Abstract
While many viruses of wild mammals are capable of infecting humans, our understanding of zoonotic potential is incomplete. Viruses vary in their degree of generalism, characterized by the phylogenetic relationships of their hosts. Among the dimensions of this phylogenetic landscape, phylogenetic aggregation, which is largely overlooked in studies of parasite host range, emerges in this study as a key predictor of zoonotic status of viruses. Plausibly, viruses that exhibit aggregation, typified by discrete clusters of related host species, may (i) have been able to close the phylogenetic distance to humans, (ii) have subsequently acquired an epidemiologically relevant host and (iii) exhibit relatively high fitness in realized host communities, which are frequently phylogenetically aggregated. These mechanisms associated with phylogenetic aggregation may help explain why correlated fundamental traits, such as the ability of viruses to replicate in the cytoplasm, are associated with zoonoses.
Keywords: virus, mammal, zoonotic, phylogeny, macroecology
1. Introduction
Given that the majority of emerging infectious diseases are caused by zoonotic pathogens [1], much attention has been given to characterizing sources of risk, uncovering disproportionate roles played by particular host and pathogen species [2–4] as well as identifying environmental and anthropogenic influences [5,6]. An outstanding challenge is determining if particular traits (e.g. transmission mode) or conditions (e.g. geographical location) simply elevate the probability of an otherwise rare event, or rather if some viruses have acquired a set of host species that mechanistically increases their zoonotic potential [7].
Viruses originating in mammals are of particular concern, having caused several recent human pandemics [4,5] and exhibiting traits, such as high mutation rates, that are associated with multi-host generalism [8]. Among primate species, viruses are more phylogenetically generalist than other pathogens [9], and at larger host taxonomic scales, many mammalian viruses show a tendency to be phylogenetically aggregated [10], meaning they are dispersed in multiple clusters of unrelated hosts. Unlike other phylogenetic metrics, this pattern is characterized by a ratio of distances rather than distance itself, and is consequently less assertive about putative donor host species. By contrast, minimum and mean phylogenetic distances from a set of host species to humans are only meaningful measures of zoonotic potential if the donor host species is either the closest related host or representative of the central tendency of relatedness to humans, when in fact the donor host could be any of the host species of the pathogen. Similarly, the phylogenetic span of a pathogen in the host phylogeny is a good predictor of zoonotic potential if the phylogenetic distance to humans is the primary barrier, but will be less informative if other factors limit cross-species transmission to humans.
While phylogenetic aggregation is fairly common among mammalian viruses, the underlying reason for the pattern is difficult to determine. A plausible explanation for aggregation is the ability of certain viruses to jump and creep through a host phylogeny; occasionally establishing in a phylogenetically novel host species and subsequently acquiring related host species [10]. This trait can increase zoonotic potential in three ways. First, establishing in a new clade within a host phylogeny may close the phylogenetic distance to humans [11]. Second, the ‘creep’ acquisition of related hosts following a jump can increase the potential of human–wildlife contact, which may be stronger with a subsequently acquired host species than with the original clade host [12]. Lastly, viruses with this trait may benefit from community assemblies, which themselves are often phylogenetically aggregated [13]. This benefit of associating with hosts dispersed in the phylogeny could increase community prevalence [14,15], buffer against local pathogen extinction [16] and increase human–wildlife contact rates [17].
Here, I build a case for phylogenetic aggregation as a trait that increases zoonotic potential of mammalian viruses using published sources of mammal–virus associations and a host phylogeny. The analysis uses distinct features of the phylogenetic landscape as predictors of zoonotic status: mean pairwise phylogenetic distance between non-human hosts, mean phylogenetic distance between host species and humans, phylogenetic span of viruses among non-human hosts and phylogenetic aggregation. Statistical modelling consistently identifies aggregation as a predictor of zoonotic status compared with all other features of the phylogenetic landscape. This predictor is then characterized by underlying viral trait data, with limited data hinting that RNA viruses capable of replicating in the cytoplasm and non-vector-borne DNA viruses are more likely to exhibit phylogenetic aggregation, and therefore harbour zoonotic potential, than their counterparts.
2. Material and methods
As an overview, data on phylogenetic traits of viruses were used to predict the zoonotic status of viruses using logistic regression. Models were compared using the Bayesian information criterion (BIC) to assess which models parsimoniously explained zoonotic status. The best models included phylogenetic aggregation, meaning the viruses whose hosts were overdispersed in the phylogeny were more likely to be zoonotic. This key phylogenetic trait was then treated as a response variable predicted by fundamental viral traits in a simple linear model, as the standard effect size for phylogenetic aggregation was approximately normally distributed.
Host–virus association data were obtained by combining three published data sources [4,18,19], and filtering to mammalian hosts, and viruses that were represented in the publication [4] containing metadata for zoonotic status, research effort, captured by log10(number of PubMed citations) (hereafter log10(cites)), along with viral trait data (mean genome length, single- versus double-stranded, RNA versus DNA, ability to replicate in cytoplasm, enveloped versus non-enveloped, segmented versus non-segmented genome, vector-borne versus other transmission). Phylogenetic measures of specificity were obtained using a mammalian host phylogeny [20]. For each virus, phylogenetic standard effect sizes for the mean pairwise distance between hosts (z.mpd), the maximum distance between hosts (z.max) and phylogenetic aggregation (z.agg) were calculated using methods previously described [10]. Mean pairwise distance and maximum distance are reasonably intuitive metrics. Phylogenetic aggregation, z.agg, was calculated as the mean nearest neighbour distance across all hosts divided by the maximum distance [10]. This metric takes small values if hosts are frequently clumped together in the phylogeny, with the denominator acting to scale by the ultimate span of a virus in the phylogeny (captured separately by z.max). The scaling is important as it allows a per-span metric of aggregation; without the scaling, the metric would essentially only recapitulate z.max.
For each virus, the mean phylogenetic distance between host species and humans was also calculated (avDist). In order to calculate phylogenetic measures of viral specialism, viruses known only to infect a single host species were excluded from the analysis (n = 229). The resulting data comprised 227 virus species infecting 791 mammal species (excluding humans) from 14 mammalian host orders (Rodentia, Carnivora, Artiodactyla, Primates, Chiroptera, Eulipotyphla, Pilosa, Cingulata, Didelphimorphia, Perissodactyla, Proboscidea, Peramelemorphia, Lagomorpha, Diprotodontia).
The phylogenetic landscape variables (z.mpd, z.max, z.agg and avDist) were used in conjunction with sampling effort, log10 (cites) to predict viral zoonotic status using logistic regression in a set of related generalized linear models assessed by the BIC. BIC is useful for model selection in cases where there is high unobserved between-sample heterogeneity, as it balances likelihood and parsimony with a heavier penalty for parameter-rich models compared with Akaike information criterion (AIC) [21]. Phylogenetic aggregation, the most promising predictor from this analysis, was then treated as a response variable predicted by viral trait data using a linear model. Since viral traits that may promote zoonoses are likely to be different in RNA versus DNA viruses [22], the model was constructed to assess only the interaction between nucleic acid type and other viral traits. Genome size was excluded as sizes are extremely bimodal between RNA and DNA viruses [23], hence genome size is effectively approximated by nucleic acid type. The predictor log10(cites) was included as a separate predictor to capture research effort. Significant and near-significant viral trait predictors of phylogenetic aggregation revealed in this analysis were then visualized to clarify how they interact with nucleic acid type.
3. Results
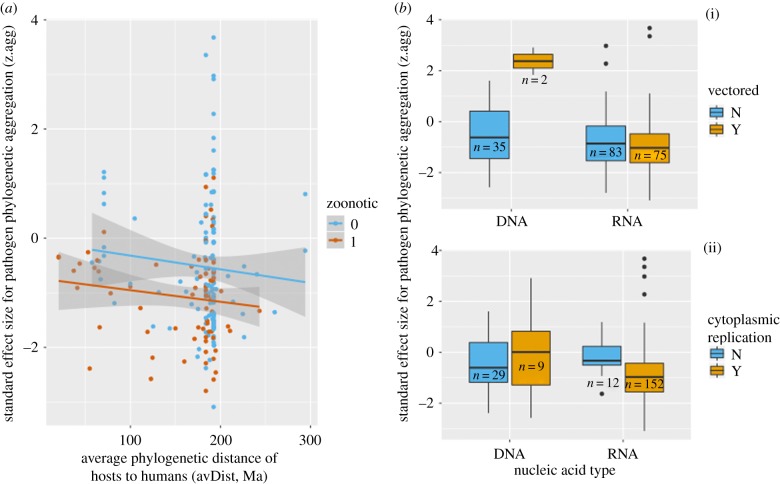
From a set of phylogenetic landscape models, zoonotic status of viruses was best predicted by accounting for both the average phylogenetic distance between host species and humans, avDist, and the degree of phylogenetic aggregation of the virus in the host phylogeny, z.agg (table 1 and figure 1a). In particular, phylogenetic aggregation was the only predictor variable, other than research effort, retained in the top two statistical models ranked by BIC (table 1). Unlike other phylogenetic predictors, it was consistently estimated to have the same (negative) sign, meaning that zoonotic viruses are more phylogenetically aggregated than other viruses (figure 1a). Lastly, it was scored as significant (p-value < 0.05) in seven of the eight models that included it. Average phylogenetic distance to humans from hosts was included in the first ranked model, although it was not a significant predictor; viruses of zoonotic status have a tendency to use hosts that are more related to humans (figure 1a). Notably, when phylogenetic aggregation is omitted from models, metrics associated with both viral specialism and generalism emerge as alternative predictors of zoonotic status. For example, the third highest ranked model suggests that zoonotic status is associated with viruses with large phylogenetic span and short mean pairwise distances between hosts. Similarly, large phylogenetic span is predictive of zoonotic status in the fourth highest ranked model, consistent with previously published work [4].
Table 1.
Summary of statistical models predicting zoonotic status of viruses by phylogenetic landscape variables (defined in methods) and sampling effort. Blank cells indicate that the predictor was omitted from the model and values in bold type refer to parameter estimates significant at the 95% confidence level. The final two columns show the Bayesian information criterion relative to the most parsimonious model (ΔBIC = 0.00), and the proportion of deviance explained. PD, Phylogenetic distance.
| interaction between average PD to humans (avDist) and standardized mean PD among all hosts (z.mpd) | interaction between average PD to humans (avDist) and standardized max PD among all hosts (z.max) | interaction between average PD to humans (avDist) and standardized PD aggregation (z.agg) | average PD to humans (avDist) | standardized mean PD among all hosts (z.mpd) | standardized max. PD among all hosts (z.max) | standardized PD aggregation (z.agg) | log10(cites) | ΔBIC | deviance explained |
|---|---|---|---|---|---|---|---|---|---|
| 0.01 | −0.01 | −2.03 | 0.22 | 0.00 | 0.10 | ||||
| −0.46 | 0.33 | 0.15 | 0.06 | ||||||
| −0.26 | 0.46 | 0.26 | 1.58 | 0.08 | |||||
| 0.27 | 0.37 | 2.64 | 0.06 | ||||||
| 0.0004 | −0.01 | 0.32 | 0.29 | 2.67 | 0.09 | ||||
| 0.15 | −0.35 | 0.33 | 4.00 | 0.07 | |||||
| −0.01 | 0.36 | 4.20 | 0.05 | ||||||
| 0.40 | 4.39 | 0.03 | |||||||
| 0.0043 | 0.01 | 0.00 | −0.48 | −2.66 | 0.21 | 4.76 | 0.12 | ||
| −0.05 | −0.46 | 0.31 | 5.26 | 0.06 | |||||
| −0.002 | 0.0024 | −0.01 | −0.01 | 0.22 | 0.18 | 5.90 | 0.12 | ||
| −0.21 | 0.35 | −0.19 | 0.26 | 6.04 | 0.08 | ||||
| −0.004 | 0.0096 | 0.02 | 0.006 | 0.50 | −1.10 | −3.48 | 0.11 | 8.17 | 0.15 |
| −0.05 | 0.38 | 9.39 | 0.03 | ||||||
| −0.001 | 0.01 | −0.01 | 0.11 | −2.05 | 0.21 | 10.71 | 0.10 | ||
| −0.0001 | −0.01 | −0.01 | 0.35 | 14.95 | 0.05 |
Figure 1.
(a) Relationship between phylogenetic aggregation (z.agg) and average phylogenetic distance between host species and humans (avDist) for both zoonotic viruses (red) and other viruses (blue). Linear regression lines are also shown fitted separately to each virus group (note, these are illustrative and not derived from the main statistical models reported in table 1). (b) Degree of phylogenetic aggregation (z.agg, y-axis) of viruses as a function of nucleic acid type (RNA versus DNA) and further structured into vector-borne versus other transmission (i) and ability versus inability to replicate in the cytoplasm (ii).
Phylogenetic aggregation was associated with certain viral traits (figure 1b) and with research effort. A significant interaction was found between nucleic acid type and vector-borne transmission (p-value = 0.001) and a near-significant interaction was found between nucleic acid type and ability to replicate in the cytoplasm (p-value = 0.084). Detecting such interactions is complicated by small group sizes (figure 1b), but tentatively these data suggest that DNA viruses that are not vector-borne are more likely to be phylogenetically aggregated than their vector-borne counterparts, and that RNA viruses that can replicate in the cytoplasm are more predisposed to become phylogenetically aggregated.
4. Discussion
Phylogenetic aggregation is a trait of pathogens that may increase zoonotic potential by closing the phylogenetic distance to humans, by acquiring a related set of host species of which one may be likely to exhibit epidemiological contact with humans, and by thriving in realized host communities which are frequently phylogenetically aggregated. Unlike phylogenetic traits expressing distance between host species and humans, it is less assertive about the known donor host and rather describes a pattern that can be linked to cross-species transmission potential. Further, it may help to explain why certain fundamental traits, such as cytoplasmic replication [4,24], are associated with zoonotic events.
As with any macroecological study of host–pathogen associations, it is important to consider sources of bias. While the underlying association data were compiled from well-documented sources [4,18,19], there will certainly be missing associations which are likely to be non-random in terms of the geographical region [25,26], host species [4], viral traits [7] and infection traits, such as virulence [27]. Further, a documented association does not indicate how commonly or rarely a given parasite infects a host species. These limitations in underlying data underscore the importance of continued virus surveillance at the human–wildlife interface [7]. Additionally, the mutability of viruses means that a trait such as phylogenetic aggregation is itself dynamic, and likely interacting with extrinsic features such as changes in host density [28]. However, linking fundamental traits to zoonotic potential via secondary traits such as phylogenetic aggregation can open lines of mechanistic inquiry, such as contact structure between humans and other clade hosts, which may improve our understanding of the risk posed by cross-species transmission.
Acknowledgements
The Macroecology of Infectious Disease Research Coordination Network provided useful discussions and support for this work.
Data accessibility
All data used in this study are available from published sources detailed in the manuscript text. Code to reproduce the analyses and figures is provided on Figshare at https://doi.org/10.6084/m9.figshare.c.4658018.v1 [29].
Competing interests
I declare I have no competing interests.
Funding
This material is based upon work supported by the National Science Foundation under grant no. NSF DEB 1754255 and by the Macroecology of Infectious Disease Research Coordination Network (grant no. NSF DEB 1316223).
References
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451, 990–993. ( 10.1038/nature06536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han BA, Schmidt JP, Bowden SE, Drake JM. 2015. Rodent reservoirs of future zoonotic diseases. Proc. Natl Acad. Sci. USA 112, 7039–7044. ( 10.1073/pnas.1501598112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han BA, Schmidt JP, Alexander LW, Bowden SE, Hayman DTS, Drake JM. 2016. Undiscovered bat hosts of filoviruses. PLoS Negl. Trop. Dis. 10, e0004815 ( 10.1371/journal.pntd.0004815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. 2017. Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650. ( 10.1038/nature22975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morse SS, Mazet JAK, Woolhouse M, Parrish CR, Carroll D, Karesh WB, Zambrana-Torrelio C, Lipkin WI, Daszak P. 2012. Prediction and prevention of the next pandemic zoonosis. Lancet 380, 1956–1965. ( 10.1016/S0140-6736(12)61684-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones BA, et al. 2013. Zoonosis emergence linked to agricultural intensification and environmental change. Proc. Natl Acad. Sci. USA 110, 8399–8404. ( 10.1073/pnas.1208059110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geoghegan JL, Holmes EC. 2017. Predicting virus emergence amid evolutionary noise. Open Biol. 7, 170189 ( 10.1098/rsob.170189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolhouse MEJ, Taylor LH, Haydon DT. 2001. Population biology of multihost pathogens. Science 292, 1109–1112. ( 10.1126/science.1059026) [DOI] [PubMed] [Google Scholar]
- 9.Cooper N, Griffin R, Franz M, Omotayo M, Nunn CL, Fryxell J. 2012. Phylogenetic host specificity and understanding parasite sharing in primates. Ecol. Lett. 15, 1370–1377. ( 10.1111/j.1461-0248.2012.01858.x) [DOI] [PubMed] [Google Scholar]
- 10.Park AW, et al. 2018. Characterizing the phylogenetic specialism–generalism spectrum of mammal parasites. Proc. R. Soc. B 285, 20172613 ( 10.1098/rspb.2017.2613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoppe E, et al. 2015. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol. Biol. Evol. 32, 2072–2084. ( 10.1093/molbev/msv090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caron A, Cappelle J, Cumming GS, de Garine-Wichatitsky M, Gaidet N.. 2015. Bridge hosts, a missing link for disease ecology in multi-host systems. Vet. Res. 46, 83 ( 10.1186/s13567-015-0217-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper N, Rodríguez J, Purvis A. 2008. A common tendency for phylogenetic overdispersion in mammalian assemblages. Proc. R. Soc. B 275, 2031–2037. ( 10.1098/rspb.2008.0420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mihaljevic JR, Joseph MB, Orlofske SA, Paull SH. 2014. The scaling of host density with richness affects the direction, shape, and detectability of diversity-disease relationships. PLoS ONE 9, e97812 ( 10.1371/journal.pone.0097812) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche B, Dobson AP, Guégan J-F, Rohani P. 2012. Linking community and disease ecology: the impact of biodiversity on pathogen transmission. Phil. Trans. R. Soc. B 367, 2807–2813. ( 10.1098/rstb.2011.0364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobson A. 2004. Population dynamics of pathogens with multiple host species. Am. Nat. 164(Suppl. 5), S64–S78. ( 10.1086/424681) [DOI] [PubMed] [Google Scholar]
- 17.Pedersen AB, Davies TJ. 2009. Cross-species pathogen transmission and disease emergence in primates. EcoHealth 6, 496–508. ( 10.1007/s10393-010-0284-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephens PR, et al. 2017. Global Mammal Parasite Database version 2.0. Ecology 98, 1476 ( 10.1002/ecy.1799) [DOI] [PubMed] [Google Scholar]
- 19.Wardeh M, Risley C, McIntyre MK, Setzkorn C, Baylis M. 2015. Database of host–pathogen and related species interactions, and their global distribution. Scient. Data 2, 150049 ( 10.1038/sdata.2015.49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritz SA, Bininda-Emonds ORP, Purvis A. 2009. Geographical variation in predictors of mammalian extinction risk: big is bad, but only in the tropics. Ecol. Lett. 12, 538–549. ( 10.1111/j.1461-0248.2009.01307.x) [DOI] [PubMed] [Google Scholar]
- 21.Brewer MJ, Butler A, Cooksley SL. 2016. The relative performance of AIC, AICC and BIC in the presence of unobserved heterogeneity. Methods Ecol. Evol. 7, 679–692. ( 10.1111/2041-210X.12541) [DOI] [Google Scholar]
- 22.Woolhouse MEJ, Brierley L, McCaffery C, Lycett S. 2016. Assessing the epidemic potential of RNA and DNA viruses. Emerg. Infect. Dis. 22, 2037–2044. ( 10.3201/eid2212.160123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campillo-Balderas JA, Lazcano A, Becerra A. 2015. Viral genome size distribution does not correlate with the antiquity of the host lineages. Front. Ecol. Evol. 3, 143 ( 10.3389/fevo.2015.00143) [DOI] [Google Scholar]
- 24.Pulliam JRC, Dushoff J. 2009. Ability to replicate in the cytoplasm predicts zoonotic transmission of livestock viruses. J. Infect. Dis. 199, 565–568. ( 10.1086/596510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen T, Murray KA, Zambrana-Torrelio C, Morse SS, Rondinini C, Di Marco M, Breit N, Olival KJ, Daszak P.. 2017. Global hotspots and correlates of emerging zoonotic diseases. Nat. Commun. 8, 1124 ( 10.1038/s41467-017-00923-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han BA, Kramer AM, Drake JM. 2016. Global patterns of zoonotic disease in mammals. Trends Parasitol. 32, 565–577. ( 10.1016/j.pt.2016.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leggett HC, Buckling A, Long GH, Boots M. 2013. Generalism and the evolution of parasite virulence. Trends Ecol. Evol. 28, 592–596. ( 10.1016/j.tree.2013.07.002) [DOI] [PubMed] [Google Scholar]
- 28.Holmes EC, Rambaut A, Andersen KG. 2018. Pandemics: spend on surveillance, not prediction. Nature 558, 180–182. ( 10.1038/d41586-018-05373-w) [DOI] [PubMed] [Google Scholar]
- 29.Park AW. 2019. Code to support ‘Phylogenetic aggregation increases zoonotic potential of mammalian viruses'. Figshare. ( 10.6084/m9.figshare.c.4658018.v1) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Park AW. 2019. Code to support ‘Phylogenetic aggregation increases zoonotic potential of mammalian viruses'. Figshare. ( 10.6084/m9.figshare.c.4658018.v1) [DOI] [PMC free article] [PubMed]
Data Availability Statement
All data used in this study are available from published sources detailed in the manuscript text. Code to reproduce the analyses and figures is provided on Figshare at https://doi.org/10.6084/m9.figshare.c.4658018.v1 [29].