Abstract
The classical model of tissue renewal posits that small numbers of quiescent stem cells (SCs) give rise to proliferating transit-amplifying cells before terminal differentiation. However, many organs house pools of SCs with proliferative and differentiation potentials that diverge from this template. Resolving SC identity and organization is therefore central to understanding tissue renewal. Here, using a combination of single-cell RNA sequencing (scRNA-seq), mouse genetics and tissue injury approaches, we uncover cellular hierarchies and mechanisms that underlie the maintenance and repair of the continuously growing mouse incisor. Our results reveal that, during homeostasis, a group of actively cycling epithelial progenitors generates enamel-producing ameloblasts and adjacent layers of non-ameloblast cells. After injury, tissue repair was achieved through transient increases in progenitor-cell proliferation and through direct conversion of Notch1-expressing cells to ameloblasts. We elucidate epithelial SC identity, position and function, providing a mechanistic basis for the homeostasis and repair of a fast-turnover ectodermal appendage.
Ectodermal appendages—such as teeth, hair follicles, mammary glands and nails—share many aspects of their development and adult renewal1–3. The maintenance and repair of these appendages is enabled by resident adult SCs that have the capacity for both prolonged self-renewal and generation of differentiated cells. Adult tissue homeostasis and repair in these organs have been thought to involve a few long-lived quiescent SCs that generate rapidly dividing, short-lived transit-amplifying proliferating progenitors, which then produce differentiated cells4–6. However, recent studies in several organs have used lineage tracing in combination with mathematical modelling and single-cell approaches to demonstrate that renewal capacity can be distributed over a large population of actively dividing progenitors or SCs7. Importantly, seemingly homogeneous SC and progenitor populations are in fact highly heterogeneous8,9. Therefore, how SCs are utilized during tissue renewal and how such heterogeneous populations are regulated remain central questions in SC biology.
The mouse incisor provides a model system for understanding the renewal and regeneration of adult tissues. This organ continuously replaces materials that are lost as a result of abrasion from gnawing, and the entire tooth is replaced every 4 weeks10 (Supplementary Fig. 1a,b), making the mouse incisor one of the most rapidly renewing mineralized tissues in mammals. This continuous growth is driven by epithelial and mesenchymal SCs that give rise to cells that produce calcified enamel or dentin and cementum, respectively11,12.
Earlier studies proposed that the incisor follows a classical SC paradigm, similar to that proposed for the haematopoietic system13, with a few slow-cycling SCs residing in the proximal portion of the outer enamel epithelium (OEE) or stellate reticulum (SR) of the labial cervical loop (laCL) that give rise to transit-amplifying inner enamel epithelium (IEE) cells, which subsequently differentiate into all of the epithelial lineages14 (Fig. 1a–c). This model was based on the observation that a few putative SC markers chosen from a candidate-based approach, such as Sox2, Bmi1, Gli1 and Lrig1, are expressed by long-lived (label-retaining) cells in the OEE of the laCL and can be lineage-traced15–18. However, owing to the broad expression of these markers throughout both the quiescent and proliferating regions, as well as the limitations in the available genetic tools, our knowledge regarding the identity, hierarchies, heterogeneity and kinetics of the SCs remain poorly understood.
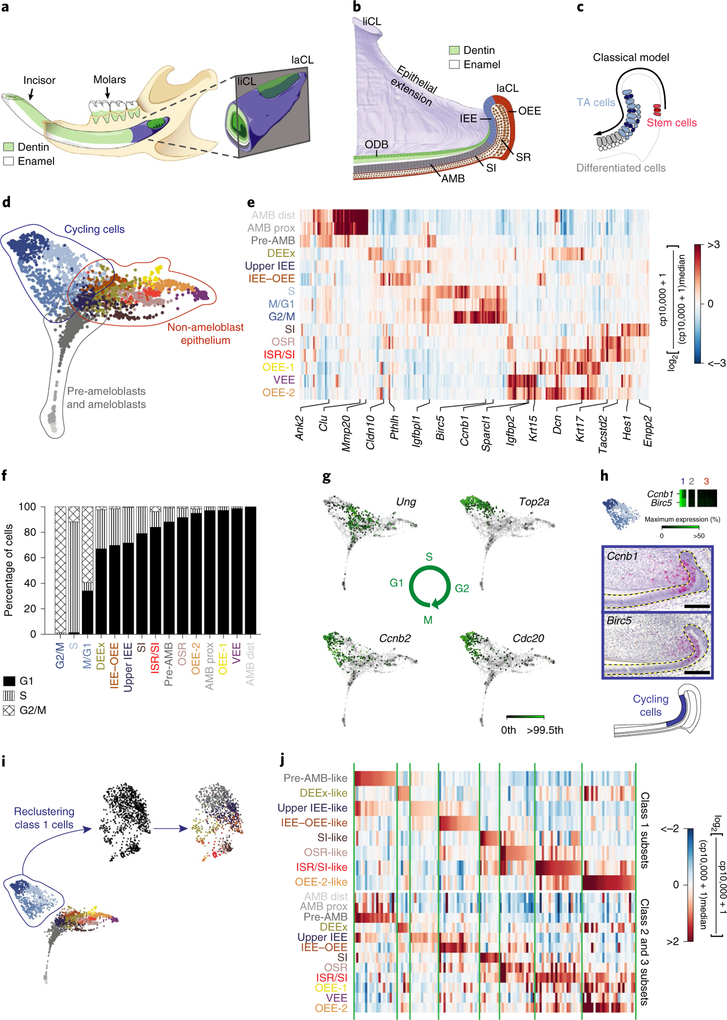
Fig. 1 |. Identification of distinct cell types by single-cell transcriptomic analysis and delineation of their spatial organization in the incisor epithelium.
a, Schematic of the mouse lower jaw and magnification of the incisor growth region showing enamel (white) and dentin (green). b, An illustration of a sagittal section through the incisor growth region showing epithelial regions. AMB, ameloblasts; ODB, odontoblasts. c, The current model posits that SCs in the OEE give rise to transit-amplifying (TA) cells in the IEE, which then differentiate into enamel-secreting ameloblasts. d, SPRING representation ofthe incisor epithelium dataset with 15 spectral clusters, grouped into 3 main classes as follows: cycling cells, pre-ameloblasts and ameloblasts, and non-ameloblast epithelial cells. e, A heat map showing the average expression of the top 20 enriched genes per cluster. Highlighted genes were used to further assess spatial expression patterns. AMB dist, distal ameloblasts; AMB prox, proximal ameloblasts; DEEx, dental epithelial extensions; IEE–OEE, junction between the IEE and OEE; ISR, inner stellate reticulum; OSR, outer stellate reticulum; VEE, ventral enamel epithelium; cp, counts per. f, The fraction of cells per cluster in each cell-cycle phase. g, SPRING plots showing gene expression of cell-cycle markers (Ung, Top2a, Cdc20 and Ccnb2). Gene expression is plotted on a linear scale; the colour scale indicates levels of expression from grey (zero counts) to dark green (color saturated at 99.5th expression percentile). h, Expression of the cell-cycle markers Ccnb1 and Birc5 in the 15 spectral clusters (top) and their spatial localization using RNAscope (middle two panels). Bottom, a schematic highlighting the cycling region. i, Class 1 cells before (left) and after regressing out the cell cycle (middle). Right, class1 cells are coloured according to the most similar profile among class 2 and 3 populations. j, The top 20 enriched genes for each of the cell populations identified in class 1 after regressing out the cell-cycle effect (top) show similar expression patterns (annotated in green vertical lines) to their counterparts in the class 2 and class 3 populations (bottom). For h, scale bars, 100 μm. Dashed lines outline the epithelium.
To address these questions, we examined cell identities using an unbiased scRNA-seq approach and then studied the kinetics and dynamics of the dental epithelial populations in incisors. By combining this information with genetic-lineage tracing and injury-repair studies, we have uncovered a highly dynamic SC model that is distinct from the traditional view of SCs in the mouse incisor and that explains the tissue homeostasis and injury repair of a fast-turnover ectodermal appendage.
Results
An unbiased analysis of cellular heterogeneity in the incisor epithelium.
As previous experiments examined mice at various stages of development12,16,18, we first determined when the incisor is in steady state (Supplementary Fig. 1c–h, Supplementary Note) and then reassessed the expression of the marker genes using high-resolution single-molecule in situ hybridization and genetic-lineage tracing. We discovered that cells of the posterior OEE, the presumed SC location, were not proliferative despite rapid epithelial turnover (Supplementary Fig. 1d,e), and that SC marker genes were more broadly expressed within the incisor growth region than previously appreciated (Supplementary Fig. 2), suggesting that the label-retaining OEE cells may not function as SCs during homeostasis. This prompted us to revisit the cellular hierarchies that underlie the homeostasis and repair of adult mouse incisors.
To address this question in an unbiased manner, we performed scRNA-seq of sorted epithelial cells (Supplementary Fig. 3a). We sequenced a total of 3,173 cells from 5 incisors of adult male mice and visualized the high-dimensional, whole-transcriptome data using SPRING19. This graph-based method is well suited for studying differentiation trajectories as it preserves the relationships between transcriptionally similar cells. Gene expression analysis identified 690 immune and 134 mesenchymal cell transcriptomes (Supplementary Fig. 3c–e), which were removed from the dataset to focus on the epithelium. The remaining cells were divided into 15 clusters that were characterized by distinct gene expression signatures (Fig. 1d); the clustering was unbiased and independent of any established markers of the incisor epithelium.
To assign putative biological identities to each cluster, we computed a heat map of the 20 most enriched genes per cluster (Fig. 1e, Supplementary Table 1), which revealed the existence of three major categories of cells (Fig. 1d). Class 1 was enriched in cycling cells (Fig. 1d, blue), class 2 contained pre-ameloblasts and ameloblasts (Fig. 1d, grey), and class 3 consisted of the remaining incisor epithelium or the non-ameloblast epithelium (Fig. 1d, brown and orange).
We next assessed the overall distribution of dividing cells and found that the vast majority were in class 1 (Fig. 1f). The transcriptomes of class 1 cells reflected successive phases of the cell cycle and formed a visible loop on the SPRING plot, implying that some cells at the end of the cycle return to their original state, reflecting self-renewal. We identified domains that are characteristic of S, G2/M and M/G1 phases, as exemplified by expression of Ung (G1 and S), Top2a (end of S phase to G2), Cdc20 (end of G2 phase to M entry) and Ccnb2 (end of M phase to G1 entry; Fig. 1g). Spatially, class 1 cells were localized to the IEE and the neighbouring stratum intermedium (SI) region of the incisor epithelium (Fig. 1h, Supplementary Fig. 3f). Given the dominance of the cell-cycle signature among the transcriptomes of class 1 cells, we further visualized class 1 cells using SPRING after regressing out the cell-cycle effect20. We found that class 1 cells had signatures reflecting class 2 and 3 populations, presumably reflecting progenitors that are cycling but beginning to upregulate differentiation genes (Fig. 1i,j, Supplementary Table 2). Interestingly, even with the cell cycle regressed out, the expression of the putative SC marker genes (Sox2, Gli1, Bmi1 and Lrig1) did not localize exclusively to any sub-region of the SPRING plot (Supplementary Fig. 2c,f,i,l).
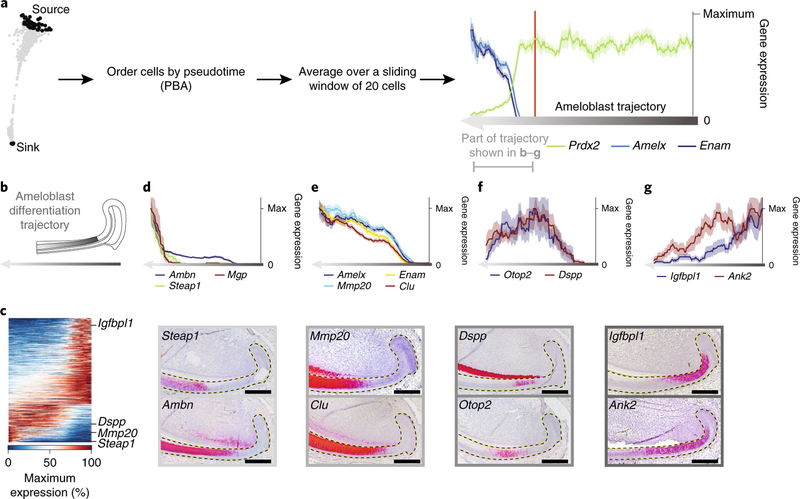
Class 2 cells (Fig. 1d), which formed a simple linear continuum in the SPRING graph, were ordered in pseudotime using population-balance analysis21 (PBA; Fig. 2a). Changes in gene expression through pseudotime (Fig. 2b–g, grey arrow) revealed groups of genes, such as Amelx, Ambn, Enam and Mmp20, that are expressed by secretory-stage ameloblasts22 (Fig. 2d,e). Furthermore, we identified groups of genes that were upregulated (Clu and Steap; Fig. 2d,e), downregulated (Igfbpl1 and Ank2; Fig. 2g), or transiently upregulated and then downregulated (Otop2 and Dspp; Fig. 2f), most of which have not previously been linked to the differentiation of ameloblasts (Supplementary Table 3). Insitu hybridization validated the expected correlation between the spatial and pseudo-temporal organization of cells undergoing amelogenesis (Fig. 2c–g, Supplementary Fig. 3g).
Fig. 2 |. An unbiased catalogue of cell types during amelogenesis.
a, PBA was used to order 431 cells through the pseudotime of their differentiation trajectory from pre-ameloblast to ameloblast. Black regions define the source and sink cells of the trajectory. Gene expression along the trajectorywas smoothed by averaging over a sliding window of 20 cells along the trajectory. We centred the exploration of gene-expression changes along the ameloblast trajectory around the onset of the expression of known mature ameloblast genes by excluding the first 300 pre-ameloblast cells. The resulting ordering of averaged cells is referred to as the ameloblast differentiation trajectory. b, Schematic of the ameloblast differentiation trajectory; dark grey represents earlier cells in the trajectory. c, A heat map of 439 genes that showed statistically significant variability (false discovery rate (FDR)-adjusted P < 0.05) along the ameloblast differentiation trajectory. Rows are genes, columns are single cells ordered by pseudotime. d–g, Expression of selected markers of ameloblast differentiation along pseudotime (scRNA-seq; top) and insitu (RNAscope; bottom). The y axis shows the average expression of a gene within the window of 20 ± s.e.m. cells (or +1/(window size) for mean values of zero), normalized to the maximum value. Dashed lines outline the epithelium. Scale bars, 100 μm.
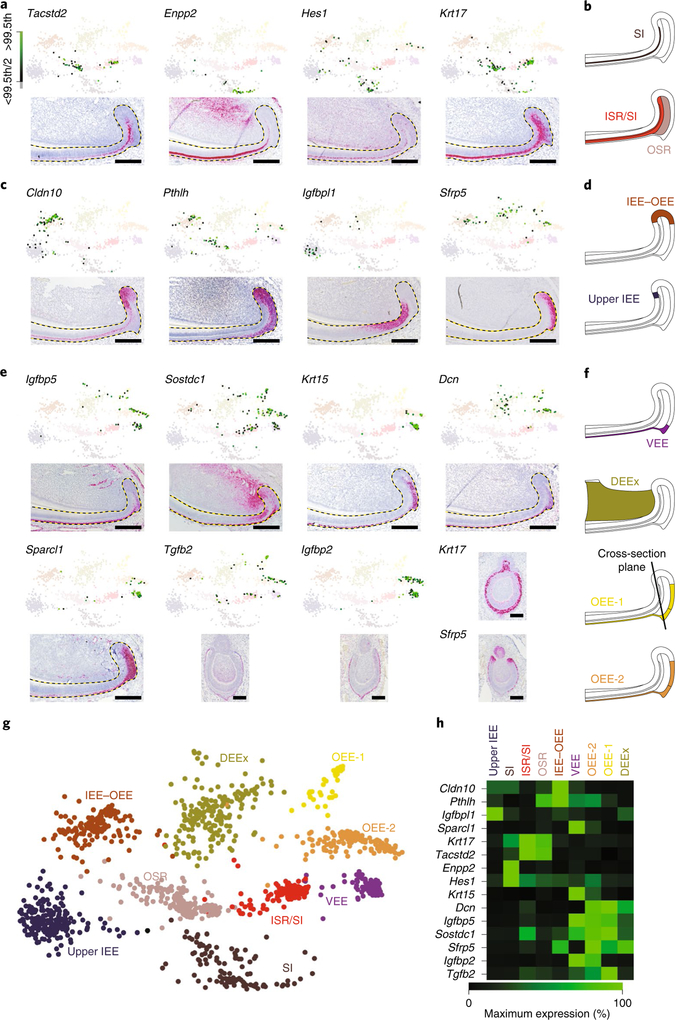
The third major class of cells was composed of 9 clusters that formed a continuum of transcriptional states in the two-dimensional (2D) representation of the SPRING k-nearest neighbour (kNN) graph without displaying clear boundaries (Fig. 1d). Enriched genes in each cell type labelled distinct compartments within the non-ameloblast region of the incisor epithelium (Fig. 3a–h, Supplementary Fig. 3h, Supplementary Table 4). Gene set enrichment analysis (GSEA) highlighted active transcription and translation in the upper IEE and at the IEE–OEE junction, suggesting that these populations may be immediate descendants of the progenitors (Supplementary Table 5), as well as significant enrichment of the Notch pathway in the SI.
Fig. 3 |. Mapping of class 3 cell types to spatial regions of the incisor-epithelium growth region.
a,c,e, SPRING plots (top) and insitu detection by RNAscope (bottom) of marker transcript combinations for each cell type in class 3. Dashed lines outline the epithelium. Scale bars, 100 μm. Color scale shows expression percentile. b,d,f, Schematics of the expression pattern for the cluster are shown. Candidate genes were selected from the top ten enriched genes per cluster. Ranking is provided in Supplementary Table 1. g, An expanded SPRING view of the 9 clusters composing class 3. h, A heat map showing the expression profile of genes used to define each cell type in class 3.
Proliferation dynamics of the incisor epithelium.
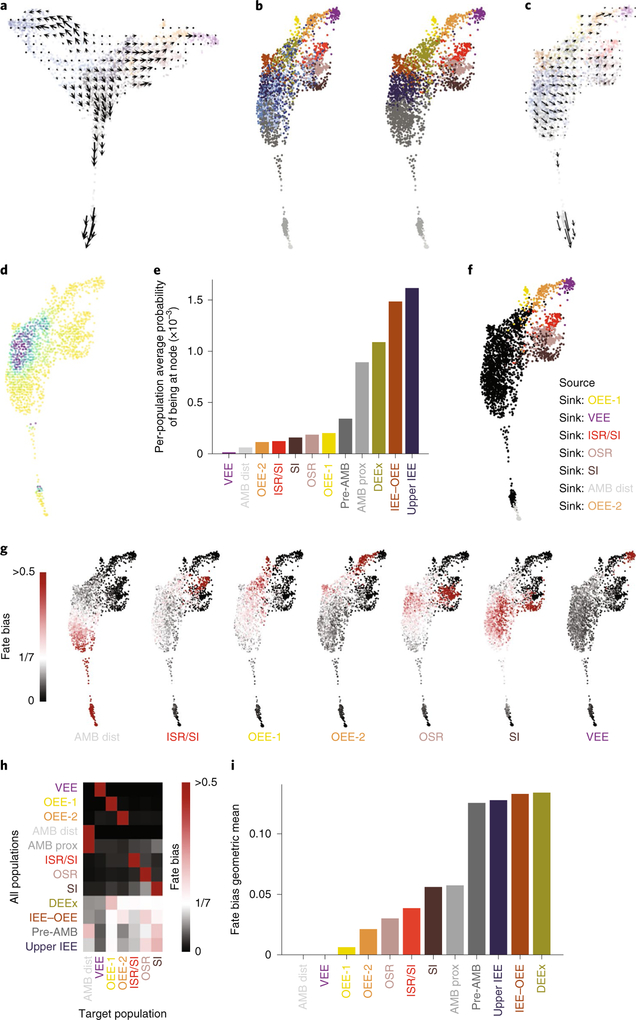
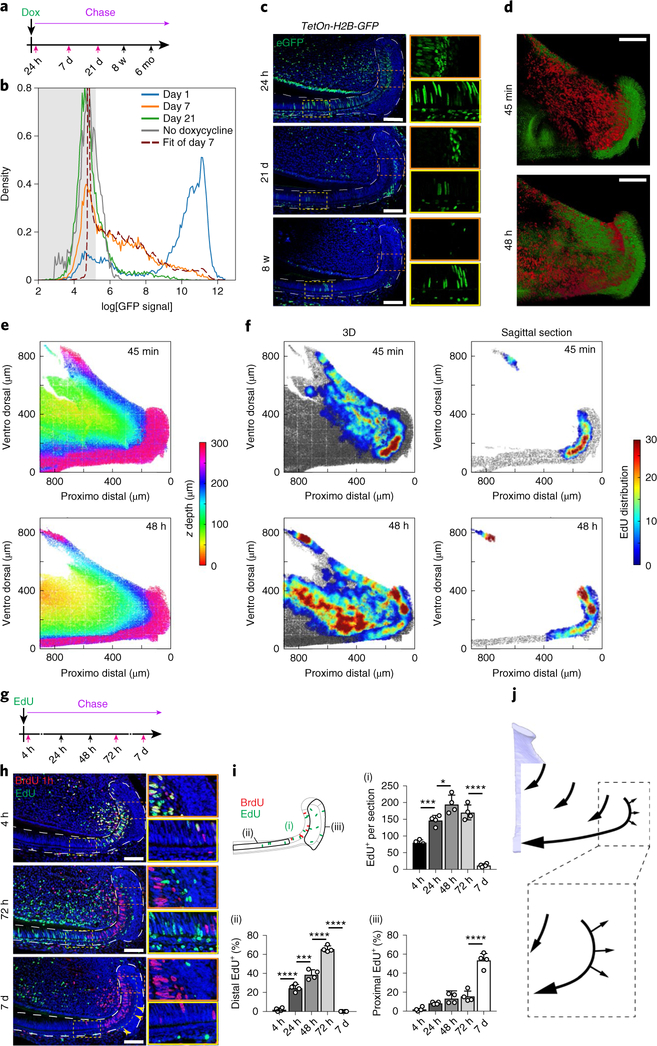
Together, these observations suggested that the IEE (class 1) houses progenitor cells that give rise to the two other classes (classes 2 and 3) of cells. Indeed, RNA velocity analysis23 confirmed these relationships (Fig. 4a), indicating class 1 cells as the root (Fig. 4b–e). We further computed the fate biases using FateID24, which also suggested that certain class 1 cells may differentiate toward specific class 2 and/or class 3 cell states (Fig. 4f–i). As such a lineage hierarchy is inconsistent with the currently accepted model (Fig. 1c), we further examined the dynamics of the incisor growth region using green fluorescent protein (GFP)-tagged H2B label dilution (Supplementary Fig. 4a–e, Supplementary Note). Epithelial cells were isolated from the incisor growth region of adult TetOn-H2B-GFP mice25, which were pulse treated with doxycycline, after various days of chase to dilute out the GFP label; GFP densities were measured using fluorescence-activated cell sorting (FACS; Fig. 5a,b). A simple Poisson model was used to fit the distribution of the GFP signal at day 7, with day 1 as the initial condition. The data indicated that, on average, the proliferating population divided 2.98 ± 0.20 times between day 1 and 7, and that 60 ± 15% of the cells were non-dividing (Fig. 5b).
Fig. 4 |. Class 1 cells house progenitor cells that give rise to class 2 and class 3 populations.
a, Velocity vectors projected onto the SPRING plot shown in Fig. 1d and smoothed over a regular grid. b, SPRING plot of all of the cells shown in Fig. 1d after regressing out the cell cycle (left). Right, the same plot as on the left after merging the cycling class 2–3-like cells with their non-cycling counterparts. c, Velocity vectors projected onto the SPRING plot from b and smoothed over a regular grid. d, The differentiation start point is represented as high-density regions on the 2D SPRING plot after a Markov process with transition probabilities on the basis of reversed velocity vectors. e, The data from d are shown as average density for each population shown in b, right. f, The seven target populations (fate) used to infer the fate biases of each cell using FateID. g, SPRING plots coloured by FateID with a fate bias towardsa given target population. The colour ranges from white (equal probability of each fate) to red (>0.5 probability of a certain fate). h, The data from g with FateID fate biases averaged over the populations shown in b, right. i, Uniformity of the rows in h measured by calculating the geometric mean; the highest bars represent populations with the most uniform fate biases and, therefore, candidate multipotent states.
Fig. 5 |. Proliferation dynamics of the incisor epithelium growth region.
a, Schematic of doxycycline pulse treatment and timepoints of sample collection during the chase; five samples were collected per timepoint. Red arrows indicate timepoints of samples that were analysed by FACS in b. b, FACS histograms showing H2B–GFP dilution patterns at chase days 1, 7 and 21, and the best fit of the distribution of the GFP signal at day 7 using the distribution at day 1 as the initial condition and the proportion of non-dividing cells (r) and the number of divisions (λ) as the parameters. The best fit was for r = 0.60 ± 0.15 and λ = 2.98 ± 0.20, indicating that the proliferating population has divided three times on average between day 1 and 7 and the proportion of cycling cells is 1 − 0.60 = 0.40. The experiment was performed once. c, GFP immunostaining in the incisor growth region of H2B–GFP expressing miceat the indicated time after pulse treatment with doxycycline. Magnifications of the areas indicated by orange and yellow dashed boxes are shown on the right. d–f, A whole-mount proliferation assay 45 min (top) and 48 h (bottom) after EdU injection. d, Two-photon micrographs of K14CreER; R26mT/mG mice. The membrane of epithelial cells is labelled green and EdU is labelled red. e, A z-depth view of the incisor growth region calculated from the two-photon images. f, Heat maps showing the local average percentage of EdU+ cells in three dimensions (3D; left) and in sagittal section (right) of the incisor growth region. The experiment was performed once. g, EdU/BrdU labelling assay and timepoints of sample collection during the chase. The red arrows mark samples used in h; four samples were collected per timepoint. h, Immunostaining of mice killed at the indicated timepoints after EdU injection; BrdU was injected 1 h before organs were collected. i, EdU+ cells were counted in the entire incisor growth region (i), distal to the BrdU+ region (ii) and proximal to the BrdU+ region (iii). The percentage of total EdU+ cells present in distal or proximal region is shown in (ii) and (iii); n = 4 animals. For the bar charts, data are mean ± s.d. Normally distributed data were analysed using one-way ANOVA with Tukey–Kramer post hoc test. Significance was taken as P < 0.05 with a confidence interval of 95%. ****P < 0.0001. j, Schematic of cell dynamics (outline by arrows) in the incisor growth region. For c and h, dashed lines outline the epithlium; magnifications (×2.6) of the areas indicated by orange and yellow dashed boxes are shown on the right; scale bars, 100 μm.
Histological analysis revealed that H2B–GFP, which was initially present in proliferating IEE and adjacent SR cells (Fig. 5c, top), was completely diluted after 21 d, consistent with the rapidly cycling nature of this region and indicating that non-dividing cells were post-mitotic cells that left the IEE. By contrast, several GFP+ cells were detected in the SR–OEE region and the ameloblast layer at 21 d chase (Fig. 5c, middle), corresponding to a GFP peak (Fig. 5b, green line); these remained for up to 12 months (Supplementary Fig. 4f). Importantly, H2B–GFP labelling was absent in the SR–OEE region after a short pulse (Fig. 5c, top), suggesting that, similar to previous studies in the ear epidermis26, the label-retaining cells in this region are post-mitotic.
To challenge these findings using an independent assay, we quantitatively analysed the spatial distribution of proliferating cells within the growth region at different timepoints (Fig. 5d–f, Supplementary Note). We focused on the initial 48 h following 5-ethynyl-2′-deoxyuridine (EdU) injection to study the first cell cycle, before labelled cells are lost due to label dilution (more than four cell cycles27) or distal displacement. After 45 min, the highest concentration of EdU+ cells was in the centre of the IEE, with labelling also present in the immediately adjacent portion of the SR and the epithelial extension from the laCL towards the lingual cervical loop (liCL; Fig. 5f), confirming that these compartments are sites of active cell division. By 48 h, higher levels of EdU+ cells were found at the liCL and the ridge of the adjacent epithelium, reflecting a doubling in the proliferating cells, and in the more distal portion of the IEE, corresponding to distal displacement of cells. Lastly, similar to the H2B–GFP results, EdU+ cells were observed in the SR–OEE region, away from the IEE (Fig. 5f). We verified these patterns of cell proliferation and movement by counting EdU-labelled and 5-bromodeoxyuridine (BrdU)-labelled cells in incisor growth-region compartments following a double-pulse analysis (Fig. 5g–i).
Taken together, the kinetics results support the hypothesis generated from the scRNA-seq data that proliferating IEE progenitors give rise to two groups of cells—a larger population that populates the ameloblast layer and a smaller population that exits the IEE to become SR and OEE cells (Fig. 5j).
scRNA-seq reveals changes in cell cycle and differentiation program during recovery.
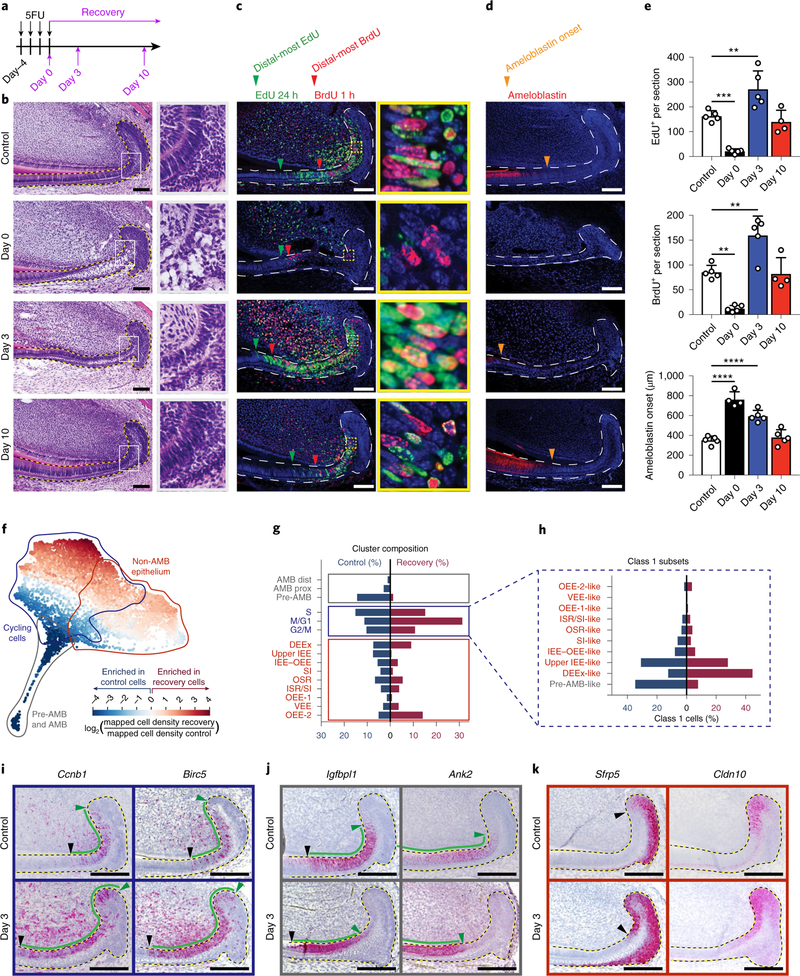
Having elucidated the steady-state population hierarchy and kinetics, we next analysed the system under non-homeostatic conditions by treating wild-type mice with 5-fluorouracil (5FU)28,29 for 4 d. This eliminated the majority of proliferative epithelial cells, which resulted in tissue loss and severely perturbed the shape of the cervical loop, whereas the non-cycling regions were relatively unaffected (Fig. 6a–c, Supplementary Fig. 5a). Moreover, 5FU-treated mice exhibited abnormal ameloblast organization and delayed and disorganized enamel matrix formation (Fig. 6b,d,e). Then, 3 d after treatment with 5FU (that is, recovery day 3), the proliferating cell population burgeoned, without concomitant proliferation in the OEE (Fig. 6c,e). Furthermore, cell-cycle time in the IEE was shortened, as evidenced by the increase in cells that re-entered the cell cycle after a 24 h chase (23–47 BrdU+EdU+ cells persection; Fig. 6c, yellow box). The regeneration process peaked between 3–5 d after treatment with 5FU and, by recovery day 10, treated incisors were indistinguishable from the controls (Fig. 6b–e).
Fig. 6 |. Single-cell transcriptomic analysis reveals changes in cell cycle and differentiation program after injury.
a, Schematic of 5FU treatment and sample collection timepoints during recovery. b–d, Control and 5FU-treated mice were analysed at the indicated timepoints using haematoxylin and eosin (H&E) staining (b), EdU and BrdU labelling (c), or ameloblastin immunostaining (d). Green and red arrowheads indicate the most distal EdU-labelled and BrdU-labelled cells, respectively, and orange arrowheads indicate the onset of ameloblastin expression. Boxed areas (grey in b, yellow in c) are magnified (b, ×3.7; c, ×9.1) on the right. e, Quantification of EdU+ and BrdU+ cells persection and the distance of ameloblastin expression onset from the proximal edge of the laCL are quantified from control or 5FU-treated mice; n = 5 animals, except for ameloblastin at day 0 and EdU and BrdU at day 10, for which n = 4 mice. For the bar charts, data are mean ± s.d. Normally distributed data were analysed using parametric tests including one-way ANOVA with Tukey–Kramer post hoc test. Significance was taken as P < 0.05 with a confidence interval of 95%. f, SPRING plot of injury and control cells coloured by the fold change in density between the two conditions. g, The proportion of cells in each population defined in control (blue) and recovery (red) conditions. h, Cycling-cell subset size for control and recovery conditions. i–k, RNAscope staining for markers of class 1 cycling cells (i), class 2 pre-ameloblasts (j), and markers of class 3 cells in control (top) and recovery day 3 (bottom) samples (k). For b and i–k, dashed lines outline the epitehlium. For b–d and i–k, scale bars, 100 μm.
To determine the transcriptional signature during recovery, we performed scRNA-seq analysis on sorted incisor epithelial cells from 5FU-treated mice at recovery day 3 (Supplementary Fig. 3b). SPRING visualization of transcriptomes from the recovery conditions retained the three-class layout observed in the control (Supplementary Fig. 5b,c). We next visualized the relative density of cells in recovery versus control conditions (Fig. 6f). We also calculated the relative abundance of each of the 15 populations after annotating cells in recovery by the closest expression profile that was observed in the control (Fig. 6g). In the recovery sample, we detected counterparts to all of the control clusters with similar gene-expression profiles, except for the proximal ameloblast cluster (Supplementary Fig. 5d, Supplementary Table 6). We found an increase in class 1 cells at recovery day 3 (57.1% of the recovery cells versus 36.7% in control; Fig. 6g), which was also shown by increased and expanded expression domains of Ccnb1 and Birc5 (Fig. 6i, Supplementary Fig. 5e). The transcriptomic signature of class 1 cells remained very similar to their control counterparts as depicted by comparable GSEA profiles (Supplementary Table 7). By contrast, the class 2 population (pre-ameloblasts and ameloblasts) was rarely observed in the recovering epithelium (1.3% of the recovery cells versus 18.3% in control; Fig. 6g). This was also demonstrated by the distal shift in the expression of two pre-ameloblast markers, Igfbpl1 and Ank2 (Fig. 6j, Supplementary Fig. 5f), as well as ameloblastin (Fig. 6d), implying delayed differentiation of ameloblasts.
Owing to the large increase in proliferative cells, we next investigated the underlying subsets of class 1 cells using SPRING after regressing out the cell-cycle effect. Similar to what we observed in the control cells, class 1 cells showed a substructure that reflected the class 2 and 3 populations (Supplementary Fig. 5h,i, Supplementary Table 8). In class 1, there was a large decrease in the pre-ameloblast signature (from around 35% to 4% of cells), a marked increase in the epithelial extension signature (from 12% of cells in control to 43% of cells in injury) and a decrease in SI-like cells (from 8% to 4%; Fig. 6h). Recovery was also associated with the upregulation of several genes that are typically expressed in class 3, including Sfrp5 and Cldn10, the expression domains of which expanded towards the proliferating regions (Fig. 6k, Supplementary Fig. 5g). The data from the recovery studies indicate that, after injury, the epithelium is repaired by recruiting more progenitors to cycle and by triggering changes in the proportions of progenitor cells, as well as by increasing the number of times progenitor cells replicate owing to a shortened cell cycle and a delayed onset of differentiation.
Injury induces Notch1-mediated conversion of SI cells into ameloblasts.
To further study the mechanisms that underlie recovery, we focused on the SI cells. SI cells have been postulated to descend from the OEE–SR16 and to act as differentiated supporting cells for ameloblasts30. However, our kinetic analyses indicated that SI cells are an intermediate population derived from the IEE, and that they are in fact progenitors of SR and OEE cells. To determine the role of the SI during homeostasis and injury, we studied our scRNA-seq results to identify markers that would enable the labelling of these cells and found that several components of the Notch signalling pathway were highly enriched in SI cells (Supplementary Fig. 6b, left panels). This was consistent with our observation of active Notch1 signalling in SI cells by immunostaining (Fig. 7a, Supplementary Fig. 6a) as well as with previous reports of Notch1 expression in SI cells12,31,32. Interestingly, during the initial days after injury, Notch1 expression and activity were downregulated (Supplementary Fig. 6a–d, Supplementary Table 9).
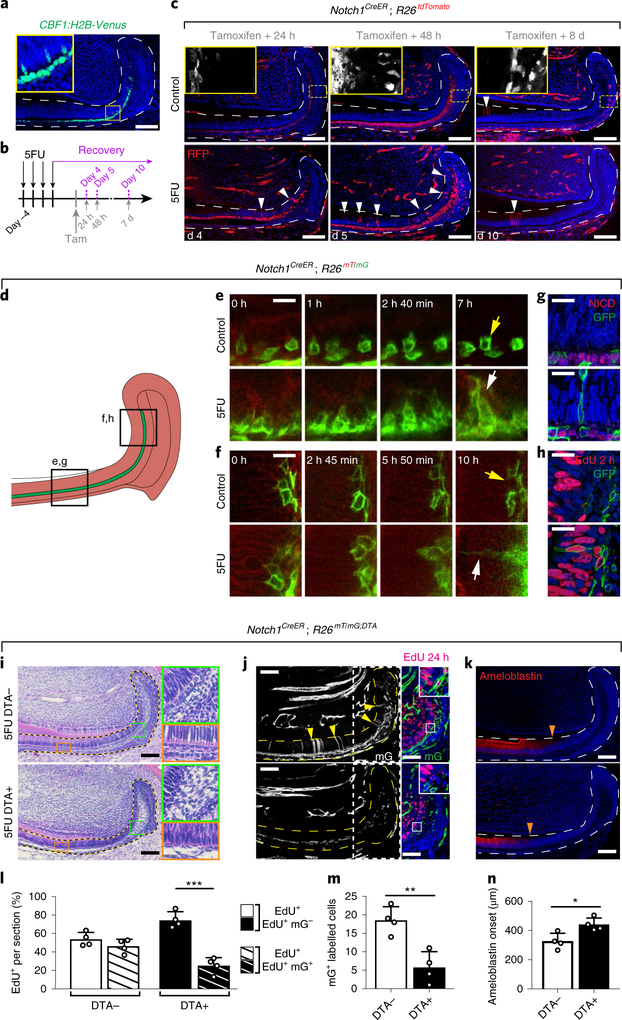
Fig. 7 |. Si cells undergo direct conversion to ameloblasts to contribute to the recovery of injured incisor epithelium.
a, Notch1 pathway activity was assessed using the CBF1:H2B-Venus reporter. b, Schematic of 5FU treatment, tamoxifen induction and sample collection timepoints of Notch1CreER; R26tdTomato mice. c, During recovery, Notch1-expressing cells contribute to the regeneration of wounded dental epithelium. Incisors from control (top row) and 5FU-treated (bottom row) mice were analysed at the indicated timepoints after tamoxifen injection. The white arrowheads indicate traced cells that have entered the proliferating region of the IEE–ameloblast layer. Areas indicated by yellow boxed areas are magnified (×4.1) on the top-left, in which tdTomato+ cells are shown in white without DAPI staining. d, Schematic of explant time-lapse imaging. e,f, Two-photon time-lapse live imaging shows the movement of Notch1-expressing cells during 8 h (ameloblast region; e), or 10 h (IEE region; f) of culture (Supplementary Videos 1–4). g, NICD (red) and GFP (green) immunostaining in control Notch1CreER; R26mT/mG mice (top) and Notch1CreER; R26mT/mG mice at recovery day 5 (bottom), 48 h after tamoxifen induction. h, EdU (red) and GFP (green) immunostaining in control Notch1CreER; R26mT/mG mice (top) and in Notch1CreER; R26mT/mG mice at recovery day 5 (bottom), 48 h after tamoxifen induction. i–n, Ablation of Notch1-expressing cells impaired normal recovery after injury with 5FU. DTA+ (Notch1CreER; R26RmT/mG; RosaDTA/–) and DTA– mice were examined at recovery day 7 using H&E (i), EdU and GFP (j), and ameloblastin immunostaining (k). Areas indicated by green (×4) orange (×4) and yellow (×3.5) boxes are magnified. Areas indicated by white boxed on the left panels in j, are shown with EdU and GFP staining on the right panels. The yellow arrowheads indicate traced cells that have entered the proliferating region of the IEE or the ameloblast layer. The orange arrowheads indicate the onset of ameloblastin expression. The number of proliferating cells (i), cells descended from Notch1-expressing progenitors in the ameloblast or IEE layers (m) and the distance of ameloblastin expression onset from the proximal edge of the laCL (n) are quantified from DTA+ or DTA− mice injured by treatment with 5FU; n = 4 animals. For bar charts, data are mean ± s.d. Normally distributed data were analysed using parametric tests including one-way ANOVA with Tukey–Kramer post hoc test. Significance was taken as P < 0.05 with a confidence interval of 95%. For a, c and i-k, dashed lines outline the epitehlium. For a, c and g–k, scale bars, 100 μm. For e and f, scale bars, 10 μm.
To fate-map and examine the clonal capacity of SI cells during both homeostasis and repair, we generated Notch1CreER; R26tdTomato mice (Fig. 7b). Marked cells were present exclusively in the SI layer 24 h after a single dose of tamoxifen (Fig. 7c, top row). Then, 48 h after CreER activation, labelled cells were found in the SR and the OEE, confirming that SR and OEE cells are derived from SI cells. Notably, 8 d after induction, scattered labelled cells were observed among the overlying ameloblasts, suggesting that descendants of SI cells infrequently contribute to normal ameloblast turnover (Fig. 7c, top row). This prompted us to investigate whether SI cells could act as reserve progenitors; lineage tracing of Notch1CreER;R26tdTomato cells after treatment with 5FU led to labelling in the proliferative region of the IEE and ameloblast layer as early as 24 h after Cre induction (Fig. 7c, top row). When a higher dosage of tamoxifen was used, we observed labelling of almost every cell in the IEE–ameloblast layer of 5FU-treated mice; by contrast, in untreated mice, the IEE–ameloblast layer remained sparsely labelled (Supplementary Fig. 7a,b). In injured mice, the number of labelled IEE and ameloblast cells continued to increase in the first 5 d after CreER induction but then decreased thereafter as labelled cells moved distally while being displaced proximally by newly formed unlabelled cells (Fig. 7c, Supplementary Fig. 7b).
To distinguish between upregulated Notch1 expression in injured IEE cells or ameloblasts and contribution to the IEE–ameloblast layer by Notch1-expressing SI cells, we performed live imaging of control and 5FU-treated incisor explants (Fig. 7d). In control explants, SI cells frequently sent small transient protrusions into the adjacent IEE–ameloblast layer; by contrast, in 5FU-treated explants, the persistent protrusions of SI cells extended further into the injured IEE–ameloblast layer, and the SI cells frequently migrated completely into these damaged layers (Fig. 7e,f, Supplementary Videos 1–4). The descendants of the Notch1-expressing cells in the IEE-ameloblast layer did not maintain Notch activity, as indicated by negative staining of the Notch1 intracellular domain (NICD; Fig. 7g), such that they lost their original SI fate and few of the descendants were actively proliferating (Fig. 7h). Importantly, the contribution of SI cells to injury repair is not limited to 5FU-induced injury, as an increase in labelled ameloblasts derived from Notch1-expressing cells was similarly observed after mechanically severing the incisor tip (Supplementary Fig. 7c,d).
Lastly, we tested the functional importance of Notch1-expressing SI cells during regeneration by eliminating Notch1+ cells through diphtheria toxin expression33 (Supplementary Fig. 7e). Elimination of Notch1+ cells resulted in marked reduction in proliferative Notch1+ (mGFP+) cells in the IEE–ameloblast layer, impaired repair and delayed ameloblast differentiation at recovery day 7, compared with 5FU-treated samples in which the Notch1-expressing cells were not ablated (Fig. 7i–n). Together, these experiments indicate that Notch1+ SI cells are critical for timely and proper recovery from injury.
Discussion
The continuously growing mouse incisor model has enabled the identification of a number of fundamental mechanisms in this renewing tissue34–36, but the underlying SC biology has remained unclear. Here, through an unbiased interrogation of the incisor epithelium, we pinpointed the identity of the various populations that comprise the mouse incisor, and we uncovered the cellular hierarchy and division kinetics of this renewing structure. Together, our data highlight a mechanism by which incisor progenitors generate both the ameloblasts and the non-ameloblast epithelium (Supplementary Fig. 7f). Although homeostatic renewal from highly proliferative progenitors has been observed in other self-renewing tissues26,37,38, it has not yet been documented in epithelial appendages, such as the hair follicle39 or the nail40. Our findings that the epithelial SCs give rise to the non-ameloblast cell layers also suggest that the SCs may produce components of their own niche.
SCs and their descendants exhibit a diverse repertoire of responses to meet the demands of injury-induced recovery41,42. Over the past several years, the transcriptional programs that underlie these cellular behaviours have been dissected using scRNA-seq, revealing transcriptional events and cell states that are unique to injury-induced recovery, such as activation of progenitor reservoirs43,44, transition of one cell state to another45,46 or initiation of a developmental program47. Our findings indicate that the incisor epithelium recovers from cytotoxic injury using the same cell types that enable homeostatic renewal without the emergence of a cell type that is unique to injury. It is still possible that a distinct repair program exists and was not detected, either because we probed the injured epithelium at a single timepoint or owing to the type of injury that we used. Thus, future studies comparing diverse injury and damage models will be useful for determining the range of approaches that tissues can utilize for repair. Furthermore, the finding that the progenitor cells in the incisor epithelium are highly heterogeneous in their differentiation potentials suggests that they are biased to differentiate either into ameloblast or into one of the non-ameloblast cell subtypes. The mechanisms that control such lineage biases are becoming more recognized9,48, but how such specification occurs in the incisor remains to be elucidated.
Our results provide an unbiased catalogue of cell types in the mouse dental epithelium at the single-cell level and offer a resource for functional studies of specific cell populations and genes during steady state and injury. Although the current study revealed the function of one of the non-ameloblast cell-layer populations, it will be important to determine the functional role of the other cell types of this class. Similarly, it will be of interest to use similar techniques to elucidate SC identity and position and the function of the mesenchymal compartment of the incisor niche to generate a broad and complete understanding of incisor renewal as a model for organ homeostasis.
Methods
Mice.
Mice were maintained in the University of California San Francisco (UCSF) specific pathogen-free animal facility in accordance with the guidelines established by the Institutional Animal Care and Use Committee and Laboratory Animal Resource Center. All of the experimental procedures were approvedby the Laboratory Animal Resource Center at UCSF. The incisor injury and kidney-capsule transplantation experiments were performed according tothe Swiss Animal Welfare Law and in compliance with the regulations of the Cantonal Veterinary Office, Zurich. Male mice aged 8–12 weeks were used for all of the experiments, except those in which mice were analysed at various ages to determine the steady state (as described in the main text). The following mouse lines were used: wild-type (C57BL/6J, Jax: 000664); Gli1CreER (Gli1tm3(cre/ERT2)Alj, Jax: 007913); Sox2CreER (Sox2tm1(cre/ERT2)Hoch, Jax: 017593); Bmi1CreER (Bmi1tm1(cre/ESR1)Mrc, Jax: 010531); Lrig1CreER (Lrig1tm1.1(cre/ERT2)Rjc, Jax: 018418); Notch1CreER (Notch1tm5(cre/ERT2)Rko, Jax: 027235); K14cre (Tg(KRT14-cre)1Amc, MGI: 2445832); R26RmT/mG (Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo, Jax: 007576); R26RlacZ (Gt(ROSA)26Sortm1Sor, Jax: 003474); R26RtdTomato (Gt(ROSA)26Sortm14(CAG-tdTomato)Hze, Jax: 007914); Rag1–/– (Rag1tm1Mom; Jax: 002216); TetOff-H2B-GFP (Tg(tetOHIST1H2BJ/GFP)47Efu, Jax: 005104); K5tTA (Tg(KRT5-tTA)1216Glk, MGI: 3575755); TetOn-H2B-GFP (Gt(ROSA)26Sortm1(rtTA*M2)Jae::Col1a1tm7(tetO-HIST1H2BJ/GFP)Jae, Jax: 016836); rtTA (Gt(ROSA)26Sortm1(rtTA*M2)Jae::Col1a1tm1(tetO-cre)Haho, Jax: 021025); CBF1:H2B-Venus (Tg(Cp-HIST1H2BB/Venus)47Hadj, Jax: 020942); and Rosa-DTA (B6.129P2-Gt(ROSA)26Sortm1(DTA)Lky/J, Jax: 009669).
Tissue preparation and histological analysis.
Euthanized mice were perfused using PBS and then 4% paraformaldehyde (PFA) in PBS. Lower mandibles were dissected away from the rest of the cranium and fixed with 4% PFA in PBS overnight at 4 °C. Mandibles were then decalcified, paraffin sectioned and H&E stained as described previously15.
Immunofluorescence staining.
For immunofluorescence, paraffin sections were rehydrated and antigen retrieval was performed by sub-boiling slides ina microwave for 15 min in a citrate buffer (pH 6.2) containing 10 mM citric acid, 2 mM EDTA and 0.05% Tween-20. For BrdU immunostaining, samples were additionally washed with 2 N HCl for 30 min. The following primaryand secondary antibodies were used: anti-ameloblastin (sc50534, Santa Cruz Biotechnology); rat anti-BrdU (ab6326, Abcam); mouse anti-BrdU (B35128, Invitrogen); anti-GFP (GFP-1020, Aves; ab13790, Abcam); anti-NICD (cleaved Notch1, D388, Cell Signaling); anti-RFP (600-401-379, Rockland). EdU was detected using a Click-iT Plus EdU Alexa Fluor 488 Assay Kit (Invitrogen, C10637). Samples were blocked in 1× animal-free blocker (Vector Laboratories, SP-5030) supplemented with 2.5% heat-inactivated goat serum, 0.02% SDS and 0.1% Triton X-100. All of the antibodies were diluted in the same blocking solution without serum. For the detection of NICD, primary antibodies were first detected using biotinylated secondary antibodies, and then sequentially amplified using VECTASTAIN Elite ABC HRP Kit (Vector Laboratories, PK-6100) and Tyramide Signal Amplification (PerkinElmer, SAT704B001EA). TUNEL staining (Roche, 12156792910) was performed according to the manufacturer’s instructions. DAPI (Invitrogen, D1306) was used for nuclear staining and all of the images were acquired using a Leica-TCS SP5 confocal microscope.
X-gal staining.
β-Galactosidase expression from Gli1CreER; R26lacZ transgenic mice was detected by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) staining as described previously18.
In situ hybridization.
An RNAscope 2.5 HD Red (ACD, 310036, 322350) detection kit was used according to the manufacturer’s instructions. Sections were boiled in the target retrieval solution at ~100 °C for 15 min and incubated in the Protease Plus solution at 40 °C for 15 min. DapB (ACD, 310043) was usedas a negative control along with appropriate positive controls. Negative controls showed little to no background staining. Probes for the following genes in Mus musculus were used: Ambn (508241), Ank2 (413221), Birc5 (422701), Bmi1 (312181), Ccnb1 (316241, custom-ordered against E5-E7), Cldn10 (320269 custom-ordered from 2–806 bp of NM_001160099.1), Clu (427191), Dspp (448301), Dcn (413281), Enpp2 (402441), Gli1 (311001), Hes1 (417701), Hey1 (319021), Igfbp2 (405951), Igfbp5 (425731), Igfbpl1 (488851), Krt15 (319091), Krt17 (479911), Lrig1 (310521), Mmp20 (479051), Notch1 (404641), Nrarp (411771), Otop2 (534911), Pthlh (456521), Sfrp5 (405001), Sostdc1 (313151), Sox2 (401041), Sparcl1 (424641), Steap1 (531061), Tacstd2 (471751) and Tgfb2 (406181).
Dual EdU and BrdU pulse–chase labelling.
Mice were given an intraperitoneal injection of 1 mg/25 g body weight of EdU (Invitrogen, C10637) and then analysed at various timepoints after treatment (exact chase timepoints are provided in the main text). Mice were injected with 1 mg/25 g body weight BrdU 1 h before analysis.
H2B–GFP label dilution experiments.
For the short H2B–GFP pulse experiments, mice were injected once intraperitoneally with 200 μl of doxycycline (2 mg ml−1, Enzo Life Sciences, ALX-380–273-G005) and then analysed at various timepoints after treatment (exact chase timepoints are provided in the main text). For the long pulse experiments (Supplementary Fig. 4f), mice were treated once with 200 μl of doxycycline (2 mg ml−1) by intraperitoneal injection. Simultaneously, doxycycline was added continually to their drinking water (1 mg ml−1 supplemented with 5% sucrose) for 3 weeks.
Incisor injury and kidney capsule transplantation.
Notch1CreER; R26mT/mG adult mice were injected intraperitoneally with 4-hydroxytamoxifen (4-OHT, Sigma-Aldrich, H7904) dissolved in corn oil (100 mg kg−1 body weight). Then, 24 h later, lower incisors were carefully dissected and the posterior part of the incisors was separated by cutting immediately anterior to the lingual cervical loop after the onset of mineralization using a surgical scalpel. The dissected incisors were transplanted under the kidney capsule of immunocompromized mice (Rag1−/−). Intact uncut incisors were used as controls. To minimize postoperative pain, 1 h before intervention, Rag1−/− mice were injected subcutaneously with buprenorphine (0.1 mg kg−1 body weight, Temgesic, Reckitt Bencklser, 3002981). The recipient mice were then anaesthetized by isoflurane inhalation (Attane Isoflurane, Piramal Healthcare, NDC 66794-014-25), laid on one side and an incision was made to enable the exposure of one kidney. The kidney capsule was then incised using forceps. The capsule was lifted and the incisors were grafted under the capsule. The mice were then sutured with an absorbable polyglycolic acid suture (Resorba, PA10242) and the skin incision was closed with a metal clip (Fine Science Tools, 12040–01). Buprenorphine was supplied in the drinking water (10 μg ml−1) for 3 d. Then, 10 d after transplantation, mice were killed and kidneys were collected and dissected, and the regions hosting the grafted incisors were isolated. The samples were then fixed in paraformaldehyde 4% for 1 h, washed with PBS and cleared for whole-mount confocal imaging by immersion in Focus Clear (CelExplorer, FC-101) overnight at 4 °C.
Micro-computed tomography.
For ex vivo imaging of the entire incisor, mandibles were collected and dehydrated through an ethanol series up to 70% ethanol. Samples were scanned using MicroXCT-200 (Carl Zeiss Microscopy) at40 kV and 160 μA. We obtained 800 projection images, taken at a total integration time of 4 s with linear magnification of ×2 and a pixel size of 10 μm. To image the incisor growth region, samples were also soaked in phosphotungstic acid overnight to differentially stain soft tissues as described previously49 and scanned at 60 kV and 200 μA. We obtained 1,200 projection images, taken at a total integration time of 6 s with a linear magnification of ×10 and a pixel size of 2.4 μm. Micro-computed tomography calibration and mineral-density determination were performed as described previously50.
For in vivo assessment of the enamel eruption rate (Supplementary Fig. 1a,b), mice were anaesthetized and the cranial region spanning the length of the incisors was scanned using Scanco vivaCT 40 (Scanco Medical AG). Images were taken at 55 kV with an integration time of 250 ms and a pixel size of 10.5 μm. Images were analysed using Avizo (FEI).
Cell suspension.
Proximal incisors (roughly 3 mm in length) were first severed away using a pair of scissors after removal of the surrounding jaw bones. The bulbous portion, as well as the lateral wing-shaped epithelium, and the surrounding mesenchyme were subsequently dissected from the rest of theincisor and collected in cold Ca2+- and Mg2+-free HBSS (pH 8.0, UCSF Cell Culture Facility, CCFAJ005–16CT01) and digested enzymatically at 37 °C in collagenase type II (2 mg ml−1; Gibco/BRL type II in DMEM) for 30 min, followed by collagenase inactivation with 2% FBS. The digested tissue was mechanically disrupted using a 1 ml pipette tip and then centrifuged at 400g for 3 min. Cells were resuspended in HBSS (pH 8.0) containing 2% FBS.
FACS analysis.
To sort epithelial cells, proximal-incisor cell suspensions were stained with monoclonal antibodies that specifically recognize CD326 (G8.8, AF647 conjugate, Biolegend, AB_1134104) for 30 min on ice followed by centrifugationat 400g for 3 min. Before FACS analysis (BD FACSAria2 SORP), cells were filtered through a 40 μm mesh (BD Falcon, 22363547) and stained with DAPI.
Time-lapse live imaging.
Time-lapse live imaging was performed as previously described51. In brief, mandibular incisors were dissected in PBS with 0.5% glucose52 and embedded in 0.65% low-melting agarose supplemented with 0.5% glucose and 1% penicillin–streptomycin in organ culture dishes (Becton Dickinson, 353037). Culturing medium (50% DMEM/F12, 50% rat serum, 0.5% glucose, 250 μg ml−1 ascorbic acid and 1% penicillin–streptomycin) was then added and agarose was carefully removed from the top of the sample to expose the dental epithelium for imaging. Samples were maintained under constant medium perfusion using a Delta T pump (Bioptechs, 60319131616) at 37 °C, 95% O2 and 5% CO2, and were imaged at a wavelength of 920 nm using a Nikon A1R two-photon microscope equipped with a ×25 1.1 NA water-immersion lens to capture the movement of GFP+ Notch-expressing SI cells. Images were taken every 5 min for 8 h. Time-lapse videos were generated using ImageJ.
Single-cell RNA barcoding and sequencing.
Epithelial cells from five controls and five 5FU-treated mice were isolated as described above. The resulting cell suspensions (~20,000 cells each) were submitted as separate samples with targeted cell recovery of 3,000 per sample to be barcoded for scRNA-seq using a Chromium Controller (10X Genomics, 120263) and a Single Cell 3′ Library Kit v2 (10X Genomics, PN-120236/37/62). The resulting libraries were sequenced on a HiSeq 4000 (Illumina, SY-401–4001) using a HiSeq 4000 PE Cluster Kit (Illumina, PE-410–1001) with a HiSeq 4000 SBS Kit (150 cycles, Illumina, FC-410–1002); one sample was loaded into each sequencing lane. About 3,500 control cells (~89,000 reads per cell) and about 5,000 5FU-treated cells (~62,000 reads per cell) were successfully barcoded and their transcriptomes sequenced. Raw sequencing data were processed for initial quality control analysis and alignment by the sequencing core of the Institute of Human Genetics (UCSF) using the Cell Ranger (v.2.0.0) pipeline.
scRNA-seq data clean-up and normalization.
We performed extra steps to clean and normalize our data. We opted for thresholds of less than 1,000 total counts or more than 10% of counts coming from mitochondrial genes to filter out cells from the analysis, leaving 3,173 and 4,645 high-quality transcriptomes for control and recovery conditions, respectively. After initial visualization of the two conditions individually, we identified a fraction of cells (824 in control and 301 in recovery) that formed well-separated clusters and were characterized by distinct expression of either immune genes or mesenchymal cell markers (Supplementary Fig. 3d). These cells were excluded from further analysis. Data were rescaled to 10,000 total counts per cell.
Visualization of scRNA-seq data in 2D.
The high-dimensional scRNA-seq data were visualized in 2D as a force-directed layout of an undirected kNN graph using SPRING19. Interactive explorers of resulting SPRING plots can be accessed using the hyperlinks provided in Supplementary Table 10.
Classification of cells by cell-cycle phase.
Each transcriptome in the scRNA-seq data was classified as G1, G2/M, S (Fig. 1f) using cell-cycle scores calculated as part of the procedure for regressing out the cell cycle described in the next section.
Regressing out the cell-cycle effect.
The cell-cycle effect was regressed out from the count data by using simple linear regression (implemention in Scanpy53, which is equivalent to Seurat19). The procedure was performed solely for the purpose of SPRING visualization (Figs. 1i and 4b, Supplementary Fig. 5h).
Defining the cell populations observed in scRNA-seq data.
In control epithelial cells, populations were defined as belonging to spectral clusters of the kNN graph shown (Fig. 1d). Spectral clustering requires predefining into how many clusters a kNN graph is to be divided. Given the continuous nature of the epithelial-cell data with no clear boundaries visible in the SPRING graph, clusters are a convenient but not entirely natural way of describing the data. Therefore, the number of clusters reported for control epithelial cells in this study reflectsa chosen degree of granularity. We confirmed that clusters represented distinct states (at least 20 significantly differentially expressed genes between any pairof clusters). The SPRING graph in Fig. 1d was first divided into 12 clusters, a configuration that well-captured 3 clusters representing different phases of thecell cycle (among class 1 cells) as well as 3 clusters with distinct gene expression along the ameloblast differentiation trajectory (class 2). To gain a better resolution for the class 3 cells, which seemed to be more complex than class 1 and class 2 from analysis of the data, we built a separate SPRING graph for class 3 cells only and found that 9 spectral clusters best captured the complexity of these cells. The resulting 15 populations were given names on the basis of RNAscope staining patterns of population-enriched genes (Fig. 3, Supplementary Table 1). To define populations in the injury condition (Fig. 6h, Supplementary Tables 6 and 8), we used a classification-based approach, for which each cell in the injury conditions was given the label of the population in the control condition with the most similar expression profile. Likelihoods from the Bayesian classifier described in the next section were used as a similarity measure.
Bayesian classifier of cell populations.
To annotate single-cell transcriptomes using a chosen set of reference expression profiles, we applied a Bayesian cell classifier on the basis of a multinomial model that was reported previously54,55 and adapted from ref.56; we describe the model here for completeness.
Given (1) raw counts uj for gene j = 1,..N from a single cell and (2) a reference expression matrix Xi,j (reference population i = 1,..,M; gene j = 1,…,N), the classifier N returns the likelihood of cell type i given u, defined as where Z is a constant ensuring and P0(i) is a prior expectation on the abundance of reference cell population i(set to = constant for all applications in this paper, that is, a naive prior was used) and is the fraction of transcripts from gene j in reference cell population i.
To classify each cell in the recovery conditions as one of the 15populations defined in the control condition (Fig. 1d), the reference matrix X contained the per-population average expression of all genes. To classify each class 1 cell by its most similar profile among class 2 and class 3 populations, we used a reduced list of genes. For a gene to be included, it had to be among the top 100 most enriched genes in any class 2 or 3 population but not within the top 100 enriched genesin class 1 populations. Population-enriched genes were identified as described below. Such gene selection was used to exclude cell-cycle-related genes, which are uninformative in distinguishing between class 2 and class 3 subsets but could, in principle, confound the classification.
Identification of population-enriched genes.
Population-enriched genes were assessed by considering only genes that were (1) detected in at least 3 cells and with at least 6 counts after normalization across all cells, and (2) statistically significantly higher in cluster i compared with all other cells (FDR-adjustedP < 0.05, Mann–Whitney U-test). The heat map in Fig. 1e was generated using genes filtered, as described above, that were found within the top 20 ordered by fold change in a cluster of i versus all other cells. Furthermore, a pseudovalue of 1 count per 10,000 was added to buffer against low expression and avoid division by zero. The table containing the top 20 per control cluster is provided in Supplementary Table 1.
RNA velocity analysis.
RNA velocity analysis was performed as described previously23 using the velocyto.py analysis pipeline (http://www.velocyto.org). In brief, the method distinguished between unspliced and spliced transcripts in scRNA-seq and uses this information to extrapolate the future transcriptional state of each cell. Extrapolated states are vectors in the high-dimensional gene-expression space, and Fig. 4a,c shows their projection onto the 2D SPRING representation.
FateID analysis.
Fate biases were calculated using FateID24; the analysis was based on the vignette provided at https://github.com/dgrun/FateID. The method requires the selection of target states that represent differentiated populations. Figure 4f shows the target states that were chosen. These target states served as the training set in an iterative classification in which the test set is composed of cells in the neighbourhood of the training set. Neighbours are determined on the basis of a distance matrix, which can be defined by the user. Here we used the same distance matrix that was used to generate the SPRING graph shown in Fig. 4b.
Ameloblast trajectory analysis.
Class 2 cells, which form a simple linear continuum in the SPRING graph (Fig. 1d), were ordered through pseudotime using PBA21 as described previously46. The method requires manually selecting source and sink cells of the trajectory, which—in our case—were the pre-ameloblast and distal-ameloblast tips, respectively (Fig. 2a). Our choice of the source and sink was based on extensive previous knowledge of several key molecular markers of amelogenesis17,57,58, and was supported by our RNA velocity analysis (Fig. 4a). Cells were ordered by the diffusion ‘potential’ parameter of PBA. Gene expression along the trajectory was smoothed by averaging over a sliding window of 20 cells along the trajectory. We centred the exploration of changes in gene expression along the ameloblast trajectory around the onset of the expression of known mature ameloblast genes by excluding the first 300 pre-ameloblast cells.
Genes variable along the ameloblast trajectory were identified as described previously59 with minor changes (Supplementary Table 2). Before statistical testing, we prefiltered genes to be expressed at a rate of at least 3 counts per 10,000 in at least 2 cells, and have a variability statistic of V > mode(V) where V is the corrected Fano Factor described previously60. For every gene i among the surviving 2,453 genes, a statistic t was calculated as ti, observed = ai,max − ai,min where ai is a vector with the smoothed expression of i along the ameloblast trajectory. The procedure was repeated in a random ordering on class 2 cells for multiple rounds, each time yielding ti, random. The one-sided P value of this permutation test was defined as the fraction of times that ti,observed > ti,random. Multiple-hypothesis testing was accounted for by controlling the FDR at 5% using the Benjamini–Hochberg procedure.
Cell-density changes on the kNN graph.
The following was performed to estimate the density of cells from the control condition on the graph combining the control and recovery conditions (Fig. 6f). Every cell from the control condition was asked to vote for its five nearest neighbours (Euclidean distance in the same principal-component space as used for the SPRING visualization). Votes were counted for every cell resulting in a raw vote vector vraw = (v1,v2,…,vn) and smoothed votes were obtained as vsmoothed = Osvraw where the smoothing operator is Os = eLβ where L is the Laplacian matrix of the kNN graph visualized using SPRING and β is the parameter that determines the extent of smoothing. In physical terms, this parameter is equivalent to diffusion time, with larger values reflecting broader diffusion. We used β = 1. Smoothed votes were normalized to a total count of 1 million. The same voting was applied on the recovering cells, resulting in a cell density map for which the enrichment in recovery versus control cells can be assessed.
GSEA.
For the GSEA, we used the GSEA PreRanked tool61,62 on genes ranked by an enrichment score Ei obtained from comparing cells within a given population i with all other epithelial cells. The Ei was defined as:
where Pi and ri are the P value and fold change, respectively, obtained from comparing cells in population i to cells outside of i as described in the ‘Identification of population-enriched genes’ section;
The c2.cp.reactome.v6.2.symbols.gmt curated Molecular Signatures Database was probed and pathways for which P < 0.05 and FDR-adjusted P < 0.25 were considered to be significant.
Statistics and reproducibility.
All of the data points are biological replicates that were randomly assigned without investigator blinding. All experiments, exceptfor the scRNA-seq and the FACS histograms showing H2B–GFP dilution patterns (Fig. 5b), were replicated at least three times. Images in Fig. 5d,f were derived from two independent biological experiments that showed similar results. All of the other images are representative of at least three independent biological samples. No data were excluded. No statistical methods were used to predetermine sample size and differences in intrasample variances were present. Statistical significance was determined using Prism (GraphPad Software). Normally distributed data were analysed using parametric tests, including one-way ANOVA with Tukey–Kramer post hoc test, except in Supplementary Fig. 4b in which an unpaired t-test with Welch’s correction was used. Bar charts indicate the mean of samples and error bars represent ± s.d. of the mean. Significance was taken as P < 0.05 with a confidence interval of 95%. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The scRNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE131204. Source data for figures are provided in Supplementary Table 11. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Code availability
The custom code developed for the whole-mount proliferation assay (Fig. 5d–f) is provided at https://github.com/kephale/incisor-cell-segmentation. The code for selected single cell RNAseq data analyses including SPRING visualizations is available as Jupyter notebooks at https:///www.github.com/rapolaszilionis/Sharir_et_al_2019.
Supplementary Material
Acknowledgements
We thank A. Rathnayake, R. D’Urso, N. Wang, B. Hoehn and S. Alto for technical assistance, L. Jimenez-Rojo for mouse surgeries, K. Lindquist for consulting on GSEA analyses and J. Bush as well as members of the O.D.K. and A.M.K. laboratories for helpful discussion. This work was supported by the NIH (R35-DE026602 and R01-DE027620 to O.D.K). A.S. was funded by the NIH (K08-DE026219). J.H. was funded by the NIH (K99-DE025874). A.M.K. was funded by the NIH (5R33CA212697). UCSF Parnassus Flow Cytometry Core is supported by the NIH (P30DK063720 and 1S10OD021822-01). UCSF Core Center for Musculoskeletal Biology and Medicine is supported by the NIH (P30AR066262).
Footnotes
Competing interests
A.M.K. is a founder of 1CellBio. C.W.S. is employed by Genentech.
Supplementary information is available for this paper at https://doi.org/10.1038/s41556-019-0378-2.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of code and data availability and associated accession codes are available at https://doi.org/10.1038/s41556-019-0378-2.
References
- 1.Ahn Y Signaling in tooth, hair, and mammary placodes. Curr. Top. Dev. Biol 111, 421–459 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Naveau A, Seidel K & Klein OD Tooth, hair and claw: comparing epithelial stem cell niches of ectodermal appendages. Exp. Cell Res 325, 96–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thesleff I, Vaahtokari A & Partanen AM Regulation of organogenesis. Common molecular mechanisms regulating the development of teeth and other organs. Int. J. Dev. Biol 39, 35–50 (1995). [PubMed] [Google Scholar]
- 4.Lajtha LG Stem cell concepts. Differentiation 14, 23–33 (1979). [DOI] [PubMed] [Google Scholar]
- 5.Cotsarelis G, Sun T-T & Lavker RM Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61, 1329–1337 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Mackenzie IC & Bickenbach JR Label-retaining keratinocytes and langerhans cells in mouse epithelia. Cell Tissue Res 242, 551–556 (1985). [DOI] [PubMed] [Google Scholar]
- 7.Clevers H & Watt FM Defining adult stem cells by function, not by phenotype. Annu. Rev. Biochem 87, 1015–1027 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Paul F et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Yang H, Adam RC, Ge Y, Hua ZL & Fuchs E Epithelial-mesenchymal micro-niches govern stem cell lineage choices. Cell 169, 483–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ness AR Eruption rates of impeded and unimpeded mandibular incisors of the adult laboratory mouse. Arch. Oral Biol 10, 439–451 (1965). [DOI] [PubMed] [Google Scholar]
- 11.Warshawsky H & Smith CE Morphological classification of rat incisor ameloblasts. Anat. Rec 179, 423–445 (1974). [DOI] [PubMed] [Google Scholar]
- 12.Harada H et al. Localization of putative stem cells in dental epithelium and their association with notch and FGF signaling. J. Cell Biol 147, 105–120 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison SJ, Uchida N & Weissman IL The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol 11, 35–71 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Hu JK-H, Mushegyan V & Klein OD On the cutting edge of organ renewal: identification, regulation, and evolution of incisor stem cells. Genesis 52, 79–92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biehs B et al. BMI1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat. Cell Biol 15, 846–852 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juuri E et al. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev. Cell 23, 317–328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidel K et al. Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. eLife 6, e24712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seidel K et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Dev. Camb. Engl 137,3753–3761 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinreb C, Wolock S & Klein AM SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinreb C, Wolock S, Tusi BK, Socolovsky M & Klein AM Fundamental limits on dynamic inference from single-cell snapshots. Proc. Natl Acad. Sci. USA 115, E2467–E2476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nanci A Ten Cate’s Oral Histology 8th edn (Elsevier, 2012). [Google Scholar]
- 23.La Manno G et al. RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herman JS, Sagar & Grün, D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods 15, 379–386 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Foudi A et al. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol 27, 84–90 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doupé DP, Klein AM, Simons BD & Jones PH The ordered architecture of murine ear epidermis is maintained by progenitor cells with random fate. Dev. Cell 18, 317–323 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Wilson A et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Lerner C & Harrison DE 5-Fluorouracil spares hemopoietic stem cells responsible for long-term repopulation. Exp. Hematol 18, 114–118 (1990). [PubMed] [Google Scholar]
- 29.Stange DE et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell 155, 357–368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Yan X, Pandya M, Luan X & Diekwisch TGH Daughters of the enamel organ: development, fate, and function of the stratum intermedium, stellate reticulum, and outer enamel epithelium. Stem Cells Dev 25, 1580–1590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harada H et al. Stratum intermedium lineage diverges from ameloblast lineage via notch signaling. Biochem. Biophys. Res. Commun 340, 611–616 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Jheon AH et al. Inhibition of notch signaling during mouse incisor renewal leads to enamel defects. J. Bone Miner. Res 31, 152–162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voehringer D, Liang H-E & Locksley RM Homeostasis and effector function of lymphopenia-induced ‘memory-like’ Tcells in constitutively Tcell-depleted mice. J. Immunol 180, 4742–4753 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaukua N et al. Glial origin of mesenchymal stem cells in a tooth model system. Nature 513, 551–554 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Hu JK-H et al. An FAK-YAP-mTOR signaling axis regulates stem cell-based tissue renewal in mice. Cell Stem Cell 21, 91–106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao H et al. Secretion of Shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell 14, 160–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klein AM, Nakagawa T, Ichikawa R, Yoshida S & Simons BD Mouse germ line stem cells undergo rapid and stochastic turnover. Cell Stem Cell 7, 214–224 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Snippert HJ et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Tumbar T et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leung Y et al. Bifunctional ectodermal stem cells around the nail display dual fate homeostasis and adaptive wounding response toward nail regeneration. Proc. Natl Acad. Sci. USA 111, 15114–15119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wabik A & Jones PH Switching roles: the functional plasticity of adult tissue stem cells. EMBO J 34, 1164–1179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanpain C & Fuchs E Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dell’Orso S et al. Single cell analysis of adult skeletal muscle stem cells in homeostatic and regenerative conditions. Development 146, dev174177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gadye L et al. Injury activates transient olfactory stem cell states with diverse lineage capacities. Cell Stem Cell 21, 775–790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joost S et al. Single-cell transcriptomics of traced epidermal and hair follicle stem cells reveals rapid adaptations during wound healing. Cell Rep 25, 585–597 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Plasschaert LW et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nusse YM et al. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 559, 109–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Velten L et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol 19, 271–281 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metscher BD MicroCT for comparative morphology: simple staining methods allow high-contrast 3D imaging of diverse non-mineralized animal tissues. BMC Physiol 9, 11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Djomehri SI et al. Mineral density volume gradients in normal and diseased human tissues. PloS ONE 10, e0121611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu JK-H, McGlinn E, Harfe BD, Kardon G & Tabin CJ Autonomous and nonautonomous roles of hedgehog signaling in regulating limb muscle formation. Genes Dev 26, 2088–2102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chavez MG et al. Isolation and culture of dental epithelial stem cells from the adult mouse incisor. J. Vis. Exp 87, e51266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolf FA, Angerer P & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zemmour D et al. Single-cell gene expression reveals a landscape of regulatory Tcell phenotypes shaped by the TCR. Nat. Immunol 19, 291–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zilionis R et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaitin DA et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu JC-C et al. Enamel defects and ameloblast-specific expression in Enam knock-out/lacZ knock-in mice. J. Biol. Chem 283, 10858–10871 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith CE, Hu Y, Hu JC-C & Simmer JP Ultrastructure of early amelogenesis in wild-type, Amelx−/−, and Enam−/− mice: enamel ribbon initiation on dentin mineral and ribbon orientation by ameloblasts. Mol. Genet. Genomic Med 4, 662–683 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tusi BK et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klein AM et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mootha VK et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE131204. Source data for figures are provided in Supplementary Table 11. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.