Abstract
Background:
Microgravity (μG) negatively influences bone metabolism by affecting normal osteoblast and osteoclast function. μG effects on bone metabolism has been an extensive field of study in recent years, due to the challenges presented by space flight.
Methods:
We systematically reviewed research data from genomic studies performed in real or simulat-ed μG, on osteoblast and osteoclast cells. Our search yielded 50 studies, of which 39 concerned cells of the osteoblast family and 11 osteoclast precursors.
Results:
Osteoblastic cells under μG show a decreased differentiation phenotype, proved by diminished expression levels of Alkaline Phosphatase (ALP) and Osteocalcin (OCN) but no apoptosis. Receptor Activator of NF-κB Ligand (RANKL)/ Osteoprotegerine (OPG) ratio is elevated in favor of RANKL in a time-dependent manner, and further RANKL production is caused by upregulation of Interleukin-6 (IL-6) and the inflammation pathway. Extracellular signals and changes in the gravitational environment are perceived by mechanosensitive proteins of the cytoskeleton and converted to intracellular signals through the Mitogen Activated Protein Kinase pathway (MAPK). This is followed by changes in the ex-pression of nuclear transcription factors of the Activator Protein-1 (AP-1) family and in turn of the NF-κB, thus affecting osteoblast differentiation, cell cycle, proliferation and maturation. Pre-osteoclastic cells show increased expression of the marker proteins such as Tryptophan Regulated Attenuation Protein (TRAP), cathepsin K, Matrix Metalloproteinase-9 (MMP-9) under μG conditions and become sensitized to RANKL.
Conclusion:
Suppressing the expression of fusion genes such as syncytine-A which acts independently of RANKL, could be possible future therapeutic targets for microgravity side effects.
Keywords: Osteoblasts, Osteoclasts, Microgravity, Gene expression, Microarrays, Space
1. INTRODUCTION
All living organisms have evolved under the effect of Earth’s inherent gravity of 1g. Since the first manned mission in space, research has focused on the impact of reduced gravitational force in the biological processes of organisms. Astronauts taking part in space missions suffer the effects of gravitational alterations, varying from increased short-term accelerations at launch, to long-term decreased gravity in orbit, known as microgravity (μG)1. Among the various systems of the human organism that are affected by altered
gravity is the skeletal system. Skeletal unloading due to μG results in 1-2% loss of bone mass per month, mainly in the pelvis and the lower extremities. Microgravity influences bone structure macroscopically and in the cellular level [1-3]. Human bone homeostasis is regulated by the coupled and synergic actions of specialized cells called osteoblasts and osteoclasts [4]. Until recently, scientists could study the influence of microgravity on gene expression, only on small groups of genes per experiment. Advances in engineering, biotechnology and information technology have facilitated the invention of high throughput methods such as the microarray and Next Generation Sequencing (NGS) technologies, which in turn enabled the complete genomic study of cells, in a way that is easily reproducible [5, 6]. Experiments in space flights are scarce mainly because of the high cost of space flights. Systems that simulate microgravitational conditions on earth have been used to perform experiments in cell cultures and organisms using different principles. Hind-Limb Suspension (HLS), Head Down Bed Rest (HDBR), the NASA-developed Rotary Cell Culture System (RCCS), Three Dimension Clinostat (3D Clinostat), Random Positioning Machine (RPM), Large Gradient High Magnetic Field (LG-HMF) are the methods most widely used in this field of research [7-9].
2. METHODS
The current study was performed according to the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statements [10]. Two independent reviewers performed the literature search as well as the evaluation of the identified studies to determine the eligibility for inclusion. A third reviewer was included in order to resolve any possible differences derived from the above-mentioned process.
2.1. Data Sources
The studies included in the current systematic review were retrieved from an independent literature search performed in the PubMed and Cochrane databases as well as other sources like Google Scholar and NASA Technical Reports Server. Independent keywords along with their combinations were applied to both databases. The specific keywords were the following: “microgravity”, “weightlessness”, “spaceflight”, “osteoblast”, “osteoclast” and “shuttle”. The above-mentioned keywords were generated through evaluation of the MeSH (Medical Subject Headings) database. Specifically, the search string applied to PubMed was: osteoblast [Title/Abstract]) OR osteoclast [Title/Abstract]) AND weightlessness [Title/Abstract] OR osteoblast [Title/Abstract]) OR osteoclast [Title/Abstract]) AND microgravity [Title/Abstract] OR osteoblast [Title/Abstract]) OR osteoclast [Title/Abstract]) AND spaceflight [Title/Abstract]. The search string applied to the Cochrane database was: microgravity AND osteoblast OR microgravity AND osteoclast.
2.2. Study Selection Criteria
In order to be included in the current review, a study is needed to adhere to specific inclusion and exclusion criteria. Particularly, a study should have provided data regarding gene expression of cells from the osteoblast or osteoclast cell lines incubated in microgravity or simulated microgravity conditions. Specifically, the study material included the a) MC3T3-E1 osteoblast precursor cell line derived from Mus musculus (mouse) calvaria, b) the MG-63 homo sapiens (human) bone osteosarcoma cells (fibroblasts), c) human osteoblast cells, d) chicken calvaria osteoblast cells, e) rat pre-osteoblasts, f) 2T3 immortalized murine osteoblast cell line, osteoblasts and osteoclasts derived from pharyngeal bone of medaka fish and g) the RAW 264.7 mus musculus (mouse) macrophage cells, murine (mouse) macrophages from bone marrow as well as mature osteoclasts.
As “microgravity” and “simulated microgravity” was considered the environment of reduced gravitational pull that is present in space shuttles, in orbit around the earth or when performing short parabolic flights and in ground facilities with devices that simulate weightlessness. The latter are the NASA developed Rotary Cell Culture System (RCCS) or Rotating Wall Vessel (RWV), the 2D or 3D Clinostat, the Random Positioning Machine (RPM), the Large Gradient High Magnetic Field (LG-HMF), Hind limb suspension (HLS) and Head Down Bed Rest (HDBR). Prior reviews that focused on a similar subject, were not considered as eligible for inclusion. Moreover, studies involving ancestral bone mesenchymal cells or mature osteocytes were also excluded, as well as studies that included only proteomic data with no adherent genomic data. Articles that the full text was not available were also excluded. Time or country of origin restrictions were not applied during the identification of eligible studies, whereas studies that were published in languages other than English were not considered as eligible for inclusion.
2.3. Data Extraction
Two authors independently reviewed the full texts of all studies that were considered eligible for inclusion and extracted the individual study data. Any discrepancies were resolved by discussion with a third author to reach a final consensus. Specifically, the data extracted included the following: study characteristics (first author, year of publication, study design), organism and cell line characteristics, location and duration of the experiment (ground based or space), method used to simulate microgravity in ground based studies, methodology of gene expression (microarray, RT-PCR, Hs DNA) and type of statistical analysis performed.
3. RESULTS
Our search in databases PubMed and Cochrane produced 176 results and additional 157 from other sources. After duplicates were removed the remaining study number was 257. After detailed screening 50 studies were eligible for qualitative synthesis in the current systematic review. The process of study selection is depicted in (Fig. 1), which consists of the actual flow diagram we followed.
Fig. (1).
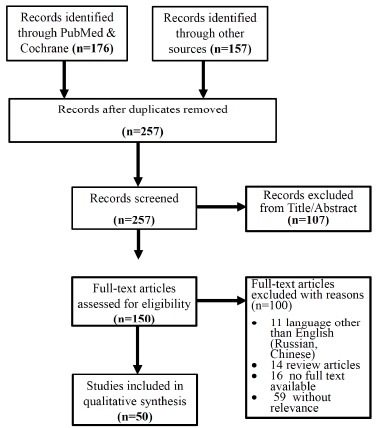
PRISMA results and literature search workflow.
3.1. Study Characteristics
A total of 50 studies met the predefined criteria and were included in the systematic review. Eleven of 50 referred to osteoclast gene expression in microgravity (Ethiraj et al. 2018 [11], Shanmugarajan et al. 2017 [12], Sambandam et al. 2016 [13], Chatani et al. 2016 [14], Chatani et al. 2015 [15], Sun et al. 2015 [16], Saxena et al. 2011 [17], Sambandam et al. 2010 [18], Sambandam et al. 2014 [19], Tamma et al. 2009 [20], Makihira et al. 2008 [21]) [11-21] and 39 out of 50 to osteoblast (Wang et al. 2016 [22], Sun et al. 2015 [23], Hu et al. 2015 [24], Makihira et al. 2008 [21], Bucaro et al. 2007 [25], Hughes et al. 2006 [26], Bucaro et al. 2004 [27], Saito et al. 2003 [28], Ontiveros et al. 2003 [29], Kumei et al. 2002 [30], Sato et al. 1999 [31], Hughes et al. 1998 [32], Z Hu et al. 2017 [33], Goyden et al. 2015 [34], Bikle et al. 1994 [35], Qian et al. 2009 [36], Shuang et al. 2013 [37], Dai et al. 2013 [38], Kapitonova et al. 2013 [39], Guignandon et al. 2014 [40], Kumei et al. 1996 [41], Carmeliet et al. 1999 [42], Landis et al. 2000 [43], Rucci et al. 2002 [44], Kumei et al. 2003 [45, 46], Nakamura et al. 2003 [47], Kumei et al. 2004 [48, 49], Kumei et al. 2006 [50], Pardo et al. 2005 [51], Rucci et al. 2007 [52], Patel et al. 2007 [53], Kumei et al. 2007 [54], Capulli et al. 2009 [55], Blaber et al. 2013 [56], Rucci et al. 2015 [57], Chatani et al. 2016 [14], Hu et al. 2015 [58]). With regard to the location that the experiments took place 21 were performed in space (shuttles, International Space Station (ISS), rockets) and 39 in ground simulators [59]. There were some comparative studies that duplicated the experiments in space and ground based microgravity models. The duration of microgravity in ground simulations varied from 1-7 days for RCCS, 2D or 3D Clinostat and LG-HMF and from 2-8 weeks for HLS or HDBR where exposure in space was 1-60 days. Detailed characteristics are depicted in Table 1 and 2.
Table 1. Study characteristics for osteoblastic cell lines.
| References | Study Design | Genomic Data | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Organism | Cell Type | Type of μG | Location | Duration | |||||||||
| Hu et al. 2017 [33] | mouse | MC3T3-E1 | RWV | EARTH | 3h | Microarray(Agilent) | |||||||
| Wang 2016 et al. [22] |
mouse | MC3T3-E1 | clinorotation | EARTH | 2d | Total RNA-cDNA-qRT-PCR | |||||||
| Chatani et al. 2016 [14] | medaka fish | OB cells | ISS | SPACE | 1-8d | HiSeq | |||||||
| Sun et al. 2015 [16] | mouse | MC3T3-E1 | clinorotation | EARTH | 2d | Total RNA-cDNA-qRT-PCR | |||||||
| Sun et al. 2015 [23] | mouse | MC3T3-E1 | clinorotation | EARTH | 2d, 3d | qPCR | |||||||
| Goyden et al. 2015 [34] | mouse | MC3T3-E1 | RCCS | EARTH | 7d | Total RNA-cDNA-qRT-PCR | |||||||
| Rucci et al. 2015 [57] | human | OB cells | HDBR | EARTH | 14d | Total RNA-cDNA | |||||||
| mouse | OB cells | HLS | EARTH | 21d | Total RNA-cDNA | ||||||||
| mouse | OB cells | BOTOx(quad, soleus, gastro, Plantaris) | EARTH | 21d | Total RNA-cDNA | ||||||||
| Hu et al. 2015 [58] | rat | OB cells(femur) | HLU | EARTH | 3w | Microarray(Agilent) | |||||||
| - | prOB | 2D-RWV | EARTH | 2d | Microarray(Agilent) | ||||||||
| Hu Li et al. 2015 [24] | Mouse | MC3T3-E1 | 3D-RPM | EARTH | 1d | RT-PCR & Real time PCR | |||||||
| Guignandon 2014 et al. [40] | human | MG-63 | FOTON-M3 | SPACE | 69h | Total RNa & real time PCR | |||||||
| Dai et al. 2013 [38] | human | OSE-MG-63 | clinostat | EARTH | 2d | Total RNA-qPCR | |||||||
| Blaber et al. 2013 [56] | mouse | OB cells | STS-131 | SPACE | 15d | qRT-PCR arrays (Qiagen) | |||||||
| prOB | RWV | EARTH | 2d | qRT-PCR arrays (Qiagen) | |||||||||
| Kapitonova et al. 2013 [39] |
human | OB cells | ISS | SPACE | 10d | Total RNA-cDNA-RT PCR(Qiagen) | |||||||
| Shuang et al. 2012 [37] | mouse | femur | HLS | EARTH | 4w & 8w | qRT -PCR | |||||||
| mouse | 2T3 | RCCS | EARTH | 2d | qRT -PCR | ||||||||
| Qian et al. 2009 [36] | human | MG-63 | LG-HMF | EARTH | 1d | Microarray(Affymetrix) | |||||||
| Capulli et al. 2009 [55] | mouse | OB cells | RWV | EARTH | 5d | Microarray(Affymetrix)-real time PCR | |||||||
| Makihira et al. 2008 [21] | mouse | MC3T3-E1 | RPM | EARTH | 3d & 7d | qRT-PCR | |||||||
| Kumei et al. 2007 [54] | rat | OB cells | space shuttle | SPACE | 4d & 5d | RT-PCR | |||||||
| Bucaro et al. 2007 [25] | mouse | MC3T3-E1 | HARV(CLINOSTAT) | EARTH | 1d & 5d & 14d | RT-PCR | |||||||
| Rucci et al. 2007 [52] | mouse | OB cells | RWV | EARTH | 1d | RT-PCR | |||||||
| Patel et al. 2007 [53] | mouse | 2T3 | RWV | EARTH | 3d | Microarray(Affymetrix) | |||||||
| Hughes et al. 2006 [26] | mouse | MC3T3-E1 | STS-76 | SPACE | 24h | RT-PCR | |||||||
| Kumei et al. 2006 [50] | rat | OB cells | space shuttle | SPACE | 4d & 5d | RT-PCR | |||||||
| Pardo et al. 2005 [51] | mouse | 2T3 | RPM | EARTH | 3d | Microarray(Amersham) | |||||||
| Bucaro et al. 2004 [27] | mouse | MC3T3-E1 | RCCS | EARTH | 5d | RT-PCR | |||||||
| Kumei et al. 2004 [48] | rat | OB cells | space shuttle | SPACE | 4d & 5d | RT-PCR | |||||||
| Saito et al. 2003 [28] | mouse | MC3T3-E1 | clinostat | EARTH | 3d | RT-PCR | |||||||
| Nakamura et al. 2003 [47] | human | OB cells | clinostat | EARTH | 12-24-48-96h | RT-PCR | |||||||
| Kumei et al. 2003 [46] | rat | OB cells | space flight | SPACE | 4d & 5d | qRT-PCR | |||||||
| Kumei et al. 2003 [45] | rat | OB cells | space flight | SPACE | 4d & 5d | qRT-PCR | |||||||
| Ontiveros and McCabe 2003 [29] | mouse | MC3T3-E1 | RWV | EARTH | 1d | Total RNA-Real time PCR | |||||||
| Kumei et al. 2002 [60] | rat | OB cells | space flight | SPACE | 5d +6d | qRT-PCR | |||||||
| Rucci et al. 2002 [44] | rat | ROS.SMER# 14 | RWV | EARTH | 2d | PT-PCR | |||||||
| Landis et al. 2000 [43] | chicken calvaria | OB cells | STS-59 | SPACE | 3d & 5d | Total RNA-cDNA(northern blot) | |||||||
| Sato et al. 1999 [31] | rat | MC3T3-E1/ HeLa cells | clinostat | EARTH | 2d | NORTHERN BLOT | |||||||
| - | TR-1A6 (rocket) | SPACE | 6min | RT-PCR | |||||||||
| Hughes et al. 1998 [32] | rat | MC3T3-E1 | STS-76 | SPACE | 29h | RT-PCR | |||||||
| Carmeliet et al. 1997 [42] | human | MG-63 | FOTON-10 | SPACE | 9d | RT-PCR, Northern blot | |||||||
| Kumei et al. 1996 [41] | rat | OB cells | STS-65 | SPACE | 5d | qRT-PCR | |||||||
| Bikle et al. 1994 [35] | rat | OB cells | HLS | EARTH | 2w | Northern blot | |||||||
| - | OB cells(tibia) | STS-54 | SPACE | 6d | Northern blot | ||||||||
(RWV: Rotating Wall Vessel, HARV: High Aspect Rotating Vessel, HLS: Hind-Limb Suspension, ISS: International Space Station, RCCS: Rotary Cell Culture System, HDBR: Head Down Bed Rest, LG-HMF: Large Gradient High Magnetic Field, RPM: Random Positioning Machine, OB: Osteoblast, RT-PCR: Reverse Transcription Polymerase Chain Reaction, HiSeq DNA: High Sequencing DNA, HLU: Hind-Limb Unloading, 2D: two Dimension, 3D: three Dimension, d: Days, h: Hours, w: Weeks, min: Minutes, qRT-PCR: Quantative: RT-PCR, cDNA: Complementary DNA).
Table 2. Study characteristics for osteoclastic cell lines.
| References | Study Design | Genomic Data | ||||
|---|---|---|---|---|---|---|
| Organism | Cell Type | Type of μG | Location | Duration | ||
| Ethiraj et al. 2018 [11] | mouse | RAW264.7 | RCCS | EARTH | 24h | RT-PCR |
| Shanmugarajan et al. 2017 [12] | mouse | RAW264.7 | RWV | EARTH | 5d | RT-PCR |
| Sambandam et al. 2016 [13] | mouse | RAW264.7 | RCCS | EARTH | 24h | RT-PCR |
| Chatani et al. 2016 [14] | medaka fish | OSC cells | ISS | SPACE | 2d | HiSeq |
| Chatani et al. 2015 [15] | medaka fish | OSC cells | ISS | SPACE | 60d | Whole transcriptome analysis |
| Yu-Long et al. 2015 [16] | mouse | RAW264.7 | LG-HMF | EARTH | 48h | RT-PCR |
| Sambandam et al. 2014 [19] | mouse | RAW264.7 | RCCS | EARTH | 24h | RT-PCR |
| Saxena et al. 2011 [17] | mouse | RAW264.7 | RCCS | EARTH | 24h | RT-PCR |
| Sambandam et al. 2010 [18] | mouse | RAW264.7 | RCCS | EARTH | 24h | Microarray(Agilent) |
| Tamma et al. 2009 [20] | mouse | (marrow macrophages) OSTEO | FOTON -M3 | SPACE | 10d | RT-PCR |
| mouse | PITS (mature OSC) | - | SPACE | 4d | RT-PCR | |
| Makihira et al. 2008 [21] | mouse | RAW264.7 | RPM | EARTH | 7d | RT-PCR |
(RCCS: Rotary Cell Culture System, RWV: Rotating Wall Vessel, ISS: International Space Station, LG-HMF: Large Gradient High Magnetic Field, HARV: High Aspect Rotating Vessel, HLS: Hind-Limb Suspension, RPM: Random Positioning Machine, OB: Osteoblast, RT-PCR: Reverse Transcription Polymerase Chain Reaction, d: days, h: hours, HiSeq: High Sequencing).
3.2. Microgravity’s Effect on Osteoblast Differentiation and Matrix Mineralization
Twenty of 50 studies included genomic data on the expression of marker genes involved in the process of osteoblastic cell differentiation and matrix mineralization. Runt related transcription factor 1 (Runx2) and Osterix (Ostx) are key factors that are implicated early in the maturation of osteoblasts. Runx2 was downregulated in 8 studies when no difference in expression levels was detected in 2. Furthermore, one study showed a gradual increase in expression from the 3rd day, but overall levels were diminished compared to ground controls. Ostx was also downregulated in one study (minimum on 2nd day) whereas in another that had a 7-day duration a gradual increase was noted up to the 7th day. Interestingly high levels were detected throughout the duration of a study performed in space, that included living organisms but with less statistical significance. RANKL was found upregulated early when exposed to μG, while OPG showed a gradual and slower increase. Maximum expression of RANKL was reached on the 2nd day and osteoprotegerine (OPG) on the 7th day but the results were not significant compared to ground controls. Notably when the ratio of RANKL/OPG was calculated, it reached significant levels, which was a common finding in all three studies. Alkaline phosphatase (ALP) and collagen I (CollI) mRNA levels manifested a somewhat diversity. Four short-duration earth studies show downregulation of ALP when five others, conducted on earth simulators as well as in space, conclude that ALP is upregulated but in lower levels than on earth gravity. One study that lasted a day showed no difference in ALP expression. CollI is expressed in a time-dependent manner with maximum levels on the 7th day of exposure. Osteocalcin (OCN), a protein implicated in matrix mineralization was upregulated in four studies, showed no difference in two and was downregulated in four. Details are depicted in Table 3.
Table 3. Functional annotations of genes reported with respect to osteoblastogenesis (OBgenesis), osteoblast (OBS) differentiation maturation and osteoblast mineralization.
| Function | OBgenesis | OBS Differentiation-maturation | OBS Mineralization | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | RunX2 | Osterix | ALP | RANKL | OPG | ColIaI | OCN | Pthr1r | Omd | OP | ON |
| Studies | |||||||||||
| Makihira et al. 2008 [21] | U (max d3) | U (max 7d) |
- | U (d1) D (d3+d7) |
U (d7 little) | U (steady) | - | - | - | - | - |
| Bucaro et al. 2007 [25] | (ND) | - | D | - | - | (ND) | (ND)- | - | - | (ND) | - |
| Bucaro et al. 2004 [27] | D | - | D | - | - | D | D | - | - | - | - |
| Ontiveros and McCabe 2003 [29] | D | - | D | - | - | - | D | - | - | - | - |
| Hu et al. 2017 [33] | D (until d3) | D (max d2) |
D (max d2) |
- | - | - | - | - | - | - | - |
| Shuang et al. 2012 [37] | D | - | D | - | - | - | D | - | - | - | - |
| Pardo et al. 2005 [51] | D | - | D | - | - | - | - | D | D | - | - |
| Patel et al. 2007 [53] | D | - | D | - | - | - | - | D | D | - | - |
| Capuli et al. 2009 [55] | D | - | - | U | D | - | - | - | - | - | - |
| Bikle et al. 1994 [35] | - | - | U (max2d) | - | - | - | D (max5d) | - | - | - | - |
| Kapitonova et al. 2013 [39] | - | - | U | - | - | U (NS) | U | - | - | NS | NS |
| Carmeliet et al. 1997 [42] | - | - | D | - | - | (D) | D min answer to stimulus. | - | - | - | - |
| Rucci et al. 2002 [44] | - | - | U | - | - | - | U | - | - | U | NA |
| Rucci et al. 2007 [52] | NA | NA | NA | U | D | NA | NA | - | - | NA | - |
| Landis et al. 2000 [43] | - | - | - | - | - | D | D | - | - | - | - |
| Dai et al. 2013 [38] | D (+after BMP2 stimulus) | - | - | - | - | - | - | - | - | - | - |
| Saito et al. 2003 [28] | - | - | - | - | - | D | - | - | - | - | - |
| Chatani et al. 2016 [14] | - | U | - | - | - | U | U | - | - | - | - |
| Hughes et al. 2006 [26] | - | - | - | - | - | - | D | - | - | - | - |
| Hughes et al. 1998 [32] | - | - | - | - | - | - | U | - | - | - | - |
| Kumei et al. 2006 [50] | - | - | - | - | - | - | - | - | - | D | U (little) |
(d: day, NS: Not Significant, ND: No Difference, NA: Not Affected, max: Maximum, min: Minimum, D: Down, U: Up, Omd: Osteomodulin, OP: Osteopontin, ON: Osteonectin, Pth1r: Parathyroid Hormone Receptor 1, CollaI: Collagen Ia, ALP: Alkaline Phospahtase, OPG: Osteoprotegerine, RANKL: Receptor Activator of NF-κB Ligant, OCN: Osteocalcin).
3.3. Microgravity’s Effect on Osteoblast Cytoskeleton, Growth, Proliferation and Apoptosis
A great number of proteins and molecules are involved in osteoblast mechanosensation and cell survival metabolic pathways. The RNA expression of cytoskeleton related genes was found increased for tallin, paxillin, supervillin, Wiskott-Aldrich Syndrome Protein Family Member 2 (ASF2), WAS Interacting Protein family member 1(WIPF1) while actin, a-tubulin and heat shock protein 73(HSP73) were decreased (Table 4). Apoptotic molecules like Bax and p53 showed upregulation that was countered by a concomitant increase in the anti-apoptotic Bcl, p21 and X-linked inhibitor of apoptosis(XIAP). Transcription factors c-fos and c-jun, that respond early in stressful stimuli and convert extracellular signals into changes in gene expression in the nucleus, where upregulated (Table 5). A number of cytokines and growth factors produced by osteoblast contribute to regulating bone homeostasis. Prostaglandin E2 (PGE-2) which has a positive effect on bone formation was upregulated in μG. Interestingly cyclooxygenase 2 (cox2), an enzyme that normally increases PGE-2 production by a feedback mechanism, was found downregulated. Interleukin-6 (IL-6) that supports and enhances osteoclastic activity was upregulated in three studies and downregulated in one (Table 6).
Table 4. Expression of cytoskeletal genes.
| - | Cytoskeleton | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SPTBN1 | WASF2 | WIPF1 | Supervillin | Destrin | Tallin | Paxillin | Vimentin | Actin | alpha-tubulin | fibronectin | HSP73 | VEGF (+) | |
| Dai et al. 2013 [38] | - | - | - | - | - | - | - | - | D | - | - | - | - |
| Qian et al. 2009 [36] |
U | U | U | U | - | U | U | - | - | - | - | - | - |
| Shuang et al. 2012 [37] | - | - | - | - | U (max 8d) |
- | - | - | - | - | - | - | - |
| Guignandon et al. 2014 [40] |
- | - | - | - | - | - | - | - | - | - | - | - | D |
| Kumei et al. 2003 [46] | - | - | - | - | - | - | - | - | - | - | - | D | - |
| Kumei et al. 2006 [50] | - | - | - | - | - | - | - | - | - | D (5d) | - | - | - |
(U: Up, D: Down, d: Days, max: Maximum).
Table 5. Expression of apoptosis- and proliferation-related genes.
| Function | Apoptosis |
Proliferation
Apoptosis |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | Bax(+)/ Bcl2(-) |
P53(+)/ p21(-) |
ERK ½ (-) | iNOS(+)/GTPCH | Caspase8 | Caspase3 | XIAp (-) |
HSP70 (-) |
PAF-R(-) | CD44 (-) |
Akt(-) | c-fos | c-jun | c-myc | MAPK |
| Buccaro et al. 2007 [25] | ND | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Hughes et al. 2006 [26] | D/D | - | - | - | - | - | - | - | - | - | - | - | - | D | - |
| Rucci et al. 2002 [44] | ND | ND | - | - | ND | - | - | - | - | - | - | - | - | - | - |
| Rucci et al. 2007 [52] | - | - | U | - | - | - | - | - | - | - | - | - | - | - | - |
| Nakamura et al. 2003 [47] |
U/U | - | - | - | ND | ND | U | - | - | - | - | - | - | - | - |
| Blaber et al. 2013 [56] | - | D/U | - | - | - | - | - | - | - | - | ND | D | - | - | D |
| Kumei et al. 2003 [45, 46] | - | - | - | U/U | - | - | - | D | U | - | - | - | - | - | - |
| Sato et al. 1999 [31] | - | - | - | - | - | - | - | - | - | - | - | D | ND | - | ND |
| Chatani et al. 2016 [14] |
- | - | - | - | - | - | - | - | - | - | - | U | U | - | - |
| Kumei et al. 2007 [54] | - | - | U | - | - | - | - | - | - | - | - | - | - | - | U |
| Kumei et al. 2006 [50] | - | - | U | - | - | - | - | - | U | U | - | - | - | - | U |
| Buccaro et al. 2004 [27] |
D | - | - | - | - | - | - | - | - | - | D | - | - | - | - |
(ND: No Difference, U: Up, D: Down).
Table 6. Expression of cytokine- and morphogenesis-related genes.
| Cytokines |
Morphogenic
Proteins |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | IL-6 | Lcn2 | MMP10 | Cox 2 | Cox 1 | PGE-2 | BMP4 | BMP2 | |||||||
| Blaber et al. 2013 [56] | - | - | U | - | - | - | - | - | |||||||
| Hughes et al. 2006 [26] | - | - | - | D | - | U | - | - | |||||||
| Kumei et al. 1996 [41] | U | - | - | - | - | U | - | - | |||||||
| Rucci et al. 2002 [44] | U | - | - | - | - | - | U | ND | |||||||
| Capulli et al. 2009 [55] | U (max 5d) |
U (max 5d) |
- | - | - | - | - | - | |||||||
| Rucci et al. 2015 [57] | - | U (12d) In humans |
- | - | - | - | - | - | |||||||
| Rucci et al. 2015 [57] | - | U | - | - | - | - | - | - | |||||||
| Patel et al. 2007 [53] | - | - | - | - | - | - | D | - | |||||||
| Saito et al. 2003 [28] | - | - | - | - | - | - | - | - | |||||||
| Kumei et al. 2007 [54] | - | - | - | - | - | U | - | - | |||||||
| Kumei et al. 2003 [46] | - | - | - | - | - | - | - | - | |||||||
| Kapitonova et al. 2013 [39] | D | - | - | - | - | - | - | - | |||||||
| Hughes et al. 1998 [32] | - | - | D | D | ND | - | - | - | |||||||
| Pardo et al. 2005 [51] | - | - | - | - | - | - | D | - | |||||||
(d: Days, ND: No Difference, U: Up, D: Down, IL: Interleukin, BMP: Bone Morphogenetic Protein, Cox: Cyclooxigenase, Lcn2: Lipocalin 2).
3.4. Microgravity’s Effect on Osteoclasts
The compilation of eleven studies concerning osteoclast genomic activity in microgravity conditions revealed upregulation in genes involved in differentiation, maturation and metabolic function. Specifically c-fos, jun-b-like and ddit4, that are involved in nuclear and mitochondrial signaling, where overexpressed as were fusion and proliferation molecules like syncytin-A, dendrocyte expressed transmembrane protein(DCSTAMP), osteoclast stimulator transmembrane protein(OCSTAMP), TNF receptor associated factor(TRAF) and TNF related apoptosis-inducing factor(TRAIL). Pre-osteoclastic cells showed increased expression of TRAP, cathepsin K and matrix metalloproteinases (MMPs) indicating osteoclast maturation. Adhesion protein β-integrin and receptor activator of NF-κB (RANK) levels were increased in response to RANKL, as well as autophagy proteins Atg5 and LC3 independent of RANKL (Table 7).
Table 7. Expression of cytokine- and morphogenesis-related genes.
| Function |
Study
Gene (Protein) |
Ethiraj
2018 [ 11 ] |
Sanmugarajan 2017 [ 12 ] |
Sambandam
2016 [ 13 ] |
Chatani
2016 [ 14 ] |
Chatani
2015 [ 15 ] |
Yu-Long 2015 [ 16 ] | Saxena 2011 [ 17 ] |
Sambandam
2010 [ 18 ] |
Tamma
2009 [ 20 ] |
Makihira
2008 [ 21 ] |
Sambandam
2014 [ 19 ] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Osteoclast Markers | RANK | - | - | - | - | - | - | U (with RANKL) | - | - | - | - |
| TRAP | - | U | - | U | - | Suppr | - | - | U | - | - | |
| MMP-9 | - | - | - | U | - | U | - | U | U | - | - | |
| Cathepsin-K | - | - | - | U | - | Suppr | U | U | U | - | - | |
| CaIR | - | - | - | - | - | - | U | - | U | - | - | |
| Fusion- Differentiation-Proliferation | Syncytin-A | U | - | - | - | - | - | - | - | - | - | - |
| Syncytin-B | ND | - | - | - | - | - | - | - | - | - | - | |
| Dcstamp | - | U | U | - | - | - | - | - | - | U | - | |
| Ocstamp | - | U | U | - | - | - | - | - | - | - | - | |
| CCN2/CTGF | - | U | - | - | - | - | - | - | - | - | - | |
| TRAF6 | - | - | U | - | - | - | - | - | - | - | - | |
| TRAIL (TNF related) | - | - | U | - | - | - | - | - | - | - | - | |
| Transcription Factors | Runx2 | - | - | - | - | - | Suppr | - | - | - | - | - |
| NFATc1 | - | - | - | - | - | U | U(without RANKL) | - | - | - | - | |
| Pcreb | - | - | - | - | - | - | - | U | - | - | - | |
| Adhesion | Podoplanin | - | - | - | - | - | - | - | U | - | - | - |
| Β-integrin | - | - | - | - | - | - | - | U | U | U (without RANKL) Down (with RANKL) |
- | |
| Signal Transduction |
S100A8 | - | - | - | - | - | - | - | U | - | - | - |
| Nucleus & mitochondrial Signaling |
c-fos | - | - | - | U | - | - | - | - | - | - | - |
| Jun-b-like | - | - | - | U | - | - | - | - | - | - | - | |
| Pai-1 | - | - | - | U | - | - | - | - | - | - | - | |
| ddit4 | - | - | - | U | U | - | - | - | - | - | - | |
| Fkbp5 | - | - | - | - | U | - | - | - | - | - | - | |
| Tsc22d3 | - | - | - | U | - | - | - | - | - | - | - | |
| Autophagy | LC3 | - | - | - | - | - | - | - | - | - | - | U |
| Atg5 | - | - | - | - | - | - | - | - | - | - | U |
(Suppr: Suppressed, ND: No Difference, U: Up).
4. DISCUSSION
Bone regulation is determined by the synergic action of specific cells that are the osteoblasts and osteoclasts. The coupled functions of these cells, which respond to environmental stimuli, determine bone production and resorption [60, 61]. In conditions of decreased gravity, the change in the environment which the aforementioned cells were normally adept, cause alterations in gene expression and protein production in an effort to accommodate for the new conditions, and results in loss of bone mass during space flights [1, 5, 35]. The mechanism of this phenomenon, that is not yet fully elucidated, involves alterations in the process of genesis, differentiation, proliferation and maturation of osteoblasts and osteoclasts.
Most research has focused on the effect of μG on osteoblasts and not as many studies have been performed that are addressing the osteoclasts. In order to understand the cellular processes that induce osteopenia, when there is exposure to microgravity, experimentation in ground based facilities as well as in space have been conducted. Different devices that employ different principles, have been used to simulate microgravity, like magnetic levitation using high magnetic field, hind limb suspension, rotation around one or two axis and head down bed rest. Unfortunately due to the high cost of launching a space shuttle, experiments
conducted in real μG are limited and sparse. The duration of exposure to μG among studies varies from 1 day to 8 weeks and in most cases cell cultures are used, with less studies being conducted in living organisms. Other concomitant factors that can affect as well the response to μG, are the hypergravity induced at launch and cosmic radiation exposure.
4.1. Microgravity and Osteoblasts
Multipotential mesenchymal cells differentiate into osteoblastic lineage cells under the effect of transcription factors runx2 and ostx. Condensed progenitor osteochondrocytes divert to pre-osteoblasts that express high level of runx2 and under the effect of osx mature to functional osteoblastic cells that produce specific marker proteins ALP, CollI, OCN of the osteoblastic cell line [62]. All ground based and space studies agree that there is a downregulation of runx2 when pre-osteoblastic cells are exposed to microgravity conditions. Runx2 is expressed by pre-osteoblastic cells exposed to μG, yet there is a delay of onset in the production as shown by Makihira et al. [21], suggesting that cell differentiation is a slower but ongoing process. In the process of maturation, ostx’s input is important for creating functional osteoblastic cells. Both in real and simulated conditions of μG, expression of the later was detected with a time depended rhythm, but overall levels had less statistical importance. Thus, the results indicate that differentiation is inhibited initially but recovers in the long run. Osteoblast-osteoclast interaction is mediated by the production of soluble and intermembrane RANKL by the osteoblasts. RANKL fuses with RANK receptor on the surface of the osteoclasts and therefore activating them. The action of RANKL is only nullified by connecting with OPG, a protein that is also produced by the osteoblasts [60]. In microgravity RANKL levels show upregulation early when exposed (day 1), while OPG follows a slow pattern of progressive production lower than in normal gravity. Even though the levels of RANKL and OPG are not altered in significant degree, the overall ratio of RANKL/OPG reaches significance. The early upregulation of RANKL and the overall downregulation of OPG, can be interpreted as a cellular response to the sudden change in gravitational conditions, diverting homeostasis towards bone resorption. Maturated osteoblasts are capable of producing large quantities of ALP, ColI and OCN so as to form bone matrix which in turn will be mineralized. This process is inhibited in μG, as studies indicate downregulation of ALP and ColI, as well as OCN, indicating an inhibition in the maturation of osteoblasts. Interestingly there are also reports of OCN upregulation as early as the 2nd day of exposure [44]. This can be explained if we consider that high levels of OCN create an anionic environment, which is necessary for osteoclastic activity [39]. Furthermore, the endocrine role of OCN is related to glucose regulation, as it increases insulin release and targets cell sensitivity towards insulin [63]. Even though it appears that μG does not affect the production of insulin growth factor (IGF), cells develop insulin resistance. The increased OCN as well as insulin growth factor receptor(IGF-R) levels can be explained as an effort to counteract the former phenomenon [35].
Osteoblasts are mechanosensitive cells that respond to gravitational alterations. Extracellular signals are perceived by transmembrane molecules and converted into intracellular signals, by a complex mechanism, causing changes in gene expression in the nucleus. Proteins of the cytoskeleton as well as cytokines and growth factors contribute significantly to this intricate metabolic pathway, via signal transduction. Growth factors and cytokines attach to their intermembrane receptors thus activating intracellular responses [26, 64]. Proteins of the cytoskeleton talin, paxillin, supervillin, WASF2, WIPF1 were upregulated when exposed to altered gravitational conditions 0g versus 2g, via large gradient diamagnetic field for 24 hours [36]. WASF2, which is a downstream effector of cell division cycle 42(Cdc42) that is implicated in actin polymerization and cytoskeletal organization as well as WIPF1that encodes a protein responsible for actin polymerization [65], were more sensitive in alterations of gravity than the magnetic field [36]. Kumei et al., 2006 found no alteration in actin or β1-integrin expression levels in rat osteoblasts cultured for 4-5days in space. In the same
study osteopontin levels decreased while CD44 increased. Integrins and CD44 are transmembrane adhesion molecules that are interconnected with actin filament, and the interaction between osteopontin (extracellular) and CD44 is mediated by integrins [66]. Disruption of the actin cytoskeleton alters cellular response to morfogenetic proteins. Dai et al. 2013 demonstrated that BMP2 proliferative effect is inhibited when actin microfilament is disrupted [38], which supports the mechanosensation role of the cytoskeleton. Changes in gravity perceived by proteins of the cytoskeleton alter cellular response to the effect of morphogenic proteins, such as BMP2, thus inhibiting osteoblast differentiation [38].
Interactions between osteoblasts and osteoclast mediated by soluble molecules such as cytokines contribute to bone homeostasis and regulation of bone production and resorption. IL-6 is a pleiotropic cytokine that is implicated in the regulation of bone turnover, by regulating osteoblast and osteoclast differentiation and function [67]. It is produced by T lymphocytes, monocytes, osteoblast/stromal cells, fibroblasts, synovial cells and cancer cells and consist of a family of 10 factors, IL6, IL11, IL27, IL31, Leukemia Inhibitory Factor (LIF), Oncostatin M (OSM), Ciliary Neurotrophic Factor (CNTF), Cardiotrophin-1 (CT-1), Cardiorophin Like Cytokine (CLC) and Neuropoietin (NP) [68-72]. IL-6 binds to a specific subunit IL-6Ra, which is either membrane or soluble (sIL-6Ra) and that complex interacts with two gp-130 molecules and forms a hexameric complex [73, 74]. In bone, soluble IL-6R is required for IL-6 effects on osteoclast development and osteoblast function but little is known of its origin. IL-6 might be produced by other cell types, like liver cells or by osteoblasts [75, 76]. All IL-6 cytokines use the transducing receptor β-subunit gp 130, which further activate Janus Protein-Tyrosin Kinases (JAKs). In turn this allows the activation of the Signal Transducer and Activator of Transcription (STATs) or of the MAPKs [77]. Activation of STAT3 is necessary for osteoblast differentiation and bone formation induced by IL-6, but it can also promote the expression of cell cycle inhibitor p21, which has shown to confer resistance to apoptosis in osteoblastic cells [78, 79]. On the other hand other in vitro reports have shown that IL-6 complex has inhibitory effects on bone formation via extracellular signal regulated kinase 1/2(ERK1/2) and PKCδ kinases (MAPKs pathway). Furthermore the transcription factor STAT5a/b and p53 act in synergy to enhance bax/bcl ration and sensitize osteoblastic cells to apoptosis [80, 81]. These dual effects of IL-6 could depend on the differentiation stage of osteoblasts, but considering that osteocytes, the final form of osteoblasts, are characterized by decreased production of osteoblastic marker proteins and high apoptotic levels, it is possible that IL-6 complex contributes throughout the osteoblastic cell life cycle [82, 83]. IL-6 role in osteoclast differentiation is performed by increasing interactions between osteoblasts and osteoclasts. The effect of IL-6 complex induces production of RANKL, PGE2, IL-1 by osteoblasts inducing osteoclast differentiation. Recent data suggest that STAT3 has a key role in RANKL production by the osteoblasts, under the effect of IL-6 [84-86]. In contrast, other studies report that IL-6 has inhibitory effects on osteoclast formation by diverting cells into the macrophage lineage. That action is mediated by inhibiting RANKL pathways through activation of NF-κB and MAPKs [86, 87]. It appears that the activity of the receptor complex of IL-6 is the result of the antagonistic action of STAT versus MAPK pathways, where STAT exerts apoptotic and anti -proliferative effects on osteoblast and osteoclasts and MAPK has mitogenic and anti-apoptotic [68, 70, 86]. In studies of simulated or real μG, IL-6 was found upregulated in three and downregulated in one. All experiments were in vitro cell cultures of the osteoblast lineage and it was interesting that down regulation of IL-6 was noted in real μG in space, while upregulation in simulated conditions. The complex actions of this cytokine as well as the fact that it can be produced by various cell types and contribute to a great number of biologic processes, precludes the necessity of in vivo study designs, in order to elucidate the whole spectrum of IL-6 complex effects on bone metabolism in microgravity environment.
Cellular response to stress induced by μG, leads to changes in the expression of apoptotic and anti-apoptotic genes. Concerning cell survival, studies concluded that microgravity does not directly induce apoptosis. In in vitro and in vivo experiments under real or simulated μG, expression of apoptosis- related genes and anti-apoptotic genes were concomitantly downregulated or showed no difference in expression with statistical importance [25, 27, 44, 46, 56]. Interestingly osteoblastic cells that were exposed to μG, became sensitized to apoptotic agents like staurosporin [27, 56] but no sensitivity was detected to the apoptotic agent sodium nitroprusside [25]. Bax mitochondrial apoptotic gene and its counterpart Bcl were increased concomitantly in one study [47], while under real microgravity on board the space shuttle, downregulation was noted which was normalized in 1g [26]. The aforementioned genes are expressed and regulate mitochondrial function. Bax releases cytochrome c and induces apoptosis through p53 tumor suppressor gene, while Bcl-2 blocks the apoptosis mechanism by stabilizing mitochondrial membrane, thus inhibiting the release of cytochrome c [88, 89]. Several anabolic signals are gravity dependent and can be downregulated in microgravity. The MAPK pathway is the central controlling point for signal transduction from the extracellular environment to the nucleus of osteoblasts [90]. The extracellular signal-regulated kinase (ERK)-mitogen-activated protein kinase (MAPK) pathway provides a major link between the cell surface and nucleus to control proliferation and differentiation [61]. Mechanical stress regulates Runx2 activation and favors osteoblast differentiation through the activation of MAPK signal transduction pathways and Ras/Raf‐dependent ERK1/2 activation, but is independent of p38 MAPK signaling [91]. Apoptosis of osteoblast mouse cells did not increase after 24 hours of clinorotation but it was increased after the introduction of MEK inhibitor PD98059, suggesting a protective role of ERK1/2 [52]. MAPK activity is induced by gravity and causes upregulation of c-fos within 30 minutes of stress. This activation is inhibited by MEK kinase inhibitor but not a p38 inhibitor. Blocking the MAPK signaling pathway and MEK1 also promotes differentiation of MC3T3-E1 pre-osteoblastic cells to more mature forms, that produce ALP and collagen I, suggesting that this pathway may influence the differentiation process in osteoblasts [92]. ERK ½ phosphorylation causes translocation of ERK to the nucleus and NF-κB mediated c-fos gene activation [90]. Members of the Fos, Jun and ATF family of proteins form the complex of transcription factors AP-1 that are implicated in bone regulation. Various members of the AP-1 complex are differentially expressed during osteoblast maturation, with high levels of c-fos and a-jun being expressed in vitro during osteoblast maturation. Chimeric mice obtained from c-Fos overexpressing embryonic stem cells develop chondrogenic tumors implying a function of c-Fos in chondrogenesis in vivo [93]. Furthermore, C-fos knock out mice showed an osteopetrotic phenotype with a shift of osteoclasts to bone marrow macrophages [93, 94]. C-fos expression is more marked during the proliferation phase of osteoblasts, and diminishes during mineralization. Over- or under- expression of this transcription factor is consistent with abnormalities in bone development [95]. In space conducted in vivo studies c-fos was upregulated early (2 days) [14], as well as ERK ½ in real or simulated μG experiments, while p38 and JNK expression did not significantly change [52, 54]. On the other hand, EGF(epidermal growth factor) -induced c-fos expression was restrained about 30% compared to ground controls, after short-term simulated microgravity and rocket flight, when no difference in MAPK phosphorylation was noted [31]. The potential role of the intricate metabolic pathway of MAPKs, in bone regulation under stressful conditions such as microgravity is far from being elucidated. Yet it is clear that it plays an important part in moderating osteoblast cell cycle, differentiation, proliferation and maturation processes.
4.2. Microgravity and Osteoclasts
In contrast to the plethora of experimental data concerning the effect of microgravity in cells of the osteoblastic lineage, osteoclast data is limited. Few short-term studies (8 in total) of simulated μG and two performed in real μG (Table 2) have provided insight to the changes in osteoclast expression in weightlessness conditions. Early exposure to μG induces expression of nucleus genes c-jun, c-fos, jun-B-like as well as proteins of the MAPK pathway (ERK, p38, JNK) which under the effect of RAKL result in the formation of gigantic multinucleated osteoclast cells [14, 17, 18]. These cells express high level of osteoclast marker proteins TRAP, MMP9, cathepsin K and Calcium receptor. Fusion process in premature osteoclasts is a complex process that still remains unclear. It has been suggested that osteoclasts chose their partners selectively according to their maturation stage and organization of fusion factors [96]. Mensah et al. [97] supported that fusion is determined by the presence or absence of DCSTAMP on extracellular membrane, while in another report syncytin-1 formed a concentrated pattern in the area facing the fusion cell [98]. It is possible that osteoclasts do not randomly choose their partner but this process is based on selectivity among a heterogeneous population based on complementarity and maturation stage [98, 99]. In different stages of osteoclast nuclearity, different fusion proteins are the prime orchestrators. Syncytin-1 promotes fusion of multi-nucleated cells and reduces the number of fusions between mono-nucleated cells of pre-osteoclasts [99]. In microgravity studies the levels of adhesion proteins participating in cell fusion DCSTAMP and OCSTAMP were increased. Microgravity was the main contributor rather than radiation and under the effect of RANKL cell fusion was noted as early as 24 hours after exposure. This resulted in the formation of mature osteoclasts presenting larger numbers of nucleuses [12]. Β-integrin’s action was also RANKL dependent and was enhanced in microgravity [21]. Another contributor to the process of osteoclast fusion is syncytin-A, which was also upregulated in μG, independent of RANKL (Table 7). When syncytin-A was blocked, cell population expressing TRAP protein was diminished and mechanism of autophagy was negatively influenced even at the presence of RANKL [11]. Interestingly OCSTAMP and DCSTAMP were found upregulated through another pathway independent of RANKL. Sambandam et al. [13], noted that through the inflammatory pathway of TNF family the upregulation of TRAIL is able to increase OS and DC STAMP levels independent of RANKL. This family of inflammatory proteins may also participate in autophagy mechanisms. Autophagy is a cellular self- consumption process that is involved in cell survival, nutrient supplementation under starvation, antigen presentation and defense against harmful agents [100]. In osteoclasts autophagy proteins (Atgs/LC3) regulate the formation of autophagosomes. In addition, the same proteins regulate the secretory lysosomes that are directed towards the ruffled area of the osteoclasts where osteoid degradation takes place [100]. Disruption of autophagy was shown to delocalize cathepsin K and reduce bone resorption [101]. Tnfs10 mRNA levels were markedly elevated in RAW 264.7 cells that were cultured for 24 hours in clinorotation. Recent studies have shown implication of TNF inflammatory cytokines in the induction of autophagy mechanisms [102] In the same study Atg5 and LC3 autophagy markers were upregulated without the effect of RANKL. The introduction of 3-MA, an autophagy inhibitor, reduced cathepsin K levels, Atg5 and LC3, therefore debilitating osteoclastic activity, without affecting their viability [19].
CONCLUSION
Our understanding of microgravity’s effect on bone metabolism has increased, with the accumulation of data from many experiments, in real or simulated conditions. Osteoblasts appear as the main orchestrators of bone metabolism in microgravity and the intricate metabolic pathway of MAPK seems to play protagonistic role in microgravity induced alterations in the osteoblastic lineage cells. Osteoclasts become sensitized in microgravity and their activity is augmented by RANKL produced by the osteoblasts as well as through inflammatory cytokines. Still we are far from fully elucidating the mechanism that causes space osteopenia. We are now at a turning point for humanity’s future regarding space exploration. Advances in robotics and biotechnology offer possibilities of performing studies in space that could improve life on Earth. Unraveling the pathophysiologic mechanisms that cause μG-induced changes in the skeletal, immune, cardiovascular and other systems of human organism may lead to the development of more efficient therapies for a variety of diseases. Furthermore, the dream of reaching to distant parts of our galaxy can become a possibility, only after the great conundrum of space-induced osteopenia is solved.
ACKNOWLEDGEMENTS
Declared none.
LIST OF ABBREVIATIONS
- ALP
Alkaline Phosphatase
- HARV
High Aspect Rotating Vessel
- HDBR
Head Down Bed Rest
- HiSeq DNA
High Sequencing DNA
- HLS
Hind-Limb Suspension
- HLU
Hind-Limb Unloading
- IL
Interleukin
- ISS
International Space Station
- LG-HMF
Large Gradient-High Magnetic Field
- NGS
Next Generation Sequencing
- OB
Osteoblast
- OCN
Osteocalcin
- OPG
Osteoprotegerin (TNF receptor superfamily member 11b)
- RANKL
TNF superfamily member 11
- RCCS
Rotary Cell Culture System
- RPM
Random Positioning Machine
- RT-PCR
Reverse Transcription Polymerase Chain Reaction
- RWV
Rotating Wall Vessel
- μG
Microgravity
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Carmeliet G., Bouillon R. The effect of microgravity on morphology and gene expression of osteoblasts in vitro. FASEB J. 1999;13(Suppl. 8):S129–S134. doi: 10.1096/fasebj.13.9001.s129. [DOI] [PubMed] [Google Scholar]
- 2.Hughes-Fulford M. Physiological effects of microgravity on osteoblast morphology and cell biology. Adv. Space Biol. Med. 2002;8:129–157. doi: 10.1016/s1569-2574(02)08017-6. [DOI] [PubMed] [Google Scholar]
- 3.Vico L., Collet P., Guignandon A., Lafage-Proust M.H., Thomas T., Rehaillia M., Alexandre C. Effects of long-term microgravity exposure on cancellous and cortical weight-bearing bones of cosmonauts. Lancet. 2000;355(9215):1607–1611. doi: 10.1016/s0140-6736(00)02217-0. [DOI] [PubMed] [Google Scholar]
- 4.Rutkovskiy A., Stenslokken K.O., Vaage I.J. Osteoblast differentiation at a glance. Med. Sci. Monit. Basic Res. 2016;22:95–106. doi: 10.12659/MSMBR.901142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clement J.Q. Innovations in biotechnology. Croatia: InTech; 2012. Gene expression microarrays in microgravity research: toward the identification of major space genes. pp. 321–348. [Google Scholar]
- 6.Trevino V., Falciani F., Barrera-Saldana H.A. DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol. Med. 2007;13(9-10):527–541. doi: 10.2119/2006-00107.Trevino. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herranz R., Anken R., Boonstra J., Braun M., Christianen P.C., de Geest M., Hauslage J., Hilbig R., Hill R.J., Lebert M., Medina F.J., Vagt N., Ullrich O., van Loon J.J., Hemmersbach R. Ground-based facilities for simulation of microgravity: organism-specific recommendations for their use, and recommended terminology. Astrobiol. 2013;13(1):1–17. doi: 10.1089/ast.2012.0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Globus R.K., Morey-Holton E. 2016.
- 9.Pavy-Le Traon A., Heer M., Narici M.V., Rittweger J., Vernikos J. From space to earth: advances in human physiology from 20 years of bed rest studies (1986-2006). Eur. J. Appl. Physiol. 2007;101(2):143–194. doi: 10.1007/s00421-007-0474-z. [DOI] [PubMed] [Google Scholar]
- 10.Moher D., Liberati A., Tetzlaff J., Altman D.G., Group P. Preferred reporting items for systematic reviews and meta-analyses: the prisma statement. J. Clin. Epidemiol. 2009;62(10):1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Ethiraj P., Link J.R., Sinkway J.M., Brown G.D., Parler W.A., Reddy S.V. Microgravity modulation of syncytin-a expression enhance osteoclast formation. J. Cell. Biochem. 2018;119(7):5696–5703. doi: 10.1002/jcb.26750. [DOI] [PubMed] [Google Scholar]
- 12.Shanmugarajan S., Zhang Y., Moreno-Villanueva M., Clanton R., Rohde L.H., Ramesh G.T., Sibonga J.D., Wu H. Combined effects of simulated microgravity and radiation exposure on osteoclast cell fusion. Int. J. Mol. Sci. 2017;18(11):2443. doi: 10.3390/ijms18112443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambandam Y., Baird K.L., Stroebel M., Kowal E., Balasubramanian S., Reddy S.V. Microgravity induction of trail expression in preosteoclast cells enhances osteoclast differentiation. Sci. Rep. 2016;6:25143. doi: 10.1038/srep25143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatani M., Morimoto H., Takeyama K., Mantoku A., Tanigawa N., Kubota K., Suzuki H., Uchida S., Tanigaki F., Shirakawa M., Gusev O., Sychev V., Takano Y., Itoh T., Kudo A. Acute transcriptional up-regulation specific to osteoblasts/osteoclasts in medaka fish immediately after exposure to microgravity. Sci. Rep. 2016;6:39545. doi: 10.1038/srep39545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatani M., Mantoku A., Takeyama K., Abduweli D., Sugamori Y., Aoki K., Ohya K., Suzuki H., Uchida S., Sakimura T., Kono Y., Tanigaki F., Shirakawa M., Takano Y., Kudo A. Microgravity promotes osteoclast activity in medaka fish reared at the international space station. Sci. Rep. 2015;5:14172. doi: 10.1038/srep14172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y.L., Chen Z.H., Chen X.H., Yin C., Li D.J., Ma X.L., Zhao F., Zhang G., Shang P., Qian A.R. Diamagnetic levitation promotes osteoclast differentiation from raw264.7 cells. IEEE Trans. Biomed. Eng. 2015;62(3):900–908. doi: 10.1109/TBME.2014.2370039. [DOI] [PubMed] [Google Scholar]
- 17.Saxena R., Pan G., Dohm E.D., McDonald J.M. Modeled microgravity and hindlimb unloading sensitize osteoclast precursors to rankl-mediated osteoclastogenesis. J. Bone Miner. Metab. 2011;29(1):111–122. doi: 10.1007/s00774-010-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambandam Y., Blanchard J.J., Daughtridge G., Kolb R.J., Shanmugarajan S., Pandruvada S.N., Bateman T.A., Reddy S.V. Microarray profile of gene expression during osteoclast differentiation in modelled microgravity. J. Cell. Biochem. 2010;111(5):1179–1187. doi: 10.1002/jcb.22840. [DOI] [PubMed] [Google Scholar]
- 19.Sambandam Y., Townsend M.T., Pierce J.J., Lipman C.M., Haque A., Bateman T.A., Reddy S.V. Microgravity control of autophagy modulates osteoclastogenesis. Bone. 2014;61:125–131. doi: 10.1016/j.bone.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamma R., Colaianni G., Camerino C., Di Benedetto A., Greco G., Strippoli M., Vergari R., Grano A., Mancini L., Mori G., Colucci S., Grano M., Zallone A. Microgravity during spaceflight directly affects in vitro osteoclastogenesis and bone resorption. FASEB J. 2009;23(8):2549–2554. doi: 10.1096/fj.08-127951. [DOI] [PubMed] [Google Scholar]
- 21.Makihira S., Kawahara Y., Yuge L., Mine Y., Nikawa H. Impact of the microgravity environment in a 3-dimensional clinostat on osteoblast- and osteoclast-like cells. Cell Biol. Int. 2008;32(9):1176–1181. doi: 10.1016/j.cellbi.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Wang H., Sun Z., Wang Y., Hu Z., Zhou H., Zhang L., Hong B., Zhang S., Cao X. MiR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting HMGA2. Sci. Rep. 2016;6:23170. doi: 10.1038/srep23170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Z., Cao X., Zhang Z., Hu Z., Zhang L., Wang H., Zhou H., Li D., Zhang S., Xie M. Simulated microgravity inhibits l-type calcium channel currents partially by the up-regulation of mir-103 in MC3T3-E1 osteoblasts. Sci. Rep. 2015;5:8077. doi: 10.1038/srep08077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu L.F., Li J.B., Qian A.R., Wang F., Shang P. Mineralization initiation of MC3T3-E1 preosteoblast is suppressed under simulated microgravity condition. Cell Biol. Int. 2015;39(4):364–372. doi: 10.1002/cbin.10391. [DOI] [PubMed] [Google Scholar]
- 25.Bucaro M.A., Zahm A.M., Risbud M.V., Ayyaswamy P.S., Mukundakrishnan K., Steinbeck M.J., Shapiro I.M., Adams C.S. The effect of simulated microgravity on osteoblasts is independent of the induction of apoptosis. J. Cell. Biochem. 2007;102(2):483–495. doi: 10.1002/jcb.21310. [DOI] [PubMed] [Google Scholar]
- 26.Hughes F.J., Turner W., Belibasakis G., Martuscelli G. Effects of growth factors and cytokines on osteoblast differentiation. Periodontol. 2006;41:48–72. doi: 10.1111/j.1600-0757.2006.00161.x. [DOI] [PubMed] [Google Scholar]
- 27.Bucaro M.A., Fertala J., Adams C.S., Steinbeck M., Ayyaswamy P., Mukundakrishnan K., Shapiro I.M., Risbud M.V. Bone cell survival in microgravity: evidence that modeled microgravity increases osteoblast sensitivity to apoptogens. Ann. N. Y. Acad. Sci. 2004;1027:64–73. doi: 10.1196/annals.1324.007. [DOI] [PubMed] [Google Scholar]
- 28.Saito M., Soshi S., Fujii K. Effect of hyper- and microgravity on collagen post-translational controls of MC3T3-E1 osteoblasts. J. Bone Miner. Res. 2003;18(9):1695–1705. doi: 10.1359/jbmr.2003.18.9.1695. [DOI] [PubMed] [Google Scholar]
- 29.Ontiveros C., McCabe L.R. Simulated microgravity suppresses osteoblast phenotype, runx2 levels and AP-1 transactivation. J. Cell. Biochem. 2003;88(3):427–437. doi: 10.1002/jcb.10410. [DOI] [PubMed] [Google Scholar]
- 30.Kumei Y., Morita S., Nakamura H., Shinomiya K., Ohya K., Shimokawa H. Does microgravity induce apoptotic signal in rat osteoblasts via cJUN-n-terminal kinase? J. Gravit. Physiol. 2002;9(1):263–P264. [PubMed] [Google Scholar]
- 31.Sato A., Hamazaki T., Oomura T., Osada H., Kakeya M., Watanabe M., Nakamura T., Nakamura Y., Koshikawa N., Yoshizaki I., Aizawa S., Yoda S., Ogiso A., Takaoki M., Kohno Y., Tanaka H. Effects of microgravity on c-FOS gene expression in osteoblast-like MC3T3-E1 cells. Adv. Space Res. 1999;24(6):807–813. doi: 10.1016/s0273-1177(99)00076-9. [DOI] [PubMed] [Google Scholar]
- 32.Hughes-Fulford M., Tjandrawinata R., Fitzgerald J., Gasuad K., Gilbertson V. Effects of microgravity on osteoblast growth. Gravit. Space Biol. Bull. 1998;11(2):51–60. [PubMed] [Google Scholar]
- 33.Hu Z., Wang H., Wang Y., Zhou H., Shi F., Zhao J., Zhang S., Cao X. Genome wide analysis and prediction of functional long noncoding RNAs in osteoblast differentiation under simulated microgravity. Mol. Med. Rep. 2017;16(6):8180–8188. doi: 10.3892/mmr.2017.7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goyden J., Tawara K., Hedeen D., Willey J.S., Oxford J.T., Jorcyk C.L. The effect of OSM on MC3T3-E1 osteoblastic cells in simulated microgravity with radiation. PLoS One. 2015;10(6):e0127230. doi: 10.1371/journal.pone.0127230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bikle D.D., Harris J., Halloran B.P., Morey-Holton E. Altered skeletal pattern of gene expression in response to spaceflight and hindlimb elevation. Am. J. Physiol. 1994;267(6 Pt 1):E822–E827. doi: 10.1152/ajpendo.1994.267.6.E822. [DOI] [PubMed] [Google Scholar]
- 36.Qian A., Di S., Gao X., Zhang W., Tian Z., Li J., Hu L., Yang P., Yin D., Shang P. cDNA microarray reveals the alterations of cytoskeleton-related genes in osteoblast under high magneto-gravitational environment. Acta Biochim. Biophys. Sin. (Shanghai) 2009;41(7):561–577. doi: 10.1093/abbs/gmp041. [DOI] [PubMed] [Google Scholar]
- 37.Shuang F., Sun Y., Yang H.H., Shao Y.C., Li H., Hu W., Zhong J., Zou H.X. Destrin deletion enhances the bone loss in hindlimb suspended mice. Eur. J. Appl. Physiol. 2013;113(2):403–410. doi: 10.1007/s00421-012-2451-4. [DOI] [PubMed] [Google Scholar]
- 38.Dai Z., Wu F., Chen J., Xu H., Wang H., Guo F., Tan Y., Ding B., Wang J., Wan Y., Li Y. Actin microfilament mediates osteoblast CBFA1 responsiveness to BMP2 under simulated microgravity. PLoS One. 2013;8(5):e63661. doi: 10.1371/journal.pone.0063661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapitonova M.Y., Salim N., Othman S., Muhd Kamauzaman T.M., Ali A.M., Nawawi H.M., Froemming G.R. Alteration of cell cytoskeleton and functions of cell recovery of normal human osteoblast cells caused by factors associated with real space flight. Malays. J. Pathol. 2013;35(2):153–163. [PubMed] [Google Scholar]
- 40.Guignandon A., Faure C., Neutelings T., Rattner A., Mineur P., Linossier M.T., Laroche N., Lambert C., Deroanne C., Nusgens B., Demets R., Colige A., Vico L. Rac1 GTPase silencing counteracts microgravity-induced effects on osteoblastic cells. FASEB J. 2014;28(9):4077–4087. doi: 10.1096/fj.14-249714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumei Y., Shimokawa H., Katano H., Hara E., Akiyama H., Hirano M., Mukai C., Nagaoka S., Whitson P.A., Sams C.F. Microgravity induces prostaglandin E2 and interleukin-6 production in normal rat osteoblasts: role in bone demineralization. J. Biotechnol. 1996;47(2-3):313–324. doi: 10.1016/0168-1656(96)01412-5. [DOI] [PubMed] [Google Scholar]
- 42.Carmeliet G., Nys G., Bouillon R. Microgravity reduces the differentiation of human osteoblastic MG-63 cells. J. Bone Miner. Res. 1997;12(5):786–794. doi: 10.1359/jbmr.1997.12.5.786. [DOI] [PubMed] [Google Scholar]
- 43.Landis W.J., Hodgens K.J., Block D., Toma C.D., Gerstenfeld L.C. Spaceflight effects on cultured embryonic chick bone cells. J. Bone Miner. Res. 2000;15(6):1099–1112. doi: 10.1359/jbmr.2000.15.6.1099. [DOI] [PubMed] [Google Scholar]
- 44.Rucci N., Migliaccio S., Zani B.M., Taranta A., Teti A. Characterization of the osteoblast-like cell phenotype under microgravity conditions in the NASA-approved rotating wall vessel bioreactor (RWV). J. Cell. Biochem. 2002;85(1):167–179. [PubMed] [Google Scholar]
- 45.Kumei Y., Morita S., Nakamura H., Akiyama H., Hirano M., Shimokawa H., Ohya K. Coinduction of GTP cyclohydrolase I and inducible no synthase in rat osteoblasts during space flight: apoptotic and self-protective response? Ann. N. Y. Acad. Sci. 2003;1010:481–485. doi: 10.1196/annals.1299.087. [DOI] [PubMed] [Google Scholar]
- 46.Kumei Y., Morita S., Shimokawa H., Ohya K., Akiyama H., Hirano M., Sams C.F., Whitson P.A. Inhibition of hsp70 and a collagen-specific molecular chaperone (hsp47) expression in rat osteoblasts by microgravity. Ann. N. Y. Acad. Sci. 2003;1010:476–480. doi: 10.1196/annals.1299.086. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura H., Kumei Y., Morita S., Shimokawa H., Ohya K., Shinomiya K. Antagonism between apoptotic (bax/bcl-2) and anti-apoptotic (IAP) signals in human osteoblastic cells under vector-averaged gravity condition. Ann. N. Y. Acad. Sci. 2003;1010:143–147. doi: 10.1196/annals.1299.023. [DOI] [PubMed] [Google Scholar]
- 48.Kumei Y., Morita S., Nakamura H., Akiyama H., Katano H., Shimokawa H., Ohya K. Platelet-activating factor receptor signals in rat osteoblasts during spaceflight. Ann. N. Y. Acad. Sci. 2004;1030:116–120. doi: 10.1196/annals.1329.014. [DOI] [PubMed] [Google Scholar]
- 49.Kumei Y., Morita S., Nakamura H., Katano H., Ohya K., Shimokawa H., Sams C.F., Whitson P.A. Osteoblast responsiveness to 1alpha,25-dihydroxyvitamin D3 during spaceflight. Ann. N. Y. Acad. Sci. 2004;1030:121–124. doi: 10.1196/annals.1329.015. [DOI] [PubMed] [Google Scholar]
- 50.Kumei Y., Morita S., Katano H., Akiyama H., Hirano M., Oyha K., Shimokawa H. Microgravity signal ensnarls cell adhesion, cytoskeleton, and matrix proteins of rat osteoblasts: osteopontin, CD44, osteonectin, and alpha-tubulin. Ann. N. Y. Acad. Sci. 2006;1090:311–317. doi: 10.1196/annals.1378.034. [DOI] [PubMed] [Google Scholar]
- 51.Pardo S.J., Patel M.J., Sykes M.C., Platt M.O., Boyd N.L., Sorescu G.P., Xu M., van Loon J.J., Wang M.D., Jo H. Simulated microgravity using the random positioning machine inhibits differentiation and alters gene expression profiles of 2T3 preosteoblasts. Am. J. Physiol. Cell Physiol. 2005;288(6):C1211–C1221. doi: 10.1152/ajpcell.00222.2004. [DOI] [PubMed] [Google Scholar]
- 52.Rucci N., Rufo A., Alamanou M., Teti A. Modeled microgravity stimulates osteoclastogenesis and bone resorption by increasing osteoblast RANKL/OPG ratio. J. Cell. Biochem. 2007;100(2):464–473. doi: 10.1002/jcb.21059. [DOI] [PubMed] [Google Scholar]
- 53.Patel M.J., Liu W., Sykes M.C., Ward N.E., Risin S.A., Risin D., Jo H. Identification of mechanosensitive genes in osteoblasts by comparative microarray studies using the rotating wall vessel and the random positioning machine. J. Cell. Biochem. 2007;101(3):587–599. doi: 10.1002/jcb.21218. [DOI] [PubMed] [Google Scholar]
- 54.Kumei Y., Shimokawa H., Ohya K., Katano H., Akiyama H., Hirano M., Morita S. Small GTPase Ras and Rho expression in rat osteoblasts during spaceflight. Ann. N. Y. Acad. Sci. 2007;1095:292–299. doi: 10.1196/annals.1397.032. [DOI] [PubMed] [Google Scholar]
- 55.Capulli M., Rufo A., Teti A., Rucci N. Global transcriptome analysis in mouse calvarial osteoblasts highlights sets of genes regulated by modeled microgravity and identifies a mechanoresponsive osteoblast gene signature. J. Cell. Biochem. 2009;107(2):240–252. doi: 10.1002/jcb.22120. [DOI] [PubMed] [Google Scholar]
- 56.Blaber E.A., Dvorochkin N., Lee C., Alwood J.S., Yousuf R., Pianetta P., Globus R.K., Burns B.P., Almeida E.A. Microgravity induces pelvic bone loss through osteoclastic activity, osteocytic osteolysis, and osteoblastic cell cycle inhibition by CDKn1a/p21. PLoS One. 2013;8(4):e61372. doi: 10.1371/journal.pone.0061372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rucci N., Capulli M., Piperni S.G., Cappariello A., Lau P., Frings-Meuthen P., Heer M., Teti A. Lipocalin 2: a new mechanoresponding gene regulating bone homeostasis. J. Bone Miner. Res. 2015;30(2):357–368. doi: 10.1002/jbmr.2341. [DOI] [PubMed] [Google Scholar]
- 58.Hu Z., Wang Y., Sun Z., Wang H., Zhou H., Zhang L., Zhang S., Cao X. miRNA-132-3p inhibits osteoblast differentiation by targeting Ep300 in simulated microgravity. Sci. Rep. 2015;5:18655. doi: 10.1038/srep18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumei Y., Nakamura H., Morita S., Akiyama H., Hirano M., Ohya K., Shinomiya K., Shimokawa H. Space flight and insulin-like growth factor-I signaling in rat osteoblasts. Ann. N. Y. Acad. Sci. 2002;973(1):75–78. doi: 10.1111/j.1749-6632.2002.tb04609.x. [DOI] [PubMed] [Google Scholar]
- 60.Boyce B.F., Xing L. The RANKL/RANK/OPG pathway. Curr. Osteoporos. Rep. 2007;5(3):98–104. doi: 10.1007/s11914-007-0024-y. [DOI] [PubMed] [Google Scholar]
- 61.Ge C., Xiao G., Jiang D., Franceschi R.T. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J. Cell Biol. 2007;176(5):709–718. doi: 10.1083/jcb.200610046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sinha K.M., Zhou X. Genetic and molecular control of osterix in skeletal formation. J. Cell. Biochem. 2013;114(5):975–984. doi: 10.1002/jcb.24439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei J., Karsenty G. An overview of the metabolic functions of osteocalcin. Rev. Endocr. Metab. Disord. 2015;16(2):93–98. doi: 10.1007/s11154-014-9307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Horowitz M.C. The role of cytokines in bone remodeling. J. Clin. Densitom. 1998;1(2):187–198. [Google Scholar]
- 65.Miki H., Suetsugu S., Takenawa T. Wave, a novel wasp-family protein involved in actin reorganization induced by RAC. EMBO J. 1998;17(23):6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katagiri Y.U., Sleeman J., Fujii H., Herrlich P., Hotta H., Tanaka K., Chikuma S., Yagita H., Okumura K., Murakami M., Saiki I., Chambers A.F., Uede T. CD44 variants but not CD44s cooperate with β1-containing integrins to permit cells to bind to osteopontin independently of arginine-glycine-aspartic acid, thereby stimulating cell motility and chemotaxis. Cancer Res. 1999;59(1):219. [PubMed] [Google Scholar]
- 67.Manolagas S.C. The role of Il-6 type cytokines and their receptors in bone. Ann. N. Y. Acad. Sci. 1998;840:194–204. doi: 10.1111/j.1749-6632.1998.tb09563.x. [DOI] [PubMed] [Google Scholar]
- 68.Franchimont N., Wertz S., Malaise M. Interleukin-6: an osteotropic factor influencing bone formation? Bone. 2005;37(5):601–606. doi: 10.1016/j.bone.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 69.Steeve K.T., Marc P., Sandrine T., Dominique H., Yannick F. Il-6, RANKL, TNF-alpha/Il-1: interrelations in bone resorption pathophysiology. Cytokine Growth Factor Rev. 2004;15(1):49–60. doi: 10.1016/j.cytogfr.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 70.Wong P.K., Campbell I.K., Egan P.J., Ernst M., Wicks I.P. The role of the interleukin-6 family of cytokines in inflammatory arthritis and bone turnover. Arthritis Rheum. 2003;48(5):1177–1189. doi: 10.1002/art.10943. [DOI] [PubMed] [Google Scholar]
- 71.Liu X.H., Kirschenbaum A., Yao S., Levine A.C. The role of the interleukin-6/GP130 signaling pathway in bone metabolism. Vitam. Horm. 2006;74:341–355. doi: 10.1016/S0083-6729(06)74014-6. [DOI] [PubMed] [Google Scholar]
- 72.Heymann D., Rousselle A.V. GP130 cytokine family and bone cells. Cytokine. 2000;12(10):1455–1468. doi: 10.1006/cyto.2000.0747. [DOI] [PubMed] [Google Scholar]
- 73.Kamimura D., Ishihara K., Hirano T. Il-6 signal transduction and its physiological roles: the signal orchestration model. Rev. Physiol. Biochem. Pharmacol. 2003;149:1–38. doi: 10.1007/s10254-003-0012-2. [DOI] [PubMed] [Google Scholar]
- 74.Ernst M., Jenkins B.J. Acquiring signalling specificity from the cytokine receptor GP130. Trends Genet. 2004;20(1):23–32. doi: 10.1016/j.tig.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 75.Tamura T., Udagawa N., Takahashi N., Miyaura C., Tanaka S., Yamada Y., Koishihara Y., Ohsugi Y., Kumaki K., Taga T. Soluble interleukin-6 receptor triggers osteoclast formation by interleukin 6. Proc. Natl. Acad. Sci. USA. 1993;90(24):11924–11928. doi: 10.1073/pnas.90.24.11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones S.A., Horiuchi S., Topley N., Yamamoto N., Fuller G.M. The soluble interleukin 6 receptor: mechanisms of production and implications in disease. FASEB J. 2001;15(1):43–58. doi: 10.1096/fj.99-1003rev. [DOI] [PubMed] [Google Scholar]
- 77.Heinrich P.C., Behrmann I., Haan S., Hermanns H.M., Muller-Newen G., Schaper F. Principles of interleukin (Il)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374(Pt 1):1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Itoh S., Udagawa N., Takahashi N., Yoshitake F., Narita H., Ebisu S., Ishihara K. A critical role for interleukin-6 family-mediated stat3 activation in osteoblast differentiation and bone formation. Bone. 2006;39(3):505–512. doi: 10.1016/j.bone.2006.02.074. [DOI] [PubMed] [Google Scholar]
- 79.Bellido T., O’Brien C.A., Roberson P.K., Manolagas S.C. Transcriptional activation of the p21WAF1,CIP1,SDI1 gene by interleukin-6 type cytokines. A prerequisite for their pro-differentiating and anti-apoptotic effects on human osteoblastic cells. J. Biol. Chem. 1998;273(33):21137–21144. doi: 10.1074/jbc.273.33.21137. [DOI] [PubMed] [Google Scholar]
- 80.Chipoy C., Brounais B., Trichet V., Battaglia S., Berreur M., Oliver L., Juin P., Redini F., Heymann D., Blanchard F. Sensitization of osteosarcoma cells to apoptosis by oncostatin M depends on stat5 and p53. Oncogene. 2007;26(46):6653–6664. doi: 10.1038/sj.onc.1210492. [DOI] [PubMed] [Google Scholar]
- 81.Brounais B., Chipoy C., Mori K., Charrier C., Battaglia S., Pilet P., Richards C.D., Heymann D., Redini F., Blanchard F. Oncostatin M induces bone loss and sensitizes rat osteosarcoma to the antitumor effect of midostaurin in vivo. Clin. Cancer Res. 2008;14(17):5400–5409. doi: 10.1158/1078-0432.CCR-07-4781. [DOI] [PubMed] [Google Scholar]
- 82.Malaval L., Aubin J.E. Biphasic effects of leukemia inhibitory factor on osteoblastic differentiation. J. Cell. Biochem. Suppl. 2001;(Suppl. 36):63–70. doi: 10.1002/jcb.1086. [DOI] [PubMed] [Google Scholar]
- 83.Chipoy C., Berreur M., Couillaud S., Pradal G., Vallette F., Colombeix C., Redini F., Heymann D., Blanchard F. Downregulation of osteoblast markers and induction of the glial fibrillary acidic protein by oncostatin M in osteosarcoma cells require PKCdelta and STAT3. J. Bone Miner. Res. 2004;19(11):1850–1861. doi: 10.1359/JBMR.040817. [DOI] [PubMed] [Google Scholar]
- 84.Palmqvist P., Persson E., Conaway H.H., Lerner U.H. Il-6, leukemia inhibitory factor, and oncostatin M stimulate bone resorption and regulate the expression of receptor activator of NF-kappa B ligand, osteoprotegerin, and receptor activator of NF-kappa B in mouse calvariae. J. Immunol. 2002;169(6):3353–3362. doi: 10.4049/jimmunol.169.6.3353. [DOI] [PubMed] [Google Scholar]
- 85.O’Brien C.A., Gubrij I., Lin S.C., Saylors R.L., Manolagas S.C. STAT3 activation in stromal/osteoblastic cells is required for induction of the receptor activator of NF-kappa B ligand and stimulation of osteoclastogenesis by GP130-utilizing cytokines or interleukin-1 but not 1,25-dihydroxyvitamin D3 or parathyroid hormone. J. Biol. Chem. 1999;274(27):19301–19308. doi: 10.1074/jbc.274.27.19301. [DOI] [PubMed] [Google Scholar]
- 86.Duplomb L. Baud’Huin, M.; Charrier, C.; Trichet, V.; Blanchard, F.; Heymann, D. Il-6 inhibits RANKL-induced osteoclastogenesis by diverting cells into the macrophage lineage: key role of serine<sup>727</sup> phosphorylation of STAT3. Bone. 2008;42:S36. doi: 10.1210/en.2007-1719. [DOI] [PubMed] [Google Scholar]
- 87.Yoshitake F., Itoh S., Narita H., Ishihara K., Ebisu S. Interleukin-6 directly inhibits osteoclast differentiation by suppressing receptor activator of NF-kappaB signaling pathways. J. Biol. Chem. 2008;283(17):11535–11540. doi: 10.1074/jbc.M607999200. [DOI] [PubMed] [Google Scholar]
- 88.Schuler M., Green D.R. Mechanisms of p53-dependent apoptosis. Biochem. Soc. Trans. 2001;29(Pt 6):684–688. doi: 10.1042/0300-5127:0290684. [DOI] [PubMed] [Google Scholar]
- 89.Schuler M., Bossy-Wetzel E., Goldstein J.C., Fitzgerald P., Green D.R. P53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J. Biol. Chem. 2000;275(10):7337–7342. doi: 10.1074/jbc.275.10.7337. [DOI] [PubMed] [Google Scholar]
- 90.Hughes-Fulford M. In: Warmbein B., editor. 23rd Annual International Gravitational Physiology Meeting; Stockholm, Sweden. June 2-7, 2002; Noordwijk, Netherlands: Publications Division; 2002. pp. 43–46. [Google Scholar]
- 91.Kanno T., Takahashi T., Tsujisawa T., Ariyoshi W., Nishihara T. Mechanical stress-mediated runx2 activation is dependent on RAS/ERK1/2 MAPK signaling in osteoblasts. J. Cell. Biochem. 2007;101(5):1266–1277. doi: 10.1002/jcb.21249. [DOI] [PubMed] [Google Scholar]
- 92.Higuchi C., Myoui A., Hashimoto N., Kuriyama K., Yoshioka K., Yoshikawa H., Itoh K. Continuous inhibition of MAPK signaling promotes the early osteoblastic differentiation and mineralization of the extracellular matrix. J. Bone Miner. Res. 2002;17(10):1785–1794. doi: 10.1359/jbmr.2002.17.10.1785. [DOI] [PubMed] [Google Scholar]
- 93.Wagner E.F. Functions of AP1 (FOS/JUN) in bone development. Ann. Rheum. Dis. 2002;61(Suppl. 2):40–42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grigoriadis A.E., Wang Z.Q., Cecchini M.G., Hofstetter W., Felix R., Fleisch H.A., Wagner E.F. C-FOS: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266(5184):443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 95.McCabe L.R., Banerjee C., Kundu R., Harrison R.J., Dobner P.R., Stein J.L., Lian J.B., Stein G.S. Developmental expression and activities of specific FOS and JUN proteins are functionally related to osteoblast maturation: role of FRA-2 and JUN D during differentiation. Endocrinology. 1996;137(10):4398–4408. doi: 10.1210/endo.137.10.8828501. [DOI] [PubMed] [Google Scholar]
- 96.Hobolt-Pedersen A.S., Delaisse J.M., Soe K. Osteoclast fusion is based on heterogeneity between fusion partners. Calcif. Tissue Int. 2014;95(1):73–82. doi: 10.1007/s00223-014-9864-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mensah K.A., Ritchlin C.T., Schwarz E.M. RANKL induces heterogeneous DC-STAMP(lo) and DC-STAMP(hi) osteoclast precursors of which the DC-STAMP(lo) precursors are the master fusogens. J. Cell. Physiol. 2010;223(1):76–83. doi: 10.1002/jcp.22012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soe K., Andersen T.L., Hobolt-Pedersen A.S., Bjerregaard B., Larsson L.I., Delaisse J.M. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone. 2011;48(4):837–846. doi: 10.1016/j.bone.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 99.Moller A.M., Delaisse J.M., Soe K. Osteoclast fusion: time-lapse reveals involvement of CD47 and syncytin-1 at different stages of nuclearity. J. Cell. Physiol. 2017;232(6):1396–1403. doi: 10.1002/jcp.25633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.DeSelm C.J., Miller B.C., Zou W., Beatty W.L., van Meel E., Takahata Y., Klumperman J., Tooze S.A., Teitelbaum S.L., Virgin H.W. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell. 2011;21(5):966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gelman A., Elazar Z. Autophagic factors cut to the bone. Dev. Cell. 2011;21(5):808–810. doi: 10.1016/j.devcel.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 102.Lin N.Y., Stefanica A., Distler J.H. Autophagy: a key pathway of TNF-induced inflammatory bone loss. Autophagy. 2013;9(8):1253–1255. doi: 10.4161/auto.25467. [DOI] [PMC free article] [PubMed] [Google Scholar]


