancer. Oncol Rep. 2016;36(4):1939–1945. doi: 10.3892/or.2016.5059 [DOI] [PubMed] [Google Scholar]
- 8.Jeffery J, Sinha D, Srihari S, Kalimutho M, Khanna KK. Beyond cytokinesis: the emerging roles of CEP55 in tumorigenesis. Oncogene. 2016;35(6):683–690. doi: 10.1038/onc.2015.128 [DOI] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pastorkova Z, Skarda J, Andel J. The role of microRNA in metastatic processes of non-small cell lung carcinoma. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160(3):343–357. doi: 10.5507/bp.2016.021 [DOI] [PubMed] [Google Scholar]
- 11.Lin C, Xie L, Lu Y, Hu Z, Chang J. miR-133b reverses cisplatin resistance by targeting GSTP1 in cisplatin-resistant lung cancer cells. Int J Mol Med. 2018;41(4):2050–2058. doi: 10.3892/ijmm.2018.3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei J, Ma Z, Li Y, et al. miR-143 inhibits cell proliferation by targeting autophagy-related 2B in non-small cell lung cancer H1299 cells. Mol Med Rep. 2015;11(1):571–576. doi: 10.3892/mmr.2014.2675 [DOI] [PubMed] [Google Scholar]
- 13.Zheng J, Xu T, Chen F, Zhang Y. MiRNA-195-5p functions as a tumor suppressor and a predictive of poor prognosis in non-small cell lung cancer by directly targeting CIAPIN1. Pathol Oncol Res. 2019;25(3):1181–1190. doi: 10.1007/s12253-018-0552-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang T, Ren Y, Liu R, et al. miR-195-5p suppresses the proliferation, migration, and invasion of oral squamous cell carcinoma by targeting TRIM14. Biomed Res Int. 2017;2017:7378148. doi: 10.1155/2017/7378148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Ren CX, Zhang JM, et al. The effects of miR-195-5p/MMP14 on proliferation and invasion of cervical carcinoma cells through TNF signaling pathway based on bioinformatics analysis of microarray profiling. Cell Physiol Biochem. 2018;50(4):1398–1413. [DOI] [PubMed] [Google Scholar]
- 16.Riehl A, Nemeth J, Angel P, Hess J. The receptor RAGE: bridging inflammation and cancer. Cell Commun Signal. 2009;7:12. doi: 10.1186/1478-811X-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18(1):4. doi: 10.1186/s12943-018-0933-7 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Chen S, Wang L, Yao X, et al. miR-195-5p is critical in REGgamma-mediated regulation of wnt/beta-catenin pathway in renal cell carcinoma. Oncotarget. 2017;8(38):63986–64000. doi: 10.18632/oncotarget.19256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng C, Zhang L, Sun Y, et al. GDPD5, a target of miR-195-5p, is associated with metastasis and chemoresistance in colorectal cancer. Biomed Pharmacother. 2018;101:945–952. doi: 10.1016/j.biopha.2018.03.028 [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Zhang X, Zou C, et al. miR-195 inhibits tumor growth and angiogenesis through modulating IRS1 in breast cancer. Biomed Pharmacother. 2016;80:95–101. doi: 10.1016/j.biopha.2016.03.007 [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Wang H, Wang Z, Cai H. MiR-195 inhibits the proliferation of human cervical cancer cells by directly targeting cyclin D1. Tumour Biol. 2016;37(5):6457–6463. doi: 10.1007/s13277-015-4540-6 [DOI] [PubMed] [Google Scholar]
- 22.Liu B, Qu J, Xu F, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6(11):9445–9456. doi: 10.18632/oncotarget.3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Jin T, Dai X, Xu J. Lentivirus-mediated knockdown of CEP55 suppresses cell proliferation of breast cancer cells. Biosci Trends. 2016;10(1):67–73. doi: 10.5582/bst.2016.01010 [DOI] [PubMed] [Google Scholar]
- 24.Jiang W, Wang Z, Jia Y. CEP55 overexpression predicts poor prognosis in patients with locally advanced esophageal squamous cell carcinoma. Oncol Lett. 2017;13(1):236–242. doi: 10.3892/ol.2016.5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang C, Zhang Y, Li Y, et al. High CEP55 expression is associated with poor prognosis in non-small-cell lung cancer. Onco Targets Ther. 2018;11:4979–4990. doi: 10.2147/OTT [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin Y, Cai J, Meng F, Sui C, Jiang Y. MiR-144 suppresses proliferation, invasion, and migration of breast cancer cells through inhibiting CEP55. Cancer Biol Ther. 2018;19(4):306–315. doi: 10.1080/15384047.2017.1416934 [DOI] [PMC free article] [PubMed] [Google Scholar]