Abstract
Methods that enable the super-resolution imaging of intracellular proteins in live bacterial cells provide powerful tools for the study of prokaryotic cell biology. Photoswitchable organic dyes exhibit many of the photophysical properties needed for super-resolution imaging, including high brightness, photostability, and photon output, but most such dyes require organisms to be fixed and permeabilized if intracellular targets are to be labeled. We recently reported a general strategy for the chemoenzymatic labeling of bacterial proteins with azide-bearing fatty acids in live cells using the eukaryotic enzyme N-myristoyltransferase. Here we demonstrate the labeling of proteins in live Escherichia coli using cell-permeant bicyclononyne-functionalized photoswitchable rhodamine spirolactams. Single-molecule fluorescence measurements on model rhodamine spirolactam salts show that these dyes emit hundreds of photons per switching event. Super-resolution imaging was performed on bacterial chemotaxis proteins Tar and CheA and cell division proteins FtsZ and FtsA. High-resolution imaging of Tar revealed a helical pattern; imaging of FtsZ yielded banded patterns dispersed throughout the cell. The precision of radial and axial localization in reconstructed images approaches 15 and 30 nm, respectively. The simplicity of the method, which does not require redox imaging buffers, should make this approach broadly useful for imaging intracellular bacterial proteins in live cells with nanometer resolution.
Short abstract
We describe a chemoenzymatic protein labeling strategy that uses cell-permeant photoswitchable dyes to enable super-resolution imaging in live bacteria without the need for special imaging buffers.
Introduction
Imaging modalities that provide resolution beyond the diffraction limit have enabled new approaches to the exploration of complex biological phenomena.1,2 Super-resolution and single-molecule imaging methods have revealed key elements of the structures of macromolecular assemblies with nanometer resolution.3 Examples include the three-dimensional architectures of microtubules and clathrin-coated pits in BS-C-1 cells,4,5 the periodicity of actin organization in neuronal cells,6 and the localization of mitochondrial proteins enriched at cristae junctions in human skin fibroblast cells.7 Methods such as photoactivated localization microscopy (PALM),8 stochastic optical reconstruction microscopy (STORM),9 and direct stochastic optical reconstruction microscopy (dSTORM)10 overcome the Abbe diffraction limit11 by exploiting fluorophores that switch between dark (“off”) and fluorescent (“on”) states.12 Conventional small-molecule dyes, which may undergo reversible photoswitching upon excitation, include those bearing carbo-rhodamine,13 cyanine,14 and oxazine15 cores. Photoswitchable fluorescent proteins, such as EYFP16 and Dronpa,17 undergo structural rearrangements that enable transitioning between off and on states. Fluorophores may also undergo irreversible conversion through means of photoactivation with weak UV illumination.18 This behavior is characteristic of azido push–pull chromophores,19 rhodamine spiroamides,20 spirocyclic diazoketones,21 photoactivatable fluorescent proteins such as PA-CFP222 and PA-mCherry,23 and photoconvertible fluorescent proteins such as mEos24 and Dendra2.25 The development of fluorescence methods that resolve molecules in space and time with high precision remains an active area of research.26
The resolution of adjacent emitters based on their individual point spread functions (PSF) is dependent on the individual fluorophore’s photon output, duty cycle, survival fraction, and number of switching cycles.27 Organic fluorophores offer important advantages that make them attractive for use in super-resolution microscopy, including photostability, brightness,28 and high photon outputs.27 Many of the conventional STORM dyes (e.g., Cy5 and Alexa 647) require the use of oxygen-scavenging buffers and the addition of exogenous reducing agents to facilitate photoswitching.29 Targets are typically labeled by means of immunofluorescence staining in fixed cells with antibodies conjugated to dyes.30 The size of antibodies used in such labeling experiments is on the scale of 10–15 nm, which may negate the localization precision of the single-molecule imaging experiment.31 The reagents needed for fixation and permeabilization of cells require careful optimization to ensure that image quality is not affected.32 For live-cell imaging, the fusion of target proteins to the HaloTag33 has found use in achieving covalent modification with photoactivatable fluorophores in live bacterial34 and eukaryotic cells,35 although the size of the tag is of the same order of magnitude as that of fluorescent proteins. Improved methods for the selective introduction of photoswitchable organic fluorophores into target proteins in live cells are needed to advance the field of super-resolution microscopy.
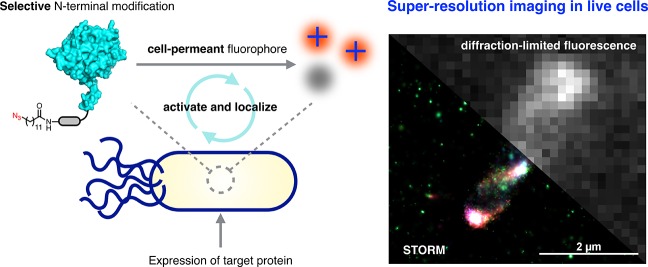
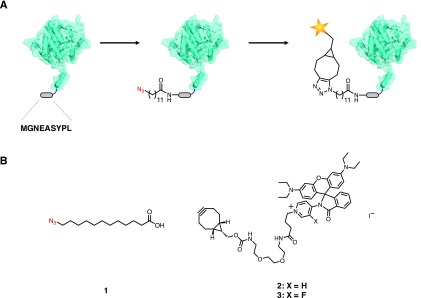
We recently reported a strategy for introducing azido fatty acids into the N-termini of intracellular proteins in E. coli for imaging with cell-permeant fluorophores (Figure 1A).36 The method uses the eukaryotic enzyme N-myristoyltransferase (NMT) to ligate the myristic acid surrogate 12-azidododecanoic acid (12-ADA, 1) to the proteins of interest.37 Target proteins are outfitted with nonapeptide sequence MGNEASYPL, an N-terminal NMT recognition sequence derived from mammalian protein calcineurin B.38 In this work, we demonstrate the labeling of chemotaxis and cell division proteins in live E. coli cells with cell-permeant photoswitchable rhodamine spirolactam dyes 2 and 3 (Figure 1B). Rhodamine spirolactams have been used as probes for the super-resolution imaging of cell-surface targets in live Caulobacter crescentus.39 Upon activation at 405 nm, the fluorophore converts from a nonfluorescent state to a fluorescent state.40 The open rhodamine isomers thermally recyclize on the order of milliseconds in polar solvents,41 allowing the molecule to undergo photoswitching. Oxygen-scavenging buffer systems and exogenous reducing agents, commonly used to facilitate the activation of STORM dyes, are not required for the photoswitching of these molecules. Other STORM dyes with similar emission characteristics include Cy3 and Atto 565,27 although such dyes require imaging buffers to facilitate photoswitching. To the best of our knowledge, these rhodamine spirolactam scaffolds have not been used for labeling and imaging specific intracellular proteins in live bacterial cells.
Figure 1.
Strategy for super-resolution imaging in live cells. (A) A bacterial protein of interest is outfitted with a short N-terminal nonapeptide NMT recognition sequence (MGNEASYPL). The treatment of cells with 1 during the expression of the target protein results in the azide-labeled protein. Subsequent strain-promoted azide–alkyne cycloaddition with cell-permeant photoswitchable dyes such as 2 and 3 tags the protein of interest for super-resolution imaging. (B) Structures of fatty acid 1 and rhodamines 2 and 3.
Inspired by the potential application of these fluorophores in live-cell imaging experiments, we examined rhodamine spirolactam salts 4 and 5 as model compounds. Bulk spectroscopic characterization was performed on these dyes in aqueous and organic solvents, and we determined each fluorophore’s photon output, duty cycle, survival fraction, and number of switching events at the single-molecule level. Finally, we used reactive dyes 2 and 3 to accomplish live-cell super-resolution imaging of chemotaxis proteins Tar and CheA42,43 and cell division proteins FtsZ and FtsA44,45 in live E. coli cells.
Results and Discussion
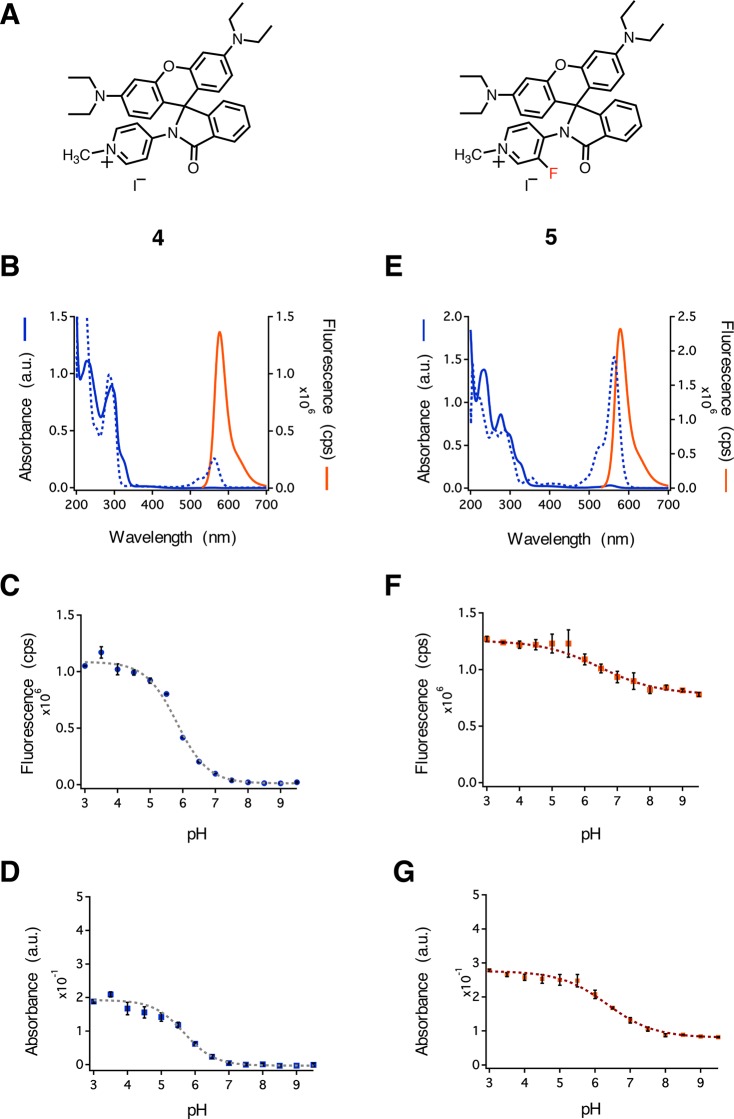
Rhodamines 4 and 5 were prepared as iodide salts from commercially available rhodamine B (Schemes S1 and S2). The activation of rhodamine B with phosphorus oxychloride in refluxing acetonitrile afforded the acid chloride, which was directly condensed with an aminopyridine at room temperature. Formation of the secondary amide induces spontaneous cyclization to form the lactam. Methylation was accomplished by treatment of the lactam with methyl iodide in refluxing acetonitrile and afforded 4 and 5 in 48 and 43% overall yields, respectively.
We characterized the spectral properties of 4 and 5 in a variety of aqueous solutions (Figure 2 and Figures S1–S6). Rhodamine 4 showed no evidence of absorption beyond 500 nm, which is typically observed in yellow-absorbing xanthene dyes in a 1:1 v/v mixture of water and acetonitrile (Figure 2B, blue solid line). The addition of acid resulted in a characteristic peak beyond 500 nm (Figure 2B, blue dashed line, λmax = 561 nm), suggesting a shift in the position of the equilibrium between the closed and open isomers. In contrast, rhodamine 5 showed a small peak at λmax = 553 nm in a 1:1 v/v mixture of water and acetonitrile (Figure 2E, blue solid line). Acidification of the solvent resulted in a strong absorption peak (λmax = 563 nm) with an intensity roughly 40-fold higher than that of the 553 nm peak in the nonacidic solvent (Figure 2E, blue dashed line). To determine the spectral properties of the open isomers of 4 and 5, we determined the molar absorptivity, quantum yield, and fluorescence emission of each compound in acidic-buffered solvents. Interestingly, rhodamine 5 had a higher molar absorptivity (ε563 nm = 55 200 M–1 cm–1) and quantum yield (φ = 0.69) when compared to 4 (ε561 nm = 10 200 M–1 cm–1 and φ = 0.54). Both 4 and 5 showed emission profiles commonly found for orange-emitting dyes (Figure 2B,E, orange solid lines). No significant shifts in emission maxima were observed for 5 when compared to 4 (λem = 578 and 577 nm, respectively). The absorption spectra of rhodamine spirolactams have been shown to respond to changes in pH.46 To test the pH sensitivity of 4 and 5, we prepared solutions in bis-tris propane at different pH values and measured both the fluorescence emission and absorption. An analysis of fluorescence emission from 4 showed a 50-fold reduction in intensity as the pH increased from 3 to 9.5 (Figure 2C,D). In contrast, emission from 5 decreased only 1.6-fold over the same range of pH (Figure 2F,G). Xanthene-based fluorophores have also been shown to respond to changes in solvent polarity.47 To examine the sensitivity of each fluorophore to the polarity of the solvent, we measured absorption and emission for 4 and 5 in mixtures of water and dioxane (Figure S7). We found 30- and 400-fold increases in fluorescence intensity for 4 and 5, respectively, as the dielectric constant increased from 2.2 to 78.5.
Figure 2.
Spectroscopic characterization of rhodamines 4 and 5. (A) Structures of rhodamines 4 and 5. (B) Absorption and fluorescence emission spectra of 4. (C) Fluorescence emission from 4 at λem = 577 nm as a function of pH. (D) Absorption by 4 at λabs = 561 nm as a function of pH. (E) Absorption and fluorescence emission spectra of 5. (F) Fluorescence emission from 5 at λem = 578 nm as a function of pH. (G) Absorption by 5 at λabs = 563 nm as a function of pH. Error bars denote the standard deviation from three independent experiments. cps = counts per second.
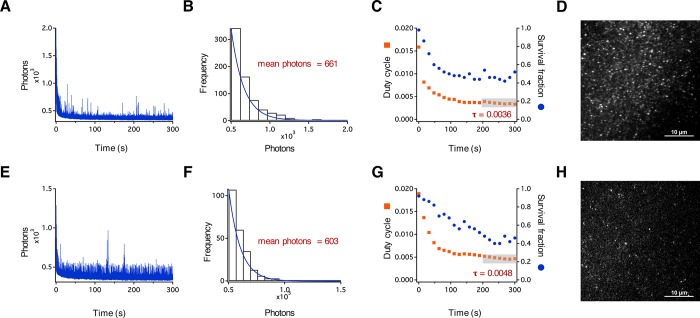
For dyes to be useful in super-resolution imaging experiments, they must exhibit photoswitching behavior on the millisecond time scale.27 To investigate the blinking behavior of 4 and 5, we analyzed their spectral properties at the single-molecule level. We determined the photon output, duty cycle, survival fraction, and number of switching cycles for each fluorophore. Dyes were prepared as 200 nM solutions in 1% w/v poly(vinyl alcohol) (PVA) and cast as films on precleaned glass slides. Immobilizing fluorescent molecules in polymeric films has been used to characterize blinking properties of single molecules.48 We imaged the cast PVA films by total internal reflection fluorescence (TIRF) microscopy using a Nikon N-STORM Ti2-E inverted microscope (Figure 3D,H). Samples of 4 and 5 were subjected to continuous activation at 405 nm (5% from a 30 mW laser source). Excitation was accomplished at 561 nm (25% from a 70 mW laser source), and emission was collected from 580 to 625 nm for 5 min at an integration time of 30 ms. Photon output profiles were plotted as a function of time (Figure 3A,E). Rhodamine 4 exhibited many photoswitching events, with a mean of 44 switching cycles during the 5 min of acquisition (Figure 3A) and an average of 661 photons per switching event (Figure 3B). Next, we calculated the duty cycle (τ)27 (i.e., the fraction of time the fluorophore spends in the on state) and the survival fraction (i.e., the fraction of molecules that are in the on state versus a dark or photobleached state) as a function of time (Figure 3C). The average duty cycle was calculated to be 0.0036 for the last 100 s of acquisition (Figure 3C, orange squares, gray box), and the survival fraction declined by roughly 60% over the course of 5 min of acquisition. The behavior of 5 was similar; we observed an average of 603 photons per switching event (Figure 3F), 56 switching events per 5 min acquisition, a duty cycle of 0.0048, and a survival fraction of approximately 40% (Figure 3G). The photon counts of 4 and 5 are comparable to those of other STORM dyes and photoactivatable fluorescent proteins27,49 and suggest that these and similar fluorophores should be useful in super-resolution imaging.
Figure 3.
Single-molecule characterization of rhodamines 4 and 5. (A) Representative single-molecule fluorescence time traces showing the number of detected photons from single molecules of 4. (B) Histogram showing the distribution of photons. The mean number of photons for 4 was calculated using a single-exponential fit (blue line). (C) The on/off duty cycle was calculated for 4 and plotted as a function of time (orange squares). The average duty cycle was calculated for the last 100 s of acquisition where the switching events reach a quasi-equilibrium state (gray box). This duty cycle was used to determine the survival fraction during image acquisition and is plotted against time (blue dots). (D) Image showing single molecules of 4 in 1% w/v PVA on glass. (E) Representative single-molecule fluorescence time traces showing the number of detected photons from single molecules of 5. (F) The mean number of photons for 5 was calculated using a single-exponential fit (blue line). (G) The on/off duty cycle was calculated for 5 and plotted as a function of time (orange squares). The average duty cycle was calculated for the last 100 s of acquisition where the switching events reach a quasi-equilibrium state (gray box). This duty cycle was used to determine the survival fraction of 5 during image acquisition and is plotted against time (blue dots). (H) Image showing single molecules of 5 in 1% w/v PVA on a glass surface.
To enable super-resolution imaging of bacterial proteins, we elaborated the rhodamine spirolactam scaffolds to generate 2 and 3 (Scheme S3) and employed the NMT labeling strategy illustrated in Figure 1. E. coli chemotaxis proteins Tar and CheA and cell division proteins FtsZ and FtsA were chosen as test substrates. Cells harboring both the pHV738-NMT1-MetAP plasmid50 for the constitutive expression of NMT and a modified pBAD24 plasmid51 for the inducible expression of the target protein downstream of the araBAD promoter were grown at 37 °C to an optical density (OD600) of 0.5 at 600 nm. The expression of each target protein was accomplished by the addition of 0.2% w/v l-arabinose for 1 h, and N-terminal labeling was achieved by the addition of 250 μM 1. Cells were collected by centrifugation, rinsed with PBS, and resuspended to an OD600 of 2 in PBS. A 100 μL aliquot of cells was labeled with 200 nM 2 at 37 °C for 1 h in the dark and then washed five times with PBS. A 10 μL aliquot of cells was pipetted onto a 1.5% w/v agarose pad and imaged using the laser conditions described previously for the characterization of single molecules of 4 and 5.
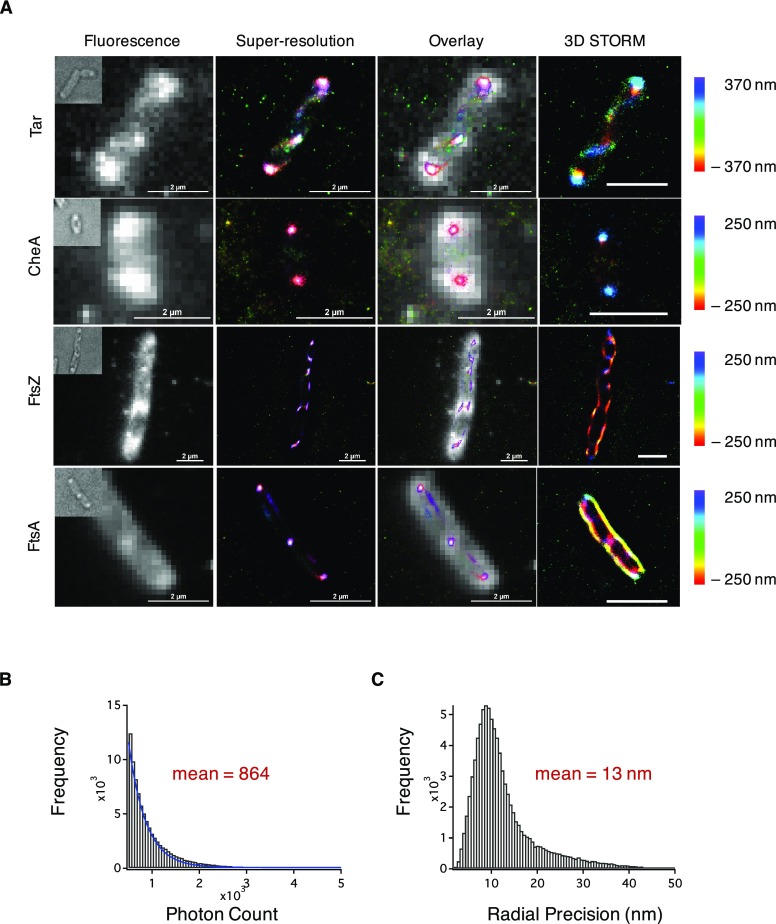
We observed the photoswitching of 2 in live cells (Supporting Information Movie S1) and reconstructed super-resolution images (Figure 4). Chemotaxis proteins Tar and CheA predominantly clustered at the cellular poles, a phenomenon believed to enhance chemotactic signaling.42 Reconstructed images of Tar also revealed smaller clusters distributed throughout the cell, in some cases in banded or helical patterns (Supporting Information Movie S2). The insertion of Tar into the polar membrane has been reported to be associated with the Sec protein–translocation pathway,52 and the Sec machinery has been suggested to form helical patterns which colocalize with expression of Tar. Greenfield and co-workers have also observed banded patterns for Tar fused to photoconvertible fluorescent protein mEos in bacterial cells53 and suggest that such patterns may reflect the spatial organization of the translocation machinery. For cell division proteins FtsZ and FtsA, localization near the septum of the cell in diffraction-limited fluorescence images was observed. Cellular division in bacteria involves the synthesis of peptidoglycan from synthases that are recruited by FtsZ and FtsA filaments. Coupling of the filaments to peptidoglycan synthases is central to the mechanism of bacterial cytokinesis. Recent studies have demonstrated that FtsZ treadmilling both recruits peptidoglycan synthases to the division plane and distributes these enzymes along the cell in the form of concentric rings.54,55 Our high-resolution image of FtsZ reveals elongated clusters of variable orientation throughout the cell, consistent with earlier reports that suggest that the Z ring forms from FtsZ protofilaments oriented both axially and longitudinally.56,57 The mean photon output of 2 measured in these live-cell imaging experiments (Figure 4B) was 864 photons, with a mean localization precision of 13 nm in the radial direction (Figure 4C) and 27 nm in the axial direction (Figure S8). Live cells labeled with 3 yielded similar results, with similar photon output and localization precision in both the radial and axial directions (Figures S9 and S10).
Figure 4.
Super-resolution imaging of bacterial proteins in live cells. Cells expressing one of four bacterial proteins were labeled with 1 and 2. (A) STORM images of bacterial proteins with polar localization (Tar and CheA) or septal localization (FtsZ and FtsA) expressed in E. coli. Bright-field images of cells are shown in the top left corner of the fluorescence images. Scale bar = 2 μm. (B) Histogram indicating the number of detected photons during image acquisition with the fit to a single exponential (blue curve). The mean number of photons is calculated from the single-exponential fit. (C) Mean radial precision for imaging in live bacterial cells.
We conducted a series of control experiments to determine the specificity of labeling, the cytocompatibility of reagents 1–3, and potential effects of the N-terminal NMT recognition sequence on protein function. Cells that did not express the target protein or were treated only with 1 did not show significant fluorescence (Figures S11 and S12), confirming the specificity of labeling. At the concentrations of the fluorophore and fatty acid used in our labeling experiments, we found no effects on cell growth (Figure S13). To determine whether the short N-terminal peptide tag affects protein function, we monitored the growth of cells that express either Tar or FtsA with or without the N-terminal NMT recognition tag. For Tar (Figure S14A–C), the growth curves were essentially unaffected by the recognition tag, although induction of the target protein (by the addition of 0.2% arabinose) reduced the growth rate and the limiting optical density of the culture (Figure S14B). Treatment with 1 caused no further changes (Figure S14C). For FtsA, the effects of induction were larger (Figure S14E), although only the limiting optical density appears to be sensitive to the presence of the recognition tag. Treatment with 1 again caused no further changes (Figure S14F). We believe that the results of these experiments are consistent with expectation in that the effects of the N-terminal peptide tag on protein function are likely to be small but should be determined for each target of interest.
There is substantial interest in the development of methods to label macromolecular targets with small-molecule fluorescent probes for super-resolution fluorescence microscopy, specifically in live cells.58 The fusion of targets to either the SNAP-Tag or the HaloTag has been successful in achieving the site-specific modification of proteins with small-molecule fluorescent probes, but both tags are large relative to the appended fluorophores. As the field progresses toward achieving the molecular resolution of new biological architectures, smaller labels will be needed. Bright organic dyes will be critical for pushing imaging modalities forward. Rhodamines 2 and 3 have photon budgets comparable to those of fluorescein, Atto 655, and Alexa 750.27 Although 2 and 3 emit fewer photons than some yellow- and red-absorbing dyes, such as Cy3 and Cy5,27 Lavis and co-workers have shown that the replacement of the diethylamino groups on rhodamines with azetidinyl substituents substantially improves the photon output.59 Furthermore, 2 and 3 can be used directly in live-cell imaging of intracellular targets without the need for fixation and permeabilization. More broadly, the strained alkyne functionality of 2 and 3 should make these fluorophores useful for labeling other azide-tagged biomolecules in both prokaryotic and eukaryotic cells. Notably, 2 and 3 do not require reducing agents or oxygen-scavenging buffers; careful optimization of the concentrations of these reagents is usually needed to facilitate the photoswitching of dyes.60 Because these dyes emit in the orange region of the spectrum, they are suited for use in two-color STORM imaging experiments with fluorophores such as Alexa 647 and Cy7.27 Although the method is limited to N-terminal labeling and is unlikely to be used on bacterial proteins bearing signal peptides,61 NMT-mediated labeling provides a simple strategy to append small, bright, photoswitchable dyes to specific target proteins. As dyes continue to be developed with chemical modifications to modulate the spectral stability,62 labeling technologies that enable site-specific covalent modification with small molecules will prove even more broadly useful.
Conclusions
The advent of super-resolution imaging methods has substantially advanced our understanding of how prokaryotic organisms orchestrate fundamental cellular processes. Here we report a new class of reactive rhodamine spirolactam dyes and demonstrate their use as cell-permeant fluorescent probes for super-resolution imaging in live bacterial cells. Super-resolution images of the bacterial chemotaxis protein Tar and the cell division protein FtsZ capture features of protein assembly that are not discernible by diffraction-limited fluorescence microscopy. New methods that continue to push the boundaries of light microscopy (e.g., cryogenic PALM) will be particularly informative for elucidating new structures with high precision.63,64 We anticipate that the results described here will expand the palette of methods for the super-resolution imaging of proteins in live cells and aid the discovery of new biological ultrastructures.
Materials and Methods
Plasmid and Strain Construction
The construction of modified pBAD24 plasmids encoding target bacterial proteins with the NMT recognition sequence has been described previously.36 Briefly, the gene encoding a bacterial protein of interest was amplified from DH10B Escherichia coli using a forward primer that contained the oligonucleotide sequence 5′-ATG GGT AAC GAA GCG TCT TAC CCG CTG-3′ to encode the NMT recognition sequence (MGNEASYPL). The amplified gene was inserted between the EcoRI and HindIII sites in pBAD24 using standard restriction enzyme digestion and ligation protocols. The modified pBAD24 plasmid and pHV738-NMT1-MetAP plasmid50 were transformed into BL21 E. coli cells and selected against ampicillin (200 μg/mL) and kanamycin (35 μg/mL).
Characterization of Spectral Properties
Spectroscopic measurements were performed in 1 cm, 3.5 mL quartz cuvettes (Starna Cells, Atascadero, CA) at ambient temperature (23 °C). Absorption spectra of rhodamine salts were recorded using a Cary 50 UV–visible spectrophotometer (Varian, Palo Alto, CA). Rhodamines were diluted to concentrations ranging from 0 to 40 μM in different solvents (e.g., 1:1 v/v acetonitrile/water, acetonitrile, toluene, methanol, and 10 mM HEPES pH 7.3). Extinction coefficients (ε) at 561 nm for 4 and at 563 nm for 5 were calculated by applying the Beer–Lambert law to the absorption spectra of diluted samples in 1:1 v/v water/acetonitrile with the addition of 1 M HCl. Measurements were made in triplicate. Fluorescence measurements were made on a PTI QuantaMaster fluorescence spectrofluorometer (Photon Technology International, Birmingham, NJ). For quantum yield determination, rhodamine salts were diluted in acidic ethanol (containing 1% v/v 1 M HCl) and adjusted to A510 < 0.1. Excitation was carried out at 510 nm, and emission was collected from 530 to 700 nm at a scan rate of 1 nm/s. The quantum yields of the rhodamine salts were calculated using the integrated fluorescence intensities at λem,max and rhodamine B as a standard with a known quantum yield (0.70).65 The reported values for φ are averages (n = 3). For pH studies, solutions of rhodamine salts were prepared at a concentration of 5 μM in 10 mM bis-tris propane at different pH values such that A510 < 0.1. Rhodamine salts were prepared in mixtures of water and dioxane with known dielectric constants66 at a concentration of 5 μM. Fluorescence emission spectra were collected using an excitation wavelength of 510 nm and emission from 530 to 700 nm with a scan rate of 1 nm/s.
Single-Molecule Fluorescence Measurements
Rhodamine salts were dissolved at a concentration of 200 nM in 1% w/v poly(vinyl alcohol) (31–50 kDa, Sigma-Aldrich, St. Louis, MO), and solutions were cast onto 75 mm × 25 mm quartz microscope slides (Electron Microscopy Sciences, Hatfield, PA). Quartz microscope slides were cleaned extensively with acetone, methanol, and doubly distilled water (ddH2O), sonicated with 1 M KOH for 60 min, and washed with acetone again before being dried under a stream of argon. Quartz coverslips (25 mm × 25 mm, Electron Microscopy Sciences, PA) were used to seal the films. Time series were recorded using a Nikon N-STORM Ti2-E inverted microscope (equipped with 405, 488, 561, and 647 nm fiber-coupled excitation lasers) with total reflection internal fluorescence (TIRF) illumination using an Apochromat TIRF 100X/1.49 NA oil-immersion objective lens, an sCMOS detector (Andor Technology, South Windsor, CT) for PSF detection, and a Perfect Focus System (PFS4) for axial stabilization. The filter cube set (TRF89902-EM-ET 405/488/561/647 nm laser band set, Chroma Technology, VT) was equipped with a quad-band-pass ZET 405/488/561/647x excitation filter, quad-band ZT405/488/561/647rpc dichroic mirror, and ZET405/488/561/647m emission filters. The excitation wavelengths were 405 nm (5% from a 30 mW laser source) and 561 nm (25% from a 70 mW laser source), and emission was collected from 580 to 625 nm. Single molecules of the rhodamine salts were identified by drawing 7 pixels × 7 pixels around areas that had integrated fluorescence intensities that were at least 5 times the standard deviation of the background fluorescence intensity. Molecules were selected such that the 7 pixel × 7 pixel areas were at least 5 pixels away from each other to ensure that molecules were not overlapping. Time series were used with an integration time of 30 ms for 5 min, resulting in 10 000 frames per acquisition. The duty cycle (DC for each single molecule was calculated as reported by Zhuang and co-workers.27 Briefly, peaks from the photon output profiles were identified as switching events if the photon count was at least 5 times above the standard deviation of the background fluctuations. A sliding window of 100 s was used in calculating the duty cycle with the following equation
where τon,i denotes the time for which the ith fluorophore is in the on state. The duty cycle is plotted against time in Figure 3 for n = 50 single molecules. The last 100 s were used to assign the DC value for determining the survival fraction.
Labeling Proteins with a ω-Azido Fatty Acid and Conjugation to the Fluorophore in Live Cells
Overnight cultures of E. coli strain BL21 harboring modified pBAD24 and pHV738-NMT1-MetAP plasmids were diluted 1:50 in LB medium supplied with 200 μg/mL ampicillin and 35 μg/mL kanamycin and labeled with azido fatty acids as previously described.36 Protein expression was carried out at 37 °C for 1 h, and cells were harvested by centrifugation, rinsed with PBS, and concentrated to OD600 = 2 in PBS. Fluorophore 2 or 3 (2 mM in DMSO) was then added to the cells to a concentration of 200 nM, and incubation proceeded at 37 °C for 1 h, after which cells were rinsed five times with PBS to remove excess fluorophore. Cells (in 10 μL aliquots) were mounted onto 1.5% w/v agarose pads in PBS for imaging.
Super-resolution Imaging (STORM) in Live Bacterial Cells
Bacterial cells labeled with 1 and either 2 or 3 were mounted on 1.5% w/v agarose pads in PBS. Imaging was performed using the Nikon N-STORM 5.0 system on an Ti2-E inverted microscope with TIRF illumination equipped with an Apochromat 100X/1.49 NA oil immersion objective lens and a sCMOS detector for PSF detection. The Perfect Focus System (PFS4) was used to stabilize and detect the axial position of the agarose pad for imaging. The filter cube set (TRF89902-EM-ET 405/488/561/647 nm Laser Band Set, Chroma Technology, VT) was equipped with a quad-band-pass ZET 405/488/561/647x excitation filter, quad-band ZT405/488/561/647rpc dichroic mirror, and ZET405/488/561/647m emission filters. The angle of the incident wave was adjusted to match the angle of the evanescent wave as close as possible to maximize the signal-to-noise ratio. Samples were continuously excited at 405 and 561 nm (5% from a 30 mW laser source and 25% from a 70 mW laser source, respectively), and images were recorded at a frame rate of 33 Hz for 10 000 frames. Drift correction was automatically implemented by NIS-Elements during image acquisition. STORM images were reconstructed using Nikon Advanced Research (AR) NIS-Elements, in which each voxel represents a three-dimensional Gaussian representing the localized centroid of each PSF. For each bacterial protein expressed and each labeling condition, at least four cells were imaged with these excitation and emission settings. Histograms of photon counts and localization precisions were determined in NIS-Elements and plotted in IGOR Pro (WaveMetrics, Portland, OR).
Acknowledgments
The authors thank members of the Tirrell laboratory for insightful discussions. S.H.H. thanks Y. Hui, A. Collazo, and R. Stanciauskas for helpful advice. This work was supported by the Joseph J. Jacobs Institute for Molecular Engineering for Medicine and the Center for Environmental Microbial Interactions at the California Institute of Technology.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b00617.
Details describing synthesis procedures, the characterization of compounds, and additional live cell super-resolution images (PDF)
Photoswitching of probe 2 in cells expressing the chemotaxis protein Tar; the movie plays at 33 fps (MOV)
Three-dimensional rendering of Tar labeled with 2 from Figure 4 (MOV)
The authors declare no competing financial interest.
Supplementary Material
References
- Klar T. A.; Jakobs S.; Dyba M.; Egner A.; Hell S. W. Fluorescence Microscopy with Diffraction Resolution Barrier Broken by Stimulated Emission. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (15), 8206–8210. 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E.; Patterson G. H.; Sougrat R.; Lindwasser O. W.; Olenych S.; Bonifacino J. S.; Davidson M. W.; Lippincott-Schwartz J.; Hess H. F. Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 2006, 313 (5793), 1642–1645. 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Gahlmann A.; Moerner W. E. Exploring Bacterial Cell Biology with Single-Molecule Tracking and Super-Resolution Imaging. Nat. Rev. Microbiol. 2014, 12 (1), 9–22. 10.1038/nrmicro3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M.; Huang B.; Dempsey G. T.; Zhuang X. Multicolor Super-Resolution Imaging with Photo-Switchable Fluorescent Probes. Science 2007, 317 (5845), 1749–1753. 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.; Wang W.; Bates M.; Zhuang X. Three-Dimensional Super-Resolution Imaging by Stochastic Optical Reconstruction Microscopy. Science 2008, 319 (5864), 810–813. 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K.; Zhong G.; Zhuang X. Actin, Spectrin, and Associated Proteins Form a Periodic Cytoskeletal Structure in Axons. Science 2013, 339 (6118), 452–456. 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans D. C.; Wurm C. A.; Riedel D.; Wenzel D.; Stagge F.; Deckers M.; Rehling P.; Jakobs S. STED Super-Resolution Microscopy Reveals an Array of MINOS Clusters along Human Mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (22), 8936–8941. 10.1073/pnas.1301820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S. T.; Girirajan T. P. K.; Mason M. D. Ultra-High Resolution Imaging by Fluorescence Photoactivation Localization Microscopy. Biophys. J. 2006, 91 (11), 4258–4272. 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M. J.; Bates M.; Zhuang X. Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat. Methods 2006, 3 (10), 793–796. 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M.; van de Linde S.; Schüttpelz M.; Kasper R.; Seefeldt B.; Mukherjee A.; Tinnefeld P.; Sauer M. Subdiffraction-Resolution Fluorescence Imaging with Conventional Fluorescent Probes. Angew. Chem., Int. Ed. 2008, 47 (33), 6172–6176. 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Abbe E. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Archiv f. mikrosk. Anatomie 1873, 9 (1), 413–418. 10.1007/BF02956173. [DOI] [Google Scholar]
- Dempsey G. T.; Bates M.; Kowtoniuk W. E.; Liu D. R.; Tsien R. Y.; Zhuang X. Photoswitching Mechanism of Cyanine Dyes. J. Am. Chem. Soc. 2009, 131 (51), 18192–18193. 10.1021/ja904588g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelsang J.; Kasper R.; Steinhauer C.; Person B.; Heilemann M.; Sauer M.; Tinnefeld P. A Reducing and Oxidizing System Minimizes Photobleaching and Blinking of Fluorescent Dyes. Angew. Chem., Int. Ed. 2008, 47 (29), 5465–5469. 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- Heilemann M.; Margeat E.; Kasper R.; Sauer M.; Tinnefeld P. Carbocyanine Dyes as Efficient Reversible Single-Molecule Optical Switch. J. Am. Chem. Soc. 2005, 127 (11), 3801–3806. 10.1021/ja044686x. [DOI] [PubMed] [Google Scholar]
- Deniz E.; Tomasulo M.; Cusido J.; Yildiz I.; Petriella M.; Bossi M. L.; Sortino S.; Raymo F. M. Photoactivatable Fluorophores for Super-Resolution Imaging Based on Oxazine Auxochromes. J. Phys. Chem. C 2012, 116 (10), 6058–6068. 10.1021/jp211796p. [DOI] [Google Scholar]
- Biteen J. S.; Thompson M. A.; Tselentis N. K.; Bowman G. R.; Shapiro L.; Moerner W. E. Super-Resolution Imaging in Live Caulobacter Crescentus Cells Using Photoswitchable EYFP. Nat. Methods 2008, 5 (11), 947–949. 10.1038/nmeth.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andresen M.; Stiel A. C.; Trowitzsch S.; Weber G.; Eggeling C.; Wahl M. C.; Hell S. W.; Jakobs S. Structural Basis for Reversible Photoswitching in Dronpa. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (32), 13005–13009. 10.1073/pnas.0700629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G. H.; Lippincott-Schwartz J. A Photoactivatable GFP for Selective Photolabeling of Proteins and Cells. Science 2002, 297 (5588), 1873–1877. 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Lord S. J.; Conley N. R.; Lee H. D.; Samuel R.; Liu N.; Twieg R. J.; Moerner W. E. A Photoactivatable Push-Pull Fluorophore for Single-Molecule Imaging in Live Cells. J. Am. Chem. Soc. 2008, 130 (29), 9204–9205. 10.1021/ja802883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi M.; Fölling J.; Belov V. N.; Boyarskiy V. P.; Medda R.; Egner A.; Eggeling C.; Schönle A.; Hell S. W. Multicolor Far-Field Fluorescence Nanoscopy through Isolated Detection of Distinct Molecular Species. Nano Lett. 2008, 8 (8), 2463–2468. 10.1021/nl801471d. [DOI] [PubMed] [Google Scholar]
- Belov V. N.; Wurm C. A.; Boyarskiy V. P.; Jakobs S.; Hell S. W. Rhodamines NN: A Novel Class of Caged Fluorescent Dyes. Angew. Chem., Int. Ed. 2010, 49 (20), 3520–3523. 10.1002/anie.201000150. [DOI] [PubMed] [Google Scholar]
- Chudakov D. M.; Verkhusha V. V.; Staroverov D. B.; Souslova E. A.; Lukyanov S.; Lukyanov K. A. Photoswitchable Cyan Fluorescent Protein for Protein Tracking. Nat. Biotechnol. 2004, 22 (11), 1435–1439. 10.1038/nbt1025. [DOI] [PubMed] [Google Scholar]
- Subach F. V.; Patterson G. H.; Manley S.; Gillette J. M.; Lippincott-Schwartz J.; Verkhusha V. V. Photoactivatable MCherry for High-Resolution Two-Color Fluorescence Microscopy. Nat. Methods 2009, 6 (2), 153–159. 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney S. A.; Murphy C. S.; Hazelwood K. L.; Davidson M. W.; Looger L. L. A Bright and Photostable Photoconvertible Fluorescent Protein for Fusion Tags. Nat. Methods 2009, 6 (2), 131–133. 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurskaya N. G.; Verkhusha V. V.; Shcheglov A. S.; Staroverov D. B.; Chepurnykh T. V.; Fradkov A. F.; Lukyanov S.; Lukyanov K. A. Engineering of a Monomeric Green-to-Red Photoactivatable Fluorescent Protein Induced by Blue Light. Nat. Biotechnol. 2006, 24 (4), 461–465. 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- Baddeley D.; Bewersdorf J. Biological Insight from Super-Resolution Microscopy: What We Can Learn from Localization-Based Images. Annu. Rev. Biochem. 2018, 87, 965–989. 10.1146/annurev-biochem-060815-014801. [DOI] [PubMed] [Google Scholar]
- Dempsey G. T.; Vaughan J. C.; Chen K. H.; Bates M.; Zhuang X. Evaluation of Fluorophores for Optimal Performance in Localization-Based Super-Resolution Imaging. Nat. Methods 2011, 8 (12), 1027–1036. 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich G.; Ziessel R.; Harriman A. The Chemistry of Fluorescent Bodipy Dyes: Versatility Unsurpassed. Angew. Chem., Int. Ed. 2008, 47 (7), 1184–1201. 10.1002/anie.200702070. [DOI] [PubMed] [Google Scholar]
- Nahidiazar L.; Agronskaia A. V.; Broertjes J.; van den Broek B.; Jalink K. Optimizing Imaging Conditions for Demanding Multi-Color Super Resolution Localization Microscopy. PLoS One 2016, 11 (7), e0158884 10.1371/journal.pone.0158884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M.; Jones S. A.; Zhuang X. Preparation of Photoswitchable Labeled Antibodies for STORM Imaging. Cold Spring Harb. Protoc. 2013, 2013 (6), 540–541. 10.1101/pdb.prot075168. [DOI] [PubMed] [Google Scholar]
- Huang B.; Babcock H.; Zhuang X. Breaking the Diffraction Barrier: Super-Resolution Imaging of Cells. Cell 2010, 143 (7), 1047–1058. 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan D. R.; Bell T. D. M. Image Artifacts in Single Molecule Localization Microscopy: Why Optimization of Sample Preparation Protocols Matters. Sci. Rep. 2015, 5, 7924. 10.1038/srep07924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los G. V.; Encell L. P.; McDougall M. G.; Hartzell D. D.; Karassina N.; Zimprich C.; Wood M. G.; Learish R.; Ohana R. F.; Urh M.; et al. HaloTag: A Novel Protein Labeling Technology for Cell Imaging and Protein Analysis. ACS Chem. Biol. 2008, 3 (6), 373–382. 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Lee H. D.; Lord S. J.; Iwanaga S.; Zhan K.; Xie H.; Williams J. C.; Wang H.; Bowman G. R.; Goley E. D.; Shapiro L.; et al. Superresolution Imaging of Targeted Proteins in Fixed and Living Cells Using Photoactivatable Organic Fluorophores. J. Am. Chem. Soc. 2010, 132 (43), 15099–15101. 10.1021/ja1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm J. B.; English B. P.; Choi H.; Muthusamy A. K.; Mehl B. P.; Dong P.; Brown T. A.; Lippincott-Schwartz J.; Liu Z.; Lionnet T.; et al. Bright Photoactivatable Fluorophores for Single-Molecule Imaging. Nat. Methods 2016, 13 (12), 985–988. 10.1038/nmeth.4034. [DOI] [PubMed] [Google Scholar]
- Ho S. H.; Tirrell D. A. Chemoenzymatic Labeling of Proteins for Imaging in Bacterial Cells. J. Am. Chem. Soc. 2016, 138 (46), 15098–15101. 10.1021/jacs.6b07067. [DOI] [PubMed] [Google Scholar]
- Kulkarni C.; Kinzer-Ursem T. L.; Tirrell D. A. Selective Functionalization of the Protein N Terminus with N-Myristoyl Transferase for Bioconjugation in Cell Lysate. ChemBioChem 2013, 14 (15), 1958–1962. 10.1002/cbic.201300453. [DOI] [PubMed] [Google Scholar]
- Aitken A.; Cohen P.; Santikarn S.; Williams D. H.; Calder A. G.; Smith A.; Klee C. B. Identification of the NH2-Terminal Blocking Group of Calcineurin B as Myristic Acid. FEBS Lett. 1982, 150 (2), 314–318. 10.1016/0014-5793(82)80759-X. [DOI] [PubMed] [Google Scholar]
- Lee M. K.; Rai P.; Williams J.; Twieg R. J.; Moerner W. E. Small-Molecule Labeling of Live Cell Surfaces for Three-Dimensional Super-Resolution Microscopy. J. Am. Chem. Soc. 2014, 136 (40), 14003–14006. 10.1021/ja508028h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer K.-H.; Gleiter R. Photochromism of Rhodamine Derivatives. Angew. Chem., Int. Ed. Engl. 1977, 16 (2), 113–113. 10.1002/anie.197701131. [DOI] [Google Scholar]
- Willwohl H.; Wolfrum J.; Gleiter R. Kinetics and Mechanism of the Photochromism of N-Phenyl-Rhodaminelactame. Laser Chem. 1989, 10, 63–72. 10.1155/1989/69709. [DOI] [Google Scholar]
- Shiomi D.; Banno S.; Homma M.; Kawagishi I. Stabilization of Polar Localization of a Chemoreceptor via Its Covalent Modifications and Its Communication with a Different Chemoreceptor. J. Bacteriol. 2005, 187 (22), 7647–7654. 10.1128/JB.187.22.7647-7654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddock J. R.; Shapiro L. Polar Location of the Chemoreceptor Complex in the Escherichia Coli Cell. Science 1993, 259 (5102), 1717–1723. 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- Sun Q.; Margolin W. FtsZ Dynamics during the Division Cycle of Live Escherichia Coli Cells. J. Bacteriol. 1998, 180 (8), 2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinall S. G.; Lutkenhaus J. FtsA Is Localized to the Septum in an FtsZ-Dependent Manner. J. Bacteriol. 1996, 178 (24), 7167–7172. 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov V. N.; Bossi M. L.; Fölling J.; Boyarskiy V. P.; Hell S. W. Rhodamine Spiroamides for Multicolor Single-Molecule Switching Fluorescent Nanoscopy. Chem. - Eur. J. 2009, 15 (41), 10762–10776. 10.1002/chem.200901333. [DOI] [PubMed] [Google Scholar]
- Grimm J. B.; Sung A. J.; Legant W. R.; Hulamm P.; Matlosz S. M.; Betzig E.; Lavis L. D. Carbofluoresceins and Carborhodamines as Scaffolds for High-Contrast Fluorogenic Probes. ACS Chem. Biol. 2013, 8 (6), 1303–1310. 10.1021/cb4000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. M.; Cubitt A. B.; Tsien R. Y.; Moerner W. E. On/off Blinking and Switching Behaviour of Single Molecules of Green Fluorescent Protein. Nature 1997, 388 (6640), 355–358. 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- Wang S.; Moffitt J. R.; Dempsey G. T.; Xie X. S.; Zhuang X. Characterization and Development of Photoactivatable Fluorescent Proteins for Single-Molecule-Based Superresolution Imaging. Proc. Natl. Acad. Sci. U. S. A. 2014, 111 (23), 8452–8457. 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valkenburgh H. A.; Kahn R. A. Coexpression of Proteins with Methionine Aminopeptidase/or N-Myristoyltransferase in Escherichia Coli to Increase Acylation Homogeneity of Protein Preparations. Methods Enzymol. 2002, 344, 186–193. 10.1016/S0076-6879(02)44715-5. [DOI] [PubMed] [Google Scholar]
- Guzman L. M.; Belin D.; Carson M. J.; Beckwith J. Tight Regulation, Modulation, and High-Level Expression by Vectors Containing the Arabinose PBAD Promoter. J. Bacteriol. 1995, 177 (14), 4121–4130. 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D.; Yoshimoto M.; Homma M.; Kawagishi I. Helical Distribution of the Bacterial Chemoreceptor via Colocalization with the Sec Protein Translocation Machinery. Mol. Microbiol. 2006, 60 (4), 894–906. 10.1111/j.1365-2958.2006.05145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield D.; McEvoy A. L.; Shroff H.; Crooks G. E.; Wingreen N. S.; Betzig E.; Liphardt J. Self-Organization of the Escherichia Coli Chemotaxis Network Imaged with Super-Resolution Light Microscopy. PLoS Biol. 2009, 7 (6), e1000137 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Lyu Z.; Miguel A.; McQuillen R.; Huang K. C.; Xiao J. GTPase Activity-Coupled Treadmilling of the Bacterial Tubulin FtsZ Organizes Septal Cell Wall Synthesis. Science 2017, 355 (6326), 744–747. 10.1126/science.aak9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson-Filho A. W.; Hsu Y.-P.; Squyres G. R.; Kuru E.; Wu F.; Jukes C.; Sun Y.; Dekker C.; Holden S.; VanNieuwenhze M. S.; et al. Treadmilling by FtsZ Filaments Drives Peptidoglycan Synthesis and Bacterial Cell Division. Science 2017, 355 (6326), 739–743. 10.1126/science.aak9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlett V. W.; Margolin W. 3D-SIM Super-Resolution of FtsZ and Its Membrane Tethers in Escherichia Coli Cells. Biophys. J. 2014, 107 (8), L17–L20. 10.1016/j.bpj.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G.; Huang T.; Buss J.; Coltharp C.; Hensel Z.; Xiao J. In Vivo Structure of the E. Coli FtsZ-Ring Revealed by Photoactivated Localization Microscopy (PALM). PLoS One 2010, 5 (9), e12680 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Frei M. S.; Salim A.; Johnsson K. Small-Molecule Fluorescent Probes for Live-Cell Super-Resolution Microscopy. J. Am. Chem. Soc. 2019, 141 (7), 2770–2781. 10.1021/jacs.8b11134. [DOI] [PubMed] [Google Scholar]
- Grimm J. B.; English B. P.; Chen J.; Slaughter J. P.; Zhang Z.; Revyakin A.; Patel R.; Macklin J. J.; Normanno D.; Singer R. H.; et al. A General Method to Improve Fluorophores for Live-Cell and Single-Molecule Microscopy. Nat. Methods 2015, 12 (3), 244–250. 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin A. G.; Lounis B.; Cognet L. Super-Resolution Microscopy Approaches for Live Cell Imaging. Biophys. J. 2014, 107 (8), 1777–1784. 10.1016/j.bpj.2014.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivankov D. N.; Payne S. H.; Galperin M. Y.; Bonissone S.; Pevzner P. A.; Frishman D. How Many Signal Peptides Are There in Bacteria?. Environ. Microbiol. 2013, 15 (4), 983–990. 10.1111/1462-2920.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butkevich A. N.; Bossi M. L.; Lukinavicius G.; Hell S. W. Triarylmethane Fluorophores Resistant to Oxidative Photobluing. J. Am. Chem. Soc. 2019, 141 (2), 981–989. 10.1021/jacs.8b11036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-W.; Chen S.; Tocheva E. I.; Treuner-Lange A.; Löbach S.; Søgaard-Andersen L.; Jensen G. J. Correlated Cryogenic Photoactivated Localization Microscopy and Cryo-Electron Tomography. Nat. Methods 2014, 11 (7), 737–739. 10.1038/nmeth.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg P. D.; Sartor A. M.; Wang J.; Saurabh S.; Shapiro L.; Moerner W. E. Identification of PAmKate as a Red Photoactivatable Fluorescent Protein for Cryogenic Super-Resolution Imaging. J. Am. Chem. Soc. 2018, 140 (39), 12310–12313. 10.1021/jacs.8b05960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeloa F. L.; Ojeda P. R.; Arbeloa I. L. Flourescence Self-Quenching of the Molecular Forms of Rhodamine B in Aqueous and Ethanolic Solutions. J. Lumin. 1989, 44 (1), 105–112. 10.1016/0022-2313(89)90027-6. [DOI] [Google Scholar]
- Critchfield F. E.; Gibson J. A.; Hall J. L. Dielectric Constant for the Dioxane—Water System from 20 to 35°. J. Am. Chem. Soc. 1953, 75 (8), 1991–1992. 10.1021/ja01104a506. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.