Microglia are activated in Huntington’s disease prior to symptom onset. Crapser et al. show that eliminating microglia from the R6/2 mouse model of Huntington’s disease reduces motor and cognitive impairments, accumulation of mutant huntingtin, and striatal volume loss. It also prevents the loss of perineuronal nets.
Keywords: microglia, Huntington's disease, CSF1R, extracellular matrix, perineuronal nets
Abstract
Huntington’s disease is associated with a reactive microglial response and consequent inflammation. To address the role of these cells in disease pathogenesis, we depleted microglia from R6/2 mice, a rapidly progressing model of Huntington’s disease marked by behavioural impairment, mutant huntingtin (mHTT) accumulation, and early death, through colony-stimulating factor 1 receptor inhibition (CSF1Ri) with pexidartinib (PLX3397) for the duration of disease. Although we observed an interferon gene signature in addition to downregulated neuritogenic and synaptic gene pathways with disease, overt inflammation was not evident by microglial morphology or cytokine transcript levels in R6/2 mice. Nonetheless, CSF1Ri-induced microglial elimination reduced or prevented disease-related grip strength and object recognition deficits, mHTT accumulation, astrogliosis, and striatal volume loss, the latter of which was not associated with reductions in cell number but with the extracellular accumulation of chondroitin sulphate proteoglycans (CSPGs)—a primary component of glial scars. A concurrent loss of proteoglycan-containing perineuronal nets was also evident in R6/2 mice, and microglial elimination not only prevented this but also strikingly increased perineuronal nets in the brains of naïve littermates, suggesting a new role for microglia as homeostatic regulators of perineuronal net formation and integrity.
Introduction
The cause of Huntington’s disease, an autosomal dominant neurological disorder, is well established as an abnormal expansion of CAG triplet repeats in exon 1 of the ubiquitously expressed huntingtin (HTT) gene, resulting in an elongated polyglutamine (Q) tract in the mutant huntingtin protein (mHTT) (MacDonald et al., 1993). While the unaffected population ranges on average 16–20 triplet repeats at this locus, affected patients contain ≥40 CAGs, with the age of symptom onset in general inversely correlating with repeat length (Duyao et al., 1993; Labbadia and Morimoto, 2013). Disease symptoms include progressive movement abnormalities (chorea, dystonia) and cognitive-behavioural impairments, accompanied by selective striatal medium spiny neuron death and cortical atrophy (Ross and Tabrizi, 2011). Pathologically, mHTT accumulates in many cells types, and in multiple forms, due at least in part to the failure of protein quality control networks, as reflected by an accumulation of ubiquitin and other post-translationally-modified proteins (Ochaba et al., 2016) and disruptions in autophagy (Kurosawa et al., 2015). Despite Huntington’s disease’s deceivingly simple monogenic origin, the contributions of specific cell types in the brain are only beginning to be elucidated.
Microglia, resident myeloid phagocytes comprising ∼12% of cells in the brain, sample the entire parenchymal milieu every 24 h for deviations from homeostasis (Nimmerjahn et al., 2005). That microglia are potent modulators of neurodegeneration and injury is by now clear (Li and Barres, 2017), and investigation into the full functional spectrum and uniqueness of microglia among related immune cells continues to elucidate critical roles in development, ageing, and disease (Bennett et al., 2018; Mrdjen et al., 2018). Previously, we developed a pharmacological method to eliminate virtually all (∼99%) microglia from the murine brain for several months at a time (Elmore et al., 2014; Dagher et al., 2015), based on inhibition of colony-stimulating factor 1 receptor (CSF1Ri). Microglial CSF1R signalling is essential to their survival and has become a thoroughly tested and highly replicated model for targeted elimination of microglia (Elmore et al., 2014; Asai et al., 2015; Dagher et al., 2015; Rice et al., 2015; Acharya et al., 2016; Feng et al., 2016; Spangenberg et al., 2016; Stafford et al., 2016; Szalay et al., 2016; Li et al., 2017; Bennett et al., 2018). By formulating inhibitors in chow, we can ensure non-invasive, uninterrupted drug delivery and elimination of these cells for the duration of treatment.
In Huntington’s disease, microglial activation is detected years before the onset of clinical symptoms and correlates with striatal neuron dysfunction in subclinical (Tai et al., 2007) as well as clinical stages of the disease (Pavese et al., 2006; Politis et al., 2011). Post-mortem Huntington’s disease brains contain elevated densities of reactive microglia in regions of neuronal loss (Sapp et al., 2001) and this microgliosis is mirrored in mouse models of the disease (Simmons et al., 2007). While microglia conventionally react to stress signals and exogenous toxins in neurodegeneration, the presence of endogenous mHTT alone alters their transcriptional landscape through increased expression of the myeloid lineage-determining factor Pu.1 (Crotti et al., 2014), resulting in enhanced production of Il6 and Tnfα transcripts as well as increased neurotoxicity in the context of a secondary immune challenge. Furthermore, intrastriatal engraftment of mHTT-expressing human glial progenitor cells in immunodeficient mice results in neuronal hyperexcitability in the striatum along with motor impairments (Benraiss et al., 2016; Osipovitch et al., 2019). We aimed to further extend these findings by eliminating microglia from a mouse model of Huntington’s disease via sustained CSF1Ri and assessing the impact on disease pathogenesis.
In this study we treated transgenic R6/2 mice, expressing exon 1 of the human huntingtin (HTT) gene containing ∼120 CAG repeats, or non-transgenic littermates with the clinically utilized and FDA-approved CSF1R inhibitor, PLX3397 (pexidartinib) (Tap et al., 2014), or vehicle. The R6/2 line is a short-lived and rapidly progressing mouse model of Huntington’s disease that displays motor and behavioural deficits beginning around 7 weeks of age and progressing until an early death at 11–13 weeks (Mangiarini et al., 1996). We found that CSF1Ri and resultant microglial depletion in R6/2 mice is accompanied by ameliorated grip strength and memory deficits, mHTT species accumulation, and protein clearance system pathway dysfunction. At the transcriptional level, we report impaired neuronal and neurite development pathways in addition to an interferon (IFN) signature in the R6/2 striatum that is similarly overrepresented in human Huntington’s disease, aspects of which are diminished following microglial depletion. Additionally, despite a reduction in R6/2 caudoputamen volume, we observed increased neuronal and neurite cytoskeletal element densities in the R6/2 striatum at 11 weeks. This was concomitant with astrogliosis and massive brain-wide deposition of extracellular matrix (ECM) chondroitin sulphate proteoglycans (CSPGs), a key component of glial scars (Hsu et al., 2008) and a negative axon guidance cue (Ohtake et al., 2016). In parallel, we observed a disease-related loss of CSPG-containing perineuronal nets (PNNs) (Fawcett et al., 2019), specialized interneuron-associated ECM structures the degradation of which contributes to seizures (Tewari et al., 2018), dampened inhibitory tone, and changes in plasticity (Banerjee et al., 2017). CSF1Ri and the depletion of microglia abrogated each of these changes, including disease-associated striatal volume loss, underscoring their role as coordinators of brain ECM remodelling that in turn appears to contribute to neurodegeneration in Huntington’s disease. Interestingly, microglial elimination in non-transgenic littermates also resulted in brain-wide increases in PNN density, suggesting that microglia tightly regulate PNN formation in health as well as disease.
Materials and methods
Compounds
CSF1R inhibitor PLX3397 (pexidartinib) was provided by Plexxikon Inc. and formulated in AIN-76A standard chow by Research Diets Inc. at 275 mg/kg.
Animals
All animal experiments were performed in accordance with animal protocols approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, Irvine, an American Association for Accreditation of Laboratory Animal Care (AAALAC)-accredited institution. R6/2 mice have been described elsewhere in detail (Mangiarini et al., 1996; Ochaba et al., 2016). For this study, 5-week-old R6/2 male mice were purchased from Jackson Laboratories and housed in groups of up to five animals/cage under a 12-h light/dark cycle with ad libitum access to control chow and water during baseline testing, and then AIN-76A chow (containing vehicle or PLX3397) starting at Week 6. Brain tissue from 16-week-old mixed sex wild-type mice on a C57BL/6J congenic background treated for 10 weeks (from Week 6) with the more selective CSF1R inhibitor PLX5622 (Plexxikon Inc.) formulated in standard chow by Research Diets Inc. at 1200 mg/kg was used for additional investigation of PNNs. Assignment of animals to treatment groups was conducted in a random fashion. Researchers were blinded to genotype and treatment group for behavioural tasks and in the analysis of behavioural, histological, stereological, and biochemical assays. CAG repeat sizing of tail samples was performed by Laragen.
Behavioural testing
Baseline behavioural characterization was performed at 5 weeks of age via Rotarod and grip strength as described previously (Ochaba et al., 2016; Morozko et al., 2018) and again during treatment at regular intervals. Briefly, animals were tested for latency to fall from an accelerating rotarod over three 5-min interval trials averaged together on Weeks 5, 7 and 9. Forelimb grip strength was assessed with a meshed force gauge that records peak force applied (IITC Life Science instrument) over five trials averaged together on Weeks 5, 7, 9, 10 and 11. Body weights were obtained daily from 5 to 11 weeks of age. Open field and novel object recognition tests were assessed on Week 11 and were analysed with EthoVision. For the open field test, mice were placed in an empty chamber and recorded while roaming freely for 5 min. For the novel object recognition test, mice were placed in a chamber for 5 min with two identical objects on Day 1, and one familiar and one novel object on Day 2, and memory for the familiar object was measured as the time spent investigating the novel object as percentage of total time exploring (i.e. discrimination index). Exclusionary criteria were as follows: if mice did not spend time investigating both objects during training, or either object during testing, the testing data were excluded from analysis, as it cannot be confirmed they spent enough time exploring to learn/discriminate. All behavioural tests were conducted and analysed in a blinded fashion.
Tissue collection
After final behavioural measurements on Week 11, mice were euthanized with administration of phenytoin/pentobarbital (Euthasol®) followed by transcardial perfusion with ice-cold phosphate-buffered saline. Brains were isolated and divided hemispherically; one hemisphere per animal was drop-fixed in 4% paraformaldehyde (Thermo Fisher Scientific) for 48 h, cryopreserved in 30% sucrose, and sectioned at 40 μm on a freezing microtome for immunohistochemistry (IHC), while the other hemisphere was used to extract, flash freeze, and store striatal tissue at −80°C for later biochemical analyses. Frozen striatal tissue was split into protein and RNA fractions.
Immunohistochemistry and confocal microscopy
Fluorescent immunolabelling followed a standard indirect technique as described previously (Elmore et al., 2014; Spangenberg et al., 2016). Brain sections were stained with antibody to IBA1 (1:1000; 019-19741, Wako and ab5076, Abcam) (Smith et al., 2013; Elmore et al., 2014), HTT (EM48) (1:500; MAB5374, Millipore), DARPP32 (1:500; SC-11365, Santa Cruz) (Morozko et al., 2018), CD68 (1:200; MCA1957GA, Bio-Rad), NeuN (1:500; MAB377, Millipore), IB4 (1:200; 121414, ThermoFisher) (Elmore et al., 2018), Olig2 (1:200; MABN50, Millipore), MAP2 (1:200; ab32454, Abcam) (Elmore et al., 2014), Tau (1:500; A0024, DAKO) (Julien et al., 2012), p62 (1:500; BML-PW9860, Enzo) (Paine et al., 2013), MBP (1:100; MAB386, Millipore) (Cardoso et al., 2015), S100β (1:200; ab52642, Abcam) (Illouz et al., 2019), CD44 (1:200; ab157107, Abcam) (Koga et al., 2018), CSPG (1:200; ab11570, Abcam) (Park et al., 2017), Wisteria floribunda agglutinin (WFA) lectin (1:1000; B-1355, Vector Labs) (Yutsudo and Kitagawa, 2015), parvalbumin (1:500; MAB1572, Millipore) (McNamara et al., 2014), c-Kit (1:100; ab25022, Abcam) (Trias et al., 2018), and Flt3 (1:100; ab73019, Abcam) (Svensson et al., 2015). Immunostained sections were mounted and coverslipped with Fluoromount-G® with DAPI (00-4959-52, SouthernBiotech). High resolution fluorescent images were obtained using a Leica TCS SPE-II Confocal microscope, and whole-brain stitches using a ZEISS Axio Scan.Z1. Cell and EM48+ puncta quantities, area, and intensity were determined using the spots module in Imaris v9.2. Per cent area coverage and integrated density measurements were determined in ImageJ (NIH) at a constant threshold for each stain using ×20 confocal images for striatal and dentate gyrus quantifications and regions of interest in whole-brain stitches for cortical analyses.
Western blotting
Striatal tissue fractionation, gel electrophoresis, and immunoblotting were performed as previously described (Ochaba et al., 2016, 2018; Morozko et al., 2018). Briefly, striatal tissue was first homogenized in lysis buffer containing 10 mM Tris (pH 7.4), 1% Triton™ X-100, 150 mM NaCl, 10% glycerol, and 0.2 mM PMSF. Tissue was lysed on ice for 60 min before centrifugation at 15 000g for 20 min at 4°C. Supernatant was collected as the detergent-soluble fraction. The pellet was washed three times with lysis buffer and centrifuged at 15 000g for 5 min each at 4°C. The pellet was resuspended in lysis buffer supplemented with 4% SDS, sonicated three times, boiled for 30 min, and collected as the detergent-insoluble fraction. Protein concentration was determined using Lowry Protein Assay (Bio-Rad).
Detergent-soluble fractions were resolved on 4–12% Bis-Tris poly-acrylamide gels (PAGE), insoluble fractions on 3–8% Tris-acetate poly-acrylamide gels, and oligomeric species determined by agarose gel electrophoresis using 1%, 375 mM Tris-HCl, pH 8.8, 1% SDS agarose gels. Insoluble mHTT fibrils were detected using a modified filter retardation assay where insoluble fractions were loaded onto a dot blot apparatus (Bio-Rad) and blotted onto a 0.2 µm cellulose acetate membrane (Morozko et al., 2018). Membranes were blocked in StartingBlock™ (Invitrogen) or 5% milk in Tris-buffered saline with Tween 20 (TBST) (for fibrils) at room temperature and incubated in primary antibody overnight at 4°C. The following primary antibodies were used: anti-HTT (1:1000; MAB5492, Millipore), anti-Ubiquitin (1:1000; sc-8017, Santa Cruz Biotechnology) (Ochaba et al., 2016), anti-p62 (1:1000; BML-PW9860, Enzo) (Wong et al., 2015), and anti-GAPDH (1:2500; 1D4, Novus Biologicals) (Zhang et al., 2018). Insoluble fibrils were labelled with anti-HTT (1:1000; VB3130, Viva Biosciences) (Ochaba et al., 2016). Western blot and agarose gel electrophoresis data were quantified by measuring mean pixel intensity using Scion Image processing software. Detergent-soluble fraction lysates were normalized to house-keeping protein loading control (GAPDH) prior to statistical analysis. Detergent-insoluble and oligomeric protein were quantified as relative protein abundance as before (Morozko et al., 2018).
Quantitative PCR and NanoString
RNA was extracted and purified from frozen striatal tissue using a RNeasy® Plus Universal Mini Kit (73404, Qiagen). Quantitative PCR was performed as before (Morozko et al., 2018) with reverse transcription using the SuperScript® III First-strand synthesis system (Invitrogen) and the following primers for cDNA amplification: R6/2 transgene (F: CCGCTCAGGTTCTGCTTTTA, R: TGGAGGGACTTGAGGGACTC), Rplp0 (F: GCTTCGTGTTCACCAAGGAGGA, R: GTCCTAGACCAGTGTTCTGAGC). For nCounter© analysis, total RNA was diluted to 20 ng/µl and probed using a mouse nCounter© PanCancer Immune Profiling Panel (NS_Mm_CancerImm_C3400, NanoString Technologies) profiling ∼700 immunology-related mouse genes. Counts for target genes were normalized to the best fitting house-keeping genes as determined by nSolver software. The WGCNA package was used to evaluate the quality of reads, as well as identify and remove appropriate outliers, based on standard deviation within normalized expression values. Principal component analysis (PCA) plots were generated using plot3D. Negative binomial linear regression analysis was performed using EdgeR, DESeq, and Limma packages to generate false discovery rate (FDR) and log-fold change values. Top significant genes are displayed as a volcano plot constructed using GLimma, ggplot2, and EnhancedVolcano (FDR < 0.10, LogFC > 1).
RNA-sequencing and analysis
For mRNA-seq, RNA from the same striatal samples used for NanoString was analysed, and as such was previously extracted using the RNeasy® Plus Universal Mini Kit (73404, Qiagen). Total RNA was monitored for quality control using the Agilent Bioanalyzer Nano RNA chip (Agilent) and NanoDrop™ absorbance ratios for 260/280 nm and 260/230 nm. Library construction was performed according to the Illumina TruSeq mRNA stranded protocol. The input quantity for total RNA was 700 ng and mRNA was enriched using oligo dT magnetic beads. The enriched mRNA was chemically fragmented for 3 min. First strand synthesis used random primers and reverse transcriptase to make cDNA. After second strand synthesis the double stranded cDNA was cleaned using AMPure XP beads (Beckman Coulter) and the cDNA was end repaired and then the 3′ ends were adenylated. Illumina barcoded adapters were ligated on the ends and the adapter ligated fragments were enriched by nine cycles of PCR. The resulting libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high sensitivity chip (Agilent). The concentrations for the libraries were normalized and then multiplexed together. The concentration used for clustering the flowcell was 200 pM. The multiplexed libraries were sequenced on one lane using PE 100 cycles chemistry for the HiSeq 4000 (Illumina). The version of HiSeq control software was HCS 3.3.76 with real time analysis software, RTA 2.7.6.
For read alignment and expression quantification, pair-end RNA-seq reads were first filtered for ribosomal RNA using SortMeRNA v.2.1b followed by trimmomatic v.0.35 to correct for overrepresented sequences via quality trimming and adapter removal. Paired-end reads were then aligned using STAR v.2.5.1b (Dobin et al., 2013) with the options (–outFilterMismatchNmax 10 –outFilterMismatchNoverReadLmax 1 –outFilterMultimapNmax 10). Rsubread was used to generate feature counts. Gene expression was measured using Limma, edgeR, and org.Mm.eg.db packages with expression values normalized into RPKM (reads per kilobase of transcript per million mapped reads). Differential expression analysis: libraries with uniquely mapping percentages higher than 80% were considered to be of good quality and kept for downstream analysis. Transcripts with expression RPKM ≥1 in at least three samples were collected for subsequent analysis. Differential expression analysis was performed by using Limma, edgeR, and org.Mm.eg.dbt (Robinson et al., 2010). Differentially expressed genes (DEGs) were selected by using FDR ≤ 0.05. Top significant genes are displayed as a volcano plot constructed using GLimma, ggplot2, and EnhancedVolcano (FDR ≤ 0.05, LogFC >1). PCA plots were generated using plot3D. Gene enrichment pathway analysis was performed using Ingenuity Pathway Analysis (IPA, Qiagen) of DEGs detected at FDR ≤ 0.10, and predicted upstream regulators are reported with activation z-scores and P-values.
Data analysis and statistics
Statistical analysis was performed with Prism GraphPad (v.8.0.1). To assess linear relationships, the two-tailed Pearson correlation test was used. To compare one variable, the two-tailed unpaired Student’s t-test was used. To compare two variables, two-way ANOVA with Tukey’s post hoc test was used. To compare two variables over multiple time points (Rotarod, grip strength), the MIXED procedure of the Statistical Analysis Systems software (SAS Institute Inc.) and subsequent post hoc contrasts were used to examine interactions as before (Elmore et al., 2015, 2018). All bar graphs are represented as means ± standard error of the mean (SEM) with individual sample values overlain. Significance is expressed as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001
Data availability
Gene expression data are deposited in GEO (ascension number GSE136158), and all RPKM values can be viewed and searched for at http://rnaseq.mind.uci.edu/green/R62_PLX/gene_search.php. All other data are available upon reasonable request.
Results
CSF1Ri via PLX3397 depletes microglia in R6/2 mice
To evaluate the dependency of microglia on CSF1R signalling in R6/2 mice—a rapidly progressing mouse model expressing an expanded repeat exon 1 transgene (Mangiarini et al., 1996)—the CSF1R/c-Kit inhibitor PLX3397 (275 mg/kg) or vehicle was provided in chow to R6/2 mice and non-transgenic littermates for 7 days to generate four experimental groups: non-transgenic + vehicle (control), non-transgenic + PLX3397 (7d PLX3397), R6/2 + vehicle (R6/2), and R6/2 + PLX3397 (R6/2 + 7d PLX3397). IHC with the myeloid marker IBA1 revealed a significant increase in striatal R6/2 microglial densities at 7 weeks of age (P < 0.001) that was not seen in the diseased somatosensory cortex at this time point (Supplementary Fig. 1). Importantly, 7 days of PLX3397 delivered at 6 weeks significantly depleted striatal and cortical microglia from R6/2 (P < 0.0001, P < 0.0001, respectively) and non-transgenic mice (P < 0.001, P < 0.001, respectively) to comparable densities of surviving cells (P > 0.05), suggesting that microglial mHTT expression does not appear to alter their dependence on CSF1R signalling for survival.
CSF1Ri via PLX3397 ameliorates grip strength and novel object recognition deficits in R6/2 mice
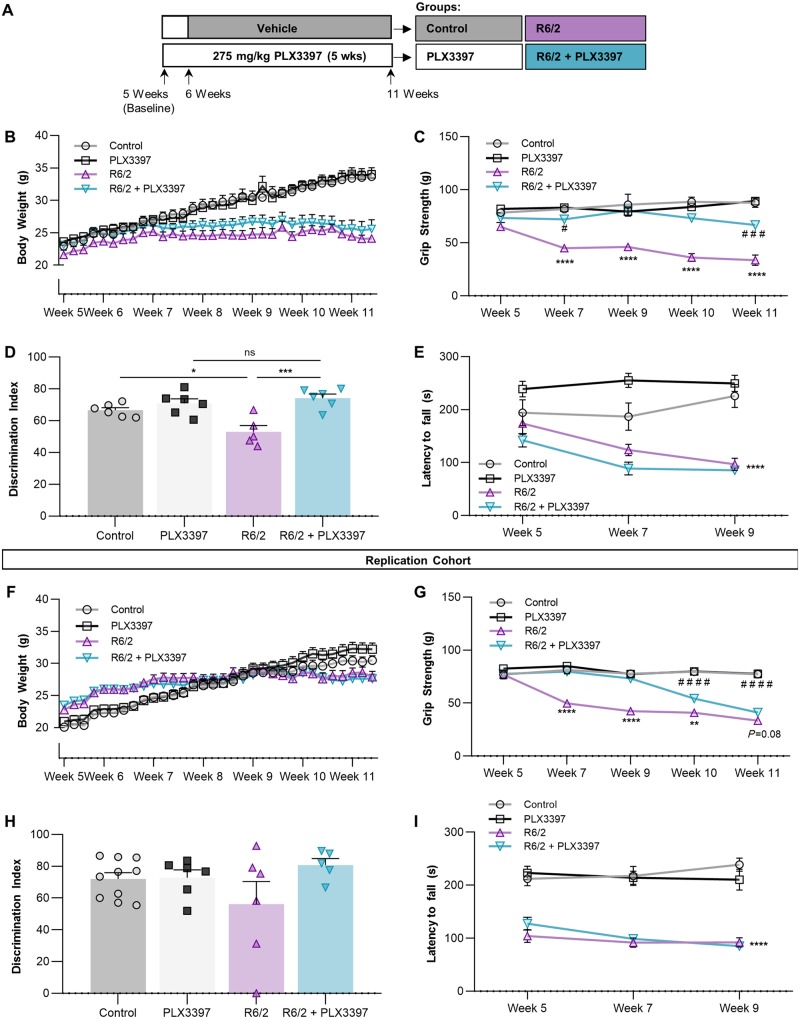
Having demonstrated that microglia could be eliminated in R6/2 mice, we next set out to investigate the roles these cells play in disease progression. Following baseline behavioural measurements, 6-week-old R6/2 mice and non-transgenic littermates were placed on diet containing 275 mg/kg PLX3397 or vehicle (n = 6/group) until sacrifice at 11 weeks (Fig. 1A). CAG repeats were confirmed at a range of 126–130 repeats in R6/2 mice. While treatment had no effect on body weight, which displayed the stereotypical plateauing in R6/2 mice at ∼7 weeks (Fig. 1B), we found that over the duration of CSF1Ri, Huntington’s disease-related grip strength deficits were prevented up to Week 11, with significant improvement remaining even at that time point (Fig. 1C). While inhibitor treatment could potentially affect muscle macrophages in a way that reduces skeletal muscle mHTT inclusions (Moffitt et al., 2009) or transcriptional alterations (Strand et al., 2005), PLX3397 has not been reported to reduce peripheral macrophage numbers (Elmore et al., 2014; Mok et al., 2014; Szalay et al., 2016). Cognitively, treatment prevented R6/2 deficits in novel object recognition at 11 weeks (Fig. 1D) while having no effect on controls, consistent with previous measures of cognition following CSF1Ri in healthy mice (Elmore et al., 2014). No groups differed statistically in average velocity or distance travelled by open field analysis (data not shown) indicating that these differences are due to effects on cognition. We did not observe any effect of treatment on Huntington’s disease-related Rotarod performance deficits (Fig. 1E). Finally, to verify cognitive and motor improvements with CSF1Ri in this model, these behavioural tasks were replicated in a second cohort of n = 10/group (Fig. 1F–I), producing similar results. The failure to achieve statistical significance in novel object recognition with this second cohort may be attributed to the rapid progression of the model, as many R6/2 mice were largely immobile/lethargic at Week 11 just prior to sacrifice, increasing the number of animals excluded during novel object training and consequently decreasing statistical power. Nonetheless, the similar cognitive and grip strength effects observed between cohorts suggests that PLX3397 may have therapeutic potential in Huntington’s disease, particularly in the case of grip strength.
Figure 1.
PLX3397 (CSF1Ri) ameliorates grip strength and object memory deficits in R6/2 mice. Following baseline measurements, 6-week-old non-transgenic or R6/2 mice were treated with vehicle (Control, R6/2) or 275 mg/kg PLX3397 (PLX3397, R6/2+PLX3397) for 5 weeks to attain microglial depletion through CSF1Ri. (A) Experimental design (n = 5–6/group). (B) Body weight from baseline to end of experiment displayed stereotypical R6/2 plateauing (n = 5–6/group). (C) R6/2+PLX3397 forelimb grip strength deficits were significantly reduced compared to R6/2 at every time point tested (P < 0.0001; MIXED procedure with post hoc contrasts; n = 5–6/group). (D) Performance in novel object recognition (NOR) test by R6/2+PLX3397 mice at Week 11 was significantly better than R6/2 mice (P < 0.001) and not different from PLX3397 controls (two-way ANOVA with Tukey’s post hoc test; n = 5–6/group). (E) Latency to fall from an accelerating Rotarod was not altered with treatment and displayed the stereotypical R6/2 genotype effect (P < 0.0001; MIXED procedure; n = 6/group). (F–I) Replication cohort for validation of behavioural outcomes using the same statistical analyses. (n = 8–10/group, except for novel object recognition where n = 5–10/group due to exclusion criteria from Day 1 acquisition period or Day 2 testing period.) Statistical significance is denoted by *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant. For grip strength, asterisk indicates R6/2 versus R6/2+PLX3397 and hash symbol indicates PLX3397 versus R6/2+PLX3397. Error bars indicate SEM.
R6/2 microglia display regional differences but are ubiquitously depleted with CSF1Ri
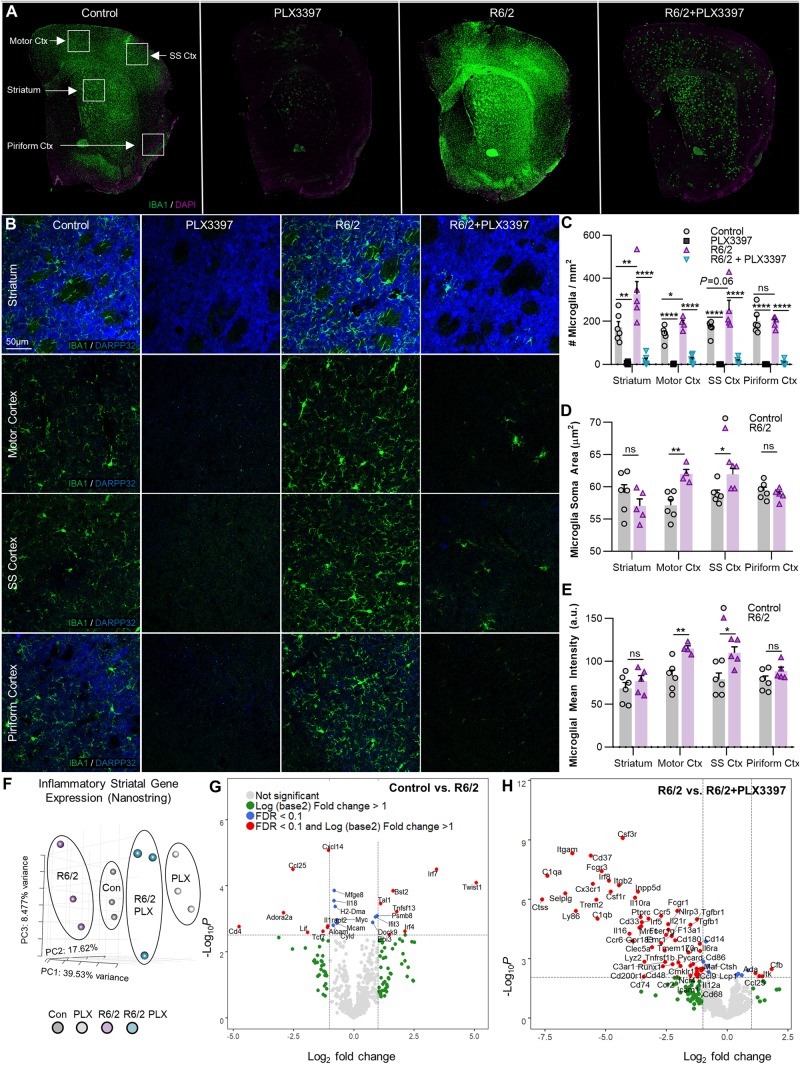
To assess the microglial response to disease, IHC with the myeloid marker IBA1 was performed on coronal brain sections from each group. Microglial number, cell body area, and IBA1 intensity were measured in the disease-relevant striatum as well as in multiple cortical regions (motor, somatosensory, and piriform cortex) (Fig. 2A). R6/2 mice displayed elevated microglial densities in the striatum (P < 0.01) and motor cortex (P < 0.05), in addition to a trending increase in the somatosensory cortex (P = 0.06; Fig. 2B and C). Microglial cell body area (Fig. 2D) and IBA1 intensity (Fig. 2E) were significantly increased only in the motor (P < 0.01, P < 0.01) and somatosensory cortices (P < 0.05, P < 0.05). Striatal microglia, although at greater densities, did not appear activated morphologically or by IBA1+ intensity, and were negative for the activation marker IB4 (data not shown). Together, this suggests that ‘classical’ microglial activation is not an overt feature of the R6/2 brain. Consistent with the data from 7 days of treatment, CSF1Ri significantly depleted microglial numbers in all tissue regions and in both genotypes as expected (Fig. 2C).
Figure 2.
CSF1Ri depletes striatal and cortical R6/2 microglia, which are increased in number but do not appear ‘classically’ activated. (A) Representative IBA1+ whole-brain and (B) Striatal and motor, somatosensory (SS), and piriform cortex (Ctx) images (×20) from non-transgenic and R6/2 mice treated with vehicle or PLX3397 confirmed R6/2 microgliosis and microglial depletion (n = 5–6/group). (C) Quantification of IBA1+ microglia revealed significant increases in the R6/2 striatum (P < 0.01) and motor cortex (P < 0.05), with a trending increase in the somatosensory cortex (P = 0.06) but no change in the piriform cortex compared to control. PLX3397 significantly depleted microglia in both genotypes and in all regions (two-way ANOVAs with Tukey’s post hoc test; n = 4–6/group). (D) Microglia increased in size with disease in the motor (P < 0.01) and somatosensory (P < 0.05) cortices but remained ramified and did not adopt an amoeboid morphology (two-tailed unpaired t-tests; n = 4–6/group). (E) IBA1+ intensity mirrored changes in cell size, with significant increases in motor (P < 0.01) and somatosensory (P < 0.05) cortices in R6/2 mice compared to control (two-tailed unpaired t-tests; n = 4–6/group). (F) Striatal inflammatory gene expression assessed with a NanoString immune panel revealed non-overlapping clustering of samples in each group by principal component analysis (PCA; n = 3/group). (G and H) Volcano plots of differentially expressed genes (DEGs) in R6/2 versus control striatum (G) indicated a lack of inflammatory transcription at an FDR of < 0.1, but (H) confirmed loss of the microglial gene signature with PLX3397 in R6/2 striatum (n = 3/group). Statistical significance is denoted by *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, ns = not significant. Error bars indicate SEM.
R6/2 striatal mRNA expression reveals dysregulated interferon signalling that is mitigated by CSF1Ri
A NanoString inflammatory panel (NS_Mm_CancerImm_C3400) was used to assess endpoint DEGs in striatal tissue from three mice per group. PCA of normalized gene counts revealed separate clustering of control and R6/2 tissue both prior to and following CSF1Ri (Fig. 2F). However, even at FDR < 0.1, few inflammation-related DEGs were found in R6/2 striatal tissue (Fig. 2G). In addition to a marked decrease in striatal adenosine A2a receptor (Adora2a), canonically downregulated in Huntington’s disease (Blum et al., 2018), we noted altered T-cell and interferon signalling (increased: Twist1, Ebi3, Tal1, Ccl19, Irf7, Irf4, Bst2, Ifit3, Psmb8; decreased: CD4, Alcam, Tcf7, Mcam, Ccl25, Il18), similar to recent reports of peripheral T-cell suppression in R6/2 mice (Lee et al., 2018). Surprisingly, we found no evidence of disease-related inflammation in terms of cytokine transcript expression (Il6, Il1b, Ifng, Tnfa) at the 11-week time point using this panel. DEGs in CSF1Ri-treated R6/2 mice largely reflected microglial depletion, in addition to partial reversals in T-cell transcript levels (Tal1, Ccl25) (Fig. 2H). As before (Najafi et al., 2018), CSF1Ri treatment in non-transgenic mice induced the expected loss of microglial gene signature from the brain transcriptome (data not shown).
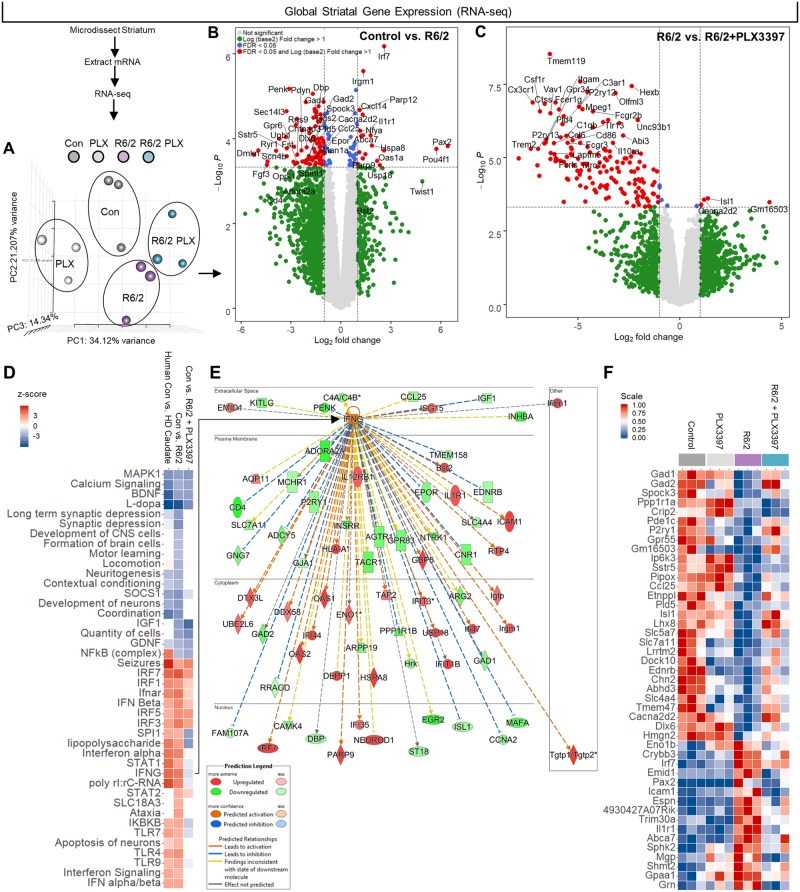
To broaden our scope of gene expression analysis, we performed RNA-seq on the same striatal extracts used for NanoString. All RPKM values can be searched and visualized at http://rnaseq.mind.uci.edu/green/R62_PLX/gene_search.php. We again observed distinct clustering of genotypes, but this time divergent effects of CSF1Ri on the R6/2 transcriptome were detected (Fig. 3A). At Week 11, striatal R6/2 transcription was considerably suppressed relative to controls (Fig. 3B) as has been described previously (Luthi-Carter et al., 2003; Hodges et al., 2006; Runne et al., 2008; Vashishtha et al., 2013). Among the downregulated genes were Gad1 and Gad2, which encode the enzymes that catalyse GABA synthesis (Lim et al., 2017), as well as the MSN voltage-gated sodium channel β4 subunit Scn4b (Oyama et al., 2006; Miyazaki et al., 2014), and the astrocyte glutamate transporter Slc7a11, all indicative of dysregulated excitatory/inhibitory balance. Microglial elimination was followed by tell-tale loss of microglial gene expression in control (not shown) and R6/2 mice (Fig. 3C). Among other genes, CSF1Ri prevented the disease-associated suppression of Isl1, a gene involved in motor neuron development (Liang et al., 2011) the conditional deletion of which results in striatal cholinergic interneuron loss (Elshatory and Gan, 2008), as well as Cacna2d2, mutations in which decrease Ca2+ channel subunit expression and drive epileptic seizures (Barclay et al., 2001; Pippucci et al., 2013). Importantly, this differential expression occurred in the absence of detectable changes in endogenous wild-type Htt transcript levels across groups. RNAseq DEGs were validated by and mirrored in the prior results from the more sensitive NanoString assay covering a subset of immune genes.
Figure 3.
RNA-seq and pathway analysis confirm lack of inflammatory transcriptional polarization in R6/2 striatum, except for a dysregulated interferon signature resembling the human Huntington’s disease caudate that is partially resolved following CSF1Ri. (A) PCA of striatal gene expression as determined by mRNA-seq confirmed similar non-overlapping clustering of control and R6/2 samples but a divergent effect of PLX3397 (n = 3/group). (B and C) Volcano plots of control versus R6/2 DEGs (B) confirmed an absence of inflammatory transcript upregulation in the R6/2 striatum and (C) a loss of microglial gene expression with PLX3397 (FDR ≤ 0.05; n = 3/group). (D) Pathway analysis (IPA) of 860 control versus R6/2 DEGs and 1380 control versus R6/2+PLX3397 DEGs (FDR ≤ 0.1; n = 3/group) revealed a distinct interferon signature and suppressed neuronal development and neuritogenesis pathways in the R6/2 striatum that closely resembled the human Huntington’s disease caudate, and which were partially resolved following PLX3397. Top significant predicted upstream regulators included HTT, IFNγ, l-DOPA, IRF7, and TRIM24, with associated activation z-scores of 2.151 (P < 8.31 × 10−22), 3.727 (P < 1.01 × 10−15), −5.654 (P < 1.19 × 10−20), 4.589 (P < 7.47 × 10−13), and −3.970 (P < 3.58 × 10−12), respectively. (E) IPA predicted IFNγ as the major cytokine upstream regulator in the R6/2 striatum, signalling that, along with IFNα, is no longer enriched in R6/2+PLX3397 striatum. (F) A heat map of control versus R6/2 DEGs (FDR < 0.1) that were reversed with treatment by unadjusted P-value (P < 0.05) to indicate potential mediators of beneficial PLX3397 effects (n = 3/group). All RPKM values can be searched and visualized at http://rnaseq.mind.uci.edu/green/R62_PLX/gene_search.php.
A growing number of non-viral diseases are characterized by the presence of a similar IFN transcriptional signature (Mostafavi et al., 2016) evident within the microglial transcriptome itself as upregulated interferon signalling gene sets in cells isolated from both an Alzheimer's disease-like model of neurodegeneration as well as ageing humans (Galatro et al., 2017; Mathys et al., 2017). We compared the activation z-scores of significantly overrepresented pathways in human control versus Huntington’s disease caudate [z-score overall = 41.23; gene expression data taken from Hodges et al., (2006)] to those in control versus R6/2 striatal tissue and found concordance between the mouse model and the human disease (Fig. 3D). A dysregulated IFN signature was found in both the murine and human manifestation of disease (Fig. 3D), facets of which were ameliorated with CSF1Ri-mediated microglial elimination as indicated by a resolution of IFNα and IFNγ, but not IFNβ, gene enrichment in control versus R6/2 + PLX3397 samples. This resolution was accompanied by a concurrent loss of IFN-involved toll-like receptor signalling (Tlr4, Tlr7, Tlr9) as well as downstream IFN effector pathway deactivation (Stat1). Pathway enrichment predicted HTT as the most significant upstream regulator of R6/2 DEGs (z-score: 2.151, P < 8.31 × 10−22), in addition to IFNγ (z-score: 3.727, P < 1.01 × 10−15), l-DOPA (z-score: −5.654, P < 1.19 × 10−20), the type I IFN regulator IRF7 (z-score: 4.589, P < 7.47 × 10−13), and TRIM24 (z-score: −3.970, P < 3.58 × 10−12). Thus, pathway analysis of the R6/2 versus control brain predicted IFNγ as the primary cytokine regulator of disease DEGs (Fig. 3E), a potent stimulator of microglial activation (Monteiro et al., 2017). Interestingly, Usp18 was markedly increased in Huntington’s disease, which inhibits microglial activation by specifically blocking IFN-induced transcription (Goldmann et al., 2015), suggesting the mobilization of relevant compensatory mechanisms in the R6/2 striatum.
Microglial elimination normalized several additional overrepresented human/murine Huntington’s disease pathways, i.e. impaired neuronal development, neuritogenesis, contextual conditioning, and coordination. These impairments occurred in the absence of an extensive inflammatory transcriptional signature. Indeed, the pro-inflammatory NFκβ pathway was upregulated in the human Huntington’s disease caudate, but not R6/2, where inflammatory transcripts such as Il6 and Il1b were in fact below detection limits, indicating that this model likely does not fully recapitulate the neuroinflammation observed in end-stage human Huntington’s disease (Crotti and Glass, 2015; Rodrigues et al., 2016; Yang et al., 2017). Accordingly, FDR and unadjusted P-values (FDR < 0.1 for control versus R6/2 plus P < 0.05 for R6/2 versus R6/2 + PLX3397) in addition to hierarchical clustering were used to identify genes that were changed in R6/2 mice but prevented with treatment to identify potential non-immune targets of CSF1Ri-mediated microglial elimination (Fig. 3F).
Microglial depletion reduces mHTT accumulation and disease-related protein clearance pathway dysregulation
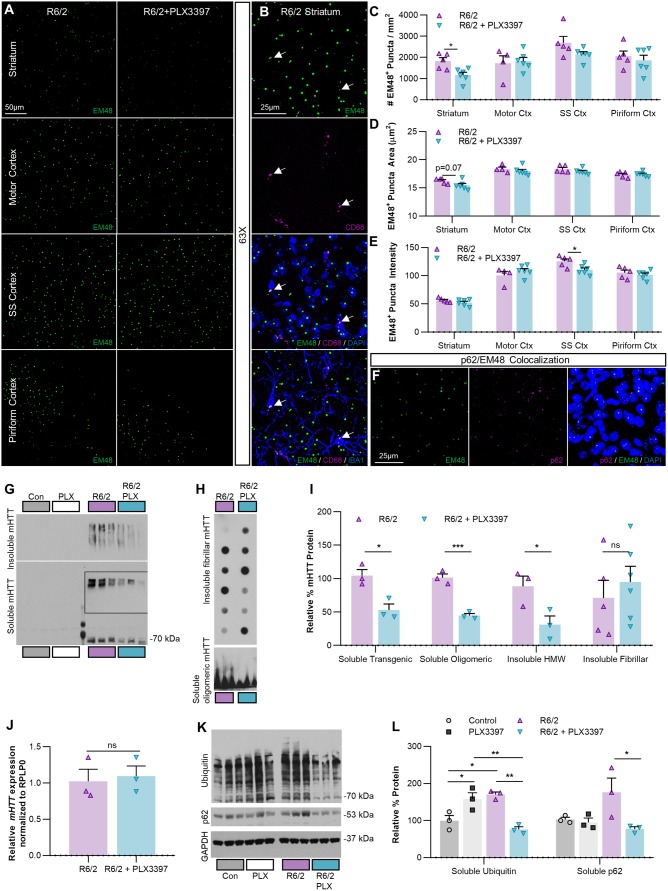
To investigate whether sustained CSF1R inhibition altered the accumulation of mHTT, we first examined intranuclear mHTT inclusion bodies (EM48) in the same regions assessed for microgliosis (Fig. 4A). Nuclear inclusion bodies were rarely seen in microglia, consistent with previous studies finding that they are predominantly neuronal (Jansen et al., 2017). Instead, when we found EM48+ staining in microglia it was localized to cytoplasmic CD68+ lysosomal granules (Fig. 4B, arrows). Following CSF1Ri the total number of EM48-labelled inclusion bodies was modestly reduced in the striatum (∼25%; P < 0.05), consistent with the results from a recent study suppressing microglial galectin 3 (Lgals3) in Huntington’s disease models (Siew et al., 2019), while EM48 intensity was significantly reduced in the somatosensory cortex (P < 0.05) (Fig. 4C–E). We also confirmed previous reports of co-localization of the autophagic protein p62 with EM48+ inclusion bodies in R6/2 mice (Kurosawa et al., 2015) (Fig. 4F). Finally, although PLX3397 is more selective for CSF1R, it can also inhibit c-Kit and, at higher concentrations, Flt3 receptor tyrosine kinases [IC50 of 0.013, 0.027, and 0.16 µM, respectively (Tap et al., 2015)]. Staining for c-Kit and Flt3 revealed no co-localization with neurons or microglia (Supplementary Figs 2 and 3), and no significant differences were detected in their transcript levels (Kit, Flt3) by RNAseq across groups, supporting the notion that the effects seen here are due specifically to the inhibition of microglial CSF1R and subsequent microglial depletion.
Figure 4.
CSF1Ri-mediated microglial depletion reduces striatal R6/2 mHTT accumulation and protein clearance system dysregulation. (A) Representative ×20 images of R6/2 and R6/2+PLX3397 striatal and cortical EM48+ nuclear mHTT inclusions in 11-week-old R6/2 mice (n = 4–6/group). (B) Representative ×63 images of microglial EM48+ mHTT. When it occurs, it is non-nuclear and co-localizes with CD68+ lysosomes. (C–E) Quantification of R6/2 EM48+ nuclear puncta, which are (C) significantly reduced in number in the striatum with PLX3397 (P < 0.05), where they (D) also display a trending decrease in area (P = 0.07). (E) Treatment also significantly reduced mean EM48+ intensity (P < 0.05) in the somatosensory cortex (two-tailed unpaired t-tests; n = 4–6/group). (F) Representative ×63 images of p62+/EM48+ nuclear co-localization in R6/2 striatum. (G and H) Western blots of striatal homogenates separated into detergent-soluble and insoluble fractions and probed for mHTT protein species (n = 3–6/group). (I) Quantification of immunoblotted mHTT species revealed that R6/2+PLX3397 mice had significantly reduced soluble transgenic (P < 0.05), soluble oligomeric (P < 0.001), and insoluble high molecular weight (HMW) mHTT (P < 0.05) in the striatum compared to R6/2 (two-tailed unpaired t-tests; n = 3–6/group). (J) Quantitative PCR of striatal R6/2 RNA indicates that PLX3397 did not change mHTT expression (two-tailed unpaired t-test; n = 3/group). (K) Western blots of striatal homogenates immunoblotted for soluble ubiquitinated proteins, p62, and the housekeeping protein GAPDH (n = 3/group). (L) The accumulation of ubiquitinated species was significantly increased in R6/2 striatum compared to control (P < 0.05) and this was prevented with PLX3397 (P < 0.01); interestingly, PLX3397 also significantly increased ubiquitination in control mice (P < 0.05). PLX3397 also induced a significant reduction in R6/2 striatal p62 levels (P < 0.05) (two-way ANOVAs with Tukey’s post hoc test; n = 3/group). Statistical significance is denoted by *P < 0.05, **P < 0.01, ***P < 0.001, ns = not significant. Error bars indicate SEM. Con = control; Ctx = cortex; SS = somatosensory.
In addition to nuclear inclusions, mHTT fragments build up in multiple detergent-soluble and -insoluble biochemical species of increasing complexity that can be resolved by fractionation (Ochaba et al., 2018). To assess mHTT flux accompanying the observed changes in inclusion body number following CSF1Ri, we immunoblotted for detergent-soluble and insoluble mHTT in striatal homogenates from R6/2 mice as described (Ochaba et al., 2016) (Fig. 4G–I). CSF1Ri and microglial depletion significantly reduced both monomeric (P < 0.05) and oligomeric (P < 0.001) mHTT in the detergent-soluble fraction, consisting of largely cytoplasmic proteins (O'Rourke et al., 2013). In the detergent-insoluble fraction, containing high molecular weight (HMW) mHTT multimers, insoluble fibrils, and nuclear proteins, we found that CSF1R reduced HMW (P < 0.05) but not fibrillar mHTT species. While microglia do express mutant HTT (mHTT), the paucity of nuclear inclusions in these cells (Jansen et al., 2017) compared to the more abundant neurons—potentially due to the former’s greater proteosomal and autophagic capacity (Yang et al., 2017)—suggests that elimination of microglia reduces neuronal mHTT accumulation. To confirm that mHTT changes did not stem from transcriptional suppression or ‘gene knockdown’ via microglial elimination, we performed qPCR of the mutant transgene in the striatum and found that CSF1Ri did not alter mHTT expression (Fig. 4J), thus suggesting that changes in protein level were independent of transcript abundance.
Ubiquitin- and autophagy-based protein clearance systems are highly impaired in Huntington’s disease, as evidenced by a striking build-up of ubiquitinated species in patient brain and mouse models (Bennett et al., 2007; Ochaba et al., 2016) and an upregulation of the autophagy marker p62 (Lee et al., 2012). Immunoblotting for ubiquitinated proteins in the detergent-soluble striatal fraction confirmed significantly greater ubiquitination in R6/2 mice (P < 0.05), but also, surprisingly, that CSF1Ri completely prevented this phenotype (Fig. 4K and L). Striatal p62 was significantly reduced in CSF1Ri-treated R6/2 mice (P < 0.05), although levels were not statistically higher in these compared to controls (Fig. 4L). Together, these data suggest that microglial depletion via CSF1Ri slows the accumulation of multiple species of mHTT in the R6/2 striatum by at least partially reversing dysfunctional protein clearance systems. Interestingly, the removal of microglia from control mice enhanced ubiquitination (P < 0.05), suggesting a potential role for these myeloid cells in homeostatic non-microglial protein clearance.
Microglial depletion with CSF1Ri prevents R6/2 striatal atrophy
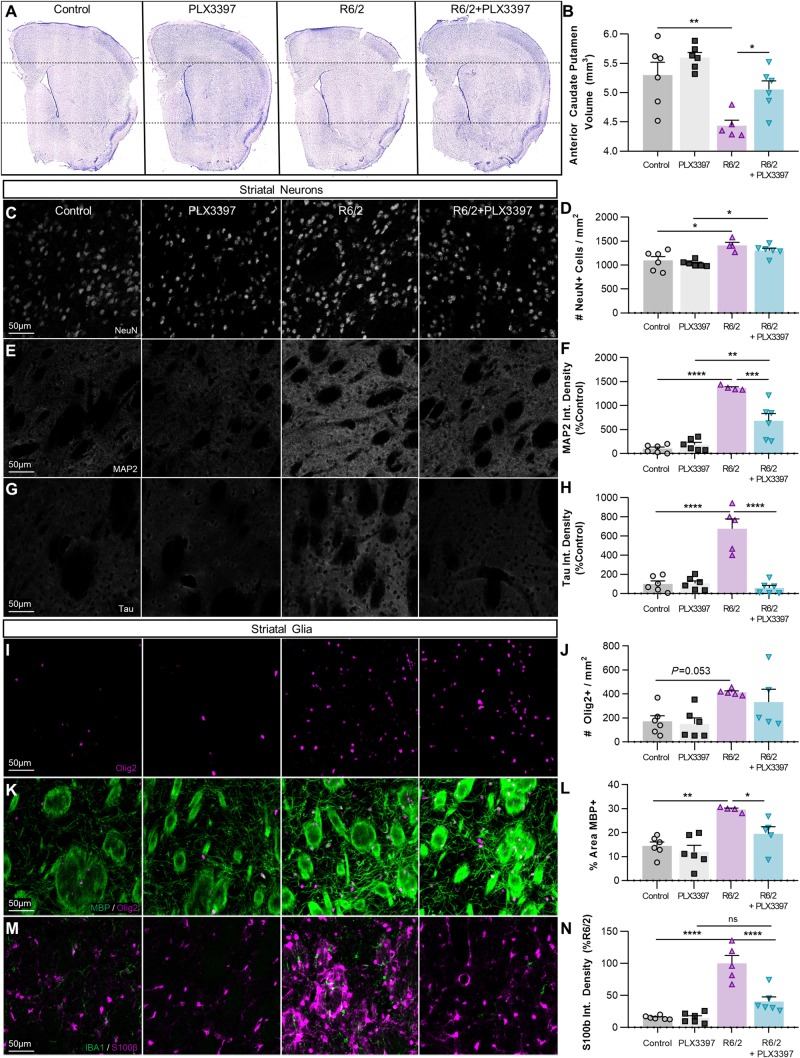
Despite minimal neuronal loss even in late stages of the disease (Dodds et al., 2014), R6/2 mice display marked striatal atrophy (Hansson et al., 1999; Li et al., 2005) similar to humans. Mutant HTT expression in R6/2 microglia effectively ‘primes’ the cell by increasing expression of the myeloid transcription factor Pu.1 (Crotti et al., 2014), a positive regulator of microglial phagocytosis (Smith et al., 2013). We hypothesized that enhanced phagocytic capability of mHTT-containing microglia could mediate remodelling of striatal neurites and/or ECM in Huntington’s disease. We performed double-blinded stereology of the anterior caudoputamen, representing the dorsal striatum, using six coronal sections collected at 240-µm intervals from CSF1Ri- and vehicle-treated control and R6/2 mice (Fig. 5A). Surprisingly, we found that microglial elimination via CSF1Ri completely prevented the decrease in striatal volume observed in R6/2 mice (Fig. 5B) independent of changes in neuronal density (Fig. 5C and D). In fact, we found significantly greater NeuN+ cell density in the Huntington’s disease striatum (P < 0.05) that did not change with treatment.
Figure 5.
PLX3397 prevents R6/2 striatal volume loss, neurite abnormalities, and astrogliosis. (A) Representative images of cresyl-violet stained coronal sections used for stereological assessment of striatal volume (n = 5–6/group, six sections/mouse separated by 240-μm intervals, spanning the anterior caudoputamen). (B) Quantification of stereological analysis reveals a significant reduction in R6/2 striatal volume versus control (P < 0.01) that is prevented with PLX3397 (P < 0.01) (two-way ANOVA with Tukey’s post hoc test; n = 5–6/group). (C–H) Investigation of striatal neurons revealed increased NeuN+ density (×20; C and D) in R6/2 and R6/2+PLX3397 mice compared to respective controls (P < 0.05) (two-way ANOVA with Tukey’s post hoc test; n = 4–6/group). Integrated MAP2+ (×20; E and F) and tau+ (×20; G and H) signal density, staining for microtubule-associated proteins localized to the dendrites and axons respectively, were significantly increased in R6/2 striatum versus control (P < 0.0001, P < 0.0001) and this was significantly prevented with PLX3397 (P < 0.001, P < 0.0001) (two-way ANOVAs with Tukey’s post hoc test; n = 4–6/group). (I–N) Analysis of striatal glia revealed a significant R6/2 genotype effect (P < 0.01) on Olig2+ number (×20; I and J) coincident with a significant increase in R6/2 myelinated (MBP+ ×20; K and L) white matter area coverage (P < 0.01) that is normalized with PLX3397 (P < 0.05) (two-way ANOVAs with Tukey’s post hoc test; n = 4–6/group). Quantification of integrated S100β+ signal density (×20; M and N) revealed extensive astrogliosis in the R6/2 striatum (P < 0.0001) that was prevented with PLX3397 (P < 0.0001) (two-way ANOVA with Tukey’s post hoc test; n = 5–6/group). Significance is denoted by *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Error bars indicate SEM.
To understand the structural relationship between the neural components of the diseased striatum, we next examined neuropil by staining for the type II microtubule-associated proteins MAP2 and tau, localized to neuronal dendrites and axons, respectively (Zhang and Dong, 2012). Both proteins stabilize microtubules critical for autophagosome transport (Kast and Dominguez, 2017) especially in neurons where distal autophagosomes travel long distances to fuse with soma-localized lysosomes (La Spada and Taylor, 2010). Furthermore, faulty splicing in Huntington’s disease increases total tau and the juvenile form of MAP2 (Fernandez-Nogales et al., 2016), which retracts from the dendrites towards the neuronal cell body in Huntington’s disease patients (Cabrera and Lucas, 2017), together promoting dendritic and striatal atrophy. We found significant elevations in the integrated densities of MAP2 (P < 0.0001) and tau (P < 0.0001) fluorescent signal in the R6/2 striatum (Fig 5E–H), alterations that were either partially (MAP2) or entirely (tau) normalized with CSF1Ri.
Staining for MBP revealed increased striatal white matter density with disease (P < 0.01), concordant with a significant effect of the Huntington’s disease genotype on Olig2+ oligodendrocyte lineage cell number (P < 0.01; Fig. 5I–L), which are reported to be uniquely proliferative in the R6/2 striatum (McCollum et al., 2013). While oligodendrocyte lineage numbers were not attenuated, CSF1Ri returned white matter densities to control levels in R6/2 mice. Furthermore, analysis of striatal astrocytes revealed marked disease-related astrogliosis as measured by integrated S100β signal density (P < 0.0001; Fig. 5M and N). This was prevented with CSF1Ri and microglial depletion, in line with the increasingly reported role of microglia in directing astrocyte reactivity (Liddelow et al., 2017; Rothhammer et al., 2018; Yun et al., 2018; Gibson et al., 2019). Although seemingly in juxtaposition to one another, the enhanced white matter and neuropil density coincident with striatal volume loss in R6/2 mice may stem from impaired neurite outgrowth (Mehta et al., 2018) or disrupted synaptic connections (Ferrante et al., 1991; Lim et al., 2017) in Huntington’s disease, consistent with the downregulated neuronal and synaptic development pathways seen in this study. However, ECM degradation could also contribute to volume loss, and so we aimed next to extend our structural findings beyond neuronal cell bodies and processes to the extracellular milieu.
CSF1Ri reduces global ECM proteoglycan deposition in the R6/2 brain
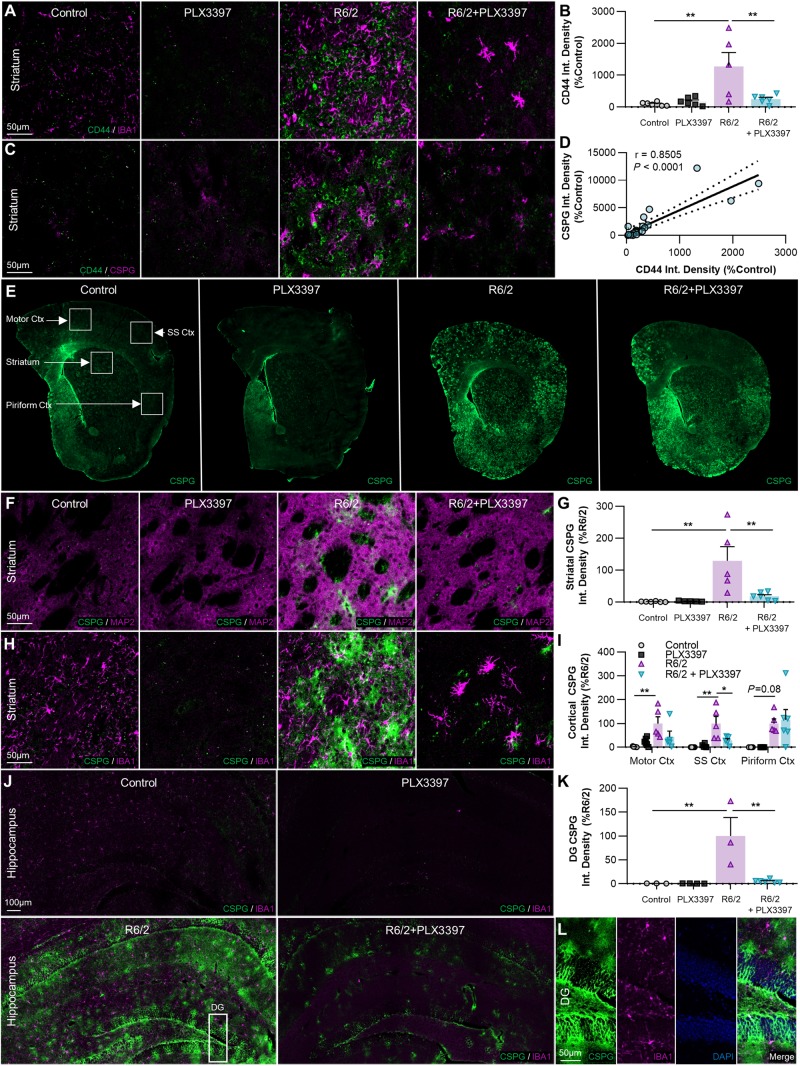
Cell-matrix adhesion in the brain is mediated largely by the transmembrane receptor CD44, which binds the primary component of parenchymal ECM, the glycosaminoglycan (GAG; polysaccharides consisting of a repeating disaccharide unit) hyaluronan, as well as other matrix constituents such as glycoproteins and proteoglycans (Dzwonek and Wilczynski, 2015). CD44 immunoreactivity was robustly enhanced in the R6/2 striatum (P < 0.01) and this was prevented with treatment (Fig. 6A and B). CD44 expression in the adult brain is primarily confined to astrocytes and activated microglia (Matsumoto et al., 2012), and while we did not observe co-localization of CD44 with the latter, microglia have been shown to regulate astrocytic expression of the receptor (Liddelow et al., 2017).
Figure 6.
Extracellular CSPGs accumulate in the R6/2 striatum, cortex, and hippocampus, but are reduced with PLX3397. (A–D) Non-microglial expression of the cell-ECM receptor CSPG8/CD44 (×20 with IBA1+A, same images with pan-CSPG+C) is significantly increased in R6/2 striatum (B) compared to control and R6/2+PLX3397 (P < 0.01, P < 0.01) (two-way ANOVA with Tukey’s post hoc test; n = 5–6/group), and correlated with CSPG accumulation (D) across all samples (P < 0.0001, r = 0.8505) (Pearson correlation test; n = 23). (E–K) Representative whole-brain (E), ×20 striatal (with MAP2+F, same images with IBA1+H), and ×20 hippocampal stitched (J) images of CSPG+ accumulation, quantified in the striatum (G), cortex (I) and dentate gyrus (DG) of the hippocampus (K) as integrated signal density (n = 5–6/group, except dentate gyrus where n = 3–6/group). Significant increases compared to control were found in R6/2 striatum (P < 0.01), motor cortex (P < 0.01), somatosensory cortex (P < 0.01), and dentate gyrus (P < 0.01), along with a trending increase in the piriform cortex (P = 0.08) (two-way ANOVAs with Tukey’s post hoc test). Interestingly, disease-related accumulation was significantly reduced with PLX3397 in the striatum (P < 0.01), somatosensory cortex (P < 0.05), and dentate gyrus (P < 0.01) (two-way ANOVAs with Tukey’s post hoc test; n = 5–6/group, except dentate gyrus where n = 3–6/group). (L) Inset of dentate gyrus from R6/2 hippocampus (J) showing CSPG+ staining in all layers. Statistical significance is denoted by *P < 0.05, **P < 0.01. Error bars indicate SEM.
CD44 is a one of a family of mostly secreted molecules, termed CSPGs, which are involved in neural development and glial scar formation, due largely in part to their role as negative guidance cues in axon growth (Ohtake et al., 2016) as well as their ability to form specialized ECM structures critical to plasticity (Yutsudo and Kitagawa, 2015; Tewari et al., 2018). CSPGs consist of a core protein covalently bound to one or more sulphated GAG sidechains, structurally related to the unsulphated GAG hyaluronan coexisting in the ECM, and are primarily produced by reactive astrocytes (Silver and Miller, 2004; Yu et al., 2012).
We therefore wondered whether other members of the CSPG family followed a similar expression pattern. Using the CS-56 antibody targeting CSPG-GAG sidechains, we performed a pan-CSPG stain and found that global CSPG signal varied directly with CD44 expression, regardless of experimental group (Pearson correlation coefficient r = 0.8505, P < 0.0001; Fig. 6C and D). Furthermore, we observed a striking and ubiquitous accumulation of extracellular CSPG throughout the R6/2 brain, which we quantified in the striatum, cortex, and dentate gyrus of the hippocampus (Fig. 6E–K). There was a significant (P < 0.01; striatum, motor cortex, somatosensory cortex, dentate gyrus) or trending increase (P = 0.08; piriform cortex) in CSPG signal in every region examined. In appearance, the diffuse patches of elevated CS-56 staining often resembled ‘dandelion clock-like structure’ (DACS), extracellular CSPG lattices reported to form around neurons at the end of the developmental critical period (Hayashi et al., 2007) as well as following kainic acid-induced seizure (Yutsudo and Kitagawa, 2015), and these deposits sometimes localized to MAP2-negative striatal fibre tracts (Fig. 6F, R6/2). The depletion of microglia with CSF1Ri significantly reduced CSPG accumulation to control levels in the striatum (P < 0.01), somatosensory cortex (P < 0.05), and dentate gyrus (P < 0.01), supporting the idea that microglia can directly or indirectly (i.e. through regulation of astrocyte reactivity) mediate the aberrant deposition of CSPGs into the extracellular space.
CSF1Ri-driven microglial elimination ubiquitously increases the density of ECM PNNs, which are degraded in the R6/2 brain
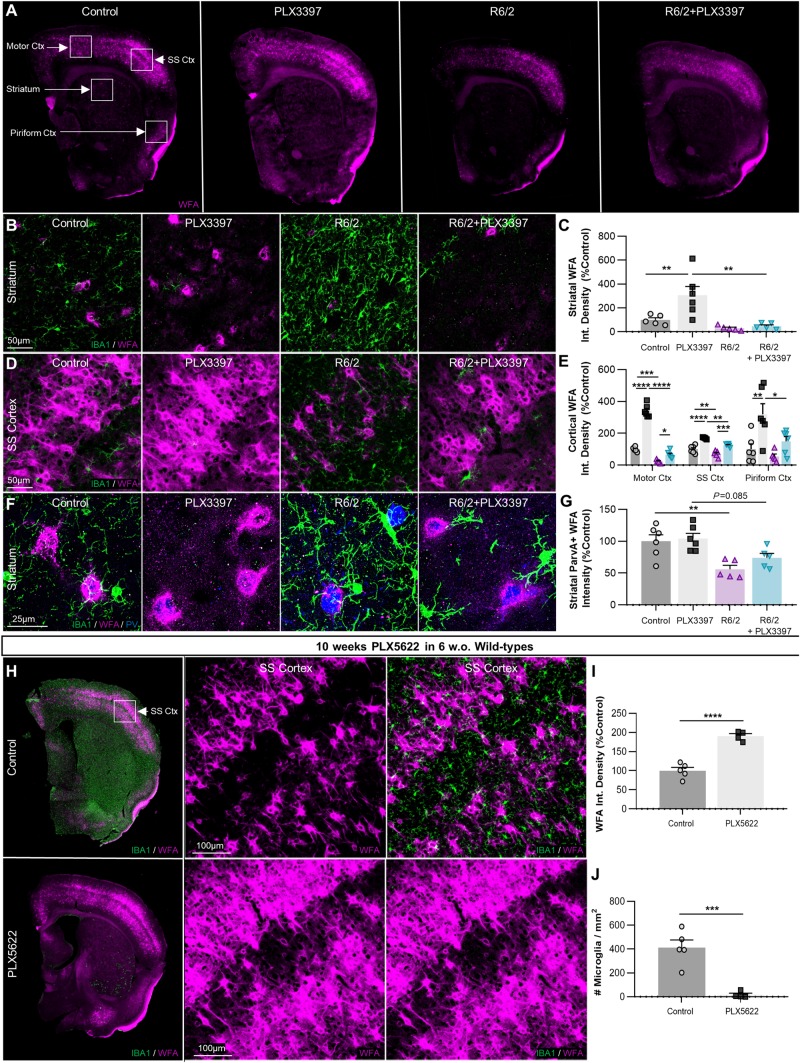
Depending on side chain sulphation pattern, CSPGs can also be preferentially incorporated into conventional PNNs (Fawcett et al., 2019) where they form a reticular dendrosomatic structure around largely parvalbumin-expressing interneurons (Miyata et al., 2012; Yutsudo and Kitagawa, 2015) that is thought to ‘lock’ synapses in place during development. Parvalbumin interneurons are reduced in number by ∼75% in the anterior caudate of patients with Huntington’s disease (Reiner et al., 2013). Furthermore, it is now known that PNN degradation impairs the inhibitory firing of GABAergic interneurons, thus contributing to tumour-associated seizures (Tewari et al., 2018) akin to those observed in R6/2 mice. Therefore, to investigate disease-associated changes in another relevant CSPG-based ECM structure, PNNs, we used the canonical lectin marker WFA to examine the same striatal and cortical regions where we observed increased CS-56 immunolabelling in the R6/2 brain (Fig. 7A). Of the regions examined, PNNs were always most abundant in the somatosensory cortex, where, along with the motor cortex, they were significantly decreased in R6/2 mice (P < 0.01 and 0.001, respectively; Fig. 7D and E), with trends to reduction seen in regions of lower PNN density and intensity (striatum, piriform cortex; Fig. 7B, C and E). Importantly, parvalbumin-expressing interneurons in the Huntington’s disease striatum displayed a significant reduction (P < 0.01) in mean WFA+ co-staining intensity (Fig. 7F and G) suggesting degradation of encapsulating PNNs.
Figure 7.
The density of CSPG-containing PNNs is reduced in R6/2 mice and this is prevented with microglial depletion, which also ubiquitously enhances PNN formation in healthy controls. (A) Representative whole-brain images of Wisteria floribunda agglutinin (WFA) lectin staining of PNNs (n = 5–6/group). (B–E) Striatal and cortical images (×20) of WFA+ PNN staining (with IBA1+, B and D) quantified as integrated density (C and E) revealed significant reductions in the motor (P < 0.001) and somatosensory (P < 0.01) cortices, which were restored by microglial depletion with PLX3397 (P < 0.05, P < 0.001) (two-way ANOVAs with Tukey’s post hoc test; n = 5–6/group). Surprisingly, PLX3397 in healthy non-transgenic controls significantly increased WFA+ PNN density in every region examined: striatum (P < 0.01), motor cortex (P < 0.0001), somatosensory cortex (P < 0.0001), and piriform cortex (P < 0.01) (two-way ANOVAs with Tukey’s post hoc test; n = 5–6/group). Quantification of mean WFA+ intensity co-localizing with striatal parvalbumin-expressing GABAergic interneurons (×63; F and G) revealed significant (P < 0.01) and trending (P = 0.085) reductions in R6/2 and R6/2+PLX3397 mice respectively, indicating loss of interneuron-associated PNN integrity (two-way ANOVA with Tukey's post hoc test; n = 5–6/group). (H) Representative whole-brain and ×20 cortical images of WFA+ PNN staining in wild-type mice treated for 10 weeks (from 6 weeks) with 1200 mg/kg PLX5622 in chow (n = 4–5/group). Analysis of PNN density as integrated WFA+ signal in the somatosensory cortex (I) revealed a significant elevation with PLX5622 (P < 0.0001) that occurred alongside the CSF1Ri-induced loss of microglia (J) (P < 0.001) (two-tailed unpaired t-tests; n = 4–5/group). Statistical significance is denoted by *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Error bars indicate SEM. Ctx = cortex; SS = somatosensory.
IBA1+ microglial processes were frequently observed proximal to and in contact with the surface of PNNs in both control and R6/2 brains (Fig. 7F). Indeed, it has been postulated that microglia mediate the degradation of PNNs in certain disease states, likely through enhanced production of extracellular proteases (Franklin et al., 2008; Sandvig et al., 2018). Microglial elimination with CSF1Ri prevented the disease-associated PNN loss observed in the somatosensory and motor cortices (P < 0.001 and 0.05, respectively), regions where microglia had adopted a more activated phenotype in R6/2 mice as measured by cell size and IBA1+ intensity. Remarkably, the depletion of microglia from naïve non-transgenic mice significantly and dramatically elevated PNN densities in every brain region examined.
Finally, to confirm that the robust enhancement of PNNs in wild-type mice are due solely to the loss of microglia, we performed IHC on previously generated brain tissue from 6-week-old congenic C57BL/6J mixed-sex mice treated for 10 weeks with the highly selective CSF1Ri PLX5622 (Spangenberg et al., 2019) at 1200 mg/kg in chow (Fig. 7H). WFA+ PNNs in the somatosensory cortex were significantly enhanced with PLX5622 treatment compared to vehicle (P < 0.0001; Fig. 7I) and this was accompanied by the depletion of IBA1+ microglia (P < 0.001; Fig. 7J). Unlike CSPGs secreted by glia, PNNs and their proteoglycan constituents are synthesized by neurons (Miyata et al., 2005). Taken together, this indicates that microglia tightly regulate basal PNN formation through sustained levels of homeostatic degradation, a function that can be pathologically altered in the context of brain injury or disease.
Discussion
The contribution of microglia to Huntington’s disease pathogenesis has been a topic of recent investigation. Although neuroinflammation is characteristic of Huntington’s disease (Crotti and Glass, 2015), treatment of R6/2 mice with the tetracycline derivative minocycline, an inhibitor of inflammatory microglial activation (Kobayashi et al., 2013), has produced mixed results (Chen et al., 2000; Smith et al., 2003). Furthermore, mHTT-expressing microglia generate exacerbated responses to inflammatory stimuli and consequently greater neuronal damage in vivo (Crotti et al., 2014), but depletion of mHTT from myeloid cells does not significantly rescue disease phenotype in the BACHD mouse model (Petkau et al., 2019). To clarify microglial function in disease further, we used the targeted and sustained inhibition of CSF1R to pharmacologically deplete microglia from the R6/2 brain and assessed the behavioural, transcriptional, and pathological consequences.
Microgliosis is a hallmark of the Huntington’s disease brain, in patients (Sapp et al., 2001; Tai et al., 2007), and in cell (Kraft et al., 2012) and mouse models (Simmons et al., 2007; Siew et al., 2019). Further, inflammatory cytokines have been detected in plasma, myeloid cells and CSF from Huntington’s disease subjects (Chang et al., 2015; Miller et al., 2016; Rodrigues et al., 2016). Accordingly, we observed elevated densities of cortical and striatal microglia, which displayed several morphological features of immune reactivity (larger soma, higher IBA1 intensity) in the motor and somatosensory cortices specifically. Beyond this, however, R6/2 microglia did not appear overtly activated in vivo, at least to the extent seen in other neurodegenerative disease models (Liberatore et al., 1999; Spangenberg et al., 2016): their processes remained ramified and thin, they did not adopt an amoeboid morphology, and the striatal transcriptome did not reflect broad pro-inflammatory polarization. However, microglia exist within a spectrum of functional states, and their dynamic developmental (Lehrman et al., 2018) and homeostatic (Bennett et al., 2018) roles could also be affected by proteinopathy.
In this study, we began CSF1R-mediated microglial depletion at Week 6, after microglial development but before overt R6/2 symptomology, attaining successful microglial elimination in all brain regions examined. We reported beneficial effects of treatment on both grip strength and novel object recognition deficits in R6/2 mice. Of clinical relevance, it is possible that peripheral actions of PLX3397 may contribute to delayed grip strength deficits, e.g. by altering CSF1R signalling in skeletal muscle macrophages. Polyglutamine inclusions accumulate in skeletal muscle from Huntington’s disease models (R6/2, HdhQ150) and patients, which display myopathic structural abnormalities (Ciammola et al., 2006; Moffitt et al., 2009; Zielonka et al., 2014). Furthermore, similar downregulations of macrophage-related transcripts we observed in the striatum here have also been reported in R6/2 skeletal muscle (Igf1, Ccl25, H2-DMa) (Strand et al., 2005) and peripheral Huntington’s disease monocytes/macrophages show dysregulated transcriptional polarization as well as impaired migration to stimuli (Crotti and Glass, 2015). Although PLX3397 has modest to no effects on homeostatic peripheral tissue macrophage numbers (Mok et al., 2014) including at doses virtually equivalent to that used in this study (Szalay et al., 2016), disruption of CSF1R signalling may alter the functional phenotype of skeletal muscle macrophages in a manner that slows disease-related muscle weakness. Future studies should investigate the effects of more translationally amenable doses of PLX3397 that do not cross the BBB on R6/2 grip strength and muscle tissue changes. Additionally, the effects seen here should be validated in longer-lived models of Huntington’s disease [e.g. BACHD (Gray et al., 2008) or zQ175 mice (Menalled et al., 2012)], where greater behavioural readout and post-symptomatic treatment opportunities are feasible than with the more aggressive R6/2 model.
Transcript analysis of the R6/2 striatum revealed a robust upregulation of IFN pathway signalling that closely resembled the affected human caudate. This is suggestive of the existence of an ‘interferon signature’ in Huntington’s disease, increasingly reported in other diseases (Mostafavi et al., 2016; Mathys et al., 2017), aspects of which were partially resolved upon microglial elimination here. Of note, microglia were recently reported to accelerate neurodegeneration in a model of prion disease via reactive upregulation of type I interferon (Nazmi et al., 2019). Consistent with a secondary rather than initiating role in Huntington’s disease, microglial depletion with CSF1Ri reduced the striatal accumulation of several pathologic mHTT species in R6/2 mice, even though microglia rarely contain nuclear mHTT inclusions themselves (Jansen et al., 2017).
While investigating the cellular integrity of the Huntington’s disease striatum, we found markedly increased expression of the cell-matrix adhesion receptor CD44, the expression of which by microglia and astrocytes is associated with ALS progression (Matsumoto et al., 2012). While we did not observe microglial co-localization, its upregulation in astrocytes is microglia-dependent (Liddelow et al., 2017), consistent with the observed astrogliosis that is abrogated alongside striatal CD44 expression with microglial elimination. CD44 (CSPG8) belongs to a family of mostly secreted proteoglycans that are fundamental components of glial scars (Anderson et al., 2016) and the brain ECM (Yutsudo and Kitagawa, 2015; Tewari et al., 2018), and staining for CSPG with CS-56 revealed a strong correlation with CD44. Indeed, Huntington’s disease mice displayed a remarkable accumulation of CSPG in the cerebral cortex, striatum, and hippocampus, consistent with reports of astrocytic CSPG expression in human Huntington’s disease striatum (DeWitt et al., 1994). Interestingly, kainic acid-induced seizures (Yutsudo and Kitagawa, 2015), which directly enhance glutamate signalling (Iadarola et al., 2015), are followed by similar CS-56+ deposition.
CSPGs generally exert inhibitory effects on axon growth (Fidler et al., 1999; Properzi et al., 2005; Fisher et al., 2011; Ohtake et al., 2016; Pearson et al., 2018). Consistent with this, transcriptomic pathway analysis confirmed impairments in striatal neuritogenesis and neuronal development, which were restored alongside CSPG levels following microglial depletion. Importantly, the same human striatal samples used for pathway analysis also displayed elevated astrocyte and CSPG mRNA with disease (Hodges et al., 2006). It is possible that the loss of microglia represses astrocyte activation and CSPG secretion (Silver and Miller, 2004; Yu et al., 2012; Liddelow et al., 2017), thereby permitting neuropil and axonal outgrowth in the striatum and restored striatal volume. Consistent with this, astrocyte reactivity and CSPG production is stimulated by TGFβ (Yu et al., 2012; Ohtake et al., 2016), which is primarily produced by microglia (Zhang et al., 2014) and disease astrocytes (Endo et al., 2015), and which was significantly reduced in R6/2 striatum following CSF1Ri (Tgfb1, LogFC = −2.0535, FDR<0.01) alongside astrogliosis. Additionally, the depletion of mHTT from astrocytes alone (Wood et al., 2018), but not microglia (Petkau et al., 2019), reduces mHTT accumulation and striatal volume loss in the BACHD mouse model. Furthermore, bilateral intrastriatal AAV-RNAi delivery of HTT-suppressing miRNA targeted to neurons and astrocytes (AAV1), but not neurons alone (AAV2-HBKO), rescued motor deficits in the YAC128 mouse model of Huntington’s disease, suggesting that astrocyte mHTT directly alters reactivity in a manner detrimental to disease (Stanek et al., 2019). In light of the current study, these reports suggest that astrocytes directly contribute to neurodegeneration, while microglia promote secondary damage through modulation of astrocyte reactivity and/or other pathways, thereby accelerating pathogenesis. The loss of the homeostatic microglial gene P2ry12 (Bennett et al., 2018) from the R6/2 striatum in our dataset (FDR < 0.1, LogFC = −0.7083 versus control) further suggests microglial dysfunction in the context of Huntington’s disease.
CSPGs are also core structural elements of conventional PNNs, reticular structures that form along the soma and proximal dendrites of primarily GABAergic parvalbumin interneurons soon after synaptic pruning is completed by microglia (Paolicelli et al., 2011), stabilizing the neuronal circuitry and inhibiting plasticity (Wang and Fawcett, 2012; Fawcett et al., 2019). CSPGs contained in PNNs are labelled by the plant lectin WFA in a manner distinct from CS-56 (Hayashi et al., 2007; Pantazopoulos et al., 2015; Yutsudo and Kitagawa, 2015), potentially due to differences in CSPG sidechain sulphation status (Sorg et al., 2016) or epitope availability. PNNs are autonomously constructed by neurons (Miyata et al., 2005; Geissler et al., 2013; Fowke et al., 2017), and in stark contrast to CS-56 staining, we found a loss of WFA+ PNN density with disease in motor and somatosensory cortices. R6/2 parvalbumin striatal interneurons also displayed a reduction in WFA+ intensity and an apparent loss of PNN structural integrity, which has before been shown to contribute to tumour-associated seizures (Tewari et al., 2018) due to impaired GABAergic inhibitory firing, as PNNs are thought to enhance GABAergic electrical transmission akin to a myelin sheath. As such, their loss may further contribute to the excitatory/inhibitory imbalance observed in the R6/2 brain. Surprisingly, the depletion of microglia from healthy, non-transgenic mice produced a dramatic and ubiquitous accumulation of PNNs, which we verified in brain tissue from adult wild-type mice treated with the selective CSF1R inhibitor PLX5622, underscoring a yet unstudied role of microglia as homeostatic regulators of PNN formation in the healthy brain.
In conclusion, we show that sustained CSF1R inhibition in the R6/2 mouse model of Huntington’s disease, which exhibited elevated microglial numbers but not broad inflammation at the transcriptional or morphological level, mitigated grip strength and object memory deficits. Microglial depletion was accompanied by a reduction in striatal mHTT accumulation and partial or full reversals of dysregulated interferon signalling, neuritogenesis, and neuronal development pathways. Furthermore, we report a massive accumulation of extracellular CSPG with disease, which is associated with striatal volume loss, astrogliosis, and neuropil alterations in the R6/2 brain, and microglial depletion with CSF1Ri partially or completely prevented each of these phenotypes. Finally, we show that a related but distinct CSPG-containing structure, the PNN, is disrupted in a region-specific manner with disease, and that the elimination of microglial not only ameliorates this, but greatly enhances PNN density in the naïve adult brain.
Supplementary Material
Acknowledgements
We thank Dr Elizabeth Dominguez for intellectual contributions regarding PNNs, Dan Hoang, and Karla Abad-Torrez for the setting up of gene expression data, and Miguel Arreola, Eva Morozko, Sylvia Yeung, and Alice Lau for technical assistance. The RNA-seq data has been deposited in GEO (ascension number GSE136158). This work was made possible, in part, through access to the Genomics High Throughput Facility Shared Resource of the Cancer Center Support Grant (P30CA-062203) at the University of California, Irvine and NIH shared instrumentation grants 1S10RR025496-01, 1S10OD010794-01, and 1S10OD021718-01.
Funding
This work was supported by the National Institutes of Health under awards R01NS083801 (NINDS), R01AG056768 (NIA), and P50AG016573 (NIA) to K.N.G., R01NS090390 (NINDS) to L.M.T., F31NS108611 (NINDS) to J.C. and NSF fellowship (to J.O.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests
The authors report no competing interests.
Glossary
Abbreviations
- CSF1Ri =
colony-stimulating factor 1 receptor inhibition
- CSPG =
chondroitin sulphate proteoglycan
- DEGs =
differentially expressed genes
- ECM =
extracellular matrix
- mHTT =
mutant huntingtin
- PNN =
perineuronal net
References
- Acharya MM, Green KN, Allen BD, Najafi AR, Syage A, Minasyan H, et al. Elimination of microglia improves cognitive function following cranial irradiation. Sci Rep 2016; 6: 31545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MA, Burda JE, Ren Y, Ao Y, O'Shea TM, Kawaguchi R, et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature 2016; 532: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T, et al. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci 2015; 18: 1584–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee SB, Gutzeit VA, Baman J, Aoued HS, Doshi NK, Liu RC, et al. Perineuronal nets in the adult sensory cortex are necessary for fear learning. Neuron 2017; 95: 169–79.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay J, Balaguero N, Mione M, Ackerman SL, Letts VA, Brodbeck J, et al. Ducky mouse phenotype of epilepsy and ataxia is associated with mutations in the Cacna2d2 gene and decreased calcium channel current in cerebellar Purkinje cells. J Neurosci 2001; 21: 6095–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett FC, Bennett ML, Yaqoob F, Mulinyawe SB, Grant GA, Hayden Gephart M, et al. A combination of ontogeny and CNS environment establishes microglial identity. Neuron 2018; 98: 1170–83.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, et al. Global changes to the ubiquitin system in Huntington's disease. Nature 2007; 448: 704–8. [DOI] [PubMed] [Google Scholar]
- Benraiss A, Wang S, Herrlinger S, Li X, Chandler-Militello D, Mauceri J, et al. Human glia can both induce and rescue aspects of disease phenotype in Huntington disease. Nat Commun 2016; 7: 11758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum D, Chern Y, Domenici MR, Buee L, Lin CY, Rea W, et al. The role of adenosine tone and adenosine receptors in Huntington's disease. J Caffeine Adenosine Res 2018; 8: 43–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera JR, Lucas JJ. MAP2 Splicing is altered in Huntington's disease. Brain Pathol (Zurich, Switzerland) 2017; 27: 181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso FL, Herz J, Fernandes A, Rocha J, Sepodes B, Brito MA, et al. Systemic inflammation in early neonatal mice induces transient and lasting neurodegenerative effects. J Neuroinflamm 2015; 12: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Wu YR, Chen YC, Chen CM. Plasma inflammatory biomarkers for Huntington's disease patients and mouse model. Brain Behav Immun 2015; 44: 121–7. [DOI] [PubMed] [Google Scholar]
- Chen M, Ona VO, Li M, Ferrante RJ, Fink KB, Zhu S, et al. Minocycline inhibits caspase-1 and caspase-3 expression and delays mortality in a transgenic mouse model of Huntington disease. Nat Med 2000; 6: 797. [DOI] [PubMed] [Google Scholar]
- Ciammola A, Sassone J, Alberti L, Meola G, Mancinelli E, Russo MA, et al. Increased apoptosis, huntingtin inclusions and altered differentiation in muscle cell cultures from Huntington's disease subjects. Cell Death Differ 2006; 13: 2068–78. [DOI] [PubMed] [Google Scholar]
- Crotti A, Benner C, Kerman B, Gosselin D, Lagier-Tourenne C, Zuccato C, et al. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci 2014; 17: 513–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti A, Glass CK. The choreography of neuroinflammation in Huntington's disease. Trends Immunol 2015; 36: 364–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagher NN,, Najafi AR, Kayala KM, Elmore MR, White TE, Medeiros R, et al. Colony-stimulating factor 1 receptor inhibition prevents microglial plaque association and improves cognition in 3xTg-AD mice. J Neuroinflamm 2015; 12: 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt DA, Richey PL, Praprotnik D, Silver J, Perry G. Chondroitin sulfate proteoglycans are a common component of neuronal inclusions and astrocytic reaction in neurodegenerative diseases. Brain Res 1994; 656: 205–9. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds L, Chen J, Berggren K, Fox J. Characterization of striatal neuronal loss and atrophy in the R6/2 mouse model of Huntington's disease. PLoS Curr 2014; 6. doi: 10.1371/currents.hd.48727b68b39b82d5fe350f753984bcf9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, Frontali M, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet 1993; 4: 387–92. [DOI] [PubMed] [Google Scholar]
- Dzwonek J, Wilczynski GM. CD44: molecular interactions, signaling and functions in the nervous system. Front Cell Neurosci 2015; 9: 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore MRP, Hohsfield LA, Kramár EA, Soreq L, Lee RJ, Pham ST, et al. Replacement of microglia in the aged brain reverses cognitive, synaptic, and neuronal deficits in mice. Aging Cell 2018; 17: e12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore MR, Lee RJ, West BL, Green KN. Characterizing newly repopulated microglia in the adult mouse: impacts on animal behavior, cell morphology, and neuroinflammation. PLoS One 2015; 10: e0122912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore MRP, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, et al. CSF1 receptor signaling is necessary for microglia viability, which unmasks a cell that rapidly repopulates the microglia-depleted adult brain. Neuron 2014; 82: 380–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y, Gan L. The LIM-homeobox gene Islet-1 is required for the development of restricted forebrain cholinergic neurons. J Neurosci 2008; 28: 3291–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo F, Komine O, Fujimori-Tonou N, Katsuno M, Jin S, Watanabe S, et al. Astrocyte-derived TGF-beta1 accelerates disease progression in ALS mice by interfering with the neuroprotective functions of microglia and T cells. Cell Rep 2015; 11: 592–604. [DOI] [PubMed] [Google Scholar]
- Fawcett JW, Oohashi T, Pizzorusso T. The roles of perineuronal nets and the perinodal extracellular matrix in neuronal function. Nat Rev Neurosci 2019; 20: 451–65. [DOI] [PubMed] [Google Scholar]
- Feng X, Jopson TD, Paladini MS, Liu S, West BL, Gupta N, et al. Colony-stimulating factor 1 receptor blockade prevents fractionated whole-brain irradiation-induced memory deficits. J Neuroinflamm 2016; 13: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Nogales M, Santos-Galindo M, Hernandez IH, Cabrera JR, Lucas JJ. Faulty splicing and cytoskeleton abnormalities in Huntington's disease. Brain Pathol (Zurich, Switzerland) 2016; 26: 772–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante R, Kowall N, Richardson E. Proliferative and degenerative changes in striatal spiny neurons in Huntington's disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J Neurosci 1991; 11: 3877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler PS, Schuette K, Asher RA, Dobbertin A, Thornton SR, Calle-Patino Y, et al. Comparing astrocytic cell lines that are inhibitory or permissive for axon growth: the major axon-inhibitory proteoglycan is NG2. J Neurosci 1999; 19: 8778–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher D, Xing B, Dill J, Li H,, Hoang HH, Zhao Z, et al. Leukocyte common antigen-related phosphatase is a functional receptor for chondroitin sulfate proteoglycan axon growth inhibitors. J Neurosci 2011; 31: 14051–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowke TM, Karunasinghe RN, Bai JZ, Jordan S, Gunn AJ, Dean JM. Hyaluronan synthesis by developing cortical neurons in vitro. Sci Rep 2017; 7: 44135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin SL, Love S, Greene JR, Betmouni S. Loss of perineuronal Net in ME7 Prion disease. J Neuropathol Exp Neurol 2008; 67: 189–99. [DOI] [PubMed] [Google Scholar]
- Galatro TF, Holtman IR, Lerario AM, Vainchtein ID, Brouwer N, Sola PR, et al. Transcriptomic analysis of purified human cortical microglia reveals age-associated changes. Nat Neurosci 2017; 20: 1162. [DOI] [PubMed] [Google Scholar]
- Geissler M, Gottschling C, Aguado A, Rauch U, Wetzel CH, Hatt H, et al. Primary hippocampal neurons, which lack four crucial extracellular matrix molecules, display abnormalities of synaptic structure and function and severe deficits in perineuronal net formation. J Neurosci 2013; 33: 7742–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson EM, Nagaraja S, Ocampo A, Tam LT, Wood LS, Pallegar PN, et al. Methotrexate chemotherapy induces persistent tri-glial dysregulation that underlies chemotherapy-related cognitive impairment. Cell 2019; 176: 43–55.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T, Zeller N, Raasch J, Kierdorf K, Frenzel K, Ketscher L, et al. USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J 2015; 34: 1612–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH, et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci 2008; 28: 6182–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson O, Petersen A, Leist M, Nicotera P, Castilho RF, Brundin P. Transgenic mice expressing a Huntington's disease mutation are resistant to quinolinic acid-induced striatal excitotoxicity. Proc Natl Acad Sci U S A 1999; 96: 8727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi N, Tatsumi K, Okuda H, Yoshikawa M, Ishizaka S, Miyata S, et al. DACS, novel matrix structure composed of chondroitin sulfate proteoglycan in the brain. Biochem Biophys Res Commun 2007; 364: 410–5. [DOI] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, et al. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet 2006; 15: 965–77. [DOI] [PubMed] [Google Scholar]
- Hsu J-YC, Bourguignon LYW, Adams CM, Peyrollier K, Zhang H, Fandel T, et al. Matrix metalloproteinase-9 facilitates glial scar formation in the injured spinal cord. The J Neurosci 2008; 28: 13467–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadarola ND, Niciu MJ, Richards EM, Vande Voort JL, Ballard ED, Lundin NB, et al. Ketamine and other N-methyl-D-aspartate receptor antagonists in the treatment of depression: a perspective review. Ther Adv Chronic Dis 2015; 6: 97–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illouz T, Madar R, Biragyn A, Okun E. Restoring microglial and astroglial homeostasis using DNA immunization in a Down syndrome mouse model. Brain Behav Immun 2019; 75: 163–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AH, van Hal M, Op den Kelder IC, Meier RT, de Ruiter AA, Schut MH, et al. Frequency of nuclear mutant huntingtin inclusion formation in neurons and glia is cell-type-specific. Glia 2017; 65: 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien C, Marcouiller F, Bretteville A, El Khoury NB, Baillargeon J, Hebert SS, et al. Dimethyl sulfoxide induces both direct and indirect tau hyperphosphorylation. PLoS One 2012; 7: e40020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast DJ, Dominguez R. The cytoskeleton-autophagy connection. Curr Biol 2017; 27: R318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Imagama S, Ohgomori T, Hirano K, Uchimura K, Sakamoto K, et al. Minocycline selectively inhibits M1 polarization of microglia. Cell Death Dis 2013; 4: e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga S, Kouri N, Walton RL, Ebbert MTW, Josephs KA, Litvan I, et al. Corticobasal degeneration with TDP-43 pathology presenting with progressive supranuclear palsy syndrome: a distinct clinicopathologic subtype. Acta Neuropathol 2018; 136: 389–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft AD, Kaltenbach LS, Lo DC, Harry GJ. Activated microglia proliferate at neurites of mutant huntingtin-expressing neurons. Neurobiol Aging 2012; 33: 621.e17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa M, Matsumoto G, Kino Y, Okuno M, Kurosawa-Yamada M, Washizu C, et al. Depletion of p62 reduces nuclear inclusions and paradoxically ameliorates disease phenotypes in Huntington's model mice. Hum Mol Genet 2015; 24: 1092–105. [DOI] [PubMed] [Google Scholar]
- La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet 2010; 11: 247–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI. Huntington's disease: underlying molecular mechanisms and emerging concepts. Trends Biochem Sci 2013; 38: 378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Noh JY, Oh Y, Kim Y, Chang JW, Chung CW, et al. IRE1 plays an essential role in ER stress-mediated aggregation of mutant huntingtin via the inhibition of autophagy flux. Hum Mol Genet 2012; 21: 101–14. [DOI] [PubMed] [Google Scholar]
- Lee SW, Park HJ, Im W, Kim M, Hong S. Repeated immune activation with low-dose lipopolysaccharide attenuates the severity of Huntington's disease in R6/2 transgenic mice. Anim Cells Syst 2018; 22: 219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman EK, Wilton DK, Litvina EY, Welsh CA, Chang ST, Frouin A, et al. CD47 protects synapses from excess microglia-mediated pruning during development. Neuron 2018; 100: 120–34.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol 2017; 18: 225. [DOI] [PubMed] [Google Scholar]
- Li M, Li Z, Ren H, Jin WN, Wood K, Liu Q, et al. Colony stimulating factor 1 receptor inhibition eliminates microglia and attenuates brain injury after intracerebral hemorrhage. J Cereb Blood Flow Metab 2017; 37: 2383–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Popovic N, Brundin P. The use of the R6 transgenic mouse models of Huntington's disease in attempts to develop novel therapeutic strategies. NeuroRx 2005; 2: 447–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Song MR, Xu Z, Lanuza GM, Liu Y, Zhuang T, et al. Isl1 is required for multiple aspects of motor neuron development. Mol Cell Neurosci 2011; 47: 215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberatore GT, Jackson-Lewis V, Vukosavic S, Mandir AS, Vila M, McAuliffe WG, et al. Inducible nitric oxide synthase stimulates dopaminergic neurodegeneration in the MPTP model of Parkinson disease. Nat Med 1999; 5: 1403. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017; 541: 481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RG, Salazar LL, Wilton DK, King AR, Stocksdale JT, Sharifabad D, et al. HD iPSC Consortium. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat Neurosci 2017; 20: 648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi-Carter R, Apostol BL, Dunah AW, DeJohn MM, Farrell LA, Bates GP, et al. Complex alteration of NMDA receptors in transgenic Huntington's disease mouse brain: analysis of mRNA and protein expression, plasma membrane association, interacting proteins, and phosphorylation. Neurobiol Dis 2003; 14: 624–36. [DOI] [PubMed] [Google Scholar]
- MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 1993; 72: 971–83. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 1996; 87: 493–506. [DOI] [PubMed] [Google Scholar]
- Mathys H, Adaikkan C, Gao F, Young JZ, Manet E, Hemberg M, et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep 2017; 21: 366–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Imagama S, Hirano K, Ohgomori T, Natori T, Kobayashi K, et al. CD44 expression in astrocytes and microglia is associated with ALS progression in a mouse model. Neurosci Lett 2012; 520: 115–20. [DOI] [PubMed] [Google Scholar]
- McCollum MH, Leon RT, Rush DB, Guthrie KM, Wei J. Striatal oligodendrogliogenesis and neuroblast recruitment are increased in the R6/2 mouse model of Huntington's disease. Brain Res 2013; 1518: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara CG, Tejero-Cantero Á, Trouche S, Campo-Urriza N, Dupret D. Dopaminergic neurons promote hippocampal reactivation and spatial memory persistence. Nat Neurosci 2014; 17: 1658–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SR, Tom CM, Wang Y, Bresee C, Rushton D, Mathkar PP, et al. Human Huntington's disease iPSC-derived cortical neurons display altered transcriptomics, morphology, and maturation. Cell Rep 2018; 25: 1081–96.e6. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Kudwa AE, Miller S, Fitzpatrick J, Watson-Johnson J, Keating N, et al. Comprehensive behavioral and molecular characterization of a new knock-in mouse model of Huntington’s disease: zQ175. PloS one 2012; 7: e49838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JR, Lo KK, Andre R, Hensman Moss DJ, Trager U, Stone TC, et al. RNA-Seq of Huntington's disease patient myeloid cells reveals innate transcriptional dysregulation associated with proinflammatory pathway activation. Hum Mol Genet 2016; 25: 2893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Komatsu Y, Yoshimura Y, Taya C, Kitagawa H. Persistent cortical plasticity by upregulation of chondroitin 6-sulfation. Nat Neurosci 2012; 15: 414. [DOI] [PubMed] [Google Scholar]
- Miyata S, Nishimura Y, Hayashi N, Oohira A. Construction of perineuronal net-like structure by cortical neurons in culture. Neuroscience 2005; 136: 95–104. [DOI] [PubMed] [Google Scholar]
- Miyazaki H, Oyama F, Inoue R, Aosaki T, Abe T, Kiyonari H, et al. Singular localization of sodium channel β4 subunit in unmyelinated fibres and its role in the striatum. Nat Commun 2014; 5: 5525. [DOI] [PubMed] [Google Scholar]
- Moffitt H, McPhail GD, Woodman B, Hobbs C, Bates GP. Formation of polyglutamine inclusions in a wide range of non-CNS tissues in the HdhQ150 knock-in mouse model of Huntington's disease. PLoS One 2009; 4: e8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S, Koya RC, Tsui C, Xu J, Robert L, Wu L, et al. Inhibition of CSF-1 receptor improves the antitumor efficacy of adoptive cell transfer immunotherapy. Cancer Res 2014; 74: 153–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro S, Roque S, Marques F, Correia-Neves M, Cerqueira JJ. Brain interference: revisiting the role of IFNgamma in the central nervous system. Prog Neurobiol 2017; 156: 149–63. [DOI] [PubMed] [Google Scholar]
- Morozko EL, Ochaba J, Hernandez SJ, Lau A, Sanchez I, Orellana I, et al. Longitudinal biochemical assay analysis of mutant huntingtin exon 1 protein in R6/2 mice. J Huntington's Dis 2018; 7: 321–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, Yoshida H, Moodley D, LeBoité H, Rothamel K, Raj T, et al. Parsing the interferon transcriptional network and its disease associations. Cell 2016; 164: 564–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrdjen D, Pavlovic A, Hartmann FJ, Schreiner B, Utz SG, Leung BP, et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 2018; 48: 380–95.e6. [DOI] [PubMed] [Google Scholar]
- Najafi AR, Crapser J, Jiang S, Ng W, Mortazavi A, West BL, et al. A limited capacity for microglial repopulation in the adult brain. Glia 2018; 66: 2385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazmi A, Field RH, Griffin EW, Haugh O, Hennessy E, Cox D, et al. Chronic neurodegeneration induces type I interferon synthesis via STING, shaping microglial phenotype and accelerating disease progression. Glia 2019; 67: 1254–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science (New York, NY) 2005; 308: 1314–8. [DOI] [PubMed] [Google Scholar]
- Ochaba J, Monteys AM, O’Rourke JG, Reidling JC, Steffan JS, Davidson BL, et al. PIAS1 regulates mutant Huntingtin accumulation and Huntington’s disease-associated phenotypes in vivo. Neuron 2016; 90: 507–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochaba J, Morozko EL, O’Rourke JG, Thompson LM. Fractionation for resolution of soluble and insoluble huntingtin species. JoVE 2018; 132: e57082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake Y, Wong D, Abdul-Muneer PM, Selzer ME, Li S. Two PTP receptors mediate CSPG inhibition by convergent and divergent signaling pathways in neurons. Sci Rep 2016; 6: 37152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke JG, Gareau JR, Ochaba J, Song W, Rasko T, Reverter D, et al. SUMO-2 and PIAS1 modulate insoluble mutant huntingtin protein accumulation. Cell Rep 2013; 4: 362–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipovitch M, Asenjo Martinez A, Mariani JN, Cornwell A, Dhaliwal S, Zou L, et al. Human ESC-derived chimeric mouse models of Huntington's disease reveal cell-intrinsic defects in glial progenitor cell differentiation. Cell Stem Cell 2019; 24: 107–22.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama F, Miyazaki H, Sakamoto N, Becquet C, Machida Y, Kaneko K, et al. Sodium channel beta4 subunit: down-regulation and possible involvement in neuritic degeneration in Huntington's disease transgenic mice. J Neurochem 2006; 98: 518–29. [DOI] [PubMed] [Google Scholar]
- Paine SM, Anderson G, Bedford K, Lawler K, Mayer RJ, Lowe J, et al. Pale body-like inclusion formation and neurodegeneration following depletion of 26S proteasomes in mouse brain neurones are independent of alpha-synuclein. PLoS One 2013; 8: e54711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantazopoulos H, Markota M, Jaquet F, Ghosh D, Wallin A, Santos A, et al. Aggrecan and chondroitin-6-sulfate abnormalities in schizophrenia and bipolar disorder: a postmortem study on the amygdala. Transl Psychiatry 2015; 5: e496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science (New York, NY) 2011; 333: 1456–8. [DOI] [PubMed] [Google Scholar]
- Park H, Kim M, Kim HJ, Lee Y, Seo Y, Pham CD, et al. Heparan sulfate proteoglycans (HSPGs) and chondroitin sulfate proteoglycans (CSPGs) function as endocytic receptors for an internalizing anti-nucleic acid antibody. Sci Rep 2017; 7: 14373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavese N, Gerhard A, Tai YF, Ho AK, Turkheimer F, Barker RA, et al. Microglial activation correlates with severity in Huntington disease: a clinical and PET study. Neurology 2006; 66: 1638–43. [DOI] [PubMed] [Google Scholar]
- Pearson CS, Mencio CP, Barber AC, Martin KR, Geller HM. Identification of a critical sulfation in chondroitin that inhibits axonal regeneration. eLife 2018; 7: e37139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkau TL, Hill A, Connolly C, Lu G, Wagner P, Kosior N, et al. Mutant huntingtin expression in microglia is neither required nor sufficient to cause the Huntington's disease-like phenotype in BACHD mice. Hum Mol Genet 2019; 28: 1661–70. [DOI] [PubMed] [Google Scholar]
- Pippucci T, Parmeggiani A, Palombo F, Maresca A, Angius A, Crisponi L, et al. A novel null homozygous mutation confirms CACNA2D2 as a gene mutated in epileptic encephalopathy. PLoS One 2013; 8: e82154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis M, Pavese N, Tai YF, Kiferle L, Mason SL, Brooks DJ, et al. Microglial activation in regions related to cognitive function predicts disease onset in Huntington's disease: a multimodal imaging study. Hum Brain Mapp 2011; 32: 258–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Properzi F, Carulli D, Asher RA, Muir E, Camargo LM, van Kuppevelt TH, et al. Chondroitin 6-sulphate synthesis is up-regulated in injured CNS, induced by injury-related cytokines and enhanced in axon-growth inhibitory glia. Eur J Neurosci 2005; 21: 378–90. [DOI] [PubMed] [Google Scholar]
- Reiner A, Shelby E, Wang H, Demarch Z, Deng Y, Guley NH, et al. Striatal parvalbuminergic neurons are lost in Huntington's disease: implications for dystonia. Mov Disord 2013; 28: 1691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice RA, Spangenberg EE, Yamate-Morgan H, Lee RJ, Arora RP, Hernandez MX, et al. Elimination of microglia improves functional outcomes following extensive neuronal loss in the hippocampus. J Neurosci 2015; 35: 9977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 2010; 26: 139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues FB, Byrne LM, McColgan P, Robertson N, Tabrizi SJ, Zetterberg H, et al. Cerebrospinal fluid inflammatory biomarkers reflect clinical severity in Huntington's disease. PLoS One 2016; 11: e0163479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Tabrizi SJ. Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol 2011; 10: 83–98. [DOI] [PubMed] [Google Scholar]
- Rothhammer V, Borucki DM, Tjon EC, Takenaka MC, Chao C-C, Ardura-Fabregat A, et al. Microglial control of astrocytes in response to microbial metabolites. Nature 2018; 557: 724–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runne H, Regulier E, Kuhn A, Zala D, Gokce O, Perrin V, et al. Dysregulation of gene expression in primary neuron models of Huntington's disease shows that polyglutamine-related effects on the striatal transcriptome may not be dependent on brain circuitry. J Neurosci 2008; 28: 9723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig I, Augestad IL, Haberg AK, Sandvig A. Neuroplasticity in stroke recovery. The role of microglia in engaging and modifying synapses and networks. Eur J Neurosci 2018; 47: 1414–28. [DOI] [PubMed] [Google Scholar]
- Sapp E, Kegel KB, Aronin N, Hashikawa T, Uchiyama Y, Tohyama K, et al. Early and progressive accumulation of reactive microglia in the Huntington disease brain. J Neuropathol Exp Neurol 2001; 60: 161–72. [DOI] [PubMed] [Google Scholar]
- Siew JJ, Chen H-M, Chen H-Y, Chen H-L, Chen C-M, Soong B-W, et al. Galectin-3 is required for the microglia-mediated brain inflammation in a model of Huntington’s disease. Nat Commun 2019; 10: 3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci 2004; 5: 146. [DOI] [PubMed] [Google Scholar]
- Simmons DA, Casale M, Alcon B, Pham N, Narayan N, Lynch G. Ferritin accumulation in dystrophic microglia is an early event in the development of Huntington's disease. Glia 2007; 55: 1074–84. [DOI] [PubMed] [Google Scholar]
- Smith AM, Gibbons HM, Oldfield RL, Bergin PM, Mee EW, Faull RL, et al. The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia 2013; 61: 929–42. [DOI] [PubMed] [Google Scholar]
- Smith DL, Woodman B, Mahal A, Sathasivam K, Ghazi-Noori S, Lowden PA, et al. Minocycline and doxycycline are not beneficial in a model of Huntington's disease. Ann Neurol 2003; 54: 186–96. [DOI] [PubMed] [Google Scholar]
- Sorg BA, Berretta S, Blacktop JM, Fawcett JW, Kitagawa H, Kwok JC, et al. Casting a wide net: role of perineuronal nets in neural plasticity. J Neurosci 2016; 36: 11459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg EE, Lee RJ, Najafi AR, Rice RA, Elmore MR, Blurton-Jones M, et al. Eliminating microglia in Alzheimer's mice prevents neuronal loss without modulating amyloid-beta pathology. Brain 2016; 139: 1265–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenberg EE, Severson P, Hohsfield LA, Crapser J, Zhang J, Burton EA, et al. Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model. Nat Commun 2019; 10: 3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford JH, Hirai T, Deng L, Chernikova SB, Urata K, West BL, et al. Colony stimulating factor 1 receptor inhibition delays recurrence of glioblastoma after radiation by altering myeloid cell recruitment and polarization. Neuro-oncology 2016; 18: 797–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek LM, Bu J, Shihabuddin LS. Astrocyte transduction is required for rescue of behavioral phenotypes in the YAC128 mouse model with AAV-RNAi mediated HTT lowering therapeutics. Neurobiol Dis 2019; 129: 29–37. [DOI] [PubMed] [Google Scholar]
- Strand AD, Aragaki AK, Shaw D, Bird T, Holton J, Turner C, et al. Gene expression in Huntington's disease skeletal muscle: a potential biomarker. Hum Mol Genet 2005; 14: 1863–76. [DOI] [PubMed] [Google Scholar]
- Svensson MN, Andersson KM, Wasén C, Erlandsson MC, Nurkkala-Karlsson M, Jonsson IM, et al. Murine germinal center B cells require functional Fms-like tyrosine kinase 3 signaling for IgG1 class-switch recombination. Proc Natl Acad Sci U S A 2015; 112: E6644–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalay G, Martinecz B, Lenart N, Kornyei Z, Orsolits B, Judak L, et al. Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat Commun 2016; 7: 11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai YF, Pavese N, Gerhard A, Tabrizi SJ, Barker RA, Brooks DJ, et al. Microglial activation in presymptomatic Huntington's disease gene carriers. Brain 2007; 130: 1759–66. [DOI] [PubMed] [Google Scholar]
- Tap WD, Anthony SP, Chmielowski B, Staddon AP, Cohn AL, Shapiro G, et al. A pilot study of PLX3397, a selective colony-stimulating factor 1 receptor (CSF1R) kinase inhibitor, in pigmented villonodular synovitis (PVNS). J Clin Oncol 2014; 32: 10503. [Google Scholar]
- Tap WD, Wainberg ZA, Anthony SP, Ibrahim PN, Zhang C, Healey JH, et al. Structure-guided blockade of CSF1R kinase in tenosynovial giant-cell tumor. N Engl J Med 2015; 373: 428–37. [DOI] [PubMed] [Google Scholar]
- Tewari BP, Chaunsali L, Campbell SL, Patel DC, Goode AE, Sontheimer H. Perineuronal nets decrease membrane capacitance of peritumoral fast spiking interneurons in a model of epilepsy. Nat Commun 2018; 9: 4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trias E, King PH, Si Y, Kwon Y, Varela V, Ibarburu S, et al. Mast cells and neutrophils mediate peripheral motor pathway degeneration in ALS. JCI Insight 2018; 3: e123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashishtha M, Ng CW, Yildirim F, Gipson TA, Kratter IH, Bodai L, et al. Targeting H3K4 trimethylation in Huntington disease. Proc Natl Acad Sci U S A 2013; 110: E3027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Fawcett J. The perineuronal net and the control of CNS plasticity. Cell Tissue Res 2012; 349: 147–60. [DOI] [PubMed] [Google Scholar]
- Wong BKY, Ehrnhoefer DE, Graham RK, Martin DDO, Ladha S, Uribe V, et al. Partial rescue of some features of Huntington disease in the genetic absence of caspase-6 in YAC128 mice. Neurobiol Dis 2015; 76: 24–36. [DOI] [PubMed] [Google Scholar]
- Wood TE, Yang Z, Gray M, Cepeda C, Barry J, Levine MS. Mutant huntingtin reduction in astrocytes slows disease progression in the BACHD conditional Huntington’s disease mouse model. Hum Mol Genet 2018; 28: 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HM, Yang S, Huang SS, Tang BS, Guo JF. Microglial activation in the pathogenesis of Huntington's disease. Front Aging Neurosci 2017; 9: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P, Wang H, Katagiri Y, Geller HM. An in vitro model of reactive astrogliosis and its effect on neuronal growth. Methods Mol Biol (Clifton, NJ) 2012; 814: 327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun SP, Kam TI, Panicker N, Kim S, Oh Y, Park JS, et al. Block of A1 astrocyte conversion by microglia is neuroprotective in models of Parkinson's disease. Nat Med 2018; 24: 931–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutsudo N, Kitagawa H. Involvement of chondroitin 6-sulfation in temporal lobe epilepsy. Exp Neurol 2015; 274: 126–33. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O'Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 2014; 34: 11929–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Dong XP. Dysfunction of microtubule-associated proteins of MAP2/tau family in Prion disease. Prion 2012; 6: 334–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yan S, Khambu B, Ma F, Li Y, Chen X, et al. Dynamic MTORC1-TFEB feedback signaling regulates hepatic autophagy, steatosis and liver injury in long-term nutrient oversupply. Autophagy 2018; 14: 1779–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka D, Piotrowska I, Marcinkowski JT, Mielcarek M. Skeletal muscle pathology in Huntington's disease. Front Physiol 2014; 5: 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data are deposited in GEO (ascension number GSE136158), and all RPKM values can be viewed and searched for at http://rnaseq.mind.uci.edu/green/R62_PLX/gene_search.php. All other data are available upon reasonable request.