See Hensel et al. (doi:10.1093/brain/awz394) for a scientific commentary on this article.
In spinal muscular atrophy (SMA), chronic low levels of SMN lead to neurodegeneration via accumulation of R-loops and DNA damage. Kannan, Jiang et al. show that overexpression of ZPR1 prevents R-loops and rescues DNA damage in SMA mice and patient cells. ZPR1 enhances SMN2 expression and rescues the SMA phenotype.
Keywords: ZPR1, R-loops, SMN, neurodegeneration, spinal muscular atrophy
Abstract
Spinal muscular atrophy (SMA) is a neuromuscular disorder caused by homozygous mutation or deletion of the survival motor neuron 1 (SMN1) gene. A second copy, SMN2, is similar to SMN1 but produces ∼10% SMN protein because of a single-point mutation that causes splicing defects. Chronic low levels of SMN cause accumulation of co-transcriptional R-loops and DNA damage leading to genomic instability and neurodegeneration in SMA. Severity of SMA disease correlates inversely with SMN levels. SMN2 is a promising target to produce higher levels of SMN by enhancing its expression. Mechanisms that regulate expression of SMN genes are largely unknown. We report that zinc finger protein ZPR1 binds to RNA polymerase II, interacts in vivo with SMN locus and upregulates SMN2 expression in SMA mice and patient cells. Modulation of ZPR1 levels directly correlates and influences SMN2 expression levels in SMA patient cells. ZPR1 overexpression in vivo results in a systemic increase of SMN levels and rescues severe to moderate disease in SMA mice. ZPR1-dependent rescue improves growth and motor function and increases the lifespan of male and female SMA mice. ZPR1 reduces neurodegeneration in SMA mice and prevents degeneration of cultured primary spinal cord neurons derived from SMA mice. Further, we show that the low levels of ZPR1 associated with SMA pathogenesis cause accumulation of co-transcriptional RNA-DNA hybrids (R-loops) and DNA damage leading to genomic instability in SMA mice and patient cells. Complementation with ZPR1 elevates senataxin levels, reduces R-loop accumulation and rescues DNA damage in SMA mice, motor neurons and patient cells. In conclusion, ZPR1 is critical for preventing accumulation of co-transcriptional R-loops and DNA damage to avert genomic instability and neurodegeneration in SMA. ZPR1 enhances SMN2 expression and leads to SMN-dependent rescue of SMA. ZPR1 represents a protective modifier and a therapeutic target for developing a new method for the treatment of SMA.
Introduction
Mutation or deletion of the survival motor neuron 1 (SMN1) gene in the presence of the SMN2 gene results in an autosomal recessive neurodegenerative disorder, spinal muscular atrophy (SMA) (Lefebvre et al., 1995). The SMN1 and inverted copy SMN2 are located on 5q13, the SMA locus. The two genes are similar, but differ by a critical single nucleotide in coding exon 7 that alters splicing and results in the majority of transcript from SMN2 lacking exon 7 thus producing ∼90% of truncated protein SMNΔ7 and 10% of full-length SMN protein (Lorson et al., 1999). SMA is caused by chronic low levels of SMN and is characterized by degeneration of the spinal cord motor neurons leading to muscle atrophy followed by respiratory failure and death (Burghes and Beattie, 2009).
The severity of SMA disease inversely correlates with the levels of SMN protein produced by varying numbers of SMN2 copies present in affected individuals (Wirth et al., 2013). Clinical reports show that SMA families with siblings that have homozygous SMN1 deletion, identical SMN2 copy number and inherited a haploidentical region of chromosome 5q13 display discordant phenotypes (Hahnen et al., 1995). These findings suggest that genes located outside of the 5q13 locus might be able to modify the SMA phenotype. Two classes of modifier genes may influence SMA phenotype: (i) SMN-independent and (ii) SMN-dependent modifiers. The SMN-independent modifier genes are those that do not influence SMN protein levels but their increased or reduced expression may modify SMA disease phenotype. The overexpression of plastin 3 (PLS3) and coronin 1C (CORO1C) genes, and the lack or reduction in expression of c-Jun NH2-terminal kinase 3 (JNK3), neurocalcin delta (NCALD) and calcineurin-like EF-hand protein 1 (CHP1) genes have been identified as SMN-independent modifiers that reduce disease severity and improve SMA phenotype (Oprea et al., 2008; Genabai et al., 2015; Hosseinibarkooie et al., 2016; Riessland et al., 2017; Janzen et al., 2018). The SMN-dependent modifier genes have potential to modulate the levels of SMN by (i) increasing full-length SMN transcript by different mechanisms, including genes encoding (ii) transcription factors that alter expression of SMN2 gene; (iii) splicing factors that enhance inclusion of exon 7; and (iv) proteins that may help stabilize protein-protein complexes and increase steady state levels of SMN protein (Burnett et al., 2009; Ahmad et al., 2012, 2016). The SMN2 gene is the most characterized and viable modifier of SMA severity and an attractive target for identification of new SMN-dependent modifiers such as transcription and splicing factors that may increase full-length SMN transcripts and protein levels (Germain-Desprez et al., 2001; Wirth et al., 2013). However, the molecular mechanisms that regulate expression of SMN genes are largely unknown.
In this study, we investigated the function of zinc finger protein ZPR1 as a potential regulator of SMN gene expression. ZPR1 is essential for cell viability in yeast and mice but its biochemical function is unclear (Galcheva-Gargova et al., 1998; Gangwani et al., 1998, 2005). ZPR1 is present in the cytoplasm and the nucleus, and is required for accumulation of SMN in nuclear gems and Cajal bodies (Gangwani et al., 2001, 2005). ZPR1 is a potential candidate modifier gene that is downregulated in SMA patients and may contribute to the severity of disease (Helmken et al., 2003; Ahmad et al., 2012). However, whether ZPR1 overexpression will rescue SMA disease and emerge as a protective modifier of SMA remains to be examined. We investigated the effect of in vivo genetic overexpression of ZPR1 on the rescue of SMA using the SMAΔ7 mouse model (Le et al., 2005). We also investigated the molecular mechanism of ZPR1-dependent prevention of R-loop and DNA damage accumulation and the rescue of SMA phenotype.
Materials and methods
Mice
Creation of transgenic mice with ZPR1 overexpression (TFZP)
To create transgenic mice overexpressing ZPR1, Flag-mZPR1 cDNA (Gangwani et al., 2001) was cloned into pBroad3 vector using AgeI/SmaI restriction sites downstream of the Rosa26 promoter (InvivoGen) and vector was linearized by digestion with PacI. Transgenic mice expressing recombinant Flag-mZPR1 gene under the control of the mouse Rosa26 promoter (TFZP) were created on FVB/N genetic background by injection of linearized vector DNA into male pronucleus at the Transgenic Animal Modeling Core at the University of Massachusetts Medical School, Worcester, MA. Thirteen positive mice were detected in the F0 generation and bred for germline transmission. Four positive F1 lines (Lines 0, 1, 4 and 8) were positive for transgene cDNA. Lines 4 and 8 were found to express recombinant Flag-ZPR1 protein. Line 8 was characterized for copy number integration using genomic DNA and real-time quantitative PCR copy number assay (Applied Biosystems). Transgenic mice were genotyped for the presence of the Flag-mZPR1 gene using PCR primers forward (5′-AGCGCCGAAGATGAGGAGCA-3′) and reverse (5′-ATCCAGCTCGGGGATCCTTG-3′).
Generation of SMA mice with ZPR1 overexpression (Z-SMA)
SMA carrier mouse line (4299) (Smn+/−; SMN2+/+; SMNΔ7+/+) (Le et al., 2005) on FVB/N background and wild-type FVB/N were purchased from the Jackson Laboratory and maintained in our laboratory. SMA carrier mice were crossed with TFZP (Flag-Zpr1+/−) mice on a pure FVB/N background and the first generations positive for the transgene were backcrossed three to four times to generate Z-SMA carrier mice with Flag-Zpr1+/− (Smn+/−; SMN2+/+; SMNΔ7+/+; Flag-Zpr1+/−). Z-SMA carrier mice were bred to generate SMA (Smn+/−; SMN2+/+; SMNΔ7+/+) and Z-SMA (Smn−/−; SMN2+/+; SMNΔ7+/+; Flag-Zpr1+/−) littermates. The homozygous state of transgenes, SMN2+/+ and SMNΔ7+/+, was confirmed by breeding Z-SMA carrier mice with wild-type mice, and litters were examined by PCR (Le et al., 2005) for the presence of SMN2 and SMNΔ7 in all pups (Genabai et al., 2015). Phenotypic analyses were blinded and littermates were genotyped for presence of Flag-ZPR1 and gender after collection of data by PCR using tail DNA. Any combination of two or three pups with genotypes normal, SMA (Smn−/−; SMN2+/+; SMNΔ7+/+) and Z-SMA (Smn−/−; SMN2+/+; SMNΔ7+/+; Flag-Zpr1+/−) in a litter were considered littermates. All experiments and procedures were approved and performed according to the guidelines and policies set by the Institutional Biosafety Committee. All animals were housed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Texas Tech University Health Sciences Center El Paso. Animals were treated and euthanasia was performed humanely using methods approved by the American Veterinary Medical Association.
Immunohistochemistry
Mouse tissues, spinal cord and hind leg skeletal muscle from 7-day-old normal, SMA and Z-SMA littermates, were isolated, fixed in 4% paraformaldehyde (PFA) and processed for frozen sections as described previously (Genabai et al., 2015). Thin serial sections (10 µm) were cut from the lumbar region of spinal cords and stained with haematoxylin and eosin or processed for immunohistochemical (IHC) staining. Motor neurons were counted in every fifth section (20 sections) of the lumbar (L1–L5) region of the spinal cord (Ahmad et al., 2012; Genabai et al., 2015). For IHC staining, sections were processed for antigen retrieval, blocked in 3% bovine serum albumin (BSA) with 0.1% Triton™ X-100, incubated overnight with antibodies against SMN (610647, BD biosciences) and choline acetyl transferase (ChAT) (ab6168, Abcam) diluted (1:100) in 3% BSA. SMN was detected by using biotinylated secondary antibody, streptavidin-conjugated horseradish peroxidase and the substrate 3,3′-diaminobenzene followed by brief counterstaining with haematoxylin. ChAT was detected with secondary antibody coupled with Alexa 594-conjugated secondary antibody. Muscle sections (16 μm) from hind limb were blocked with 3% BSA with 0.1% Triton™ X-100 incubated with primary antibodies to β-actin (Sigma) and dystrophin (ab15277, Abcam) overnight. The next day, sections were washed, incubated with secondary antibody (Alexa 488) followed by washing with phosphate-buffered saline-Triton (PBST) (5 × 5 min). Double staining was performed using anti-neurofilament (NF-M, 145 kDa) (MAB1621, Millipore), anti-synaptophysin antibodies (ab14692, Abcam) and Alexa 594-conjugated α-bungarotoxin (BTX) for 60 min using sequential staining (Genabai et al., 2015). Microscopic slides or coverslips were mounted using Vectashield with DAPI. Immunofluorescence was examined by confocal microscopy. Muscle fibre transverse sections stained with dystrophin were used to measure diameter of fibres. Fibre diameter was measured at three different points, including longest and shortest along the irregular circular shape of each fibre and averaged. One hundred fibres per mouse (three mice per group) were measured to determine myofibre diameter.
Primary spinal cord motor neuron culture
Mouse spinal cord explants from 7-day-old normal, SMA and Z-SMA mice were cultured in vitro for 12–14 days in 8-well chamber microscope slides, coated with poly-d-lysine/laminin using serum-free Neurobasal™ medium supplemented with B-27 (Genabai et al., 2015; Kannan et al., 2018). The identity and morphology of the spinal cord motor neurons and glial cells were established by staining with specific markers, including ChAT and Hlxb9 (Hb9) (Genabai et al., 2015; Kannan et al., 2018). Cultured neurons were infected with Ad5CMV-GFP or Ad5CMV-FlagZPR1-GFP (Viral Vector Core Facility, University of Iowa) at 100 MOI (multiplicity of infection) in a volume of 200 μl/well of 8-well chamber and 400 µl of medium added after 4 h and medium was replaced after 24 h. Neurons were either fixed (4% PFA) or harvested for protein extraction at 48 h post-infection for immunofluorescence and immunoblotting analysis, respectively.
SMA patient primary fibroblast and HeLa cell culture
Primary fibroblast derived from SMA type I patients, GM03813 and GM09677 were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 15% foetal bovine serum (FBS) and HeLa cells were cultured in DMEM with 10% FBS (Gangwani et al., 2001; Kannan et al., 2018). Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Cells were cultured on glass coverslips (105 cells/well) in 6-well plates and transfected with phrGFPIIc (GFP) or phrGFPIIc-FlagZPR1 (ZPR1-GFP) (1 µg/well) using Lipofectamine® 2000 according to the manufacturer’s protocol (Gangwani et al., 2001; Kannan et al., 2018). Cells were either harvested for immunoblot analysis or fixed for immunofluorescence analysis after 30 h post-transfection. Cells were also transfected with pcDNA3 or pcDNA3-FlagZPR1 and harvested after 30 h post transfection to isolate total RNA for analysis of SMN expression using real time PCR analysis (Gangwani et al., 2001; Kannan et al., 2018). HeLa cells were mock-transfected (Control), transfected with antisense oligonucleotide against ZPR1 (As-ZPR1) and scrambled oligonucleotide (Scramble).
Immunoblot analysis
Protein extracts for immunoblot analysis were prepared from mouse spinal cord, brain, heart, lung, liver and muscle tissues collected from 7-day-old normal, SMA and Z-SMA littermates, SMA patient cell lines GM03813 and GM09677 transfected with phrGFPIIc (GFP) or phrGFPIIc-FlagZPR1 (ZPR1-GFP), cultured primary spinal cord SMA neurons infected with adenovirus Ad5CMV-GFP (GFP) or Ad5CMV-FlagZPR1-GFP (ZPR1-GFP) (Kannan et al., 2018) and HeLa cells co-transfected with human pGL3/SMN1-Luc or pGL3/SMN2-Luc with pcDNA3-FlagZPR1 or with antisense (As-ZPR1) and scramble (Scram) oligos (Genabai et al., 2017). Cell and tissue lysates were prepared using Triton™ lysis buffer (Gangwani et al., 1998). Specific proteins were detected by automated capillary western blot system, Wes™ System (ProteinSimple), which uses capillary based electrophoretic separation and detection of proteins using antibodies as described previously (Kannan et al., 2018). Signal intensity (area) of the protein was normalized to the peak area of loading control α-tubulin. The following primary phospho and non-phospho antibodies were used for immunoblot analysis, SMN (610647, BD Biosciences), ZPR1 (Clone LG-C61) (Genabai et al., 2017), RNA Polymerase II (clone #8WG16, BioLegend), recombinant Flag-ZPR1 was detected by Flag M2 (F1804, Sigma), γH2AX (phospho Ser 139) (ab26350), p-DNA-PKcs (Ser2056) (ab18192), total DNA-PKcs (ab53701), and senataxin (ab220827, Abcam), GFP (A11122, Invitrogen) and α-tubulin (T8203, Sigma). Data analysis and quantitation of protein levels were performed using Compass Software (ProteinSimple) (Kannan et al., 2018). The relative levels of proteins [mean ± standard error of the mean (SEM)] normalized to tubulin, are presented.
Immunofluorescence analysis
Mammalian cells cultured on glass were washed with PBS and fixed in pre-chilled methanol (−20°C) for 5 min followed by 2 min in pre-chilled acetone at −20°C. Cell were stained with primary antibody against SMN, γH2AX, RNA-DNA hybrid (R-loops), 53BP1 or SETX for 1 h followed by secondary antibody Alexa 594-conjugated IgG (1 h) and coverslips were mounted on microscope slides with mounting medium containing DAPI (Gangwani et al., 2001; Kannan et al., 2018). PFA fixed primary spinal cord motor neurons were washed with PBS, permeabilized with 0.1% Triton™ X-100 for 5 min, washed 3 × 5 min with PBS with 0.2% Tween-20 (PBS-T), blocked with 3% BSA in PBS-T for 30 min and double-labelled using sequential incubation (1 h each) with primary antibodies [SMN, γH2AX, p-DNA-PKcs, RNA-DNA hybrid (R-loops) (S9.6) or SETX] followed by secondary antibody Alexa 633-conjugated IgG, washed 3 × 5 min with PBS-T and incubated with second primary antibody, mouse anti-β-tubulin class-III neuron-specific antibody (clone TUJ1) or rabbit anti-tubulin β-III (TUBB3), washed 3 × 5min with PBS-T followed by incubation with Alexa 594-conjugated anti-mouse or anti-rabbit IgG secondary antibody (Kannan et al., 2018). Immunofluorescence of stained cells was examined using a confocal microscope.
Real-time quantitative PCR analysis
Total RNA was isolated from (i) mouse spinal cord from 7-day-old normal, SMA and Z-SMA littermates; and (ii) SMA patient fibroblast cell lines, GM03813 and GM09677 transfected with pcDNA3 or pcDNA3-FlagZPR1 using RNeasy® Mini Kit (Qiagen). Total RNA (100 ng) per sample was reverse-transcribed using SuperScript™ VILO cDNA synthesis Kit (Invitrogen). Real-time quantitative PCR (qPCR) amplification for full-length SMN and truncated SMNΔ7 transcripts was performed using SYBR® Green Master Mix. Relative mRNA levels normalized to GAPDH were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001; Genabai et al., 2017; Kannan et al., 2018). The primer sequences were as follows: mouse Gapdh primers: forward (5′-AAGGTCATCCCAGAGCTGAA-3′), reverse (5′-CTGCTTCACCACCTTCTTGA-3), human GAPDH primers: forward (5′-ATAGGCGAGATCCCTCCAA-3′), reverse (5′-TGAAGACGCCAGTGGAC-3′), SMN2 Jxn E5/E6 forward F2 (5′-TTCCTTCTGGACCACCAATAA-3′), SMN2 Jxn E7/E8 reverse R2 (5′-TCTATGCCAGCATTTCTCCTTAATTTAAG-3′) and SMN2 Jxn E6/E8 Reverse R3 (5′-TGCTCTATGCCAGCATTTCCATAT-3′). Full-length SMN and SMNΔ7 transcripts were amplified using F2+R2 and F2+R3 primers, respectively (Seo et al., 2016).
Chromatin immunoprecipitation-quantitative PCR assay
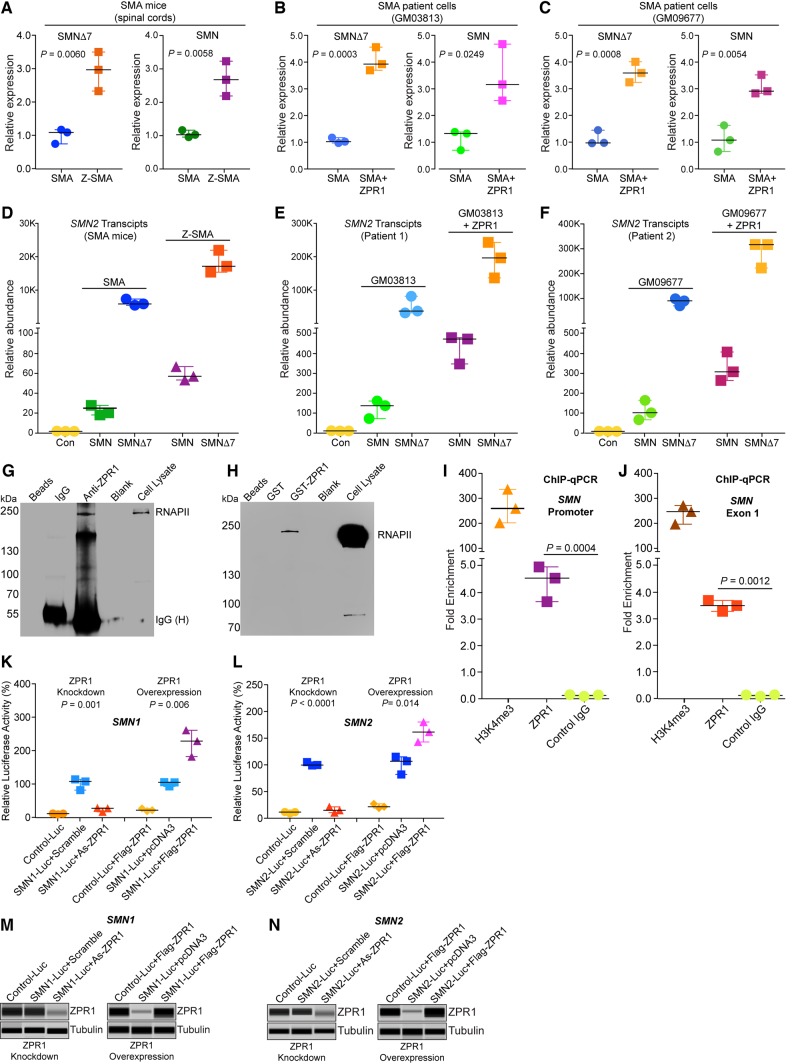
Chromatin samples were prepared from HeLa cells treated with 3.7% PFA to crosslink DNA and protein complexes. Chromatin was sheared by sonication to generate fragments ranging between 200 and 1000 bp. Chromatin immunoprecipitation (ChIP) was performed using antibodies against ZPR1 (Clone: LG-C61), H3K4me3 as a positive control and anti-Flag M2 as a negative control (IgG) according to manufacturer’s protocol using Magna ChIP™ HiSens Kit. Real-time quantitative PCR amplification was performed using SYBR® Green reagents and human SMN promoter region, SMN1&2-F5: 5′-GATCTGCCGCCTTCCTTCCTG-3′, SMN1&2-R5: 5′-CTTAGGCCTCGTCTCGAACTC-3′ and SMN exon 1 region, SMN1&2-F3: 5′-CAGTGCAGTCTCCCTATTAGCG-3′ and SMN1&2-R3: 5′-CACAACTCCAGTGAGCGGATCG-3′. Levels of SMN genomic DNA present in ChIP with each antibody were measured using the comparative CT (ΔΔCT) method for fold enrichment (Genabai et al., 2017).
Effect of ZPR1 levels modulation on SMN1 and SMN2 gene expression
To examine the effect of change in the levels of ZPR1 on alteration of the SMN gene expression, we used HeLa cells co-transfected with human SMN1 or SMN2 promoter driven luciferase reporter vector pGL3/SMN1-P750-Luc and SMN2-P750-Luc (provided by Dr Arthur Burghes) (Monani et al., 1999) and either pcDNA3-FlagZPR1 (ZPR1 overexpression) (Ahmad et al., 2012) or with ZPR1-antisense oligonucleotides (As-ZPR1) (ZPR1 knockdown) (Gangwani, 2006). To examine the effect of ZPR1 knockdown, cultured HeLa cells were transfected with either human SMN-Luc or empty control reporter vectors (Con-Luc) using Lipofectamine® 2000. Transfected cells were retransfected after 24 h with scrambled or ZPR1 antisense oligonucleotides to knockdown ZPR1 levels. Cells were harvested after 24 h post second transfection for determination of luciferase activity or immunoblot analysis. To examine the effect of ZPR1 overexpression, HeLa cells were transfected with combinations of two plasmids: (i) Con-Luc + pcDNA3-FlagZPR1; (ii) SMN1-Luc + pcDNA3 (empty); (iii) SMN1-Luc + pcDNA3-FlagZPR1; (iv) SMN2-Luc + pcDNA3 (empty); and (v) SMN2-Luc + pcDNA3-FlagZPR1 using the Lipofectamine® reagent. After 30 h post transfection, cells were harvested for determining luciferase activity or immunoblot analysis. Luciferase activity was measured using the luciferase assay system in the synergy H1 hybrid microplate reader. Relative levels of luciferase activity (mean ± SEM) are presented.
Statistical analyses
The quantitative analysis of continuously distributed data is presented as scattered plots or box-and-whisker plots with quantitative elements, including, median with interquartile interval, minimum and maximum ranges. Statistical analysis performed using mean ± SEM using either Kaplan-Meier survival analysis, Log-rank (Mantel-Cox) test, one-way ANOVA or Student’s t-test (unpaired, two-tailed) with GraphPad Prism (version 5.0d). A value of P ≤ 0.05 was considered significant. In all experiments with mice, ‘n’ represents the number of mice used per group; and in experiments with tissues or cells, ‘n’ represents the number of times experiment was performed. A minimum of n = 3 mice or number of times experiment performed was used in all the experiments, unless otherwise specified in an experiment.
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Results
Generation of SMA mice with ZPR1 overexpression
To examine the in vivo effect of ZPR1 overexpression on modulation of SMA disease severity, we generated transgenic mice expressing the recombinant Flag-Zpr1 gene under the control of the murine Rosa26 promoter on an FVB genetic background to match with the SMAΔ7 model (Supplementary Fig. 1A). Transgene copy number analysis using Real-Time PCR copy number analysis (Applied Biosystems) revealed single copy integration into mouse genome. Heterozygous Flag-Zpr1+/− mice were normal and fertile. Analysis of phenotype of homozygous Flag-Zpr1+/+ mice showed that female mice were normal and fertile but male mice developed inflammation of the genitals, and breeding potential, ability or interest to mate were compromised. However, male mice showed normal development and lifespan. We focused our analysis on heterozygous mice that provided an advantage to generate littermates with/without transgene for comparison. Next, we tested the functionality of recombinant ZPR1. Mutation of the Zpr1−/− gene causes early (E3.5) embryonic lethality in mice (Gangwani et al., 2005). To test whether recombinant Flag-ZPR1 is a functional protein, we bred TFZP mice with Zpr1+/− mice to rescue embryonic lethality of Zpr1−/− mice. Embryonic lethality of Zpr1−/− mice was rescued by recombinant Flag-ZPR1, rescued mice (Zpr1−/−; Flag-Zpr1+/−) were fertile and displayed normal phenotype and lifespan suggesting that the recombinant Flag-ZPR1 expresses during early embryonic development, retains biological activity and can complement for ZPR1 essential function (Supplementary Fig. 1B and C). We crossed Flag-Zpr1+/− mice with SMAΔ7 mice and generated SMA carrier mice with Flag-Zpr1 (Smn+/−; SMN2+/+; SMNΔ7+/+; Flag-Zpr1+/−). Homozygosity of transgenes, SMN2 and SMNΔ7, in Z-SMA carrier mice was confirmed by PCR and breeding methods (Genabai et al., 2015). SMA mice overexpressing ZPR1 (Smn−/−; SMN2+/+; SMNΔ7+/+; Flag-Zpr1+/−) are referred to as Z-SMA.
ZPR1 improves overall growth of SMA mice
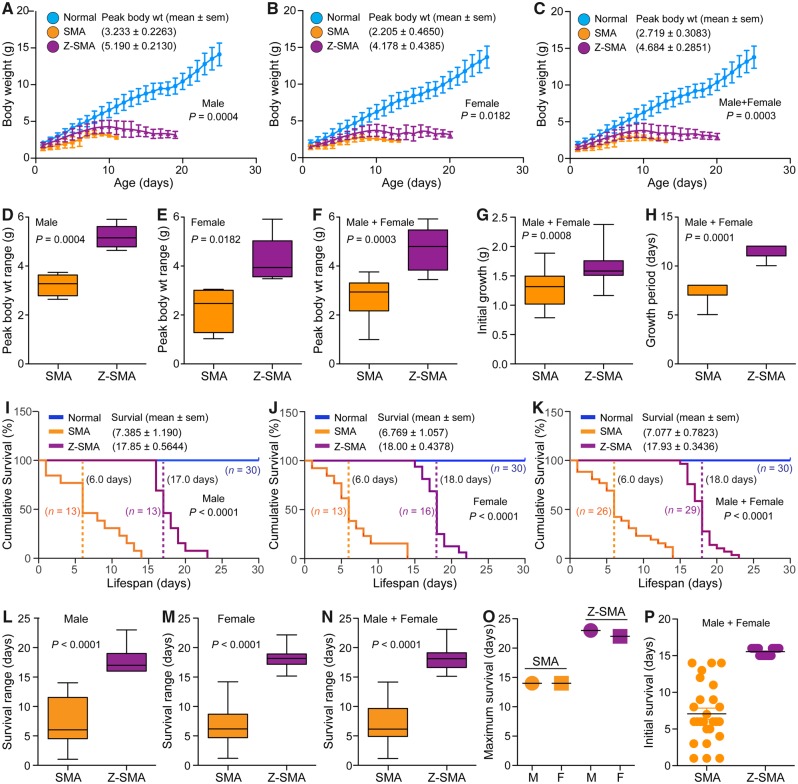
SMA mice with ZPR1 overexpression (Z-SMA) were healthier, and were able to stand and walk compared to SMA mice (SMA) between the ages of 6 and 14 days. Initial observations suggested that ZPR1 overexpression might be beneficial in improving the overall health and gross motor function of SMA mice as shown in a photograph and a video clip of littermates with normal, SMA and Z-SMA genotypes (Supplementary Fig. 1D, E and Supplementary Video 1). Analysis of growth of littermates shows that ZPR1 improved average peak (mean ± SEM, n = 5) body weight (g) in Z-SMA males (5.19 ± 0.21, P = 0.0004) compared to SMA males (3.23 ± 0.22) and in Z-SMA females (4.17 ± 0.43, P = 0.0182) compared to SMA females (2.20 ± 0.46) (Fig. 1A and B) . Comparison of combined (male + female) peak body weight of Z-SMA (4.684 ± 0.285) with SMA (2.719 ± 0.308) mice show statistically significant (P = 0.0003) improvement in growth of Z-SMA mice (Fig. 1C). Gender-based analysis shows overall growth increase in Z-SMA females was ∼89% and in Z-SMA males was ∼61% compared to SMA females and males, respectively (Fig. 1D–F). These data show better improvement in Z-SMA females with ∼28% higher body weight compared to Z-SMA males. Chronic SMN deficiency results in reduced embryonic growth and newborn human SMA babies have lower body weight compared to normal babies. Comparison of body weights at birth shows statistically significant (P = 0.0008) improvement in Z-SMA (1.642 ± 0.065, n = 20) compared to SMA (1.294 ± 0.069, n = 20) mice (Fig. 1G). Comparison of body weight between normal (1.800 ± 0.064, n = 20), SMA and Z-SMA mice shows reduced weight loss in newborn pups of Z-SMA (0.16 g) compared to SMA (0.51 g), which shows 3.19-fold improvement in embryonic growth of Z-SMA mice (ANOVA, P < 0.0001). ZPR1 overexpression also increased growth period from (7.00 ± 0.378, n = 7) (SMA) to (11.29 ± 0.285, n = 7) (Z-SMA) days, and shows ∼60% increase (P = 0.0001) in postnatal growth period (Fig. 1H). These data suggest that in vivo ZPR1 overexpression improves embryonic and postnatal growth of Z-SMA mice, which may contribute to reduction of disease severity and increase the lifespan of SMA mice.
Figure 1.
Genetic overexpression of the Zpr1 gene improves overall growth and survival of SMA mice. Overexpression of recombinant Flag-Zpr1 gene under the control of mouse Rosa26 promoter increases growth and the lifespan of mice with SMA (SMAΔ7). (A–C) ZPR1 overexpression improves growth of mice with SMA. Overall growth (body weight in grams) curves of normal (blue), Z-SMA (purple), and SMA (orange) mice littermates. Body weights recorded every day are presented as growth curves with mean ± SEM, n = minimum 3 mice/group for combined (male and female) group and for individual male and female groups. (D–F) Box and whisker plots and median with interquartile range (IQR) (minimum and maximum). (D) Average peak body weight (median, min, max) for males, SMA (3.27, 2.64, 3.74) and Z-SMA (5.15, 4.64, 5.91). (E) Average peak body weight for females, SMA (2.42, 0.98, 3.0) and Z-SMA (3.89, 3.43, 5.87). (F) Average peak body weight for combined males and females, SMA (2.92, 0.98, 3.74) and Z-SMA (4.78, 3.43, 5.91). These data show statistically significant increase in average peak body weight (g) of Z-SMA males (5.19 ± 0.21, n = 5) compared to SMA males (3.23 ± 0.22, P = 0.0004, unpaired t-test, two tailed), which represents ∼61% increase in body weight of Z-SMA males. Z-SMA females (4.17 ± 0.43, n = 5) compared to SMA females (2.20 ± 0.46, P = 0.0182) show ∼89% increase in peak body weight. Comparison of combined (male + female) peak body weight in Z-SMA (4.684 ± 0.285, n = 10) with SMA (2.719 ± 0.308, P = 0.0003) shows an average peak body weight increase of ∼72% in Z-SMA mice. (G) ZPR1 improves initial (embryonic) growth. Combined male and female growth (g) at birth for SMA (1.30, 0.77, 1.87) and Z-SMA (1.56, 1.15, 2.36). Statistical analysis shows significant improvement in initial growth of Z-SMA (1.642 ± 0.065, n = 20, P = 0.0008) compared to SMA (1.294 ± 0.069, n = 20) mice. (H) ZPR1 improves overall postnatal growth period of Z-SMA (11.0, 10.0, 12.0) compared to SMA (7.0, 5.0, 8.0) mice that increases from 7.00 ± 0.378 days (SMA, n = 7) to 11.29 ± 0.285 days (Z-SMA, n = 7), which shows ∼60% (P = 0.0001) increase in postnatal growth period. Growth analysis was blinded with colour coding. Gender and genotypes were confirmed by PCR-based method after euthanasia of pups. (I–P) ZPR1 overexpression improves survival of SMA mice. (I–K) Kaplan-Meier survival curves of normal (blue), Z-SMA (purple), and SMA (orange) mice littermates. Dotted lines show median survival. (L–N) Box-and-whisker plots with IQR show increase in (median, min, max) survival (days) of Z-SMA mice. (L) Z-SMA males (17.0, 16.0, 23.0) compared to SMA males (6.0, 1.0, 14.0). (M) Z-SMA females (18.0, 15.0, 22.0) compared to SMA females (6.0, 1.0, 14.0). (N) Z-SMA males and females (18.0, 15.0, 23.0) compared to SMA males and females (6.0, 1.0, 14.0). These data show ∼3-fold increase in median survival of Z-SMA mice compared to SMA mice. Statistical analysis of male and female survival show increase in average survival of Z-SMA (17.93 ± 0.34 days, n = 29) (Log-rank test, P < 0.0001) compared to SMA (7.07 ± 0.78 days, n = 26) mice. Gender-based analysis show average survival of Z-SMA males (17.85 ± 0.56, n = 13) is higher than SMA males (7.38 ± 1.19, n = 13). Average survival of Z-SMA females (18.00 ± 0.43 days, n = 16) is also higher compared to SMA females (6.76 ± 1.05, n = 13). (O) Maximum survival of SMA males (M) and females (F) is 14 days and Z-SMA males is 23 days and females is 22 days. Increase in the lifespan among Z-SMA male + female (combined) is 2.53-fold (P < 0.0001, t-test) compared to SMA mice. Increase in Z-SMA versus SMA males is 2.42-fold (P < 0.0001) and Z-SMA versus SMA females is 2.66-fold (P < 0.0001). (P) Scatter plot shows increase (15.5-fold) in initial survival of Z-SMA mice compared to SMA mice. All Z-SMA males and females survived at least 16 and 15 days, respectively, compared to SMA males and females that survived at least 1 day. Survival analysis was blinded with colour coding. Gender and genotypes were confirmed using a PCR-based method after euthanasia of pups.
ZPR1 improves the lifespan of SMA mice
To determine whether ZPR1 overexpression improves the lifespan of SMA mice, we examined survival of SMA and Z-SMA littermates using Kaplan-Meier analysis. ZPR1 overexpression resulted in 2.53-fold increase in average (male and female) survival of Z-SMA mice (17.93 ± 0.34, n = 29) days compared to SMA (7.07 ± 0.78, n = 26) (Log-rank test, P < 0.0001) (Fig. 1I–K). Comparison of median survival of combined males and females shows 3.0-fold increase in the lifespan of Z-SMA mice (Fig. 1K). Gender-based analysis shows average survival of Z-SMA males (17.85 ± 0.56, n = 13) and median survival (17.0 days) is higher than SMA males average (7.38 ± 1.19, n = 13) and median (6.0 days) survival (Fig. 1I and L). Average survival of Z-SMA females (18.00 ± 0.43 days, n = 16) and median survival (18.0 days) is also higher than SMA females average (6.76 ± 1.05, n = 13) and median (6.0 days) survival (Fig. 1J and M). Comparison of the average lifespan of Z-SMA males (2.42-fold) and females (2.66-fold) showed a ∼24% higher improvement in Z-SMA females than Z-SMA males (Fig. 1I–K). These data are consistent with overall growth in Z-SMA females that is ∼28% higher compared to Z-SMA males (Fig. 1A–C). Notably, 9 days increase in maximum survival of Z-SMA mice (∼64% increase in max lifespan) includes 6 days of growth (reduced severity) that shows about 67% of the total increase in the lifespan is with reduced severity of illness (Fig. 1O). Analysis of initial survival shows a marked 15.5-fold increase in the initial survival of Z-SMA compared to SMA mice suggesting that every Z-SMA pup (n = 29), male or female, survived for at least 15 days after birth (Fig. 1P). These data suggest that ZPR1 overexpression reduces postnatal disease severity, markedly decreases early mortality and increases the lifespan of Z-SMA mice.
ZPR1 improves gross motor function and muscle strength of SMA mice
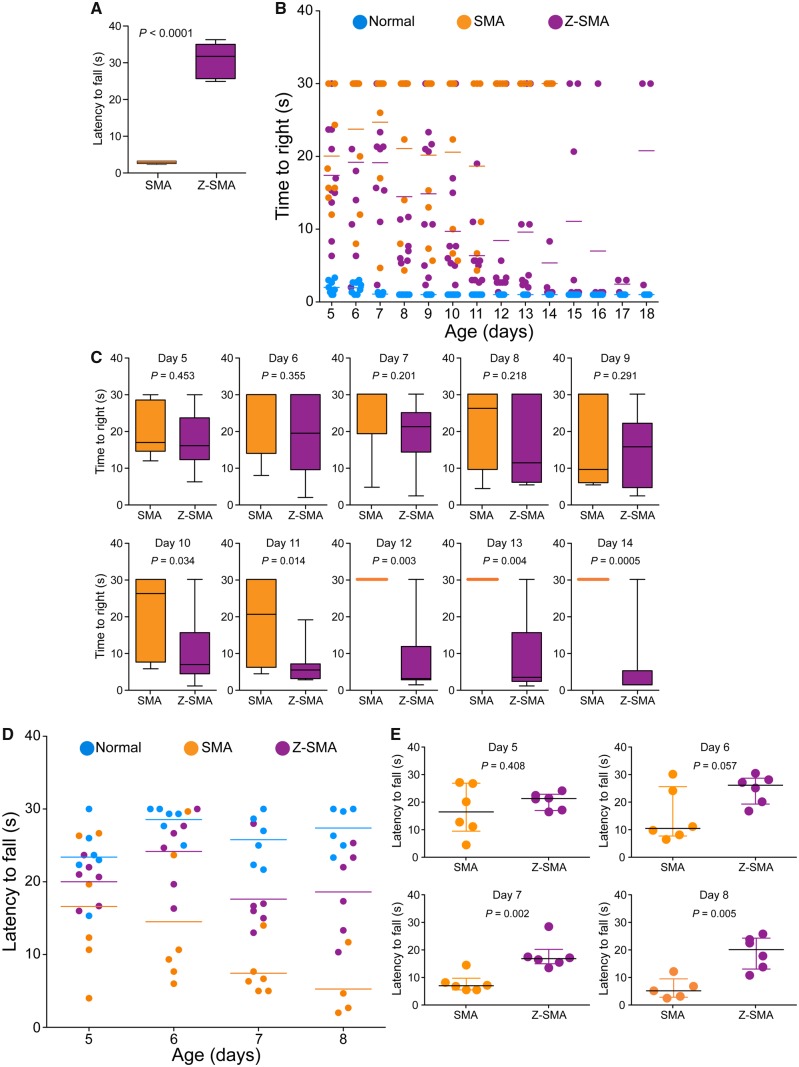
Impaired gross motor functions and the loss of ability to walk are clinical features of SMA. To evaluate beneficial effects of ZPR1 overexpression, we examined gross motor functions, ability to right, stand on paws and walk. The ability to stand on paws and walk (time in seconds) was improved in Z-SMA mice (30.61 ± 1.82, P < 0.0001) compared to SMA mice (2.55 ± 0.15) (Fig. 2A). Video clip of 12-day-old three littermates [normal, SMA (smallest), Z-SMA (black mark on the tail)] shows SMA pup has severe defects in ability to right and stand on paws and unable to walk. In contrast, the Z-SMA pup was able to walk and right itself within a few seconds and continues to walk (Supplementary Video 1). The representative video clip shows that the Z-SMA mouse is able to right itself at the age of 12 days. These data show ZPR1 overexpression results in marked improvement in the ability of Z-SMA mice to walk.
Figure 2.
ZPR1 overexpression improves gross motor function and muscle strength of SMA mice. (A) ZPR1 improves ability to stand on paws and walk. Ability to stand on paws and walk was determined by measuring the time to fall off in an effort to walk, and was recorded in 12-day-old SMA and Z-SMA littermates. Comparison of latency to fall (s) from paws presented as a box-and-whisker plot with IQR (median, min, max) for SMA (2.49, 2.0, 3.0) and Z-SMA (31.50, 24.66, 36.00) showing all Z-SMA pups were able to stand and walk for a median time of ∼30 s, suggesting improvement in gross motor function. Statistical analysis of (mean ± SEM, n = 6 mice/group) (s) between SMA (2.55 ± 0.15) and Z-SMA (30.61 ± 1.82), P < 0.0001 (unpaired t-test, two-tailed) for 12-day-old mice shows marked improvement in the ability of Z-SMA mice to stand on paws and walk (see Supplementary Video 1). (B) Ability of mice to right was recorded for 5–18-day-old normal (blue), SMA (orange), and Z-SMA (purple) littermates with all three genotypes present in the same litter. Time-to-right (TTR) with a time limit of 30 s for test and average of three recordings per pup are plotted. Data were collected using n = 10 (normal), 8 (SMA), and 10 (Z-SMA) mice groups are presented as a scatter plot. (C) Improvement in the motor function is demonstrated by increase in the ability of mice to right faster in Z-SMA mice compared to SMA mice is shown as box-and-whisker plots (median, min, max) starting from PND10 [SMA (26.17, 5.67, 30.0) and Z-SMA (6.83, 1.0, 30.0)], PND11 [SMA (20.50, 4.33, 30.0) and Z-SMA (5.33, 2.67, 19.0)], PND12 [SMA (30.0, 30.0, 30.0) and Z-SMA (3.0, 1.33, 30.0)], PND13 [SMA (30.0, 30.0, 30.0) and Z-SMA (3.33, 1.0, 30.0)], PND14 [SMA (30.0, 30.0, 30.0) and Z-SMA (1.33, 1.33, 30.0)]. Statistical analysis of (mean ± SEM) shows significant improvement in Z-SMA mice from PND10 [SMA (20.58 ± 3.97, n = 8) and Z-SMA (9.70 ± 2.76, n = 10), t-test, P = 0.034), PND11 [SMA (18.67 ± 5.14, n = 6) and Z-SMA (6.36 ± 1.60, n = 10), P = 0.014), PND12 [SMA (30.00 ± 1.00, n = 4) and Z-SMA (8.43 ± 3.61, n = 10), P = 0.003], PND13 [SMA (30.00 ± 1.00, n = 4) and Z-SMA (9.60 ± 3.56, n = 10), P = 0.004] to PND14 [SMA (30.00 ± 1.00, n = 4) and Z-SMA (5.36 ± 3.1, n = 10), P = 0.0005]. (D) The hind-limb suspension test (HLST) shows ZPR1 improves muscle strength in mice with SMA. Littermates aged 5–8 days were hung by both hind legs on the edge of a 50 ml plastic conical tube and time (s) was recorded until fall from the edge of the tube. Latency to fall (mean ± SEM, 6 mice/group) shown as a scatter plot and (E) as scatter plots with median with IQR (median, min, max) for each time point (day), PND5 [SMA (16.0, 4.0, 26.67) and Z-SMA (20.84, 16.0, 23.67)], PND6 [SMA (10.0, 6.0, 29.67) and Z-SMA (25.67, 16.33, 30.0)], PND7 [SMA (6.5, 5.0, 14.0) and Z-SMA (16.34, 13.0, 28.0)], PND8 [SMA (4.67, 2.0, 11.70) and Z-SMA (19.67, 10.33, 25.33)]. Statistical analysis (t-test, unpaired) shows marked increase (P = 0.0301) in hanging time for Z-SMA compared to SMA with increasing age PND5 [Z-SMA (20.00 ± 1.23) and SMA (16.61 ± 3.73), P = 0.408], PND6 [Z-SMA (24.16 ± 2.11) and SMA (14.50 ± 3.97), P = 0.057], PND7 [Z-SMA (17.61 ± 2.15) and SMA (7.445 ± 1.3762), P = 0.002], PND8 [Z-SMA (18.60 ± 2.42) and SMA (5.26 ± 2.22), P = 0.005] shows gradual increase in muscle strength of Z-SMA mice compared to SMA mice.
To gain insight into improvement in gross motor function during postnatal development, we examined time-to-right (TTR) for 5–18-day-old pups with a test time of 30 s (Genabai et al., 2015). Analysis of TTR shows statistically significant improvement in the motor function of Z-SMA compared to SMA mice starting from postnatal day (PND)10 [SMA (20.58 ± 3.97, n = 8) and Z-SMA (9.70 ± 2.76, n = 10), P = 0.0342] to PND14 [SMA (30.00 ± 1.0, n = 4) and Z-SMA (5.36 ± 3.1, n = 10), P = 0.0005] (Fig. 2B and C). All SMA pups in this assay were dead by Day 14. However, Z-SMA pups showed continued improvement in their ability to right until PND17 (2.44 ± 0.55, n = 3) (Fig. 2B). The ability to right for Z-SMA pups began declining from PND18 and mice were unable to right by PND20. Increase in inability to right with reduction in TTR of Z-SMA compared to SMA mice with increasing age suggest gradual improvement in the gross motor function and reduction in disease severity of Z-SMA mice during postnatal development.
Improvement in motor function of Z-SMA mice suggest possibility of increase in muscle strength, to test whether ZPR1 also increases muscle strength, we used a hind-limb suspension test to evaluate improvement in the proximal hind-limb muscle strength (El-Khodor et al., 2008; Ahmad et al., 2012; Genabai et al., 2015). Five to eight-day-old mice were hung by both hind legs on the edge of a 50 ml plastic conical tube fitted with soft cotton pad at the bottom, and time was recorded until fall from the edge of tube. Comparison of latency to fall (seconds) between Z-SMA (20.00 ± 1.23, n = 6) and SMA (16.61 ± 3.73, n = 6) at PND5 did not show statistically significant difference (P = 0.4087) (Fig. 2D and E). However, a marked increase in hanging time of Z-SMA compared to SMA mice was found at PND6 [SMA (14.50 ± 3.97) and Z-SMA (24.16 ± 2.11, P = 0.057)], PND7 (SMA (7.445 ± 1.3762) and Z-SMA (17.61 ± 2.15, P = 0.0026) and PND8 (SMA (5.26 ± 2.22) and Z-SMA (18.60 ± 2.42, P = 0.0052)]. An increase in hind-limb suspension test time suggests improvement in hind-limb muscle strength of Z-SMA mice compared to SMA mice. Together, these data suggest that ZPR1 overexpression improves muscle strength and gross motor function of Z-SMA mice.
ZPR1 reduces neuron degeneration, improves neuromuscular junction innervation and muscle fibre size in SMA mice
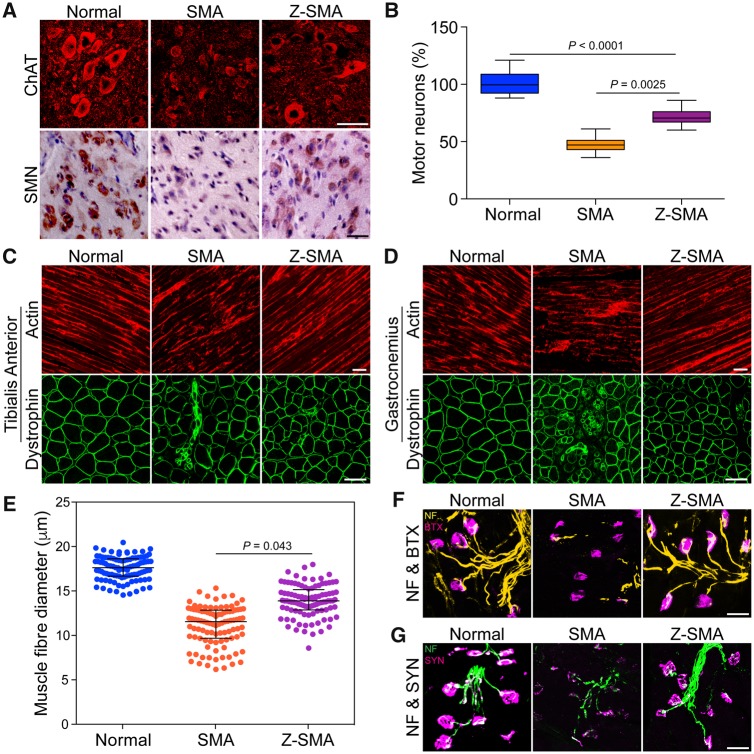
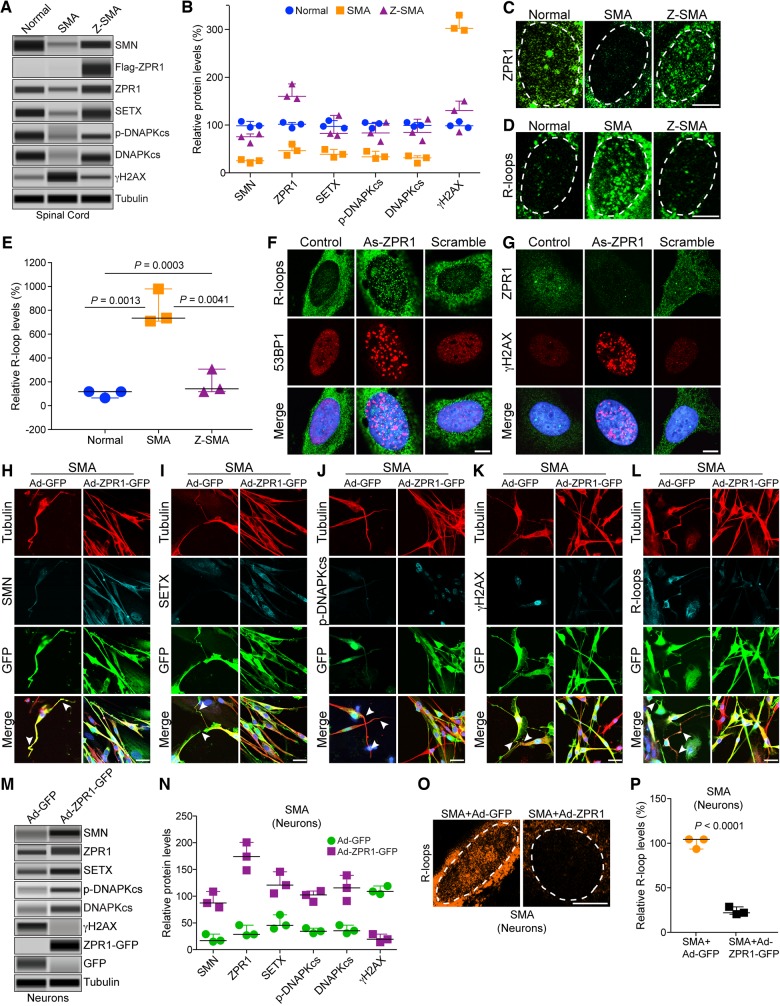
Downregulation of ZPR1 has been shown to contribute to neurodegeneration and severity of SMA disease in mice (Doran et al., 2006; Ahmad et al., 2012). To test whether in vivo ZPR1 overexpression will help reduce neurodegeneration and contribute to the rescue of SMA phenotype, we examined motor neurons in the spinal cords of normal, SMA and Z-SMA mice. Immunohistochemical staining of the spinal cord sections with antibodies against ChAT and SMN showed reduced loss of motor neurons in Z-SMA compared to SMA mice (Fig. 3A). Comparison of number of motor neurons in the lumbar (L1–L5) region of spinal cords (mean ± SEM%, three mice per group) of 7-day-old normal (101.20 ± 4.03%, n = 3), SMA (47.03 ± 2.85%) and Z-SMA (71.55 ± 2.24%) mice shows that ZPR1 overexpression reduced the loss of motor neurons by (24.52 ± 3.63%, P = 0.0025)% in Z-SMA compared to SMA mice and provided neuroprotection by increasing survival of neurons up to (71.55 ± 2.24%, P < 0.0001) in Z-SMA compared to normal and SMA mice (Fig. 3B).
Figure 3.
ZPR1 overexpression reduces neuron degeneration, improves NMJ innervation and muscle fibre size in SMA mice. (A) ZPR1 reduces loss of spinal cord motor neurons in mice with SMA. Immunohistochemical staining of the lumbar region spinal cord sections from 7-day-old normal, SMA, and Z-SMA littermates with anti-ChAT and SMN antibodies. Scale bar = 50 µm. (B) ZPR1 increases survival of SMN-deficient spinal cord motor neurons from SMA mice. Box-and-whisker plot with median and IQR (median, min, max) shows increase in the relative number of motor neurons (20 sections/mice, 3 mice/group) in the lumbar region of the spinal cords from Z-SMA (70.50, 60.0, 86.0) compared to SMA (47.0, 36.0, 61.0). Statistical analysis using mean ± SEM and comparison between SMA (47.03 ± 2.85%, n = 3) and Z-SMA (71.55 ± 2.24%) compared with normal (non-SMA) mice (littermates) as a reference point (101.20 ± 4.03%) shows statistically significant increase (24.52 ± 3.63%) in the number of motor neurons in Z-SMA mice, SMA versus Z-SMA (P = 0.0025, t-test, unpaired) and normal versus SMA versus Z-SMA (P < 0.0001, ANOVA). (C) ZPR1 reduces muscle degeneration in mice with SMA. Immunohistochemical staining of hind limb muscles, tibialis anterior longitudinal sections with β-actin (top) and transverse sections with dystrophin (bottom) from normal, SMA, and Z-SMA littermates. Scale bar = 50 µm (top) and 25 µm (bottom). (D) Immunohistochemical staining of hind limb muscles, gastrocnemius longitudinal sections with β-actin (top) and transverse sections with dystrophin (bottom) from normal, SMA, and Z-SMA littermates. Scale bar = 50 µm (top) and 25 µm (bottom). (E) ZPR1 improves muscle fibre diameter of SMA mice. The diameter of individual muscle fibres (μm) was measuring using transverse sections of gastrocnemius muscle from normal (blue), SMA (orange), and Z-SMA (purple) littermates stained with dystrophin. Diameter was measured at three different points, including longest and shortest along the irregular circular shape of each fibre and averaged. One hundred fibres per mouse (three mice per group) were measured to determine myofibres diameter. Scatter plots with median, min, max range for normal (17.61, 14.53, 20.44), SMA (11.53, 6.16, 15.30) and Z-SMA (13.87, 8.55, 17.97) show distribution of myofibre diameters. Statistical analysis shows increase in mean diameter (µm) for Z-SMA (13.88 ± 0.91) compared to SMA (11.06 ± 0.31), P = 0.043. (F) Immunohistochemical staining of gastrocnemius skeletal muscle with antibody to neurofilament M protein (NF, green) and BTX coupled with Alexa 594 (red) from normal, SMA, and Z-SMA littermates. Scale bars = 25 µm. (G) Immunohistochemical staining of gastrocnemius skeletal muscle with antibody to synaptophysin (SYN, red) and neurofilament (NF, green) from normal, SMA, and Z-SMA littermates. Scale bars = 25 µm. Note that littermates for all three genotypes were present in the same litter.
Improvement in gross motor functions, increased hind limb strength (Fig. 2) and reduced loss of motor neurons in Z-SMA mice suggest the possibility of improvement in muscle growth and function. Immunohistochemical analysis of hind limb muscles, tibialis anterior and gastrocnemius skeletal muscles, performed by staining longitudinal and transverse sections with antibodies against dystrophin and β-actin (markers for muscle morphology) shows marked improvement with reduced muscle atrophy in Z-SMA compared to SMA mice (Fig. 3C and D). Analysis of muscle fibre diameter using scatter plots shows reduced variation in myofibre size and increased mean diameter (μm) in Z-SMA mice (13.88 ± 0.91) compared to SMA mice (11.06 ± 0.31) with P = 0.043 (Fig. 3E). Comparison of muscle fibre thickness (μm) between normal (17.52 ± 0.90), SMA (11.06 ± 0.31) and Z-SMA (13.88 ± 0.91) show increase (∼21%, P = 0.043) in muscle fibre diameter of Z-SMA compared to SMA mice (Fig. 3E).
Neuron degeneration, including axonal retraction results in poor innervations of neuromuscular junctions (NMJs) leading to defects in maturation of NMJs in SMA patients and SMA mice (Torres-Benito et al., 2012; Genabai et al., 2015). To test whether neuroprotection provided by ZPR1 overexpression will improve NMJs, we examined hind-leg gastrocnemius skeletal muscle stained with BTX and antibody against NF-M protein to visualize acetylcholine receptors (AChRs) and nerves, respectively. Comparison of NMJs innervation, indicated by co-localization of AChRs and NF staining, shows improved innervations of NMJs in Z-SMA compared to SMA mice that have NMJs with partial or full denervation (Fig. 3F). Neuron degeneration coupled with NMJs denervation causes defects in synapse formation and contributes to synaptopathy in SMA. Defects in synaptic maturation and reduced levels of synapse formation have been shown to be present in SMA animal models (Torres-Benito et al., 2012; Genabai et al., 2015). To determine whether ZPR1 also improves synaptic maturation in Z-SMA mice, we examined gastrocnemius muscle stained with antibodies against NF-M and synaptophysin. SMA mice show NMJs with reduced innervation, retracting nerve fibres and marked reduction in synaptophysin staining compared to Z-SMA mice (Fig. 3G). ZPR1 overexpression results in increased synaptophysin staining of NMJs in Z-SMA and show marked improvement in synapse formation in Z-SMA compared to SMA mice (Fig. 3G). Together, these data show ZPR1 overexpression results in improvement of motor neuron survival, NMJs innervation, gross motor function, overall growth and the lifespan of Z-SMA mice, which suggest amelioration of severe SMA phenotype to moderate SMA phenotype.
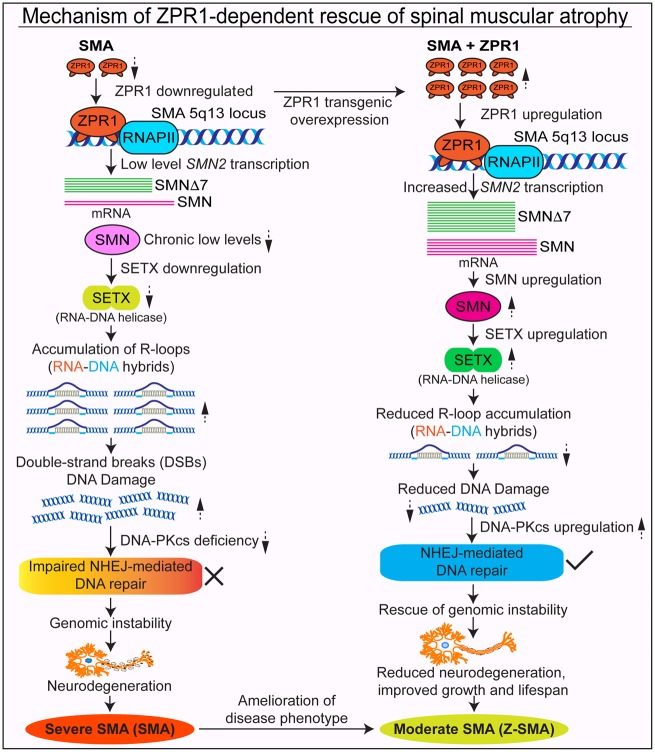
Mechanism of improvement in SMA phenotype by ZPR1
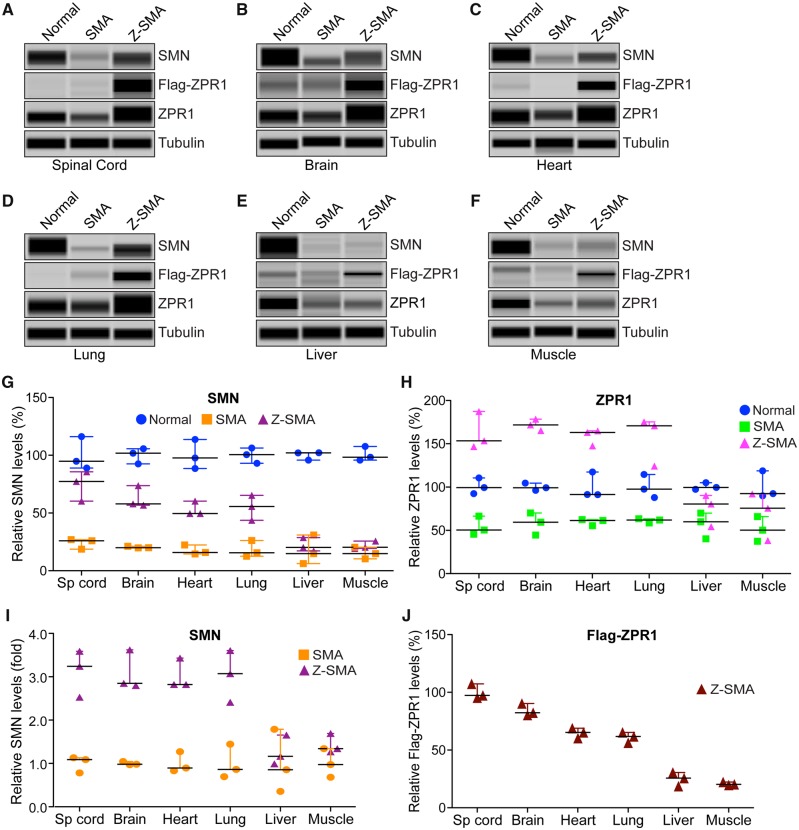
The immunohistochemical staining of spinal cord sections with SMN antibodies reproducibly showed increased staining for SMN in Z-SMA compared to SMA mice, which suggests an increase in SMN levels in Z-SMA mice (Fig. 3A, bottom). To determine whether improvement in phenotype of Z-SMA mice is SMN-dependent, we examined effect of ZPR1 overexpression on SMN levels in neuronal, brain and spinal cord and non-neuronal, heart, lung, liver and muscle tissues that are affected in SMA (Shababi et al., 2014) using automated capillary western blot analysis (Genabai et al., 2017; Kannan et al., 2018). Quantitative (mean ± SEM, n = 3 mice/group) and statistical analysis of relative protein levels normalized to tubulin with reference to normal mice (100%) show marked increase in levels of SMN protein in Z-SMA mice spinal cord (74.42 ± 7.44%, P = 0.0031) and brain (62.82 ± 5.43%, P = 0.0015) compared to SMA spinal cord (23.86 ± 2.62%) and brain (20.33 ± 0.45%) tissues, respectively (Fig. 4A, B and G). Analysis of ZPR1 shows downregulation of endogenous ZPR1 levels (∼40–45%) in the brain and spinal cord, and in non-neuronal tissues, heart, lung, liver and muscle from SMA mice (Fig. 4A–F, H, and Supplementary Figs 2 and 3) suggesting systemic downregulation of ZPR1 (∼40–50%) in SMA mice, which is consistent with downregulation of ZPR1 reported in SMA patients (Helmken et al., 2003; Ahmad et al., 2012). Analysis of total ZPR1 and recombinant Flag-ZPR1 levels shows a large increase (∼3-fold) in total ZPR1 levels in the spinal cord and brain tissues of Z-SMA compared to SMA mice (Fig. 4A, B and H). Analysis of SMN levels shows similar increase in the spinal cord [3.12-fold (P = 0.0031)] and brain [3.09-fold (P = 0.0015)] tissues of Z-SMA compared to SMA mice (Fig. 4I). These data suggest that increase in SMN levels by ZPR1 overexpression in vivo may have a direct correlation between ZPR1 and SMN levels in the CNS of SMA mice.
Figure 4.
ZPR1 overexpression increases SMN levels in neuronal and non-neuronal tissues and leads to SMN-dependent amelioration of SMA phenotype. Proteins levels of SMN, ZPR1, Flag-ZPR1 and tubulin were examined by immunoblot analysis of mouse tissues. (A) spinal cord, (B) brain, (C) heart, (D) lung, (E) liver and (F) muscle from 7-day-old normal, SMA and Z-SMA mice using automated capillary Wes™ System (ProteinSimple) and quantification was performed using Compass software. Representative capillary-blot images of proteins are shown (full-length blots are provided in Supplementary Figs 2 and 3). Quantitative data are shown as a scatter plot with median, min and max; (median, min, max) range shows relative increase in SMN levels (%) in different tissues by ZPR1 overexpression in the: (G) spinal cord [SMA (25.96, 18.65, 26.98) and Z-SMA (77.32, 60.32, 85.63)], brain [SMA (19.96, 19.79, 21.24) and Z-SMA (57.93, 56.87, 73.67)], heart [SMA (15.76, 14.62, 22.36) and Z-SMA (49.61, 49.60, 60.32)], lung [SMA (15.61, 12.58, 26.19) and Z-SMA (55.69, 43.72, 65.36)], liver [SMA (14.82, 6.13, 30.98) and Z-SMA (20.20, 17.18, 28.65)] and muscle [SMA (14.78, 10.32, 20.36) and Z-SMA (20.36, 19.32, 25.63)] and statistical analysis (unpaired t-test) of protein levels (mean ± SEM, n = 3 mice/group) shows that ZPR1 overexpression resulted in statistically significant increase of SMN levels in the spinal cord of Z-SMA (74.42 ± 7.44, P = 0.0031) compared to SMA (23.86 ± 2.62), brain of Z-SMA (62.82 ± 5.43, P = 0.0015) compared to SMA (20.33 ± 0.45), heart of Z-SMA (53.18 ± 3.57, P = 0.0012) compared to SMA (17.58 ± 2.41), lung of Z-SMA (54.92 ± 6.25, P = 0.0080) compared to SMA (18.13 ± 4.12) but not in the liver of Z-SMA (22.01 ± 3.43, P = 0.5907) compared to SMA (17.31 ± 7.27) and muscle of Z-SMA (21.77 ± 1.95, P = 0.1317) compared to SMA (15.15 ± 2.90) relative to respective normal mice tissues protein levels (100%) normalized to tubulin. (H) Quantitative analysis of increase in relative levels (%) of total ZPR1 expression in the: spinal cord [SMA (50.36, 45.63, 66.36) and Z-SMA (153.47, 146.73, 187.32)], brain [SMA (59.34, 44.50, 70.12) and Z-SMA (171.80, 165.32, 178.36)], heart [SMA (61.36, 55.48, 62.36) and Z-SMA (163.06, 147.51, 164.91)], lung [SMA (61.89, 58.50, 63.35) and Z-SMA (170.76, 124.33, 175.32)], liver [SMA (59.99, 40.32, 69.97) and Z-SMA (80.36, 54.32, 90.32)] and muscle [SMA (50.32, 37.39, 65.95) and Z-SMA (75.66, 38.25, 92.04)]. Statistical analysis of increase in levels of total ZPR1 protein using antibody against ZPR1 shows increase in ZPR1 levels in the spinal cord of Z-SMA (162.50 ± 12.56, P = 0.0015) compared to SMA (54.12 ± 6.27), brain of Z-SMA (171.80 ± 3.76, P = 0.0002) compared to SMA (57.99 ± 7.42), heart of Z-SMA (158.50 ± 5.51, P = 0.0001) compared to SMA (59.73 ± 2.14), lung of Z-SMA (156.80 ± 16.29, P = 0.0043) compared to SMA (61.25 ± 1.43), liver of Z-SMA (75.00 ± 10.73, P = 0.2574) compared to SMA (56.76 ± 8.71) and muscle of Z-SMA (68.65 ± 15.92, P = 0.3861) compared to SMA (51.22 ± 8.25) relative to respective normal mouse tissue protein levels (100%) normalized to tubulin. (I) Quantitative data are presented as a scatter plot with median, min and max range shows relative increase in SMN levels in different tissues as fold change in Z-SMA mice compared to SMA mice; spinal cord (3.11 ± 0.31, P = 0.0031), brain (3.08 ± 0.26, P = 0.0015), heart (3.02 ± 0.20, P = 0.0012), lung (3.029 ± 0.34, P = 0.0080), liver (1.27 ± 0.19, P = 0.5911) and muscle (1.43 ± 0.13, P = 0.1333) relative to respective SMA mice tissues protein levels (100% or 1.0-fold) normalized to tubulin. (J) Quantitative data are shown as a scatter plot with median, min and max range shows relative expression (%) of recombinant Flag-ZPR1 in Z-SMA mice; spinal cord (100.027 ± 3.71), brain (84.36 ± 3.05), heart (64.86 ± 2.50), lung (61.17 ± 2.62), liver (24.98 ± 3.47) and muscle (20.78 ± 0.80) relative to Z-SMA spinal cord Flag-ZPR1 levels (100%) normalized to tubulin.
To gain insight into ZPR1-dependent systemic increase in SMN levels, we examined levels of SMN in non-neuronal tissues, heart, lung, liver and skeletal muscle that are also affected by chronic low levels of SMN in SMA (Shababi et al., 2014). Quantitation shows an increase in SMN levels (∼3-fold) in the heart (P = 0.0012) and lung (P = 0.0080) tissues of Z-SMA compared to SMA mice (Fig. 4C, D, G and I). Analysis of total ZPR1 levels shows increase in the heart (2.65-fold, P < 0.0001) and lung (2.56-fold, P = 0.0043) in Z-SMA compared to SMA mice. In contrast, liver and muscle show a small and statistically non-significant increase in SMN [1.27-fold (P = 0.5911) and [1.43-fold (P = 0.1333), respectively] in Z-SMA compared to SMA mice (Fig. 4E–G). ZPR1 levels also shows a small and non-significant increase in liver (1.32-fold, P = 0.2577) and muscle (1.34-fold, P = 0.3876) of Z-SMA mice compared to SMA mice (Fig. 4H). Analysis of Flag-ZPR1 levels in all examined tissues shows the lowest levels in the liver (24.98 ± 3.47%) and muscle (20.78 ± 0.80%) tissues relative to the spinal cord (100%) (Fig. 4J). These data suggest that low levels of Flag-ZPR1 in liver and muscle tissues may be because of reduced expression of the Rosa26 promoter in these tissues. Analysis of data from different tissues shows that a 2–3-fold increase in ZPR1 is required for a corresponding 2–3-fold increase in SMN, suggesting a direct correlation between ZPR1 and SMN levels. Together, these data suggest that ZPR1-mediated improvement in SMA phenotype is SMN-dependent.
ZPR1 clears R-loops and rescues DNA damage in SMN-deficient spinal cord neurons and SMA mice
Chronic low levels of SMN causes molecular defects that include downregulation of SETX and DNA-activated protein kinase catalytic subunit (DNA-PKcs), which lead to DNA damage accumulation and neurodegeneration in SMA (Kannan et al., 2018). SETX is an RNA/DNA helicase required for resolution of RNA-DNA hybrids (R-loops) (Skourti-Stathaki et al., 2011). SETX deficiency causes accumulation of R-loops, which results in double-stranded DNA breaks leading to genomic instability. DNA-PKcs is essential for non-homologous end joining (NHEJ)-mediated double strand break repair (Davis et al., 2014). Neurons predominantly use NHEJ for DNA repair (Rass et al., 2007). Therefore, combined deficiency of SETX and DNA-PKcs results in gradual accumulation of DNA damage because of inefficient NHEJ-mediated DNA repair leading to genomic instability and predominant degeneration of motor neurons in SMA (Kannan et al., 2018). Notably, ectopic increase in SMN levels restores SETX and DNA-PKcs levels and rescues DNA damage in SMA motor neurons and SMA patient cells (Kannan et al., 2018). To test whether ZPR1-dependent increase in SMN will restore levels of SETX and DNA-PKcs and reduce DNA damage in SMA, we examined spinal cords and quantitation shows that ZPR1 overexpression increases SETX levels in Z-SMA (93.66 ± 13.39%, P = 0.0199) compared to (40.30 ± 4.80%) in SMA (Fig. 5A, B and Supplementary Fig. 4). ZPR1 also increases levels of DNA-PKcs in Z-SMA (88.11 ± 13.12%, P = 0.0133) compared to (29.36 ± 4.48%) in SMA (Fig. 5A and B), which results in increased levels of phospho (p)-DNA-PKcs in Z-SMA (85.13 ± 11.62%, P = 0.0175) compared to (36.41 ± 4.58%) in SMA (Fig. 5A and B) suggesting an increase in DNA-PKcs activity to enhance NHEJ-mediated DNA repair. Examination of activation of DNA damage response response marker, γH2AX (phospho-H2AX), shows marked reduction (2.5-fold, P = 0.0009) in γH2AX levels in Z-SMA compared to SMA mice suggesting decrease in DNA damage in Z-SMA mice (Fig. 5A and B). Together, these data show that ZPR1 overexpression increases SETX and p-DNA-PKcs levels, which downregulates DNA damage response and rescue DNA damage in Z-SMA mice.
Figure 5.
ZPR1 rescues molecular defects and DNA damage associated with SMA pathogenesis in SMA motor neurons and SMA mice. Protein extracts were prepared from the spinal cords isolated from 7-day-old normal, SMAΔ7 (SMA) and Z-SMA mice, and examined using an automated capillary-based western blot system. Representative capillary-blot images of proteins are shown (full-length blots are provided in Supplementary Fig. 4). (A) Immunoblot analysis of spinal cord protein extracts. (B) Quantitative immunoblot data are presented as a scatter plot with median, min and max (median, min, max) range shows relative change (%) in levels of SMN and DNA damage markers caused by in vivo ZPR1 overexpression in the spinal cords of mice, SMN [SMA (25.64, 20.65, 27.98) and Z-SMA (75.69, 62.65, 75.69)], SETX [SMA (38.95, 32.74, 49.21) and Z-SMA (82.32, 78.32, 120.33)], p-DNA-PKcs [SMA (33.65, 30.21, 45.36) and Z-SMA (83.65, 65.79, 105.96)], total DNA-PKcs [SMA (31.70, 20.69, 35.68) and Z-SMA (84.65, 67.32, 112.36)] and γH2AX [SMA (302.69, 298.63, 330.25) and Z-SMA (130.65, 85.63, 150.32)]. Statistical analysis (mean ± SEM, n = 3 mice/group) using t-test (unpaired, two-tailed) of proteins shows increase in ZPR1 (167.40 ± 9.55%, P = 0.0005) in Z-SMA compared to ZPR1 (47.99 ± 6.78%) levels in SMA mice results in increase of SMN (73.33 ± 5.61%, P = 0.0013) in Z-SMA compared to SMN (24.76 ± 2.16%) levels in SMA mice, SETX (93.66 ± 13.39%, P = 0.0199) in Z-SMA compared to SETX (40.30 ± 4.80%) levels in SMA mice, phospho-DNA-PKcs (p-DNAPKcs) (85.13 ± 11.62%, P = 0.0175) in Z-SMA compared to p-DNA-PKcs (36.41 ± 4.58%) levels in SMA mice and total DNA-PKcs (88.11 ± 13.12%, P = 0.0133) in Z-SMA compared to DNA-PKcs (29.36 ± 4.48%) levels in SMA mice. Analysis of DNA damage response marker shows decrease in γH2AX (122.20 ± 19.15%, P = 0.0009) in Z-SMA compared to γH2AX (310.5 ± 9.93%) levels in SMA mice suggesting the rescue of DNA damage in vivo by ZPR1 overexpression. (C and D) Primary spinal cord neurons were cultured from 7-day-old normal, SMA and Z-SMA mice. (C) Neurons were stained with ZPR1 and (D) R-loops (S9.6) antibodies and high magnification images of nuclei are presented. Dotted ellipses represent nuclei. Scale bar = 5.0 μm. (E) Quantitative analysis of relative levels of accumulation of nuclear R-loops in motor neurons from normal, SMA and Z-SMA mice is presented as a scatter plot with IQR (median, min, max), normal (118.0, 65.25, 118.7), SMA (734.90, 709.40, 978.9) and Z-SMA (141.8, 118.20, 306.90). Statistical analyses (mean ± SEM, n = 3 experiments, 50 neurons/experiment) using t-test (unpaired, two-tailed) of accumulation of R-loops in SMA (807.7 ± 85.90%, P = 0.0013) and normal neurons (106 ± 17.69%) shows large (∼8-fold) increase in accumulation of R-loops in SMA compared to normal mice. Comparison of R-loop accumulation between SMA (807.7 ± 85.90) and Z-SMA (189.0 ± 59.36) using t-test (P = 0.0041) and comparison between normal, SMA and Z-SMA using ANOVA (P = 0.0003) shows statistically significant decrease in in vivo R-loop accumulation by ZPR1 overexpression in Z-SMA mice. (F and G) HeLa cells were transfected with mock (Control), antisense ZPR1 oligonucleotide (As-ZPR1) and scrambled oligonucleotides (Scramble) (100 nM). (F) Knockdown of ZPR1 results in accumulation of R-loops (green) in the nucleus and formation 53BP1 foci (red) suggesting DNA double-strand breaks (DSBs). (G) ZPR1 (green) knockdown results in loss ZPR1 nuclear foci and causes accumulation of γH2AX foci (red) suggesting activation of DNA damage response in response to DNA damage caused by ZPR1 deficiency. Scale bar = 5 μm. (H–L) Cultured primary spinal cord neurons from SMA mice were infected with adenovirus (100 MOI) expressing green fluorescent protein (GFP) (Ad-GFP) and ZPR1-GFP fusion protein (Ad-ZPR1-GFP) and stained with antibodies against neuron-specific β-tubulin-III (red), SMN, SETX, p-DNA-PKcs, R-loops and γH2AX, and immunofluorescence was examined by confocal microscopy. GFP and ZPR1-GFP (green) were detected by GFP fluorescence. Axonal defects include retraction, bending, folding of axons (arrowheads) that indicate degeneration of SMN-deficient neurons. (H) Staining of neurons with SMN (cyan) and β-tubulin (red), (I) SETX (cyan) and β-tubulin (red), (J) p-DNA-PKcs (cyan) and β-tubulin (red), (K) γH2AX (cyan) and β-tubulin (red) and (L) R-loops (cyan) and β-tubulin (red). SMA neurons with ZPR1 ectopic expression (Ad-ZPR1-GFP panels) show reduction in neuron degenerative features. Nuclei were stained with DAPI (blue). Scale bar = 25 μm. Enlarged images of merged panels (H–L) are included in Supplementary Fig. 5 to show features of axonal degeneration such as loosening, bending retraction and ballooning in SMA neurons. (M) Immunoblot analysis of in vitro cultured motor neurons from SMA mice expressing GFP (Ad-GFP) and ZPR1-GFP (Ad-ZPR1-GFP) for changes in levels of SMN and DNA damage markers, SETX, p-DNA-PKcs, total DNA-PKcs and γH2AX. (N) Quantitative immunoblot data are presented as a scatter plot with median, min and max range shows relative change (%) in levels of SMN and DNA damage markers caused by in vitro ZPR1 overexpression in cultured motor neurons from SMA mice, SMN [SMA+GFP (17.06, 15.02, 29.0) and SMA+ZPR1-GFP (87.32, 79.06, 108.63)], SETX [SMA+GFP (45.62, 39.52, 65.32) and SMA+ZPR1-GFP (120.69, 105.36, 145.63)], p-DNA-PKcs [SMA+GFP (34.56, 30.65, 40.31) and SMA+ZPR1-GFP (102.36, 89.65, 109.65)], total DNA-PKcs [SMA+GFP (35.62, 30.65, 45.98) and SMA+ZPR1-GFP (115.47, 91.20, 138.97)] and γH2AX [SMA+GFP (108.90, 104.04, 118.99) and SMA+ZPR1-GFP (19.45, 14.13, 28.96)]. Statistical analysis using unpaired t-test of quantitative data (mean ± SEM, n = 3 mice/group) from spinal cord neuron immunoblots shows increase in ZPR1 levels (5.14 ± 0.44, P = 0.0010)-fold results in marked increase in levels of SMN (4.50 ± 0.43, P = 0.0019)-fold, SETX (2.47 ± 0.23, P = 0.0075)-fold, p-DNA-PKcs (2.85 ± 0.16, P = 0.0006)-fold and total DNA-PKcs (3.07 ± 0.36, P = 0.0059)-fold leading to decrease in γH2AX levels (5.04 ± 0.03, P = 0.0002)-fold. These data suggest the rescue of DNA damage in SMA spinal cord neurons (full-length blots are provided in Supplementary Fig. 6). (O) Enlarged images of nuclei of neurons stained with S9.6 antibody (R-loops, pseudocoloured orange) from SMA+Ad-GFP (SMA) and SMA+Ad-ZPR1-GFP (SMA+ZPR1) groups of SMA neurons. (P) Quantitative and statistical analysis of R-loop accumulation in cultured SMA motor neurons, SMA+GFP (103.5, 92.80, 103.7) and SMA+ZPR1-GFP (21.36, 19.65, 27.89) shows reduced accumulation (22.97 ± 2.51%, P < 0.0001) in neurons overexpressing ZPR1 (SMA+ZPR1-GFP) compared SMA+ GFP neurons.
Genomic instability is caused by R-loop accumulation during transcription that leads to selective degeneration of motor neurons in SMA (Kannan et al., 2018). To gain insight into the effect of ZPR1 overexpression on R-loop accumulation, we examined cultured primary neurons isolated from the spinal cords of 7-day-old, normal, SMA and Z-SMA mice and stained with antibodies against ZPR1 and R-loops (S9.6). We have previously shown that cultured primary spinal cord neurons from postnatal mice stain positive for known motor neuron markers, such as homeobox Hlxb9 (Hb9) and ChAT, suggesting that cultured spinal cord neurons may have characteristics similar to motor neurons (Ahmad et al., 2012; Genabai et al., 2015; Kannan et al., 2018). Immunofluorescence analysis of neurons stained with neuron-specific β-tubulin shows that neurons from Z-SMA mice were healthy and were rescued of axonal degeneration defects compared to neurons from SMA mice. SMA neurons show reduced accumulation of ZPR1 in the nucleus compared to normal neurons (Fig. 5C) and is consistent with data from patient cells (Gangwani et al., 2001). Neurons from Z-SMA show increased accumulation of ZPR1 in the nucleus compared to SMA suggesting that deficiency of ZPR1 in the nucleus of SMA (SMN-deficient) neurons is rescued by ZPR1 overexpression (Fig. 5C). Comparison of R-loops shows ∼8-fold higher accumulation in SMA compared to normal (P = 0.0013). Interestingly, ZPR1 overexpression results in ∼4.3-fold (P = 0.0041) reduction in R-loop accumulation in Z-SMA (Fig. 5D and E) suggesting that ZPR1 can complement and decrease R-loops in vivo in SMN-deficient neurons. These data raise an interesting question: does ZPR1-deficiency causes R-loop accumulation and contributes to genomic instability associated with SMA pathogenesis (Kannan et al., 2018)? To address this, we examined the effect of ZPR1 knockdown on HeLa cells, which showed that ZPR1-deficiency causes accumulation of R-loops and DNA damage as suggested by activation of DNA damage response markers, γH2AX and 53BP1 (Fig. 5F and G). These data suggest that ZPR1-deficiency may contribute to R-loop accumulation and DNA damage associated with SMA. ZPR1 overexpression and reduction in R-loop accumulation in Z-SMA neurons suggest that ZPR1 may contribute to resolution of R-loops generated during transcription and supports putative function of ZPR1 in transcription (Gangwani, 2006).
To test whether ZPR1 can also prevent DNA damage in vitro in SMA neurons, we performed a rescue experiment by complementation with adenovirus expressing ZPR1-GFP using in vitro cultured primary spinal cords neurons from SMA mice (Genabai et al., 2015; Kannan et al., 2018). We have recently shown that an increase in SETX levels alone can rescue molecular defects associated with DNA damage in SMA neurons and SMA patients cells. Therefore, we focused our analysis of ZPR1 overexpression on increase in SETX levels, reduction in R-loop accumulation and the rescue of DNA damage (Kannan et al., 2018). SMA neurons were infected with adenovirus containing GFP (Ad-GFP) and recombinant human ZPR1-GFP (Ad-ZPR1-GFP), stained with antibodies against tubulin, SMN, SETX, p-DNA-PKcs, R-loops (S9.6), and γH2AX. Control SMA neurons expressing GFP show axonal degeneration, bending, retraction and folding that is consistent with previous findings (Genabai et al., 2015; Kannan et al., 2018), suggesting that GFP did not alter phenotype of SMN-deficient spinal cord neurons from SMA mice (Fig. 5H–L, Ad-GFP panels). Neurons expressing ZPR1-GFP (Ad-ZPR1-GFP) show increase in SMN staining and the rescue of neurodegenerative features such as axonal bending, swelling, ballooning, retraction and folding compared to GFP expressing SMA neurons (Fig. 5H–L, Ad-ZPR1-GFP panels). Protein analysis show that Ad-ZPR1-GFP expression increases total ZPR1 levels (5.14-fold, P = 0.0010) compared control neurons with Ad-GFP (Fig. 5M and N). Notably, ZPR1-GFP overexpression increases SMN levels by 4.5-fold (P = 0.0019) (Fig. 5H, M and N), increases levels of SETX (2.47-fold, P = 0.0075) (Fig. 5I, M and N), p-DNA-PKcs (2.85 ± 0.16, P = 0.0006)-fold and total DNA-PKcs (3.07 ± 0.36, P = 0.0059)-fold (Fig. 5J, M and N), and decreases γH2AX levels by 5.04-fold (P = 0.0002) (Fig. 5K, M and N) suggesting reduction in double strand break accumulation and downregulation of DNA damage response. Further, examination of neurons stained with an antibody (S9.6) against R-loops shows marked reduction (4.35-fold, P < 0.0001) in R-loops suggesting that increase in SETX levels by ZPR1 results in reduced accumulation of R-loops in SMA neurons (Fig. 5L, O and P). These data show that ZPR1-dependent restoration of SMN levels in SMA neurons increase SETX levels, which reduce R-loop accumulation, and increase DNA-PKcs levels, which repairs DNA double-strand breaks, and together rescue DNA damage in SMA neurons. Thus, these results suggest an important role for ZPR1 in maintaining optimal levels of SMN and its downstream targets SETX and DNA-PKcs critical for resolving R-loops and DNA repair, respectively, to avert genomic instability and prevent degeneration of motor neurons in SMA.
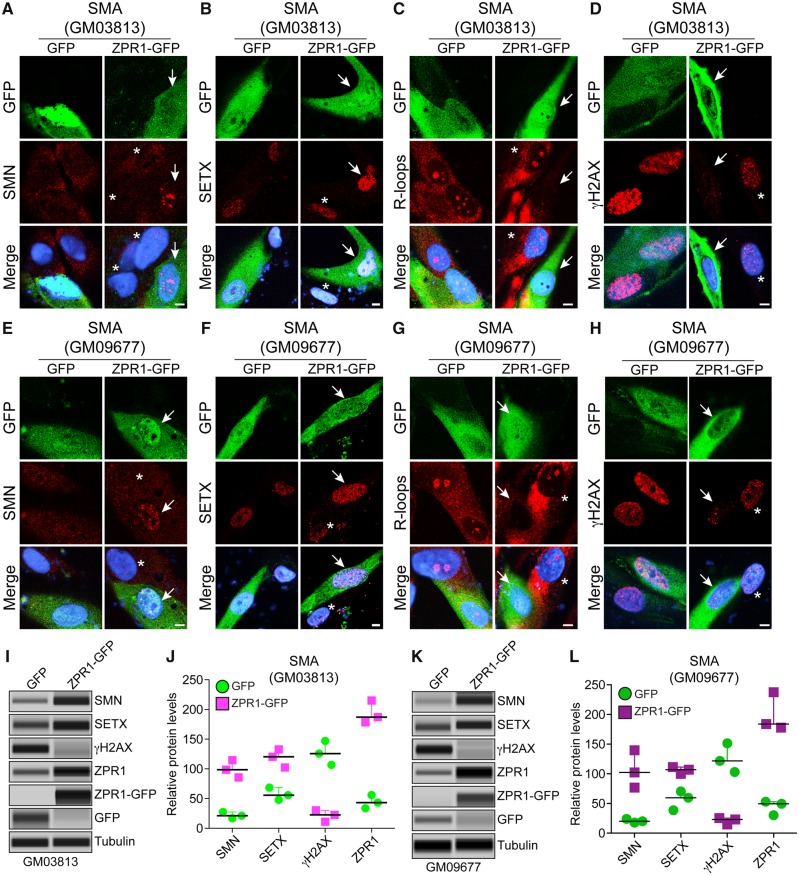
ZPR1 rescues molecular defects and DNA damage in SMA patient cells
To test whether the molecular defects associated with DNA damage can be rescued by ZPR1 in SMA type I patient-derived primary fibroblasts, we performed a rescue experiment with ectopic expression of recombinant ZPR1-GFP protein. Comparison of control GFP transfected and non-transfected cells show that GFP expression did not alter the phenotype of SMA patient cells and is consistent with previous findings (Fig. 6A–H) (Kannan et al., 2018). Ectopic expression of ZPR1-GFP induces accumulation of SMN in the nucleus of transfected cells compared to non-transfected in GM03813 and GM09677 cells as shown in Fig. 6A and E. Quantitative analysis shows an increase in ZPR1 levels (∼4.5-fold) elevated SMN levels in GM03813 (4.5-fold, P = 0.0009) (Fig. 6A, I and J) and GM09677 (5.1-fold, P = 0.0095) cells (Fig. 6E, K, L and Supplementary Figs 7 and 8). Notably, ZPR1 also increased SETX levels 2.0-fold (P = 0.0049) in GM03813 (Fig. 6B, I and J) and 1.9-fold (P = 0.0061) in GM09677 cells (Fig. 6F, K and L). Cells stained with antibody against R-loops show reduced accumulation of R-loops in the nucleus of cells expressing ZPR1-GFP compared to non-transfected or GFP transfected cells (Fig. 6C and G). Quantitation of nuclear R-loop shows decrease in GM03813 (4.55-fold, P = 0.00015) and GM09677 (4.0-fold, P = 0.00008) cells expressing ZPR1-GFP compared control cells suggesting that increase in SETX levels by ZPR1 reduces R-loop accumulation in SMA patient cells (Fig. 6B, C, F and G).
Figure 6.
Complementation with ZPR1 rescues senataxin levels and DNA damage in SMA patient cells. SMA patient primary fibroblast cell lines, GM03813 (A–D) and GM09677 (E–H) were transfected with phrGFP (GFP) or phrZPR1-GFP (ZPR1-GFP), fixed and stained with antibodies against SMN, SETX, R-loops and γH2AX. Ectopic ZPR1 expression elevates levels of SMN and SETX and reduces R-loop accumulation and rescues DNA damage in SMA patient cells. (A and E) SMN (red) and ZPR1-GFP (green), (B and F) SETX (red) and ZPR1-GFP, (C and G) R-loops (red) and ZPR1-GFP, (D and H) γH2AX (red) and ZPR1-GFP. Nuclei were stained with DAPI (blue). Scale bar = 5.0 μm. Arrows show transfected cells and asterisks indicate non-transfected cells. (I–L) Effect of ZPR1 overexpression on SMN, SETX, and γH2AX levels in patient cells GM03813 (I-J) and GM09677 (K-L) examined by IB (full-length blots are provided in Supplementary Figs 7 and 8). (I) Immunoblot analysis of patient cells (GM03813) expressing GFP and ZPR1-GFP. (J) Quantitative immunoblot data are presented as a scatter plot with IQR (median, min, max) shows relative change (%) in levels of SMN, SETX and γH2AX caused by in vitro ZPR1 overexpression in SMA patient cells (GM03813), SMN [GFP (21.28, 16.83, 27.44) and ZPR1-GFP (98.65, 85.69, 114.67)], SETX [GFP (55.69, 47.96, 68.52) and ZPR1-GFP (120.36, 102.36, 132.65)] and γH2AX [GFP (125.69, 106.59, 146.96) and ZPR1-GFP (22.65, 10.96, 30.32)]. Statistical analysis of quantitative data from GM03813 (GFP) and GM03813 (ZPR1-GFP) cells show ZPR1 overexpression (4.37 ± 0.25, P = 0.0003)-fold increases levels of SMN (4.56 ± 0.38, P = 0.0009)-fold and SETX (2.06 ± 0.15, P = 0.0049)-fold and result in marked reduction of γH2AX levels (6.0 ± 0.04, P = 0.0012)-fold in (GM03813+ZPR1-GFP) compared to control GM03813+GFP cells suggesting rescue of DNA damage in patient cells by ZPR1 complementation. (K) Immunoblot analysis of patient cells (GM09677) expressing GFP and ZPR1-GFP. (L) Quantitative immunoblot data are presented as a scatter plot with IQR (median, min, max) shows relative change (%) in levels of SMN, SETX and γH2AX caused by in vitro ZPR1 overexpression in SMA patient cells (GM09677), SMN [GFP (20.05, 17.65, 24.08) and ZPR1-GFP (102.32, 76.92, 139.52)], SETX [GFP (59.68, 38.69, 70.26) and ZPR1-GFP (106.98, 99.87, 111.36)] and γH2AX [GFP (121.8, 103.04, 151.52) and ZPR1-GFP (22.98, 14.65, 25.36)]. Statistical analysis of quantitative data from GM09677+GFP and GM09677+ZPR1-GFP cells shows (4.51 ± 0.42, P = 0.0015)-fold increase in ZPR1, which results in marked reduction of γH2AX levels (6.08 ± 0.026, P = 0.0018)-fold compared to GM09677+GFP suggesting the rescue of DNA damage that is supported by increase in levels of SMN (5.15 ± 0.88%, P = 0.0095)-fold and SETX (1.88 ± 0.059, P = 0.0061)-fold in ZPR1-GFP complemented patient cells compared to patient cells with GFP.
Furthermore, immunofluorescence analysis of ZPR1 overexpressing cells stained for γH2AX shows reduced γH2AX staining and foci in transfected compared to non-transfected GM03813 and GM09677 cells (Fig. 6D and H). Quantitation shows marked reduction in the levels of γH2AX in GM03813 (6.0-fold, P = 0.0012) (Fig. 6I and J) and GM09677 (6.08-fold, P = 0.0018) cells complemented with ZPR1 suggesting that ZPR1 is able to rescue DNA damage in SMA patient cells (Fig. 6K and L). Together, these data on ZPR1-mediated increase in SMN and SETX levels leads to reduction in R-loop accumulation and the rescue of DNA damage in patient cells suggest that ZPR1 has potential to rescue the molecular defects associated with SMA pathogenesis, in SMA neurons and patient cells and represents an SMN-dependent protective modifier of SMA.
ZPR1 transcriptionally upregulates SMN2 expression in SMA patient cells and SMA mice
ZPR1 may have functions in mRNA biogenesis, including transcription and may be a putative transcription factor (Gangwani, 2006; Kielbowicz-Matuk et al., 2016). To test the function of ZPR1 as a transcriptional regulator and unravel the molecular mechanism of ZPR1-mediated increase in SMN protein levels, we examined in vitro and in vivo effects of ZPR1 overexpression on SMN2 transcripts, full-length SMN and SMNΔ7, in SMA patient cells and Z-SMA mice using qPCR. Comparative analysis of in vivo transcription data from the spinal cords show increase (2.93 ± 0.33-fold, P = 0.0060) in SMNΔ7 and (2.65 ± 0.30-fold, n = 3, P = 0.0058) in full-length SMN transcripts in Z-SMA compared to SMA mice (Fig. 7A). Analysis of SMA patient fibroblast with ectopic expression of recombinant Flag-ZPR1 shows increase in SMNΔ7 (4.00 ± 0.25-fold, P = 0.0003) and full-length SMN transcripts (3.42 ± 0.62-fold, P = 0.0249) in GM03813+ZPR1 (SMA+ZPR1) compared to control (transfected with empty vector) GM03813 cells (SMA) (Fig. 7B). Examination of second SMA patient fibroblast cell line (GM09677) also shows increase in SMNΔ7 (3.58 ± 0.22-fold, P = 0.0008) and in full-length SMN transcripts (3.04 ± 0.21-fold, P = 0.0054) in GM09677+ZPR1 (SMA+ZPR1) compared to control GM09677 cells (SMA) (Fig. 7C). We also examined the effect of ZPR1 on change in relative abundance of SMN and SMNΔ7 transcripts in SMA and Z-SMA mice that shows proportionate ∼3-fold increase in both transcripts in Z-SMA mice (Fig. 7D). Similar increase (>3-fold) in both transcripts was found in SMA patient cell lines complemented with recombinant ZPR1 (Fig. 7E and F). The higher levels of SMNΔ7 compared to SMN transcript in mice and patient cells is because of one copy each of SMN2 and SMNΔ7 cDNA in mice and three copies of SMN2 gene in both cell lines, respectively, and is consistent with the published data that SMN2 produces majority of SMNΔ7 transcript (Fig. 7D–F) (Lorson et al., 1999; Stabley et al., 2015). These data show that ZPR1 overexpression increases overall transcription of SMN2 gene in vitro (patient cells) and in vivo (SMA mice) under pathophysiological conditions of SMA suggesting that ZPR1 may be a positive transcriptional regulator of the SMN2 gene.
Figure 7.
ZPR1 overexpression upregulates SMN2 gene transcription in SMA patient cells and SMA mice. (A) ZPR1 overexpression in vivo upregulates total SMN2 transcription under SMA conditions. Total RNA was isolated from the spinal cords of 7-day-old SMA and Z-SMA littermates and examined by qPCR using specific primers for amplification of SMNΔ7 and full-length SMN transcripts generated from SMN2. The relative mRNA levels were calculated using fold-enrichment (2−ΔΔCT) method and presented as a scatter plot with median, min and max range. Quantitative data for SMNΔ7 [SMA (1.08, 0.74, 1.16) and Z-SMA (2.96, 2.32, 3.50)] and SMN [SMA (0.98, 0.91, 1.12) and Z-SMA (2.63, 2.14, 3.18)]. ZPR1 overexpression increases full-length SMN (2.65 ± 0.30-fold, n = 3, P = 0.0058) and SMNΔ7 (2.93 ± 0.33-fold, P = 0.0060) transcript levels in Z-SMA mice compared to SMA mice. (B) SMA patient cell lines GM03813 was transfected with pcDNA3 or pcDNA3/Flag-ZPR1. The scatter plot shows quantitative data for SMNΔ7 [SMA (0.96, 0.92, 1.12) and SMA+ZPR1 (3.87, 3.64, 4.5)] and SMN [SMA (1.28, 0.65, 1.34) and SMA+ZPR1 (3.12, 2.51, 4.62)]. Statistical analysis shows ectopic ZPR1 overexpression increases full-length SMN (3.42 ± 0.62-fold, P = 0.0249) and SMNΔ7 (4.00 ± 0.25-fold, P = 0.0003) transcript levels in GM03813+ZPR1 (SMA+ZPR1) compared to control GM03813 (SMA) patient cells. (C) SMA patient cell line GM09677 was transfected with pcDNA3 or pcDNA3/Flag-ZPR1. Quantitative data for SMNΔ7 [SMA (0.94, 0.93, 1.42) and SMA+ZPR1 (3.57, 3.21, 3.98)] and SMN [SMA (1.03, 0.60, 1.59) and SMA+ZPR1 (2.86, 2.79, 3.48)]. Statistical analysis shows that ZPR1 increases full-length SMN (3.04 ± 0.21-fold, P = 0.0054) and SMNΔ7 (3.58 ± 0.22-fold, P = 0.0008) transcript levels in GM09677+ZPR1 (SMA+ZPR1) compared to control GM09677 (SMA) patient cells. (D–F) Analysis of relative abundance of SMN and SMNΔ7 transcripts in (D) SMA and Z-SMA mice, and (E) SMA patient cell line (GM03813) and (F) SMA patient cell line (GM09677) without and with ZPR1 overexpression using qPCR. (G–N) ZPR1 interacts with RNA polymerase and in vivo associates with genomic SMN locus. Modulation of ZPR1 levels influences SMN expression. (G) Immunoprecipitation with anti-ZPR1 antibody shows pulldown of RNAPII from wild-type mouse brain protein extract suggesting in vivo interaction of ZPR1 with RNAPII. (H) GST-ZPR1 pulldown of RNAPII from HeLa cell lysate shows in vitro direct interaction of ZPR1 with RNAPII. (I–J) ZPR1 interacts with genomic SMN locus in vivo. Chromatin immunoprecipitation (ChIP) was performed using antibodies against ZPR1, H3K4me3 (positive control) and FLAG (M2) antibody (negative control) and chromatin prepared from human HeLa cells. Presence of SMN genomic locus in ChIP was detected by real-time qPCR using primers in the human SMN promoter region and SMN exon 1 region. ChIP assay shows ZPR1 associates with SMN locus and result in 4.34 ± 0.38-fold (n = 3, P = 0.0004) (promoter) and 3.66 ± 0.43-fold (P = 0.0012) (SMN exon 1) regions amplifications compared to control (IgG). (K–L) Modulation of ZPR1 levels influences expression of SMN1-Luc and SMN2-Luc reporter genes. (K) Effect of ZPR1 knockdown on ectopic SMN1 and SMN2 genes expression in cultured HeLa cells. Cells were transfected with either SMN1-Luc or SMN2-Luc or empty control reporter vector (Con-Luc). Transfected cells were retransfected after 24 h with scrambled or ZPR1 antisense oligonucleotides (As-ZPR1) to knockdown the levels of ZPR1. Cells were harvested after 24 h post-second transfection for determination of luciferase activity or (M) immunoblot analysis for ZPR1 expression. To examine the effect of ZPR1 overexpression, HeLa cells were transfected with combination of two plasmids: (i) Con-Luc + pcDNA3-FlagZPR1, (ii) SMN1-Luc or SMN2-Luc + pcDNA3 (empty); and (iii) SMN1-Luc or SMN2-Luc + pcDNA3-FlagZPR1. After 30 h post-transfection, cells were harvested for determining luciferase activity or (N) immunoblot analysis for ZPR1 expression. Quantification of luciferase activity shows ZPR1 knockdown causes decreases and ZPR1 overexpression increases levels of SMN1 and SMN2 promoters driven luciferase expression. Scatter plot with median, min and max range shows modulation of luciferase activity for SMN1-Luc (106.70, 80.48, 112.9), SMN1-Luc+As-ZPR1 (26.14, 15.59, 27.23) and SMN1-Luc+Flag-ZPR1 (227.6, 180.9, 260.0). SMN2-Luc (98.41, 97.67, 103.90), SMN2-Luc+As-ZPR1 (13.72, 9.28, 20.19) and SMN2-Luc+Flag-ZPR1 (160.10, 141.6, 178.9). Statistical analysis (mean ± SEM; n = 3) shows ZPR1 knockdown reduced levels of luciferase activity of SMN1-Luc to 22.99 ± 3.71% (P = 0.001) and SMN2-Luc to 14.40 ± 3.16% (P < 0.0001) in cells treated with antisense oligonucleotides compared to control cells with scrambled oligo. ZPR1 overexpression results in increase of luciferase activity for SMN1-Luc to 222.8 ± 22.95% (P = 0.0062) and SMN2-Luc to 160.2 ± 10.76% (P = 0.014) compared to cells without ZPR1 overexpression (full-length blots are provided in Supplementary Fig. 9).
ZPR1 interacts with RNA polymerase, associates in vivo with SMN genomic locus and regulates SMN transcription
The above data on enhancement of SMN2 transcription by ZPR1 suggest that ZPR1 may be a componenet of RNA transcription complexes; therefore, we examined the interaction of ZPR1 with RNA polymerase II (RNAPII). Immunoprecipitation with anti-ZPR1 antibody and pulldown with GST-ZPR1 followed by western blot analysis shows that ZPR1 interacts in vivo with RNAPII (Fig. 7G) and ZPR1 may interact directly with RNAPII (Fig. 7H) suggesting that ZPR1 is a part of the basal transcription complex containing RNAPII. These data support the idea that downregulation of ZPR1 in SMA patients may contribute to reduced transcription of SMN2 and the low levels of SMN, which is further supported by our current data that ZPR1 overexpression enhances SMN2 transcription and levels of SMN protein in SMA mice and patient cells.
To strengthen the idea that ZPR1 is a transcriptional regulator of SMN genes, we tested whether ZPR1 interacts in vivo with SMN genomic locus, we performed ChIP with antibodies against H3K4me3 (positive control) and ZPR1, and control antibody (negative control) followed by qPCR to measure binding of ZPR1 with SMN genes using promoter and exon 1 primers with sequences identical in SMN1 and SMN2 genes. The ChIP-PCR assay using primers, which amplify promoter region (−622 to −430) of SMN1 and SMN2 genes, show that ZPR1 antibody pulldown results in 4.34 ± 0.38-fold (P = 0.0004) amplification compared to IgG control (Fig. 7I). Similarly, PCR with exon 1 region primers shows 3.66 ± 0.43-fold (P = 0.0012) amplification of SMN genes compared to IgG control (Fig. 7J). Together, these data show that ZPR1 interacts in vivo with RNAPII and associates with genomic SMN locus on chromosome 5q13 suggesting that ZPR1 is a part of the core transcription complex and may be involved in transcriptional regulation of SMN expression.
Furthermore, to test whether modulation of ZPR1 levels influence transcription of both SMN1 and SMN2 gene and ZPR1 may be a transcriptional regulator of SMN genes, we examined the effect of change in ZPR1 levels using in vitro biochemical experiments. To examine the effect of ZPR1 on transcription, we used plasmids containing the luciferase (Luc) gene under the control of human SMN1 and SMN2 promoter regions (750 bp DNA sequences upstream of start codon) (Monani et al., 1999) and transfected HeLa cells to measure the luciferase enzyme activity. ZPR1 levels were: (i) downregulated by knockdown using antisense oligonucleotide (Gangwani, 2006); and (ii) upregulated by ectopic expression of recombinant human Flag-ZPR1 cDNA (Ahmad et al., 2012; Genabai et al., 2017). Knockdown of ZPR1 levels (As-ZPR1) causes marked reduction in luciferase activity in cells expressing SMN1-Luc (22.99 ± 3.71%, P = 0.001) (Fig. 7K) and SMN2-Luc (14.40 ± 3.16%, P < 0.0001) (Fig. 7L) compared to cells treated with scrambled oligonucleotide. Notably, overexpression of ZPR1 (Flag-ZPR1) results in increase of luciferase activity in cells expressing SMN1-Luc (222.80 ± 22.95%, P = 0.006) (Fig. 7K) and SMN2-Luc (160.20 ± 10.76%, P = 0.014) (Fig. 7L) compared to control cells (pcDNA3) without ZPR1 overexpression (Fig. 7M, N and Supplementary Fig. 9). The increase in SMN2 promoter activity is consistent with in vivo increase in the levels of transcription and SMN protein levels, which are upregulated by Zpr1 gene overexpression in Z-SMA mice and SMA patient cells. These findings suggest that modulation of ZPR1 levels directly correlates and influences levels of SMN genes transcription and ZPR1 is a transcriptional regulator of the SMN1 and SMN2 genes.
Discussion
Chronic low levels of SMN result in motor neuron degeneration causing skeletal muscle atrophy leading to respiratory failure and death in SMA. The severity of SMA disease ranges from SMA type I (severe) to type IV (mild) and is inversely correlated with SMN levels, which are dependent on copy number of the SMN2 gene in SMA patients that have homozygous SMN1 deletion or mutation (Lefebvre et al., 1997; Wirth et al., 2013). Each copy of the SMN2 gene produces ∼10% SMN protein and variation in SMN2 copy number causes proportionate changes in SMN levels and result in a broad clinical spectrum of SMA patients with varying severity of disease (Lorson et al., 1999; Wirth et al., 2013). These findings have established SMN2 as a naturally occurring modifier of disease severity with tremendous therapeutic potential for SMN-dependent rescue of SMA. Two methods can be used for producing higher amounts of SMN from SMN2: (i) correction of pre-mRNA splicing; and (ii) upregulation of transcription of the SMN2 gene. Excellent progress has been made in the past decade to develop a therapeutic method using antisense oligonucleotides to correct splicing of the SMN2 gene and approved by FDA as the first drug (nusinersen; SPINRAZA®) to treat SMA (Singh et al., 2017). However, there are challenges that need to be addressed, including side effects and complications in patients, which are discussed in recent papers (Sumner and Crawford, 2018). In contrast, only modest progress has been made towards understanding the transcriptional regulation of the SMN2 gene. Cis regulatory elements have been identified in the promotor region of the SMN2 gene but the transcription factors that may regulate expression of the SMN2 gene have not been reported (Ottesen et al., 2017). In this study, we demonstrate that ZPR1 is a positive transcriptional regulator of both SMN1 and SMN2 genes. ZPR1 overexpression in SMA patient cells (in vitro) and SMA mice (in vivo) enhances SMN2 transcription, which increases SMN protein levels and results in an SMN-dependent amelioration of severe SMA phenotype to moderate SMA phenotype (Fig. 8).
Figure 8.
Graphical summary: the molecular mechanism of ZPR1-dependent rescue of spinal muscular atrophy. ZPR1 interacts with RNAPII and associates in vivo with SMA 5q13 genomic locus. ZPR1 levels are downregulated in SMA. Low levels of ZPR1 result in reduced transcription of SMN2 gene leading to chronic low levels of SMN in SMA. Low levels of SMN cause deficiency of senataxin (SETX), which results in accumulation of RNA-DNA hybrids (R-loops). Accumulation of R-loops causes DNA double-strand breaks (DSBs) and activation of DNA damage response (DDR). In dividing cells, double strand breaks are repaired by homologous recombination (HR) and NHEJ, but neurons predominantly use NHEJ that requires DNA-PKcs activity. Chronic low levels of SMN cause deficiency of DNA-PKcs that causes defects in NHEJ-mediated double strand break repair leading to genome instability and predominant degeneration of motor neurons in SMA. ZPR1 overexpression in SMA increases transcription of SMN2 gene resulting in higher levels of SMN. Increase in SMN elevates SETX and DNA-PKcs levels that result in reduction of R-loops and the rescue of DNA damage leading to neuroprotection and amelioration of severe to moderate SMA disease phenotype.
Mechanism of ZPR1-mediated transcriptional upregulation of the SMN2 gene in SMA
Transcriptional regulation of SMN1 and SMN2 genes is poorly understood. Initial studies have identified a promoter region within 3.4 kb and a core promoter within 750 bp upstream of the start codon. Several common consensus DNA cis elements, including HNF-3beta, HiNF, IRF-E/ISRE, Elk-1, CREB, Sp1, MAZ, AhR, HNF-3 and N-Oct3 that are known to regulate gene expression are present in the core promoter regions of SMN1 and SMN2 genes (Echaniz-Laguna et al., 1999; Germain-Desprez et al., 2001). Preliminary studies suggest that transcriptional regulation modulates SMN genes expression during development and cellular differentiation. However, specific trans-acting/transcription factors that may regulate expression of SMN genes remain to be identified.
The human ZPR1 gene is located on Chr11q23.2 and is evolutionary conserved in eukaryotes and encodes a protein with two C4-type Zn2+ fingers with novel helix-loop-helix motifs that may mediate protein-protein and protein-nucleic acid interactions (Mishra et al., 2007). Genetic studies in yeast and mice have shown that the ZPR1 is an essential gene (Galcheva-Gargova et al., 1998; Gangwani et al., 1998, 2005). However, the essential biological function of ZPR1 is unknown. Biochemical studies have provided insight that ZPR1 interacts with eEF1A and the SMN protein (Gangwani et al., 1998, 2001). ZPR1 mediates receptor tyrosine kinases (RTKs) signalling by binding to the cytoplasmic domain of epidermal growth factor receptor (EGFR) and platelet-derived growth factor receptor (PDGFR), which is highly conserved among family of RTKs (Galcheva-Gargova et al., 1996). In quiescent mammalian cells, ZPR1 binds to inactive EGFR and PDGFR, upon mitogen or serum treatment, ZPR1 translocates from the cytoplasm to the nucleus and triggers mitosis suggesting that ZPR1 may play an important role in nuclear processes involving expression of genes required for the cell growth and proliferation (Galcheva-Gargova et al., 1996, 1998). The precise function of ZPR1 in the nucleus is unclear. However, ZPR1 deficiency causes defects in splicing and transcription (Gangwani et al., 2001, 2005; Gangwani, 2006). Recent studies showed that ZPR1 may be a putative transcription factor that regulate light regulated genes in plants and developmentally regulated Hox genes in mammals (Kielbowicz-Matuk et al., 2016; Genabai et al., 2017). ZPR1 in vivo interacts with SMN and is required for accumulation of SMN in sub-nuclear bodies, including gems and Cajal bodies. Interaction of ZPR1 with SMN is indirect (Gangwani et al., 2001, 2005). Our data demonstrate that ZPR1 interacts in vivo and in vitro with RNAPII and is a part of transcription complexes. A recent study demonstrated that SMN interacts with RNAPII and is a part of transcription complexes and the disruption of RNAPII-SMN complexes by mutation in RNAPII causes defects in transcription termination (Zhao et al., 2016). Direct interaction of ZPR1 with RNAPII suggests that ZPR1 and SMN interaction may be mediated in vivo by RNAPII. ZPR1 and SMN may play distinct roles in transcription. SMN has been shown to play a critical role in pre-mRNA splicing (Pellizzoni et al., 1998) and our data show that the ZPR1 may play a role in regulation of pre-mRNA transcription by contributing to resolution of co-transcriptional R-loops. Accumulation of R-loops in cells is a major cause of DNA damage and genomic instability leading to human diseases, including neurodegenerative disorders (Sollier and Cimprich, 2015; Kannan et al., 2018).
Binding of ZPR1 with RNAPII and its in vivo association with genomic SMA (5q13) locus suggest that ZPR1 may be a part of core transcription complexes. However, whether ZPR1 is a specific trans-acting factor or transcriptional enhancer that has a specific DNA binding site or has broad specificity and interacts with nucleic acids as a part of core transcription complexes, and regulates transcription is unclear and requires further studies. Our current data demonstrate that ZPR1 may play a role in resolution of R-loops that are formed during transcription and regulates transcription but the molecular mechanism of ZPR1-dependent R-loop resolution and transcriptional regulation is unclear. However, knockdown of ZPR1 in HeLa cells results in accumulation of R-loops and DNA damage, which is consistent with data on accumulation of R-loops in SMA patient cells that have low levels of ZPR1, suggesting that ZPR1 deficiency causes R-loop accumulation and contributes to genomic instability that leads to neurodegeneration in SMA (Ahmad et al., 2012; Kannan et al., 2018). Because ZPR1 interacts with RNAPII and is a part of core transcription complexes, it is possible that ZPR1 may be required for resolution of co-transcriptional R-loops.
Modulation of ZPR1 levels in vitro by knockdown or overexpression in cultured cells causes downregulation or upregulation of recombinant SMN1-Luc and SMN2-Luc expression levels, respectively, suggesting a direct correlation between ZPR1 and SMN levels. This is supported by in vivo data of ZPR1 overexpression in Z-SMA mice on upregulation of SMN2 gene transcription and increase in SMN levels in Z-SMA mice that show ∼3-fold increase in ZPR1 expression results in ∼3-fold increase in SMN levels, which is further supported by in vitro data on upregulation of endogenous SMN2 transcription and SMN levels by ZPR1 in SMA patient fibroblast that show ∼4–5-fold increase in ZPR1 expression results in ∼4–5-fold increase in SMN transcript and protein levels. The higher levels of transcripts and SMN protein in patient cells complemented with Flag-ZPR1 compared to Z-SMA mice could be because of three copies of SMN2 present in GM03813 and GM09677 cell lines (Stabley et al., 2015). Together, our current data demonstrate that ZPR1 is a positive transcriptional regulator of SMN2 gene and supports the idea that it could be exploited for its therapeutic potential to treat SMA using pharmacological method to upregulate ZPR1 expression to elevate SMN levels.
SMN-dependent systemic improvement of SMA phenotype by ZPR1
We show that genetic in vivo overexpression of ZPR1 increases SMN protein levels and results in SMN-dependent improvements, including gross motor function, increase in overall growth, decrease in neurodegeneration and muscle atrophy, and increase in the lifespan of SMA mice. Together, these improvements show a systemic decrease in the severity of disease in SMA mice, which is conceptually in-line and consistent with findings that increase in SMN levels would result in amelioration of disease severity and the rescue of SMA phenotype. ZPR1 overexpression resulted in partial but systemic SMN-dependent rescue of SMA mice.
ZPR1 overexpression reduces severity of disease, increases lifespan and improves SMA phenotype. Comparison of survival of SMNΔ7 mice with other published studies shows broad range of average survival from ∼7 to 12 days reported in different studies with SMNΔ7 mouse model (Butchbach et al., 2010; Cherry et al., 2013; Genabai et al., 2015; Kim et al., 2017). The SMAΔ7 mice without Flag-ZPR1 (SMA) show an average survival of SMA (male and female) (7.07 ± 0.78, n = 26) days under our laboratory conditions and is consistent with our and others’ findings (Genabai et al., 2015; Kim et al., 2017). Z-SMA mice (littermates) show significant increase in average survival (17.93 ± 0.34, P < 0.0001) days. The decrease in average survival of the original SMA line (4299) (Le et al., 2005) from ∼10 to ∼7 days was stabilized in our laboratory and this might be because of several factors, including diet, local environment and genetic modifications such as addition of a transgene to create new strain. It is established that the changes in the genetic content of SMAΔ7 model causes alteration in phenotype, including average and max survival (Monani et al., 2000; Genabai et al., 2015). To eliminate the possibility of genetic variation, we analysed and compared data from littermates produced by breeding of Z-SMA carrier strain (Smn+/−; SMN2+/+; SMNΔ7+/+; Flag-Zpr1+/−). The increase in the median (3-fold), average (2.53-fold) and maximum (9 days) survival time of Z-SMA mice compared to SMA mice suggests that ZPR1 overexpression markedly reduces severity of disease resulting in an increase of survival of Z-SMA mice. Notably, marked increase in the initial (15.5-fold) suggest minimization of variation in disease severity present in SMA mice that leads to striking increase in minimal survival after birth for at least 15 days in male and female Z-SMA mice. Gender-based analysis shows that ZPR1 improves phenotypes of male and female mice. The normal mouse lifespan of ∼8–9 days is equivalent to ∼1 year of human life (Dutta and Sengupta, 2016); however, similar increases in lifespan under disease conditions may have a profound effect on the increase in lifespan of SMA patients and such correlation remains to be studied.
Our data show systemic partial rescue and the reasons for this may include moderate to low levels increase in SMN amount in different neuronal and non-neuronal tissues. Low level increases in SMN might be because of insufficient expression of the Zpr1 gene under the control of mouse Rosa26 promoter in liver and muscle tissues. Our data show that ∼3-fold increase in total ZPR1 levels in the spinal cord, brain, heart and lung tissues results in ∼3-fold increase in SMN levels suggesting a direct correlation between ZPR1 and SMN levels. Analysis of other tissues affected in SMA show a small increase in total ZPR1expression in liver (∼18%) that result in a very small and non-significant increase (∼5%) in SMN. SMN deficiency in liver has been shown to causes defects in liver development that result in late embryonic lethality in mice, suggesting that low levels of SMN in the liver may partially impair its function and contribute to severity during the course of disease progression (Vitte et al., 2004). Skeletal muscle is the second most affected tissue in SMA and downregulation of SMN in skeletal muscle results in severe muscle dystrophy, suggesting that the normal SMN levels are critical for maintenance and function of skeletal muscle (Cifuentes-Diaz et al., 2001). A small increase (∼7%) in SMN in muscle may be because of low level expression of Rosa26 promoter in muscle tissue resulting in a non-significant increase (∼17%, P = 0.386) in ZPR1 levels that maybe insufficient for full rescue of skeletal muscle phenotype in Z-SMA. However, improvements found in skeletal muscle, including reduced degeneration and increase in myofibre size might be because of reduced neurodegeneration, which is consistent with previous findings that SMN-independent reduction in neurodegeneration improves muscle growth, strength and myofibre size in the SMAΔ7 model (Genabai et al., 2015). Together, these findings suggest that restoration of SMN levels in the liver and muscle tissues maybe critical for full rescue of SMA disease phenotype.
The moderate improvement in the overall disease phenotype with modest increase in the median survival (3-fold) of Z-SMA mice compared to other reported splicing modifiers and gene therapy methods used in SMAΔ7 mice raise the possibility of contribution of unanticipated toxic and off target effects of ZPR1 overexpression (Singh et al., 2009, 2017; Naryshkin et al., 2014; Meyer et al., 2015). The inflammation of genitals noted in males homozygous for the transgene (Flag-Zpr1+/+) compared to males heterozygous for the transgene (Flag-Zpr1+/−) indicates a possibility of a toxic effect of ZPR1 overexpression. However, the inflammation in homozygous males might not be because of the toxic effect of increased ZPR1 levels because females with the homozygous Flag-Zpr1+/+ genotype did not show any adverse effect, were fertile and lived the normal lifespan, suggesting undetectable or no toxicity on doubling of ZPR1 expression, and homozygous males did not show any other complication and lived a normal lifespan. Therefore, it is possible that inflammation in males might be due to the disruption of a non-essential gene, specific to male genitals, at the site of integration of transgene in the mouse genomic locus.
Our previous studies with stable and transient ZPR1 overexpression in single cell models (Saccharomyces cerevisiae and S. pombe) and mammalian cells did not show any detectable cellular toxicity (Galcheva-Gargova et al., 1998; Gangwani et al., 1998; Ahmad et al., 2012). The probability of toxic or off target effects may be low because ZPR1 forms complexes with SMN and is required for translocation of SMN to the nucleus. Cells derived from SMA patients have very low levels of ZPR1-SMN complexes (Gangwani et al., 2001). Several studies have demonstrated that deficiency of either ZPR1 or SMN cause similar defects in mRNA splicing and transcription (Pellizzoni et al., 1998; Gangwani et al., 2001; Zhao et al., 2016) leading to neurodegeneration mediated by the JNK signalling pathway involving neuron-specific isoform JNK3 (Genabai et al., 2015; Jiang et al., 2019) suggesting that ZPR1-SMN complexes might be involved in common cellular functions, including mRNA biogenesis. Therefore, the majority of ZPR1 may form protein-protein complexes with SMN that might be critical for restoring cellular functions of SMA cells.
ZPR1 is downregulated in SMA patients and SMA mice (Gangwani et al., 2001; Helmken et al., 2003; Ahmad et al., 2012). Upregulation of SMN levels by ZPR1 overexpression may help restore levels of critical molecular targets stemming downstream of SMN deficiency that are reported to be affected in SMA (Zhang et al., 2008). Our data show that ZPR1 overexpression increases levels of two critical targets—SETX and DNA-PKcs—and rescues DNA damage, a leading cause of genomic instability and neurodegeneration in SMA (Kannan et al., 2018). However, the effects of ZPR1 overexpression on systemic changes in the levels of potential ZPR1 targets, toxicity and off targets in Z-SMA mouse model remains to be examined. Moreover, the development of an actual therapeutic strategy for the treatment of SMA may utilize two different methods: (i) a pharmacological method using small compounds to upregulate the levels of ZPR1; and (ii) a gene therapy method using AAV9 vector to deliver AAV9-ZPR1 intravenously. These therapeutic approaches will have their respective toxic or off target effects and will require preclinical studies to determine the efficacy and safety of drug compounds and gene therapy method before initiating human clinical trials.
Together, these data suggest that moderate increase in median survival (3-fold) of Z-SMA mice correlates with ∼3-fold increase in SMN levels by ZPR1, which may be equivalent to the amount of SMN produced by about three copies of SMN2 in humans that may lead to moderate reduction in severity and amelioration of disease phenotype from severe (type I) to moderate (type II) in SMA patients. However, ZPR1-dependent rescue may not represent the best stand-alone strategy for the treatment of severe SMA (type I) but could be very useful in a combination therapy. Nevertheless, our data suggest that similar increase in ZPR1 expression in moderate SMA (type II) and mild SMA (type III and IV) patients that have higher SMN2 copy numbers may show profound improvements with amelioration of type II to types III or IV phenotypes, and the rescue of types III and IV to the normal phenotype. In conclusion, this study shows ZPR1 is a transcriptional regulator of SMN2 gene and an SMN-dependent protective modifier of SMA. ZPR1 represents a viable therapeutic target for developing a new method for the treatment of SMA.
Supplementary Material
Acknowledgements
We thank Dr Arthur Burghes of Ohio State University for plasmids with human SMN1 and SMN2 promoters fused to luciferase. We thank Dr Olof Sundin, TTUHSC EP and Charlotte Sumner, John Hopkins Medical Institute, for reading and helpful comments on the manuscript. We thank graduate student Kabir Masih for assisting Dr Saif Ahmad with histological analysis and confocal microscopy.
Funding
This study was supported by research grants to L.G. from the US National Institutes of Health (R01 NS064224), Cure SMA (GAN1920) and Muscular Dystrophy Association (MDA 480210).
Competing interests
The authors report no competing interests.
Glossary
Abbreviations
- ChIP =
chromatin immunoprecipitation
- DNA-PKcs =
DNA-activated protein kinase-catalytic subunit
- GFP =
green fluorescent protein
- NHEJ =
non-homologous end joining
- NMJ =
neuromuscular junction
- PND =
postnatal day
- RNAPII =
RNA polymerase II
- SMA =
spinal muscular atrophy
See Hensel et al. (doi:10.1093/brain/awz394) for a scientific commentary on this article.
References
- Ahmad S, Bhatia K, Kannan A, Gangwani L. Molecular mechanisms of neurodegeneration in spinal muscular atrophy. J Exp Neurosci 2016; 10: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S, Wang Y, Shaik GM, Burghes AH, Gangwani L. The zinc finger protein ZPR1 is a potential modifier of spinal muscular atrophy. Hum Mol Genet 2012; 21: 2745–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 2009; 10: 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol 2009; 29: 1107–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchbach ME, Singh J, Thorsteinsdottir M, Saieva L, Slominski E, Thurmond J, et al. Effects of 2,4-diaminoquinazoline derivatives on SMN expression and phenotype in a mouse model for spinal muscular atrophy. Hum Mol Genet 2010; 19: 454–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JJ, Osman EY, Evans MC, Choi S, Xing X, Cuny GD, et al. Enhancement of SMN protein levels in a mouse model of spinal muscular atrophy using novel drug-like compounds. EMBO Mol Med 2013; 5: 1035–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes-Diaz C, Frugier T, Tiziano FD, Lacene E, Roblot N, Joshi V, et al. Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J Cell Biol 2001; 152: 1107–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AJ, Chen BP, Chen DJ. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair 2014; 17: 21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran B, Gherbesi N, Hendricks G, Flavell RA, Davis RJ, Gangwani L. Deficiency of the zinc finger protein ZPR1 causes neurodegeneration. Proc Natl Acad Sci U S A 2006; 103: 7471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Sengupta P. Men and mice: relating their ages. Life Sci 2016; 152: 244–8. [DOI] [PubMed] [Google Scholar]
- Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J. The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am J Hum Genet 1999; 64: 1365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khodor BF, Edgar N, Chen A, Winberg ML, Joyce C, Brunner D, et al. Identification of a battery of tests for drug candidate evaluation in the SMNDelta7 neonate model of spinal muscular atrophy. Exp Neurol 2008; 212: 29–43. [DOI] [PubMed] [Google Scholar]
- Galcheva-Gargova Z, Gangwani L, Konstantinov KN, Mikrut M, Theroux SJ, Enoch T, et al. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol Biol Cell 1998; 9: 2963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galcheva-Gargova Z, Konstantinov KN, Wu IH, Klier FG, Barrett T, Davis RJ. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science 1996; 272: 1797–802. [DOI] [PubMed] [Google Scholar]
- Gangwani L. Deficiency of the zinc finger protein ZPR1 causes defects in transcription and cell cycle progression. J Biol Chem 2006; 281: 40330–40. [DOI] [PubMed] [Google Scholar]
- Gangwani L, Flavell RA, Davis RJ. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol Cell Biol 2005; 25: 2744–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwani L, Mikrut M, Galcheva-Gargova Z, Davis RJ. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J Cell Biol 1998; 143: 1471–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat Cell Biol 2001; 3: 376–83. [DOI] [PubMed] [Google Scholar]
- Genabai NK, Ahmad S, Zhang Z, Jiang X, Gabaldon CA, Gangwani L. Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum Mol Genet 2015; 24: 6986–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genabai NK, Kannan A, Ahmad S, Jiang X, Bhatia K, Gangwani L. Deregulation of ZPR1 causes respiratory failure in spinal muscular atrophy. Sci Rep 2017; 7: 8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain-Desprez D, Brun T, Rochette C, Semionov A, Rouget R, Simard LR. The SMN genes are subject to transcriptional regulation during cellular differentiation. Gene 2001; 279: 109–17. [DOI] [PubMed] [Google Scholar]
- Hahnen E, Forkert R, Marke C, Rudnik-Schoneborn S, Schonling J, Zerres K, et al. Molecular analysis of candidate genes on chromosome 5q13 in autosomal recessive spinal muscular atrophy: evidence of homozygous deletions of the SMN gene in unaffected individuals. Hum Mol Genet 1995; 4: 1927–33. [DOI] [PubMed] [Google Scholar]
- Helmken C, Hofmann Y, Schoenen F, Oprea G, Raschke H, Rudnik-Schoneborn S, et al. Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1. Hum Genet 2003; 114: 11–21. [DOI] [PubMed] [Google Scholar]
- Hosseinibarkooie S, Peters M, Torres-Benito L, Rastetter RH, Hupperich K, Hoffmann A, et al. The power of human protective modifiers: PLS3 and CORO1C unravel impaired endocytosis in spinal muscular atrophy and rescue SMA phenotype. Am J Hum Genet 2016; 99: 647–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen E, Mendoza-Ferreira N, Hosseinibarkooie S, Schneider S, Hupperich K, Tschanz T, et al. CHP1 reduction ameliorates spinal muscular atrophy pathology by restoring calcineurin activity and endocytosis. Brain 2018; 141: 2343–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kannan A, Gangwani L. ZPR1-dependent neurodegeneration is mediated by the JNK signaling pathway. J Exp Neurosci 2019; 13: 1179069519867915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan A, Bhatia K, Branzei D, Gangwani L. Combined deficiency of Senataxin and DNA-PKcs causes DNA damage accumulation and neurodegeneration in spinal muscular atrophy. Nucleic Acids Res 2018; 46: 8326–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielbowicz-Matuk A, Czarnecka J, Banachowicz E, Rey P, Rorat T Solanum tuberosum ZPR1 encodes a light-regulated nuclear DNA-binding protein adjusting the circadian expression of StBBX24 to light cycle. Plant Cell Environ 2016. [DOI] [PubMed]
- Kim JK, Caine C, Awano T, Herbst R, Monani UR. Motor neuronal repletion of the NMJ organizer, Agrin, modulates the severity of the spinal muscular atrophy disease phenotype in model mice. Hum Mol Genet 2017; 26: 2377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet 2005; 14: 845–57. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80: 155–65. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 1997; 16: 265–9. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)] method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A 1999; 96: 6307–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Ferraiuolo L, Schmelzer L, Braun L, McGovern V, Likhite S, et al. Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: a dose-response study in mice and nonhuman primates. Mol Ther 2015; 23: 477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra AK, Gangwani L, Davis RJ, Lambright DG. Structural insights into the interaction of the evolutionarily conserved ZPR1 domain tandem with eukaryotic EF1A, receptors, and SMN complexes. Proc Natl Acad Sci U S A 2007; 104: 13930–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Coovert DD, Burghes AH. Animal models of spinal muscular atrophy. Hum Mol Genet 2000; 9: 2451–7. [DOI] [PubMed] [Google Scholar]
- Monani UR, McPherson JD, Burghes AH. Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT). Biochim Biophys Acta 1999; 1445: 330–6. [DOI] [PubMed] [Google Scholar]
- Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, et al. Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 2014; 345: 688–93. [DOI] [PubMed] [Google Scholar]
- Oprea GE, Krober S, McWhorter ML, Rossoll W, Muller S, Krawczak M, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science 2008; 320: 524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen EW, Seo J, Singh NN, Singh RN. A multilayered control of the human survival motor neuron gene expression by Alu elements. Front Microbiol 2017; 8: 2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 1998; 95: 615–24. [DOI] [PubMed] [Google Scholar]
- Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell 2007; 130: 991–1004. [DOI] [PubMed] [Google Scholar]
- Riessland M, Kaczmarek A, Schneider S, Swoboda KJ, Lohr H, Bradler C, et al. Neurocalcin delta suppression protects against spinal muscular atrophy in humans and across species by restoring impaired endocytosis. Am J Hum Genet 2017; 100: 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J, Singh NN, Ottesen EW, Sivanesan S, Shishimorova M, Singh RN. Oxidative stress triggers body-wide skipping of multiple exons of the spinal muscular atrophy gene. PLoS One 2016; 11: e0154390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shababi M, Lorson CL, Rudnik-Schoneborn SS. Spinal muscular atrophy: a motor neuron disorder or a multi-organ disease? J Anat 2014; 224: 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Howell MD, Androphy EJ, Singh RN. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther 2017; 24: 520–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol 2009; 6: 341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 2011; 42: 794–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Cimprich KA. Breaking bad: R-loops and genome integrity. Trends Cell Biol 2015; 25: 514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabley DL, Harris AW, Holbrook J, Chubbs NJ, Lozo KW, Crawford TO, et al. SMN1 and SMN2 copy numbers in cell lines derived from patients with spinal muscular atrophy as measured by array digital PCR. Mol Genet Genomic Med 2015; 3: 248–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner CJ, Crawford TO. Two breakthrough gene-targeted treatments for spinal muscular atrophy: challenges remain. J Clin Invest 2018; 128: 3219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Benito L, Ruiz R, Tabares L. Synaptic defects in spinal muscular atrophy animal models. Dev Neurobiol 2012; 72: 126–33. [DOI] [PubMed] [Google Scholar]
- Vitte JM, Davoult B, Roblot N, Mayer M, Joshi V, Courageot S, et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am J Pathol 2004; 165: 1731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B, Garbes L, Riessland M. How genetic modifiers influence the phenotype of spinal muscular atrophy and suggest future therapeutic approaches. Curr Opin Genet Dev 2013; 23: 330–8. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008; 133: 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 2016; 529: 48–53. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.