Abstract
The mammalian gut harbors a vast community of microorganisms — termed the microbiota — whose composition and dynamics are considered to be critical drivers of host health. These factors depend, in part, upon the manner in which microbes interact with one another. Microbes are known to engage in a myriad of different ways, ranging from unprovoked aggression to actively feeding each other. However, the relative extent to which these different interactions occur between microbes within the gut is unclear. In this minireview we assess our current knowledge of microbe–microbe interactions within the mammalian gut microbiota, and the array of methods used to uncover them. In particular, we highlight the discrepancies between different methodologies: some studies have revealed rich networks of cross-feeding interactions between microbes, whereas others suggest that microbes are more typically locked in conflict and actively cooperate only rarely. We argue that to reconcile these contradictions we must recognize that interactions between members of the microbiota can vary across condition, space, and time — and that only through embracing this dynamism will we be able to comprehensively understand the ecology of our gut communities.
Introduction
The mammalian intestine harbors a vast community of microbes, collectively known as the gut microbiota [1], that plays a critical role in host health [2]. The composition of the mammalian microbiota demonstrates both remarkable dynamism and striking stability: on the one hand, diversifying after the initial colonization that follows birth and fluctuating in response to diet and disease, yet maintaining core taxa over periods of years [3]. Determining the forces that shape our gut communities is fundamental to our understanding of the functioning of the human microbiota and will also offer a window into many of the ecological and evolutionary forces that shape microbial communities more broadly. Central to this goal is establishing an understanding of the ecology of our microbiota. How does each organism survive in the gut, what niche does it occupy, and how does it interact with those around it? Specifically, with which other organisms does each compete, which organisms do they each rely upon, and how do these interactions change with environmental conditions? Box 1 and Figure 1 each introduce common ecological terms that describe the nature of interactions between organisms and how they are applied in the context of microbial ecology.
Box 1. An ecologist’s guide to microbe–microbe interactions in the host gut.
Positive interactions: cooperation, commensalism, and cross-feeding
Positive interactions between microbial species range from commensal — wherein one species derives a benefit from another that is itself unaffected — to true cooperation — where one species has evolved specifically to increase the fitness of another. Cooperation may or may not be reciprocal; though if there is a cost to one species, which there tends to be, indirect fitness benefits must be received in return, in order for the cooperation to be evolutionarily stable. Positive interactions within the microbiome are typically cases of cross-feeding or syntrophy — commensal interactions in which one species grows on waste products produced by another. Very few examples of true interspecies cooperation have been identified within the gut microbiome, perhaps unsurprising given cooperation is often both ecologically and evolutionarily unstable [37,38].
Negative interactions: competition, ammensalism, exploitation, and interference
Negative interactions may be symmetric (−/− competition) or one-sided (-/0 ammensalism), though often both are loosely referred to as competition, which itself falls into two categories. Exploitative competition is an indirect interaction wherein two species compete for a common resource, reducing one another’s fitness as a consequence. In contrast, interference competition typically involves one species directly affecting the fitness of another. In the mammalian microbiome, exploitative competition commonly occurs over shared nutrients; however, microbes may also compete for space. Meanwhile, a range of weapons for interference competition between microbes have been identified, ranging from type VI secretion systems and bacteriocins, to mechanisms that induce host immune responses that in turn inhibit other microbes.
Asymmetric interactions: exploitation, predation and parasitism
Asymmetric interactions (+/−, confusingly also often termed exploitation) are those in which one species gains a fitness benefit at a cost to another. In macro-ecology this is often classified as predation or parasitism but conceptualizing such interactions for microbes can be challenging. The predatory bacterium Bdellovibrio bacteriovorus actively feeds on other bacteria [49], whereas parasitic bacteriophages multiply inside and often eventually kill their bacterial hosts [48]. However, in the gut microbiome asymmetric interactions are more likely to manifest when one species receives a growth boost from another (for example, via cross-feeding) and in response alters the environment in a manner that harms its partner, for example through producing a toxic waste product or changing the environmental pH [50].
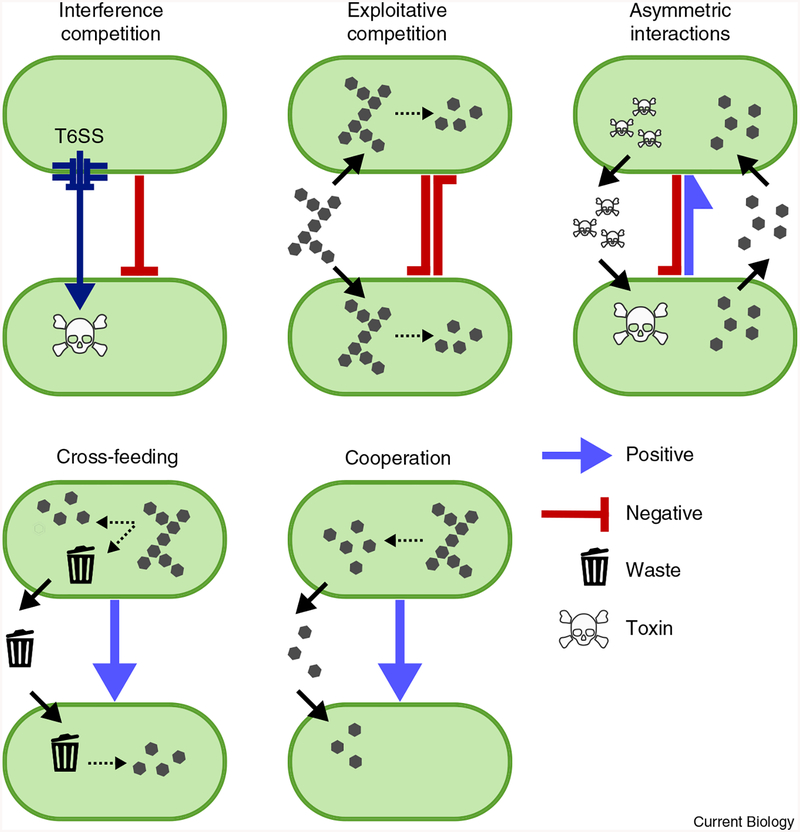
Figure 1. Types of interactions by fitness effect and mechanism.
Pairwise interactions between species are defined based on the effect each microbe has on the other’s fitness, and on the mechanism by which that effect is achieved. Interactions in which one microbe is negatively affected and the other is either unaffected (-/0) or harmed (−/−) are defined as ammensal or competitive, respectively. When these interactions are mediated by competition for a nutrient they are termed exploitation, and interference refers to when the interaction is direct (for example, type VI killing). Interactions can also be asymmetric, when one species gains a fitness benefit at the expense of another (+/−). Positive interactions within the gut are typically either defined as cross-feeding when microbes grow on by-products, such as fermentation products, amino acids or digested sugars produced by others, or cooperation when one microbe has evolved an adaptation specifically to increase the fitness of another. These interactions can either be reciprocal (+/+) or not, in which case they are termed commensalism (+/0).
Determining how species interact with one another is a challenge in any diverse ecosystem. The behavior of any given species results from the integration of its interactions with all other community members and the environment — rendering it hard to disentangle the nature of any individual interaction. Moreover, unlike in macro-ecological communities, rarely can we directly observe microbes in situ within their mammalian hosts (although some non-mammalian hosts offer the exciting opportunity to do so [4]). Quantifying microbial interactions within the mammalian microbiome is, therefore, a formidable task.
Methods for measuring interactions within host-associated microbiomes can broadly be classified into those used to study members within their natural environment (that is, within their host) and those that characterize members within a controlled experimental setting (Figure 2). However, different approaches often give contrasting results. Some computational models have predicted the possibility of rich cross-feeding networks within the mammalian microbiome, with numerous microbes supplementing one another’s growth [5]; yet some in vitro experiments have suggested that the human microbiota is dominated by competition [6]. What then is the overarching picture of interactions within the mammalian microbiome, and moreover, what drives these differences between methods? To what extent are differences driven by technical biases, and to what extent do they tell us about the biology of the microbiota itself? In this minireview we discuss the diverse methods for quantifying interactions between microbes within the gut and assess what each can tell us about the ecology of the mammalian microbiota. Crucially though, we also highlight that interactions between members of the microbiota can vary across condition, space, and potentially evolutionary time — and that only through embracing this dynamism will we be able to comprehensively understand microbe–microbe interactions within the gut.
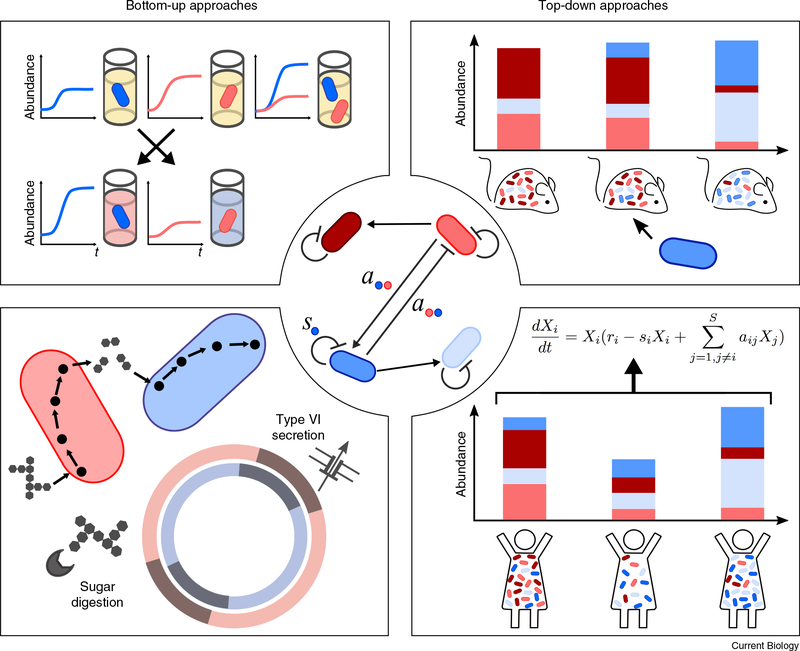
Figure 2. Bottom-up and top-down approaches to infer microbe–microbe interactions within the gut microbiome.
Approaches to inferring microbe–microbe interactions within the mammalian microbiome can largely be divided into those that build an understanding from bottom-up reductionism (left), and those that attempt to learn interactions from top-down community data (right). Experimental reductionism (top left) often involves directly growing species together or culturing microbes in one another’s supernatant in order to assess the effects they have on one another’s growth. Reductionist computational methods (bottom left) can parse a given microbe’s genome to identify mechanisms for specific interactions (for example, microbial weapons) or apply metabolic-flux modeling to predict how species may compete for nutrients or cross-feed one another. By contrast, top-down approaches include experimentally perturbing an existing microbiota community in a defined manner (for example, by introducing a new species) and determining how members of the resident microbial community respond (top right); or, tracking an individual’s microbiota composition over time or across different conditions, and then using statistical tools to infer likely interactions between microbial taxa (bottom right).
Learning from the Community In Vivo
The physical and chemical environment of the mammalian gut is likely to influence the manner in which microbes interact. As such, the ideal context to quantify microbiota interactions is within the host itself. As the microbiota cannot easily be observed directly, most approaches rely on analyzing proxies of gut composition. The most common approach by far is to amplify variable regions in the genes encoding ribosomal RNA to assign taxa within stool samples. More recently, advances in metagenomics have enabled us to determine the composition of the microbiota down to the resolution of individual strains [3]. What, though, can these data tell us about how members of the microbiota interact?
A common approach to quantifying inter-microbial interactions is to use these genomic data to build correlation networks — identifying species that occur together more often than expected by chance. A number of methods have been developed specifically for generating co-occurrence networks from microbiome data where species abundances are often described in relative terms, confounding traditional correlation analyses [7]. Although these methods identify members of the microbiota that frequently co-occur, and are thus likely to interact ecologically, they cannot determine how these microbes interact with one another (Box 1). If two microbes positively correlate, it may be tempting to conclude that they facilitate one another. However, this correlation might equally result from the two occupying a similar niche, in which common environmental conditions are selecting for increased abundances of both members. In the latter case, we would expect niche overlap to drive strong competition between the microbes in question [8].
Nonetheless, although correlation analysis alone is insufficient to infer ecological interactions, it can inform targeted experiments that test ecological interactions mechanistically. For example, by identifying taxa correlated with recovery from diarrhea following Vibrio cholera infection, a consortium of species predicted to directly inhibit V. cholera was determined. This consortium was then refined further to identify a specific inhibitory species, Ruminococcus obeum, which bacterial-genetic approaches revealed represses V. cholera via quorum sensing factors [9].
To robustly infer microbial interactions from community data alone, however, we must look beyond co-occurrence to how taxa change in response to one another. This requires data tracking the same microbiota (in the same host) over time to determine how individual taxa change between timepoints, as well as statistical tools to infer whether these changes are influenced by the presence of other taxa. A number of methods exist to achieve this, typically based on fitting microbiota next-generational sequencing time-series data with a generalized Lotka-Volterra model [10–12]. This simple mathematical model describes how the growth rate of any given taxon is affected by interactions with other members of a microbial community. Parameterizing this model enables us to infer directed relationships between microbes — for example, species A increases the growth rate of species B. When combined with machine-learning tools that disentangle true signals of interactions from false positives, this approach can build an entire interaction network of how each taxon’s growth rate is affected by each other member of the community.
Several studies have used this statistical approach to characterize the interaction networks of mouse [10] or human [11,13] gut-microbiome communities, determining all possible pairwise interactions between common taxa. In each case communities appeared to be dominated by competition and exploitation. For example, in the developing mouse microbiome, of 136 possible microbial interactions no mutualistic pairs were identified, whereas only four were identified as commensal [11] (Box 1). However, these methods are still in their infancy and are vulnerable to mistakes if dynamics are driven by unmeasured external factors (for example, antibiotics) [14]. Therefore, they are best treated as hypothesis generators from which the strongest predicted interactions can then be validated experimentally. For example, this approach was used to identify microbes that inhibit the pathogen Clostridium difficile in humans; one of the strongest inhibitory links identified (via Clostridium scindens) was confirmed and the mechanism of inhibition elucidated using in vivo and in vitro mouse models [13].
The above methods have relied on simply observing interactions in natural communities. Further insight can be gained from actively manipulating communities in vivo. One such approach is to add new members to natural communities and determine both how these new members behave and how they affect the existing microbiota. This approach revealed that Bacteroides species confer colonization resistance — whereby an invading strain cannot colonize the host if conspecifics are already present — implying strong within-species competition [15]. A simpler approach still is to manipulate small artificial communities in vivo. For example, mice mono-colonized with Bacteroides thetaiotaomicron supported higher burdens of the pathogens Salmonella enterica serovar Typhimurium and C. difficile than did germ-free mice colonized with these pathogens alone, leading to hypotheses that both pathogens benefited from the presence of this member of the gut microbiota. Tracking changes in gene expression, as well as metabolite production and consumption, revealed that the positive interactions between B. thetaiotaomicron and the pathogens were mediated by two distinct types of cross-feeding: via monosaccharides liberated by the cleaving of host glycans [16] or by fermentation of waste products [17]. Studying reduced-complexity communities in vivo in this manner has also revealed that B. thetaiotaomicron cross-feeds the common sulfur-reducing bacterium Desulfovibrio piger [18] and facilitates colonization by another common gut bacterium Faecalibacterium prausnitzii [19].
Learning from Reductionism In Vitro
Complementing these in vivo studies are ‘bottom-up’ approaches in which inter-microbial interactions are determined from examining individuals or pairs of species within controlled environments. These methods range from classic in vitro screens for interference competition (Box 1) to newer approaches harnessing metabolomics and computational modelling to infer interaction networks.
The most reductionist approach is to characterize individual members of the microbiota and subsequently infer how they may interact with other community members. This is often achieved using a combination of phenotypic screens — assaying individuals in vitro to determine the presence of a trait (for example, ability to utilize a particular nutrient) — and bioinformatic screens that identify homologous genes in one organism associated with specific interaction mechanisms in another.
These approaches have identified a vast array of mechanisms for interference competition within the microbiota [20]. A classic example is the type VI secretion system — a molecular apparatus used by Gram-negative bacteria to deliver a toxic payload to another bacterium, or in some cases a host organism [20]. Early work used phenotypic screens of mutant libraries growing on target cells to reveal both the presence of, and the genes associated with, the type VI secretion system machinery in several species [21,22]. Later, bioinformatics screens uncovered homologous type VI secretion system genes in more than half of the Bacteroidales strains within human microbiota samples [23,24]. Subsequent functional analyses confirmed the role of these genes in intraspecies competition within the gut [25,26].
Reductionist methods have also predicted large amounts of exploitative competition (Box 1) within the microbiota. Typically this has been achieved by identifying cases in which pairs of microbes each possess the ability to digest a specific polysaccharide — implying that they will compete for this nutrient [27]. For example, in vitro characterization revealed that many members of the Bacteroides genus share the machinery for digesting a range of dietary and host glycans [28], predicting a resource competition that was subsequently validated in vivo [29]. However, it is crucial to identify not only whether a microbe utilizes a nutrient, but also how. Some dietary glycans are digested extracellularly, providing breakdown products that benefit not only the focal strain but also others in the surrounding area. This is in contrast to ‘selfish’ mechanisms whereby glycan utilization is privatized [30–33]. Very rarely, such as in the case of Bacteroides ovatus, microbes have even been found to engage in non-essential costly ‘public’ polysaccharide digestion in order to help other species — displaying true, evolved interspecies cooperation [31] (Box 1).
An alternative to studying individual strains in isolation is to directly assess interactions between pairs of species in a simplified environment, either experimentally or computationally. Experimental approaches typically involve either directly growing species together or culturing species in one another’s supernatant. Meanwhile, computational approaches to inferring interactions have typically relied on metabolic-network modelling. Here annotated genomes are used to construct networks of all the metabolic reactions within a given set of microbes. These networks are then combined alongside information about metabolic constraints to simulate the flow of metabolites within and between each of the species using flux balance analyses [5,34]. In doing so, one can identify potential competition for nutrients or metabolite cross-feeding. Regardless of the specific method, these approaches give a more direct quantification of interactions than do techniques that rely on characterizing individual strains or analyzing natural communities in vivo. However, the downside is that these approaches quickly become unfeasible for natural communities — a community with 100 species contains over 5000 pairs. As such, these approaches are typically applied to small synthetic consortia of microbes designed to capture the dominant taxa within mammalian microbiomes.
Experimental assessments of interactions suggest that the mammalian microbiota is dominated by competitive and exploitative interactions [6,35,36]. For example, directly coculturing a 12-species synthetic community determined that competitive and ammensal (Box 1) interactions together comprised 68% of all interactions, with cooperative and commensal interactions representing around 5% [6]. By contrast, computational approaches have produced mixed results regarding the nature of interactions within the microbiome [5,34]. A metabolic network model of an 11-species synthetic community predicted mutualism and commensalism to be rare, together representing approximately 10% of all interactions [34]. However, a more extensive model containing 773 members of the human microbiome identified commensalism as the second most dominant interaction type after exploitation, suggesting that a rich network of beneficial cross-feeding exists within the microbiota [5]. Importantly, however, the almost 300,000 interactions classified in this study were not filtered for whether the taxa in question were known to commonly co-occur. When interaction pairs were classified based upon whether the focal species commonly co-occurred, microbiota members were on average more competitive against species with whom they commonly co-occurred compared to species with which they rarely associated [27].
Interactions within the Microbiota — Discrepancies and Dynamism
Taken together, what can we conclude about interactions within the mammalian gut microbiome? Longitudinal community data, pairwise cultivation, and the presence of a diverse array of molecular weaponry suggest that the microbiota is dominated by exploitation and competition. This is perhaps unsurprising — theory suggests that communities dominated by cooperation are likely to be unstable on both ecological and evolutionary timescales [37,38]. However, reduced complexity ecosystems in vivo and metabolic models have each demonstrated rich potential for beneficial cross-feeding between microbes. What then drives these discrepancies?
Technical Drivers of Discrepancies
The intrinsic technical biases and blind-spots of any approach will, inevitably, influence the conclusions we draw about microbiota ecology. Supernatant-based growth experiments and metabolic modeling will fail to capture contact-dependent interference competition — leading to an overrepresentation of positive interactions. Conversely, co-culturing organisms in batch cultures may drive competition for nutrients that would never be in demand in vivo, rendering microbes more competitive in vitro than they would be within their host. More broadly, most methods for inferring interactions require strong simplifications and assumptions about the underlying environment in which communities reside, and any ex-vivo approach will inevitably remove many of the physical and chemical complexities of the gut environment. For example, both in vitro approaches and metabolic modelling require defined growth media; however, this media will itself influence the interactions that occur [5,34]. Microbes cannot compete for nutrients that are not there; nor can they cross-feed if the necessary substrates are missing. Meanwhile, though in vivo systems capture communities in their natural settings, unmeasured variability between hosts may confound any attempts to infer microbial interactions [14]. Another important consideration is that top-down approaches may be unable to capture interactions between strains, due to limitations in sequencing resolution or depth.
Biological Drivers of Discrepancies — Dynamism in Interactions
More fundamentally however, these technical limitations highlight a key principle of microbial ecology in the gut: that the balance between competition and cooperation is both dynamic and context dependent. Although we are trying to define static interactions, in reality, the chemical, social, and physical environment will all dynamically influence how a microbe interacts with others at any given time.
Microbes change their gene expression depending on context, potentially altering how they interact with the environment around them. For example, many members of the gut microbiota regulate their nutrient utilization depending on environmental availability. The Bacteroides demonstrate hierarchies of glycan preference [39,40], shifting their nutrient utilization over time and potentially reducing competition for some nutrients while increasing competition for others [40–42]. Meanwhile, model systems have demonstrated how changing nutrient availability can reverse interactions entirely; for example, from obligate mutualism at low nutrients to competition when nutrients are abundant [43]. Similarly, many mechanisms of bacterial warfare are under tight regulation [20,44,45]. The type VI secretion system, for example, is deployed very differently across species. The pathogen Pseudomonas aeruginosa exhibits a ‘tit-for-tat’ behavior, increasing the rate at which it fires its weaponry when it is attacked by other cells [44]. In contrast, Serratia marcescens employs its type VI system pre-emptively, firing regardless of whether it has already been attacked [45]. More fundamentally, this dynamism in gene expression also highlights a key limitation of any method that focuses on inferring pairwise interactions between species: that it will fail to capture higher-order interactions — whereby the presence of one species may alter how others interact.
Physical forces too can influence microbial interactions. Although the extent of spatial organization in the mammalian gut is relatively unknown, certain microbes are differentially distributed [46]. Such community structure may weaken interactions if it prevents cells from encountering one another, whereas in a spatially limited environment strong competition for space may outweigh any beneficial effects of cross-feeding [37].
Thus far we have focused solely on microbe–microbe interactions, agnostic of the presence of the host. However, the gut epithelium provides many of the nutrients for which species compete, potentially providing a means for hosts to manipulate microbial interactions [37]. Moreover, there is increasing evidence of microbe–host–microbe interactions, whereby one microbial species influences another, via inducing a host response. For example, certain strains of S. enterica employ virulence factors that induce epithelial inflammation in the gut, and the production of new electron acceptors by the host that can only be used by S. enterica. This environmental change enables S. enterica to respire in the gut, providing it with a growth advantage against other bacteria and enabling it to outcompete the resident microbiota [47]. Determining the role of the host in regulating interactions among members of the microbiota will be critical to fully understanding microbe–microbe interactions in the gut.
Outlook
If we wish to manipulate our microbiota we must account for the ecology of our gut communities. A probiotic will only be successful if it can, at minimum, colonize its host — and this colonization ability will depend critically upon how the probiotic competes or cooperates with the natural microbiota. Although we are beginning to elucidate the forces that shape microbial communities, we are still far from fully disentangling how microbes interact within the mammalian gut.
To comprehensively understand microbial interactions within the complex gut microbiota we must appreciate that they are dynamic and context-dependent. Progress will be made here if in vivo studies can begin to account for the unseen factors that shape interactions — for example, by mapping the micro-niches that exist in the gut and accounting for chemical and physical variation within a host. Reductionist approaches will benefit from better theoretical frameworks in order to map upwards and understand how individual interactions integrate to drive broader community dynamics.
Finally, although in-roads are being made towards disentangling bacterial interactions within the microbiota, far less is currently known of how fungi or viruses contribute to microbiota dynamics [48]. Elucidating the within- and cross-kingdom interactions that occur in the mammalian gut is essential if we wish to fully understand the ecology of our microbiota.
More fundamentally, here we have focused on how microbes affect one another on ecological timescales. To fully understand our gut communities, however, we must also seek to understand how microbes influence each other on evolutionary timescales. How will these selective forces shape the ecological interactions between microbes — and vice versa? Understanding this eco-evolutionary interplay will undoubtedly be a formidable task but will be pivotal in building our understanding of the microbial communities that live inside us.
ACKNOWLEDGEMENTS
We thank E. Bath, members of the Foster group, members of the S.R.-N. lab and our anonymous reviewers for helpful comments on the manuscript. K.Z.C. is funded by a Sir Henry Wellcome Postdoctoral Research Fellowship (grant 201341/Z/16/Z). S.R-N. is supported by a Career Award for Medical Scientists from the Burroughs Wellcome Fund, a Pew Biomedical Scholarship, a Basil O’Connor Starter Scholar Award from the March of Dimes and 1K08AI130392-01.
REFERENCES
- 1.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, and Relman DA (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch SV, and Pedersen O (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med 375, 2369–2379. [DOI] [PubMed] [Google Scholar]
- 3.Yassour M, Vatanen T, Siljander H, Hämälinen A-M, Härkönen T, Ryhänen ST, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, et al. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med 8, 343ra81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logan SL, Thomas J, Yan J, Baker RP, Shields DS, Xavier JB, Hammer BK, and Parthasarathy R (2018). The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc. Natl. Acad. Sci. USA 115, E3779–E3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, Greenhalgh K, Jäger C, Baginska J, Wilmes P, et al. (2016). Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol 35, 81–89. [DOI] [PubMed] [Google Scholar]
- 6.Venturelli OS, Carr AC, Fisher G, Hsu RH, Lau R, Bowen BP, Hromada S, Northen T, and Arkin AP (2018). Deciphering microbial interactions in synthetic human gut microbiome communities. Mol. Syst. Biol 14, e8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtz ZD, Müller CL, Miraldi ER, Littman DR, Blaser MJ, and Bonneau RA (2015). Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol 11, e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardin G (1960). The competitive exclusion principle. Science 131, 1292–1297. [DOI] [PubMed] [Google Scholar]
- 9.Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA, Haque R, Ahmed T, and Gordon JI (2014). Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515, 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, Sander C, and Xavier JB (2013). Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput. Biol 9, e1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faust K, Bauchinger F, Laroche B, de Buyl S, Lahti L, Washburne AD, Gonze D, and Widder S (2018). Signatures of ecological processes in microbial community time series. Microbiome 6, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bucci V, Tzen B, Li N, Simmons M, Tanoue T, Bogart E, Deng L, Yeliseyev V, Delaney ML, Liu Q, et al. (2016). MDSINE: Microbial Dynamical Systems INference Engine for microbiome time-series analyses. Genome Biol. 17, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao H-T, Gibson TE, Bashan A, and Liu Y-Y (2017). Inferring human microbial dynamics from temporal metagenomics data: Pitfalls and lessons. BioEssays 39, 1600188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, and Mazmanian SK (2013). Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. (2013). Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, and Sonnenburg JL (2014). Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16, 770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, and Gordon JI (2013). Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl. Acad. Sci. USA 110, 13582–13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrzosek L, Miquel S, Noordine M-L, Bouet S, Joncquel Chevalier-Curt M, Robert V, Philippe C, Bridonneau C, Cherbuy C, Robbe-Masselot C, et al. (2013). Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol 11, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Bayona L, and Comstock LE (2018). Bacterial antagonism in host-associated microbial communities. Science 361, eaat2456. [DOI] [PubMed] [Google Scholar]
- 21.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, and Mekalanos JJ (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. USA 103, 1528–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coyne MJ, Roelofs KG, and Comstock LE (2016). Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verster AJ, Ross BD, Radey MC, Bao Y, Goodman AL, Mougous JD, and Borenstein E (2017). The landscape of Type VI secretion across human gut microbiomes reveals its role in community composition. Cell Host Microbe 22, 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wexler AG, Bao Y, Whitney JC, Bobay L-M, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, et al. (2016). Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl. Acad. Sci. USA 113, 3639–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatzidaki-Livanis M, Geva-Zatorsky N, and Comstock LE (2016). Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl. Acad. Sci. USA 113, 3627–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy R, and Borenstein E (2013). Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl. Acad. Sci. USA 110, 12804–12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salyers AA, Vercellotti JR, West SE, and Wilkins TD (1977). Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol 33, 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, and Sonnenburg JL (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakoff-Nahoum S, Coyne MJ, and Comstock LE (2014). An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol 24, 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rakoff-Nahoum S, Foster KR, and Comstock LE (2016). The evolution of cooperation within the gut microbiota. Nature 533, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. (2015). Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517, 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Basle A, Morland C, Day AM, Zheng H, et al. (2015). Glycan complexity dictates microbial resource allocation in the large intestine. Nat. Commun 6, 7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heinken A, and Thiele I (2015). Anoxic conditions promote species-specific mutualism between gut microbes in silico. Appl. Environ. Microbiol 81, 4049–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medlock GL, Carey MA, McDuffie DG, Mundy MB, Giallourou N, Swann JR, Kolling GL, and Papin JA (2018). Inferring metabolic mechanisms of interaction within a defined gut microbiota. Cell Syst. 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biggs MB, Medlock GL, Moutinho TJ, Lees HJ, Swann JR, Kolling GL, and Papin JA (2017). Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J 11, 426–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coyte KZ, Schluter J, and Foster KR (2015). The ecology of the micro-biome: networks, competition, and stability. Science 350, 663–666. [DOI] [PubMed] [Google Scholar]
- 38.Mitri S, and Richard Foster K (2013). The genotypic view of social interactions in microbial communities. Annu. Rev. Genet 47, 247–273. [DOI] [PubMed] [Google Scholar]
- 39.Lynch JB, and Sonnenburg JL (2012). Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol. Microbiol 85, 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuncil Y, Xiao Y, Porter N, Reuhs B, Martens E, and Hamakera B Reciprocal prioritization to dietary glycans by gut bacteria in a competitive environment promotes stable coexistence. mBio 8, e01068–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. (2009). Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. USA 106, 5859–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwalm ND, and Groisman EA (2017). Navigating the gut buffet: control of polysaccharide utilization in Bacteroides spp. Trends Microbiol. 25, 1005–1015. [DOI] [PubMed] [Google Scholar]
- 43.Hoek TA, Axelrod K, Biancalani T, Yurtsev EA, Liu J, and Gore J (2016). Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol. 14, e1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basler M, Ho BT, and Mekalanos JJ (2013). Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152, 884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostrowski A, Cianfanelli FR, Porter M, Mariano G, Peltier J, Wong JJ, Swedlow JR, Trost M, and Coulthurst SJ (2018). Killing with proficiency: integrated post-translational regulation of an offensive type VI secretion system. PLoS Pathog. 14, e1007230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, and Borisy GG (2017). Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl. Acad. Sci. USA 114, E9105–E9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. (2007). Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5, e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shkoporov AN, and Hill C (2019). Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25, 195–209. [DOI] [PubMed] [Google Scholar]
- 49.Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, et al. (2004). A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303, 689–692. [DOI] [PubMed] [Google Scholar]
- 50.Ratzke C, and Gore J (2018). Modifying and reacting to the environmental pH can drive bacterial interactions. PLoS Biol. 16, e2004248. [DOI] [PMC free article] [PubMed] [Google Scholar]