Abstract
Epicardial adipose tissue (EAT) inflammation is thought to potentiate the development of coronary artery disease (CAD). Overall diet quality and statin therapy are important modulators of inflammation and CAD progression. Our objective was to examine the effects and interaction of dietary patterns and statin therapy on EAT gene expression in the Ossabaw pig. Pigs were randomized to 1 of 4 groups; Heart Healthy diet (high in unsaturated fat, unrefined grain, fruits/vegetables [HHD]) or Western diet (high in saturated fat, cholesterol, refined grain [WD]), with or without atorvastatin. Diets were fed in isocaloric amounts for 6 months. A two-factor edge R analysis identified the differential expression of 21 genes. Relative to the HHD, the WD resulted in a significant 12-fold increase of radical s-adenosyl methionine domain containing 2 (RSAD2), a gene induced by interferon signaling. Atorvastatin led to the significant differential expression of 17 genes predominately involved in interferon signaling. Results were similar using the Porcine Translational Research Database. Pathway analysis confirmed the up-regulation of interferon signaling in response to the WD and atorvastatin independently. An expression signature of the largely interferon related differentially expressed genes had no predictive capability on a histological assessment of atherosclerosis in the underlying coronary artery. These results suggest that a WD and atorvastatin evoke an interferon mediated immune response in EAT of the Ossabaw pig, which is not associated with the presence of atherosclerosis.
Keywords: Ossabaw pig, Epicardial adipose tissue, Transcriptomics, Dietary patterns, Atorvastatin, Interferon signaling
1. Introduction
Epicardial adipose tissue (EAT) is the perivascular adipose tissue surrounding the coronary arteries and in direct contact with the myocardium [1]. Continuity between EAT and the coronary arteries is characterized by lack of fascial layer separating the tissues and the vascularization of EAT by the coronary vasa vasorum [1]. As a readily available source of fatty acids EAT may serve as an energy store for heart and provide thermal insulation [2]. Like abdominal visceral adipose tissue (VAT), EAT originates from the splanchnopleuric mesoderm and when compared to subcutaneous adipose tissue has higher expression of pro-inflammatory cytokines such as interleukin 6 (IL-6), interleukin 1 beta (IL-1β), tumor necrosis factor alpha (TNFα.), and the chemokine monocyte chemotactic protein (MCP-1) [2,3].
In humans, EAT volume is thought to associate with its inflammatory status, with larger EAT volume representing increases in adipocyte size and number, and infiltration of immune cells [4,5]. It is hypothesized that inflamed EAT may secrete inflammatory cytokines to the underlying coronary arteries and potentiate the development of coronary atherosclerosis [1]. Concurrently, EAT volume, independent of body mass index and/or abdominal VAT mass, has been positively associated with coronary artery calcification, plaque progression, unstable plaques, and future incidence of coronary artery disease (CAD) [6–11]. Consistent with these observations, EAT from individuals with obesity or established CAD have lower expression and secretion of adiponectin, and higher expression of inflammatory cytokines (IL-6, IL-1β, and TNFα), and macrophage infiltration (CD68, CD11c, CD206) [3,12–14]. Together, these data imply that EAT may be an independent risk factor in the development of CAD. Accordingly, EAT may represent a therapeutic target for the prevention and mitigation of CAD.
Lifestyle modifications including weight loss and increases in physical activity are standard recommendations for the prevention of cardiovascular diseases (CVD), including CAD [15,16]. Weight loss from caloric restriction and physical activity has been associated with a reduction in EAT volume [17]. In addition to weight loss, current CVD risk reduction recommendations call for improvements in diet quality [18]. The Dietary Guidelines for Americans recommends improvements in diet quality by focusing on complete dietary patterns and highlight the replacement of saturated fat with poly- and monoun-saturated fats and a shift from refined to whole grain products [19]. Previous work has demonstrated that under conditions of weight loss a Mediterranean style diet provided greater reduction in intrapericardial adipose tissue when compared to a lower-fat control [17]. This suggests that distinct dietary patterns have the ability to differentially influence EAT.
Statin medications, standard pharmacotherapy for individuals with elevated low-density lipoprotein cholesterol (LDL-C), has also been associated with EAT regression [20–22]. Simvastatin has been reported to decrease expression of inflammatory markers in EAT, suggesting statins may reduce EAT volume through their pleiotropic anti-inflammatory effects [21,23]. How statin medications may interact with diet to modulate EAT inflammation is unknown. Due to the limited opportunistic nature of collecting EAT in humans, and lack of EAT in standard rodent models [24], there is little data examining the effect of dietary patterns and atorvastatin, independent of weight loss, on EAT gene expression.
We have previously established the Ossabaw miniature pig as a translational model of diet-induced atherosclerosis by feeding food-based diets, designed to reflect human dietary patterns, with or without statin therapy [25]. Compared to pigs fed a Heart Healthy diet (HHD), pigs fed a Western diet (WD) exhibited significantly higher LDL-C concentrations, systemic inflammation, and incidence and severity of early coronary atherosclerotic lesions [25]. Statin therapy led to a significant reduction in LDL-C and triglycerides [25]. Our objective was to characterize the effect of dietary patterns and statin therapy on the EAT transcriptome in the Ossabaw miniature pig and to determine if differentially expressed genes were associated with the presence of atherosclerosis. We hypothesized that, relative to the HHD, EAT from pigs fed the WD would have a significant increase in inflammatory gene expression, which will be attenuated by atorvastatin.
2. Materials and methods
2.1. Study design, animals, and diets
The present investigation reports results ancillary to prior work designed to assess the effect of diet patterns and statin therapy on atherosclerosis in the Ossabaw miniature pig [25]. Thirty-two Ossabaw miniature pigs (16 boars, 16 gilts) were randomized using a 2×2 factorial design into four groups: Hearty Healthy diet (HHD); Heart Healthy diet + atorvastatin (HHD + S); Western diet (WD); and Western diet + atorvastatin (WD + S). One pig assigned to the HHD group died during the acclimatizing period and another pig in the WD group died during the baseline blood collection resulting in a final sample size of 30 pigs. Diets were designed to reflect human dietary patterns and were fed in isocaloric amounts for 6 months following a 2-month acclimatization period. A detailed diet composition has been reported previously [25]. Briefly, diets were composed of 38% energy (E) from fat, 47% E from carbohydrate and 15% E from protein but differed in the type of dietary fat and carbohydrate, and the amount of fiber and cholesterol [25]. The HHD contained 29% E as fat from canola, soybean, corn, and fish oils; 9% E as fat from anhydrous milk fat; 47% E from whole wheat flour and oats; a freeze-dried fruit and vegetable mix; 13 g of fiber per 100 g of diet; 0.1% wet weight of cholesterol; and 2.5% wet weight of a vitamin and mineral mix. The WD contained 16% E as fat from canola, soybean, and corn oils; 22% E as fat from anhydrous milk fat; 47% E from sugar and white flour; 7 g of fiber per 100 g of diet, 1.5% wet weight of cholesterol; and 2.5% wet weight of a vitamin and mineral mix. HHD fed pigs were supplemented with fish oil capsules (Epanova 1 g, 550 mg eicosapentaenoic acid [EPA] + 200 mg docosahexaenoic acid [DHA]) three times per week. Statin treated pigs were given atorvastatin (Lipitor), daily at a dose of 20 mg/day during months 1–3, and 40 mg/day during months 4–6.
2.2. Sample collection
The collection of blood and fixation of samples for histology has been previously described [25]. EAT adjacent to the proximal left anterior descending artery was collected at necropsy and flash frozen in liquid nitrogen. Samples were stored at−80 °C until processing.
2.3. Sample processing
2.3.1. Blood samples
Serum total cholesterol, high-density lipoprotein cholesterol (HDL-C), LDL-C, triglyceride, and high sensitivity C-reactive protein (hsCRP) measurements were assessed and reported as previously described [25]. Data on LDL-C concentrations was estimated using the Friedewald equation [26]. Plasma hsCRP concentrations were measured by a two-site enzyme-linked immunoassay procedure, (Pig High-Sensitive CRP ELISA Cat. No. KT-184, Kamiya Biomedical Company, Seattle, WA, USA).
2.3.2. Histopathology
As part of the original study histological presence of atherosclerosis in the proximal left anterior descending coronary artery was determined by a blinded board-certified veterinary cardiovascular pathologist using the previously established Stary system of classification [25,27].
2.3.3. EAT RNA isolation
EAT was homogenized and RNA was isolated using RNeasy Universal Midi kit (Qiagen, Valencia, CA, USA) per the manufacturer’s instructions for lipid rich tissue. Isolated RNA was treated with Turbo DNase (Ambion, Waltham, MA) to minimize genomic DNA contamination. RNA quality was assessed using the Experion RNA analysis electrophoresis kit (Bio-Rad, Hercules, CA, USA). Only samples with an RNA Quality Indicator (RQI) greater than 7 were sequenced.
2.4. TruSeq library preparation, sequencing, and processing of reads
Prior to RNA sequencing samples were processed (mRNA purification and fragmentation, cDNA synthesis, end repair, 3′ adenylation, adaptor ligation, and DNA enrichment) using the Illumina TruSeq RNA Sample Preparation Kit v2 (Illumina, San Diego, CA, USA) and AMPure XP beads (Beckman Coulter, Brea, CA, USA) per the manufacturer’s instructions. DNA fragment size was determined using Experion DNA 1 K chips (Bio-Rad, Hercules, CA, USA) and library quantification was completed using the KAPA Library Quantification kit (KAPA Biosystems, Wilmington, MA). Samples were sequenced on an Illumina NextSeq 500 sequencer (Illumina, San Diego, CA, USA) with 100 base pair single end reads. Raw data in FASTQ format was processed for quality using CLC Bio Genomic Workbench (Qiagen, Valencia, CA, USA). The EAT transcriptome was assembled using the annotated Sus scrofa10.2 as a reference genome [28].
2.5. Transcriptomic analysis
Differential expression analysis was performed on 22,862 genes using a two-factor model design matrix in edge R, a Bioconductor package based on a negative binomial distribution [29]. Three pigs (one HHD and two HHD + S) displayed abnormal high counts across all genes and were excluded from subsequent analysis, resulting in a final sample of n=27. Count data on annotated genes was filtered based on a minimum of at least one count per million across six samples in all groups and normalized using the trimmed mean of M values (TMM) method. The edge R two-factor model was designed to determine differential gene expression attributable to diet, statin, or a diet × statin interaction. Differentially expressed genes were identified using the Cox-Reid profile-adjusted likelihood method and likelihood ratio test. A Benjamini-Hochberg false discovery rate (FDR) method was used to adjust p-values for multiple comparisons [30]. Sample size and power were previously determined based on prior work assessing the primary outcome of atherosclerosis and not for a transcriptomic analysis. Thus, given the ancillary nature of this study genes were considered differentially expressed based on an FDR adjusted P<.1 and absolute fold change ≥1.5. Fold change for genes identified by the edge R two-factor model were interpreted as diet effect: fold change of WD relative to HHD and statin effect: fold change of atorvastatin relative to no atorvastatin. Differentially expressed genes were only attributed to main effects (diet and statin) if there was no significant diet × statin interaction. A significant diet × stain interaction is indicated by a FDR adjusted P<.1.
As a replication of our transcriptomic analysis we conducted a secondary differential expression analysis using the manually curated Porcine Translational Research Database [31]. The secondary transcriptomic analysis was completed using the methods described above by replacing the Sus scrofa10.2 genome database with the Porcine Translational Research Database.
2.6. Pathway analysis
Following differential expression analysis genes were analyzed using Ingenuity Pathway Analysis (IPA; v 9.0, Mountain View, CA, USA) to determine relevant biological pathways affected by the diet and statin therapy. An exploratory pathway analysis was conducted. In total 1432 genes with an absolute fold change ≥1.5 were uploaded to IPA and evaluated using the canonical pathway analysis. Z-scores were calculated for each pathway to indicate the direction of pathway regulation (activated versus inhibited) and match to IPA Knowledge Base (observed versus predicted). Pathways with an absolute Z score ≥2 and an FDR adjusted P≤.05 were considered significant.
2.7. Additional statistical analyses
Principal component analysis (PCA) was performed on genes differentially expressed by the main effects (diet and statin) (Table 1) to create a gene expression signature. Expression data (reads per kilobase million [rpkm]) for differentially expressed genes, from a pooled sample of pigs (n=27), was scaled and centered prior to PCA analysis. The first principal component explained over 78% of the variance (Appendix Fig. A.1) and was used as a signature of EAT differentially expressed genes (EATPC). Three logistic regression models were used to determine the predictive capability of the EAT expression signature on the presence of coronary atherosclerosis in the underlying left anterior descending coronary artery and to determine if the EAT expression signature would provide additional predictive capability in the presence of LDL-C and hsCRP. Three hsCRP values were more than two standard deviations beyond mean values and were replaced by median values for pigs in the respective diet +/− statin group. Model 1 evaluated the EATPC alone; model 2 evaluated LDL-C and hsCRP; and model 3 evaluated LDL-C, hsCRP, and the EATPC. Stary score was previously used for a histological assessment of the presence and degree of atherosclerosis in the proximal left anterior descending artery [25]. A score of 0 was defined as no presence of atherosclerosis and scores ≥1 were defined was presence of atherosclerosis. Likelihood ratio tests were used to determine model significance and compare models. Receiving operating characteristic (ROC) curves displaying area under the curve (AUC) and 95% confidence intervals (CI) were used to evaluate predictive capability. ROC curves and AUC were generated using the pROC package in R [32].
Table 1.
Diet, statin, and diet × statin interaction differentially expressed genes in EAT1
| Gene Symbol | Gene Name | Diet effect | Statin effect | Diet × statin | Average expression (rpkm) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| FC | FDR | FC | FDR | FDR | WD (n=7) | WD + S(n=8) | HHD (n=6) | HHD + S(n=6) | ||
| CCL5 | C-C motif chemokine 5 | 2.59 | 0.814 | 7.15 * | 0.054 | 0.563 | 0.86 | 0.77 | 0.33 | 2.40 |
| CTHRC1 | Collagen triple helix repeat-containing protein 1 | −2.21 | 0.735 | −4.20 * | 0.080 | 1.000 | 0.33 | 0.28 | 0.73 | 0.17 |
| CXCL10 | C-X-C motif chemokine 10 | 11.96 | 0.468 | 22.56 * | 0.072 | 0.313 | 3.93 | 2.28 | 0.33 | 7.47 |
| DDX60 | ATP-dependent RNA helicase DDX60 | 5.65 | 0.352 | 6.48 * | 0.077 | 1.000 | 1.35 | 1.56 | 0.24 | 1.57 |
| Unknown 12 | ENSSSCG000000145653 | 2.28 | 0.513 | 2.96 * | 0.080 | 1.000 | 10.73 | 12.96 | 4.72 | 14.05 |
| ETV7 | Transcription factor ETV7 | 9.94 | 0.352 | 14.10 * | 0.059 | 0.385 | 0.47 | 0.38 | 0.05 | 0.68 |
| GBP1 | Guanylate-binding protein 1 | 6.05 | 0.468 | 10.64 * | 0.054 | 0.385 | 17.55 | 14.63 | 2.91 | 31.22 |
| GBP6 | Guanylate-binding protein 6 | 7.56 | 0.352 | 14.41 ** | 0.025 | 0.167 | 2.54 | 1.99 | 0.33 | 4.90 |
| IDO1 | Indoleamine 2,3-dioxygenase 1 | 7.87 | 0.352 | 14.41 ** | 0.028 | 0.064 * | 0.31 | 0.16 | 0.04 | 0.57 |
| IFIT3 | Interferon-induced protein with tetratricopeptide | 3.51 | 0.352 | 4.09 * | 0.077 | 0.962 | 5.61 | 5.87 | 1.60 | 6.62 |
| ISG12(A) | Interferon alpha-inducible protein 27 | 3.65 | 0.493 | 5.91 * | 0.054 | 0.925 | 24.95 | 28.38 | 6.82 | 40.66 |
| MYL4 | Myosin light chain 4 | 0.09 | 0.353 | −8.99 | 0.460 | 0.002 ** | 0.07 | 1.67 | 0.83 | 0.10 |
| MYL7 | Myosin light chain 7 | 0.12 | 0.473 | −8.09 | 0.358 | 0.002 ** | 0.07 | 1.19 | 0.55 | 0.06 |
| NKL | Antimicrobial peptide NK-lysin | 4.63 | 0.618 | 12.14** | 0.048 | 0.083 * | 1.42 | 0.68 | 0.30 | 3.75 |
| OAS1 | 2′−5′-oligoadenylate synthase 1 | 3.11 | 0.352 | 3.86** | 0.028 | 0.791 | 12.58 | 14.62 | 4.07 | 15.82 |
| OAS2 | 2′−5′-oligoadenylate synthase 2 | 3.27 | 0.473 | 5.17 ** | 0.048 | 1.000 | 10.56 | 14.25 | 3.24 | 16.91 |
| PTN | Pleiotrophin | −3.61 | 0.513 | −10.88 ** | 0.005 | 0.364 | 0.97 | 0.78 | 3.56 | 0.33 |
| RSAD2 | Radical S-adenosyl methionine domain-containing protein 2 | 12.27 ** | 0.015 | 13.63 ** | 0.005 | 0.313 | 3.10 | 3.72 | 0.25 | 3.49 |
| SLPI | Secretory leukocyte peptidase inhibitor | 1.38 | 0.814 | 1.97 ** | 0.041 | 0.318 | 5.32 | 4.75 | 3.86 | 7.70 |
| UBD | Ubiquitin D | 3.2 | 0.605 | 6.00 ** | 0.048 | 0.364 | 3.54 | 2.88 | 1.10 | 6.68 |
| XAF1 | XIAP associated factor 1 | 3.98 | 0.358 | 5.81 ** | 0.041 | 0.840 | 3.80 | 4.55 | 0.96 | 5.63 |
FDR adjusted P<.1.
FDR adjusted P<.05.
EAT: epicardial adipose tissue, FC: fold change, rpkm: reads per kilobase per million mapped reads, HHD: Heart Healthy diet, WD: Western diet, HHD+S: Heart Healthy diet with atorvastatin, WD+S: Western diet with atorvastatin, Diet effect: WD relative to HHD, Statin effect: atorvastatin relative to no atorvastatin.
Unknown transcript.
Ensembl ID.
3. Results
3.1. Differential expression analysis
With the FDR adjusted p-value cutoff of b0.1 our two-factor edge R analysis identified a total of 21 genes with differential expression. There was 1 significant gene with differential expression attributable to the diet effect (WD relative to HHD), 17 significant genes with differential expression attributable to the statin effect (atorvastatin relative to no atorvastatin), and an additional four genes with a significant diet × statin interaction. All genes designated as significantly different had an absolute fold change value >1.5. Fold change and average expression values for differentially expressed genes are displayed in Table 1 and values for all genes are included in Appendix Table A.1.
3.1.1. Diet effect
Radical s-adenosyl methionine domain containing 2 (viperin [RSAD2]), an antiviral gene induced by interferon signaling, and involved in immune response, had a more than 12-fold increase in expression in EAT of pigs fed the WD relative to the HHD.
3.1.2. Statin effect
Atorvastatin resulted in increased expression of 15 genes and decreased expression of 2 genes (Table 1). Genes with increased expression due to atorvastatin were largely involved in immune response and inflammation. Whereas, pleiotrophin (PTN), a gene involved in cell proliferation, and collagen triple helix repeat-containing protein (CTHRC1), a gene involved in cell migration, had decreased expression.
3.1.3. Diet × statin interaction
Four genes had a significant diet × statin interaction, indicating the effect of atorvastatin on EAT gene expression was differentially modified by the respective dietary patterns (Table 1). The addition of atorvastatin to the WD resulted in a decrease in expression of indoleamine 2,3-dioxygenase 1 (IDO1) and antimicrobial peptide NK-lysin (NKL), whereas IDO1 and NKL expression were increased with the addition of atorvastatin to the HHD. Additionally, the addition of atorvastatin led to relatively large increases in expression of two genes, myosin light chain 4 (MYL4) and myosin light chain 7 (MLY7), in EAT of pigs fed the WD but decreases in expression of pigs fed the HHD.
3.2. Pathway and secondary analyses
To further determine biological relevance of the diet and statin effects an IPA canonical pathway analysis was conducted. This analysis was exploratory in nature and included genes with an absolute fold change >1.5. Five pathways were identified as significantly regulated by the diet effect and 11 by the statin effect (Appendix Table A.2). The 3 top scoring pathways and associated genes for the diet and statin effects are displayed in Table 2.
Table 2.
Top biological pathways of EAT differentially expressed genes with a fold change of±1.51
| Top pathways | Activation | Molecules | FDR |
|---|---|---|---|
| Diet effect | |||
| Interferon signaling | Up | IFIT1, IFIT3, IFITM1, IRF1, IRF9, MX1,OAS1, STAT1 | 0.007 |
| Neuropathic pain Signaling in dorsal horn neurons | Up | CAMK4, CAMK2B, FGFR3, FOS, GPR37, GRIN3A, GRM4, KCNH2, KL, PLCD4, PLCH1, PRKCB, PRKCZ | 0.009 |
| Role of pattern recognition receptors in recognition of bacteria and viruses | Up | CASP1,CCL5, IL8, FGFR3, IFIH1, IL25, IRF7, KL, MAPK10, OAS1, OAS2, PRKCB, PRKCZ | 0.027 |
| Statin effect | |||
| Interferon signaling | Up | IFI35, IFIT1, IFIT3, IFITM1, IRF1, IRF9, MX1,OAS1, SOCS1, STAT1, STAT2 | <0.0001 |
| IL-8 signaling | Down | ANGPT2, CCND2, CR2, IL8, CYBB, BEK, FGFR3, FRS2, GNA13, TGAM, MMP9, MYL7, NCF2, PIK3C2G, PTGS2, RHOU | 0.003 |
| Leukocyte extravasation signaling | Down | CLDN4, CLDN12, CLDN19, CXCL12, CYBB, EZR, BEK, FGFR3, FRS2, ITGAM, MMP9, MMP25, NCF1, NCF2, PIK3C2G, RASGRP1 | 0.004 |
EAT: epicardial adipose tissue, Diet effect: WD relative to HHD, HHD: Heart Healthy diet, WD: Western diet, Statin effect: atorvastatin relative to no atorvastatin.
3.2.1. Diet effect
Top pathways in EAT of pigs fed the WD compared to the HHD included and up-regulation of (1) Interferon signaling, (2) Neuropathic Pain Signaling in Dorsal Horn Neurons, and (3) Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses.
3.2.2. Statin effect
Top pathway regulated by atorvastatin included an up-regulation of (1) Interferon Signaling and the down-regulation of (2) IL-8 Signaling and (3) Leukocyte Extravasation Signaling.
Differential expression analysis using the Porcine Translational Research Database as the referent yielded similar results for the diet and statin effects (Appendix Table A.3). In contrast, no significant genes were identified a diet × statin interaction. Pathway analysis of genes with an absolute fold change >1.5 from this analysis confirmed an up-regulation of interferon signaling as the primary pathway regulated by both diet (FDR<0.0001) and statin (FDR<0.0001) effects.
3.3. EAT gene expression signature and presence of atherosclerosis
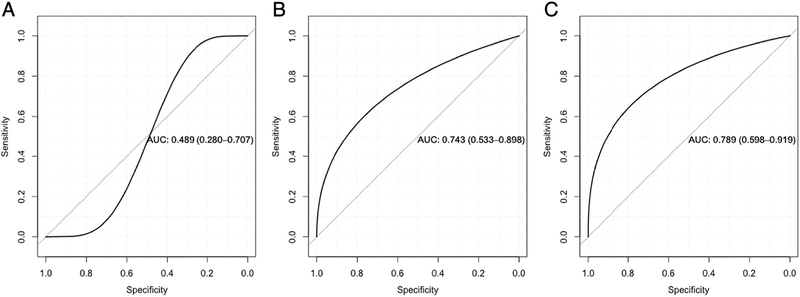
We have previously reported the effects of the dietary patterns and atorvastatin on LDL-C, hsCRP concentrations, and the degree of atherosclerosis [25]. Here we sought to examine the relationship between the expression of differentially expressed genes in EAT and the presence of atherosclerosis independent of dietary pattern or atorvastatin (see Methods 2.7). Additionally, we examined if a EAT gene expression signature added predictive capability to a model that included LDL-C and hsCRP concentrations. Using the pooled group of pigs (n=27) LDL-C concentrations ranged from 0.83 to 19.27 mmol/L with a median value of 2.97 mmol/L and hsCRP concentrations ranged from 42 to 163 mg/L with a median value 79 mg/L. The EATPC contained all differentially expressed genes attributable to the main effects (diet and statin). Interferon stimulated genes (ISGs) had relatively high factor loading scores, while unrelated genes such as PTN and CTHRC1 had negative scores (Appendix Fig. A.1). Thus, the EATPC may be considered representative of differential ISG expression. The AUC of model 1 (EATPC alone) was 0.49 (95% CI: 0.28, 0.71) indicating that the EAT signature was not a significantly better predictor of atherosclerosis than random chance (Fig. 1). In contrast, the AUCs for models 2 (LDL-C + hsCRP) and 3 (LDL-C + hsCRP + EATPC) were 0.74 (95% CI: 0.53, 0.90) and 0.79 (95% CI: 0.60, 0.92), respectively, indicating moderate capability to predict presence of atherosclerosis (Fig. 1). Concurrently, model 1 was not a significant classifier of atherosclerosis (X2=1.74, P=.187), while models 2 (LDL-C + hsCRP) and 3 (LDL-C + hsCRP + EATPC) were both significant classifiers (X2=6.82, P=.033 and X2=7.98, P=.047 respectively). However, there was no significant difference between models 2 and 3 (X2=1.15, P=.283). Together these data indicate that the EAT gene expression signature alone is not an independent predictor of atherosclerosis. Although the EAT gene expression signature provided a small increase in predictive capability to a model with LDL-C and hsCRP, this increase did not translate to a statistically significant improvement in predictive capacity.
Fig. 1.
Receiving operating characteristic curves display AUC and 95% CI, and demonstrate predictive performance of three different models on the classification of coronary atherosclerosis determined by histological assessment using the Stary Score Classification System. (A) Model 1: EATPC, (B) Model 2: LDL-C and hsCRP, (C) Model 3: LDL-C, hsCRP, and EATPC. AUC: area under the curve, CI: confidence interval, EAT: epicardial adipose tissue, EATPC: gene expression signature of EAT differentially expressed genes, LDL-C: low-density lipoprotein cholesterol, hsCRP: high sensitive C-reactive protein.
4. Discussion
Promising data suggests weight loss and statin therapy reduce EAT volume and inflammation in humans. Little data is available on the effects of dietary patterns and statin therapy, independent of weight loss, on alterations in EAT gene expression. The present study was designed to address this gap using the Ossabaw miniature pig model of diet-induced atherosclerosis.
We found that a WD, relative to a HHD, led to a significant increase in expression of RSAD2, an antiviral ISG involved in immune response. Atorvastatin resulted in significant increases in expression of genes predominately related to interferon signaling, including ISGs. Interferon signaling was identified as the top biological pathway for both the diet and statin effects. We determined that a gene expression signature of our largely interferon related gene expression phenotype was not significantly associated with the presence of atherosclerosis in the underlying artery and added no significant predictive value to established risk factors.
4.1. Diet effect
RSAD2 was the only gene with expression significantly affected by the WD, relative to the HHD. However, pathway analysis indicated the up-regulation of two pathways related to type 1 interferon signaling (interferon signaling and role of pattern recognition receptors in recognition of bacteria and viruses). These results may have been mediated in part by the effects of saturated fatty acids on toll-like receptor (TLR) signaling [33]. Saturated fatty acids act as agonists for TLR2 and TLR4 which initiate type I interferon signaling [34–36]. While the HHD and WD were matched for total fat content (38% of energy), 22% of energy in the WD was from anhydrous milk fat (rich in saturated fatty acids) compared to 9% of energy in the HHD [25]. It has previously been demonstrated that palmitic acid, a predominant fatty acid in anhydrous milk fat, induces type I interferon expression in both hepatocytes and macrophages [37]. The higher saturated fat content of the WD may have evoked interferon signaling by the direct induction of TLRs in EAT or indirectly via mechanisms such as an unfavorable modulation of the gut microbiome. Adipose tissue is the major site of de novo lipogenesis in the pig [38]. Consequently, endogenous synthesis of saturated fatty acids in EAT may have in part mitigated significant differences in saturated fatty acid induced gene expression between diets, resulting in only a modest effect of the WD.
4.2. Statin effect
The differential expression analysis revealed a more pronounced effect on ISGs by atorvastatin and our pathway analysis indicated that atorvastatin led to an up-regulation in type I interferon signaling. These results were unexpected as statins are recognized to have pleiotropic anti-inflammatory effects, which are thought to mediate the association between statin therapy and EAT regression [20–22]. Though few studies have examined the effect of statins directly on ISGs, crosstalk between interferon signaling and sterol biosynthesis has been documented [39]. In vitro studies have demonstrated that type I interferon signaling leads to reductions in cellular sterol biosynthesis which further potentiates interferon signaling [39,40]. Evidence suggests that interferon mediated reductions in sterol biosynthesis are in part due to inhibition of sterol regulatory binding protein-2 (SREBP2) and mevalonate activity attributed to increased concentrations of the oxysterol 25-hydroxycholesterol [39,41,42]. Statin medications have been reported to have antiviral activity and an in vitro study has demonstrated up-regulation of gene and protein expression of ISGs by the targeted down-regulation of 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) reductase or treatment with simvastatin [39,43,44]. Hence, antiviral effects of statins are likely due to suppression of sterol and isoprenoid biosynthesis pathways [39,44]. Our results here corroborate interferon-sterol crosstalk with the induction of ISGs by atorvastatin in EAT of the Ossabaw miniature pig.
4.3. Diet × statin interaction
Differential expression analysis indicated a diet × statin interaction for four genes. Expression of MYL4 and MYL7 likely reflect the myocardium associated with EAT and are not of biological importance. The addition of atorvastatin to the WD resulted in lower expression of two immune response genes, ISG IDO1, relative to addition of atorvastatin to the HHD. Reasons for the potential interaction are unclear. Of note, these results were not substantiated in our secondary analysis and should be interpreted with caution.
The EAT gene expression signature was representative of all differentially expressed genes attributable to the diet and statin effects. In particular ISGs had high factor loadings scores. This gene expression signature was not associated with the presence of atherosclerosis and had no independent predictive value. Though ROC curves demonstrated that adding this gene expression signature to a model with LDL-C and hsCRP slightly improved the predictive capability, there was no significant difference between this model and a model including only LDL-C and hsCRP. These results imply that the specific EAT gene expression phenotype we observed had limited, if any, local effects on the development of atherosclerosis in the underlying coronary artery.
Interferon signaling is integral in the response and regulation of the innate immune system [45]. Additionally, interferons, and the proteins that regulate their signaling (interferon regulatory factors [IRFs]), have been positively associated with obesity, adipose tissue inflammation and atherosclerosis [35,46–52]. In contrast, type I interferon signaling, and the expression of ISGs, in adipose tissue may protect against metabolic dysfunction [37,53,54]. The modest up-regulation of interferon signaling in EAT of pigs fed a WD and a more pronounced up-regulation in EAT of pigs receiving atorvastatin was predominately attributable to the number of differentially expressed genes that were ISGs. ISGs are induced by interferons binding to their respective receptors, ultimately leading to the phosphorylation and translocation of IRFs into the nucleus where they bind to a interferon stimulated response element (ISRE) [35,55,56]. We cannot determine if pro-longed interferon signaling may induce inflammation or if the expression of ISGs is protective against adipose tissue inflammation. However, we did not detect differential expression of interferons (IFNα, IFNβ, and IFNγ) or inflammatory cytokines typically induced by interferon signaling. This may explain why the EAT gene expression signature had no significant association with atherosclerosis.
The present study has several strengths but is not without limitations. The differential gene expression we detected was likely attributable to cells in the stromal vascular fraction of adipose tissue. As RNA was isolated from adipose tissue homogenates, reflective of multiple cell types, we expect gene expression from this fraction was diluted. We did not detect changes in the expression of inflammatory cytokines that have previously been associated with an inflamed EAT and are thought to mediate the association between EAT and CAD in humans. This could be due to the aforementioned dilution of the stromal vascular fraction or may in part be due to feeding the two diets in isocaloric amounts, making it unlikely that changes in gene expression more closely associated with obesity would be detected here. The results presented in this experimental study require replication and should be interpreted in the context of the identified biological pathway(s) and not individual genes. We did not detect changes in the expression of inflammatory cytokines that have previously been associated with an inflamed EAT and are thought to mediate the association between EAT and CAD in humans. This could be due to the aforementioned dilution of the stromal vascular fraction or may in part be due to feeding the two diets in isocaloric amounts, making it unlikely that changes in gene expression more closely associated with obesity would be detected here. A strength of our study is the secondary differential expression analysis using the Porcine Translational Research Database [31]. As the porcine genome may not be as well annotated as the genomes of more typical experimental models (e.g. rodents) this secondary analysis helps validate our results. An additional strength is the translational value of the study, particularly the use of the food-based diets designed to replicate human dietary patterns [25]. Typical control and disease inducing diets fed to experimental models frequently focus on the quantity of dietary fat (low- vs. high-fat) and not on the macronutrient quality. Hence, such diets do not reflect current dietary guidance and are of limited translational value. Our approach was intended to replicate human dietary patterns and determine how these patterns affect EAT gene expression. Lastly, porcine models, like the Ossabaw miniature pig, represent an innovative approach to study the effects of diet on EAT as they provide the benefit of controlled feeding and have a EAT depot, similar to humans.
In conclusion, we investigated the effects of dietary patterns and atorvastatin on EAT in the Ossabaw miniature pig. Our transcriptomic and pathway analyses indicated that relative to an HHD, a WD resulted in an upregulation of interferon signaling. Additionally, atorvastatin resulted in a more pronounced up-regulation of interferon signaling. This interferon related gene expression profile exhibited no predictive capability on a histological determination of atherosclerosis. These results indicate that in the Ossabaw miniature pig a WD pattern and statin therapy play a role in inducing interferon signaling in EAT, which could in turn influence the regulation of EAT inflammatory status.
Supplementary Material
Funding source:
This work was supported by the Tufts HNRCA Cardiovascular Research Cluster, NHLBI T32 Nutrition and Cardiometabolic Disorders Pre-doctoral Research Training Grant [T32HL069772-15], NIH Multidisciplinary Training Program in Cardiovascular Epidemiology [5T32HL125232], USDA agreement 588-1950-9-001 and USDA-ARS project 8040-51530-056-00-D. Any opinions, findings, conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the USDA.
Footnotes
Declaration of interest: None.
Data statement
All raw RNA sequencing data from this manuscript will be deposited in the Gene Expression Omnibus (GEO) repository for public access (GSE129301).
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jnutbio.2019.02.003.
References
- [1].Iacobellis G. Local and systemic effects of the multifaceted epicardial adipose tissue depot. Nat Rev Endocrinol 2015;11:363–71. 10.1038/nrendo.2015.58. [DOI] [PubMed] [Google Scholar]
- [2].Fitzgibbons TP, Czech MP. Epicardial and perivascular adipose tissues and their influence on cardiovascular disease: basic mechanisms and clinical associations. J Am Heart Assoc 2014;3:e000582 10.1161/JAHA.113.000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mazurek T, Zhang L, Zalewski A, Mannion JD, Diehl JT, Arafat H, et al. Human epicardial adipose tissue is a source of inflammatory mediators. Circulation 2003; 108:2460–6. 10.1161/01.CIR.0000099542.57313.C5. [DOI] [PubMed] [Google Scholar]
- [4].Salazar J, Luzardo E, Mejías JC, Rojas J, Ferreira A, Rivas-Ríos JR, et al. Epicardial fat: physiological, pathological, and Therapeutic Implications. Cardiol Res Pract 2016; 2016:1291537 10.1155/2016/1291537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chistiakov DA, Grechko AV, Myasoedova VA, Melnichenko AA, Orekhov AN. Impact of the cardiovascular system-associated adipose tissue on atherosclerotic pathology. Atherosclerosis 2017;263:361–8. 10.1016/j.atheroscle-rosis.2017.06.017. [DOI] [PubMed] [Google Scholar]
- [6].Nerlekar N, Brown AJ, Muthalaly RG, Talman A, Hettige T, Cameron JD, et al. Association of epicardial adipose tissue and high-risk plaque characteristics: a systematic review and meta-analysis. J Am Heart Assoc 2017;6 10.1161/JAHA.117.006379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kunita E, Yamamoto H, Kitagawa T, Ohashi N, Oka T, Utsunomiya H, et al. Prognostic value of coronary artery calcium and epicardial adipose tissue assessed by non-contrast cardiac computed tomography. Atherosclerosis 2014;233: 447–53. 10.1016/j.atherosclerosis.2014.01.038. [DOI] [PubMed] [Google Scholar]
- [8].Liu J, Fox CS, Hickson D, Sarpong D, Ekunwe L, May WD, et al. Pericardial adipose tissue, atherosclerosis, and cardiovascular disease risk factors: the Jackson heart study. Diabetes Care 2010;33:1635–9. 10.2337/dc10-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rosito GA, Massaro JM, Hoffmann U, Ruberg FL, Mahabadi AA, Vasan RS, et al. Pericardial fat, visceral abdominal fat, cardiovascular disease risk factors, and vascular calcification in a community-based sample: the Framingham heart study. Circulation 2008;117:605–13. 10.1161/CIRCULATIONAHA.107.743062. [DOI] [PubMed] [Google Scholar]
- [10].Goeller M, Achenbach S, Marwan M, Doris MK, Cadet S, Commandeur F, et al. Epicardial adipose tissue density and volume are related to subclinical atherosclerosis, inflammation and major adverse cardiac events in asymptomatic subjects. J Cardiovasc Comput Tomogr 2017. 10.1016/j.jcct.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gaborit B, Abdesselam I, Dutour A. Epicardial fat: more than just an “epi” phenomenon? Horm Metab Res 2013;45:991–1001. 10.1055/s-0033-1358669. [DOI] [PubMed] [Google Scholar]
- [12].Karastergiou K, Evans I, Ogston N, Miheisi N, Nair D, Kaski J-C, et al. Epicardial adipokines in obesity and coronary artery disease induce atherogenic changes in monocytes and endothelial cells. Arterioscler Thromb Vasc Biol 2010;30:1340–6. 10.1161/ATVBAHA.110.204719. [DOI] [PubMed] [Google Scholar]
- [13].Baker AR, da Silva NF, Quinn DW, Harte AL, Pagano D, Bonser RS, et al. Human epicardial adipose tissue expresses a pathogenic profile of adipocytokines in patients with cardiovascular disease. Cardiovasc Diabetol 2006;5:1 10.1186/1475-2840-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hirata Y, Tabata M, Kurobe H, Motoki T, Akaike M, Nishio C, et al. Coronary atherosclerosis is associated with macrophage polarization in epicardial adipose tissue. J Am Coll Cardiol 2011;58:248–55. 10.1016/j.jacc.2011.01.048. [DOI] [PubMed] [Google Scholar]
- [15].Eckel RH, Jakicic JM, Ard JD, de Jesus JM, Miller NH, Hubbard VS, et al. 2013 AHA/ACC guideline on lifestyle management to reduce cardiovascular risk: a report of the American College of Cardiology/American Heart Association task force on practice guidelines. Circulation 2014;129:S76–99. 10.1161/01.cir.0000437740.48606.d1. [DOI] [PubMed] [Google Scholar]
- [16].Jensen MD, Ryan DH, Apovian CM, Ard JD, Comuzzie AG, Donato KA, et al. 2013 AHA/ACC/TOS guideline for the management of overweight and obesity in adults: a report of the American College of Cardiology/American Heart Association task force on practice guidelines and the Obesity Society. Circulation 2014;129: S102–38. 10.1161/01.cir.0000437739.71477.ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tsaban G, Wolak A, Avni-Hassid H, Gepner Y, Shelef I, Henkin Y, et al. Dynamics of intrapericardial and extrapericardial fat tissues during long-term, dietary-induced, moderate weight loss. Am J Clin Nutr 2017;106:984–95. 10.3945/ajcn.117.157115. [DOI] [PubMed] [Google Scholar]
- [18].Smith SC, Benjamin EJ, Bonow RO, Braun LT, Creager MA, Franklin BA, et al. AHA/ACCF secondary prevention and risk reduction therapy for patients with coronary and other atherosclerotic vascular disease: 2011 update: a guideline from the American Heart Association and American College of Cardiology Foundation endorsed by the World Heart Federation and the Preventive Cardiovascular Nurses Association. J Am Coll Cardiol 2011;58:2432–46. 10.1016/j.jacc.2011.10.824. [DOI] [PubMed] [Google Scholar]
- [19].Millen BE, Abrams S, Adams-Campbell L, Anderson CA, Brenna JT, Campbell WW, et al. The 2015 dietary guidelines advisory committee scientific report: development and major conclusions. Adv Nutr 2016;7:438–44. 10.3945/an.116.012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Alexopoulos N, Melek BH, Arepalli CD, Hartlage G-R, Chen Z, Kim S, et al. Effect of intensive versus moderate lipid-lowering therapy on epicardial adipose tissue in hyperlipidemic post-menopausal women: a substudy of the BELLES trial (Beyond Endorsed Lipid Lowering with EBT Scanning). J Am Coll Cardiol 2013;61:1956–61. 10.1016/j.jacc.2012.12.051. [DOI] [PubMed] [Google Scholar]
- [21].Iacobellis G. Epicardial fat: a new cardiovascular therapeutic target. Curr Opin Pharmacol 2016;27:13–8. 10.1016/j.coph.2016.01.004. [DOI] [PubMed] [Google Scholar]
- [22].Soucek F, Covassin N, Singh P, Ruzek L, Kara T, Suleiman M, et al. Effects of atorvastatin (80 mg) therapy on quantity of Epicardial adipose tissue in patients undergoing pulmonary vein isolation for atrial fibrillation. Am J Cardiol 2015;116: 1443–6. 10.1016/j.amjcard.2015.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Grosso AF, de Oliveira SF, Higuchi M de L, Favarato D, Dallan LA de O, da Luz PL. Synergistic anti-inflammatory effect: simvastatin and pioglitazone reduce inflammatory markers of plasma and epicardial adipose tissue of coronary patients with metabolic syndrome. Diabetol Metab Syndr 2014;6:47 10.1186/1758-5996-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yamaguchi Y, Cavallero S, Patterson M, Shen H, Xu J, Kumar SR, et al. Adipogenesis and epicardial adipose tissue: a novel fate of the epicardium induced by mesenchymal transformation and PPARγ activation. Proc Natl Acad Sci U S A 2015;112:2070–5. 10.1073/pnas.1417232112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Matthan NR. The Ossabaw pig is a suitable translational model to evaluate dietary patterns and coronary artery disease risk. J Nutr 2018. 10.1093/jn/nxy002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972;18:499–502. [PubMed] [Google Scholar]
- [27].Stary HC, Chandler AB, Glagov S, Guyton JR, Insull W, Rosenfeld ME, et al. A definition of initial, fatty streak, and intermediate lesions of atherosclerosis. A report from the committee on vascular lesions of the council on arteriosclerosis, American Heart Association. Circulation 1994;89:2462–78. [DOI] [PubMed] [Google Scholar]
- [28].Sus scrofa - Ensembl genome browser 91. n.d. https://useast.ensembl.org/Sus_scrofa/Info/Index.
- [29].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. 10.1093/bioinformatics/btp6.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125:279–84. 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- [31].Dawson HD, Chen C, Gaynor B, Shao J, Urban JF. The porcine translational research database: a manually curated, genomics and proteomics-based research resource. BMC Genomics 2017;18:643 10.1186/s12864-017-4009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Fessler MB, Rudel LL, Brown JM. Toll-like receptor signaling links dietary fatty acids to the metabolic syndrome. Curr Opin Lipidol 2009;20:379–85. 10.1097/MOL.0b013e32832fa5c4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Uematsu S, Akira S. Toll-like receptors and type I interferons. J Biol Chem 2007; 282:15319–23. 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- [35].Zhang X-J, Zhang P, Li H. Interferon regulatory factor Signalings in Cardiometabolic diseases. Hypertension 2015;66:222–47. 10.1161/HYPER-TENSIONAHA.115.04898. [DOI] [PubMed] [Google Scholar]
- [36].Hwang DH, Kim J-A, Lee JY. Mechanisms for the activation of toll-like receptor 2/4 by saturated fatty acids and inhibition by docosahexaenoic acid. Eur J Pharmacol 2016;785:24–35. 10.1016/j.ejphar.2016.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wieser V, Adolph TE, Grander C, Grabherr F, Enrich B, Moser P, et al. Adipose type I interferon signalling protects against metabolic dysfunction. Gut 2018;67: 157–65. 10.1136/gutjnl-2016-313155. [DOI] [PubMed] [Google Scholar]
- [38].O’Hea EK, Leveille GA. Significance of adipose tissue and liver as sites of fatty acid synthesis in the pig and the efficiency of utilization of various substrates for lipogenesis. J Nutr 1969;99:338–44. [DOI] [PubMed] [Google Scholar]
- [39].Blanc M, Hsieh WY, Robertson KA, Watterson S, Shui G, Lacaze P, et al. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol 2011;9:e1000598 10.1371/journal.pbio.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].York AG, Williams KJ, Argus JP, Zhou QD, Brar G, Vergnes L, et al. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell 2015;163:1716–29. 10.1016/j.cell.2015.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Blanc M, Hsieh WY, Robertson KA, Kropp KA, Forster T, Shui G, et al. The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 2013;38: 106–18. 10.1016/j.immuni.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu S-Y, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 2013;38:92–105. 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yang X, Ouyang H, Chen F, Ma T, Dong M, Wang F, et al. Inhibition of 3-hydroxy-3-methylglutaryl-coenzyme a reductase increases the expression of interferon-responsive genes. Clin Exp Pharmacol Physiol 2014;41:950–5. 10.1111/1440-1681.12299. [DOI] [PubMed] [Google Scholar]
- [44].Ye J, Wang C, Sumpter R, Brown MS, Goldstein JL, Gale M. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc Natl Acad Sci U S A 2003;100:15865–70. 10.1073/pnas.2237238100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Rauch I, Müller M, Decker T. The regulation of inflammation by interferons and their STATs. JAK-STAT 2013;2:e23820 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang Y, Li H. Reprogramming interferon regulatory factor signaling in cardiometabolic diseases. Physiology (Bethesda) 2017;32:210–23. 10.1152/physiol.00038.2016. [DOI] [PubMed] [Google Scholar]
- [47].Boshuizen MCS, de Winther MPJ. Interferons as essential modulators of atherosclerosis. Arterioscler Thromb Vasc Biol 2015;35:1579–88. 10.1161/ATVBAHA.115.305464. [DOI] [PubMed] [Google Scholar]
- [48].McLaughlin T, Ackerman SE, Shen L, Engleman E. Role of innate and adaptive immunity in obesity-associated metabolic disease. J Clin Invest 2017;127:5–13. 10.1172/JCI88876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ghosh AR, Bhattacharya R, Bhattacharya S, Nargis T, Rahaman O, Duttagupta P, et al. Adipose recruitment and activation of plasmacytoid dendritic cells fuel metaflammation. Diabetes 2016;65:3440–52. 10.2337/db16-0331. [DOI] [PubMed] [Google Scholar]
- [50].Kumari M, Wang X, Lantier L, Lyubetskaya A, Eguchi J, Kang S, et al. IRF3 promotes adipose inflammation and insulin resistance and represses browning. J Clin Invest 2016;126:2839–54. 10.1172/JCI86080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Eguchi J, Kong X, Tenta M, Wang X, Kang S, Rosen ED. Interferon regulatory factor 4 regulates obesity-induced inflammation through regulation of adipose tissue macrophage polarization. Diabetes 2013;62:3394–403. 10.2337/db12-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].King KR, Aguirre AD, Ye Y-X, Sun Y, Roh JD, Ng RP, et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat Med 2017;23:1481–7. 10.1038/nm.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Alsaggar M, Mills M, Liu D. Interferon beta overexpression attenuates adipose tissue inflammation and high-fat diet-induced obesity and maintains glucose homeostasis. Gene Ther 2017;24:60 10.1038/gt.2016.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Qi Z, Xia J, Xue X, Liu J, Liu W, Ding S. Targeting viperin improves diet-induced glucose intolerance but not adipose tissue inflammation. Oncotarget 2017;8: 101418–36. 10.18632/oncotarget.20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 2014;32:513–45. 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat Rev Immunol 2008;8:559–68. 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.