Difficulties in confirming and discriminating human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2 infections by serological Western blot (WB) assays (HTLV Blot 2.4; MP Biomedicals) have been reported in Brazil, mainly in HIV/AIDS patients, with a large number of WB-indeterminate and WB-positive but HTLV-untypeable results. Nonetheless, a line immunoassay (LIA) (INNO-LIA HTLV-I/II; Fujirebio) provided enhanced specificity and sensitivity for confirming HTLV-1/2 infections.
KEYWORDS: diagnostic, HTLV, confirmatory serological tests, LIA, WB
ABSTRACT
Difficulties in confirming and discriminating human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2 infections by serological Western blot (WB) assays (HTLV Blot 2.4; MP Biomedicals) have been reported in Brazil, mainly in HIV/AIDS patients, with a large number of WB-indeterminate and WB-positive but HTLV-untypeable results. Nonetheless, a line immunoassay (LIA) (INNO-LIA HTLV-I/II; Fujirebio) provided enhanced specificity and sensitivity for confirming HTLV-1/2 infections. To add information concerning the improved ability of the LIA in relation to WB when applied to samples of individuals from different risk groups from Brazil, we performed the present study. Three groups were analyzed: group 1 (G1), with 62 samples from HIV/AIDS patients from São Paulo, SP (48 WB indeterminate and 14 HTLV untypeable); group 2 (G2), with 24 samples from patients with hepatitis B or hepatitis C from São Paulo (21 WB indeterminate and 3 HTLV untypeable; 17 HIV seropositive); and group 3 (G3), with 25 samples from an HTLV outpatient clinic in Salvador, Bahia (16 WB indeterminate and 9 HTLV untypeable; all HIV seronegative). Overall, the LIA confirmed HTLV-1/2 infection (HTLV-1, HTLV-2, or HTLV) in 66.1% (G1), 83.3% (G2), and 76.0% (G3) of samples. Interestingly, the majority of WB-indeterminate results were confirmed by the LIA as being HTLV-2 positive in G1 and G2 but not in G3, in which the samples were defined as being HTLV-1 or HTLV positive. These results agree with the virus types that circulate in such patients of different regions in Brazil and emphasize that the LIA is the best serological test for confirming HTLV-1 and HTLV-2 infections, independently of being applied in HTLV-monoinfected or HTLV-coinfected individuals.
INTRODUCTION
Despite regional variations, Brazil is one of the largest areas of the world where human T-cell lymphotropic virus type 1 (HTLV-1) is endemic (1). Higher rates of HTLV-1 infection have been detected in general populations in the north and northeast regions, while human T-cell lymphotropic virus type 2 (HTLV-2) is endemic among indigenous communities in the north as well as in injectable drug users in urban areas, mostly in southeastern in Brazil (2).
Difficulties in confirming the diagnosis of HTLV-1 and HTLV-2 by serological assays (Western blot [WB]) (HTLV Blot 2.3 and 2.4) have been reported in Brazil, due to a large number of WB-indeterminate and HTLV-positive but untypeable results, mainly in patients truly infected with HTLV-2 and/or HIV (3–5). Consequently, molecular assays have been employed to detect proviral DNA segments of HTLV-1 and HTLV-2 (pol, long terminal repeat [LTR], env, and tax) in peripheral blood mononuclear cells using nested PCR, PCR hybridization, and/or PCR-restriction fragment length polymorphism (RFLP) assays (6–11). Nonetheless, there was no consensus regarding the criteria to consider a blood sample truly infected by HTLV-1/2 using these techniques if positive for one or at least two DNA proviral segments of HTLV-1/2 (6, 10, 11). Subsequently, another molecular assay, real-time PCR or quantitative PCR (qPCR), was proposed as a confirmatory HTLV-1/2 molecular assay; however, low sensitivity was found when applied on HIV blood samples and on those from Brazilian patients infected with HTLV-2, which could be due to low HTLV-2 proviral loads (12–15).
In 1998, a new HTLV-1/2 serological confirmatory assay employed a line immunoassay (LIA) with a nylon membrane sensitized with the most relevant antigens, recombinant proteins, or synthetic peptides of HTLV-1 and HTLV-2 (16). This new immunoassay (INNO-LIA HTLV) demonstrated better results than WB, with improved sensitivity for the confirmation of HTLV-1 and HTLV-2 infections, thereby reducing numbers of WB-indeterminate results (16). However, the high cost of INNO-LIA prevented its routine adoption in Brazil. Consequently, few studies have compared the performances of LIA and WB in Brazil, one employing blood bank samples (17) and two using serum samples from HIV/AIDS patients (14, 15). These studies indicated that the LIA was the best assay to confirm or rule out HTLV-1/2 infection.
In Brazil, molecular biology laboratories are not widely available due to differences in socioeconomic conditions. Thus, the use of a confirmatory serological assay with high performance is essential and necessary for HTLV-1/2 diagnosis. Accordingly, the present study aimed to evaluate the use of INNO-LIA for clarifying WB-indeterminate and WB-HTLV-untypeable serum samples.
MATERIALS AND METHODS
Samples.
The serum samples employed in the present study were obtained from the biorepositories of the HTLV Research Laboratory (LPHTLV), Department of Immunology, Adolfo Lutz Institute (IAL), located in São Paulo, Brazil, and the Integrated and Multidisciplinary HTLV Center (CHTLV), located at the Bahiana School of Medicine and Public Health (EBMSP) in Salvador, Bahia, Brazil. Briefly, the samples from São Paulo were collected between 2012 and 2016 in the course of previous studies designed to detect the prevalence of HTLV-1/2 in HIV-infected individuals as well as in patients with hepatitis B virus (HBV) or hepatitis C virus (HCV) infection in the state of São Paulo, Brazil (14, 15, 18–20). The samples from Salvador, Bahia, were obtained by routine diagnostic procedures at an outpatient clinic in Salvador (CHTLV) from 2015 to 2017; these samples were additionally used to assess the performances of four commercially available HTLV serological screening tests in Brazil (21). Table 1 lists the characteristics of the samples collected from these two biorepositories (number of samples per sex and age) as well as HTLV-1/2 screening results and confirmatory WB assay results (HTLV Blot 2.4; MP Biomedicals). Only samples that were WB inconclusive (WB indeterminate or HTLV untypeable) were selected for analysis in the present investigation.
TABLE 1.
Characteristics of the study groups whose serum samples were analyzed for the presence of HTLV-1/2 antibodies and results of HTLV-1/2 screening and Western blot confirmatory assays
| Groupa | Study population | No. of individuals | No. of samples | % of samples | Age (yr) |
No. of screen-positive samplesb | No. of WB-positive samples |
No. of WB-inconclusive samples |
% WB-inconclusive samplesc |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Min–max | HTLV-1 | HTLV-2 | HTLV | IND | HTLV | IND | ||||||
| G1 | HIV | 4,395 | 311 | 97 | 65 | 16 | 66 | 5.14 | 21.22 | ||||
| Male | 2,935 | 66.78 | 39.12 | 16–84 | |||||||||
| Female | 1,459 | 33.20 | 40.36 | 16–83 | |||||||||
| Total | 4,395 | 100 | 39.74 | 16–84 | |||||||||
| G2 | HBV/HCV | 3,228 | 99 | 45 | 29 | 3 | 21 | 3.03 | 21.21 | ||||
| Male | 1,749 | 54.18 | 48.40 | 14–88 | |||||||||
| Female | 1,479 | 45.82 | 48.50 | 13–94 | |||||||||
| Total | 3,228 | 100 | 48.70 | 13–94 | |||||||||
| G3d | Monoinfection | 362 | 207 | 127e | 36e | 11 | 27 | 5.31 | 13.04 | ||||
| Male | 105 | 29.01 | 39.00 | 24–60 | |||||||||
| Female | 257 | 70.99 | 42.20 | 16–73 | |||||||||
| Total | 362 | 100 | 41.04 | 16–73 | |||||||||
G1, group of HIV/AIDS patients from São Paulo, SP, Brazil; G2, group of patients with hepatitis B virus (HBV) and hepatitis C virus (HCV) from São Paulo, SP; G3, group of individuals from HTLV outpatient clinics in Salvador, BA, Brazil.
Screening assays as described in Materials and Methods.
Western blot results according to the manufacturer’s criteria for HTLV Blot 2.4 (MP Biomedicals). IND, indeterminate.
Age data obtained only from patients with WB-inconclusive results.
Five samples were positive for both HTLV-1 and HTLV-2.
Of the 145 samples with WB-inconclusive results, a volume sufficient for immunoassaying was present in 111; these samples were used to determine the ability of the LIA (INNO-LIA HTLV; Fujirebio) to confirm and discriminate HTLV-1/2-specific antibodies. The evaluated samples originated from three groups of patients: group 1 (G1), with 62 samples (48 WB indeterminate and 14 HTLV untypeable) obtained in the course of routine HTLV diagnosis at the Adolfo Lutz Institute or from the São Paulo Sexually Transmitted Disease/AIDS Reference and Training Center (CRT DST/AIDS-São Paulo), all of which were obtained from individuals who were known to be HIV seropositive; group 2 (G2), with 24 samples (21 WB indeterminate and 3 HTLV untypeable) that were collected from patients who were initially seen at gastroenterology centers in São Paulo, 14 of whom had HBV and 10 of whom had HCV infection, with 17 of these being HIV seropositive (10 with HBV and 7 with HCV); and group 3 (G3), with 25 HTLV-monoinfected samples from patients seen at the CHTLV outpatient clinic in Salvador, Bahia (16 WB indeterminate and 9 HTLV untypeable).
Screening assays.
The samples from São Paulo were screened for the presence of HTLV-1/2 antibodies by two enzyme immunoassays (EIAs): Murex HTLV-I+II (DiaSorin S.p.A., Dartford, UK) and Gold ELISA (enzyme-linked immunosorbent assay) HTLV-1/2 (REM Indústria e Comércio Ltd., São Paulo, Brazil). The samples from Salvador were initially screened by the Ortho HTLV-1/HTLV-2 Ab-Capture ELISA system (Ortho Clinical Diagnostics, Raritan, NJ, USA) as well as by four other HTLV-1/2 screening tests commercially available in Brazil: three EIAs (Murex HTLV-I+II [DiaSorin S.p.A., Dartford, UK], anti-HTLV-1/2 Sym solution [Symbiosis Diagnóstica Ltd., Leme, Brazil], and Gold ELISA HTLV-1/2 [REM Indústria e Comércio Ltd., São Paulo, Brazil]) and one chemiluminescence assay (CLIA) kit (Architect rHTLV-1/2; Abbott Diagnostics Division, Wiesbaden, Germany). All assays were performed according to the manufacturers’ instructions, which were also used to interpret results. Cutoff values and gray zones were calculated for each assay, and samples considered reactive or inconclusive in screening were submitted for a confirmatory assay.
Confirmatory assays.
A WB assay (HTLV Blot 2.4; MP Biomedicals Asia Pacific Pte. Ltd., Singapore) was used to confirm HTLV-1 and HTLV-2 infection for all the previous studies that generated the samples analyzed here, and these results were interpreted according to the stringent criteria provided by the manufacturer. Briefly, HTLV-1-positive serum samples were defined as having the presence of Gag (p19 with or without p24) and two Env (GD21 and rgp46-I) bands. HTLV-2-positive samples were defined as demonstrating reactivity to Gag (p24 with or without p19) and two Env (GD21 and rgp46-II) bands. Samples that showed the presence of antibodies to both Gag (p19 and p24) and Env (GD21) were defined as being HTLV positive but were considered untypeable. Any other pattern of bands was deemed to be indeterminate.
The present study employed an LIA (INNO-LIA HTLV I/II; Fujirebio, Europe NV, Belgium) in an attempt to confirm and/or discriminate samples with inconclusive results by WB (i.e., WB indeterminate or HTLV positive but untypeable). The strips used in the LIA contain antigens for validation, confirmation, and discrimination. For validation, the line marked by each sample was compared to the control line, and a score ranging from +/− to +3 was assigned. The confirmatory antigens included Gag p19-I/II, Gag p24-I/II, Env gp46-I/II, and Env gp21-I/II. No bands or the occurrence of a single band (Gag p19-I/II, Gag p24-I/II, or Env gp46-I/II) denoted a negative result. The presence of one band (Env gp21-I/II) or two bands (except Env gp21-I/II) indicated indeterminate results, while two bands (Env gp21-I/II and Gag p19-I/II, Gag p24-I/II, or Env gp46-I/II) indicated HTLV positivity. Three discriminatory bands (Gag p19-I, Env gp46-I, and Env gp46-II) were considered as follows: HTLV-1 positivity was indicated by reactivity to Gag p19-I and/or Env gp46-I, while HTLV-2 positivity was found when samples showed Env gp46-II or a higher intensity of the Env gp46-II band than the Gag p19-I and Env gp46-I bands.
Statistical analyses.
Differences in the numbers of males and females in each group were evaluated statistically using the chi-square test. GraphPad Prism software version 5.03 (GraphPad, San Diego, CA, USA) was used for age comparisons between groups using Kruskal-Wallis analysis of variance (ANOVA), complemented with Dunn’s multiple-comparison test. Results with a P value of ≤0.05 were considered statistically significant.
Ethical considerations.
The present research protocol was approved by the Institutional Review Board of the IAL in São Paulo, Brazil (protocol no. 106D/2012, 62H/2015, and 21I/2016) and by the Institutional Research Board (IRB) of the EBMSP in Salvador, Bahia, Brazil (protocol no. 464.286). All procedures were performed in accordance with the principles established in the Declaration of Helsinki and its subsequent revisions.
RESULTS
The characteristics of the patients (sex and age) and the distribution of WB-inconclusive results in each study group are presented in Table 2. More males were found among the HBV/HCV-infected patients in G2, with significant differences in relation to HIV/AIDS patients in G1 and HTLV patients in G3 (P = 0.0048). A comparative analysis of age among the groups showed no significant differences, although the individuals in G2 were older overall. Concerning the distribution of WB-inconclusive samples, the three groups contained more WB-indeterminate than HTLV-untypeable samples: 77.4% in G1, 87.5% in G2, and 64.0% in G3.
TABLE 2.
Characteristics (sex and age) and numbers of individuals whose serum samples yielded HTLV-inconclusive WB results and were available for the LIA in each study group
| Parameter | Value for groupa |
||
|---|---|---|---|
| G1 (n = 62) | G2 (n = 24) | G3 (n = 25) | |
| No. (%) of individuals of sexb | |||
| Male | 31 (50.00) | 20 (83.30) | 10 (40.00) |
| Female | 31 (50.00) | 4 (16.70) | 15 (60.00) |
| Mean age (yr) (min–max) | 44.06 (18–68) | 49.50 (35–76) | 41.08 (16–73) |
| No. of WB-indeterminate resultsc | 48 | 21 | 16 |
| No. of WB HTLV-untypeable resultsc | 14 | 3 | 9 |
G1, group of HIV/AIDS patients from São Paulo, SP, Brazil; G2, group of patients with HBV and HCV infection from São Paulo, SP; G3, group of individuals from an HTLV outpatient clinic in Salvador, BA, Brazil.
The P value for sex was 0.0048, which was statistically significant using a chi-square test.
Indeterminate and HTLV-untypeable results according to the manufacturer’s criteria for HTLV Blot 2.4 (MP Biomedicals).
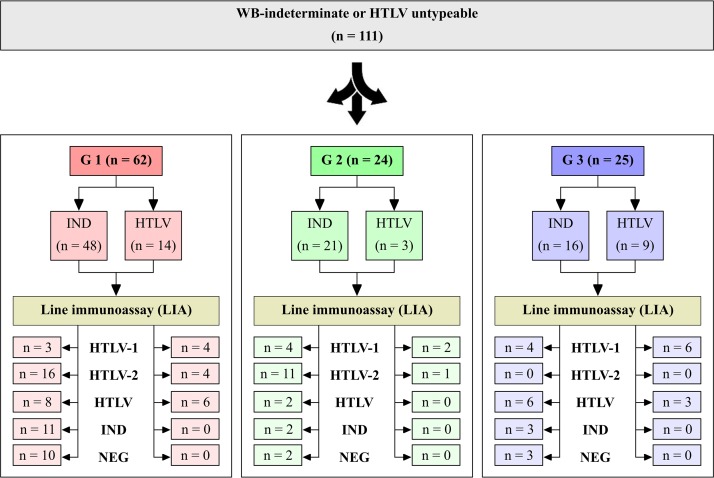
The LIA provided confirmation of HTLV-1/2 infection (HTLV-1, HTLV-2, or HTLV) in 66.1% (G1), 83.3% (G2), and 76.0% (G3) of the samples analyzed. Interestingly, most WB-indeterminate results in G1 and G2 were confirmed to be HTLV-2 positive by the LIA, but this was not the case in G3. In G3, only HTLV-1 (40.0%)- and HTLV (36.0%)-positive samples were detected among both WB-indeterminate and HTLV-untypeable samples (Fig. 1).
FIG 1.
Numbers of serum samples in each study group that scored WB indeterminate or HTLV untypeable in previous analyses and HTLV infections confirmed or excluded by a Western blot (WB) assay (HTLV Blot 2.4; MP Biomedicals) and a line immunoassay (LIA) (INNO-LIA HTLV-I/II; Fujirebio). IND, indeterminate; NEG, negative.
Table 3 shows the WB-indeterminate profiles detected in the present study, the numbers of samples that presented each profile, and the numbers and percentages of samples with confirmed HTLV-1/2 infection. These results indicate that the WB patterns showing Env bands (GD21 and/or rgp46-I or -II) plus p19 or p24 were confirmed as being positive for HTLV-1/2 infection by the LIA. The overall patterns from the WB-inconclusive samples (n = 111) and the patterns returned by the LIA are presented in Table 4, revealing that the majority of indeterminate WB profiles that were not confirmed as being positive for HTLV-1/2 infection by the LIA presented only Gag bands in G1, only GD21 in G2, and one of three bands (GD21, rgp46-II, and p24) in G3. It is noteworthy that among 26 HTLV-positive but untypeable samples (14 in G1, 3 in G2, and 9 in G3), after the LIA, 28.6% were confirmed as being HTLV-1 positive, 28.6% were confirmed as being HTLV-2 positive, and 42.8% remained HTLV untypeable in G1. In G2, 66.7% were confirmed as being HTLV-1 positive and 33.3% were confirmed as being HTLV-2 positive, and in G3, 66.7% were confirmed as being HTLV-1 positive and 33.3% remained HTLV untypeable (Fig. 1 and Table 4).
TABLE 3.
WB-indeterminate profiles detected in serum samples from each study group and numbers and percentages of serum samples with HTLV-1/2 infection confirmed by LIA
| WB-indeterminate profilea | No. of samples with profile |
No. of LIA-positive samples (%)b | |||
|---|---|---|---|---|---|
| G1 (n = 48) | G2 (n = 21) | G3 (n = 16) | Total | ||
| GD21, p24 | 10 | 3 | 13 | 12 (92.30) | |
| rgp46 only (I or II) | 5 | 1 | 6 | 5 (83.33) | |
| GD21, p19 | 3 | 3 | 3 | 9 | 8 (88.89) |
| p24, rgp46-II | 4 | 3 | 7 | 7 (100.00) | |
| GD21, rgp46 (I and/or II) | 7 | 1 | 4 | 12 | 9 (75.00) |
| GD21 | 4 | 4 | 6 | 14 | 3 (21.43) |
| p19 | 5 | 5 | 1 (20.00) | ||
| p24 | 4 | 1 | 1 | 6 | 1 (16.67) |
| GD21, p24, rgp46-I | 3 | 3 | 1 (33.33) | ||
| p19, rgp46-I | 1 | 1 | 2 | 2 (100.00) | |
| GD21, gp21, rgp46-II | 1 | 1 | 1 (100.00) | ||
| GD21, p19, p26 | 1 | 1 | 1 (100.00) | ||
| GD21, p24, p32, p36 | 1 | 1 | 1 (100.00) | ||
| p19, p24, rgp46-II | 1 | 1 | 1 (100.00) | ||
| p24, p36, rgp46-II | 1 | 1 | 1 (100.00) | ||
| p19, p24 | 1 | 1 | |||
| p19, p26, p28, p53 | 2 | 2 | |||
According to criteria established by the manufacturer of HTLV Blot 2.4 (MP Biomedicals).
According to the requirements established by the manufacturer of INNO-LIA HTLV-I/II (Fujirebio).
TABLE 4.
Final results and profiles obtained with confirmatory serological assays for detecting HTLV-1/2 antibodies in 111 serum samples that yielded WB-inconclusive results and were tested by LIA
| Group and WB result for each samplea | WB profile | LIA resultb | LIA profile | Final result |
|---|---|---|---|---|
| G1 | ||||
| Indeterminate | GD21, p24 | HTLV | p19-I/II, p24-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24 | HTLV | p19-I/II, p24-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24, p26, p28, p32, p36, p53 | HTLV | p19-I/II, gp21-I/II | HTLV |
| Indeterminate | p19, p26, p28, p53 | Negative | p19-I/II | Negative |
| Indeterminate | GD21,c p24c | HTLV | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II | HTLV |
| Indeterminate | GD21, p24 | HTLV | p19-I/II, p24-I/II, gp21-I/II | HTLV |
| Indeterminate | p24, rgp46-II | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21 | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | p24 | Indeterminate | p19-I/II, p24-I/II | Indeterminate |
| Indeterminate | GD21, rgp46-II | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p19 | HTLV-1 | p19-I/II, gp21-I/II, p19-I | HTLV-1 |
| Indeterminate | p24, p36, rgp46-II | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21 | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | p19, p26, p28, p53 | Negative | p19-I/II | Negative |
| Indeterminate | p24c | Indeterminate | gp21-I/II | Indeterminate |
| HTLV | GD21,c p19,c p24 | HTLV-1 | p19-I/II, p24-I/II, gp21-I/II, p19-I | HTLV-1 |
| Indeterminate | p19 | Negative | p19-I/II, p19-I | Negative |
| Indeterminate | p19, p24 | Negative | p24-I/II | Negative |
| Indeterminate | GD21,c p24c | Indeterminate | gp21-I/II | Indeterminate |
| HTLV | GD21, p19, p24 | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, gp21, rgp46-II | HTLV-2 | p19-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p24 | HTLV | p19-I/II, p24-I/II, gp46,-I/II, gp21-I/II | HTLV |
| Indeterminate | GD21, rgp46-II, rgp46-I | Negative | No bands | Negative |
| Indeterminate | GD21, p24,c rgp46-I | HTLV-1 | p19-I/II, gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| Indeterminate | GD21c | Negative | p19-I/II | Negative |
| HTLV | GD21, p19, p24, p26, p28, p32 | HTLV | p19-I/II, p24-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24 | HTLV | p19-I/II, gp21-I/II | HTLV |
| Indeterminate | p19,c rgp46-I | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| Indeterminate | p24,c rgp46-II | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| HTLV | GD21, p19, p24, p26, p32, p36, p53 | HTLV | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24 | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p24 | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p19 | HTLV | p19-I/II, gp46-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24 | HTLV | p19-I/II, p24-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24 | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21,c p24 | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p24, p32, p36 | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | p24,c rgp46-II | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | p24, rgp46-II | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p24 | HTLV | p19-I/II, p24-I/II, gp21-I/II | HTLV |
| Indeterminate | p24c | Negative | No bands | Negative |
| Indeterminate | GD21 | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | GD21, rgp46-II | HTLV-2 | gp46-I/II, gp21-I/II, gp46II | HTLV-2 |
| Indeterminate | p19 | Negative | p19-I/II | Negative |
| Indeterminate | GD21, p24,c rgp46-Ic | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | GD21, p19, p26 | HTLV | p19-I/II, gp46-I/II, gp21-I/II | HTLV |
| Indeterminate | GD21, rgp46-II | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | GD21, p24,c rgp46-Ic | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | GD21, rgp46-I | Indeterminate | gp21-I/II | Indeterminate |
| HTLV | GD21, p19, p24 | HTLV-2 | p19-I/II, p24-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, rgp46-II, rgp46-Ic | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p24 | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | p24 | Indeterminate | p19-I/II, p24-I/II | Indeterminate |
| Indeterminate | p19 | Negative | p19-I/II | Negative |
| Indeterminate | p19c | HTLV | p19-I/II, gp21-I/II | HTLV |
| Indeterminate | GD21, rgp46-II | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21,c p19 | HTLV-2 | p19-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| HTLV | GD21, p19, p24, p26, p32, p36 | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, p19-I, gp46-I | HTLV-1 |
| Indeterminate | GD21, p24 | HTLV-2 | p19-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| HTLV | GD21, p19, p24 | HTLV-1 | p19-I/II, p24-I/II, gp21-I/II, p19-I | HTLV-1 |
| Indeterminate | p19 | Negative | p19-I/II | Negative |
| HTLV | GD21, p19, p24 | HTLV-1 | p19-I/II, p24-I/II, gp21-I/II, p19-I | HTLV-1 |
| G2 | ||||
| Indeterminate | rgp46-I | HTLV-1 | p24-I/II, gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| Indeterminate | p24, rgp46-II | HTLV-2 | p24-I/II, gp46-I/II, gp21-I/II, gp46-I,c gp46-II | HTLV-2 |
| HTLV | GD21, p19, p24, p26,c p28,c p32,c p36c | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, p19-I, gp46-I | HTLV-1 |
| Indeterminate | GD21c | Negative | No bands | Negative |
| Indeterminate | rgp46-Ic | HTLV-1 | gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| Indeterminate | GD21,c rgp46-I | HTLV-1 | gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| Indeterminate | GD21, p19c | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | p24, rgp46-II | HTLV-2 | gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | rgp46-II | HTLV-2 | p19-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21c | HTLV | p19-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24 | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| Indeterminate | GD21 | Negative | No bands | Negative |
| Indeterminate | GD21, p19 | HTLV | p19-I/II, p24-I/II, gp21-I/II | HTLV |
| Indeterminate | p24, rgp46-II | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21c | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | p24 | HTLV-2 | gp46-I/II, gp21-I/II, gp46IIc | HTLV-2 |
| Indeterminate | GD21, p24 | HTLV-2 | p19-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | p19, p24, rgp46-II | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p24 | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | rgp46-II | HTLV-2 | p19-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | rgp46-II | HTLV-2 | p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21, p24 | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-II | HTLV-2 |
| Indeterminate | GD21,c p19c | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, p19-I,c gp46-I | HTLV-1 |
| HTLV | GD21,c p19,c p24 | HTLV-2 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-IIc | HTLV-2 |
| G3 | ||||
| HTLV | GD21, p19, p24, p26,c p28,c p36c | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, p19-I, gp46-I | HTLV-1 |
| HTLV | GD21, p19, p24, p26,c p28,c p32,c p36,c gp46c | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| HTLV | GD21,c p19, p24c | HTLV | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24, p26c, p28c, p36, gp46 | HTLV | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24, p26, p28, p32, p36, gp46, p53 | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, p19-I | HTLV-1 |
| HTLV | GD21, p19, p24, p26, p28, p32, p36 | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, p19-I | HTLV-1 |
| Indeterminate | GD21,c p19c | HTLV | p19-I/II, gp46-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24c | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, p19-I | HTLV-1 |
| Indeterminate | GD21, rgp46-II | HTLV | p19-I/II, gp21-I/II | HTLV |
| HTLV | GD21, p19, p24,c p26, p28, p32, p36 | HTLV-1 | p19-I/II, p24-I/II, gp46-I/II, gp21-I/II, p19-I, gp46-I | HTLV-1 |
| Indeterminate | GD21, rgp46-I | HTLV-1 | p19-I/II, gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| Indeterminate | GD21, p19 | HTLV | p19-I/II, gp21-I/II | HTLV |
| Indeterminate | GD21c | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | GD21 | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | GD21 | HTLV-1 | gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| Indeterminate | GD21 | Negative | No bands | Negative |
| Indeterminate | p24c | Negative | No bands | Negative |
| Indeterminate | rgp46-IIc | Negative | No bands | Negative |
| Indeterminate | GD21, rgp46-I | HTLV-1 | p19-I/II, gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
| HTLV | GD21, p19, p24 | HTLV | p19-I/II, gp21-I/II | HTLV |
| Indeterminate | GD21 | HTLV | p19-I/II, gp21-I/II | HTLV |
| Indeterminate | GD21, rgp46-IIc | HTLV | p19-I/II, gp21-I/II | HTLV |
| Indeterminate | GD21 | Indeterminate | gp21-I/II | Indeterminate |
| Indeterminate | GD21, p19 | HTLV | p19-I/II, gp46-I/II, gp21-I/II | HTLV |
| Indeterminate | p19, rgp46-I | HTLV-1 | p19-I/II, gp46-I/II, gp21-I/II, gp46-I | HTLV-1 |
WB, Western blot (HTLV Blot 2.4; MP Biomedicals).
LIA, line immunoassay (INNO-LIA HTLV-I/II; Fujirebio).
Faint bands.
DISCUSSION
HTLV-1- and HTLV-2-seroindeterminate WB results are prevalent worldwide, with rates fluctuating according to country and study group (geographic areas and populations where the disease is or is not endemic). Several attempts have been made to improve WB sensitivity and specificity, such as adding HTLV-1 and HTLV-2 recombinant envelope proteins and transmembrane protein to the HTLV-1 viral lysate. These include rgp46-I, rgp46-II, and GD21, the latter of which blocks the cross-reactivity of gp21 with Plasmodium falciparum in regions where malaria is endemic (22, 23). In spite of these efforts, Blot 2.4 continues to yield high rates of WB-indeterminate and/or untypeable HTLV results (4–8, 10, 11, 13–17, 21). The present study found similar results and disclosed that in populations presenting a high risk of acquiring viral infections (G1 and G2) as well in the general population (G3) of Brazil, a large number of WB-inconclusive results were detected. Several hypotheses were taken into consideration for these WB-inconclusive results, such as low HTLV-1 and HTLV-2 proviral loads, mutations in the provirus (defective particles), low-level production of viral antigens consequently leading to a low level of specific antibody production, the seroconversion period, cross-reactivity with other antigens or viruses, coinfection with HIV, and the use of antiretroviral therapy, among others (5, 12–15, 24–27).
Interestingly, the majority of WB-indeterminate results that were not confirmed by seroconversion or PCR assays were due to cross-reactivity with Gag antigens. For instance, the WB-indeterminate pattern exhibiting p19, p26, p28, p32, p36, and p53, termed the HTLV-1 Gag indeterminate profile (HGIP), has been detected in epidemiological studies, mostly in Cameroon and in the Caribbean, but was not associated with true HTLV-1 infection (23, 24). This pattern was not frequently described in Brazilian WB-indeterminate samples (5, 13, 15, 26) and was also not observed in any samples analyzed in the present study. Although two samples here presented an incomplete HGIP pattern (without p32 and p36 bands), only the p19-I/II band was detected by the LIA. Thus, these samples were considered HTLV negative according to the manufacturer’s criteria for the LIA. Corroborating the LIA result, the same profile (only the p19-I/II band) was detected when we analyzed samples from five patients who attended HTLV outpatient clinics in São Paulo during the years 2000 to 2006, which exhibited p19, p26, p28, and p36 WB bands (5).
Moreover, we conducted a retrospective analysis of 108 well-characterized blood samples from patients from São Paulo (tested by two serological assays [WB and LIA] and two molecular assays [qPCR for pol and PCR-RFLP analysis for tax]) and confirmed a series of more than 10 HTLV-1 and 10 HTLV-2 samples as being positive by both WB and LIA criteria, and they were concordant. In addition, we revised data for two samples that were HTLV-1 and -2 positive by WB, one of which was confirmed to be HTLV-1 and HTLV-2 positive by the LIA and molecular assays (15). Additionally, we tested two other HTLV-1 and -2 WB-positive samples by the LIA, and only HTLV-1 was confirmed, taking into account the intensity of the discriminatory bands (p19-I, gp46-I, and gp46-II). Of note, the WB-indeterminate profile (strong GD21 and p28 bands) described in Pygmies and Bantus living in the southern Cameroonian rainforest (28) has not been detected in any study conducted in São Paulo (5, 13, 15) and here.
Although PCR assays presented lower sensitivity than WB in detecting true HTLV-1/2 infection in HIV-HTLV-coinfected individuals in São Paulo, Brazil, the molecular assays were able to confirm and discriminate between HTLV-1 and HTLV-2 in some WB-indeterminate and HTLV-untypeable cases, indicating that both serological and molecular assays are useful for HTLV diagnosis (13, 15). Due to the presence of large numbers of WB-indeterminate samples, coupled with the high cost of obtaining WB assays and LIAs in Brazil, we recently proposed an algorithm that employs qPCR to confirm HTLV infection, followed by testing any PCR-negative samples by WB or an LIA. This strategy was shown to reduce costs and improve the diagnostic accuracy of HTLV-1/2 detection (13, 15). Nonetheless, due to highly divergent socioeconomic conditions among different regions in Brazil, in laboratories without the means to perform molecular assays, high-performance serological testing presents an acceptable alternative.
Some studies of HTLV diagnosis conducted in blood donors in Latin America (considered an area where HTLV-1/2 is endemic) have reported differing numbers of WB-indeterminate samples, which were subsequently confirmed to be positive by PCR (29–31). Also, in blood donors from another area where the disease is endemic, northeast Iran, WB-indeterminate samples were found to be positive by PCR, and the most prevalent WB bands presented various combinations of rgp46-I, GD21, and gp21 (32).
In corroboration with these findings, the majority of serum samples here that presented WB patterns of GD21 and/or rgp46-I or -II plus p19 or p24 were subsequently confirmed to be positive by the LIA. These types of WB patterns were observed in G1, and PCR assays demonstrated HTLV-1 or HTLV-2 positivity (data not shown) (15). In addition, the majority of serum samples that presented only Gag bands in the WB analysis were negative for HTLV-1/2 infection by the LIA. Of note, one blood sample from G1 that showed a faint GD21 band in the WB analysis tested negative for HTLV-1/2 by both the LIA and PCR. Another serum sample presenting a GD21, rgp46-I, and rgp46-II WB pattern was found to be negative by the LIA; unfortunately, this sample could not be analyzed by PCR because only serum was sent to the laboratory for analysis. However, retesting of this serum sample by WB and the LIA confirmed the discrepant results. It is interesting to note that in serum samples (n = 14) that tested HTLV untypeable by the LIA, PCR confirmed the presence of HTLV-1 in 5 samples and HTLV-2 in another 2 samples from G1 (data not shown), emphasizing the need for employing molecular assays to confirm an HTLV diagnosis in patients with HIV-HTLV coinfection.
In G2, the majority of WB-indeterminate patterns presented either GD21 alone or this protein in association with one Gag or envelope band. The LIA confirmed the presence of HTLV-2 in 11/21 (52.4%) of the WB-indeterminate samples. The high number of HTLV-2-positive samples in G2 leads us to suppose that these patients acquired HBV and HCV, as well HTLV-1/2 and HIV, at the same time, probably by the parenteral route and prior to the time when serological testing for HIV and HBV (1989), and subsequently for HTLV and HCV (1993), became mandatory in blood banks throughout Brazil; in addition, intravenous drug addiction was more frequent in this country, as previously described (18–20). Corroborating this hypothesis, older age and male sex predominated in G2. Regarding the lack of WB detection of truly HTLV-2-infected samples, we have described this difficulty since 2000 (8, 13, 15) and hypothesized that the rgp46-II (K55) present in the WB strip is not as sensitive for the detection antibodies to the HTLV-2 strains that circulate in Brazil (HTLV-2a subtype, variant 2c) (33). This seems not to be the case for the gp46-II present in the LIA strip.
Concerning the two WB-indeterminate samples in G2 with negative HTLV results by the LIA, both presented reactivity for GD21 in the WB analysis, and one of the samples showed a faint band. Curiously, the LIA demonstrates the best performance in this group of patients, with 20/24 (83.3%) of HTLV-positive samples being detected among the WB-inconclusive samples. Unfortunately, only plasma/serum samples from these patients were available for analysis, which did not allow the use of PCR to perform a comparative analysis of serological and molecular results. Nonetheless, associations between HTLV-1/2 and hepatitis B and C have been reported in several studies conducted in Brazil and elsewhere (18–20, 24, 29, 34).
The WB-inconclusive patterns in G3 were quite different from those in the other groups analyzed. Several of the HTLV-untypeable samples demonstrated the presence of almost all bands corresponding to the HTLV-1 viral lysate, without reactivity to rgp46-I, and six of nine were subsequently confirmed to be HTLV-1 positive by the LIA. Twelve out of the 25 WB-inconclusive samples that could be submitted for PCR (9 WB indeterminate and 3 HTLV indeterminate), 11 were confirmed to be HTLV-1 infected, 6 of which were HTLV untypeable by LIAs (data not shown). In addition, the serum samples that tested negative by LIAs presented three different WB-indeterminate patterns: (i) GD21, (ii) p24 (faint band), and (iii) rgp46-II (faint band). Only one of these samples could be tested by PCR and presented an HTLV-negative result (data not shown). In summary, the samples in G3 were confirmed to be positive for HTLV-1 or HTLV but not HTLV-2 infection. This finding could be partially related to the ethnic origin of the included individuals (African descendants), the lack of HIV infection in this group, and the characteristics of the patients seen at HTLV outpatient clinics in Salvador, BA (35).
Of note, the reasons described above to explain WB-inconclusive results could also be applied to the PCR-negative results in truly HTLV-1/2-infected individuals, including the low proviral loads in HIV/AIDS patients undergoing antiretroviral therapy in G1 and, in some cases, in G2 (13–15); the observation that HTLV-2 infection is known to show low proviral loads (11, 12, 25); the presence of defective provirus not detected by the primers employed in the PCR assays (27); and infection with other viruses, such as HTLV-3 or HTLV-4, which can be detected only by using specific primers (36, 37).
It is noteworthy that since the discovery of HTLV-3 and HTLV-4 in central Africa (36, 37), studies have been conducted to ascertain whether assays that are commercially available and employed for HTLV-1/2 diagnosis are able to detect these new HTLVs (38–40). The results obtained confirmed that HTLV-1/2 screening assays are sensitive for the detection of antibodies to HTLV-3 and HTLV-4 (38) and disclosed discordant and misclassified results in relation to confirmatory serological assays (WB and LIA) (36–41).
In fact, we could not rule out HTLV-3 and HTLV-4 infections in Brazil, since populations in central Africa migrated from Africa and Australia to the American continent before Asiatic population migration, and their descendants, such as the Amerindians, could maintain such viruses or spread them to the general population, which could justify the frequent presence of WB-indeterminate results in Amerindians, as previously described (42).
In relation to the LIA, although it presented the best performance in detecting HTLV-1 and HTLV-2, we could not exclude misclassified positive results, as occurred in HTLV-3- and HTLV-4-infected individuals, who could be erroneously diagnosed as being infected with HTLV-2 (36–38).
Of note, despite the fact that the LIA demonstrated better performance than WB in the serological diagnosis of both HTLV-1 and HTLV-2, additional considerations are warranted for both assays. With respect to WB, we consider the lack of an ability to score the intensity of a positive band to be a problem, since it is not known when a faint band should be considered truly positive. The criteria (band profiles) established by the manufacturer to confirm HTLV-1 and HTLV-2 infections in WB assays are excessively stringent and deserve a review. Taking into account the results obtained here, we suggest that samples presenting only one Gag band (p19 or p24) plus GD21 and rgp46-I or rgp-46-II should be considered HTLV-1 and HTLV-2 positive, respectively, since samples that demonstrated p24, GD21, and rgp46-I bands were confirmed to be HTLV-1 positive by the LIA and PCR. In contrast, we detected true HTLV-2 positivity in samples that showed p19, GD21, and rgp46-II bands. In addition, when Gag bands were undetectable but both envelope bands (GD21 and rgp46-I or rgp46-II) were present, it was impossible to rule out true HTLV-1 or HTLV-2 infection, since seroconversion could be taking place. Indeed, when all bands showed reactivity to HTLV-1 viral lysate antigens, even in the absence of rgp46-I, it was possible to confirm HTLV-1 infection.
In conclusion, the LIA was shown to be the best serological test for confirming HTLV-1 and HTLV-2 infections, regardless of whether individuals were HTLV monoinfected or coinfected. We further highlight the need to review some WB criteria based on our results and those reported previously by others. It remains to be determined whether the superior performance of the LIA was due to the less stringent criteria employed than for WB. Further studies are necessary to confirm these results in a variety of risk populations from Brazil and elsewhere.
ACKNOWLEDGMENTS
This study was supported in São Paulo by grants from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (2012/51220-8 and 2016/03654-0), and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (302661/2015-8) and in Salvador by grants from the CNPq (473667/2012 and 311054/2014-50), the Fundação de Amparo à Pesquisa do Estado da Bahia (FAPESB) (2574/2013), and the Fundação Nacional para o Desenvolvimento do Ensino Superior (FUNDADESP) (9600113). F.L.N.S. and K.R.C. are fellows supported by CAPES (financial code 001). The publication fee was covered by the Oswaldo Cruz Foundation.
We thank Andris K. Walter for providing English language revision and manuscript copyediting assistance.
We declare that we have no commercial or other associations which might pose a conflict of interest to the manufacturers of the kits reported in the present study.
REFERENCES
- 1.Gessain A, Cassar O. 2012. Epidemiological aspects and world distribution of HTLV-1 infection. Front Microbiol 3:338. doi: 10.3389/fmicb.2012.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paiva A, Casseb J. 2015. Origin and prevalence of human T-lymphotropic virus type 1 (HTLV-1) and type 2 (HTLV-2) among indigenous populations in the Americas. Rev Inst Med Trop Sao Paulo 57:1–13. doi: 10.1590/S0036-46652015000100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caterino-de-Araujo A, Simao do Rosario Casseb J, Neitzert E, Xavier de Souza ML, Mammano F, Del Mistro A, De Rossi A, Chieco-Bianchi L. 1994. HTLV-I and HTLV-II infections among HIV-1 seropositive patients in São Paulo, Brazil. Eur J Epidemiol 10:165–171. doi: 10.1007/bf01730366. [DOI] [PubMed] [Google Scholar]
- 4.Jacob F, Santos-Fortuna E, Azevedo RS, Caterino-de-Araujo A. 2007. Performances of HTLV serological tests in diagnosing HTLV infection in high-risk population of São Paulo, Brazil. Rev Inst Med Trop Sao Paulo 49:361–364. doi: 10.1590/s0036-46652007000600005. [DOI] [PubMed] [Google Scholar]
- 5.Jacob F, Santos-Fortuna E, Azevedo RS, Caterino-de-Araujo A. 2008. Serological patterns for HTLV-I/II and its temporal trend in high-risk populations attended at Public Health Units of São Paulo, Brazil. J Clin Virol 42:149–155. doi: 10.1016/j.jcv.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Caterino-de-Araujo A, de los Santos-Fortuna E, Zandoná Meleiro MC, Suleiman J, Calabrò ML, Favero A, De Rossi A, Chieco-Bianchi L. 1998. Sensitivity of two ELISA tests in relation to Western blot in detecting HTLV-I and HTLV-II infections among HIV-1-infected patients from São Paulo, Brazil. Diagn Microbiol Infect Dis 30:173–182. doi: 10.1016/S0732-8893(97)00236-8. [DOI] [PubMed] [Google Scholar]
- 7.Ishak R, Vallinoto AC, Azevedo VN, Vicente AC, Hall WW, Ishak MO. 2007. Molecular evidence for infection by HTLV-2 among individuals with negative serological screening tests for HTLV antibodies. Epidemiol Infect 135:604–609. doi: 10.1017/S0950268806006984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morimoto HK, Morimoto AA, Reiche EMV, Ueda LT, Matsuo T, Reiche FV, Caterino-de-Araujo A. 2007. Difficulties in the diagnosis of HTLV-2 infection in HIV/AIDS patients from Brazil. Comparative performances of serologic and molecular assays, and detection of HTLV-2b subtype. Rev Inst Med Trop Sao Paulo 49:225–230. doi: 10.1590/s0036-46652007000400006. [DOI] [PubMed] [Google Scholar]
- 9.Novoa P, Penalva de Oliveira AC, Posada Vergara MP, da Silva Duarte AJ, Casseb J. 2007. Molecular characterization of human T-cell lymphotropic virus type 2 (HTLV-II) from people living in urban areas of Sao Paulo city: evidence of multiple subtypes circulation. J Med Virol 79:182–187. doi: 10.1002/jmv.20775. [DOI] [PubMed] [Google Scholar]
- 10.Costa JMP, Segurado AC. 2009. Molecular evidence of human T-cell lymphotropic virus types 1 and 2 (HTLV-1 and HTLV-2) infections in HTLV seroindeterminate individuals from São Paulo, Brazil. J Clin Virol 44:185–189. doi: 10.1016/j.jcv.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Olah I, Fukumori LM, Smid J, de Oliveira AC, Duarte AJ, Casseb J. 2010. Neither molecular diversity of the envelope, immunosuppression status, nor proviral load causes indeterminate HTLV Western blot profiles in samples from human T-cell lymphotropic virus type 2 (HTLV-2)-infected individuals. J Med Virol 82:837–842. doi: 10.1002/jmv.21718. [DOI] [PubMed] [Google Scholar]
- 12.Montanheiro P, Olah I, Fukumori LM, Smid J, Oliveira AC, Kanzaki LI, Fonseca LA, Duarte AJ, Casseb J. 2008. Low DNA HTLV-2 proviral load among women in São Paulo City. Virus Res 135:22–25. doi: 10.1016/j.virusres.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Costa EAS, Magri MC, Caterino-Araujo A. 2011. The best algorithm to confirm the diagnosis of HTLV-1 and HTLV-2 in at-risk individuals from São Paulo, Brazil. J Virol Methods 173:280–283. doi: 10.1016/j.jviromet.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 14.Campos KR, Gonçalves MG, Caterino-de-Araujo A. 2017. Failures in detecting HTLV-1 and HTLV-2 in patients infected with HIV-1. AIDS Res Hum Retroviruses 33:382–385. doi: 10.1089/AID.2016.0191. [DOI] [PubMed] [Google Scholar]
- 15.Campos KR, Gonçalves MG, Costa NA, Caterino-de-Araujo A. 2017. Comparative performances of serologic and molecular assays for detecting HTLV-1 and HTLV-2 in patients infected with HIV-1. Braz J Infect Dis 21:297–305. doi: 10.1016/j.bjid.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zrein M, Louwagie J, Boeykens H, Govers L, Hendrickx G, Bosman F, Sablon E, Demarquilly C, Boniface M, Saman E. 1998. Assessment of a new immunoassay for serological confirmation and discrimination of human T-cell lymphotropic virus infections. Clin Diagn Lab Immunol 5:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabino EC, Zrein M, Taborda CP, Otani MM, Ribeiro-dos-Santos G, Sáez-Alquézae A. 1999. Evaluation of the INNO-LIA HTLV I/II assay for confirmation of human T-cell leukemia virus-reactive sera in blood bank donations. J Clin Microbiol 37:1324–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caterino-de-Araujo A, Sacchi CT, Gonçalves MG, Campos KR, Magri MC, Alencar WK, Group of Surveillance and Diagnosis of HTLV of São Paulo. 2015. Current prevalence and risk factors associated with HTLV-1 and HTLV-2 infections among HIV/AIDS patients in São Paulo, Brazil. AIDS Res Hum Retroviruses 31:543–549. doi: 10.1089/AID.2014.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caterino-de-Araujo A, Alves FA, Campos KR, Lemos MF, Moreira RC. 2018. Making the invisible visible: searching for human T-cell lymphotropic virus types 1 and 2 (HTLV-1 and HTLV-2) in Brazilian patients with viral hepatitis B and C. Mem Inst Oswaldo Cruz 113:130–134. doi: 10.1590/0074-02760170307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alves FA, Campos KR, Lemos MF, Moreira RC, Caterino-de-Araujo A. 2018. Hepatitis C viral load in HCV-mono and HCV/HIV-1-, HCV/HTLV-1/-2-, and/or HCV/HIV/HTLV-1/-2-co-infected patients from São Paulo, Brazil. Braz J Infect Dis 22:123–128. doi: 10.1016/j.bjid.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.da Silva Brito V, Santos FLN, Gonçalves NLS, Araujo TH, Nascimento DSV, Pereira FM, Boa-Sorte NCA, Grassi MFR, Caterino-de-Araujo A, Galvão-Castro B. 2018. Performance of commercially available serological screening tests for human T-cell lymphotropic virus infection in Brazil. J Clin Microbiol 56:e00961-18. doi: 10.1128/JCM.00961-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varma M, Rudolph DL, Knuchel M, Switzer WM, Hadlock KG, Velligan M, Chan L, Foung SK, Lal RB. 1995. Enhanced specificity of truncated transmembrane protein for serologic confirmation of human T-cell lymphotropic virus type 1 (HTLV-1) and HTLV-2 infections by Western blot (immunoblot) assay containing recombinant envelope glycoproteins. J Clin Microbiol 33:3239–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahieux R, Horal P, Mauclère P, Mercereau-Puijalon O, Guillotte M, Meertens L, Murphy E, Gessain A. 2000. Human T-cell lymphotropic virus type 1 Gag indeterminate Western blot patterns in Central Africa: relationship to Plasmodium falciparum infection. J Clin Microbiol 38:4049–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rouet F, Meertens L, Courouble G, Herrmann-Storck C, Pabingui R, Chancerel B, Abid A, Strobel M, Mauclere P, Gessain A. 2001. Serological, epidemiological, and molecular differences between human T-cell lymphotropic virus type 1 (HTLV-1)-seropositive healthy carriers and persons with HTLV-I Gag indeterminate Western blot patterns from the Caribbean. J Clin Microbiol 39:1247–1253. doi: 10.1128/JCM.39.4.1247-1253.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy EL, Lee TH, Chafets D, Nass CC, Wang B, Loughlin K, Smith D, HTLV Outcomes Study Investigators. 2004. Higher human T lymphotropic virus (HTLV) provirus load is associated with HTLV-I versus HTLV-II, with HTLV-II subtype A versus B, and with male sex and a history of blood transfusion. J Infect Dis 190:504–510. doi: 10.1086/422398. [DOI] [PubMed] [Google Scholar]
- 26.Martins ML, Santos AC, Namen-Lopes MS, Barbosa-Stancioli EF, Utsch DG, Carneiro-Proietti AB. 2010. Long-term serological follow-up of blood donors with an HTLV-indeterminate Western blot: antibody profile of seroconverters and individuals with false reactions. J Med Virol 82:1746–1753. doi: 10.1002/jmv.21881. [DOI] [PubMed] [Google Scholar]
- 27.Kuramitsu M, Sekizuka T, Yamochi T, Firouzi S, Sato T, Umeki K, Sasaki D, Hasegawa H, Kubota R, Sobata R, Matsumoto C, Kaneko N, Momose H, Araki K, Saito M, Nosaka K, Utsunomiya A, Koh K-R, Ogata M, Uchimaru K, Iwanaga M, Sagara Y, Yamano Y, Okayama A, Miura K, Satake M, Saito S, Itabashi K, Yamaguchi K, Kuroda M, Watanabe T, Okuma K, Hamaguchi I. 2017. Proviral features of human T cell leukemia virus type 1 in carriers with indeterminate Western blot analysis results. J Clin Microbiol 55:2838–2849. doi: 10.1128/JCM.00659-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Filippone C, Bassot S, Betsem E, Tortevoye P, Guillotte M, Mercereau-Puijalon O, Plancoulaine S, Calattini S, Gessain A. 2012. A new and frequent human T-cell leukemia virus indeterminate Western blot pattern: epidemiological determinants and PCR results in central African inhabitants. J Clin Microbiol 50:1663–1672. doi: 10.1128/JCM.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos TJT, Costa CM, Goubau P, Vandamme AM, Desmyter J, Van Doren S, Mota RM, de Castro Costa FB, Oliveira AC, Barreto V, Gomes AF, Carneiro-Proietti AB, de Bruin VM, de Sousa FC, Oriá RB. 2003. Western blot seroindeterminate individuals for human T-lymphotropic virus 1/2 (HTLV-1/2) in Fortaleza (Brazil): a serological and molecular diagnostic and epidemiological approach. Braz J Infect Dis 7:202–209. doi: 10.1590/S1413-86702003000300005. [DOI] [PubMed] [Google Scholar]
- 30.Mangano AM, Remesar M, del Pozo A, Sen L. 2004. Human T lymphotropic virus types I and II proviral sequences in Argentinian blood donors with indeterminate Western blot patterns. J Med Virol 74:323–327. doi: 10.1002/jmv.20172. [DOI] [PubMed] [Google Scholar]
- 31.Berini CA, Eirin ME, Pando MA, Biglione MM. 2007. Human T-cell lymphotropic virus types I and II (HTLV-I and -II) infection among seroindeterminate cases in Argentina. J Med Virol 79:69–73. doi: 10.1002/jmv.20731. [DOI] [PubMed] [Google Scholar]
- 32.Zanjani DS, Shahabi M, Talaei N, Afzalaghaee M, Tehranian F, Bazargani R. 2011. Molecular analysis of human T cell lymphotropic virus type 1 and 2 (HTLV-1/2) seroindeterminate blood donors from northeast Iran: evidence of proviral tax, env, and gag sequences. AIDS Res Hum Retroviruses 27:131–135. doi: 10.1089/aid.2010.0017. [DOI] [PubMed] [Google Scholar]
- 33.Magri MC, Brigido LFM, Morimoto HK, Caterino-de-Araujo A. 2013. HTLV type 2a strains among HIV-1-coinfected patients from Brazil have originated mostly from Brazilian Amerindians. AIDS Res Hum Retroviruses 29:1010–1018. doi: 10.1089/aid.2013.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morimoto HK, Caterino-de-Araujo A, Morimoto AA, Reiche EMV, Ueda LT, Matsuo T, Stegmann JW, Reiche FV. 2005. Seroprevalence and risk factors for human T cell lymphotropic virus type 1 and 2 infection in human immunodeficiency virus-infected patients attending AIDS referral center health units in Londrina and other communities in Paraná, Brazil. AIDS Res Hum Retroviruses 21:256–262. doi: 10.1089/aid.2005.21.256. [DOI] [PubMed] [Google Scholar]
- 35.Dourado I, Alcantara LCJ, Barreto ML, da Glória Teixeira M, Galvão-Castro B. 2003. HTLV-I in the general population of Salvador, Brazil, a city with African ethnic and sociodemographic characteristics. J Acquir Immune Defic Syndr 34:527–531. doi: 10.1097/00126334-200312150-00013. [DOI] [PubMed] [Google Scholar]
- 36.Calattini S, Chevalier SA, Duprez R, Bassot S, Froment A, Mahieux R, Gessain A. 2005. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in central Africa. Retrovirology 2:30. doi: 10.1186/1742-4690-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe ND, Heneine W, Carr JK, Garcia AD, Shanmugam V, Tamoufe U, Torimiro JN, Prosser AT, Lebreton M, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Switzer WM. 2005. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc Natl Acad Sci U S A 102:7994–7999. doi: 10.1073/pnas.0501734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Switzer WM, Hewlett I, Aaron L, Wolfe ND, Burke DS, Heneine W. 2006. Serologic testing for human T-lymphotropic virus-3 and -4. Transfusion 46:1647–1648. doi: 10.1111/j.1537-2995.2006.00950.x. [DOI] [PubMed] [Google Scholar]
- 39.Calattini S, Betsem E, Bassot S, Chevalier SA, Mahieux R, Froment A, Gessain A. 2009. New strain of human T lymphotropic virus (HTLV) type 3 in a Pygmy from Cameroon with peculiar HTLV serologic results. J Infect Dis 199:561–564. doi: 10.1086/596206. [DOI] [PubMed] [Google Scholar]
- 40.Mahieux R, Gessain A. 2011. HTLV-3/STLV-3 and HTLV-4 viruses: discovery, epidemiology, serology and molecular aspects. Viruses 3:1074–1090. doi: 10.3390/v3071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng H, Wolfe ND, Sintasath DM, Tamoufe U, Lebreton M, Djoko CF, Diffo JLD, Pike BL, Heneine W, Switzer WM. 2010. Emergence of a novel and highly divergent HTLV-3 in a primate hunter in Cameroon. Virology 401:137–145. doi: 10.1016/j.virol.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanzaki LIB, Casseb J. 2008. Human T-lymphotropic viruses evolution possibly explained by primate deltaretrovirus geographical segregation. Retrovirology (Auckl) 1:1178–1238. [Google Scholar]