Abstract
Spherical nucleic acids (SNAs) are nanomaterials typically consisting of a nanoparticle core and a functional, dense, and highly oriented oligonucleotide shell with unusual biological properties that make them appealing for many applications, including sequence-specific gene silencing, mRNA quantification, and immunostimulation. When placed in biological fluids, SNAs readily interact with serum proteins, leading to the formation of ill-defined protein coronae on the surface, which can influence the targeting capabilities of the conjugate. In this work, SNAs were designed and synthesized with functional proteins, such as antibodies and serum albumin, deliberately adsorbed onto their surfaces. These particles exhibit increased resistance to protease degradation compared with native SNAs but still remain functional, as they can engage in hybridization with complementary oligonucleotides. SNAs with adsorbed targeting antibodies exhibit improved cellular selectivity within mixed cell populations. Similarly, SNAs coated with the dysopsonizing protein serum albumin show reduced macrophage uptake, providing a strategy for tailoring selective SNA delivery. Importantly, the protein coronae remain stable on the SNAs in human serum, exhibiting a less than 45% loss of protein through exchange after 12 h at 37 °C. Taken together, these results show that protein–SNA complexes and the method used to prepare them provide a new avenue for enhancing SNA stability, targeting, and biodistribution.
Short abstract
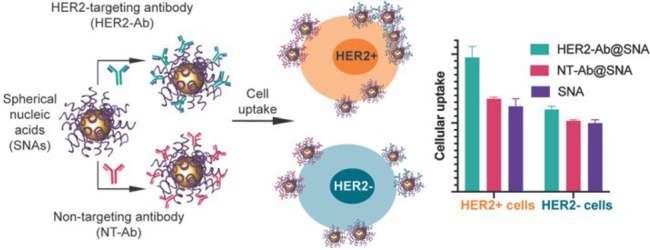
Anti-HER2-coated spherical nucleic acids selectively enter HER2-positive breast cancer cells at a higher efficiency than bare and nontargeting protein-coated SNAs.
Introduction
Certain nanomaterials can carry and present peptides, proteins, oligonucleotides, and small molecules within highly engineered structures to target tissues, making them appealing for biomedical and life science applications.1 However, many nanomaterials, when introduced to biological fluids, nonspecifically adsorb biomolecules, resulting in the formation of a protein corona around the structure.2 The protein corona alters the biological stability,3−7 biodistribution,8,9 and targeting efficiency10−13 of a nanomaterial, sometimes diminishing its therapeutic potential. Though the surface charge,14,15 size,8,16,17 and shape17 of a nanomaterial can modulate the composition of the protein corona, its formation is largely unavoidable in biological environments. Careful modification of the nanoparticle surface, however, can help dictate protein corona formation and mediate its effects on pharmacokinetics, yielding constructs with improved targeting capabilities12,18−20 that sometimes exceed covalent attachment methods12 or reduced nonspecific cellular uptake.4,21,22
Spherical nucleic acids (SNAs), a unique class of nanomaterials consisting of a spherical nanoparticle core densely functionalized with a highly oriented nucleic acid shell,23,24 have enhanced biological properties, including increased resistance to nuclease degradation compared with linear oligonucleotides of the same sequence,25 the ability to rapidly enter cells in high quantities without transfection agents,26,27 and tailorable immunogenicity.28 These properties have positioned SNAs for use in applications, such as gene silencing,29−32 immunomodulation,33−35 drug delivery,36,37 and nucleic acid detection in vitro(38−41) and in live cells.42−46 However, SNAs, like many other nanomaterials, interact with serum proteins, resulting in the formation of a protein corona that can alter their uptake properties.20 Antibody–DNA conjugates have been hybridized onto the surface of SNAs to improve their targeting capabilities and direct them to cancer cells.19 However, in this approach, the antibody densities utilized were so low (1–2 antibodies/particle) that they would be unlikely to alter protein corona formation. Alternatively, PEGylation of the nanoparticle core has shown the ability to reduce nonspecific adsorption of serum proteins, thus extending blood circulation times, but such modifications compromise SNA uptake efficiency by target cells.47
Herein we report a universal method for targeting SNAs to specific cell types. To accomplish this goal, we designed and synthesized SNAs with predefined protein coronae consisting of functional proteins immobilized on the oligonucleotide shell. We then explored the stability of these structures in buffer and human serum. In addition, we studied their ability to hybridize with complementary oligonucleotides as well as selectively target cell populations based on molecular signatures present on the cell surface. For example, SNAs with adsorbed human epithelial growth factor receptor 2 (HER2) monoclonal antibodies (mAbs) exhibited selectivity for HER2-positive breast cancer cells in mixed cell cultures with HER2-negative breast cancer cells. Taken together, our results show that this approach provides an easy, efficient, and flexible method for controlling SNA interactions within cells that has the potential to improve SNA stability, cell targeting, and biodistribution.
Results and Discussion
Particle Synthesis and Characterization
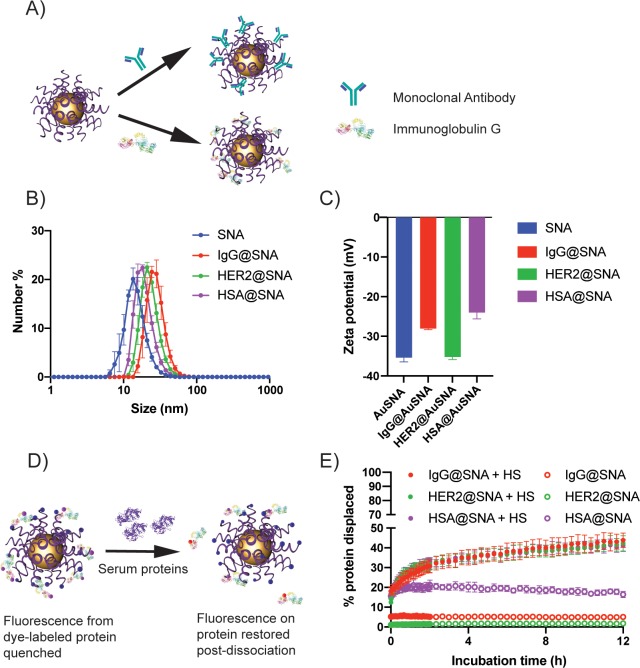
In order to generate particles with defined protein coronae, we first synthesized SNAs by functionalizing 13 nm gold nanoparticles (AuNPs) with thiolated oligodeoxynucleotides (ODNs) using established synthetic methods.23,48,49 We then incubated these SNAs with different functional proteins, such as human epithelial growth factor receptor 2 monoclonal antibodies (anti-HER2), immunoglobulin G (IgG), and human serum albumin (HSA). Each protein was selected on the basis of its potential ability to confer specific targeting properties to SNAs. Specifically, anti-HER2 was used as a model protein for cell targeting because it is a clinically relevant target for HER2-positive breast cancer.50 IgG was used because it is an immunogenic protein that attracts macrophages, while HSA was explored because it can shield nanoparticles from liver clearance and macrophage uptake.51 After incubating the individual proteins with the SNAs, we washed away the loosely bound proteins by pelleting the particles via centrifugation, leaving primarily a hard corona (defined as one with a high affinity for a particle) adsorbed on the SNA surface.52
To verify the formation of a hard corona, we measured the diameter and ζ potential of the SNAs before and after incubation with the different proteins (Figure 1). Dynamic light scattering (DLS) revealed that the average diameters of IgG-adsorbed SNAs (28.3 ± 4.1 nm), anti-HER2-adsorbed SNAs (23.5 ± 2.1 nm), and HSA-adsorbed SNAs (19.6 ± 1.4 nm) were larger than those of the bare SNAs (15.1 ± 2.2 nm) (Figure 1B). Furthermore, the SNAs with adsorbed IgG (−28.0 ± 0.6 mV) and HSA (−24.0 ± 3.6 mV) displayed a more positive ζ potential compared with bare SNAs (−35.4 ± 2.4 mV) (Figure 1C). Since IgG has an isoelectric point between 6.6 and 7.2 and that of HSA is 4.7, both proteins are negatively charged at physiological pH. The shielding of the highly negatively charged ODN by the protein coronae can contribute to the observed elevated ζ potential. Noticeably, neither IgG nor HSA undergoes significant conformational changes in the pH ranges used in this study.53,54A lack of significant change in the ζ potential of the anti-HER2-adsorbed SNAs (−35.2 ± 0.7 mV) may be due to the charge variants of the mAb obtained from the manufacturer, which could result in a less positive surface charge of the mAb (the isoelectric point of the main isoform is 8.7). Notably, charge variants of mAbs do not seem to significantly affect their binding affinity for target receptors.55 As further verification of protein corona formation, we examined the electrophoretic mobility of SNAs after incubation with proteins. All of the SNAs with protein coronae had reduced mobility in an agarose gel compared with bare SNAs (Figure S1A), further confirming that protein adsorption occurred. Notably, the number of DNA strands per particle did not change appreciably upon protein adsorption, indicating that all shifts in electrophoretic mobility were due to the protein coronae and not changes in the total number of DNA strands (Table S2).
Figure 1.
Synthesis and characterization of protein-adsorbed SNAs. (A) Schematic representation of monoclonal antibody (top) or IgG (bottom) immobilization on the ODN shell of SNAs. (B) Distributions of the hydrodynamic diameters of bare, IgG- (IgG@SNA), anti-HER2- (HER2@SNA), and HSA-immobilized SNAs (HSA@SNA). (C) ζ potentials of bare SNAs, IgG@SNAs, HER2@SNAs, and HSA@SNAs. (D) Schematic depicting the displacement of Texas Red-X-labeled proteins from the surface of Cy5-labeled SNAs by serum proteins. The fluorescence of Texas Red-X increases as protein displacement occurs. (E) Kinetic fluorescence profiles of Texas Red-X-labeled IgG, anti-HER2, and HSA during incubation with 10% serum proteins.
Furthermore, the number of proteins adsorbed onto each SNA was quantified via a fluorescence-based assay using fluorophore (i.e., Texas Red-X)-labeled proteins. For a 13 nm AuNP functionalized with ∼140 ODNs, approximately 40 ± 2, 26 ± 2, and 23 ± 2 IgG, anti-HER2, and HSA proteins/NP were found to be adsorbed, respectively. The reproducibility of this coating method was confirmed with HSA, as additional separate batches yielded similar numbers of proteins per NP (Table S3). The higher-density IgG coating is likely due to the higher affinity of IgG for the ODN shell.
The interactions driving binding of the proteins and SNAs likely consist of both electrostatic and hydrogen-bonding interactions, which have been reported for interactions between proteins and DNA; however, the exact mechanisms are still unclear.56 In our studies, increased salt concentrations (1 M NaCl, MgCl2), nonionic detergents (0.1%Triton X-100), and ethanol (1–20%) had negligible effects on the coronae. Only harsh denaturing conditions consisting of 0.1% sodium dodecyl sulfate (SDS) removed the coronae. These results suggest that there is a strong multifaceted interaction between the proteins and SNAs.
Stability of Protein Coronae in Biological Fluids
After establishing that we could adsorb an initial protein corona on the SNAs, we examined the stability of the different protein coronae by studying the exchange dynamics of proteins on the surface of SNAs under serum-rich conditions (i.e., 10% human serum (HS)). To accomplish this objective, we synthesized SNAs with Cy5-labeled ODNs, to which we adsorbed proteins labeled with Texas Red-X, a fluorophore that can transfer emitted energy to the Cy5 fluorophore on the ODN shell when attached to the SNAs. Thus, as proteins were displaced from the particle surface, an increase in Texas Red-X fluorescence was observed. We incubated the SNAs in 10% HS at 37 °C and tracked the change in fluorescence for 12 h, at which point the change in fluorescence had plateaued (Figure 1E). After 12 h, we found that more than 55% of the initial corona remained (full protein dissociation was established in 0.1% SDS), indicating that a stable hard protein corona had formed on the SNAs. Compared with IgG and anti-HER2, adsorbed HSA tends to dissociate from the SNA surface even without serum, indicating its weaker affinity for the ODN shell; the addition of serum increases the HSA desorption by about 20%. Significantly, the stable adhesion of the functional proteins ensures that active protein coronae are retained even in physiologically relevant media. To assess the composition of the protein coronae following incubation in serum, the protein-coated SNAs were incubated in 10% HS for 4 h at 37 °C, the unbound protein was then removed through centrifugation, and the bound protein was dissociated using SDS. The extracted protein was then analyzed using an SDS-PAGE gel running with Tris–glycine–SDS buffer (Figure S1B). Darker protein bands at ∼150 kDa indicate antibody enrichment for the IgG@SNAs and anti-HER2@SNAs. Enrichment of HSA was not observed for HSA@SNAs, most likely because of the abundance of albumin in HS. Albumin composes 50–60% of blood plasma proteins,57 and IgG is about one-fifth of the albumin content.58
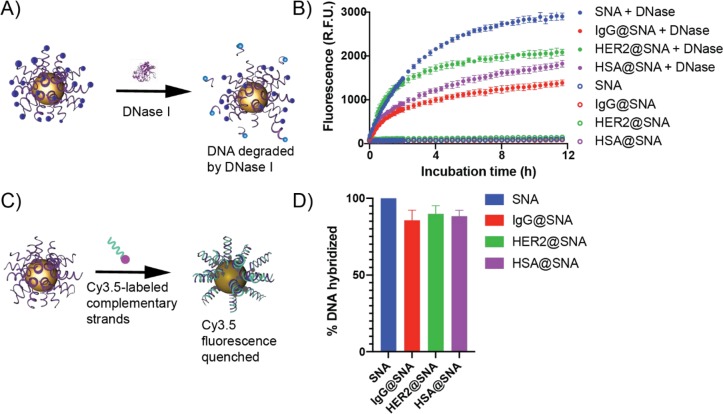
We explored the characteristics of SNAs precoated with corona proteins to ensure that they retained some of the same key biological properties as the original SNAs that make them valuable in biology and medicine. First, we examined their resistance to nucleases by quantifying the amount of ODNs degraded. To measure degradation, we synthesized SNAs with Cy5-labeled ODNs; the Cy5 dye is quenched when the labeled strand is attached to the gold core. Upon incubation with deoxyribonuclease I (DNase I), an endonuclease, ODNs are cleaved from the AuNP core, resulting in increased Cy5 emission intensity. In this experiment, preadsorption of IgG or HSA significantly reduced both the rate and efficiency of ODN degradation, and the anti-HER2 coating lessened the ODN degradation efficiency compared with the bare SNAs (Figure 2B); this result indicates that preadsorption of proteins enhances SNA resistance to nucleases. This presumably occurs because of the increased steric hindrance of the protein corona that prevents nucleases from accessing the ODNs.
Figure 2.
In vitro properties of SNAs preadsorbed with functional proteins. (A) Schematic representation of the degradation of the ODN shell in the presence of DNase I, in which the Cy5 fluorophore attached to the outer shell is no longer quenched by AuNPs following protease degradation. (B) Fluorescence kinetic profiles of the bare, IgG- (IgG@SNA), anti-HER2- (HER2@SNA), and HSA-immobilized SNAs (HSA@SNA) with and without DNase I treatment. (C) Schematic representation of the hybridization of Cy3.5-labeled complementary strands to the ODNs immobilized on AuNPs. Fluorescence is quenched as hybridization occurs. (D) Degrees of hybridization of SNAs with complementary strands for IgG@SNAs, HER2@SNAs, and HSA@SNAs compared with that for bare SNAs.
Given that the ODNs are potentially sterically hindered when a protein corona is adsorbed to the structures, we examined whether a protein corona reduced their ability to recognize complementary binding partners, a necessary step for antisense and RNA interference pathways as well as mRNA sensing. To assess this property, we designed a AuNP-based fluorescence quenching assay in which the hybridization of fluorophore-labeled (i.e., Cy3.5) strands complementary to those making up the SNA shell results in quenching due to the proximity of the Cy3.5 fluorophore to the AuNP core. The quenching of the Cy3.5 fluorescence by the AuNP core is an indicator of the amount of hybridization and therefore a measure of the surface DNA accessibility (Figure 2C). The percentage of DNA hybridized to protein-immobilized SNAs was calculated in comparison with hybridization measured for protein-free SNAs and was normalized to the hybridization of protein-free SNAs to noncomplementary (i.e., T20) strands. Surprisingly, we found that the preadsorbed protein coronae decreased DNA accessibility by only ∼10% (Figure 2D) compared with bare SNAs. Furthermore, when we assessed whether the most-dense protein corona (i.e., IgG@SNA) altered the specificity, we found no apparent increase in binding of noncomplementary strands compared to a protein-free SNA (Figure S2). Together, these experiments imply that the protein corona does not significantly alter recognition of complementary strands.
Cellular Selectivity of the SNAs with Immobilized Antibodies
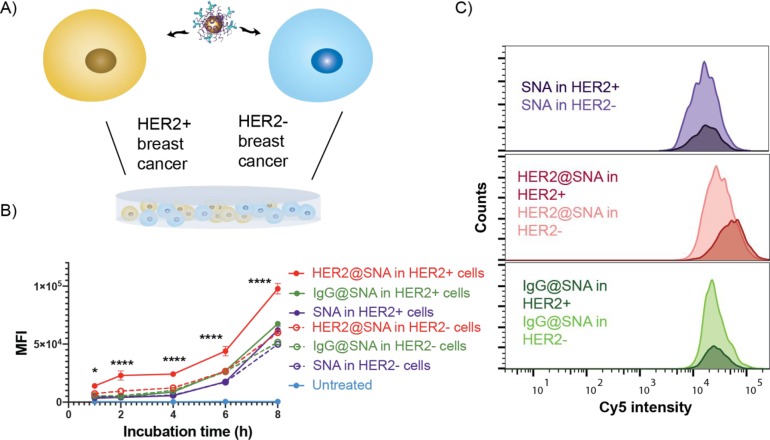
On a cellular level, we investigated whether we could use preadsorbed protein coronae on SNAs to modulate their uptake by targeted cell types. A key attribute of SNAs is that they enter nearly any cell type (over 50 to date),27 an especially powerful property for many therapeutic and diagnostic applications; however, selective targeting could impart an enhanced therapeutic effect. As a first test, we examined the targeting capabilities of SNAs with immobilized antibodies. For these experiments, SNAs were synthesized with an ODN shell containing 10% Cy5-labeled strands for flow cytometry detection, thereby minimizing potential perturbation to the uptake pathway caused by the fluorophore. We then incubated these constructs with either anti-HER2 mAbs or a nontargeting antibody, IgG. We assessed the selectivity of the SNAs by treating the cocultured breast cancer cell lines SK-BR-3 (HER2 overexpressing) and MDA-MB-231 (HER2 negative) with the particles (Figure 3A). Furthermore, the MDA-MB-231 cells were engineered to express green fluorescent protein (GFP), such that the two cell lines could be separated by flow cytometry on the basis of GFP fluorescence intensity. Significantly, the anti-HER2-adsorbed SNAs preferentially entered the HER2-positive cells compared with the HER2-negative ones over an 8 h treatment time (Figure 3B), even under conditions with greater numbers of HER2-negative cells (Figure S3). In contrast, nontargeting IgG-adsorbed SNAs and bare SNAs showed no cellular selectivity. Importantly, precoating SNAs with nontargeting proteins did not seem to reduce their cancer cell uptake efficiency compared to that for bare SNAs (Figure 3C). This finding is consistent with a previous report that mAb functionalization improves cellular selectivity,19 but the physical adsorption of mAbs demonstrated here is easier and more adaptable than the reported conjugation approach. Significantly, cells were treated with mAb-adsorbed SNAs in complete growth medium supplemented with 10% serum, and the targeting capabilities still persisted. These conditions show that mAb-adsorbed SNAs are able to retain cellular selectivity even in the presence of other serum proteins.
Figure 3.
Selective cellular uptake of the monoclonal HER2 antibody-adsorbed SNAs. (A) Schematic describing the SNA treatment of the cocultured HER2-expressing breast cancer cells, SK-BR-3, and non-HER2-expressing breast cancer cells, MDA-MB-231. (B) Uptake profiles of anti-HER2-adsorbed (HER2@SNA), IgG-adsorbed (IgG@SNA), and bare SNAs following incubation for 1–8 h with the cocultured cells (solid circles, HER2-positive cells; open circles, HER2-negative cells). MFI = median fluorescence intensity. (C) Representative overlaid cellular uptake histograms (as measured by Cy5 intensity) for cocultured breast cancer cells after treatment with HER2@SNAs, IgG@SNAs, and bare SNAs for 6 h. The distributions of Cy5 fluorescence of HER2+ cells are denoted by the darker-shaded histograms, while those for the HER2– cells are shown in a lighter shade.
Evasion of Macrophage Clearance of the Dysopsonin-Adsorbed SNAs
Lastly, we investigated the potential utility of our approach for creating SNAs that can target or avoid macrophages, which play central roles in immunomodulation and clearance. For this purpose, we preadsorbed either a recognized opsonin, IgG,59 or a dysopsonin, HSA,21 on the ODN shell of SNAs and incubated them with human macrophages (Figure 4A). Typically, an opsonin marks a construct as foreign and induces macrophage clearance, while dysopsonins do the opposite.60 To perform this experiment, human THP-1 monocytes were first differentiated into macrophages using phorbol 12-myristate 13-acetate (PMA) (Figure S4).61,62 We then treated the macrophages with bare and active protein-coated SNAs for 1–8 h at 37 °C. We hypothesized that IgG preadsorption would improve the SNA uptake efficiency by macrophages while HSA preadsorption would reduce it.21,63 Interestingly, both IgG and HSA adsorption lowered the uptake of the SNAs into the macrophages, with HSA adsorption having a more significant impact on reducing SNA clearance by macrophages (Figure 4B).
Figure 4.
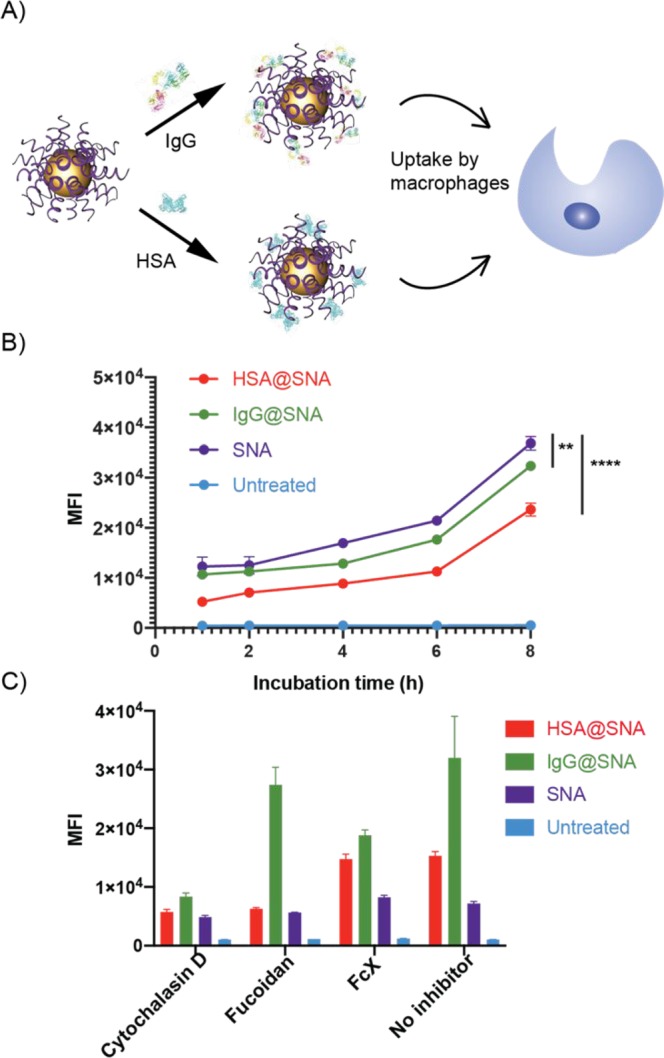
Cellular uptake of the IgG (opsonin)- and HSA (dysopsonin)-adsorbed SNAs. (A) Schematic describing the SNA treatment of THP-1-derived macrophages. (B) Uptake of HSA-adsorbed (HSA@SNA), IgG-adsorbed (IgG@SNA), and bare SNAs following 1–8 h incubation with THP-1-derived macrophages. (C) Receptor inhibited THP-1 macrophage uptake of HSA@SNA, IgG@SNA, and bare SNAs following pretreatment with cytochalasin D, fucoidan, or Fc receptor blocker (FcX).
IgG is an opsonin that can be cleared by macrophages through Fc receptor recognition,64 while non-protein-coated SNAs and HSA alone are reported to enter cells through scavenger receptor A (SR-A) recognition.65,66 Previous findings showed that changes in protein corona composition change the cellular uptake pathway for nanoparticles.67 Therefore, we speculated that coating SNAs with IgG could alter their preferred uptake pathway. Indeed, significantly diminished uptake efficiency was observed for these SNAs when the Fc receptors were blocked by FcX (Figure 4C), meaning that the IgG-coated SNAs entered cells through a different route than observed with typical SNAs. As a comparison, when SR-A was inhibited by fucoidan, the reduction in the uptake efficiency of bare SNAs and HSA-immobilized SNAs is more significant than for IgG-adsorbed SNAs. Since macrophages are phagocytic, when phagocytosis is inhibited by cytochalasin D, the uptake of all SNA types is suppressed. Taken together, these results indicate that IgG immobilization alters the major cellular uptake pathway of SNAs, which could be the reason for the overall reduction of uptake efficiency. Significantly, HSA coating of SNAs reduces nonspecific macrophage uptake compared with bare SNAs, opening new avenues to explore for increasing blood circulation half-life.
Conclusions
This work has introduced a straightforward and flexible method for incorporating active protein coronae on SNA surfaces with relatively high stability even in the presence of serum. Importantly, this method increases the cellular selectivity of SNAs and reduces nonspecific macrophage clearance without significantly affecting the accessibility of the oligonucleotide shell. Therefore, this work points toward a potential strategy for improving SNA targeting and distribution in vivo, which could impact ongoing clinical efforts68 aimed at SNA therapeutic development. Finally, looking forward, this methodology, depending upon particle surface characteristics, could be generally applied to other nanomaterials to improve cellular selectivity.
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Awards U54CA199091 and R01CA208783. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health. This work made use of the IMSERC MS Facility at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205), the State of Illinois, and the International Institute for Nanotechnology (IIN). This work was supported by the Northwestern University Flow Cytometry Core Facility supported by Cancer Center Support Grant NCI CA060553. B.M. acknowledges support from the Eden and Steven Romick Postdoctoral Fellowship through the American Committee for the Weizmann Institute of Science.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.9b01105.
Experimental procedures, tables, and supporting figures (PDF)
Author Present Address
∥ B.M.: Department of Biomedical Engineering, University of North Texas, Denton, TX 76207, United States.
Author Contributions
§ W.Z. and B.M. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Hubbell J. A.; Chilkoti A. Nanomaterials for Drug Delivery. Science 2012, 337 (6092), 303–305. 10.1126/science.1219657. [DOI] [PubMed] [Google Scholar]
- Cedervall T.; Lynch I.; Lindman S.; Berggård T.; Thulin E.; Nilsson H.; Dawson K. A.; Linse S. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2007, 104 (7), 2050–2055. 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y.; Gause K. T.; Kamphuis M. M.; Ang C. S.; O’Brien-Simpson N. M.; Lenzo J. C.; Reynolds E. C.; Nice E. C.; Caruso F. Differential roles of the protein corona in the cellular uptake of nanoporous polymer particles by monocyte and macrophage cell lines. ACS Nano 2013, 7 (12), 10960–10970. 10.1021/nn404481f. [DOI] [PubMed] [Google Scholar]
- Schöttler S.; Becker G.; Winzen S.; Steinbach T.; Mohr K.; Landfester K.; Mailänder V.; Wurm F. R. Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers. Nat. Nanotechnol. 2016, 11, 372–377. 10.1038/nnano.2015.330. [DOI] [PubMed] [Google Scholar]
- Aoyama M.; Hata K.; Higashisaka K.; Nagano K.; Yoshioka Y.; Tsutsumi Y. Clusterin in the protein corona plays a key role in the stealth effect of nanoparticles against phagocytes. Biochem. Biophys. Res. Commun. 2016, 480 (4), 690–695. 10.1016/j.bbrc.2016.10.121. [DOI] [PubMed] [Google Scholar]
- Saha K.; Rahimi M.; Yazdani M.; Kim S. T.; Moyano D. F.; Hou S.; Das R.; Mout R.; Rezaee F.; Mahmoudi M.; Rotello V. M. Regulation of Macrophage Recognition through the Interplay of Nanoparticle Surface Functionality and Protein Corona. ACS Nano 2016, 10 (4), 4421–4430. 10.1021/acsnano.6b00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Dardir K.; Lee K. B.; Fabris L. Impact of Protein Corona in Nanoflare-Based Biomolecular Detection and Quantification. Bioconjugate Chem. 2019, 30 (10), 2555–2562. 10.1021/acs.bioconjchem.9b00495. [DOI] [PubMed] [Google Scholar]
- Schaffler M.; Sousa F.; Wenk A.; Sitia L.; Hirn S.; Schleh C.; Haberl N.; Violatto M.; Canovi M.; Andreozzi P.; Salmona M.; Bigini P.; Kreyling W. G.; Krol S. Blood protein coating of gold nanoparticles as potential tool for organ targeting. Biomaterials 2014, 35 (10), 3455–3466. 10.1016/j.biomaterials.2013.12.100. [DOI] [PubMed] [Google Scholar]
- Staufenbiel S.; Weise C.; Müller R. H. Targeting of Intravenous Polymeric Nanoparticles by Differential Protein Adsorption. Macromol. Symp. 2014, 345 (1), 42–50. 10.1002/masy.201400062. [DOI] [Google Scholar]
- Salvati A.; Pitek A. S.; Monopoli M. P.; Prapainop K.; Bombelli F. B.; Hristov D. R.; Kelly P. M.; Aberg C.; Mahon E.; Dawson K. A. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 2013, 8 (2), 137–143. 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- Jiang X.; Weise S.; Hafner M.; Rocker C.; Zhang F.; Parak W. J.; Nienhaus G. U. Quantitative analysis of the protein corona on FePt nanoparticles formed by transferrin binding. J. R. Soc., Interface 2010, 7 (Suppl_1), S5–S13. 10.1098/rsif.2009.0272.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonigold M.; Simon J.; Estupinan D.; Kokkinopoulou M.; Reinholz J.; Kintzel U.; Kaltbeitzel A.; Renz P.; Domogalla M. P.; Steinbrink K.; Lieberwirth I.; Crespy D.; Landfester K.; Mailander V. Pre-adsorption of antibodies enables targeting of nanocarriers despite a biomolecular corona. Nat. Nanotechnol. 2018, 13 (9), 862–869. 10.1038/s41565-018-0171-6. [DOI] [PubMed] [Google Scholar]
- Mirshafiee V.; Mahmoudi M.; Lou K.; Cheng J.; Kraft M. L. Protein corona significantly reduces active targeting yield. Chem. Commun. 2013, 49 (25), 2557–2559. 10.1039/c3cc37307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist M.; Stigler J.; Elia G.; Lynch I.; Cedervall T.; Dawson K. A. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (38), 14265–14270. 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz S.; Schöttler S.; Kotman N.; Baier G.; Musyanovych A.; Kuharev J.; Landfester K.; Schild H.; Jahn O.; Tenzer S.; Mailänder V. Protein Corona of Nanoparticles: Distinct Proteins Regulate the Cellular Uptake. Biomacromolecules 2015, 16 (4), 1311–1321. 10.1021/acs.biomac.5b00108. [DOI] [PubMed] [Google Scholar]
- Liu X.; Huang N.; Li H.; Jin Q.; Ji J. Surface and Size Effects on Cell Interaction of Gold Nanoparticles with Both Phagocytic and Nonphagocytic Cells. Langmuir 2013, 29 (29), 9138–9148. 10.1021/la401556k. [DOI] [PubMed] [Google Scholar]
- García-Álvarez R.; Hadjidemetriou M.; Sánchez-Iglesias A.; Liz-Marzán L. M.; Kostarelos K. In vivo formation of protein corona on gold nanoparticles. The effect of their size and shape. Nanoscale 2018, 10 (3), 1256–1264. 10.1039/C7NR08322J. [DOI] [PubMed] [Google Scholar]
- Dai Q.; Yan Y.; Ang C.-S.; Kempe K.; Kamphuis M. M. J.; Dodds S. J.; Caruso F. Monoclonal Antibody-Functionalized Multilayered Particles: Targeting Cancer Cells in the Presence of Protein Coronas. ACS Nano 2015, 9 (3), 2876–2885. 10.1021/nn506929e. [DOI] [PubMed] [Google Scholar]
- Zhang K.; Hao L.; Hurst S. J.; Mirkin C. A. Antibody-linked Spherical Nucleic Acids for Cellular Targeting. J. Am. Chem. Soc. 2012, 134 (40), 16488–16491. 10.1021/ja306854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen A. B.; Guan C. M.; Ko C. H.; Mirkin C. A. The Impact of Protein Corona Formation on the Macrophage Cellular Uptake and Biodistribution of Spherical Nucleic Acids. Small 2017, 13 (16), 1603847. 10.1002/smll.201603847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawara K.-i.; Furumoto K.; Nagayama S.; Minato K.; Higaki K.; Kai T.; Kimura T. Pre-coating with serum albumin reduces receptor-mediated hepatic disposition of polystyrene nanosphere: implications for rational design of nanoparticles. J. Controlled Release 2004, 100 (3), 451–455. 10.1016/j.jconrel.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Prozeller D.; Pereira J.; Simon J.; Mailander V.; Morsbach S.; Landfester K. Prevention of Dominant IgG Adsorption on Nanocarriers in IgG-Enriched Blood Plasma by Clusterin Precoating. Adv. Sci. (Weinh) 2019, 6 (10), 1802199. 10.1002/advs.201802199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin C. A.; Letsinger R. L.; Mucic R. C.; Storhoff J. J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382 (6592), 607–609. 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- Cutler J. I.; Auyeung E.; Mirkin C. A. Spherical Nucleic Acids. J. Am. Chem. Soc. 2012, 134 (3), 1376–1391. 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- Seferos D. S.; Prigodich A. E.; Giljohann D. A.; Patel P. C.; Mirkin C. A. Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett. 2009, 9 (1), 308–311. 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giljohann D. A.; Seferos D. S.; Daniel W. L.; Massich M. D.; Patel P. C.; Mirkin C. A. Gold Nanoparticles for Biology and Medicine. Angew. Chem., Int. Ed. 2010, 49 (19), 3280–3294. 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi N. L.; Giljohann D. A.; Thaxton C. S.; Lytton-Jean A. K. R.; Han M. S.; Mirkin C. A. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science 2006, 312 (5776), 1027–1030. 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- Massich M. D.; Giljohann D. A.; Seferos D. S.; Ludlow L. E.; Horvath C. M.; Mirkin C. A. Regulating Immune Response Using Polyvalent Nucleic Acid-Gold Nanoparticle Conjugates. Mol. Pharmaceutics 2009, 6 (6), 1934–1940. 10.1021/mp900172m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giljohann D. A.; Seferos D. S.; Prigodich A. E.; Patel P. C.; Mirkin C. A. Gene Regulation with Polyvalent siRNA-Nanoparticle Conjugates. J. Am. Chem. Soc. 2009, 131 (6), 2072–2073. 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S. A.; Day E. S.; Ko C. H.; Hurley L. A.; Luciano J. P.; Kouri F. M.; Merkel T. J.; Luthi A. J.; Patel P. C.; Cutler J. I.; Daniel W. L.; Scott A. W.; Rotz M. W.; Meade T. J.; Giljohann D. A.; Mirkin C. A.; Stegh A. H. Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Sci. Transl. Med. 2013, 5 (209), 209ra152–209ra152. 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbakry A.; Zaky A.; Liebl R.; Rachel R.; Goepferich A.; Breunig M. Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano Lett. 2009, 9 (5), 2059–2064. 10.1021/nl9003865. [DOI] [PubMed] [Google Scholar]
- Ruan W.; Zheng M.; An Y.; Liu Y.; Lovejoy D. B.; Hao M.; Zou Y.; Lee A.; Yang S.; Lu Y.; Morsch M.; Chung R.; Shi B. DNA nanoclew templated spherical nucleic acids for siRNA delivery. Chem. Commun. 2018, 54 (29), 3609–3612. 10.1039/C7CC09257A. [DOI] [PubMed] [Google Scholar]
- Radovic-Moreno A. F.; Chernyak N.; Mader C. C.; Nallagatla S.; Kang R. S.; Hao L.; Walker D. A.; Halo T. L.; Merkel T. J.; Rische C. H.; Anantatmula S.; Burkhart M.; Mirkin C. A.; Gryaznov S. M. Immunomodulatory spherical nucleic acids. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (13), 3892–3897. 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Qin L.; Yamankurt G.; Skakuj K.; Huang Z.; Chen P. C.; Dominguez D.; Lee A.; Zhang B.; Mirkin C. A. Rational vaccinology with spherical nucleic acids. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (21), 10473–10481. 10.1073/pnas.1902805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molino N. M.; Neek M.; Tucker J. A.; Nelson E. L.; Wang S. W. Display of DNA on Nanoparticles for Targeting Antigen Presenting Cells. ACS Biomater. Sci. Eng. 2017, 3 (4), 496–501. 10.1021/acsbiomaterials.7b00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X.; Li Z.; Wang G.; He X.; Shen X.; Sun Q.; Wang L.; Yue R.; Ma N. MicroRNA-Catalyzed Cancer Therapeutics Based on DNA-Programmed Nanoparticle Complex. ACS Appl. Mater. Interfaces 2017, 9 (39), 33624–33631. 10.1021/acsami.7b09420. [DOI] [PubMed] [Google Scholar]
- Zhang X.-Q.; Xu X.; Lam R.; Giljohann D.; Ho D.; Mirkin C. A. Strategy for Increasing Drug Solubility and Efficacy through Covalent Attachment to Polyvalent DNA-Nanoparticle Conjugates. ACS Nano 2011, 5 (9), 6962–6970. 10.1021/nn201446c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taton T. A.; Mirkin C. A.; Letsinger R. L. Scanometric DNA Array Detection with Nanoparticle Probes. Science 2000, 289 (5485), 1757–1760. 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- Kim D.; Daniel W. L.; Mirkin C. A. Microarray-based multiplexed scanometric immunoassay for protein cancer markers using gold nanoparticle probes. Anal. Chem. 2009, 81 (21), 9183–9187. 10.1021/ac9018389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M. S.; Lytton-Jean A. K. R.; Mirkin C. A. A Gold Nanoparticle Based Approach for Screening Triplex DNA Binders. J. Am. Chem. Soc. 2006, 128 (15), 4954–4955. 10.1021/ja0606475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medley C. D.; Smith J. E.; Tang Z.; Wu Y.; Bamrungsap S.; Tan W. Gold Nanoparticle-Based Colorimetric Assay for the Direct Detection of Cancerous Cells. Anal. Chem. 2008, 80 (4), 1067–1072. 10.1021/ac702037y. [DOI] [PubMed] [Google Scholar]
- Prigodich A. E.; Seferos D. S.; Massich M. D.; Giljohann D. A.; Lane B. C.; Mirkin C. A. Nano-flares for mRNA Regulation and Detection. ACS Nano 2009, 3 (8), 2147–2152. 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seferos D. S.; Giljohann D. A.; Hill H. D.; Prigodich A. E.; Mirkin C. A. Nano-flares: Probes for Transfection and mRNA Detection in Living Cells. J. Am. Chem. Soc. 2007, 129 (50), 15477–15479. 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib M.; Mohamadi R. M.; Poudineh M.; Ahmed S. U.; Ivanov I.; Huang C.-L.; Moosavi M.; Sargent E. H.; Kelley S. O. Single-cell mRNA cytometry via sequence-specific nanoparticle clustering and trapping. Nat. Chem. 2018, 10 (5), 489–495. 10.1038/s41557-018-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X.; Zeng T.; Li Z.; Wang G.; Ma N. Catalytic Molecular Imaging of Micro RNA in Living Cells by DNA-Programmed Nanoparticle Disassembly. Angew. Chem., Int. Ed. 2016, 55 (9), 3073–3076. 10.1002/anie.201509726. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Huang J.; Yang X.; Quan K.; Wang H.; Ying L.; Xie N.; Ou M.; Wang K. FRET Nanoflares for Intracellular mRNA Detection: Avoiding False Positive Signals and Minimizing Effects of System Fluctuations. J. Am. Chem. Soc. 2015, 137 (26), 8340–8343. 10.1021/jacs.5b04007. [DOI] [PubMed] [Google Scholar]
- Chinen A. B.; Ferrer J. R.; Merkel T. J.; Mirkin C. A. Relationships between Poly(ethylene glycol) Modifications on RNA-Spherical Nucleic Acid Conjugates and Cellular Uptake and Circulation Time. Bioconjugate Chem. 2016, 27 (11), 2715–2721. 10.1021/acs.bioconjchem.6b00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Liu J. Freezing Directed Construction of Bio/Nano Interfaces: Reagentless Conjugation, Denser Spherical Nucleic Acids, and Better Nanoflares. J. Am. Chem. Soc. 2017, 139 (28), 9471–9474. 10.1021/jacs.7b04885. [DOI] [PubMed] [Google Scholar]
- Hurst S. J.; Lytton-Jean A. K. R.; Mirkin C. A. Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes. Anal. Chem. 2006, 78 (24), 8313–8318. 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina M. A.; Codony-Servat J.; Albanell J.; Rojo F.; Arribas J.; Baselga J. Trastuzumab (Herceptin), a Humanized Anti-HER2 Receptor Monoclonal Antibody, Inhibits Basal and Activated HER2 Ectodomain Cleavage in Breast Cancer Cells. Cancer Res. 2001, 61 (12), 4744–4749. [PubMed] [Google Scholar]
- Cai R.; Chen C. The Crown and the Scepter: Roles of the Protein Corona in Nanomedicine. Adv. Mater. 2019, 31 (45), 1805740 10.1002/adma.201805740. [DOI] [PubMed] [Google Scholar]
- Nguyen V. H.; Lee B.-J. Protein corona: a new approach for nanomedicine design. Int. J. Nanomed. 2017, 12, 3137–3151. 10.2147/IJN.S129300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockal M.; Carter D. C.; Rüker F. Conformational Transitions of the Three Recombinant Domains of Human Serum Albumin Depending on pH. J. Biol. Chem. 2000, 275 (5), 3042–3050. 10.1074/jbc.275.5.3042. [DOI] [PubMed] [Google Scholar]
- Calmettes P.; Cser L.; Rajnavolgyi E. Temperature and pH dependence of immunoglobulin G conformation. Arch. Biochem. Biophys. 1991, 291 (2), 277–283. 10.1016/0003-9861(91)90135-6. [DOI] [PubMed] [Google Scholar]
- Khawli L. A.; Goswami S.; Hutchinson R.; Kwong Z. W.; Yang J.; Wang X.; Yao Z.; Sreedhara A.; Cano T.; Tesar D.; Nijem I.; Allison D. E.; Wong P. Y.; Kao Y.-H.; Quan C.; Joshi A.; Harris R. J.; Motchnik P. Charge variants in IgG1: Isolation, characterization, in vitro binding properties and pharmacokinetics in rats. MAbs 2010, 2 (6), 613–624. 10.4161/mabs.2.6.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baimanov D.; Cai R.; Chen C. Understanding the Chemical Nature of Nanoparticle-Protein Interactions. Bioconjugate Chem. 2019, 30 (7), 1923–1937. 10.1021/acs.bioconjchem.9b00348. [DOI] [PubMed] [Google Scholar]
- Steel L. F.; Trotter M. G.; Nakajima P. B.; Mattu T. S.; Gonye G.; Block T. Efficient and Specific Removal of Albumin from Human Serum Samples. Mol. Cell. Proteomics 2003, 2 (4), 262–270. 10.1074/mcp.M300026-MCP200. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Quintela A.; Alende R.; Gude F.; Campos J.; Rey J.; Meijide L. M.; Fernandez-Merino C.; Vidal C. Serum levels of immunoglobulins (IgG, IgA, IgM) in a general adult population and their relationship with alcohol consumption, smoking and common metabolic abnormalities. Clin. Exp. Immunol. 2008, 151 (1), 42–50. 10.1111/j.1365-2249.2007.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter E. V.; Stollerman G. H. The opsonization of bentonite particles by γ-globulin. J. Immunol. 1961, 87, 110–118. [PubMed] [Google Scholar]
- Liu F.; Frick A.; Yuan X.; Huang L. Dysopsonin activity of serum DNA-binding proteins favorable for gene delivery. J. Pharmacol. Exp. Ther. 2010, 332 (2), 500–504. 10.1124/jpet.109.159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigneault M.; Preston J. A.; Marriott H. M.; Whyte M. K. B.; Dockrell D. H. The Identification of Markers of Macrophage Differentiation in PMA-Stimulated THP-1 Cells and Monocyte-Derived Macrophages. PLoS One 2010, 5 (1), e8668 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small A.; Lansdown N.; Al-Baghdadi M.; Quach A.; Ferrante A. Facilitating THP-1 macrophage studies by differentiating and investigating cell functions in polystyrene test tubes. J. Immunol. Methods 2018, 461, 73–77. 10.1016/j.jim.2018.06.019. [DOI] [PubMed] [Google Scholar]
- Furumoto K.; Yokoe J. I.; Ogawara K. i.; Amano S.; Takaguchi M.; Higaki K.; Kai T.; Kimura T. Effect of coupling of albumin onto surface of PEG liposome on its in vivo disposition. Int. J. Pharm. 2007, 329 (1–2), 110–116. 10.1016/j.ijpharm.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Sobota A.; Strzelecka-Kiliszek A.; Gladkowska E.; Yoshida K.; Mrozińska K.; Kwiatkowska K. Binding of IgG-opsonized particles to FcγR is an active stage of phagocytosis that involves receptor clustering and phosphorylation. J. Immunol. 2005, 175 (7), 4450–4457. 10.4049/jimmunol.175.7.4450. [DOI] [PubMed] [Google Scholar]
- Choi C. H.; Hao L.; Narayan S. P.; Auyeung E.; Mirkin C. A. Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (19), 7625–7630. 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M.; Baumgartner R.; Aldi S.; Dusart P.; Hedin U.; Gustafsson B.; Caidahl K. Human serum albumin-based probes for molecular targeting of macrophage scavenger receptors. Int. J. Nanomed. 2019, 14, 3723–3741. 10.2147/IJN.S197990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia V.; Yang K.; Deville S.; Reker-Smit C.; Nelissen I.; Salvati A. Corona Composition Can Affect the Mechanisms Cells Use to Internalize Nanoparticles. ACS Nano 2019, 13 (10), 11107–11121. 10.1021/acsnano.9b03824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NU-0129 in Treating Patients With Recurrent Glioblastoma or Gliosarcoma Undergoing Surgery. ClinicalTrials.gov, 2017. https://clinicaltrials.gov/ct2/show/NCT03020017 (accessed Oct 29, 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.