Abstract
Salmonella is an important food-borne pathogen associated with public health and high economic losses. To investigate the prevalence and the characteristics of Salmonella in a pig slaughterhouse in Yangzhou, a total of 80 Salmonella isolates were isolated from 459 (17.43%) samples in 2016–2017. S. Derby (35/80, 43.75%) was the most prevalent, followed by S. Rissen (16/80, 20.00%) and S. Newlands (11/80, 13.75%). The highest rates of susceptibility were observed to cefoxitin (80/80, 100.0%) and amikacin (80/80, 100.0%), followed by aztreonam (79/80, 98.75%) and nitrofurantoin (79/80, 98.75%). The highest resistance rate was detected for tetracycline (65/80, 81.25%), followed by ampicillin (60/80, 75.00%), bactrim (55/80, 68.75%), and sulfisoxazole (54/80, 67.50%). Overall, 91.25% (73/80) of the isolates were resistant to at least one antibiotic, while 71.25% (57/80) of the isolate strains were multidrug resistant in the antimicrobial susceptibility tested. In addition, 86.36% (19/22) of the 22 antimicrobial resistance genes in the isolates were identified. Our data indicated that the resistance to certain antimicrobials was significantly associated, in part, with antimicrobial resistance genes. Furthermore, 81.25% (65/80) isolates harbored the virulence gene of mogA, of which 2 Salmonella Typhimurium isolates carried the mogA, spvB and spvC virulence genes at the same time. The results showed that swine products in the slaughterhouse were contaminated with multidrug resistant Salmonella commonly, especially some isolates carry the spv virulence genes. The virulence genes might facilitate the dissemination of the resistance genes to consumers along the production chain, suggesting the importance of controlling Salmonella during slaughter for public health.
Keywords: Salmonella, Pig slaughterhouse, Antimicrobial susceptibility, Antimicrobial resistance genes, Virulence genes
Introduction
Salmonella has emerged as a major food-borne pathogen associated with breeding industry and public health in many countries (Eurosurveillance editorial team 2012; Majowicz et al. 2010; Kasimoglu Dogru et al. 2010). So far, more than 2600 identified serovars of Salmonella have been recorded (Guibourdenche et al. 2010). It is one of the leading causes of human gastroenteritis and causes more than 93.8 million infection cases annually (Majowicz et al. 2010). Salmonella has been estimated at least 1 million cases in the USA each year, resulting in the loss of 365 million dollars (Yang et al. 2019). In China, Salmonella infection cases are also frequently reported and accounted for 70–80% of bacterial food poisonings (Wang et al. 2006; Yang et al. 2016). Chen et al. reported that there were 134 outbreaks of food poisoning events caused by Salmonella in Guangxi from 1981 to 2003, which caused 7285 cases of salmonellosis (Chen et al. 2004). Between July 2010 and December 2011, nontyphoidal Salmonella were isolated from 316 (17.2%) of 1833 cases of acute gastroenteritis in children in Shanghai (Li et al. 2014a). After being infected with Salmonella, human and livestock can be asymptomatic carriers, which can reduce the fertility, aggravate the morbidity or mortality, and even be manifested as clinically fatal diseases. Pigs are considered to be one of the most important reservoir for many serovars of Salmonella, and most human infections are attributed to consumption of contaminated pork (Eurosurveillance editorial team 2012; Li et al. 2013; Vo et al. 2006a). Information on the distribution of different Salmonella enterica serovars in contaminated pork is important to public health.
Slaughterhouse is a main place where livestock and poultry products might be contaminated with Salmonella. In order to control the transmission of salmonella, the European Union has carried out many studies on pig slaughterhouses and processes. From these studies, Swart et al. have developed a model to evaluate the effects of different interventions (Swart et al. 2016). The swine herd population and pork production in China account for half of the world (Windhorst 2012). Therefore, it is of great public health significance to study the status of Salmonella contamination from pig slaughterhouses in China (Botteldoorn et al. 2004). As a big pork producer, China must strengthen its control of Salmonella transmission.
As of 2013, the total antibiotics usage in China was approximately 162,200 tons, of which 84,100 tons were for animals (Tang et al. 2016). The use of antibiotics in China accounts for about half of the world. These data show that China is one of the countries with the most severe abuse of antibiotics. In animals, salmonellosis is mainly treated with antibiotics for prevention and control. However, due to the abuse of antibiotics in recent years, the rate of Salmonella with drug resistance and even multidrug resistance has risen significantly resulting in the increased frequency of treatment failure in human clinical medicine (Hidalgo-Vila et al. 2008; Kariuki et al. 2015; Kingsley et al. 2009; Dahshan et al. 2011; Beyene et al. 2011; Hendriksen et al. 2009; Pan et al. 2009; Li and Liu 2005). Salmonella with antibiotic resistance in contaminated products could infect humans directly or transmit their resistance genes to human pathogens through the food chain, leading to the failure of antibiotic treatment and posing a threat to human health. Thus it is a reason that the disease caused by Salmonella is not well controlled clinically.
In this study, a total of 459 swine samples were randomly collected from a pig slaughterhouse between 2016 and 2017 in Yanzhou, China. All the isolates were examined for serotype distribution, antimicrobial resistance, major genotypes, and the relationship between drug resistance phenotype and drug resistance genes. The results could provide a reference for the epidemiological investigation of Salmonella in pigs.
Materials and methods
Sample collection and Salmonella isolation
During a period of 13 months, from October 2016 to October 2017, a total of 459 samples (distal ilea, n = 230; livers, n = 166; feces, n = 63) were collected at the start of the slaughter line from slaughtered pigs in a large scale industrialized slaughterhouse in Yangzhou, China. After collection, all samples were stored in sterilized containers with ice bags, and immediately processed to isolate Salmonella strains. Briefly, the samples of the ileum and liver were sheared, and 0.5 g fecal samples were picked. The samples mentioned above were added to 5 mL buffered peptone water (BPW; Neogen, Lansing, MI, USA) and incubated at 37 °C for 8–18 h. Subsequently, 100 μL pre-enriched culture was inoculated into in 5 mL of selenite cysteine (SC) broth at 37 °C for 12–18 h and then streaked on MacConkey and Salmonella and Shigella (SS) plates. After incubation for 18–24 h at 37 °C, suspected colonies were picked from MacConkey medium plates for purification. After purifying, the suspected Salmonella colonies were stained according to the Gram staining instruction manual. Then, the gram-negative bacteria were further confirmed by combined polymerase chain reaction (PCR) analysis using 2 pairs of primers [Salmonella enterotoxin gene (stn, 260 bp) and histidine transporter gene (hut, 495 bp)]. The wild-type S. Choleraesuis strain C78-3 (CVCC79103) was purchased from China Institute of Veterinary Drugs Control and used as a positive control. The primers used in this study are described in Table 1.
Table 1.
Primers used for PCR amplification
| Target genes | Nucleotide sequences | Size (bp) |
|---|---|---|
| Salmonella spp. detection primer sets | ||
| stn-F | 5′-CTTTGGTCGTAAAATAAGGCG-3′ | 260 |
| stn-R | 5′-TGCCCAAAGCAGAGAGATTC-3′ | |
| hut-F | 5′-ACTGGCGTTATCCCTTTCTCTGCTG-3′ | 495 |
| hut-R | 5′-ATGTTGTCCTGCCCCTGGTAAGAGA-3′ | |
| 16S rRNA primer set | ||
| 27F | 5′-AGAGTTTGATCCTGGCTCAG-3′ | 1466 |
| 1492R | 5′-TACGGTTACCTTGTTACGACTT-3′ | |
| Antimicrobial resistance gene primer sets | ||
| Quinolones | ||
| qnrA-F | 5′-TTCAGCAAGAGGATTTCTCA-3′ | 500 |
| qnrA-R | 5′-GGCAGCACTATTACTCCCAA -3′ | |
| qnrB-F | 5′-CCTGAGCGGCACTGAATTTT-3′ | 617 |
| qnrB-R | 5′-GTTTGCTGCTCGCCAGTCGA-3′ | |
| qnrC-F | 5′-GGGTTGTACATTTATTGAATC-3′ | 447 |
| qnrC-R | 5′-TCCACTTTACGAGGTTCT-3′ | |
| qnrD-F | 5′-TTACGGGGAATAGAGTTA-3′ | 468 |
| qnrD-R | 5′-AATCAGCCAAAGACCAAT-3′ | |
| qnrS-F | 5′-ACATAAAGACTTAAGTGATC-3′ | 619 |
| qnrS-R | 5′-CAATTAGTCAGGATAAAC-3′ | |
| qepA-F | 5′-CCAGCTCGGCAACTTGATAC-3′ | 570 |
| qepA-R | 5′-ATGCTCGCCTTCCAGAAAA-3′ | |
| oqxA-F | 5′-CTCGGCGCGATGATGCTC-3′ | 392 |
| oqxA-R | 5′-CACTCTTCACGGGAGACGA-3′ | |
| oqxB-F | 5′-TTCTCCCCCGGGGGGAAGTCCTCGGC-3′ | 512 |
| oqxB-R | 5′-CATTTTGGCGCGTA-3′ | |
| Aminoglycosides | ||
| aac(6′)-Ib-F | 5′-TTGCGATGCTCTATGAGTGGCTA-3′ | 482 |
| aac(6′)-Ib-R | 5′-CTCGAATGCCTGGCGTGTT-3′ | |
| aadA1-F | 5′-GCGCCATCTCGAACCGACGTT-3′ | 573 |
| aadA1-R | 5′-GCCCAGTCGGCAGCGACATC-3′ | |
| Sulfonamides | ||
| sul1-F | 5′-TGGCGTCGCGACTGCGAAAT-3′ | 813 |
| sul1-R | 5′-TGGTGACGGTGTTCGGCATTCT-3′ | |
| sul2-F | 5′-GTTTCTCCGATGGAGGCCGGT-3′ | 517 |
| sul2-R | 5′-AGCGAGGTTTCGGGAGCAGC-3′ | |
| Trimethoprim | ||
| dfrA1-F | 5′-AGTGCCAAAGGTGAACAGCTCCT-3′ | 308 |
| dfrA1-R | 5′-ACATCACCTTCCGGCTCGATGTCT-3′ | |
| β-lactamase | ||
| blaOXA-1-F | 5′-ATGAAAAACACAATACATATC-3′ | 830 |
| blaOXA-1-R | 5′-AATTTAGTGTGTTTAGAATGG-3′ | |
| blaPSE-1-F | 5′-CGCTTCCCGTTAACAAGTAC-3′ | 420 |
| blaPSE-1-R | 5′-CTGGTTCATTTCAGATAGCG-3′ | |
| blaTEM-F | 5′-ATAAAATTCTTGAAGACGAAA-3′ | 1080 |
| blaTEM-R | 5′-GACAGTTACCAATGCTTAATC-3′ | |
| BlaCMY-2-F | 5′-TGGCGGTTGCCGTTATCTAC-3′ | 210 |
| BlaCMY-2-R | 5′-CCCGTTTTATGCACCCATGA-3′ | |
| Tetracyclines | ||
| tetA-F | 5′-TGGTCCGGAGGCCAGACGTG-3′ | 867 |
| tetA-R | 5′-TTCCGAGCATGAGTGCCCGC-3′ | |
| tetB-F | 5′-GGAGCTACTGGGGCTGTCGCACC-3′ | 374 |
| tetB-R | 5′-ACCCACACCGTTGCGGGAAT-3′ | |
| tetG-F | 5′-TCTTGCAGGAGCCGCAGTCGAT-3′ | 721 |
| tetG-R | 5′-GGCCGGCATGCCAACACCC-3′ | |
| Chloramphenicols | ||
| catA1-F | 5′-TCTTGCCCGCCTGATGAATGC-3′ | 388 |
| catA1-R | 5′-AACCTGAATCGCCAGCGGCA-3′ | |
| floR-F | 5′-AACCCGCCCTCTGGATCAAGTCAA-3′ | 549 |
| floR-R | 5′-CAAATCACGGGCCACGCTGTATC-3′ | |
| Virulence gene primer sets | ||
| mogA-F | 5′-ATTGGCTTAGTTTCTATCTCCG-3′ | 419 |
| mogA-R | 5′-CCTTCCAGCGTTTCTTTGA-3′ | |
| pvB-F | 5′-CCGTAGAGCAGACGCTGTAAGC-3′ | 1856 |
| spvB-R | 5′-GTATCTATGAGTTGAGTACCCCTATG-3′ | |
| spvC-F | 5′-CCGCAAAGTAGTGCATCTAAAC-3′ | 919 |
| spvC-R | 5′-CCATACTTACTCTGTCATCAAACG-3′ | |
Salmonella serotyping
To evaluate the serotype distribution of the Salmonella isolates, the confirmed Salmonella strains were serotyped by slide agglutination test for O and H antigens of Salmonella using commercially available antiserum (Lanzhou Institute of Biological Products, China). Briefly, a single colony, picked with a loop, was evenly coated to a 10 μL of Salmonella standard antiserum on a slide. The slide was shaked gently for 1–2 min. The agglutinator which showed a uniform turbidity was determined as the serotype positive of Salmonella based on the measured antigenic formula according to GB/T 4789.4-2010. In this process, the negative control of saline group was also included.
Analysis of the 16S rRNA sequence
To determine the homology of the Salmonella isolates, 16S rRNA genes of the identified strains were sequenced by Sangon Biotech. Primers for the amplification of 16S rRNA corresponding to the universal primers 27F and 1492R are listed in Table 1. The DNA sequences of 16S rRNA were edited and assembled using the programs SeqMan and Edit Seq (DNA Star, Laser Gene 6, Madison, WI, USA). The sequences were aligned using MEGA v6.0 and MegAlign software. Genetic distances were defined using the Kimura 2-parameter model (Kumar et al. 2004). The phylogenetic tree was constructed by the neighbour-joining method in MEGA v6.0 (Saitou et al. 1987). Percent divergence and similarity were calculated by comparing sequence pairs in relation by MegAlign.
Antimicrobial susceptibility testing
Antimicrobial susceptibility test results of the Salmonella isolates to 22 kinds of antimicrobials was carried out in accordance with the standard Kirby-Bauer disk diffusion method recommended by the Clinical and Laboratory Standards Institute (CLSI 2017) (Berchieri et al. 2001). The reference strain, Escherichia coli ATCC 25922 was used as a control. The isolates were classified as susceptible, intermediate, or resistant according to the CLSI (2017) guidelines. Salmonella isolates resistant to at least 3 different antimicrobials were defined as multidrug resistance isolates (Pokharel et al. 2006). The following antibiotics were used in this study: ampicillin (AMP, 10 μg), mezlocillin (MEZ, 75 μg), amoxicillin/clavulanic acid (augmentin, AMC, 20/10 μg), cefoxitin (CFX, 30 μg), ceftriaxone (CRO, 30 μg), aztreonam (ATM, 30 μg), polymyxin B (POL, 300 IU), gentamicin (GEN, 10 μg), tobramycin (TOB, 10 μg), amikacin (AMK, 30 μg), kanamycin (KAN, 30 μg), neomycin (NEO, 30 μg), streptomycin (STR, 10 μg), tetracycline (TET, 30 μg), chloramphenicol (CHL, 30 μg), florfenicol (FFC, 30 μg), ciprofloxacin (CIP, 5 μg), enrofloxacin (ENR, 10 μg), sulfisoxazole (SUL, 300 μg), trimethoprim/sulfamethoxazole (bactrim, SXT, 1.25/23.75 μg), trimethoprim (TMP, 5 μg), and nitrofurantoin (NIT, 300 μg). The disks of 22 different antimicrobial agents were purchased from Hangzhou Microbiological Reagent Co., Ltd.
PCR amplification of antimicrobial resistance genes
DNA templates of the Salmonella isolates for PCR were prepared according to the boiled lysis method (Ahmed et al. 2010). In short, all the Salmonella isolates were maintained in Luria–Bertani (LB) broth. Then, an overnight bacterial culture (200 μL) was mixed with 800 μL of distilled water and boiled for 10 min. The mixture samples were centrifuged at 4 °C for 5 min and the supernatant were used as the DNA templates. After extraction of DNA, the antimicrobial resistance genes, including quinolones, aminoglycosides, sulfonamides, trimethoprim, β-lactamase, tetracyclines, and chloramphenicols, were examined by PCR amplification, using the previously described primers listed in Table 1 (Petermann et al. 2011). The PCR products were subjected to electrophoresis in a 1.0% agarose gel, and sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The DNA sequences were compared with data in the GenBank database using the BLAST tool available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Detection of virulence genes
The Salmonella pathogenicity island-I (SPI-1) virulence gene mogA and the virulence genes spvB and spvC were selected to detect the virulence of Salmonella through the multiplex PCR as described by Skyberg et al. (Skyberg et al. 2006). The primers of virulence genes used in this study are described in Table 1.
Results
Isolation of Salmonella
To investigate the contamination status of Salmonella in a pig slaughterhouse, 459 samples (distal ilea, n = 230; livers, n = 166; feces, n = 63) were collected randomly from the slaughtered pigs in Yangzhou between October 2016 and October 2017. After enrichment, purification, Gram stain and PCR procedure, a total of 80 Salmonella isolates were recovered and identified from 459 (17.43%) samples. The isolation rate of Salmonella spp. was 27.83% (64/230) in ileum samples, 8.43% (14/166) in liver samples and 3.17% (2/63) in feces samples, respectively (Table 2). The isolates showed higher positive rate for Salmonella in ileum samples than that in liver and feces samples.
Table 2.
Information of Salmonella isolated from a pig slaughterhouse in Yangzhou, China
| No. | Serotype | Origin | Virulence genes | Resistance genes | Date | ||
|---|---|---|---|---|---|---|---|
| mogA | spvB | spvC | |||||
| 1 | S. Sinstorf | Feces | − | − | − | oqxA, oqxB, aac(6′)-Ib, sul1, sul2, tetA, tetG, catA1, floR | 2 Oct 2016 |
| 2 | S. Typhimurium | Liver | + | + | + | blapsE-1, tetG, catA1 | 2 Oct 2016 |
| 3 | S. Typhimurium | Ileum | + | + | + | blapsE-1, tetG, catA1 | 2 Oct 2016 |
| 4 | S. Typhimurium | Ileum | + | − | − | qnrS, oqxB, blatem, tetA, floR, catA1 | 2 Oct 2016 |
| 5 | S. Rissen | Ileum | + | − | − | qepA, sul2, tetA | 2 Oct 2016 |
| 6 | S. Derby | Liver | + | − | − | qepA, oqxB, aac(6′)-Ib, blaoxA-1, tetA, tetG, floR, dfrA1 | 22 Oct 2016 |
| 7 | S. Sinstorf | Liver | + | − | − | oqxA, aac(6′)-Ib, blaCMY-2, tetA, tetG, floR, dfrA1 | 23 Oct 2016 |
| 8 | S. Rissen | Ileum | + | − | − | qepA, oqxB, blatem, tetA, catA1, dfrA1 | 23 Oct 2016 |
| 9 | S. Sinstorf | Liver | − | − | − | oqxA, oqxB, aac(6′)-Ib, tetA, tetG, catA1, floR, dfrA1 | 23 Oct 2016 |
| 10 | S. Derby | Liver | + | − | − | qepA, aac(6′)-Ib, blaoxA-1, tetA, dfrA1 | 23 Oct 2016 |
| 11 | S. Derby | Liver | + | − | − | oqxA, oqxB, tetA, dfrA1 | 4 Dec 2016 |
| 12 | S. Newlands | Ileum | − | − | − | tetA | 4 Dec 2016 |
| 13 | S. Newlands | Ileum | − | − | − | 4 Dec 2016 | |
| 14 | S. Newlands | Ileum | − | − | − | catA1 | 4 Dec 2016 |
| 15 | S. Derby | Ileum | + | − | − | aac(6′)-Ib, blatem, aadA1, floR | 4 Dec 2016 |
| 16 | S. Derby | Ileum | + | − | − | aac(6′)-Ib, aadA1 | 4 Dec 2016 |
| 17 | S. Derby | Ileum | + | − | − | aac(6′)-Ib, aadA1 | 10 Dec 2016 |
| 18 | S. Typhimurium | Ileum | + | − | − | qnrB, oqxA, oqxB, tetB, floR, catA1, aadA1 | 10 Dec 2016 |
| 19 | S. Derby | Ileum | + | − | − | aac(6′)-Ib | 10 Dec 2016 |
| 20 | S. Derby | Ileum | + | − | − | 10 Dec 2016 | |
| 21 | S. Derby | Ileum | + | − | − | tetA | 10 Dec 2016 |
| 22 | S. Newlands | Ileum | − | − | − | tetA | 10 Dec 2016 |
| 23 | S. Newlands | Ileum | − | − | − | 10 Dec 2016 | |
| 24 | S. Derby | Liver | + | − | − | tetA | 26 Dec 2016 |
| 25 | S. Newlands | Feces | − | − | − | dfrA1 | 26 Dec 2016 |
| 26 | S. Derby | Ileum | + | − | − | oqxA, aac(6′)-Ib, sul1, sul2, blaoxA-1, tetA, aadA1, floR | 26 Dec 2016 |
| 27 | S. Typhimurium | Ileum | + | − | − | qnrS, oqxB, blatem, tetA, floR, catA1 | 26 Dec 2016 |
| 28 | S. Rissen | Ileum | + | − | − | tetA, aadA1, catA1 | 26 Dec 2016 |
| 29 | S. Typhimurium | Ileum | + | − | − | qnrS, oqxB, blatem, tetA, floR, catA1 | 26 Dec 2016 |
| 30 | S. Derby | Liver | + | − | − | oqxA, sul2, blatem, tetA, aadA1, catA1, dfrA1 | 14 Jan 2017 |
| 31 | S. Derby | Liver | + | − | − | oqxA, sul2, blatem, tetA, aadA1, dfrA1 | 14 Jan 2017 |
| 32 | S. Derby | Liver | + | − | − | oqxA, tetA, aadA1, dfrA1 | 14 Jan 2017 |
| 33 | S. Newlands | Ileum | − | − | − | sul2, blapsE-1, tetA, catA1, floR | 14 Jan 2017 |
| 34 | S. Rissen | Ileum | − | − | − | sul2, catA1 | 25 Feb 2017 |
| 35 | S. Newlands | Ileum | − | − | − | sul2, tetA, aadA1, floR | 25 Feb 2017 |
| 36 | S. Derby | Ileum | + | − | − | oqxA, aac(6′)-Ib, sul1, sul2, blaoxA-1, blapsE-1, tetA, aadA1, floR | 25 Feb 2017 |
| 37 | S. Nchanga | Ileum | − | − | − | sul2, blapsE-1, catA1, floR | 25 Feb 2017 |
| 38 | S. Derby | Ileum | + | − | − | oqxA, sul1, sul2, blaoxA-1, blapsE-1, tetA, aadA1, floR | 25 Feb 2017 |
| 39 | S. Derby | Ileum | + | − | − | oqxA, aac(6′)-Ib, sul1, sul2, blaoxA-1, tetA, aadA1, floR | 25 Feb 2017 |
| 40 | S. Derby | Ileum | + | − | − | oqxA, aac(6′)-Ib, sul1, sul2, blaoxA-1, tetA, aadA1, floR | 25 Feb 2017 |
| 41 | S. Newlands | Ileum | − | − | − | 25 Feb 2017 | |
| 42 | S. Derby | Liver | + | − | − | aac(6′)-Ib, blaoxA-1, tetA | 2 Apr 2017 |
| 43 | S. Newlands | Ileum | + | − | − | sul2, tetA, aadA1, floR | 2 Apr 2017 |
| 44 | S. Derby | Liver | + | − | − | tetA, aadA1, catA1 | 22 Apr 2017 |
| 45 | S. Derby | Ileum | + | − | − | aadA1 | 22 Apr 2017 |
| 46 | S. Derby | Ileum | + | − | − | tetA | 22 Apr 2017 |
| 47 | S. Newlands | Ileum | − | − | − | 22 Apr 2017 | |
| 48 | S. Rissen | Ileum | + | − | − | sul2, blatem, tetA, aadA1, catA1 | 22 Apr 2017 |
| 49 | S. Rissen | Ileum | + | − | − | Blatem, tetA, aadA1, catA1 | 22 Apr 2017 |
| 50 | S. Rissen | Ileum | + | − | − | Blatem, tetA, aadA1, catA1 | 22 Apr 2017 |
| 51 | S. Rissen | Ileum | + | − | − | Blatem, tetA, aadA1, catA1 | 22 Apr 2017 |
| 52 | S. Rissen | Ileum | + | − | − | Blatem, tetA, aadA1, catA1 | 22 Apr 2017 |
| 53 | S. Derby | Ileum | + | − | − | Blatem, tetA, aadA1, catA1, floR | 22 Apr 2017 |
| 54 | S. Rissen | Ileum | + | − | − | Blatem, tetA, aadA1, catA1, dfrA1 | 22 Apr 2017 |
| 55 | S. Rissen | Ileum | + | − | − | Blatem, tetA, aadA1, catA1 | 22 Apr 2017 |
| 56 | S. Rissen | Ileum | + | − | − | Blatem, tetA, aadA1, catA1 | 22 Apr 2017 |
| 57 | S. Rissen | Ileum | + | − | − | Blatem, tetA, aadA1, catA1 | 22 Apr 2017 |
| 58 | S. Rissen | Ileum | + | − | − | sul2, blatem, tetA, aadA1, catA1, dfrA1 | 22 Apr 2017 |
| 59 | S. Derby | Ileum | + | − | − | sul2, tetA, aadA1, catA1 | 22 Apr 2017 |
| 60 | S. Derby | Liver | + | − | − | tetA, aadA1, catA1, floR | 20 May 2017 |
| 61 | S. Derby | Ileum | + | − | − | tetA | 20 May 2017 |
| 62 | S. Rissen | Ileum | + | − | − | qnrS, blatem, tetA | 20 May 2017 |
| 63 | Untyped | Ileum | + | − | − | blatem, tetA | 20 May 2017 |
| 64 | S. Derby | Ileum | + | − | − | tetA | 17 Jul 2017 |
| 65 | S. Nchanga | Ileum | + | − | − | aac(6′)-Ib, tetA | 17 Jul 2017 |
| 66 | S. Derby | Ileum | + | − | − | tetA | 20 Oct 2017 |
| 67 | S. Derby | Ileum | + | − | − | 20 Oct 2017 | |
| 68 | S. Derby | Ileum | + | − | − | catA1 | 20 Oct 2017 |
| 69 | S. Derby | Ileum | + | − | − | 20 Oct 2017 | |
| 70 | S. Derby | Liver | + | − | − | 22 Oct 2017 | |
| 71 | S. Derby | Ileum | + | − | − | 22 Oct 2017 | |
| 72 | S. Nchanga | Ileum | − | − | − | qnrS, sul2, blapsE-1, tetA, aadA1, catA1, floR | 22 Oct 2017 |
| 73 | S. Rissen | Ileum | + | − | − | aadA1, catA1 | 22 Oct 2017 |
| 74 | S. Derby | Ileum | + | − | − | 22 Oct 2017 | |
| 75 | S. Chester | Ileum | + | − | − | tetB, catA1 | 22 Oct 2017 |
| 76 | S. Chester | Ileum | + | − | − | tetB, catA1 | 22 Oct 2017 |
| 77 | S. Derby | Ileum | + | − | − | oqxA, aac(6′)-Ib, sul1, sul2, blaoxA-1, blapsE-1, tetA, aadA1, catA1, floR | 22 Oct 2017 |
| 78 | S. Typhimurium | Ileum | + | − | − | sul2, tetB, catA1 | 28 Oct 2017 |
| 79 | S. Typhimurium | Ileum | + | − | − | sul2, tetB, catA1 | 28 Oct 2017 |
| 80 | S. Typhimurium | Ileum | + | − | − | sul2, tetB, catA1 | 28 Oct 2017 |
Serotyping of Salmonella isolates
Of the 80 isolates, 79 (98.75%) were typable and 1 (1.25%) was non-typable (Table 3). The 79 Salmonella isolates consisted of 7 serotypes: S. Derby, S. Rissen, S. Newlands, S. Typhimurium, S. Sinstorf, S. Nchanga, and S. Chester. S. Derby (35/80, 43.75%) was the predominant one, followed by S. Rissen (16/80, 20.00%) and S. Newlands (11/80, 13.75%). The isolates contained 3 serovar groups: B (46/80, 57.50%), C1 (16/80, 20.00%), and E1 (18/80, 22.5%).
Table 3.
Serotype distribution of the Salmonella isolates
| Groups of sero-group of Salmonella | Serotype of Salmonella | Numbers of isolates |
|---|---|---|
| Group B | S. Derby | 35 |
| S. Typhimurium | 9 | |
| S. Chester | 2 | |
| Group C1 | S. Rissen | 16 |
| Group E1 | S. Newlands | 11 |
| S. Sinstorf | 3 | |
| S. Nchanga | 3 | |
| Untyped | 1 |
Phylogenetic analysis based on 16S rRNA gene sequences
To investigate the serotype homology of the Salmonella isolates, phylogenetic analysis of 16S rRNA sequences was carried out. All sequences of the isolates were submitted to GenBank (GenBank ID: MH548440–MH548519). A total of 80 16S rRNA gene sequences from isolates were compared based on differences in 16S rRNA sequence to construct an evolutionary tree. As shown in Fig. 1, the 80 isolates in the phylogenetic analysis were divided into three main clusters. We found that the same serotype can not be divided into discrete clusters, while the same cluster can contain multiple serotypes, which means that some Salmonella serotypes closely related to the gene sequences of 16S rRNA.
Fig. 1.
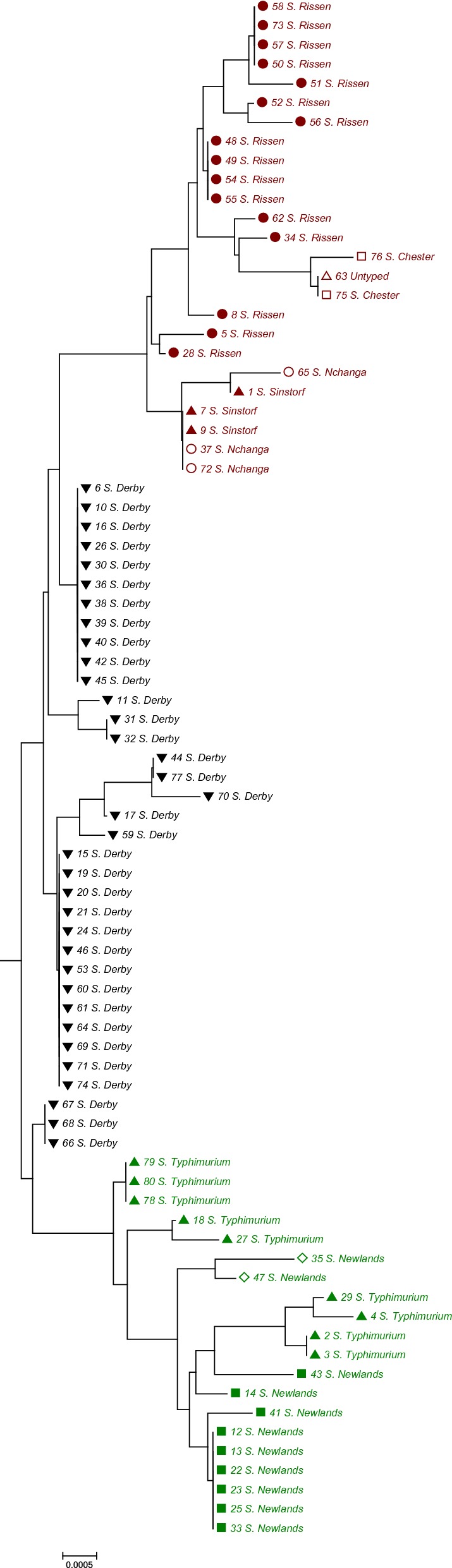
Phylogenetic tree of 80 Salmonella isolates based on 16S rRNA analysis. 16S rRNA gene sequences (1466 bp) were amplified by PCR and the nucleotide sequences determined. This is a neighbour-joining tree based on 80 Salmonella 16S rRNA sequences. The scale bar indicates one base substitution per 10,000 nt position. The number shown next to each node indicates the bootstrap value (1000 replicates)
Antimicrobial resistance of Salmonella isolates
All of the 80 Salmonella isolates were tested for antimicrobial susceptibility against 22 antimicrobial agents. The results of the antimicrobial resistance determination of the isolates are shown in Table 4. All the Salmonella isolates were susceptible to cefoxitin and amikacin and 65 (81.25%), 60 (75.00%), 55 (68.75%) and 54 (67.50%) isolates were resistant to tetracycline, ampicillin, bactrim and sulfisoxazole, respectively. Of particular note, all isolates were nonresistant against ceftriaxone, cefoxitin, azimium, polymyxin B, amikacin, and nitrofurantoin. In addition, 44 (55.00%), 31 (38.75%) and 30 (37.50%) of the isolates were moderately sensitive to enrofloxacin, polymyxin B and mezlocillin, respectively.
Table 4.
Antimicrobial resistance rates of the 80 Salmonella isolates
| Antimicrobials | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| Ampicillin | 6 (7.50%) | 14 (17.50%) | 60 (75.00%) |
| Mezlocillin | 40 (50.00%) | 30 (37.50%) | 10 (12.50%) |
| Augmentin | 52 (65.00%) | 6 (7.50%) | 22 (27.50%) |
| Ceftriaxone | 68 (85.00%) | 12 (15.00%) | 0 (0.00%) |
| Cefoxitin | 80 (100.00%) | 0 (0.00%) | 0 (0.00%) |
| Aztreonam | 79 (98.75%) | 1 (1.25%) | 0 (0.00%) |
| Polymyxin B | 49 (61.25%) | 31 (38.75%) | 0 (0.00%) |
| Gentamicin | 64 (80.00%) | 3 (3.75%) | 13 (16.25%) |
| Tobramycin | 65 (81.25%) | 3 (3.75%) | 12 (15.00%) |
| Amikacin | 80 (100.00%) | 0 (0.00%) | 0 (0.00%) |
| Kanamycin | 64 (80.00%) | 4 (5.00%) | 12 (15.00%) |
| Neomycin | 70 (87.50%) | 0 (0.00%) | 10 (12.50%) |
| Streptomycin | 31 (38.75%) | 26 (32.50%) | 23 (28.75%) |
| Tetracycline | 14 (17.50%) | 1 (1.25%) | 65 (81.25%) |
| Florfenicol | 46 (57.50%) | 4 (5.00%) | 30 (37.50%) |
| Ciprofloxacin | 55 (68.75%) | 17 (21.25%) | 8 (10.00%) |
| Enrofloxacin | 25 (31.25%) | 44 (55.00%) | 11 (13.75%) |
| Bactrim | 25 (31.25%) | 0 (0.00%) | 55 (68.75%) |
| Sulfisoxazole | 21 (26.25%) | 5 (6.25%) | 54 (67.50%) |
| Chloramphenicol | 41 (51.25%) | 2 (2.50%) | 37 (46.25%) |
| Nitrofurantoin | 79 (98.75%) | 1 (1.25%) | 0 (0.00%) |
| Trimethoprim | 49 (61.25%) | 7 (8.75%) | 24 (30.00%) |
The drug resistance profiles of the 80 isolates were constructed (Table 5). Among all of the 80 isolates, 73 (91.25%) of the isolates were resistant to at least one antibiotic, and 57 isolates showed multidrug resistance (resistant to three or more different antimicrobial agents), yielding the high rate of 71.25%. The Salmonella isolates in this study displayed a high and wide spectrum of antibiotic resistance. The most common resistance spectrums were AMP-STR-TET-SXT-SUL-TMP (n = 5) and AMP (n = 5). Totally, 46 resistance phenotypes of these isolates to 22 classes of antimicrobials were found in this study, among which most isolates were multidrug resistant to 6 and 8 classes of antimicrobials, accounted for 13.75% (11/80) and 12.50% (10/80), respectively. The highest drug resistance in the isolates were resistance to 14 antibacterial agents. Of particular note, our results showed that multidrug resistance of the isolates were frequently observed among the pig slaughterhouse.
Table 5.
Antimicrobial resistance phenotypes of the 80 Salmonella isolates
| Resistant phenotypes (numbers) | Numbers of isolates |
|---|---|
| AMP | 5 |
| STR | 1 |
| TET | 3 |
| FFC | 1 |
| AMP-TET | 2 |
| TET-SUL | 2 |
| TET-SXT | 2 |
| TET-SXT-SUL | 1 |
| AMP-TET-SXT | 1 |
| AMP-TET-SXT-SUL | 2 |
| STR-TET-SXT-TMP | 1 |
| TET-CHL-SXT-SUL | 1 |
| AMP-TET-SXT-SUL-TMP | 4 |
| AMP-MEZ-TET-SXT-SUL-TMP | 1 |
| AMP-AMC-TET-SXT | 1 |
| AMP-KAN-TET-CHL | 2 |
| AMP-AMC-TET-SXT-SUL | 1 |
| AMP-TET-CHL-SXT-SUL | 1 |
| AMP-MEZ-AMC-TET-SXT-SUL | 1 |
| AMP-STR-TET-SXT-SUL-TMP | 5 |
| AMP-TET-CHL-FFC-SXT-SUL | 2 |
| AMP-TET-CHL-SXT-SUL-TMP | 1 |
| STR-TET-CHL-FFC-SXT-SUL | 1 |
| AMP-MEZ-STR-TET-SXT-SUL-TMP | 1 |
| AMP-TET-CHL-FFC-SXT-SUL-TMP | 1 |
| AMP-AMC-TET-CHL-FFC-SXT-SUL | 3 |
| AMP-STR-TET-FFC-SXT-SUL-TMP | 1 |
| AMP-GEN-STR-TET-CHL-FFC-SXT-SUL | 1 |
| AMP-MEZ-AMC-TET-CHL-FFC-SXT-SUL | 3 |
| AMP-NEO-STR-TET-CHL-SXT-SUL-TMP | 1 |
| AMP-STR-TET-CHL-FFC-SXT-SUL-TMP | 3 |
| AMP-GEN-STR-TET-CHL-FFC-SXT-SUL-TMP | 1 |
| AMP-MEZ-STR-TET-CHL-FFC-SXT-SUL-TMP | 1 |
| AMP-AMC-GEN-TOB-KAN-STR-CHL-FFC-SXT-SUL | 1 |
| AMP-AMC-STR-TET-CHL-SXT-SUL | 1 |
| AMP-AMC-GEN-STR-TET-CHL-SXT-SUL | 1 |
| AMP-MEZ-AMC-TET-CHL-FFC-ENR-SUL | 1 |
| AMP-MEZ-GEN-TOB-STR-TET-CHL-FFC-CIP-ENR-SUL | 1 |
| AMP-GEN-TOB-KAN-NEO-STR-TET-CHL-FFC-CIP-ENR-SXT-SUL-TMP | 1 |
| AMP-AMC-GEN-TOB-STR-TET-CHL-ENR-SXT-SUL | 1 |
| AMP-AMC-TOB-KAN-NEO-TET-CHL-FFC-CIP-SXT-SUL | 1 |
| AMP-AMC-GEN-TOB-KAN-NEO-TET-CHL-FFC-ENR-SXT-SUL | 1 |
| AMP-AMC-GEN-TOB-KAN-NEO-TET-CHL-FFC-CIP-ENR-SXT-SUL | 3 |
| AMP-AMC-TOB-KAN-NEO-TET-CHL-FFC-CIP-ENR-SXT-SUL-TMP | 1 |
| AMP-MEZ-AMC-GEN-TOB-KAN-NEO-TET-CHL-FFC-ENR-SXT-SUL | 1 |
| AMP-AMC-GEN-TOB-KAN-NEO-TET-CHL-FFC-CIP-ENR-SXT-SUL-TMP | 1 |
Detection of antimicrobial resistance genes
Among the 22 resistance genes detected by PCR, 19 kinds of resistant genes in the Salmonella isolates were detected (Table 6). The highest rate was observed to the tetA gene (51/80, 63.75%), followed by catA1 (38/80, 47.50%) and aadA1 (33/80, 41.25%), which mediate the resistance to chloramphenicol and streptomycin, respectively. Only two isolates carried the qnrB gene and blaCMY-2 gene, respectively, each accounting for 1.25%. It is noteworthy that all of the aac(6′)-Ib genes harbored the -cr mutation (Trp-Arg at locus 102 and Asp-Tyr at locus 179). Three antimicrobial resistance genes qnrA, qnrC and qnrD were not detected in the isolates. Moreover, 12.50% (10/80) of the isolates did not harbored any resistance genes, of which six of them exhibited extremely weak drug resistance (intermediate). We found that the resistance genes of tetracyclines (59/80, 73.75%) displayed the highest rate, which is in general consistency with the observations of antimicrobial resistance of the isolates. Higher rates of resistance genes to chloramphenicols (48/80, 60.00%) and aminoglycosides (41/80, 51.25%) than to sulfonamides (22/80, 27.50%) and trimethoprim (12/80, 15.00%) were observed. Among the 22 resistance genes, the genes including tetA (63.75%), catA1 (47.50%), aadA1 (41.25%) and sul2 (26.25%) were the dominate genes in their corresponding resistance gene categories. Although the detection rates of quinolones and β-lactamase resistance genes were relatively high, the composition of each resistance gene was relatively dispersed.
Table 6.
Antimicrobial resistance genes of the Salmonella isolates
| Drug classes | Resistance genes | Number of isolates | Positive rates (%) |
|---|---|---|---|
|
Quinolones 40.00% |
qnrA | 0 | 0.00 |
| qnrB | 1 | 1.25 | |
| qnrC | 0 | 0.00 | |
| qnrD | 0 | 0.00 | |
| qnrS | 5 | 6.25 | |
| qepA | 4 | 5.00 | |
| oqxA | 14 | 17.50 | |
| oqxB | 9 | 11.25 | |
|
Aminoglycosides 51.25% |
aac(6′)-Ib | 16 | 20.00 |
| aadA1 | 33 | 41.25 | |
|
Sulfonamides 27.50% |
sul1 | 8 | 10.00 |
| sul2 | 21 | 26.25 | |
|
β-lactamase 43.75% |
blaOXA-1 | 9 | 11.25 |
| blaPSE-1 | 8 | 10.00 | |
| blaTEM | 20 | 25.00 | |
| blaCMY-2 | 1 | 1.25 | |
|
Tetracyclines 73.75% |
tetA | 51 | 63.75 |
| tetB | 6 | 7.50 | |
| tetG | 6 | 7.50 | |
|
Chloramphenicols 60.00% |
catA1 | 38 | 47.50 |
| floR | 22 | 27.50 | |
|
Trimethoprim 15.00% |
dfrA1 | 12 | 15.00 |
Relationship of antimicrobial resistance genes with antimicrobial susceptibility
The relationship between the antimicrobial resistance genes and the resistance phenotypes of the Salmonella isolates were analyzed by integrating the above data in this study. As shown in Table 7, the association of antimicrobial resistance genes with antimicrobial susceptibility was variable among differents Salmonella isolates. Among the six major categories of antimicrobials, the high relativity (> 80%) between the phenotypes and the antimicrobial resistance genotypes in the isolates was 33/41 (strains resistant to aminoglycosides antimicrobials/strains harbored resistant genes to aminoglycosides antimicrobials), 65/59 and 39/48, respectively for aminoglycosides, tetracyclines and chloramphenicols antimicrobials. In contrast, there were different in the quinolones, folate pathway inhibitors and β-lactamase with a coincidence rate of about 50%. For the specific antimicrobial resistance, the correlations between phenotypes and genotypes of the isolates for cephalosporins and tetracyclines were 0/1 and 65/59, respectively, much higher (> 90%) than other antimicrobials. These data indicated that the resistance to certain antimicrobials was associated, in part, with antimicrobial resistance genes.
Table 7.
Resistance genes and phenotype relationship of Salmonella isolates
| Drug classes | Quinolones | Aminoglycosides | Folate pathway inhibitors | β-lactamase | Tetracyclines | Chloramphenicols | ||
|---|---|---|---|---|---|---|---|---|
| Ciprofloxacin Enrofloxacin |
Gentamicin, Tobramycin Kanamycin, Neomycin Streptomycin |
Sulfonamides | Trimethoprim | Penicillins | Cephalosporins | Tetracycline | Chloramphenicol Florfenicol |
|
| Resistance genes | qnr/qepA/oqxA/oqxB | aadA1/aac(6′)-Ib | sul1/sul2 | dfrA1 | blaOXA-1/blaPSE-1/blaTEM | blaCMY-2 | tetA/tetB/tetG | catA1/floR |
| Number of isolates carrying drug-resistant genes | 23 | 41 | 22 | 12 | 34 | 1 | 59 | 48 |
| Number of drug-resistant isolates | 12 | 33 | 59 | 24 | 60 | 0 | 65 | 39 |
Detection of virulence genes
Among the 80 isolates, 3 virulence genes mogA, spvB and spvC were detected. We found that 81.25% (65/80) isolates at least carried the virulence gene of mogA, of which 2 Salmonella Typhimurium strains (2.50%) harbored the mogA, spvB and spvC virulence genes at the same time (Table 2). 18.75% (15/80) of the isolates did not carried any of the virulence genes. The results suggested that swine products in the slaughterhouse were commonly contaminated with mogA virulence gene. It is noteworthy that some isolates had the spv virulence genes, which is a great threat to public health safety.
Discussion
Salmonella is one of the most common food-borne pathogens, with a wide range of hazards that can cause contamination of various agricultural products (Shao et al. 2011). It has been reported that the isolation rate of swine Salmonella in China was range from 11 to 35% and the predominant serotypes of the isolates were S. Derby, S. Typhimurium, S. Enteritidis, and S. Argona (Huang et al. 2012; Wang et al. 2016; Zou et al. 2012; Song et al. 2004; Kuang et al. 2015). Salmonella strains with strong pathogenicity were widespread, among them, S. Typhimurium and S. Enteritidis are the major non-typhoid Salmonella that cause diarrhea in humans. In this study, samples were collected randomly from the slaughtered pigs in mainland China between October 2016 and October 2017. Eventually, 80 Salmonella isolates were recovered from 459 samples of the pig slaughterhouse, and the overall isolation rate of Salmonella spp. was 17.43%. Among the isolates, the prevalence of Salmonella was 27.83% (64/230) in ileum samples, 8.43% (14/166) in liver samples and 3.17% (2/63) in feces samples, respectively. The prevalence of Salmonella from ileum was slightly higher than the 23.8% observed in Brazil (da Silva et al. 2012) and lower than the 36.5% reported in Huaian in China (Zhou et al. 2017).The serotyping results indicated that S. Derby (43.75%) in B group was the predominant serovar in the slaughterhouse and processing chain, which is consistent with previous studies (Bonardi et al. 2016; Piras et al. 2011). In fact, S. Derby has been shown to be the most common serovar across the world. For example, S. Derby has been shown to represent the most significant proportion of serovars in both pork and slaughterhouse in China (Cai et al. 2016; Li et al. 2014b). In general consistency with the reports by Zhou et al. (Zhou et al. 2017), we observed that S. Rissen was the second most common serovar in the slaughterhouse. A total of 14 Salmonella strains were isolated from liver samples consisting of S. Derby (n = 11), S. Chester (n = 2) and S. Typhimurium (n = 1). It is noteworthy that all the 3 serotypes of Salmonella have been reported to infect humans in China with the clinical syndromes including diarrhoea and septicemia (Liang et al. 2016; Zhou et al. 2013; Guo et al. 2015; Sun et al. 2014). These results suggest the importance of controlling Salmonella during slaughter process and regular surveillance for public health. In addition, Salmonella was isolated from October 2016 to October 2017, no significant difference in the prevalence was observed (data not shown).
It has been shown that Salmonella is widely drug-resistant and commonly multidrug resistant (Hidalgo-Vila et al. 2008; Li and Liu 2005; Chen et al. 2008; Wang et al. 2007, 2009). In this study, our results showed that 73 Salmonella isolates were resistant to at least one antimicrobial agent and most of the isolates showed multidrug resistance, mainly to tetracycline, ampicillin, bactrim, sulfisoxazole, and chloramphenicol. Our results concerning the phenomenon of particularly severe drug resistance are consistent with previously described findings in China (Yang et al. 2019; Lu et al. 2011). In this study, multidrug resistance isolate rate of Salmonella (71.25%) was similar to another two studies (71.4% and 73.9%) in China (Yang et al. 2019; Li et al. 2013). Our results showed that multidrug resistance of the isolates were frequently observed among the pig slaughterhouse. Reducing antibiotics use in pigs is particularly important to limit the emergence of multidrug resistance bacteria and to maintain good public health. According to previous reports (Chen et al. 2008; Vo et al. 2006b; Zhao et al. 2007), Salmonella strains were highly resistant to ampicillin, chloramphenicol, kanamycin, streptomycin, sulfonamides, tetracyclines and quinolones, consistent with the results in this study. Among the 80 Salmonella isolates, 7 isolates were none drug resistant strains including 5 S. Newlands strains, one S. Derby and one S. Typhimurium. The none drug resistant isolates were mainly distribution the S. Newlands, accounted for 45.45% (5/11) of all the isolated S. Newlands. Among all of the 9 S. Typhimurium isolates, one isolate was none drug resistant, and 5 isolates were only resistant to 1 or 2 kinds of antibiotics, indicating that the resistance of S. Newlands and S. Typhimurium were relatively low. Combined with the data of the resistant phenotypes and the antimicrobial resistance genes of the Salmonella isolates, the isolates have a high correlation between the phenotypes and genotypes of aminoglycosides, cephalosporins, tetracyclines and chloramphenicols, while the relationship between the resistance genes and the resistance phenotypes of the Salmonella isolates of trimethoprim, sulfonamides, penicillins and quinolones were relatively lower. These data indicated that the resistance to certain antimicrobials was associated with antimicrobial resistance genes. Moreover, the association of antimicrobial resistance genes with antimicrobial susceptibility were variable among differents Salmonella isolates. Some isolates harboring drug resistance genes were not highly drug-resistant while resistance genes could not be amplified by PCR from some highly drug-resistant isolate strains. It might be due to untested or unknown drug resistance genes in the resistant strains, and propose that further study is necessary.
MogA is a virulence gene associated with invasiveness on Salmonella SPI-1. SpvB gene has adenosine diphosphate (ADP) ribose transferase activity which mediates the modification of G-actin and the block of F-actin, and then disrupts the cytoskeletal function of actin (Tezcan-Merdol et al. 2005; Mesa-Pereira et al. 2013). The protein encoded by the spvC gene has a phosphorylated threonine lyase activity that inhibits MAP phosphokinase (Haneda et al. 2012; Mazurkiewicz et al. 2008). Notably, spvB and spvC are required for the expression of the spv gene simultaneously. The pathogenicity of Salmonella strains will greatly increase when both spvB and spvC genes exist at the same time. In this study, we found that swine products in the slaughterhouse were commonly contaminated with the mogA virulence gene (65/80, 81.25%). In general, the high detection rate of virulence genes highlights the pathogenic potential of these isolates, which may indicate serious salmonellosis and a threat to public health (Fardsanei et al. 2017). Our results also suggested that the Salmonella isolates harboring spvB and spvC virulence genes existed in the swine slaughterhouse, which is a great threat to public health.
The work described here highlights the prevalence and antimicrobial resistance of Salmonella in a pig slaughterhouse in mainland China. Swine products in the slaughterhouse were contaminated with multidrug resistant Salmonella commonly, even a small fraction of them might carry the spv virulence genes, which suggests efficient measures to facilitate the reasonable use of antimicrobials in animal husbandry must be taken to control Salmonella during slaughter for public health, underlying strict hygiene method and HACCP (Hazard Analysis and Critical Control Points) management are vital for reducing cross-contamination. To reduce Salmonella contamination, several other interventions have proven successful. Moreover, a mature and healthy livestock system should be established to strictly control the environmental hygiene, carcass hygiene, drinking water and feed hygiene, as well as the supervision of the processing and circulation of animal products. We believe that it is necessary to extend more studies about practical interventions in pig slaughterhouses to control Salmonella in China. Collectively, nationwide regular surveillance is needed to screen any changes in antimicrobial resistance patterns in Salmonella isolates in the swine industry.
Acknowledgements
Not applicable.
Authors’ contributions
QL and JY performed the experiments, interpreted the data and wrote the manuscript; ZL and ZWL performed some experiments; YZD, WWG, MB and SFW participated in the discussion and revised the manuscript; HYS participated in experimental design interpreted the data, and supervised the research project. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 31672516, 31172300, and 30670079), Grant No. BE2016343 from Jiangsu province, the Jiangsu University and College Natural Science Foundation (No. 12KJA230002), the Natural Science Foundation of Jiangsu Province (No. BK20190886), China Postdoctoral Science Foundation [No. 2019M661953], the Doctoral Program of Higher Education of China (No. 20133250110002), supported by State Key Laboratory of Genetically Engineered Veterinary Vaccines (No. AGVSKL-ZD/ZY-201807) and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Availability of data and materials
Not applicable.
Ethical approval and consent to participate
Procedures involving the care and use of animals were approved by the Jiangsu Administrative Committee for Laboratory Animals (permission number SYXK-SU-2007-0005) and complied with the Jiangsu Laboratory Animal Welfare and Ethics guidelines of the Jiangsu Administrative Committee of Laboratory Animals.
Consent for publication
All authors gave their informed consent prior to their inclusion in the study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Quan Li and Jian Yin contributed equally to this work
Contributor Information
Quan Li, Email: liquan2018@yzu.edu.cn.
Jian Yin, Email: 987780941@qq.com.
Zheng Li, Email: 1156851868@qq.com.
Zewei Li, Email: 942282754@qq.com.
Yuanzhao Du, Email: 2325164237@qq.com.
Weiwei Guo, Email: 924214484@qq.com.
Matthew Bellefleur, Email: mbellefl@ufl.edu.
Shifeng Wang, Email: shifengwang@ufl.edu.
Huoying Shi, Email: hystg2017@163.com.
References
- Ahmed AM, Shimabukuro H, Shimamoto T. Isolation and molecular characterization of multidrug-resistant strains of Escherichia coli and Salmonella from retail chicken meat in Japan. J Food Sci. 2010;74(7):M405–M410. doi: 10.1111/j.1750-3841.2009.01291.x. [DOI] [PubMed] [Google Scholar]
- Berchieri A, Jr, Murphy CK, Marston K, Barrow PA. Observations on the persistence and vertical transmission of Salmonella enterica serovars Pullorum and Gallinarum in chickens: effect of bacterial and host genetic background. Avian Pathol. 2001;30(3):221–231. doi: 10.1080/03079450120054631. [DOI] [PubMed] [Google Scholar]
- Beyene G, Nair S, Asrat D, Mengistu Y, Engers H, Wain J. Multidrug resistant Salmonella Concord is a major cause of salmonellosis in children in Ethiopia. J Infect Dev Ctries. 2011;5(1):23–33. doi: 10.3855/jidc.906. [DOI] [PubMed] [Google Scholar]
- Bonardi S, Alpigiani I, Bruini I, Barilli E, Brindani F, Morganti M, Cavallini P, Bolzoni L, Pongolini S. Detection of Salmonella enterica in pigs at slaughter and comparison with human isolates in Italy. Int J Food Microbiol. 2016;218:44–50. doi: 10.1016/j.ijfoodmicro.2015.11.005. [DOI] [PubMed] [Google Scholar]
- Botteldoorn N, Herman L, Rijpens N, Heyndrickx M. Phenotypic and molecular typing of Salmonella strains reveals different contamination sources in two commercial pig slaughterhouses. Appl Environ Microbiol. 2004;70(9):5305–5314. doi: 10.1128/AEM.70.9.5305-5314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Tao J, Jiao Y, Fei X, Zhou L, Wang Y, Zheng H, Pan Z, Jiao X. Phenotypic characteristics and genotypic correlation between Salmonella isolates from a slaughterhouse and retail markets in Yangzhou, China. Int J Food Microbiol. 2016;222:56–64. doi: 10.1016/j.ijfoodmicro.2016.01.020. [DOI] [PubMed] [Google Scholar]
- Chen XL, Tang ZZ, Huang L. The epidemiological evaluation of the food poisoning in Guangxi from 1981 to 2003 and the strategies of intervention in future. Guangxi J Prev Med. 2004;10(4):200–204. [Google Scholar]
- Chen L, Cong-Ming WU, Shen JZ. Detection of antimicrobial resistance and class 1 integrons among Salmonella isolates from animals. Chin J Vet Med. 2008;44:6–9. [Google Scholar]
- da Silva LE, Dias V, Ferronatto A, Guerra P, Berno L, Triches N, Kich JD, Corbellini LG, Cardoso M. Longitudinal dissemination of Salmonella enterica clonal groups through the slaughter process of Salmonella-positive pig batches. J Food Prot. 2012;75(9):1580–1588. doi: 10.4315/0362-028X.JFP-11-515. [DOI] [PubMed] [Google Scholar]
- Dahshan H, Abd-El-Kader MA, Chuma T, Moriki H, Okamoto K. Re-emergence of multi-drug resistant Salmonella enterica serovar Stanley from cattle. Vet Res Commun. 2011;35(1):55. doi: 10.1007/s11259-010-9448-4. [DOI] [PubMed] [Google Scholar]
- Eurosurveillance editorial team (2012) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. Euro Surveill 17(10) [PubMed]
- Fardsanei F, Soltan Dallal MM, Douraghi M, Zahraei Salehi T, Mahmoodi M, Memariani H, Nikkhahi F. Genetic diversity and virulence genes of Salmonella enterica subspecies enterica serotype Enteritidis isolated from meats and eggs. Microb Pathog. 2017;107:451–456. doi: 10.1016/j.micpath.2017.04.026. [DOI] [PubMed] [Google Scholar]
- Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, Weill FX. Supplement 2003–2007 (No. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol. 2010;161(1):26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Guo Z, Su C, Huang J, Niu J. A food-borne outbreak of gastroenteritis caused by different Salmonella serotypes in 2 universities in Xiamen, Fujian, China, in 2012. Jpn J Infect Dis. 2015;68(3):187–191. doi: 10.7883/yoken.JJID.2014.235. [DOI] [PubMed] [Google Scholar]
- Haneda T, Ishii Y, Shimizu H, Ohshima K, Iida N, Danbara H, Okada N. Salmonella type III effector SpvC, a phosphothreonine lyase, contributes to reduction in inflammatory response during intestinal phase of infection. Cell Microbiol. 2012;14(4):485–499. doi: 10.1111/j.1462-5822.2011.01733.x. [DOI] [PubMed] [Google Scholar]
- Hendriksen RS, Mikoleit M, Kornschober C, Rickert RL, Duyne SV, Kjelsø C, Hasman H, Cormican M, Mevius D, Threlfall J. Emergence of multidrug-resistant Salmonella concord infections in Europe and the United States in children adopted from Ethiopia, 2003–2007. Pediatr Infect Dis J. 2009;28(28):814–818. doi: 10.1097/INF.0b013e3181a3aeac. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Vila J, Díaz-Paniagua C, Pérez-Santigosa N, Frutos-Escobar CD, Herrero-Herrero A. Salmonella in free-living exotic and native turtles and in pet exotic turtles from SW Spain. Res Vet Sci. 2008;85(3):449–452. doi: 10.1016/j.rvsc.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Huang FB, Bing-Xia LU, Liu L, Guo X, Zhang MX, Lan JN, Liao CQ, Zhao CQ, Huang WJ. Isolation identification, drug resistance analysis and pathogenicity tests of salmonella isolates from slaughter pigs intestine. China Anim Husbandry Vet Med. 2012;39(1):172–177. [Google Scholar]
- Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine. 2015;33(1):C21–C29. doi: 10.1016/j.vaccine.2015.03.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasimoglu Dogru A, Ayaz ND, Gencay YE. Serotype identification and antimicrobial resistance profiles of Salmonella spp. isolated from chicken carcasses. Trop Anim Health Prod. 2010;42(5):893–897. doi: 10.1007/s11250-009-9504-7. [DOI] [PubMed] [Google Scholar]
- Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, Harris D, Clarke L, Whitehead S, Sangal V. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19(12):2279. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang X, Hao H, Dai M, Wang Y, Ahmad I, Liu Z, Yuan Z. Serotypes and antimicrobial susceptibility of Salmonella spp. isolated from farm animals in China. Front Microbiol. 2015;6:602. doi: 10.3389/fmicb.2015.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Li WX, Liu CZ. Studies on the minimum inhibitory concentration of six kinds of fluoroquinolones to Salmonella. Chin J Vet Drug. 2005;39:4–8. [Google Scholar]
- Li R, Lai J, Wang Y, Liu S, Li Y, Liu K, Shen J, Wu C. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int J Food Microbiol. 2013;163(1):14–18. doi: 10.1016/j.ijfoodmicro.2013.01.020. [DOI] [PubMed] [Google Scholar]
- Li Y, Xie X, Xu X, Wang X, Chang H, Wang C, Wang A, He Y, Yu H, Wang X. Nontyphoidal salmonella infection in children with acute gastroenteritis: prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis. 2014;11(3):200. doi: 10.1089/fpd.2013.1629. [DOI] [PubMed] [Google Scholar]
- Li YC, Pan ZM, Kang XL, Geng SZ, Liu ZY, Cai YQ, Jiao XA. Prevalence, characteristics, and antimicrobial resistance patterns of Salmonella in retail pork in Jiangsu province, eastern China. J Food Prot. 2014;77(2):236–245. doi: 10.4315/0362-028X.JFP-13-269. [DOI] [PubMed] [Google Scholar]
- Liang YX, Xiao-Nan LI, Shao-Dong WUDO. Inspection, clinical analysis of 204 children infected by mouse typhus salmonella. China Pract Med. 2016;11(17):44–45. [Google Scholar]
- Lu Y, Wu CM, Wu GJ, Zhao HY, He T, Cao XY, Dai L, Xia LN, Qin SS, Shen JZ. Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog Dis. 2011;8(1):45–53. doi: 10.1089/fpd.2010.0605. [DOI] [PubMed] [Google Scholar]
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50(6):882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- Mazurkiewicz P, Thomas J, Thompson JA, Liu M, Arbibe L, Sansonetti P, Holden DW. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol Microbiol. 2008;67(6):1371–1383. doi: 10.1111/j.1365-2958.2008.06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa-Pereira B, Medina C, Camacho EM, Flores A, Santero E. Novel tools to analyze the function of Salmonella effectors show that SvpB ectopic expression induces cell cycle arrest in tumor cells. PLoS ONE. 2013;8(10):e78458. doi: 10.1371/journal.pone.0078458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan WJ, Xiang C, Wang XQ, Cong QX, Pan ZM, Song G, Jiao XA. The analysis of quinolone resistance of the avian Escherichia coli and Salmonella isolates from 1993 to 2008. Chin J Zoo. 2009;9(28):3567–3579. [Google Scholar]
- Petermann SR, Sherwood JS, Stepan RM, Logue CM. Molecular and comparative analysis of Salmonella enterica Senftenberg from humans and animals using PFGE, MLST and NARMS. BMC Microbiol. 2011;11(1):153. doi: 10.1186/1471-2180-11-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras F, Brown DJ, Meloni D, Mureddu A, Mazzette R. Investigation of Salmonella enterica in Sardinian slaughter pigs: prevalence, serotype and genotype characterization. Int J Food Microbiol. 2011;151(2):201–209. doi: 10.1016/j.ijfoodmicro.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Pokharel BM, Koirala J, Dahal RK, Mishra SK, Khadga PK, Tuladhar NR. Multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing Salmonella enterica (serotypes Typhi and Paratyphi A) from blood isolates in Nepal: surveillance of resistance and a search for newer alternatives. Int J Infect Dis. 2006;10(6):434–438. doi: 10.1016/j.ijid.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Saitou NNM, Nei MC, Saitou N, Nei M. The neighbor-joining method—a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shao D, Shi Z, Wei J, Ma Z. A brief review of foodborne zoonoses in China. Epidemiol Infect. 2011;139(10):1497–1504. doi: 10.1017/S0950268811000872. [DOI] [PubMed] [Google Scholar]
- Skyberg JA, Logue CM, Nolan LK. Virulence genotyping of Salmonella spp. with multiplex PCR. Avian Dis. 2006;50(1):77–81. doi: 10.1637/7417.1. [DOI] [PubMed] [Google Scholar]
- Song L, Ning YB, Zhang GC, Shen QC, Wang Q, Zhao Y, Feng ZW, Shi-Xin XU, Gao G, Gao YC. The isolation and serum-type identification of Escherichia coli (E. coli) and Salmonella from the east China. Chin J Prev Vet Med. 2004;26(2):142–145. [Google Scholar]
- Sun J, Ke B, Huang Y, He D, Li X, Liang Z, Ke C. The molecular epidemiological characteristics and genetic diversity of salmonella typhimurium in Guangdong, China, 2007–2011. PLoS ONE. 2014;9(11):e113145. doi: 10.1371/journal.pone.0113145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart AN, Evers EG, Simons RL, Swanenburg M. Modeling of Salmonella contamination in the pig slaughterhouse. Risk Anal. 2016;36(3):498–515. doi: 10.1111/risa.12514. [DOI] [PubMed] [Google Scholar]
- Tang Q, Song P, Li J, Kong F, Sun L, Xu L. Control of antibiotic resistance in China must not be delayed: the current state of resistance and policy suggestions for the government, medical facilities, and patients. Biosci Trends. 2016;10(1):1–6. doi: 10.5582/bst.2016.01034. [DOI] [PubMed] [Google Scholar]
- Tezcan-Merdol D, Engstrand L, Rhen M. Salmonella enterica SpvB-mediated ADP-ribosylation as an activator for host cell actin degradation. Int J Med Microbiol. 2005;295(4):201–212. doi: 10.1016/j.ijmm.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Vo AT, van Duijkeren E, Fluit AC, Heck ME, Verbruggen A, Maas HM, Gaastra W. Distribution of Salmonella enterica serovars from humans, livestock and meat in Vietnam and the dominance of Salmonella Typhimurium phage type 90. Vet Microbiol. 2006;113(1–2):153–158. doi: 10.1016/j.vetmic.2005.10.034. [DOI] [PubMed] [Google Scholar]
- Vo AT, Van DE, Fluit AC, Heck ME, Verbruggen A, Van DZK, Gaastra W. Class 1 integrons in Dutch Salmonella enterica serovar Dublin isolates from clinical cases of bovine salmonellosis. Vet Microbiol. 2006;117(2):192–200. doi: 10.1016/j.vetmic.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Wang SJ, Yang J, Shen ZQ. Analysis on 766 events of bacterial food poisoning in China from 1994 to 2003. China Prev Med. 2006;7(3):180–184. [Google Scholar]
- Wang XQ, Jiao XA, Liu XW, Chen X, Huan HX. Characterization of multidrug-resistant Salmonella serovars isolated from meats and human samples in some regions of Jiangsu. Wei Sheng Wu Xue Bao. 2007;47(2):221–227. [PubMed] [Google Scholar]
- Wang J, Wang YD, Zheng ZR, Zhao SJ, Huang XM. Study on susceptibility for avian E. coli and Salmonella. Poult Sci. 2009;6:8–10. [Google Scholar]
- Wang W, Zhang S, Zhou B, Hu X, Zhang H, Wen M, Cheng Z, Wang K. Isolation, identification and drug resistance of Salmonella from Swine. Guizhou J Anim Husbandry Vet Med. 2016;40(6):1–4. [Google Scholar]
- Windhorst HW (2012) China is the world’s biggest pork producer. A status report about the dynamics, patterns and problems of China’s steadily growing swine industry
- Yang H, Mou Y, Luo W. Research progress of food-borne Salmonella. Heilongjiang Anim Sci Vet Med. 2016;69(4):69–71. [Google Scholar]
- Yang J, Ju Z, Yang Y, Zhao X, Jiang Z, Sun S. Serotype, antimicrobial susceptibility and genotype profiles of Salmonella isolated from duck farms and a slaughterhouse in Shandong province, China. BMC Microbiol. 2019;19(1):202. doi: 10.1186/s12866-019-1570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Mcdermott PF, White DG, Qaiyumi S, Friedman SL, Abbott JW, Glenn A, Ayers SL, Post KW, Fales WH. Characterization of multidrug resistant Salmonella recovered from diseased animals. Vet Microbiol. 2007;123(1):122–132. doi: 10.1016/j.vetmic.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Pan Z, Li Y, Kang X, Wang X, Geng S, Liu Z, Jiao X, Liu X. Epidemiological analysis of Salmonella isolates recovered from food animals and humans in eastern China. Food Res Int. 2013;54(1):223–229. doi: 10.1016/j.foodres.2013.07.002. [DOI] [Google Scholar]
- Zhou ZH, Li JW, Zheng HJ, Jin XC, Shen Y, Lei TY, Sun XY, Pan ZM, Jiao XA. Diversity of Salmonella isolates and their distribution in a pig slaughterhouse in Huaian, China. Food Control. 2017;78:238–246. doi: 10.1016/j.foodcont.2017.02.064. [DOI] [Google Scholar]
- Zou LK, Yan-Jun PU, Yang L, Liu CH, Xiao P, Luo Y, Bei LI. Isolation and drug resistance analysis of Escherichia coli and Salmonella spp. Pork from Sichuan Province. Food Sci. 2012;33(13):202–206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


