Van Bergen et al. add to the growing number of ‘trappopathies’ by identifying a pathogenic variant in TRAPPC4 in seven children with early-onset seizures and developmental delay. The incompletely penetrant splicing defect reduces protein levels and TRAPP complex assembly, and impairs vesicular trafficking and autophagy in patient cells and yeast.
Keywords: autophagy, molecular genetics, whole-exome sequencing, vesicular transport, intellectual disability
Abstract
The conserved transport protein particle (TRAPP) complexes regulate key trafficking events and are required for autophagy. TRAPPC4, like its yeast Trs23 orthologue, is a core component of the TRAPP complexes and one of the essential subunits for guanine nucleotide exchange factor activity for Rab1 GTPase. Pathogenic variants in specific TRAPP subunits are associated with neurological disorders. We undertook exome sequencing in three unrelated families of Caucasian, Turkish and French-Canadian ethnicities with seven affected children that showed features of early-onset seizures, developmental delay, microcephaly, sensorineural deafness, spastic quadriparesis and progressive cortical and cerebellar atrophy in an effort to determine the genetic aetiology underlying neurodevelopmental disorders. All seven affected subjects shared the same identical rare, homozygous, potentially pathogenic variant in a non-canonical, well-conserved splice site within TRAPPC4 (hg19:chr11:g.118890966A>G; TRAPPC4: NM_016146.5; c.454+3A>G). Single nucleotide polymorphism array analysis revealed there was no haplotype shared between the tested Turkish and Caucasian families suggestive of a variant hotspot region rather than a founder effect. In silico analysis predicted the variant to cause aberrant splicing. Consistent with this, experimental evidence showed both a reduction in full-length transcript levels and an increase in levels of a shorter transcript missing exon 3, suggestive of an incompletely penetrant splice defect. TRAPPC4 protein levels were significantly reduced whilst levels of other TRAPP complex subunits remained unaffected. Native polyacrylamide gel electrophoresis and size exclusion chromatography demonstrated a defect in TRAPP complex assembly and/or stability. Intracellular trafficking through the Golgi using the marker protein VSVG-GFP-ts045 demonstrated significantly delayed entry into and exit from the Golgi in fibroblasts derived from one of the affected subjects. Lentiviral expression of wild-type TRAPPC4 in these fibroblasts restored trafficking, suggesting that the trafficking defect was due to reduced TRAPPC4 levels. Consistent with the recent association of the TRAPP complex with autophagy, we found that the fibroblasts had a basal autophagy defect and a delay in autophagic flux, possibly due to unsealed autophagosomes. These results were validated using a yeast trs23 temperature sensitive variant that exhibits constitutive and stress-induced autophagic defects at permissive temperature and a secretory defect at restrictive temperature. In summary we provide strong evidence for pathogenicity of this variant in a member of the core TRAPP subunit, TRAPPC4 that associates with vesicular trafficking and autophagy defects. This is the first report of a TRAPPC4 variant, and our findings add to the growing number of TRAPP-associated neurological disorders.
Introduction
The process of intracellular trafficking is of critical importance to cells, and this requires numerous proteins to guide cargo, contained within carrier vesicles, to specific intracellular compartments. Such proteins include those that co-ordinate vesicle formation, GTPases of the Rab family, transport and tethering factors, and soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins that mediate vesicle fusion with the target membrane (Cai et al., 2007). The TRAnsport Protein Particle (TRAPP) family of complexes act as a trafficking complex and participate in events upstream of vesicle fusion, and in some aspects of vesicle tethering to their correct intracellular target membrane.
In humans, the TRAPP complex is formed by the core proteins (TRAPPC1, TRAPPC2, two of TRAPPC3, TRAPPC4, TRAPPC5, TRAPPC6 and TRAPPC2L) that self-assemble to form a stable core (Sacher et al., 2019). This TRAPP core then interacts with a number of accessory proteins to form two distinct, yet related complexes called TRAPP II (core plus TRAPPC9 and TRAPPC10) and TRAPP III (core plus TRAPPC8, TRAPPC11, TRAPPC12 and TRAPPC13). Both TRAPP II and TRAPP III have an accepted function in the secretory pathway, and recent evidence demonstrates that TRAPP III is also involved in autophagy (Behrends et al., 2010; Lamb et al., 2016; Stanga et al., 2019). Although the exact subunit composition of the human TRAPP complexes is still being resolved, the core TRAPP proteins including human TRAPPC4 (and the yeast homologue Trs23) are essential to the stability of both TRAPP complexes. All of the core subunits of the TRAPP complex are highly conserved from yeast to mammals (Loh et al., 2005; Sacher et al., 2008, 2019), and some of the core subunits are essential for cell viability in either yeast or human cells, or both, whilst the accessory subunits are mostly conserved, but not always essential for viability (Blomen et al., 2015; Hart et al., 2015; Wang et al., 2015; Kim et al., 2016). The best characterized biochemical function of the TRAPP complex is as a guanine nucleotide exchange factor (GEF) to activate Ypt/Rab GTPases (Jones et al., 2000; Wang et al., 2000; Thomas and Fromme, 2016). The Ypt/Rab GTPases coordinate intracellular trafficking pathways by acting as molecular switches, which can be stimulated by GEFs to mediate different stages of vesicle trafficking (Segev, 2001).
Recent evidence highlights the critical importance of the TRAPP complex to a spectrum of human diseases, now collectively termed TRAPPopathies, which feature diverse but also overlapping phenotypes (reviewed by Brunet and Sacher, 2014; Kim et al., 2016; Sacher et al., 2019). Intellectual disability and brain malformation-related phenotypes are a common feature of TRAPPopathies. Variants in TRAPPC9 (MIM: 611966) have been associated with autosomal-recessive intellectual disability, thinning of the corpus callosum and white matter loss, microcephaly, dysmorphism, in some cases impaired mobility, yet rarely seizures (Mir et al., 2009; Mochida et al., 2009; Philippe et al., 2009; Kakar et al., 2012; Marangi et al., 2013). TRAPPC6A (MIM: 610396) variants have been associated with Alzheimer’s disease (Hamilton et al., 2011; Chang et al., 2015), and more recently intellectual disability, microcephaly, hypotonia and skeletal abnormalities (Mohamoud et al., 2018). TRAPPC6B (MIM: 617862) variants were identified in patients with neurodevelopmental delay, brain abnormalities, movement disorder and microcephaly and all cases experienced seizures (Marin-Valencia et al., 2018). TRAPPC11 (MIM: 615356) variants have been associated with movement disorder and neurological abnormalities including cerebral atrophy, ataxia and intellectual disability and several cases were reported with seizures and scoliosis (Bogershausen et al., 2013; Fee et al., 2017; Koehler et al., 2017; Liang et al., 2017; Matalonga et al., 2017; Larson et al., 2018). Pathogenic variants in TRAPPC12 (MIM: 617669) result in childhood encephalopathy, severe intellectual disability, microcephaly and brain abnormalities (Milev et al., 2017). Lastly, single cell expression profiling of dopaminergic neurons identified TRAPPC4 as a differentially regulated gene in Parkinson disease (Elstner et al., 2009) but no primary pathogenic variants have been reported to date in TRAPPC4. Nevertheless, collectively these studies suggest that various TRAPP subunit variants result in a wide range of clinical brain malformation-related phenotypes.
The brain-related phenotypes may be partly explained by the high neuronal expression levels of some TRAPP proteins. Although widely expressed throughout the body, TRAPPC9 expression appears highest in the cerebellum (Court et al., 2014), where it is involved in the nuclear factor-kappa B signalling pathway (Hu et al., 2005), and may be critical for neuronal differentiation (Zhang et al., 2014) and neurite outgrowth (Hu et al., 2005). TRAPPC4 (also known as synbindin) is highly expressed in the CNS, particularly in large neurons such as pyramidal neurons, Purkinje cells, and motor neurons, where it is responsible for postsynaptic membrane trafficking (Ethell et al., 2000).
In this report we demonstrate the critical importance of TRAPPC4 in humans. We report a TRAPPC4 pathogenic variant, discovered by whole exome sequencing and family-based rare variant analyses, in three unrelated families with children suffering from early-onset seizures, profound intellectual disability, microcephaly and sensorineural hearing loss, spastic quadriparesis, and with progressive cortical and cerebellar atrophy on brain MRI. All children, from different ethnicities, carried an identical homozygous pathogenic intronic variant in TRAPPC4, which was not due to a founder effect based on haplotype analysis of the families we had access to analyse. We provide evidence that a significant reduction of TRAPPC4 protein in patient fibroblasts due to impaired splicing negatively influences intracellular trafficking and autophagy. To mimic the human variant in the yeast system, we characterized a temperature-sensitive variant in TRS23, the yeast homologue of TRAPPC4. This variant results in a low level of the TRAPP subunit, affecting both secretion and autophagy. Given our genetic findings of homozygosity for the variant in seven affected subjects from three unrelated families, that TRAPPC4 appears to be highly expressed in neurons, and that pathogenic variation in genes for other TRAPP complex proteins result in neurological diseases, we suggest that the TRAPPC4 variant we have identified is the cause of the neurological disorder in these subjects.
Materials and methods
Full materials and methods are available in the online Supplementary material.
Subjects and variant analysis
All procedures followed were in accordance with the ethical standards and approved by the human research ethics committee of the respective participating institutes. Exome sequencing was used to identify candidate genes and single nucleotide polymorphism (SNP) array analysis was used to determine if there was a shared haplotype. Fibroblast cultures were established from skin biopsies and compared to control fibroblasts established from in-house controls from healthy children without suspected genetic disorders.
Protein expression and TRAPP complex stability
Fibroblast extracts were analysed by SDS-PAGE, native PAGE or size exclusion chromatography and fractions separated by SDS-PAGE followed by immunoblotting against TRAPPC4, TRAPPC8, TRAPPC12, TRAPPC2L, or TRAPPC10 as representative subunits of the TRAPP complexes.
Reverse transcription PCR analysis
Total RNA was isolated using a commercially available kit (Qiagen RNeasy® kit). cDNA was synthesized using Invitrogen SuperScript™ III first strand master mix and random hexamers. Reverse-transcription PCR (RT-PCR) reactions were performed using custom primers and PCR products were extracted from agarose gels followed by Sanger sequencing.
Quantitative PCR analysis
Total RNA was isolated using a commercially available kit (Qiagen RNeasy® kit). cDNA was synthesized (GoScript™ Reverse Transcription master mix with random hexamers, Promega). Quantitative reverse-transcription PCR (qPCR) reactions were performed using custom primers and AccuPower® 2X Greenstar™ qPCR Master Mix (Bioneer Pacific) and a Roche Real Time PCR machine.
VSVG–GFP ts045 trafficking assay
The temperature sensitive vesicular stomatitis virus glycoprotein (VSVG)–GFP ts045 construct was used as a marker for transport along the secretory pathway. Cells were transfected with a plasmid containing the VSVG–GFP ts045 construct, incubated at 40°C overnight then transferred to 32°C and left for fixed intervals of 0, 30, 60 or 120 min. At each time point cells were fixed, permeabilized and immunostained with the Golgi marker anti-GM130, Alexa-595 conjugated secondary antibody then counterstained with the nuclear marker 4′,6-diamidino-2-phenylindole (DAPI). Co-localization of VSVG–GFP ts045 and the Golgi maker GM130 was used to determine the kinetics of transport through the Golgi.
Cellular fractionation and protease protection assay
Cells were cultured in either Dulbecco’s modified Eagle medium (DMEM) with 10% foetal bovine serum (FBS; non-starved) or Earle’s Balanced Salt Solution (EBSS) containing 200 nM bafilomycin A1, homogenized and the post-nuclear supernatant (PN) was obtained by centrifugation at 300g. This was further centrifuged at 7700g to generate the low speed pellet (LP). The resulting supernatant was centrifuged at 100 000g to generate a high-speed pellet (HP) and a high speed supernatant (HS). The low and high speed pellets were resuspended in the same volume of homogenization buffer. To analyse the integrity of the membranes, the low and high speed pellets were either left untreated or incubated with proteinase K with or without Triton™ X-100. All samples (LP, HP and HS) were precipitated by incubating in 10% trichloroacetic acid, washed, resuspended in 1× Laemmli sample buffer and processed for western analysis.
Autophagic flux assay
Cells were washed twice with phosphate-buffered saline (PBS) and incubated with EBSS for 2 h. In some cases bafilomycin A1 was included during a 2 h starvation to block the fusion of autophagosomes with lysosomes. Cells starved and treated with bafilomycin A1 were returned to nutrient-rich medium by washing the cells twice with PBS and incubating in DMEM with 10% FBS for 20 or 40 min. The cells were lysed and analysed by western analysis.
Immunofluorescence microscopy
Cells were fixed, quenched with glycine, permeabilized with Triton™ X-100, blocked then incubated with primary antibodies (anti-LC3B and anti-LAMP1) followed by the appropriate cross-adsorbed secondary antibodies and counterstained with DAPI.
Yeast strains, plasmids and reagents
Reagents related to yeast strains, plasmids, antibodies, plasmid construction and yeast strain generation are described in the Supplementary material.
Yeast cell growth and analysis
Media preparation, yeast cell growth, transformation, microscopy and autophagy phenotype analyses, nitrogen starvation and cargo processing, were carried out as previously described (Lipatova et al., 2016). Endoplasmic reticulum (ER)-phagy analysis was described in Lipatova and Segev (2015), and cell viability analysis was described in Zhou et al. (2019). The general secretion assay was performed as previously published (Gaynor and Emr, 1997), with some modifications (Jedd et al., 1995).
Yeast protein analyses
Yeast lysate preparation, immunoblot analysis and quantification were performed as previously described (Lipatova et al., 2016). GST-pulldown was carried out as previously described (Lipatova et al., 2008) except that 0.2% Triton™ X-100 was added to the wash buffer.
Statistical analysis
Statistical analyses were carried out using either a two-tailed Student’s t-test or one-way ANOVA corrected for multiple comparisons as appropriate (GraphPad Prism® Software). Error bars represent the standard deviation of the mean (± SD). A P-value < 0.05 was considered to be statistically significant.
Data availability
The raw data that support the findings of this study are available from the corresponding author, upon request.
Results
Case descriptions
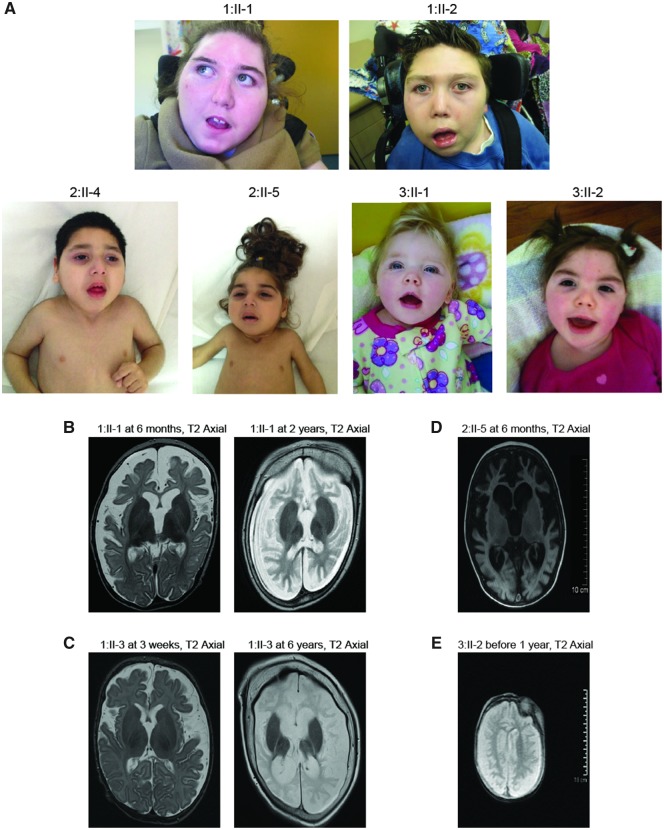
We report seven subjects from three unrelated families, the subjects originating from Caucasian, Turkish and French-Canadian backgrounds. All affected subjects carried an identical homozygous variant in TRAPPC4. The predominant clinical features of the affected subjects included normal birth followed by feeding difficulties during the neonatal period. Most subjects developed progressive microcephaly, severe to profound developmental disability, seizures, spastic quadriparesis, and progressive scoliosis with some subjects requiring surgical treatment. The majority of affected subjects had common clinical features of microcephaly, facial dysmorphism with bitemporal narrowing and a long philtrum (Fig. 1A). A shared similar gestalt for facial dymorphology is perhaps emerging; however, a clinically recognizable pattern is not yet delineated.
Figure 1.
Clinical and brain imaging features of affected subjects with TRAPPC4-related neurological disorder. (A) Frontal clinical images of affected subjects, noting the common features of microcephaly, facial dysmorphism, bitemporal narrowing and long philtrum. (B) T2-weighted axial images show that between 6 months and 2 years of age Subject 1:II-1 had progressive generalized severe cerebral atrophy, particularly affecting white matter, with relative sparing of the basal ganglia. (C) T2-weighted images showed that between 3 weeks and 6 years Subject 1:II-3 had progressive large extra-axial spaces and enlarged ventricles, plagiocephaly involving the occipital parietal region, with a thin corpus callosum and wide Sylvian fissures. (D) T2 FLAIR SE axial image of Subject 2:II-5 at 6 months of age showing severe cerebral atrophy, and relatively milder cerebellar atrophy. There was preservation of deep grey matter structures. (E) T2-weighted images of Subject 3:II-2 in the first year of life showed cerebral atrophy. Full clinical features are described in Supplementary Table 1.
Brain MRI showed that Subject 1:II-1 had progressive generalized severe cerebral atrophy, particularly affecting white matter, with relative sparing of the basal ganglia (Fig. 1B). Subject 1:II-3 had progressive large extra-axial spaces and enlarged ventricles, plagiocephaly involving the occipital parietal region, with a thin corpus callosum and wide Sylvian fissures (Fig. 1C). Subject 2:II-5 showed severe cerebral atrophy, relatively milder cerebellar atrophy, with preservation of deep grey matter structures (Fig. 1D), and Subject 3:II-2 showed cerebral atrophy in the first year of life (Fig. 1E). Hypothalamic dysfunction (hypothermia, premature adrenarche), optic atrophy and cortical visual impairment were present in some subjects. Detailed clinical reports of all cases are available in the Supplementary material and Supplementary Table 1. Fibroblasts were collected from one of the affected children (Subject 1:II-1) and both parents for ongoing studies.
Sequencing and in silico analysis
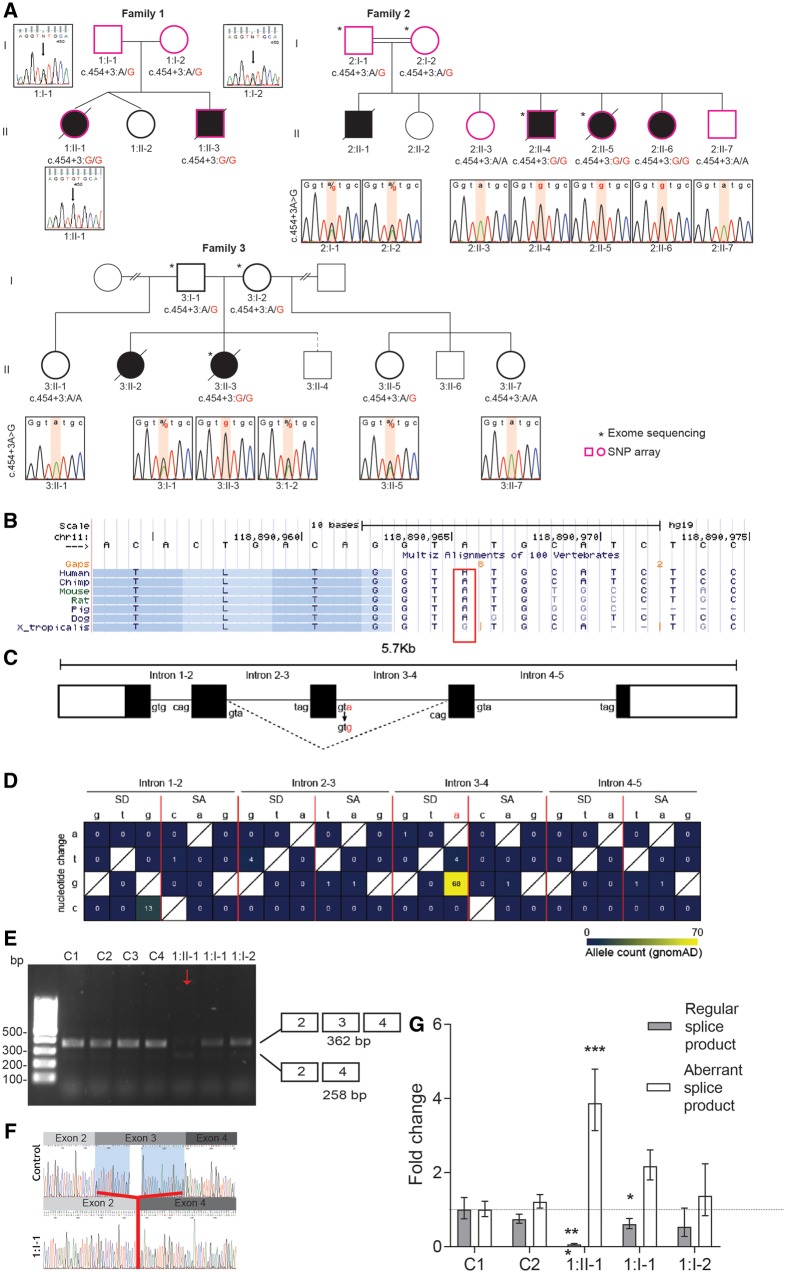
The researchers for all families were connected using GeneMatcher (Sobreira et al., 2015). Exome sequencing analysis was undertaken using DNA extracted from blood of the affected subjects and their parents (Fig. 2A). After alignment to the reference human genome (UCSC Genome Browser hg19) and variant calling, several filtering steps were applied (Supplementary material) for rare variant family based genomic analyses applied (Supplementary material). The affected subjects were confirmed to be homozygous for the trafficking protein particle complex 4 gene [TRAPPC4; hg19:11:g.118890966A->G; NM_016146.5; c.454+3A>G; p.(Leu120Aspfs*9)] by Sanger sequencing, whilst in all cases parents were heterozygous for this variant and unaffected siblings were either heterozygous for the variant, or homozygous for the wild-type variant (Fig. 2A) consistent with autosomal recessive inheritance. The TRAPPC4 variant was located in a putative, highly conserved splice site within TRAPPC4 (Fig. 2B). Human Splicing Finder (HSF v3.1) analysis (Desmet et al., 2009) predicted that the wild-type donor site would be disrupted, and most likely affect splicing (Fig. 2C). The total minor allele frequency according to gnomAD was 0.0002419, with a total of 68 heterozygotes, and no homozygotes reported. The allele count for other variants in TRAPPC4 splicing donor and/or splicing acceptor sites was very low (Fig. 2D). The heterozygote TRAPPC4 cases in gnomAD are reported in European, Latino, South Asian and African populations.
Figure 2.
Pedigrees of families and genetic findings in TRAPPC4-deficient families. (A) Pedigrees of Families 1–3 all with a recessive inherited c.454+3A>G variant in TRAPPC4. Confirmatory Sanger sequencing in the three families shows a homozygous c.454+3A>G TRAPPC4 variant in the affected subjects, a heterozygous variant in each parent and homozygous wild-type or heterozygous configuration in unaffected tested siblings. Asterisk indicates exome sequencing. (B) Multiple sequence alignment surrounding the variant was performed using the Multiz Alignment of 100 vertebrates in the UCSC browser shows that the c.454+3A>G TRAPPC4 variant is a highly conserved intronic residue (surrounded by red box). (C) The structure of TRAPPC4 (NM_016146.5) with the reference sequence of exon-intron junctions shown, and the c.454+3A>G variant indicated in red. (D) The heat map showing the allele count in gnomAD for the variants in splicing donor (SD) and splicing acceptor (SA) sites in TRAPPC4. Yellow colour shows higher frequency and blue colour shows the lower frequency allele counts. (E) RT-PCR analysis demonstrated a splicing defect leading to incomplete exon skipping and formation of an additional PCR product in an affected subject (Subject1:II-1, patient marked with a red arrow) that was not present in the affected subject’s parents (Subjects 1:I-1 and 1:I-2) or four controls (C1–C4). (F) Sanger sequencing of the RT-PCR fragments in E confirmed the sequence of the 362-bp product contained exons 2, 3 and 4 in controls, and in the affected subject (Subject 1:II-1), whilst the lower band only present in the affected subject confirmed skipping of exon 3. (G) Quantitative real-time PCR in control, parental and subject fibroblasts showed a significant decrease in the regular splice product of TRAPPC4 in Subject 1:II-1 fibroblasts and one of the parents (Subject 1:I-2) compared to controls. Additionally, there is a significant increase in levels of the aberrant transcript in fibroblasts from Subject 1:II-1 compared to controls. Statistical significance was determined using one-way ANOVA with Bonferroni correction for multiple comparisons. Data show mean, upper and lower limits of n = 3 independent biological collections. *P < 0.05; ***P < 0.001 versus C1.
Single nucleotide polymorphism array analysis
The phasing results revealed that there was no haplotype shared in this region involving contributing ‘recurrent alleles’ from Subjects 1:II-1 and 1:II-3 (affected siblings) in the first family and Subject 2:II-5 in the second family from distinct world populations. We detected absence of heterozygosity (AOH) regions from SNP array data with BafCalculator (https://github.com/BCM-Lupskilab/BafCalculator) (Karaca et al., 2018). In the first family, we identified an ∼1.51 Mb AOH region (chr11:118022107-119537683) in Subjects 1:II-1 and 1:II-3 affected siblings including the TRAPPC4 recurrent allele. In the second family, we identified an ∼4.5 Mb AOH region (chr11:117469122-121971098) in Subject 2:II-5 that included the TRAPPC4 recurrent allele.
mRNA analysis confirms altered splicing in Subject 1:II-1 fibroblasts
Because the TRAPPC4 c.454+3A>G variant was predicted to alter splicing, we determined the effect on the mature TRAPPC4 transcript using RT-PCR and primers to amplify exons 2–4, followed by Sanger sequencing of the resulting fragments. In four different control fibroblasts one product of 362 bp was observed, representing the wild-type TRAPPC4 transcript with exons 2, 3 and 4 properly spliced. In fibroblasts from Subject 1:II-1, two RT-PCR products were apparent, representing the wild-type TRAPPC4 transcript as well as a shorter 258 bp PCR amplicon containing TRAPPC4, which lacked exon 3. The shorter product was not visually detected in four paediatric control fibroblast lines, nor in the two parental lines (Fig. 2E). Sanger sequencing confirmed skipping of exon 3 (Fig. 2F). The TRAPPC4 c.454+3A>G variant was further quantitatively investigated. Quantitative PCR using primers designed specific to the wild-type splice product (forward primer spanned the exon 2–3 boundary) or to the aberrant splice product (forward primer spanned the exon 2–4 boundary) revealed that fibroblasts from Subject 1:II-1 only had 6% of the regular splice product as compared with control fibroblasts (100%). There was also a significant reduction (54% of controls) of the regular splice product in the mother (Subject 1:I-2). Additionally, there was a 387% increase in the transcript abundance of the aberrant splice product in patient fibroblast from Subject 1:II-1 compared to controls (100%, Fig. 2G). From this we can conclude that the TRAPPC4 c.454+3A>G variant leads to a splicing defect that results in skipping of exon 3, and a loss of 96% of the intact TRAPPC4 transcript that contributes to the phenotype of the patient.
TRAPPC4 protein level is reduced in fibroblasts derived from Subject 1:II-1
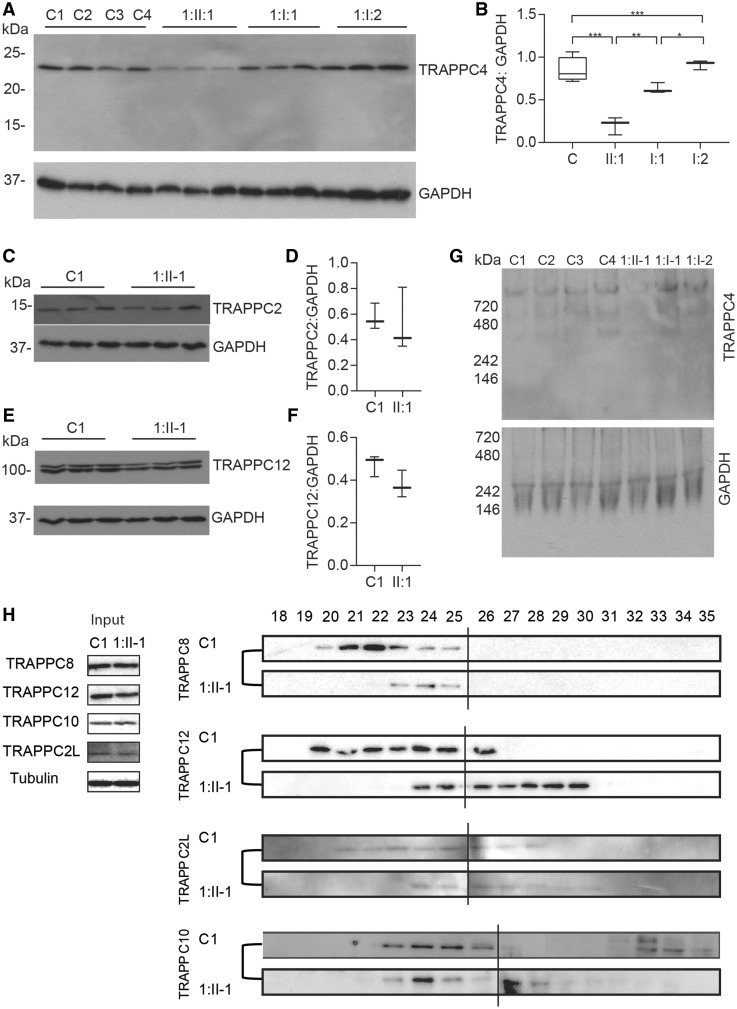
The homozygous c.454+3A>G variant leading to skipping of exon 3 is predicted to result in a frameshift and early truncation of the TRAPPC4 protein p.(Leu120Aspfs*9) likely resulting in nonsense-mediated decay. However, RT-PCR confirmed some wild-type TRAPPC4 transcript was still produced in fibroblasts from Subject 1:II-1. Western blot analysis detected full-length TRAPPC4 protein in this subject (predicted size of 24.3 kDa) (Fig. 3A), but the TRAPPC4 protein levels were significantly reduced compared to heterozygous carrier parental fibroblast samples and paediatric controls (Fig. 3B). There were no smaller protein products detected using an antibody against the N-terminal region of TRAPPC4, which would detect the protein product missing exon 3. Therefore, the shortened transcript is likely unstable and subject to nonsense-mediated decay. Further analysis of TRAPPC2 and TRAPPC12, two other TRAPP complex subunits, did not show any reduction in protein levels under denaturing conditions (Fig. 3C–F). Thus, this TRAPPC4 variant leads to a reduced level of the TRAPPC4 protein without affecting protein levels of other representative TRAPP proteins.
Figure 3.
Decreased protein levels and impaired TRAPP complex assembly in fibroblasts from an affected subject with TRAPPC4 c.454+3A>G homozygous variant. (A) Western blot of TRAPPC4 and GAPDH levels in four paediatric controls (C1–C4), as well as fibroblasts from an affected subject with c.454+3A>G homozygous variant (Subject 1:II-1) and parents (heterozygous for the same variant; Subjects1:I-1 and 1:I-2) in triplicate. (B) Quantification of the western blots in A demonstrate that there was a significant decrease in endogenous TRAPPC4 protein levels normalized to GAPDH in fibroblasts from an affected subject (Subject 1:II-1) compared to both parental fibroblasts (Subjects 1:I-1 and 1:I-2) and paediatric control fibroblasts. (C) Western blot of TRAPPC2 and GAPDH in control fibroblasts and an affected subject (Subject 1:II-1) and (D) quantification of western blots in C demonstrate no difference in protein levels of TRAPPC2 in Subject 1:II-1. (E) Western blot of TRAPPC12 and GAPDH in a control fibroblast and an affected subject (Subject 1:II-1) and (F) quantification of western blots in E demonstrate no difference in protein levels of TRAPPC12 in Subject 1:II-1. (G) Native PAGE electrophoresis revealed decreased levels of TRAPPC4-containing complexes in fibroblasts from Subject 1:II-1 relative to parental fibroblasts (Subjects 1:I-1 and 1:I-2) and paediatric control fibroblasts (C1–C4). Molecular size standards are indicated to the left. (H) Size exclusion chromatography of fibroblasts from Subject 1:II-1 and control fibroblasts. The TRAPP proteins TRAPPC8, TRAPPC12 and TRAPPC2L are shifting to a smaller molecular size in fibroblasts from the affected subject compared to control fibroblasts. The numbers at the top represent the fraction off the column, with smaller numbers indicating larger molecular size. Data in B, D and F are box-and-whisker plots showing median, interquartile interval, minimum and maximum of n > 3 per measurement from at least three biological collections of an affected subject and at least two independent experiments. Statistical significance was determined using one-way ANOVA with Bonferroni correction for multiple comparisons. *P < 0.05; **P < 0.01; ***P < 0.001. Molecular weight standards are indicated on the right (kDa).
TRAPPC4 c.454+3A>G leads to a reduction in the levels of TRAPP complexes
As the TRAPPC4 c.454+3A>G variant resulted in a significant reduction of full-length protein on denaturing PAGE, and TRAPPC4 is a core subunit of the TRAPPII and TRAPPIII complexes, we used native PAGE and size exclusion chromatography to determine whether reduced levels of the core TRAPPC4 protein in fibroblasts affected the overall levels and/or stability of assembled TRAPP complexes. Proteins were extracted and either separated by native PAGE to retain protein complex integrity or separated on size exclusion chromatography and fractions immunoblotted. Native PAGE analysis of TRAPP complexes in Subject 1:II-1 fibroblasts revealed three major bands detected with an anti-TRAPPC4 antibody, corresponding to >1000 kDa, ∼800 kDa and ∼500 kDa, which could correspond to the TRAPP III and TRAPP II complexes, and a subassembly product (Fig. 3G). The intensity of the anti-TRAPPC4-detected bands was lower in Subject 1:II-1 compared to controls and parental samples. The relative intensity of both the ponceau-stained membrane (representing total cellular protein levels, data not shown) and GAPDH protein level was comparable in all samples, and the detected GAPDH band in fibroblast lysates had an apparent molecular size of ∼720 kDa, as previously described for cultured cells (Kunjithapatham et al., 2015). When Subject 1:II-1 and a control fibroblast sample were separated by size exclusion chromatography, there was a significant shift in the migration of TRAPP-containing complexes in fibroblasts from Subject 1:II-1 compared to control fibroblasts with the proteins shifting towards a smaller molecular size [smaller molecular sizes appear in the higher numbered (later) fractions] (Fig. 3H). Collectively, using two independent methodologies, we have demonstrated that lysates from an individual with a TRAPPC4 c.454+3A>G variant have reduced levels of fully-assembled TRAPP complexes.
Altered VSVG-GFP trafficking in fibroblasts from an affected subject
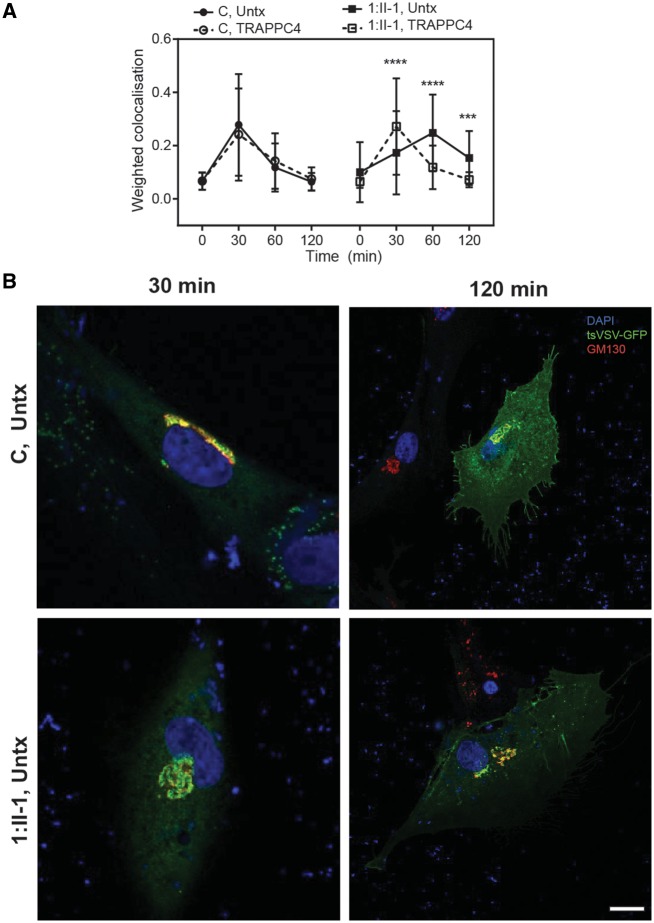
As the TRAPP complexes are required for several steps in the biosynthetic pathway, and variants in other TRAPP genes are known to affect secretion (Bogershausen et al., 2013; Koehler et al., 2017; Milev et al., 2017, 2018), we investigated whether the reduced levels of TRAPPC4 protein affected secretion in Subject 1:II-1 fibroblasts. To determine whether secretion was delayed between the ER, Golgi and plasma membrane we used a temperature sensitive form of VSVG fused to GFP (VSVG–GFP ts045) as a marker protein for movement along the secretory pathway (Bergmann and Singer, 1983; Hirschberg et al., 1998). This commonly used protein construct misfolds at higher temperatures (40°C) and accumulates in the ER. When cells are returned back to permissive temperature, the construct exits the ER and is transported via the Golgi to the cell surface (Bergmann and Singer, 1983). In as little as 25–30 min the construct appears in the Golgi, then reaches the cell surface after 90–120 min (Hirschberg et al., 1998). We determined secretion by transfecting fibroblasts with the VSVG–GFP ts045 construct, then cells were held at restrictive temperature (40°C) overnight, followed by release back to permissive temperature (32°C). Cells were fixed at regular intervals then counterstained with the Golgi marker GM130 (Supplementary Figs 1 and 2) and the degree of co-localization was examined. In control cells VSVG–GFP ts045 co-localization with the Golgi peaked at 30 min, and progressively reduced with extended time points and only 23% remained in the Golgi after 120 min (Fig. 4A and B). The trafficking dynamics we observed in control fibroblasts corresponded to previous findings where VSVG–GFP ts045 reached the Golgi at ∼30 min (Koehler et al., 2017; Milev et al., 2017, 2018) and secreted to the cell surface after 90–120 min (Hirschberg et al., 1998). Lentiviral transduction of wild-type TRAPPC4 into control cells had no obvious effect. In fibroblasts from Subject 1:II-1, VSVG-GFP ts045 co-localization with the Golgi was significantly delayed compared to control cells with a peak accumulation occurring at 60 min. There was also delayed exit from the Golgi, where after 120 min 62% of VSVG–GFP ts045 still remained in the Golgi (Fig. 4A and B). Lentiviral transduction with wild-type TRAPPC4 construct into fibroblasts from Subject 1:II-1 completely restored the trafficking defect to that of control cells. These experiments suggest that there is a secretory defect that can be explained by the reduced levels of TRAPPC4 protein.
Figure 4.
Impaired Golgi trafficking in fibroblasts from an affected subject is rescued by lentiviral transduction of wild-type TRAPPC4. Control fibroblasts and fibroblasts derived from Subject 1:II-1 were either untransduced (Untx) or stably transduced with wild-type TRAPPC4 lentivirus (TRAPP). Cells were then transfected with a temperature-sensitive vesicular stomatitis virus glycoprotein VSVG–GFP ts045, then incubated at the non-permissive temperature of 40°C overnight. Cells were then shifted to the permissive temperature of 32°C for the time intervals indicated, fixed with PFA then immunostained with the Golgi marker GM130 and counterstained with the nuclear marker DAPI. VSVG-GFP ts045 co-localization with GM130 was determined for a minimum of 60 cells per time point and treatment. (A) There was no difference in trafficking in naïve control cells or those transduced with TRAPPC4 lentivirus. However, in fibroblasts from the affected subject there was a significant delay in VSVG-GFP ts045 entering the Golgi, and a significant delay in exiting the Golgi compared to control cells. Golgi trafficking in fibroblasts from the affected subject was restored back to levels of control upon transduction with wild-type TRAPPC4 lentivirus. (B) Representative confocal images of intracellular trafficking (blue, DAPI; green, VSVG-GFP ts045; red, GM130). More representative figures can be found in Supplementary Figs 1 and 2. Data are average ± SD, n > 60 cells per treatment and time point. Significance was measured by a two-way ANOVA with Sidak multiple comparisons correction. ***P < 0.001, ****P < 0.0001. Scale bar = 20 µm.
Autophagic flux is affected in fibroblasts from an affected subject
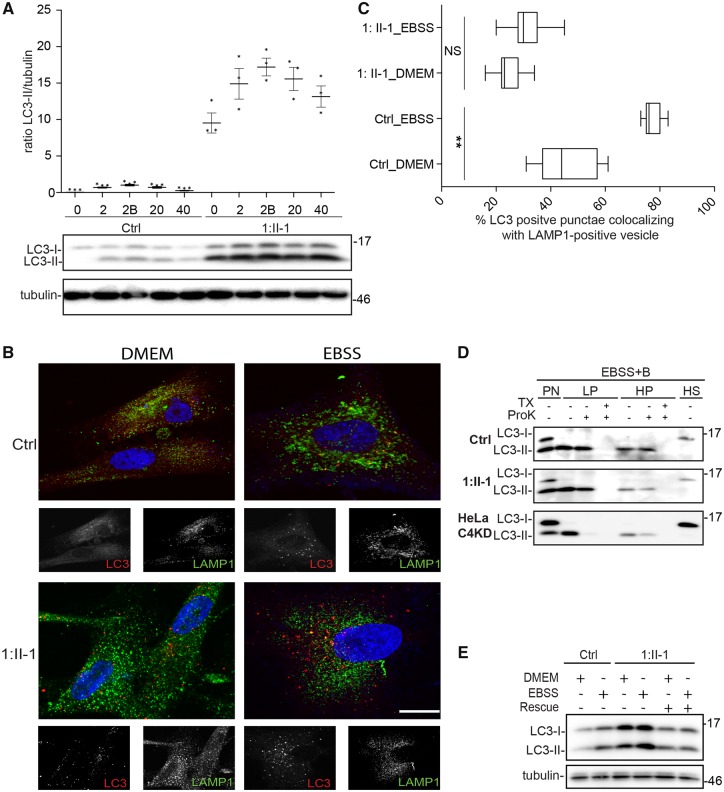
The TRAPPC4 protein is a critical subunit of the core TRAPP complex, which is central to both the TRAPP II and III complexes. As the TRAPP III complex has been implicated in starvation-induced autophagy (Behrends et al., 2010; Lamb et al., 2016; Ramirez-Peinado et al., 2017; Stanga et al., 2019) we examined whether the reduced levels of TRAPPC4 in the affected subject resulted in an autophagy defect. As shown in Fig. 5A, the levels of the autophagic marker LC3-II in the fibroblasts from the affected subject were elevated even prior to starvation. An increase in LC3-positive structures was also seen by immunofluorescence microscopy (Fig. 5B). While starvation resulted in a further increase in LC3-II, upon return to nutrient-rich medium there was no significant reduction in LC3-II compared to control, suggesting an autophagic flux defect. As LC3-positive autophagic membranes ultimately fuse with the lysosome, we examined the cells for overlap between LC3 and the lysosomal marker LAMP1. Consistent with an autophagic flux defect, the percentage of overlap between these two markers was reduced both prior to and following starvation in fibroblasts from the affected subject (Fig. 5B and C). We have recently shown that the TRAPP III protein TRAPPC11 is involved in sealing of isolation membranes into autophagosomes (Stanga et al., 2019). Given the reduced levels of assembled TRAPP III (Fig. 3), we examined if the LC3-II-positive membranes were defective in sealing into closed autophagosomes using a protease protection assay. The assay examines the resistance of LC3-II to proteinase K as an indication of sealed autophagosome closure in both high speed and low speed pellet fractions (HP and LP, respectively). As shown in Fig. 5D, while LC3-II in both the high-speed and low-speed supernatants from the control was largely resistant to degradation by proteinase K treatment, that from the affected subject was sensitive to the protease treatment. Suspecting that the incomplete sensitivity in the affected subject was due to reduced, but not absent, levels of TRAPPC4, we reduced the levels of TRAPPC4 to nearly undetectable levels in HeLa cells by RNA interference. In this case, virtually all of the LC3-II in the high speed and low speed pellet fractions was sensitive to proteinase K treatment. These results suggest that the reduced levels of TRAPPC4 in the affected subject result in a partial defect in both basal and starvation-induced autophagy, partly due to a defect in autophagosome formation.
Figure 5.
Autophagy is defective in fibroblasts from an affected subject. (A) Control fibroblasts and fibroblasts derived from a subject with the homozygous c.454+3A>G variant in TRAPPC4 (Subject 1:II-1) were either unstarved (0) or transferred into starvation medium (EBBS) for 2 h without (2) or with (2B) bafilomycin A1. The bafilomycin A1-treated cells were then transferred into nutrient-rich (DMEM) medium for 20 or 40 min (indicated as 20 and 40). Lysates were prepared and LC3 and tubulin were detected by western analysis. The ratio of LC3-II:tubulin was calculated and is plotted in the graph with a representative western analysis shown. (B) The same fibroblasts as in A were either left in nutrient-rich medium or starved for 2 h. The cells were the processed for immunofluoerescence microscopy as described in the ‘Materials and methods’ section. (C) The per cent overlap between LC3 and LAMP1 from the cells in B was calculated using Imaris for n = 10 cells in all cases. (D) The same fibroblasts as in A were grown in starvation (EBSS) medium for 2 h in the presence of bafilomycin A1. Lysates were prepared and processed for the membrane sealing assay as described in the ‘Materials and methods’ section. Prior to fractionation, some samples were treated with proteinase K (ProK) or with ProK in the presence of 1% Triton X-100 (TX). The fractions, composed of a postnuclear supernatant (PN), low-speed pellet (LP), high-speed pellet (HP) and high-speed supernatant (HS) were probed for LC3. The reduction in LC3-II following proteinase K was ∼50% in lysates from the affected subject. Data are a box-and-whisker plot showing median, interquartile interval, minimum and maximum of n > 3 per measurement from at least three biological collections of an affected subject and at least two independent experiments. Significance was measured by a one-way ANOVA with a Tukey’s post hoc HSD analysis. NS = not significant. **P < 0.01.
Low levels of Trs23 in yeast result in a reduction of the core TRAPP complex level
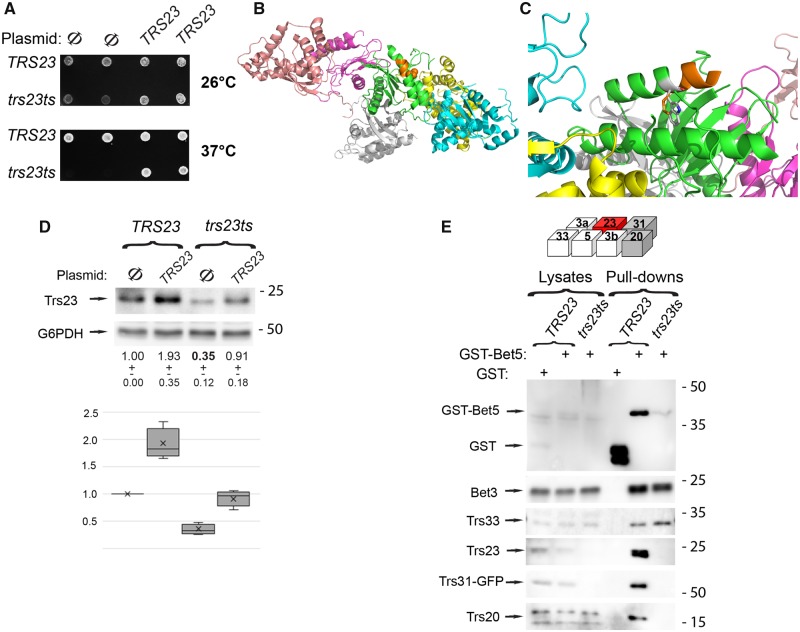
The essential role of the core TRAPP complex in yeast cell viability, secretion and autophagy is well-established (Kim et al., 2016). We wanted to extend our findings in human cells in a eukaryotic model organism. To address this, we used a temperature sensitive variant in TRS23, trs23ts. Temperature-sensitive variants of the four core-TRAPP subunits, bet3ts, bet5ts, trs23ts and trs31ts, were constructed in a high-throughput study (Ben-Aroya et al., 2008). These variants were used to show an effect on autophagy (Zou et al., 2018); however, the trs23ts variant has not been further characterized. We confirmed the temperature-sensitive growth phenotype of trs23ts mutant cells and the complementation of this defect by the wild-type TRS23 expressed from the plasmid (Fig. 6A). Sequencing of trs23ts mutant cells revealed that the five C-terminal amino acids of the protein, 215–219, are deleted and the M214 is changed to W. The yeast Trs23 and its human orthologue TRAPPC4 share 22% identity; however, with omission of the PDZL domain there is 45% sequence identity (Kim et al., 2016). We termed this allele trs23ts-1. When modelling this modification on the 3D structure of Trs23 in the context of the core-TRAPP complex, it seems that the interactions of Trs23 with the other core-TRAPP subunits, or with its substrate Ypt1 (Fig. 6B and C) would not be affected. However, the M214W replacement might affect the folding of the Trs23 protein itself and thereby affect protein stability.
Figure 6.
The yeast trs23ts-1 variant results in low level of the Trs23 protein and impaired assembly of the TRAPP core. (A) Wild-type and trs23-1 mutant cells were transformed with an empty CEN plasmid (Ø) or a plasmid expressing TRS23 with its own promoter and terminator. Cells were grown at permissive (26°C) or restrictive (37°C) temperature. The trs23ts-1 cells had a growth defect at 26°C and were non-viable at 37°C, which was rescued by introduction of the wild-type plasmid. Shown are two independent colonies for wild-type (top) and trs23-1 (bottom), transformed with empty plasmid (Ø) or plasmid expressing TRS23. (B and C). The interaction of the trs23ts-1 mutant protein with other TRAPP complex subunits is shown in 3D; Trs23 (green), Bet3 (yellow and light pink), Bet5 (magenta), and Trs31 (cyan), associated with Ypt1 (grey). In Trs23, the deleted five amino acids are highlighted in orange. The deleted five amino acids are not in a region of Trs23 that associates with other TRAPP complex proteins (Bet3 and Bet5) and is on the opposite side of its interaction surface with Ypt1. Ribbon 3D diagram was rendered using PyMOL. (C) Focuses on the mutated domain of trs23ts-1 (180° rotation from B) and shows the 214W side chain. (D) Wild-type and trs23-1 mutant cells were transformed with an empty CEN plasmid (Ø) or a plasmid expressing TRS23. Lysates were collected and probed for Trs23 and G6PDH as a loading control. The numbers beneath each lane represent the relative levels of Trs23 compared to control. Quantification of a minimum of three such immunoblots normalized to G6PDH is shown. (E) Wild-type and trs23ts cells expressing Trs31-yEGFP were transformed with a plasmid expressing GST or GST-Bet5. Cell lysates were subjected to GST pulldown followed by immunoblot analysis. The cartoon shows the architecture of core TRAPP complex. Bet5 was revealed using anti-GST, Trs31 was revealed with anti-GFP and the remainder of the subunits were revealed with subunit-specific antibodies. G6PDH was used to ensure equal loading of the lysates (not shown). Molecular size standards are indicated on the right (kDa). Results in D are presented as a box-and-whisker plot showing median, interquartile interval, minimum and maximum of n > 3 per measurement from at least three biological collections.
The level of the Trs23 protein was compared in wild-type and trs23ts-1 mutant cells using immunoblot analysis and anti-Trs23 antibody. The level of the mutant Trs23 was 3-fold lower than that of the wild-type protein, and expression of wild-type Trs23 from a plasmid resulted in an increase of the total Trs23 protein in mutant cells (Fig. 6D). Thus, even though at the protein sequence level the yeast variant is not similar to that of the human mutant protein described above, the overall effect on the protein level is similar in that both variants result in a significant reduction in Trs23/TRAPPC4 protein level.
We hypothesized that a low level of Trs23 protein in the trs23ts-1 mutant cells would result in a reduction in the level of the core TRAPP complex that would likely affect the levels of the holocomplexes. It was previously shown that when the six TRAPP subunits, Bet3, Bet5, Trs23, Trs31, Trs20 and Trs33, are expressed in the same cells, they form a stable complex (Kim et al., 2006). In addition, partial TRAPP complexes that contain Bet3-Bet5 and Trs33 can also form a stable sub-TRAPP complex in the absence of Trs23 (Kim et al., 2006). To determine whether the TRAPP core complexes were reduced in the yeast model, wild-type and mutant cells were transformed with a plasmid expressing GST-tagged Bet5 and interacting proteins were captured on glutathione-agarose beads. The level of the other TRAPP subunits in cell lysates and their co-precipitation with GST-Bet5 was compared by immunoblot analysis using antibodies against specific subunits or their tag. Similar levels of Bet3 and Trs33 were present in wild-type and trs23ts-1 mutant cells and co-precipitated with GST-Bet5. However, the levels of Trs31-GFP and Trs20 proteins that co-precipitated with GST-Bet5 were much lower in trs23ts-1 mutant cells when compared to wild-type cells. The level of Trs31-GFP was lower in trs23ts-1 mutant cells when compared to wild-type (the anti-Trs20 antibody was not clean enough to identify the protein in cell lysates) (Fig. 6E). Thus, while partial complexes that contain Bet5, Bet3 and Trs33 can form in trs23ts-1 mutant cells, the level of the complete core TRAPP complex was reduced due to the low level of Trs23 mutant protein. The low level of Trs31 in mutant cells is probably due to the fact that it is not incorporated into the core TRAPP complex, which results in its degradation. Therefore, a reduction in the level of Trs23 affects the assembly of the core TRAPP complex and likely affects assembly of TRAPP II and TRAPP III in yeast.
Reduced Trs23 levels cause secretion and autophagy defects in yeast
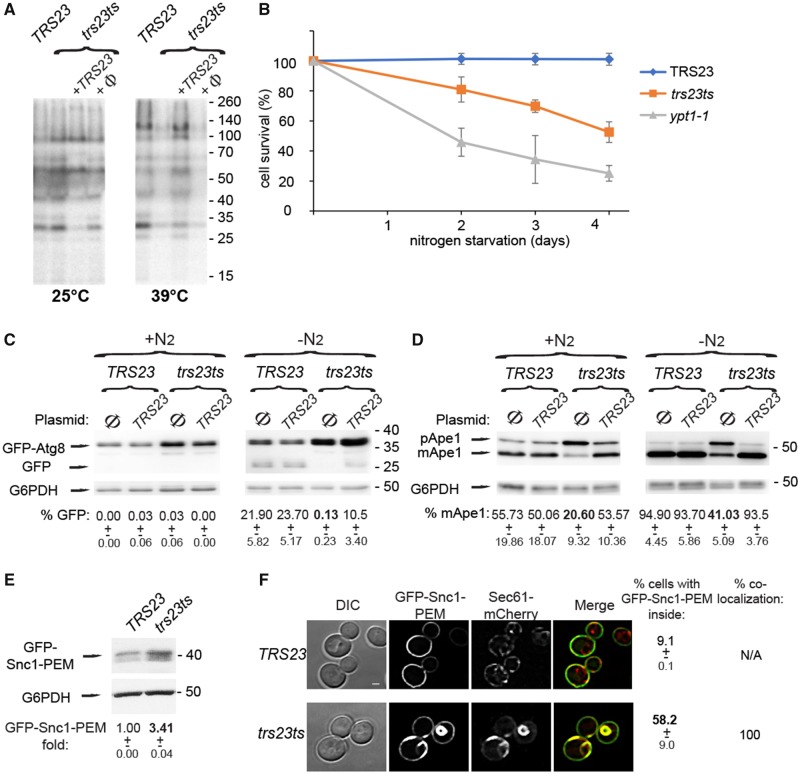
We next determined the effect of the trs23ts-1 variant on general secretion in yeast cells. Secreted proteins can be detected in the cell medium using pulse-chase followed by denaturing gel electrophoresis analysis (Gaynor and Emr, 1997). When wild-type and trs23ts-1 yeast were grown at 25°C, both showed the same pattern of secreted proteins (Fig. 7A), although some lower molecular weight secreted proteins were not fully rescued by wild-type TRS23 at 25°C but were fully complemented at 39°C. When the cells were subjected to heat stress (39°C for 1 h), trs23ts-1 mutant cells, but not wild-type, exhibited a severe block in general secretion. This block was complemented by expression of wild-type TRS23 from a plasmid (Fig. 7A).
Figure 7.
trs23ts-1 cells display secretory and autophagy defects. (A) trs23ts-1 cells exhibit a general secretion defect at 39°C (right) but not at 25°C (left). S35-labelled proteins from media of cells grown at 25°C or 39°C were precipitated with trichloroacetic acid (TCA) and analysed by SDS-PAGE and Phosphoimager (see ‘Materials and methods’ section). Shown from left to right for each temperature: wild-type, trs23ts-1, trs23ts-1 transformed with a plasmid expressing TRS23, and trs23ts-1 transformed with an empty plasmid (Ø). (B) Wild-type, trs23ts-1 and ypt1-1 were starved in medium lacking nitrogen. Cell survival was determined by vital staining with trypan blue. (C) Cells (wild-type or trs23ts-1) expressing GFP-Atg8 were transformed with an empty CEN plasmid (Ø) or a plasmid expressing TRS23. Transformants were grown in selective medium containing nitrogen (+N2, left) or incubated for 4 h in medium without nitrogen (−N2, right). Cell lysates were subjected to immunoblot analysis using anti-GFP (top) and anti-G6PDH (as a loading control). The per cent GFP detected for each condition is shown beneath each lane along with the standard deviation. (D) Cell lysates from C were subjected to an immunoblot analysis using anti-Ape1 and anti-G6PDH (as a loading control) to determine preApe1 (pApe1) and mature Ape1 (mApe1) levels. The per cent mApe1 detected in each condition is indicated beneath each lane along with the standard deviation. (E) Wild-type and trs23ts were transformed with a 2-μ plasmid for expression of GFP-Snc1-PEM and grown at 26°C. GFP-Snc1-PEM levels were determined by immunoblot analysis with anti-GFP antibody. G6PDH was included as a loading control. The level of GFP-Snc1-PEM relative to wild-type is indicated beneath each lane along with the standard deviation. (F) Wild-type (top) and trs23ts (bottom) cells expressing endogenously-tagged Sec61-mCherry and GFP-Snc1-PEM from a plasmid were grown as in E and cells were visualized by confocal fluorescence microscopy. Shown from left to right: DIC (cell contour), GFP channel, mCherry channel, merge. The percentage of cells with intracellular GFP-Snc1-PEM (±SD) and per cent co-localization of GFP-Snc1-PEM with Sec61-mCherry are indicated to the right of the fluorescence micrographs. Scale bar = 1 µm. Molecular size standards are indicated to the right of the gels and immunoblots in A, C, D and E. Results in this figure represent three independent experiments.
An autophagy defect for the trs23ts-1 mutant cells has been previously reported at the permissive temperature in a high-throughput analysis (Zou et al., 2018). Therefore, we further characterized autophagy defects in the mutant cells following two different autophagy processes. First, we showed that trs23ts-1 mutant cells exhibit a mild defect in cell viability during nitrogen starvation compared to the severe defect exhibited by ypt1-1 mutant cells as a positive control (Fig. 7B). Second, cargo-processing defects of trs23ts-1 mutant cells were also tested under nitrogen starvation at the permissive temperature. In agreement with previous observations, processing of GFP-Atg8 to GFP was defective in trs23ts-1 mutant cells, and the phenotype was partially complemented by TRS23 expressed from a plasmid (back to ∼45% of the wild-type level) (Fig. 7C). In the cytosol-to-vacuole (CVT) pathway, preApe1 is processed to mature (mm)Ape1 during normal growth. Processing of preApe1 to mApe1 was partially defective in trs23ts-1 mutant cells under nitrogen starvation (∼2-fold lower than wild-type). This phenotype was completely complemented by TRS23 expressed from a plasmid (Fig. 7D). We also tested the ER-phagy phenotype of trs23ts-1 mutant cells during normal growth at the permissive temperature. In this assay, accumulation of an overexpressed membrane protein, GFP-Snc1-PEM, in the ER is assessed using immunoblot analysis and fluorescence microscopy (Lipatova and Segev, 2015). As we have previously reported for ypt1-1 and trs85Δ mutant cells (Lipatova et al., 2016), the level of GFP-Snc1-PEM was increased in trs23ts-1 mutant cells (Fig. 7E). Moreover, ∼60% of the mutant cells (compared to ∼10% of wild-type cells) accumulated intracellular GFP-Snc1-PEM, which completely co-localized with the ER marker Sec61 (Fig. 7F).
Together, our results demonstrated that yeast cells expressing low levels of the Trs23 protein exhibit defects in autophagy, both in nutrient-rich and -depleted conditions, even at the permissive temperature. In contrast, the secretory defect of these mutant cells was only evident under extreme conditions of high temperature. Therefore, as seen in the fibroblasts with reduced levels of TRAPPC4, reduced levels of Trs23 in yeast affect secretion and autophagy.
Discussion
This work expands the increasing number of TRAPP genes associated with human disease, where we identify the first subjects with a pathogenic variant in TRAPPC4. The affected subjects manifested early onset seizures, profound intellectual disability, microcephaly, sensorineural hearing loss, spastic quadriparesis and cerebral and cerebellar atrophy revealed by brain MRI. The clinical manifestations in these subjects correlate well with subjects with pathogenic variants in other TRAPP complexes, collectively termed TRAPPopathies (Sacher et al., 2019). Intellectual disability, regression or developmental delay, brain abnormalities, microcephaly and in some cases seizures, are common features of TRAPPopathies (Table 1), with the exception of TRAPPC2 variants, which are associated with the skeletal disorder spondyloepiphyseal dysplasia tarda (SEDT; reviewed by Sacher et al., 2019).
Table 1.
Clinical summary of affected family members with TRAPPC4-related neurological disorder and comparison to other TRAPPopathy disorders
| TRAPPC4 | TRAPPC2L | TRAPPC6A | TRAPPC6B | TRAPPC9 | TRAPPC11 | TRAPPC12 | |
|---|---|---|---|---|---|---|---|
| References | This report | Milev et al., 2018 | Mohamoud et al., 2018 | Marin-Valencia et al., 2018 | Mir et al., 2009; Mochida et al., 2009; Philippe et al., 2009; Kakar et al., 2012; Marangi et al., 2013 | Bogershausen et al., 2013; Koehler et al., 2017; Matalonga et al., 2017; Larson et al., 2018 | Milev et al., 2017 |
| Seizures | 7/7 | 2/2 | n/a | 6/6 | 2/15 | 8/13a | 2/3 |
| Intellectual disability | 7/7 | 2/2 | 3/3 | 6/6 | 15/15 | 11/13a | 3/3 |
| Regression or developmental delay | 7/7 | 2/2 | n/a | 6/6 | 11/15 | 11/13a | 3/3 |
| Brain abnormalities on MRI | 4/4a | 2/2 | n/a | 4/4a | 9/11a | 9/13a | 3/3 |
| Microcephaly | 7/7 | 2/2 | 3/3 | 6/6 | 13/15 | 1/1a | 3/3 |
| Impaired mobility | 7/7 | 2/2 | n/a | 6/6 | 4/10a | 12/13 | 3/3 |
| Hearing loss | 2/6a | n/a | 0/3 | n/a | n/a | 1/1a | 3/3 |
| Vision issues | 3/6a | 2/2 | 0/3 | 6/6 | n/a | 6/11a | 3/3 |
Information not available for some cases. More detailed clinical information for all TRAPPC4 cases are provided in Supplementary Table 1 and Supplementary material, full case reports.
n/a = not available.
Brain MRIs from the subjects reported in this study revealed cortical and cerebellar atrophy, which was similar to subjects with TRAPPC6B variants, who displayed thin corpus callosum, cortical, cerebellar and/or brainstem atrophy (Marin-Valencia et al., 2018) and subjects with TRAPPC9 variants, who had a thin corpus callosum, hypoplasia of cerebellum and corpus callosum, white matter loss and microcephaly (Mir et al., 2009; Mochida et al., 2009; Philippe et al., 2009; Marangi et al., 2013). This is somewhat distinct from the broader neurological phenotype of subjects with TRAPPC11 variants, where only a subgroup of subjects had cerebral atrophy (Bogershausen et al., 2013; Koehler et al., 2017). Subjects with pathogenic TRAPPC6A variants also feature intellectual disability and microcephaly (Mohamoud et al., 2018). TRAPPC6A was also associated with Alzheimer’s disease (Hamilton et al., 2011), by forming extracellular matrix plaques in the cortex and cerebellum (Chang et al., 2015). The subjects in the present study had spastic quadraparesis and visual impairment, which has featured in other TRAPPopathies (Table 1). Collectively we provide a comprehensive analysis of some shared and unique clinical features of the TRAPPopathies, although specific genotype-phenotype associations remain to be established with larger patient cohorts.
The homozygous TRAPPC4 c.454+3A>G splicing defect was incompletely penetrant as revealed by a significant reduction in the production of the wild-type transcript as well as an increase in the levels of a mutant transcript lacking exon 3. Aberrant splicing and reduced protein levels have also been reported in other TRAPPopathies including splicing variants in TRAPPC11 (Bogershausen et al., 2013; Koehler et al., 2017), TRAPPC9 (Kakar et al., 2012) and TRAPPC6B (Marin-Valencia et al., 2018). The c.454+3A>G variant resulted in a significant reduction in full-length TRAPPC4 protein levels, but this did not affect the total levels of either TRAPPC2 (part of the core TRAPP complex) or TRAPPC12 (part of the TRAPP III complex). The reduced TRAPPC4 protein levels also affected the assembly of the TRAPP complex as might be expected, as TRAPP complex assembly is likely dependent upon TRAPPC4 levels. The mammalian TRAPP complex exists in two distinct forms, TRAPP II and TRAPP III (Bassik et al., 2013; Zhao et al., 2017). Although yeast has been reported to have an additional TRAPP I complex, its existence in vivo (Brunet et al., 2012; Thomas et al., 2018), as well as a human equivalent, have been challenged (Sacher et al., 2019). The TRAPP II complex has been reported as a complex of ∼670 kDa in humans (Yamasaki et al., 2009) and yeast (Brunet et al., 2012). The yeast TRAPP III complex was estimated to be >1000 kDa (Choi et al., 2011; Brunet et al., 2012). The TRAPP complexes can undergo homotypic or heterotypic oligomerization (Choi et al., 2011) to form higher ordered structures (Kim et al., 2016). Additionally, the method of extraction can significantly influence the molecular weight of the holocomplex (Brunet et al., 2012). Therefore, it is consistent that reduced levels of TRAPPC4, a core subunit of the TRAPP complex, affect the overall levels and/or stability of fully assembled TRAPP complexes.
To determine whether the reduced TRAPPC4 levels block membrane trafficking and secretion we monitored movement of VSVG–GFP ts045 along the secretory pathway (Bergmann and Singer, 1983). Membrane trafficking dysfunction was apparent in fibroblasts from an affected subject, and was completely restored by lentiviral transduction with wild-type TRAPPC4. To validate our findings in a model organism, a temperature-sensitive Saccharomyces cerevisiae yeast trs23ts mutant strain was developed. Because the yeast gene does not have introns, the best way to model the disease in yeast is to have a mutant that results in a low level of the Trs23 protein. This was possible with the temperature-sensitive trs23ts variant. In this mutant, both the level of the Trs23 protein and the full TRAPP complex are low. TRAPP subunits fell into two groups: the protein level of members of the first group (Bet3, Bet5 and Trs33) was similar in wild-type and the trs23ts mutant cells, and in mutant cells they collectively formed a subcomplex. In contrast, members of the second group (Trs23, Trs31 and Trs20) did not join the subcomplex in mutant cells and the protein level of Trs23 and Trs31 was lower. At restrictive temperature the yeast trs23ts strain displayed a secretory defect much like the patient fibroblasts. This defect could be complemented by reintroduction of the wild-type TRAPPC4 gene in a rescue plasmid. The trafficking delay due to decreased TRAPPC4/Trs23 levels is consistent with membrane trafficking dysfunction from reduced levels of other TRAPP proteins associated with human disease (Bogershausen et al., 2013; Koehler et al., 2017; Milev et al., 2017, 2018).
Aside from the well accepted function of TRAPP complexes in secretion, emerging evidence implicates both the yeast and mammalian TRAPP III in autophagy (Behrends et al., 2010; Lynch-Day et al., 2010; Scrivens et al., 2011; Brunet et al., 2013; Taussig et al., 2014; Lamb et al., 2016; Ramirez-Peinado et al., 2017; Zhao et al., 2017; Stanga et al., 2019). Although, currently the only evidence to connect human disorders and autophagy defects is due to pathogenic variants in TRAPPC11 (Stanga et al., 2019). Here, for the first time, we clearly demonstrate defects in autophagy due to a variant in TRAPPC4 that are likely caused by to a reduced level of one of the core TRAPP subunits, TRAPPC4. Fibroblasts from an affected subject had defects in sealing of autophagosome membranes, and an increase in LC3-II levels prior to starvation. These findings were validated using the temperature-sensitive S. cerevisiae yeast trs23ts mutant strain, which had an autophagy defect both at restrictive and permissive temperatures. The yeast autophagy defects could be complemented with the wild-type gene. Together, these results reflect a defect in basal autophagy, and a delay in autophagy flux due to reduced TRAPPC4 levels in patient fibroblasts.
In conclusion, we provide evidence that perturbations of a core TRAPP subunit are associated with a distinct neurological disorder. The TRAPPC4 c.454+3A>G splicing variant in fibroblasts from an affected subject leads to reduced TRAPPC4 protein levels, a decrease in TRAPP complex abundance, secretory defects and a delay in autophagy flux. We speculate that the reduced levels of all TRAPPC4-containing complexes (which include TRAPP II and III) account for the cellular phenotypes and the pleiotropic clinical phenotypes. Ongoing research is likely to identify other subjects with pathogenic TRAPPC4 variants, and development of animal models with reduced levels of the core TRAPP subunits including TRAPPC4 would provide important insight into the collective disease mechanisms for TRAPPopathies. Given that TRAPPC4 and other TRAPP complex subunits are highly expressed in neurons, and variants in other TRAPP complex subunits are associated with neurological disorders, we propose that the pathogenic TRAPPC4 variant is likely to be the cause of the neurological disease in the presented subjects.
Supplementary Material
Acknowledgements
We thank A. Lavie for help generating the structure models, Y. Liang for strains and Y. Ohsumi for antisera.
Funding
The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. Sequencing, data analysis and Sanger validation were done in the Center for Applied Genomics at the Children’s Hospital of Philadelphia through research funding from Aevi Genomic Medicine, Inc. Funding for M.S. was from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council. Funding for N.S. was from the National Institute of General Medical Sciences (NIGMS) grant GM-45444 and the National Institute of Neurological Disorders and Stroke (NINDS) grant NS-099556. This work was supported in part by the US National Human Genome Research Institute (NHGRI)/National Heart Lung and Blood Institute (NHLBI) UM1 HG006542 to the Baylor Hopkins Center for Mendelian Genomics (BHCMG), the US National Institute of Neurological Disorders and Stroke (NINDS) grant R35 NS105078 to J.L. and the Muscular Dystrophy Association, grant #512848 awarded to J.L. T.M. is supported by the Uehara Memorial Foundation.
Competing interests
Baylor College of Medicine (BCM) and Miraca Holdings have formed a joint venture with shared ownership and governance of the Baylor Genetics (BG), which performs clinical microarray analysis and clinical exome sequencing. P.L. is currently a full-time employee of BG, and J.R. also receives salary support from BG. J.L. serves on the Scientific Advisory Board of the BG. J.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from molecular genetic testing offered at BG. The views expressed herein are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defence, or the U.S. Government. All other authors declare that they have no conflict of interest.
Glossary
Abbreviations
- TRAPP =
transport protein particle
- VSVG =
vesicular stomatitis virus glycoprotein
References
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 2013; 152: 909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466: 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Coombes C, Kwok T, O'Donnell KA, Boeke JD, Hieter P. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell 2008; 30: 248–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JE, Singer SJ. Immunoelectron microscopic studies of the intracellular transport of the membrane glycoprotein (G) of vesicular stomatitis virus in infected Chinese hamster ovary cells. J Cell Biol 1983; 97: 1777–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen VA, Majek P, Jae LT, Bigenzahn JW, Nieuwenhuis J, Staring J, et al. Gene essentiality and synthetic lethality in haploid human cells. Science 2015; 350: 1092–6. [DOI] [PubMed] [Google Scholar]
- Bogershausen N, Shahrzad N, Chong JX, von Kleist-Retzow JC, Stanga D, Li Y, et al. Recessive TRAPPC11 mutations cause a disease spectrum of limb girdle muscular dystrophy and myopathy with movement disorder and intellectual disability. Am J Hum Genet 2013; 93: 181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S, Noueihed B, Shahrzad N, Saint-Dic D, Hasaj B, Guan TL, et al. The SMS domain of Trs23p is responsible for the in vitro appearance of the TRAPP I complex in Saccharomyces cerevisiae. Cell Logist 2012; 2: 28–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S, Sacher M. In sickness and in health: the role of TRAPP and associated proteins in disease. Traffic 2014; 15: 803–18. [DOI] [PubMed] [Google Scholar]
- Brunet S, Shahrzad N, Saint-Dic D, Dutczak H, Sacher M. A trs20 mutation that mimics an SEDT-causing mutation blocks selective and non-selective autophagy: a model for TRAPP III organization. Traffic 2013; 14: 1091–104. [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 2007; 12: 671–82. [DOI] [PubMed] [Google Scholar]
- Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, et al. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinform 2012; 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JY, Lee MH, Lin SR, Yang LY, Sun HS, Sze CI, et al. Trafficking protein particle complex 6A delta (TRAPPC6ADelta) is an extracellular plaque-forming protein in the brain. Oncotarget 2015; 6: 3578–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Davey M, Schluter C, Pandher P, Fang Y, Foster LJ, Conibear E. Organization and assembly of the TRAPPII complex. Traffic 2011; 12: 715–25. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Program NCS, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 2005; 15: 901–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court F, Camprubi C, Garcia CV, Guillaumet-Adkins A, Sparago A, Seruggia D, et al. The PEG13-DMR and brain-specific enhancers dictate imprinted expression within the 8q24 intellectual disability risk locus. Epigenetics Chromatin 2014; 7: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009; 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med 2017; 9: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstner M, Morris CM, Heim K, Lichtner P, Bender A, Mehta D, et al. Single-cell expression profiling of dopaminergic neurons combined with association analysis identifies pyridoxal kinase as Parkinson's disease gene. Ann Neurol 2009; 66: 792–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethell IM, Hagihara K, Miura Y, Irie F, Yamaguchi Y. Synbindin, A novel syndecan-2-binding protein in neuronal dendritic spines. J Cell Biol 2000; 151: 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee DB, Harmelink M, Monrad P, Pyzik E. Siblings with mutations in TRAPPC11 presenting with limb-girdle muscular dystrophy 2S. J Clin Neuromuscul Dis 2017; 19: 27–30. [DOI] [PubMed] [Google Scholar]
- Gambin T, Akdemir ZC, Yuan B, Gu S, Chiang T, Carvalho CMB, et al. Homozygous and hemizygous CNV detection from exome sequencing data in a Mendelian disease cohort. Nucleic Acids Res 2017; 45: 1633–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol 1997; 136: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton G, Harris SE, Davies G, Liewald DC, Tenesa A, Starr JM, et al. Alzheimer's disease genes are associated with measures of cognitive ageing in the lothian birth cohorts of 1921 and 1936. Int J Alzheimers Dis 2011; 2011: 505984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 2015; 163: 1515–26. [DOI] [PubMed] [Google Scholar]
- Hirschberg K, Miller CM, Ellenberg J, Presley JF, Siggia ED, Phair RD, Lippincott-Schwartz J. Kinetic analysis of secretory protein traffic and characterization of golgi to plasma membrane transport intermediates in living cells. J Cell Biol 1998; 143: 1485–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WH, Pendergast JS, Mo XM, Brambilla R, Bracchi-Ricard V, Li F, et al. NIBP, a novel NIK and IKK(beta)-binding protein that enhances NF-(kappa)B activation. J Biol Chem 2005; 280: 29233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol 1995; 131: 583–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell 2000; 11: 4403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar N, Goebel I, Daud S, Nurnberg G, Agha N, Ahmad A, et al. A homozygous splice site mutation in TRAPPC9 causes intellectual disability and microcephaly. Eur J Med Genet 2012; 55: 727–31. [DOI] [PubMed] [Google Scholar]
- Karaca E, Posey JE, Coban Akdemir Z, Pehlivan D, Harel T, Jhangiani SN, et al. Phenotypic expansion illuminates multilocus pathogenic variation. Genet Med 2018; 20: 1528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Lipatova Z, Segev N. TRAPP complexes in secretion and autophagy. Front Cell Dev Biol 2016; 4: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Raunser S, Munger C, Wagner J, Song YL, Cygler M, et al. The architecture of the multisubunit TRAPP I complex suggests a model for vesicle tethering. Cell 2006; 127: 817–30. [DOI] [PubMed] [Google Scholar]
- Koehler K, Milev MP, Prematilake K, Reschke F, Kutzner S, Juhlen R, et al. A novel TRAPPC11 mutation in two Turkish families associated with cerebral atrophy, global retardation, scoliosis, achalasia and alacrima. J Med Genet 2017; 54: 176–85. [DOI] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–81. [DOI] [PubMed] [Google Scholar]
- Kunjithapatham R, Geschwind JF, Devine L, Boronina TN, O'Meally RN, Cole RN, et al. Occurrence of a multimeric high-molecular-weight glyceraldehyde-3-phosphate dehydrogenase in human serum. J Proteome Res 2015; 14: 1645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Nuhlen S, Judith D, Frith D, Snijders AP, Behrends C, Tooze SA. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J 2016; 35: 281–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AA, Baker PR 2nd, Milev MP, Press CA, Sokol RJ, Cox MO, et al. TRAPPC11 and GOSR2 mutations associate with hypoglycosylation of alpha-dystroglycan and muscular dystrophy. Skelet Muscle 2018; 8: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WC, Tian X, Yuo CY, Chen WZ, Kan TM, Su YN, et al. Comprehensive target capture/next-generation sequencing as a second-tier diagnostic approach for congenital muscular dystrophy in Taiwan. PLoS One 2017; 12: e0170517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci U S A 2012; 109: 6981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Majumdar U, Segev N. Trs33-containing TRAPP IV: a novel autophagy-specific Ypt1 GEF. Genetics 2016; 204: 1117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Segev N. A role for macro-ER-phagy in ER quality control. PLoS Genet 2015; 11: e1005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Shah AH, Kim JJ, Mulholland JW, Segev N. Regulation of ER-phagy by a Ypt/Rab GTPase module. Mol Biol Cell 2013; 24: 3133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipatova Z, Tokarev AA, Jin Y, Mulholland J, Weisman LS, Segev N. Direct interaction between a myosin V motor and the Rab GTPases Ypt31/32 is required for polarized secretion. Mol Biol Cell 2008; 19: 4177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh E, Peter F, Subramaniam VN, Hong W. Mammalian Bet3 functions as a cytosolic factor participating in transport from the ER to the Golgi apparatus. J Cell Sci 2005; 118: 1209–22. [DOI] [PubMed] [Google Scholar]
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A 2010; 107: 7811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangi G, Leuzzi V, Manti F, Lattante S, Orteschi D, Pecile V, et al. TRAPPC9-related autosomal recessive intellectual disability: report of a new mutation and clinical phenotype. Eur J Hum Genet 2013; 21: 229–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Valencia I, Novarino G, Johansen A, Rosti B, Issa MY, Musaev D, et al. A homozygous founder mutation in TRAPPC6B associates with a neurodevelopmental disorder characterised by microcephaly, epilepsy and autistic features. J Med Genet 2018; 55: 48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matalonga L, Bravo M, Serra-Peinado C, Garcia-Pelegri E, Ugarteburu O, Vidal S, et al. Mutations in TRAPPC11 are associated with a congenital disorder of glycosylation. Hum Mutat 2017; 38: 148–51. [DOI] [PubMed] [Google Scholar]
- Milev MP, Graziano C, Karall D, Kuper WFE, Al-Deri N, Cordelli DM, et al. Bi-allelic mutations in TRAPPC2L result in a neurodevelopmental disorder and have an impact on RAB11 in fibroblasts. J Med Genet 2018; 55: 753–64. [DOI] [PubMed] [Google Scholar]
- Milev MP, Grout ME, Saint-Dic D, Cheng YH, Glass IA, Hale CJ, et al. Mutations in TRAPPC12 manifest in progressive childhood encephalopathy and golgi dysfunction. Am J Hum Genet 2017; 101: 291–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir A, Kaufman L, Noor A, Motazacker MM, Jamil T, Azam M, et al. Identification of mutations in TRAPPC9, which encodes the NIK- and IKK-beta-binding protein, in nonsyndromic autosomal-recessive mental retardation. Am J Hum Genet 2009; 85: 909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Mahajnah M, Hill AD, Basel-Vanagaite L, Gleason D, Hill RS, et al. A truncating mutation of TRAPPC9 is associated with autosomal-recessive intellectual disability and postnatal microcephaly. Am J Hum Genet 2009; 85: 897–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamoud HS, Ahmed S, Jelani M, Alrayes N, Childs K, Vadgama N, et al. A missense mutation in TRAPPC6A leads to build-up of the protein, in patients with a neurodevelopmental syndrome and dysmorphic features. Sci Rep 2018; 8: 2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe O, Rio M, Carioux A, Plaza JM, Guigue P, Molinari F, et al. Combination of linkage mapping and microarray-expression analysis identifies NF-kappaB signaling defect as a cause of autosomal-recessive mental retardation. Am J Hum Genet 2009; 85: 903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Peinado S, Ignashkova TI, van Raam BJ, Baumann J, Sennott EL, Gendarme M, et al. TRAPPC13 modulates autophagy and the response to Golgi stress. J Cell Sci 2017; 130: 2251–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Kim YG, Lavie A, Oh BH, Segev N. The TRAPP complex: insights into its architecture and function. Traffic 2008; 9: 2032–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Shahrzad N, Kamel H, Milev MP. TRAPPopathies: An emerging set of disorders linked to variations in the genes encoding transport protein particle (TRAPP)-associated proteins. Traffic 2019; 20: 5–26. [DOI] [PubMed] [Google Scholar]
- Scrivens PJ, Noueihed B, Shahrzad N, Hul S, Brunet S, Sacher M. C4orf41 and TTC-15 are mammalian TRAPP components with a role at an early stage in ER-to-Golgi trafficking. Mol Biol Cell 2011; 22: 2083–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N. Ypt and Rab GTPases: insight into functions through novel interactions. Curr Opin Cell Biol 2001; 13: 500–11. [DOI] [PubMed] [Google Scholar]
- Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat 2015; 36: 928–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga D, Zhao Q, Milev MP, Saint-Dic D, Jimenez-Mallebrera C, Sacher M. TRAPPC11 functions in autophagy by recruiting ATG2B-WIPI4/WDR45 to preautophagosomal membranes. Traffic 2019; 20: 325–45. [DOI] [PubMed] [Google Scholar]
- Taussig D, Lipatova Z, Segev N. Trs20 is required for TRAPP III complex assembly at the PAS and its function in autophagy. Traffic 2014; 15: 327–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LL, Fromme JC. GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J Cell Biol 2016; 215: 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LL, Joiner AMN, Fromme JC. The TRAPPIII complex activates the GTPase Ypt1 (Rab1) in the secretory pathway. J Cell Biol 2018; 217: 283–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, et al. Identification and characterization of essential genes in the human genome. Science 2015; 350: 1096–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol 2000; 151: 289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki A, Menon S, Yu S, Barrowman J, Meerloo T, Oorschot V, et al. mTrs130 is a component of a mammalian TRAPPII complex, a Rab1 GEF that binds to COPI-coated vesicles. Mol Biol Cell 2009; 20: 4205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bitner D, Pontes Filho AA, Li F, Liu S, Wang H, et al. Expression and function of NIK- and IKK2-binding protein (NIBP) in mouse enteric nervous system. Neurogastroenterol Motil 2014; 26: 77–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Li CM, Luo XM, Siu GK, Gan WJ, Zhang L, et al. Mammalian TRAPPIII Complex positively modulates the recruitment of Sec13/31 onto COPII vesicles. Sci Rep 2017; 7: 43207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Wu Z, Zhao M, Murtazina R, Cai J, Zhang A, et al. Rab5-dependent autophagosome closure by ESCRT. J Cell Biol 2019; 218: 1908–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S, Liu Y, Liang Y. Trs20, Trs23, Trs31 and Bet5 participate in autophagy through GTPase Ypt1 in Saccharomyces cerevisiae. Arch Biol Sci 2018; 70: 109–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data that support the findings of this study are available from the corresponding author, upon request.