Abstract
Most people are now familiar with the concepts of big data, deep learning, machine learning, and artificial intelligence (AI) and have a vague expectation that AI using medical big data can be used to improve the quality of medical care. However, the expectation that big data could change the field of medicine is inconsistent with the current reality. The clinical meaningfulness of the results of research using medical big data needs to be examined. Medical staff needs to be clear about the purpose of AI that utilizes medical big data and to focus on the quality of this data, rather than the quantity. Further, medical professionals should understand the necessary precautions for using medical big data, as well as its advantages. No doubt that someday, medical big data will play an essential role in healthcare; however, at present, it seems too early to actively use it in clinical practice. The field continues to work toward developing medical big data and making it appropriate for healthcare. Researchers should continue to engage in empirical research to ensure that appropriate processes are in place to empirically evaluate the results of its use in healthcare.
Keywords: Artificial intelligence, Big data, Data science, Medical informatics, Deep learning, Machine learning
INTRODUCTION
Artificial intelligence (AI) is one of the biggest talking points in the medical field today [1,2,3]. Big data, machine learning, deep learning, and the common data model (CDM), among others, are also commonly mentioned in the medical field [4,5,6,7,8]. In fact, most medical staff wrongfully assume that medical big data can be easily extracted from electronic medical records (EMR), and analyzed using advanced statistics—a naïve assumption driven by the proliferation of medical data. In reality, extracting good data to use for research is not an easy task [4,5]. This is due to the inherent limitations of big data, specifically claim data, that had not been collected for research purposes. Ultimately, big data is a primitive form of data that needs to be manually reviewed for the extraction of useful and meaningful knowledge. Medical staff should be able to extract correct medical information in an environment overflowing with uncertain information.
Limitations of machine learning
Machine learning (or deep learning) is used to develop AI algorithms, and requires a large amount of data for full functionality. According to some scholars [9,10], machine learning performance increases along with the amount of data; however, the sophistication of analytic models does not improve performance significantly. In one study, an outstanding prediction model was developed using simple logistic regression [11]. Here, we are faced with a very basic question: why is deep learning necessary? In some cases, if the data is well refined, simple statistical procedures, like simple logistic regression, provide excellent results or prediction models, even without the application of complicated deep learning techniques that takes time to acquire, and requires high-end equipment. Due to the increased attention on machine learning [12,13], methodological know-how is often perceived as the key for unlocking knowledge; however, the most important consideration in research is not the methodology, but rather how well the data extracted from the sample reflects the realities of the whole population. The use of complicated procedures and the development of statistical hypotheses reflect the fact that data quality cannot be guaranteed; when data is collected in a manner that reflects the unique qualities of the sample population, even simple statistics can produce sufficiently good results in some cases. This is why we continually emphasize the importance of data quality, rather than quantity. Of course, regardless of data quality, effective analysis may not be possible using traditional statistical methods, depending on the type of data. When digital data are standardized to a certain level and are well-managed in terms of quality (image or bio-signal data), existing traditional analytical methods cannot be used to elicit new interpretations. In such cases machine learning can be useful, despite limitations derived from inaccurate data or an absence of data quality management (DQM). Of great importance, however, is that medical staff are trained to look at data itself first to determine whether machine learning or traditional statistical methods would better serve the analytic purpose.
Most clinical data contain noise and missing values, necessitating repetitive and labor-intensive tasks that involve time and effort, including data handling, DQM, and data cleansing [4]. Machine learning relies on data, and the deep learning itself is less important than the quality of data. If the data is unrefined, AI will not produce any good results, as exemplified by Microsoft's Tay Chatbot [14], which learned nonsense when it was taught nonsense. Various retrospective studies further demonstrated how undesirable phenomena produced different results, depending on the type of data and the research method [15,16]. Such results support the importance of data rather than methodology, and of quality over quantity. Who can assess the quality of this data? Data scientists provide expertise in analytics, but they do not have any experience with medical practice. The role should therefore be filled by scientists working in the medical field.
HOW CAN WE ACQUIRE GOOD QUALITY DATA?
Whether we are considering EMR data or claim data, such as data generated by the National Health Insurance Service [17,18] or Health Insurance Review & Assessment Service [19,20], it is important to note that these are not data for research purposes [4]. Would it then be possible to conduct clinical research with data not intended for clinical research purposes? In order to secure good quality medical data, it is necessary to develop an operational definition and a strategy for DQM. If these are not properly considered, the reliability of the data will drop dramatically, and the results of research relying on this data will not be worth looking at.
Conceptual definition vs. operational definition
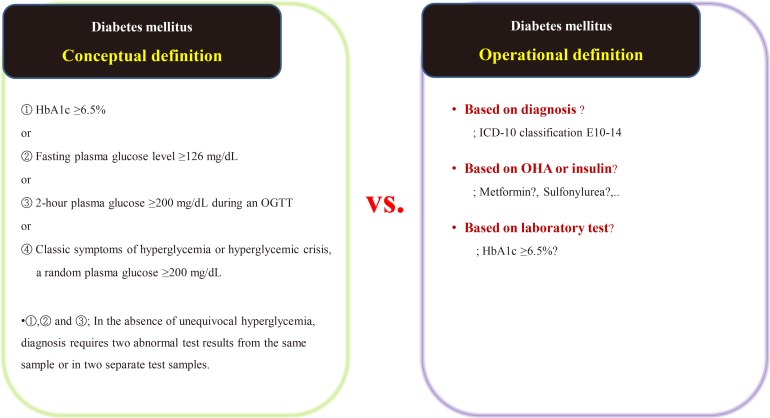
Suppose we are conducting a study on the accompanying rate of cardiovascular disease in patients with diabetes mellitus (DM). The first thing to do is to define people with DM. The widely used definition of DM involves the patient's fasting/postprandial glucose and hemoglobin A1c (HbA1c) levels [21,22]. This is a conceptual definition of DM, and most medical staff are familiar with it. In a data-driven study, however, this conceptual definition is not only impossible to extract, but also not very efficient. Therefore, we use operational rather than conceptual definition.
We can develop an operational definition of DM using the International Classification of Diseases 10th Revision (ICD-10) classification code, an oral hypoglycemia agent such as metformin or sulfonylurea, or traditional laboratory findings such as HbA1c levels (Fig. 1) [23]. Of course, operational definitions can also be developed from the intersections or combinations produced by these three axes. There is no clear, correct way of developing an operational definition, and the researcher should do so based on their research objectives. It should be noted that, in real-world research, the first diagnosis date, the date of first taking the drug, and the date of blood test when diagnosed with DM are all different, as the data are often poorly stored. The study results are informed by the development of the operational definition, and this aspect of research requires the careful attention of medical staff. Importantly, steps need to be taken to minimize the bias that can occur in retrospective research using big data.
Fig. 1. Example of the conceptual [23] and operational definitions of diabetes mellitus. HbA1c, hemoglobin A1c; OGTT, oral glucose tolerance test; ICD-10, International Classification of Diseases 10th Revision; OHA, oral hypoglycemic agents.
Continuous communication between data scientists and medical staff can reduce research-related problems. However, medical staff often struggle to clarify an operational definition, and for this reason we need deeper reflection, rather than common sense, as the same data can render different results. Active collaboration with the medical staff who know the data best is required.
Data quality management
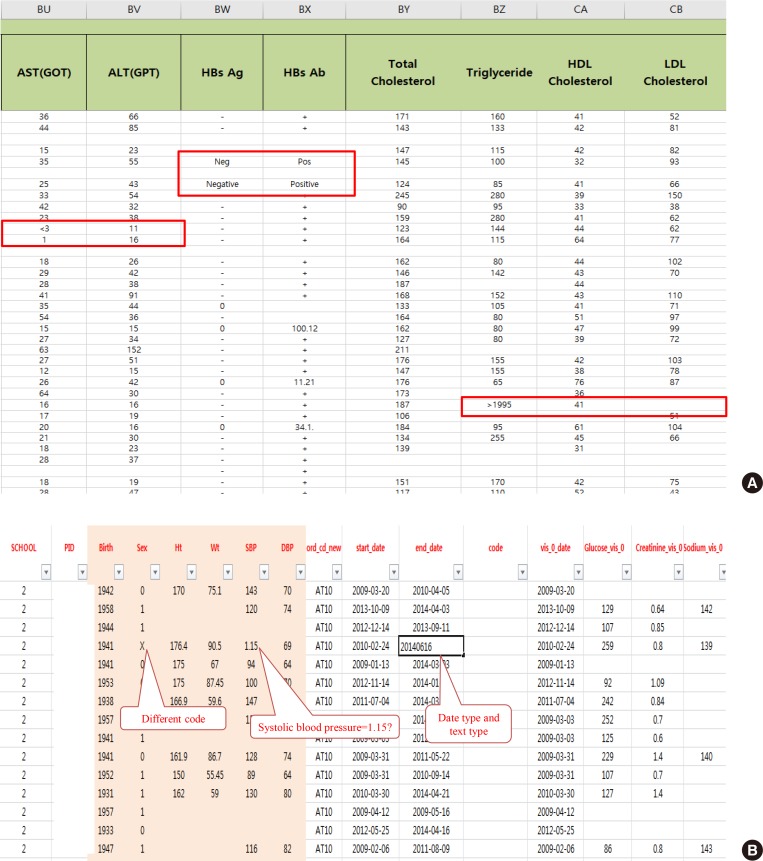
After extracting medical data for analysis, it may become clear that it is not fit for research purposes. While data for research purposes must be expressed in a structured form, that is, in numbers, most EMR data is not well organized (Fig. 2). For example, result of hepatitis B surface antigen is described in various ways such as neg, negative, 0, and (−). In some cases, height and weight or systolic and diastolic blood pressure are reversed. Most clinicians will realize intuitively that the data in such cases were mistyped, but they are unable to change the numbers arbitrarily. Although some modifications can be made through operational definitions, in most cases such data is eventually converted to missing data. After all, DQM is the most time-consuming task in the research process, and must be managed by researcher or medical staff member who clearly understands medical data [4]. It is recommended that clear protocols and guidelines are put in place before the study so that other researchers can consistently follow the same approach.
Fig. 2. Examples of real cases requiring data quality management. (A) Various written data. Even though the laboratory test result is “<3,” it is also written as “1.” The physicians' role is defining and classifying data, and staff who are most familiar with the data should do so. (B) Example of incorrectly entered data. AST, aspartate aminotransferase; GOT, glutamate oxaloacetate transaminase; ALT, aspartate aminotransferase; GPT, glutamate pyruvate transaminase; HBsAg, hepatitis B surface antigen; HBsAb, hepatitis B surface antibody; HDL, high-density lipoprotein; LDL, low-density lipoprotein.
STANDARDIZATION AND THE DEVELOPMENT OF A MULTI-CENTER REGISTRY
As mentioned above, the advantages of big data are positively correlated with the amount of available data [9]. In the case of tertiary university hospitals, large amounts of meaningful medical data are expected to be available. However, for research conducted using real EMR data, the amount of missing data presents further challenges [24,25]. Ultimately, the sample size from which the so-called big data can be extracted may contain smaller numbers than large-scaled randomized clinical trials. Given this, is the name “big data” still accurate, and how can greater amounts of medical data be collected? One answer would be to develop a multi-center registry as soon as possible to merge the available datasets from multiple hospitals [26].
Big data research in a single institution presents various challenges, including controlling various operational definitions, DQM, and numerous biases [4]. While challenges exist with single hospitals, they are even larger when in a multi-center. Nevertheless, the integration of data from multiple hospitals appears to be required for future research. One critical consideration for the development of a multi-center registry is the development of an international coding system and mapping this [27], which would enable not only domestic partnerships but also international collaboration and would ultimately lead to the creation of an international standard [28]. Such a standard would include guidelines for the interpretation of specific types of data, and its establishment should be negotiated among various stakeholders, under the auspices of a suitably qualified institution. By doing this, every participating institution would agree to abide by the standard. However, in research conducted using multi-center registries, researchers tend not to consider the future need for an international code and choose instead a domestic standard that is agreed upon by a small number of institutions, rendering international mapping impossible. Thus, international mapping is mandatory if we are to consider future work.
After the standard has been established, the role of hospitals and doctors need to be clarified. Inputting EMR data must be done according to the standardized format. Proper collection of data in the initial stages significantly decreases the time and money necessary for future research [4]. Data that is made or input without a specific standard will eventually lose its reliability and value. However, in the reality of the Korean medical environment, each patient is limited to 3 to 5 minutes when consulting with a doctor [29], during which it is realistically impossible for medical professionals to input data in the correct format. Nonetheless, the development of a standard remains important [28]; if EMR data accumulates in a standardized format, it will likely remain useful over time.
HOT ISSUES IN KOREA: THE COMMON DATA MODEL
There has been increasing interest in CDM for research in various institutions in Korea. CDM unifies data in diverse formats into one common format [30,31,32]. Among the various types of CDM, the one that is most popular in Korea is the observational health data sciences and informatics–observational medical outcomes partnership model (OHDSI-OMOP CDM), which is used in various institutions for retrospective big data collection and analysis, which makes it very useful for researchers [31,33]. Moreover, CDM does not share patient identification numbers, which is a powerful advantage of CDM in terms of privacy However, the CDM itself has not been accredited by an authorized institution. Thus, there is a need to understand whether the CDM is an organizational or institutional standard, which is why the CDM consortium promises to use only in this consortium. Another challenge of CDM is that data mapping must be done manually, step-by-step [34]. Data mapping is nearly impossible to automatize as each hospital has a unique data structure, and automatic data mapping without consideration of the discrepancies would result in definite biases. EMR data does not exist solely for academic research, but is fundamentally aimed at supporting patient treatment [35]—a fact that should not be forgotten.
CONCLUSIONS
From the perspective of medical professionals, the purpose of collecting and standardizing medical big data needs to be defined clearly. Collecting standardized data properly is the most important task, as the main goal of retrospective research using EMR data is to care for patients. As long as data is collected properly, it does not matter whether you use deep learning, machine learning, or very simple statistical methods. Medical staff are best placed to use and interpret medical data, and the key is for them to critically observe the data, identifying any underlying problems and dealing with them appropriately. Finally, results comprising the most compelling value is found when refined data is combined with medical ideas.
ACKNOWLEDGMENTS
This work was supported by the Technology development Program (S2726209) funded by the Ministry of SMEs and Startups (MSS, Korea). I am indebted Prof Shin Soo-yong, whose lecture is referenced often in this text.
Footnotes
CONFLICTS OF INTEREST: No potential conflict of interest relevant to this article was reported.
References
- 1.Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. 2017;69S:S36–S40. doi: 10.1016/j.metabol.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Miller DD, Brown EW. Artificial intelligence in medical practice: the question to the answer? Am J Med. 2018;131:129–133. doi: 10.1016/j.amjmed.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 3.Kantarjian H, Yu PP. Artificial intelligence, big data, and cancer. JAMA Oncol. 2015;1:573–574. doi: 10.1001/jamaoncol.2015.1203. [DOI] [PubMed] [Google Scholar]
- 4.Kim HS, Kim JH. Proceed with caution when using real world data and real world evidence. J Korean Med Sci. 2019;34:e28. doi: 10.3346/jkms.2019.34.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim HS, Lee S, Kim JH. Real-world evidence versus randomized controlled trial: clinical research based on electronic medical records. J Korean Med Sci. 2018;33:e213. doi: 10.3346/jkms.2018.33.e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obermeyer Z, Emanuel EJ. Predicting the future: big data, machine learning, and clinical medicine. N Engl J Med. 2016;375:1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miotto R, Wang F, Wang S, Jiang X, Dudley JT. Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform. 2018;19:1236–1246. doi: 10.1093/bib/bbx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravi D, Wong C, Deligianni F, Berthelot M, Andreu-Perez J, Lo B, et al. Deep learning for health informatics. IEEE J Biomed Health Inform. 2017;21:4–21. doi: 10.1109/JBHI.2016.2636665. [DOI] [PubMed] [Google Scholar]
- 9.Sun C, Shrivastava A, Singh S, Gupta A. Revisiting unreasonable effectiveness of data in deep learning era; 2017 IEEE International Conference on Computer Vision (ICCV); 2017 Oct 22–29; Venice, IT. Piscataway, NJ: IEEE; 1997. pp. 843–852. [Google Scholar]
- 10.Rubin DB. Estimating causal effects from large data sets using propensity scores. Ann Intern Med. 1997;127:757–763. doi: 10.7326/0003-4819-127-8_part_2-199710151-00064. [DOI] [PubMed] [Google Scholar]
- 11.Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med. 2018;1:18. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bastanlar Y, Ozuysal M. Introduction to machine learning. Methods Mol Biol. 2014;1107:105–128. doi: 10.1007/978-1-62703-748-8_7. [DOI] [PubMed] [Google Scholar]
- 14.Williams H. Microsoft's teen chatbot has gone wild [Internet] Surry Hills: Gizmodo; 2016. [cited 2019 Dec 9]. Available from: https://www.gizmodo.com.au/2016/03/microsofts-teen-chatbot-has-gone-wild. [Google Scholar]
- 15.Levesque LE, Hanley JA, Kezouh A, Suissa S. Problem of immortal time bias in cohort studies: example using statins for preventing progression of diabetes. BMJ. 2010;340:b5087. doi: 10.1136/bmj.b5087. [DOI] [PubMed] [Google Scholar]
- 16.Maugis PG. Big data uncertainties. J Forensic Leg Med. 2018;57:7–11. doi: 10.1016/j.jflm.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, Lee JS, Park SH, Shin SA, Kim K. Cohort profile: the National Health Insurance Service-National Sample Cohort (NHIS-NSC), South Korea. Int J Epidemiol. 2017;46:e15. doi: 10.1093/ije/dyv319. [DOI] [PubMed] [Google Scholar]
- 18.Noh J. The diabetes epidemic in Korea. Endocrinol Metab (Seoul) 2016;31:349–353. doi: 10.3803/EnM.2016.31.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo GH, Chung JH. Incidence and prevalence of overt hypothyroidism and causative diseases in Korea as determined using claims data provided by the Health Insurance Review and Assessment Service. Endocrinol Metab (Seoul) 2015;30:288–296. doi: 10.3803/EnM.2015.30.3.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YK, Yoon BH, Koo KH. Epidemiology of osteoporosis and osteoporotic fractures in South Korea. Endocrinol Metab (Seoul) 2013;28:90–93. doi: 10.3803/EnM.2013.28.2.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2004;27:S5–S10. doi: 10.2337/diacare.27.2007.s5. [DOI] [PubMed] [Google Scholar]
- 22.Ko SH, Hur KY, Rhee SY, Kim NH, Moon MK, Park SO, et al. Antihyperglycemic agent therapy for adult patients with type 2 diabetes mellitus 2017: a position statement of the Korean Diabetes Association. Diabetes Metab J. 2017;41:337–348. doi: 10.4093/dmj.2017.41.5.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Expert Committee. International Expert Committee report on the role of the A1C assay in the diagnosis of diabetes. Diabetes Care. 2009;32:1327–1334. doi: 10.2337/dc09-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z. Missing data exploration: highlighting graphical presentation of missing pattern. Ann Transl Med. 2015;3:356. doi: 10.3978/j.issn.2305-5839.2015.12.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim TM, Kim H, Jeong YJ, Baik SJ, Yang SJ, Lee SH, et al. The differences in the incidence of diabetes mellitus and prediabetes according to the type of HMG-CoA reductase inhibitors prescribed in Korean patients. Pharmacoepidemiol Drug Saf. 2017;26:1156–1163. doi: 10.1002/pds.4237. [DOI] [PubMed] [Google Scholar]
- 26.Chen PH, Loehfelm TW, Kamer AP, Lemmon AB, Cook TS, Kohli MD. Toward data-driven radiology education-early experience building multi-institutional academic trainee interpretation log database (MATILDA) J Digit Imaging. 2016;29:638–644. doi: 10.1007/s10278-016-9872-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matney SA, Settergren TT, Carrington JM, Richesson RL, Sheide A, Westra BL. Standardizing physiologic assessment data to enable big data analytics. West J Nurs Res. 2017;39:63–77. doi: 10.1177/0193945916659471. [DOI] [PubMed] [Google Scholar]
- 28.Kalra D. Electronic health record standards. Yearb Med Inform. 2006:136–144. [PubMed] [Google Scholar]
- 29.Lee CH, Lim H, Kim Y, Park AH, Park EC, Kang JG. Analysis of appropriate outpatient consultation time for clinical departments. Health Policy Manag. 2014;24:254–260. [Google Scholar]
- 30.Kim H, Choi J, Jang I, Quach J, Ohno-Machado L. Feasibility of representing data from published nursing research using the OMOP common data model. AMIA Annu Symp Proc. 2017;2016:715–723. [PMC free article] [PubMed] [Google Scholar]
- 31.Ceusters W, Blaisure J. A realism-based view on counts in OMOP’s common data model. Stud Health Technol Inform. 2017;237:55–62. [PubMed] [Google Scholar]
- 32.Kimura E, Suzuki H. Development of a common data model facilitating clinical decision-making and analyses. Stud Health Technol Inform. 2019;264:1514–1515. doi: 10.3233/SHTI190511. [DOI] [PubMed] [Google Scholar]
- 33.Park RW. The distributed research network, observational health data sciences and informatics, and the South Korean research network. Korean J Med. 2019;94:309–314. [Google Scholar]
- 34.FitzHenry F, Resnic FS, Robbins SL, Denton J, Nookala L, Meeker D, et al. Creating a common data model for comparative effectiveness with the observational medical outcomes partnership. Appl Clin Inform. 2015;6:536–547. doi: 10.4338/ACI-2014-12-CR-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HS. Decision-making in artificial intelligence: is it always correct? J Korean Med Sci. 2020 doi: 10.3346/jkms.2020.35.e1. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]