Trauma-related invasive fungal wound infections (IFIs) are associated with significant morbidity and mortality. Early identification and treatment are critical. Traditional identification methods (e.g., fungal cultures and histopathology) can be delayed and insensitive. We assessed a PCR-based sequencing assay for rapid identification of filamentous fungi in formalin-fixed paraffin-embedded (FFPE) specimens obtained from combat casualties injured in Afghanistan.
KEYWORDS: invasive fungal wound infection, trauma, combat, PCR-based assay, mucormycosis, PCR, invasive fungal infection
ABSTRACT
Trauma-related invasive fungal wound infections (IFIs) are associated with significant morbidity and mortality. Early identification and treatment are critical. Traditional identification methods (e.g., fungal cultures and histopathology) can be delayed and insensitive. We assessed a PCR-based sequencing assay for rapid identification of filamentous fungi in formalin-fixed paraffin-embedded (FFPE) specimens obtained from combat casualties injured in Afghanistan. Blinded FFPE specimens from cases (specimens positive on histopathology) and controls (specimens negative on histopathology) were submitted for evaluation with a panfungal PCR. The internal transcribed spacer 2 (ITS2) region of the fungal ribosomal repeat was amplified and sequenced. The PCR results were compared with findings from histopathology and/or culture. If injury sites contributed multiple specimens, findings for the site were collapsed to the site level. We included 64 case subjects (contributing 95 sites) and 102 controls (contributing 118 sites). Compared to histopathology, panfungal PCR was specific (99%), but not as sensitive (63%); however, sensitivity improved to 83% in specimens from sites with angioinvasion. Panfungal PCR identified fungi of the order Mucorales in 33 of 44 sites with angioinvasion (75%), whereas fungal culture was positive in 20 of 44 sites (45%). Saksenaea spp. were the dominant fungi identified by PCR in specimens from angioinvasion sites (57%). Panfungal PCR is specific, albeit with lower sensitivity, and performs better at identifying fungi of the order Mucorales than culture. DNA sequencing offers significant promise for the rapid identification of fungal infection in trauma-related injuries, leading to more timely and accurate diagnoses.
INTRODUCTION
Given the significant morbidity and mortality associated with trauma-related invasive fungal infections (IFIs) (1–3), to improve outcomes, early identification of the causative fungus is essential. Unfortunately, however, both culture and histopathology have limitations. Specifically, conventional cultures are delayed and insensitive, while histopathology does not offer genus level identification (4, 5). Molecular-based methods offer a high degree of precision related to species identification and rapid turnaround times compared with conventional cultures.
Recent analyses have shown that fungal DNA can be efficiently amplified from formalin-fixed paraffin-embedded (FFPE) tissue samples, with the sensitivities of assays targeting either internal transcribed spacer (ITS) units or ribosomal (or mitochondrial) DNA (rDNA) ranging from 38% to 89% (6–11). However, these studies have not been performed in the setting of blast-related trauma. Soil contamination is common in this setting, and infections are often polymicrobial, with multiple fungal species being cultured, creating a challenge. We developed and assessed the performance characteristics of a panfungal PCR-based assay for the identification of filamentous fungi in FFPE specimens obtained during surgical debridement of blast wounds, comparing it with culture and histopathology. We also compared panfungal PCR with seminested PCR assays in a preplanned protocol-specified blinded analysis.
(This work was presented in part at IDWeek, 3 to 7 October 2018, San Francisco, CA [12].)
MATERIALS AND METHODS
Study design.
Data utilized for this retrospective analysis were collected through the Trauma Infectious Disease Outcomes Study (TIDOS) (13), an observational, multicenter study focused on examining outcomes of trauma-related infections among wounded military personnel. The FFPE tissue specimens were procured through a separate protocol approved by the Institutional Review Board of the Uniformed Services University of the Health Sciences. Due to concerns that formalin fixation and the resultant DNA degradation could result in reduced yield, a prespecified pilot analysis that involved submission of 20% of blinded specimens to two laboratories (University of Texas Health Sciences Center, San Antonio, TX [UTHSCSA], and LADR GmbH, Medizinisches Versorgungszentrum Dr. Kramer und Kollegen, Geesthact, Germany) was conducted. The decision to move to the second phase of the study was based on the achievement of a concordance of 60% or higher with the diagnosis obtained by histopathology. The results are presented here.
Study population.
Patients were eligible for inclusion if they sustained a trauma-related injury in Afghanistan (June 2009 through December 2014), were admitted to a participating U.S. military hospital (Walter Reed National Military Medical Center or Brook Army Medical Center) after transition through Landstuhl Regional Medical Center, Germany (LRMC), and had surgical specimens available. Cases were included only if they had positive histopathology (i.e., fungus identified either invading the vasculature [angioinvasion] or present in tissue). A hospital-based clinical practice guideline involving the sampling of all patients at risk for IFI was implemented in 2011 (14). Controls were identified from this population if they had specimens submitted to assess for IFI that were negative for fungus on histopathology and culture. Information on demographics, injury characteristics, and trauma care were obtained from the Department of Defense Trauma Registry (15), while results of cultures and histopathology were collected from the supplemental TIDOS infectious disease module (13).
Tissue specimens.
To reduce contamination, tissue blocks were loaded in an aseptic fashion (technicians donned sterile gloves for the procedure), DNA Away (ThermoFisher Scientific, Waltham, MA) was used to wipe down the surface before sectioning and between samples, and sectioning was performed with a sterile microtome. Finally, to reduce the chances of contamination, the first 5-μm section of the block was discarded. Sections (5 to 20 μm thick) were cut and shipped to the laboratories overnight in sterile cryovials. Both laboratories were blinded to the culture and histopathology results.
PCR-based assays.
The panfungal PCR-based assay was developed by the Advanced Nucleic Acid Core Facility at UTHSCSA and was prespecified as the default assay to be used on all specimens. For DNA extraction, five paraffin sections were transferred to a 2.0-ml screw-cap tube (Sarstedt, Numbrecht, Germany), pelleted briefly in a microcentrifuge at low speed, and prepared according to the manufacturer’s instructions with modifications. Briefly, 400 μl of incubation buffer containing 2 mg/ml proteinase K (Maxwell 16 FFPE Tissue LEV DNA purification kit; Promega, Inc., Madison, WI) was added to each specimen, followed by incubation of samples in a 70°C water bath overnight. After incubation, 100 μl (vol/vol) 0.5-mm glass beads (BioSpec Products, Inc., Bartlesville, OK) was added to each tube. The tubes were agitated on a bead beater (BioSpec) for 1 min at maximum speed and then spun in a microcentrifuge at maximum speed for 1 min. The supernatants were transferred to a new, sterile 2.0-ml screw-cap tube, taking care not to carry over beads or debris. Two times the supernatant volume of Maxwell 16 FFPE Tissue LEV DNA purification kit lysis buffer was added to each tube and mixed by brief vortexing. Samples were stored overnight at room temperature, but not refrigerated, or were processed immediately. The DNA was purified from the samples using a Maxwell 16 automated DNA extraction instrument (Promega) according to the manufacturer’s instructions. After purification, samples were measured for DNA content with a NanoDrop spectrophotometer (ThermoFisher, Grand Island, NY).
For fungal identification using a panfungal PCR assay, template DNA was amplified using the primers ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) to amplify the ITS2 region in the fungal ribosomal-DNA locus, as previously described (16). These primers yield amplicons ∼350 to 390 bp in length, depending on the species. Sequences were used for a BLAST search of GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and identities of ≥97% were required for identification to the species level (16). Additional PCR and sequencing were performed when needed for species confirmation; if BLAST results yielded low identities (17); when yields, as determined by gel electrophoresis, were low; or when sequence identity was poor due to sequence quality.
LADR used accredited in-house seminested PCR assays (18) targeting the mitochondrial DNA of Aspergillus spp., the 18S ribosomal DNA of molds belonging to the order Mucorales, and the ITS/28S region of members of the genus Fusarium. The primers AM 1 (5′-CTTTGGTTGCGGGTTTAGGGATT-3′) and AM 2 (5′-GGGAGTTCAAATCTCCCTTGGG-3′) amplify a 201-bp fragment complementary to positions 938 to 738 of an Aspergillus fumigatus sequence (GenBank accession number L37095). The second round, using primers AM 1 and AM 3 (5′-GAAAGGTCAGGTGTTCGAGTCAC-3′), amplifies a 135-bp seminested product used for species identification by sequencing. Primers ZM 1 (5′-ATTACCATGAGCAAATCAGA-3′) and ZM 2 (5′-TCCGTCAATTCCTTTAAGTTTC-3′) amplify a 407- or 408-bp fragment of the 18S ribosomal DNA corresponding to positions 722 to 1129 in the genus Cunninghamella (GenBank accession number AF113421) or 711 to 1096 in Rhizopus spp. (AF113440). Using primers ZM 1 and ZM 3 (5′-CAATCCAAGAATTTCACCTCTAG-3′) in the second round amplifies a 176- or 177-bp product used for genus identification (19). Whereas the variability within the targeted region is high enough to distinguish genera, it is too low to discriminate species within a genus. For Fusarium spp., a PCR assay utilized FUS I (P58SL) and FUS II (P28SL) primers (5′-AGTATTCTGGCGGGCATGCCTGT-3′ and 5′-ACAAATTACAACTCGGGCCCGAGA-3′, respectively) to amplify a 357-bp DNA sequence within the ITS/28S region in Fusarium spp. (20). The FUS I primer is complementary to positions 329 to 351, and FUS II is complementary to positions 685 to 662 (GenBank accession number L36634). To increase the sensitivity of the Fusarium PCR assay, a third primer, FUS III (5′-CCGTTACTGAGGCAATCCCTGTT-3′), was constructed. The seminested product of FUS I and FUS III is 296 bp long. The PCR conditions and processes were identical to the ones used for Aspergillus sp.- and Mucorales-specific PCRs (19). Nested-PCR products were sequenced as previously described (19). A PCR assay was considered positive if the product obtained from the Aspergillus sp. PCR was identical to an Aspergillus sp. sequence in GenBank. In addition, a PCR assay was positive if the product amplified by Mucorales PCR showed 100% homology to an 18S rDNA sequence of a genus in the order Mucorales or if the product of the Fusarium PCR showed 100% sequence identity to a fungal genus. Except for a single nucleotide position, Apophysomyces and Saksenaea have identical nested products in the Mucorales PCR. Therefore, because the difference is too small to discriminate (we cannot exclude the possibility of an error of the polymerase of a 1-nucleotide difference out of 900 nucleotides), the results were identified as Apophysomyces/Saksenaea sp.
Statistical analysis.
Subject characteristics were compared using Fisher’s exact test or a chi-square test (as appropriate) for categorical variables and a Wilcoxon rank sum test for continuous variables. P values of <0.05 were considered statistically significant.
The PCR findings were assessed for concordance with the results of culture and histopathology. Since a subject could have more than one wound and these wounds were often debrided multiple times, each subject could contribute more than one specimen from the same or different injury sites. All analyses were performed at the injury site level; if multiple specimens were obtained from an injury site, the findings were pooled and analyzed. Specimens were defined as negatives, where the only fungi identified were common skin or environmental contaminants, such as Malassezia spp. (see Table S1 in the supplemental material) (21).
Measures of concordance/agreement between PCR findings and both conventional cultures and histopathology were calculated using Cohen’s kappa coefficient. McNemar’s test statistic was also used to assess concordance between PCR findings and histopathology. Results were considered concordant if there was a match for a fungus. Diagnostic performance parameters assessing the PCR-based assay against histopathology were calculated as previously described (22).
Results for the subset of IFI case specimens that had both seminested and panfungal PCR assays were compared and are presented. For measures of concordance between the panfungal and seminested PCR-based assays, specimens identified as Apophysomyces elegans were included with Saksenaea spp., as the target sequences of the LADR Mucorales PCR assay were identical.
Data availability.
All relevant data are provided within the paper and its supporting documentation.
RESULTS
Study population.
A total of 64 case and 102 control subjects were identified. Case subjects had a greater proportion of blast injuries (97% versus 80%; P = 0.002) and higher injury severity scores (23) (median, 42 versus 26; P < 0.0001), required more blood transfusions during the day following injury (median number of units, 32 versus 10; P < 0.0001), and had more traumatic above-knee amputations (59% versus 21%; P < 0.0001) than controls (Table 1). These characteristics are all well-described risk factors for IFI (24).
TABLE 1.
Characteristics of trauma patients with tissue specimens analyzed by PCR-based assays
| Characteristica | Case patientsb (n = 64) | Controlsb (n = 102) | P valuef |
|---|---|---|---|
| Male [no. (%)] | 64 (100) | 102 (100) | NA |
| Afghanistan operational theater [no. (%)] | 64 (100) | 102 (100) | NA |
| Blast injury [no. (%)] | 62 (96.9) | 82 (80.4) | 0.002 |
| Dismounted injuryc [no. (%)] | 49 (94.2) | 76 (85.4) | 0.168 |
| Injury severity score [median (IQR)] | 42 (33–57) | 25.5 (18–33) | <0.0001 |
| 1–9 (mild) [no. (%)] | 0 | 3 (2.9) | <0.0001 |
| 10–15 (moderate) [no. (%)] | 0 | 11 (10.8) | |
| 16–25 (severe) [no. (%)] | 2 (3.1) | 37 (36.3) | |
| ≥26 (critical) [no. (%)] | 62 (96.9) | 51 (50.0) | |
| Blood (RBC) units 24 h postinjury [median (IQR)] | 32 (23–44) | 10 (5–19) | <0.0001 |
| Zero [no. (%)] | 0 | 2 (2.2)d | <0.0001 |
| 1–9 [no. (%)] | 4 (6.3) | 40 (43.0) | |
| 10–20 [no. (%)] | 6 (9.4) | 32 (34.4) | |
| >20 [no. (%)] | 54 (84.4) | 19 (20.4) | |
| Traumatic amputationse [no. (%)] | |||
| Above knee | 38 (59.4) | 21 (20.6) | <0.0001 |
| Bilateral above knee | 9 (14.1) | 5 (4.9) | 0.039 |
| Amputation plus serious injury to opposite limb | 31 (48.4) | 16 (15.7) | <0.0001 |
| Perineal, GU injury, or abdominal injury [no. (%)] | 61 (95.3) | 63 (61.8) | <0.0001 |
| Colostomy [no. (%)] | 20 (31.3) | 3 (2.9) | <0.0001 |
| LRMC SOFA [median (IQR)] | 11 (6–13) | 3 (2–8) | <0.0001 |
| U.S. hospital SOFA [median (IQR)] | 8 (4–12) | 1 (0–2) | <0.0001 |
| Mechanical ventilation [no. (%)] | <0.0001 | ||
| None | 11 (17.2) | 63 (61.8) | |
| LRMC only | 7 (10.9) | 20 (19.6) | |
| LRMC and U.S. hospital ≤1 wk | 46 (71.9) | 17 (16.7) | |
| LRMC and U.S. hospital >1 wk | 0 | 2 (2.0) |
GU, genitourinary injury; RBC, red blood cell; SOFA, sequential organ failure assessment.
Case patients were those with histopathology positive for fungal nonvascular-tissue invasion or angioinvasion; controls were patients with histopathology that did not identify any fungal elements.
Dismounted refers to being on foot patrol. Mounted status was missing for 12 case patients and 13 controls. Percentages and P values are based on total minus missing.
Blood information was missing for nine controls. Percentages and P values are based on total minus missing.
Patients may have had more than one type of traumatic amputation, so the categories are not mutually exclusive.
NA, not applicable.
The 64 case subjects contributed 171 specimens from 95 injury sites, with fungal cultures available for 87 sites (92%). The 102 controls had 128 specimens collected from 118 injury sites.
Analysis of FFPE specimens with positive histopathology using panfungal PCR.
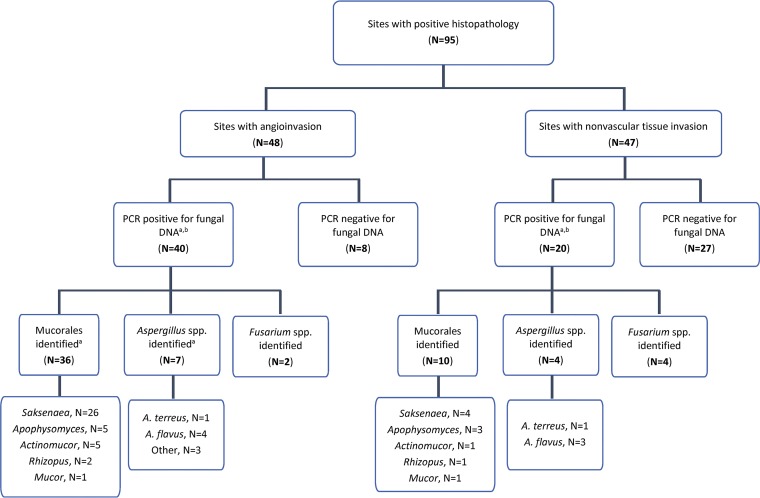
Based on histopathology, injury sites were categorized into two groups: sites exhibiting angioinvasion (n = 48) and those with fungus present in tissue but no angioinvasion (i.e., nonvascular-tissue invasion) (n = 47) (Fig. 1). PCR failed to detect fungal DNA in 35 sites (8 with angioinvasion and 27 with nonvascular-tissue invasion). When restricted to 60 sites with fungal DNA detection, fungi from the order Mucorales were identified from 36 sites with angioinvasion (90% of 40 sites) and 10 sites with nonvascular tissue invasion (50% of 20 sites), respectively. Saksenaea spp. were present either alone or in combination in specimens from 26 (65% of 40) sites with angioinvasion and 4 (20% of 20) sites with nonvascular-tissue invasion. Other fungi from the order Mucorales detected included Apophysomyces spp. (eight sites), Actinomucor spp. (six sites), Rhizopus spp. (three sites), and Mucor spp. (two sites). Aspergillus spp. were identified in specimens from seven (18% of 40) sites with angioinvasion and four (20% of 20) sites with nonvascular-tissue invasion; Aspergillus flavus was predominant (seven sites). A low proportion of Fusarium spp. were also identified (six sites). Other organisms were also detected, including Acrophialophora nainiana, Beauveria bassiana, Chaetomium murorum, Pyrenochaetopsis spp., Pythium aphanidermatum, Rasamsonia argillacea, Scedosporium spp., and Ustilago spp. (see Table S2 in the supplemental material for full species data).
FIG 1.
Flow diagram of PCR findings from sites with positive histopathology. a, sites may grow more than one fungus, so totals may sum to more than the number of sites. b, fungi other than members of the order Mucorales, Aspergillus spp., or Fusarium spp. were identified from eight sites with angioinvasion and four sites with nonvascular-tissue invasion.
Comparison of panfungal PCR with culture-based results among cases.
Of the 95 injury sites, 87 had culture data available (Table 2). Fungi grew from cultures of 68 sites (78%). Fungi belonging to the order Mucorales were grown from 31 (46%) of the 68 sites, with Saksenaea spp. the predominant fungi (8 sites). Rhizopus spp., Actinomucor spp., and Apophysomyces spp. were grown from four, three, and one site, respectively. Twenty-four sites had Aspergillus sp. growth, with A. flavus and Aspergillus terreus grown in 12 and 10 sites, respectively.
TABLE 2.
Comparison of molecular-sequencing results with fungal culture in all specimens with any positive histopathology (angioinvasion or nonvascular-tissue invasion)a
| Growth of filamentous fungi | Resultsb
|
Kappa coefficient [median (IQR)] | ||
|---|---|---|---|---|
| Culture | PCR | Concordant | ||
| Cultures collected (87 sites) [no. (%)] | ||||
| No fungi | 19 (21.8) | 30 (34.5) | 11 (12.6) | 0.248 (0.039–0.457) |
| Any fungi | 68 (78.2) | 57 (65.5) | ||
| Fungi identified at genus levelc | 63 (72.4) | 57 (65.5) | ||
| Order Mucorales | 31 (35.6) | 43 (49.4) | 21 (24.1) | 0.262 (0.067–0.457) |
| Saksenaea spp. | 8 (9.2) | 31 (35.6) | 5 (5.7) | 0.129 (−0.039–0.298) |
| Aspergillus spp. | 24 (27.6) | 12 (13.8) | 9 (10.3) | 0.387 (0.169–0.605) |
| Fusarium spp. | 15 (17.2) | 6 (6.9) | 3 (3.5) | 0.208 (−0.050–0.466) |
| Cultures collected within 24 h of histopathology specimen collection (77 sites) [no. (%)] | ||||
| No fungi | 22 (28.6) | 29 (37.7) | 14 (24.7) | 0.332 (0.115–0.549) |
| Any fungi | 55 (71.4) | 48 (62.3) | ||
| Fungi identified at genus leveld | 50 (64.9) | 48 (62.3) | ||
| Order Mucorales | 25 (32.5) | 37 (48.1) | 20 (26.0) | 0.421 (0.227–0.614) |
| Saksenaea spp. | 7 (9.1) | 26 (33.8) | 5 (6.5) | 0.187 (−0.004–0.377) |
| Aspergillus spp. | 14 (18.2) | 8 (10.4) | 5 (6.5) | 0.371 (0.093–0.650) |
| Fusarium spp. | 8 (10.4) | 3 (3.9) | 0 | −0.060 (−0.112–0.008) |
Injury sites were restricted to those that also had a culture collected. By definition, control specimens did not have any histopathology or culture evidence of a fungal infection, so data from control patients are not included.
The PCR specimens were collected only from U.S. hospitals; however, cultures were collected at various time points and at different facilities. Analysis was restricted to subsets of injury sites with tissue specimens for histopathology and cultures collected from the same injury site at Landstuhl Regional Medical Center or U.S. hospitals (n = 87) and within 24 h of histopathology specimen collection (n = 77). Categories are independent of each other. Percentage for concordance was calculated using the total number of sites (n = 87 or 77).
Excludes Mycelia sterilia and fungi classified as “not otherwise specified.” Three sites with fungal culture results classified as “zygomycetes” were also excluded.
Excludes five sites with fungal culture results classified as “zygomycetes.”
In a comparison of PCR and culture findings, PCR identified fungi of the order Mucorales in 43 of 87 sites (49%), compared to 31 of 87 sites (36%) grown from culture (Table 2). Specimens from 21 injury sites were concordant regarding detection of fungi from the order Mucorales (kappa coefficient, 0.262) compared to 9 and 3 injury sites for Aspergillus spp. and Fusarium spp. Findings were similar when the injury sites were restricted to those with a concomitant culture collected (i.e., within 24 h of specimen collection [n = 77]) (Table 2).
The PCR-based assay failed to detect fungal DNA in specimens from 30 of the sites that had cultures collected (7 sites with angioinvasion and 23 sites with nonvascular-tissue invasion), and 19 of the sites exhibited growth on culture. Specifically, nine sites (four with angioinvasion and five with nonvascular-tissue invasion) that had false-negative PCR findings had culture growth of fungi from the order Mucorales, while six and five sites grew Aspergillus spp. and Fusarium spp., respectively (three sites grew combinations of Mucorales plus Aspergillus or Fusarium, and two sites grew other fungi).
When examining the 44 injury sites with evidence of angioinvasion (excluding 4 sites where a culture was not collected), both PCR and culture identified/grew fungi from 37 sites, with 31 concordant at the genus level (Table 3). PCR detected fungi from the order Mucorales at a greater frequency than culture (75% versus 46%; kappa coefficient, 0.087) while detecting Aspergillus spp. at a lower frequency (16% versus 30%; kappa coefficient, 0.370).
TABLE 3.
Comparison of molecular-sequencing results with fungal culture in specimens with histopathologic evidence of angioinvasion onlya
| Growth of filamentous fungi | Resultsb
|
Kappa coefficient [median (IQR)] | ||
|---|---|---|---|---|
| Culture | PCR | Concordant | ||
| Cultures collected (44 sites) [no. (%)] | ||||
| No fungi | 7 (15.9) | 7 (15.9) | 1 (2.3) | −0.019 (−0.304–0.266) |
| Any fungi | 37 (84.1) | 37 (84.1) | ||
| Fungi identified at genus levelc | 35 (79.6) | 37 (84.1) | ||
| Order Mucorales | 20 (45.5) | 33 (75.0) | 16 (36.4) | 0.087 (−0.154–0.327) |
| Saksenaea spp. | 6 (13.6) | 25 (56.8) | 4 (9.1) | 0.049 (−0.130–0.228) |
| Aspergillus spp. | 13 (29.6) | 7 (15.9) | 5 (11.3) | 0.370 (0.069–0.670) |
| Fusarium spp. | 11 (25.0) | 2 (4.6) | 2 (4.5) | 0.250 (−0.039–0.539) |
| Cultures collected within 24 h of histopathology specimen collection (41 sites) [no. (%)] | ||||
| No fungi | 6 (14.6) | 8 (19.5)d | 1 (2.4) | −0.029 (−0.316–0.257) |
| Any fungi | 35 (85.4) | 33 (80.5) | ||
| Fungi identified at genus levele | 32 (78.1) | 33 (80.5) | ||
| Order Mucorales | 18 (43.9) | 28 (68.3) | 15 (35.6) | 0.253 (−0.004–0.510) |
| Saksenaea spp. | 5 (12.2) | 20 (48.8) | 4 (9.8) | 0.155 (-0.049–0.359) |
| Aspergillus spp. | 6 (14.6) | 5 (12.2) | 3 (7.3) | 0.476 (0.084–0.868) |
| Fusarium spp. | 6 (14.6) | 1 (2.4) | 0 | −0.044 (−0.119–0.032) |
Injury sites were restricted to those that also had a culture collected. By definition, control specimens did not have any histopathology or culture evidence of a fungal infection, so data from control patients are not included.
The PCR specimens were collected only from U.S. hospitals; however, cultures were collected at various time points and at different facilities. Analysis was restricted to injury sites with observed angioinvasion and cultures collected from the same injury site at Landstuhl Regional Medical Center or U.S. hospitals (n = 44) and within 24 h of histopathology specimen collection (n = 41). Categories are independent of each other. Percentage for concordance was calculated using the total number of sites (n = 44 or 41).
Excludes Mycelia sterilia and fungi classified as “not otherwise specified.” One site with fungal culture results classified as “zygomycetes” was also excluded.
Injury sites might have had more than one specimen collected, and if so, the results were collapsed to the site level. One subject with two specimens from one injury site had one of the specimens excluded after the restriction of having a culture collected within 24 h was applied. The specimen excluded was positive for fungal growth (Aspergillus spp. and Saksenaea spp.), while the included specimen was negative.
Excludes three sites with fungal culture results classified as “zygomycetes.”
(i) Performance characteristics of panfungal PCR-based assay. Performance characteristics were determined by comparing PCR findings with histopathology (see Table S3 in the supplemental material). Fungal DNA was detected in specimens from 60 histopathology-positive sites, resulting in sensitivity of 63.2% (Table 4; see Table S3). Specificity was calculated to be 99.2% based upon identification of one false-positive result. The positive predictive value of the PCR-based assay was calculated to be 98.4%, while the negative predictive value was 77.0%.
TABLE 4.
PCR-based assay performance compared to histopathology
| Parameter | PCR results for specimens from sites with: |
|
|---|---|---|
| Any histopathologya (n = 95) | Angioinvasion (n = 48) | |
| Sensitivity [% (95% CIb )] | 63.2 (52.6–72.8) | 83.3 (69.8–92.5) |
| Specificity [% (95% CI)] | 99.2 (95.4–99.9) | 99.2 (95.4–99.9) |
| Positive predictive value [% (95% CI)] | 98.4 (89.4–99.8) | 97.6 (85.0–99.7) |
| Negative predictive value [% (95% CI)] | 77.0 (72.0–81.3) | 93.6 (88.6–96.5) |
| False-positive rate (%) | 0.8 | 0.8 |
| False-negative rate (%) | 36.8 | 16.7 |
| Positive likelihood ratio | 75.8 | 98.3 |
| Negative likelihood ratio | 0.37 | 0.17 |
Includes both angioinvasion and nonvascular-tissue invasion.
CI, confidence interval.
The PCR findings restricted to sites with angioinvasion (n = 48) were examined (see Table S3). The specificity of the PCR-based assay remained at 99.2%; however, the negative predictive value increased to 93.6%, and the false-negative rate decreased to 16.7% (Table 4).
(ii) Comparison of panfungal and seminested PCR assays. Sixty-seven specimens, of which 5 were specimens from nontrauma patients (true negatives) and 62 were histopathology positive (21% of the specimens assessed by UTHSCSA), were sent to LADR for analysis with seminested assays and compared to findings of the panfungal PCR (see Table S4 in the supplemental material). While the panfungal PCR failed to detect fungal DNA in 15 (24%) of the 62 specimens, fungal DNA was not identified in 6 specimens (10%) using the seminested assays. Overall, the results were not concordant (kappa coefficient, 0.171) for identification of fungi at the genus level. Nevertheless, concordance improved with fungi of the order Mucorales (kappa coefficient, 0.494). Assessment of specimens from sites with angioinvasion produced similar results (see Table S3).
DISCUSSION
The results of our study (the largest to date in patients with trauma-associated IFIs) demonstrate that even in the setting of blast wounds, where environmental contamination is common (25–27), it is possible to amplify fungal DNA from FFPE specimens with a high degree of specificity (99%), albeit with lower sensitivity (63%). However, the sensitivity is comparable to that in studies using other PCR-based assays (6–11). Panfungal PCR performed better on FFPE specimens with documented angioinvasion (sensitivity, 83%; negative predictive value, 94%). One potential explanation for the reduced yield in specimens without angioinvasion could be related to the timing of collection. We strategically avoided inclusion of early specimens, as soil and debris introduced at the time of the injury could have resulted in identifying fungi that were contaminants. Case specimens were collected at U.S. hospitals, and wounds at that time point had generally undergone a median of two debridements, usually once in Afghanistan and once at LRMC. These debridements may have reduced the fungal burden and yield, especially in specimens without angioinvasion. Our findings may have also been impacted by the specimen age; specimens were collected a median of 5.5 (interquartile range [IQR], 4.8 to 6.1) years prior to analysis, leading to reduced yield due to DNA fragmentation (28). Historically, FFPE specimens have been challenging for sequencing-based diagnostics (8). Cutting and extraction methods in the analysis were standardized and automated, keeping contamination to a minimum and contributing to the observed high specificity.
Consistent with prior results, fungi from the order Mucorales were the primary pathogenic agents (29, 30). Saksenaea spp. were identified in over half of the cases with documented angioinvasion, and in combination with fungi of the genus Apophysomyces, they accounted for two-thirds of the cases. This outcome is consistent with data reported from the Indian subcontinent, where these injuries occurred, which frequently link fungi of the two closely related genera Apophysomyces and Saksenaea with trauma-associated IFIs (31). Further, Apophysomyces trapeziformis was identified from all patients examined following the EF-5 tornado in Joplin, MO (2). In contrast, molds causing mucormycosis in immunocompromised patients belong to (in decreasing order) Rhizopus spp. (53%), Mucor spp. (16%), Rhizomucor spp. (6%), and Lichtheimia spp. (6%) (32).
There are limitations to the use of panfungal PCR or culture as standalone diagnostics. Reliance on culture alone may have resulted in unfavorable outcomes, as it was insensitive at detecting fungi from the order Mucorales. Tissue preparation for culturing typically destroys the ribbon-like hyphal elements of fungi belonging to the order Mucorales, reducing growth (33). To improve recovery of mucormycetes, our laboratory did not grind tissue, but rather used a stomacher (Seward Laboratory Systems, Inc., Bohemia, NY) to stomach the specimens for 1 min before inoculating them into media. Further, current culture methods fail to mimic physiologic conditions required for fungal growth, thereby decreasing yield. This is especially true for Saksenaea spp. and Apophysomyces spp., which fail to sporulate well in routine fungal media (34), whereas reliance on panfungal PCR alone would have failed to identify fungi of the genera Aspergillus and Fusarium when there was growth. Specifically, A. terreus was grown from 10 sites using conventional culture, while it was identified from only 2 sites using panfungal PCR. As A. terreus is resistant to amphotericin B, identification of the fungus is important to direct treatment. Thus, despite the significant potential of the panfungal PCR, a PCR platform with high sensitivity for both fungi of the order Mucorales and Aspergillus spp. is needed to help guide treatment in trauma-related IFIs. Use of PCR on fresh tissue likely would improve sensitivity due to omission of formalin and paraffin in the specimens, which reduce yield due to DNA fragmentation.
In a random sample of 62 specimens, seminested PCR assays had improved ability to identify fungal organisms, including Fusarium spp. and Aspergillus spp. There are a few potential explanations for the varied results obtained with the seminested and panfungal PCR assays, including differences in extraction methods. Specifically, an additional boiling step to antagonize the protein-complexing effect of formalin was used with the seminested PCRs. Boiling results in release of proteins, and the amount of protein found in the supernatant correlates with the duration of boiling (35). Since histones around the DNA are complexed by formalin, the boiling step should increase the amount of DNA released for amplification. Furthermore, the seminested assays that have been developed have an increased number of amplification cycles, a total of 70 compared to 40 for panfungal PCR, resulting in improved DNA yield. Finally, as template DNA, FFPE-amplifiable regions are often shorter due to fragmentation or cross-linking. The shorter, nested product is advantageous; however, it limits identification at the species level. While use of a higher number of amplification cycles and fewer nucleotides to call a match improves the sensitivity of the seminested PCR-based method, it might reduce the specificity of the technique. For example, DNA of spores of ubiquitous molds within the paraffin used for embedding tissue samples might be amplified and indistinguishable from fungal DNA within the tissue. Accordingly, based on newly revised and updated consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium, a positive PCR result from paraffin-embedded tissue can be accepted only if it correlates with histopathology (36). However, we believe that nested PCR could be a beneficial diagnostic strategy when a narrow spectrum of fungi is suspected. In this study, nested PCRs were targeted to the Mucorales, Aspergilli, and Fusaria based on our prior results (29).
Accurate identification of fungi is essential for selection of appropriate antifungal therapy. The trade-off between specificity and sensitivity is worth considering. The decision to initiate antifungals for combat casualties is based on easily ascertainable clinical criteria (37), not laboratory findings. Based on current practice guidelines, this approach would involve the use of both amphotericin-based compounds and an azole (37). Although a clinician is unlikely to withhold antifungals when a person meets clinical criteria, a test that allows the clinician to safely withdraw antifungals is desirable (i.e., an assay with high negative predictive value, which was noted with panfungal PCR in specimens with angioinvasion). Nonetheless, there is a need to continue to work on strategies that will improve sensitivity for all specimen types (both angioinvasion and nonvascular-tissue invasion) without reducing specificity. Therefore, we intend to examine the performance characteristics of the seminested PCR assays on controls.
There are limitations of the analysis that should be considered. The selection of the study population from trauma patients at risk for IFI based on histopathological evidence could have biased results relating to the predictive value of cultures. Moreover, PCR was conducted using FFPE archived tissue specimens rather than fresh tissue. Occasionally, repeat PCRs were performed on the same sample, particularly if amplicons yielded poor sequence or a bad identity that was less than 97%. This strategy would be feasible only in a molecular laboratory with experienced personnel. To overcome these limitations, we are currently exploring the role of probe-based assays.
Overall, our findings indicate that the panfungal PCR studied had a high degree of specificity and moderate sensitivity for specimens with angioinvasion. Compared to conventional cultures, the PCR-based assay was better at identifying fungi of the order Mucorales, which is clinically relevant, as the use of culture alone to direct therapy might have resulted in failure to use amphotericin-based compounds; however, the technique did not perform as well at identifying fungi of the genus Aspergillus. Although there are limitations to the panfungal PCR-based assay assessed in this analysis, the identification of a PCR platform with high sensitivity for both fungi of the order Mucorales and Aspergillus spp. would add to the current armamentarium for the diagnosis of blast-related IFI. Due to species-specific differences in susceptibility to antifungals, the availability of such a platform could better guide antifungal selection by providing species-specific identification with rapid turnaround (38).
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to the Infectious Disease Clinical Research Program (IDCRP) TIDOS team of clinical coordinators, microbiology technicians, data managers, clinical site managers, and administrative support personnel for their tireless efforts to ensure the success of this project. We thank Brian Johnson, Denise Bennett, and Teresa Merritt of the IDCRP, as well as Jianmin Fu of the University of Texas Health Sciences Center at San Antonio, for their invaluable contributions to this project.
This work (IDCRP-077) was conducted by the Infectious Disease Clinical Research Program, a Department of Defense program executed through the Uniformed Services University of the Health Sciences, Department of Preventive Medicine and Biostatistics, through a cooperative agreement with the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF).
This project has been funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (Inter-Agency Agreement Y1-AI-5072); the Department of the Navy under the Wounded, Ill, and Injured Program (HU0001-10-1-0014); and the Defense Medical Research and Development Program (HT9404-12-1-0014).
No competing financial interests exist.
The views expressed are ours and do not reflect the official views of the Uniformed Services University of the Health Sciences; the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.; the National Institutes of Health or the Department of Health and Human Services; Brooke Army Medical Center; Walter Reed National Military Medical Center; Landstuhl Regional Medical Center; the U.S. Army Institute of Surgical Research; the U.S. Army Medical Department; the U.S. Army Office of the Surgeon General; the Department of Defense; or the Departments of the Army, Navy, and Air Force. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Warkentien T, Rodriguez C, Lloyd B, Wells J, Weintrob A, Dunne J, Ganesan A, Li P, Bradley W, Gaskins LJ, Seillier-Moiseiwitsch F, Murray CK, Millar EV, Keenan B, Paolino K, Fleming M, Hospenthal DR, Wortmann GW, Landrum ML, Kortepeter MG, Tribble DR. 2012. Invasive mold infections following combat-related injuries. Clin Infect Dis 55:1441–1449. doi: 10.1093/cid/cis749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neblett Fanfair R, Benedict K, Bos J, Bennett SD, Lo YC, Adebanjo T, Etienne K, Deak E, Derado G, Shieh WJ, Drew C, Zaki S, Sugerman D, Gade L, Thompson EH, Sutton DA, Engelthaler DM, Schupp JM, Brandt ME, Harris JR, Lockhart SR, Turabelidze G, Park BJ. 2012. Necrotizing cutaneous mucormycosis after a tornado in Joplin, Missouri, in 2011. N Engl J Med 367:2214–2225. doi: 10.1056/NEJMoa1204781. [DOI] [PubMed] [Google Scholar]
- 3.Lewandowski LR, Weintrob AC, Tribble DR, Rodriguez CJ, Petfield J, Lloyd BA, Murray CK, Stinner D, Aggarwal D, Shaikh F, Potter BK. 2016. Early complications and outcomes in combat injury related invasive fungal wound infections: a case-control analysis. J Orthop Trauma 30:e93–e99. doi: 10.1097/BOT.0000000000000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schofield CM, Murray CK, Horvath EE, Cancio LC, Kim SH, Wolf SE, Hospenthal DR. 2007. Correlation of culture with histopathology in fungal burn wound colonization and infection. Burns 33:341–346. doi: 10.1016/j.burns.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 5.Heaton SM, Weintrob AC, Downing K, Keenan B, Aggarwal D, Shaikh F, Tribble DR, Wells J. 2016. Histopathological techniques for the diagnosis of combat-related invasive fungal wound infections. BMC Clin Pathol 16:11–19. doi: 10.1186/s12907-016-0033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau A, Chen S, Sorrell T, Carter D, Malik R, Martin P, Halliday C. 2007. Development and clinical application of a panfungal PCR assay to detect and identify fungal DNA in tissue specimens. J Clin Microbiol 45:380–385. doi: 10.1128/JCM.01862-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buitrago MJ, Aguado JM, Ballen A, Bernal-Martinez L, Prieto M, Garcia-Reyne A, Garcia-Rodriguez J, Rodriguez-Tudela JL, Cuenca-Estrella M. 2013. Efficacy of DNA amplification in tissue biopsy samples to improve the detection of invasive fungal disease. Clin Microbiol Infect 19:E271–E277. doi: 10.1111/1469-0691.12110. [DOI] [PubMed] [Google Scholar]
- 8.Rickerts V. 2016. Identification of fungal pathogens in formalin-fixed, paraffin-embedded tissue samples by molecular methods. Fungal Biol 120:279–287. doi: 10.1016/j.funbio.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Babouee Flury B, Weisser M, Prince SS, Bubendorf L, Battegay M, Frei R, Goldenberger D. 2014. Performances of two different panfungal PCRs to detect mould DNA in formalin-fixed paraffin-embedded tissue: what are the limiting factors? BMC Infect Dis 14:692. doi: 10.1186/s12879-014-0692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gade L, Hurst S, Balajee SA, Lockhart SR, Litvintseva AP. 2017. Detection of mucormycetes and other pathogenic fungi in formalin fixed paraffin embedded and fresh tissues using the extended region of 28S rDNA. Med Mycol 55:385–395. doi: 10.1093/mmy/myw083. [DOI] [PubMed] [Google Scholar]
- 11.Springer J, McCormick Smith I, Hartmann S, Winkelmann R, Wilmes D, Cornely O, Kessel J, Loffler J, Rickerts V. 2019. Identification of Aspergillus and Mucorales in formalin-fixed, paraffin-embedded tissue samples: comparison of specific and broad-range fungal qPCR assays. Med Mycol 57:308–313. doi: 10.1093/mmy/myy041. [DOI] [PubMed] [Google Scholar]
- 12.Ganesan A, Shaikh F, Peterson P, Bradley W, Johnson B, Bennett D, Carson ML, Merritt T, Akers K, Wells J, Bialek R, Tribble DR, Wickes B. 2018. Tissue-based molecular diagnostics: a sensitive and specific way for the identification of invasive fungal infections in the combat related setting, poster 2053. IDWeek 2018, 3 to 7 October 2018, San Francisco, CA: https://idsa.confex.com/idsa/2018/webprogram/Paper70458.html. [Google Scholar]
- 13.Tribble D, Conger N, Fraser S, Gleeson T, Wilkins K, Antonille T, Weintrob A, Ganesan A, Gaskins L, Li P, Grandits G, Landrum M, Hospenthal D, Millar E, Blackbourne L, Dunne J, Craft D, Mende K, Wortmann G, Herlihy R, McDonald J, Murray C. 2011. Infection-associated clinical outcomes in hospitalized medical evacuees following traumatic injury: Trauma Infectious Disease Outcome Study (TIDOS). J Trauma 71:S33–S42. doi: 10.1097/TA.0b013e318221162e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lloyd B, Weintrob A, Rodriguez C, Dunne J, Weisbrod A, Hinkle M, Warkentien T, Murray C, Oh J, Millar E, Shah J, Shaikh F, Gregg S, Lloyd G, Stevens J, Carson ML, Aggarwal D, Tribble DR. 2014. Effect of early screening for invasive fungal infections in U.S. service members with explosive blast injuries. Surg Infect 15:619–626. doi: 10.1089/sur.2012.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eastridge BJ, Jenkins D, Flaherty S, Schiller H, Holcomb JB. 2006. Trauma system development in a theater of war: experiences from Operation Iraqi Freedom and Operation Enduring Freedom. J Trauma 61:1366–1372. doi: 10.1097/01.ta.0000245894.78941.90. [DOI] [PubMed] [Google Scholar]
- 16.Romanelli AM, Sutton DA, Thompson EH, Rinaldi MG, Wickes BL. 2010. Sequence-based identification of filamentous basidiomycetous fungi from clinical specimens: a cautionary note. J Clin Microbiol 48:741–752. doi: 10.1128/JCM.01948-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balajee SA, Borman AM, Brandt ME, Cano J, Cuenca-Estrella M, Dannaoui E, Guarro J, Haase G, Kibbler CC, Meyer W, O'Donnell K, Petti CA, Rodriguez-Tudela JL, Sutton D, Velegraki A, Wickes BL. 2009. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in clinical mycology laboratory: where are we and where should we go from here? J Clin Microbiol 47:877–884. doi: 10.1128/JCM.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammond SP, Bialek R, Milner DA, Petschnigg EM, Baden LR, Marty FM. 2011. Molecular methods to improve diagnosis and identification of mucormycosis. J Clin Microbiol 49:2151–2153. doi: 10.1128/JCM.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bialek R, Konrad F, Kern J, Aepinus C, Cecenas L, Gonzalez GM, Just-Nübling G, Willinger B, Presterl E, Lass-Flörl C, Rickerts V. 2005. PCR based identification and discrimination of agents of mucormycosis and aspergillosis in paraffin wax embedded tissue. J Clin Pathol 58:1180–1184. doi: 10.1136/jcp.2004.024703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hue FX, Huerre M, Rouffault MA, de Bievre C. 1999. Specific detection of Fusarium species in blood and tissues by a PCR technique. J Clin Microbiol 37:2434–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grice EA, Segre JA. 2011. The skin microbiome. Nat Rev Microbiol 9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anonymous. 2012. Appendix: test performance metrics In Chang SM, Matchar DB, Smetana GW, Umscheid CA (ed), Methods guide for medical test reviews. Agency for Healthcare Research and Quality, Rockville, MD: https://www.ncbi.nlm.nih.gov/books/NBK98249/. [PubMed] [Google Scholar]
- 23.Linn S. 1995. The injury severity score—importance and uses. Ann Epidemiol 5:440–446. doi: 10.1016/1047-2797(95)00059-3. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez C, Weintrob AC, Shah J, Malone D, Dunne JR, Weisbrod AB, Lloyd BA, Warkentien T, Murray CK, Wilkins K, Shaikh F, Carson ML, Aggarwal D, Tribble DR. 2014. Risk factors associated with invasive fungal Infections in combat trauma. Surg Infect 15:521–526. doi: 10.1089/sur.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evriviades D, Jeffery S, Cubison T, Lawton G, Gill M, Mortiboy D. 2011. Shaping the military wound: issues surrounding the reconstruction of injured servicemen at the Royal Centre for Defence Medicine. Philos Trans R Soc Lond B Biol Sci 366:219–230. doi: 10.1098/rstb.2010.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tribble DR, Rodriguez CJ, Weintrob AC, Shaikh F, Aggarwal D, Carson ML, Murray CK, Masuoka P. 2015. Environmental factors related to fungal wound contamination after combat trauma in Afghanistan, 2009–2011. Emerg Infect Dis 21:1759–1769. doi: 10.3201/eid2110.141759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheean AJ, Tintle SM, Rhee PC. 2015. Soft tissue and wound management of blast injuries. Curr Rev Musculoskelet Med 8:265–271. doi: 10.1007/s12178-015-9275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe M, Hashida S, Yamamoto H, Matsubara T, Ohtsuka T, Suzawa K, Maki Y, Soh J, Asano H, Tsukuda K, Toyooka S, Miyoshi S. 2017. Estimation of age-related DNA degradation from formalin-fixed and paraffin-embedded tissue according to the extraction methods. Exp Ther Med 14:2683–2688. doi: 10.3892/etm.2017.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warkentien TE, Shaikh F, Weintrob AC, Rodriguez CJ, Murray CK, Lloyd BA, Ganesan A, Aggarwal D, Carson ML, Tribble DR. 2015. Impact of Mucorales and other invasive molds on clinical outcomes of polymicrobial traumatic wound infections. J Clin Microbiol 53:2262–2270. doi: 10.1128/JCM.00835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kronen R, Liang SY, Bochicchio G, Bochicchio K, Powderly WG, Spec A. 2017. Invasive fungal infections secondary to traumatic injury. Int J Infect Dis 62:102–111. doi: 10.1016/j.ijid.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Chander J, Kaur M, Singla N, Punia RPS, Singhal SK, Attri AK, Alastruey-Izquierdo A, Stchigel AM, Cano-Lira JF, Guarro J. 2018. Mucormycosis: battle with the deadly enemy over a five-year period in India. J Fungi 4:E46. doi: 10.3390/jof4020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chitasombat MN, Kontoyiannis DP. 2016. Treatment of mucormycosis in transplant patients: role of surgery and of old and new antifungal agents. Curr Opin Infect Dis 29:340–345. doi: 10.1097/QCO.0000000000000277. [DOI] [PubMed] [Google Scholar]
- 33.Kontoyiannis DP, Chamilos G, Hassan SA, Lewis RE, Albert ND, Tarrand JJ. 2007. Increased culture recovery of Zygomycetes under physiologic temperature conditions. Am J Clin Pathol 127:208–212. doi: 10.1309/7KU5XWURYM0151YN. [DOI] [PubMed] [Google Scholar]
- 34.Lombardi G, Padhye AA, Standard PG, Kaufman L, Ajello L. 1989. Exoantigen tests for the rapid and specific identification of Apophysomyces elegans and Saksenaea vasiformis. J Med Vet Mycol 27:113–120. doi: 10.1080/02681218980000151. [DOI] [PubMed] [Google Scholar]
- 35.Ikeda K, Monden T, Kanoh T, Tsujie M, Izawa H, Haba A, Ohnishi T, Sekimoto M, Tomita N, Shiozaki H, Monden M. 1998. Extraction and analysis of diagnostically useful proteins from formalin-fixed, paraffin-embedded tissue sections. J Histochem Cytochem 46:397–403. doi: 10.1177/002215549804600314. [DOI] [PubMed] [Google Scholar]
- 36.Donnelly JP, Chen SC, Kauffman CA, Steinbach WJ, Baddley JW, Verweij PE, Clancy CJ, Wingard JR, Lockhart SR, Groll AH, Sorrell TC, Bassetti M, Akan H, Alexander BD, Andes B, Azoulay E, Bialek R, Bradsher RW Jr, Bretagne S, Calandra T, Caliendo AM, Castagnola E, Cruciani M, Cuenca-Estrella M, Decker CF, Desai SR, Fisher BT, Harrison TS, Heussel CP, Jensen HE, Kibbler CC, Kontoyiannis DP, Kullberg BJ, Lagrou K, Lamoth F, Lehrnbecher T, Loeffler J, Lortholary O, Maertens J, Marchetti O, Marr KA, Masur H, Meis JF, Morrissey CO, Nucci M, Ostrosky-Zeichner L, Pagano L, Patterson TF, Perfect JR, Racil Z, Roilides E, Ruhnke M, Schaefer-Prokop C, Shoham S, Slavin MA, Stevens DA, Thompson GII, Vazquez JA, Viscoli C, Walsh TJ, Warris A, Wheat LJ, White PL, Zaoutis TE, Pappas PG. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez CJ, Tribble DR, Murray CK, Jessie EM, Fleming ME, Potter BK, Gordon WT, Shackelford SA. 2016. Invasive fungal infection in war wounds (CPG: 28). https://jts.amedd.army.mil/assets/docs/cpgs/JTS_Clinical_Practice_Guidelines_(CPGs)/Invasive_Fungal_Infection_04_Aug_2016_ID28.pdf. [DOI] [PubMed]
- 38.Espinel-Ingroff A, Chakrabarti A, Chowdhary A, Cordoba S, Dannaoui E, Dufresne P, Fothergill A, Ghannoum M, Gonzalez GM, Guarro J, Kidd S, Lass-Florl C, Meis JF, Pelaez T, Tortorano AM, Turnidge J. 2015. Multicenter evaluation of MIC distributions for epidemiologic cutoff value definition to detect amphotericin B, posaconazole, and itraconazole resistance among the most clinically relevant species of Mucorales. Antimicrob Agents Chemother 59:1745–1750. doi: 10.1128/AAC.04435-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are provided within the paper and its supporting documentation.