Abstract
In the most recent years, an extraordinary research effort has emerged to disentangle osteoarthritis heterogeneity, opening new avenues for progressing with therapeutic development and unravelling the pathogenesis of this complex condition. Several phenotypes and endotypes have been proposed albeit none has been sufficiently validated for clinical or research use as yet. This review discusses the latest advances in OA phenotyping including how new modern statistical strategies based on machine learning and big data can help advance this field of research.
Keywords: Osteoarthritis, phenotype, stratification, precision medicine, big data, machine learning
INTRODUCTION
Osteoarthritis (OA) is among the most prevalent and debilitating chronic diseases worldwide, affecting predominantly older adults (1, 2). There has been significant effort to develop therapies to improve care for OA patients both from the symptomatic perspective and from the point of view of structure modification. Despite that, no therapies have been proven to modify disease progression or proven to be highly effective for symptomatic relief, other than joint replacement for advanced disease, leading to profound disappointment among researchers, patients and clinicians.
The contemporaneous evidence-based management of OA is based on non-pharmacological and pharmacological therapies, with surgical intervention reserved for patients with severe disabling symptoms who have not improved with non-surgical interventions (3). However, despite numerous treatment options being available, outcomes for patients with OA are usually suboptimal and patients remain vulnerable to the clinical consequences of the disease on pain and physical function (4). An important aspect of OA is its extraordinary interpatient variability in clinical and structural manifestations (5, 6). This heterogeneity may be one of the major factors associated with the complexity of OA and with the difficulties to identify one-fits-all therapeutic strategies.
Major advances in the physiopathology and manifestations of OA have occurred in the last decade, revealing the variety of potential molecular and cellular changes that can be involved in the joint destruction process. It has been demonstrated that all joint tissues can be affected (7), which occurs in different extents across patients and results in an array of possible structural OA manifestations (e.g. variable degrees of inflammation, meniscal lesions, bone damage, etc.). In addition, the pain experience can be caused by different factors including the peripheral joint pathology and extra-articular sources of pain such as psychosocial factors and neural mechanisms (8). New therapies have been developed that target pain but not structure including inhibitors of nerve growth factor (NGF) such as the anti-NGF monoclonal antibody tanezumab (9, 10). Interventions targeting many of these structural pathologies and pain mechanisms have been tested in trials but none to date have been approved as structural or disease modifying therapies.
One reason for the failure of clinical trials testing therapeutics intended for structure modification in OA is that it is unclear at present which patients would be most suitable for a specific therapy. For example, the failure of bisphosphonates to slow OA progression might have been due to enrolling any patient with symptomatic OA rather than selecting patients with greater subchondral bone turnover (11). In order to address the heterogeneity of OA to improve clinical research and trials, a new model of understanding OA based on a phenotype-guided approach is needed. Recently, a significant research effort has emerged aimed to define a classification of OA phenotypes for the purpose of better identifying individuals at higher risk of progression and to better delineate OA subpopulations caused by distinct risk factors and disease mechanisms that would be suitable for targeted treatment and prevention strategies.
Other medical fields are more advanced than OA when it comes to disease phenotyping such as chronic obstructive pulmonary disease (12) and heart failure (13). A classification of phenotypes has been achieved in these fields, defining disease subtypes within the spectrum of the condition. In this regard, such classifications are only relevant and clinically useful if they can inform on differences in underlying pathophysiology, clinical outcomes or management. For example, heart failure is recognized as being divided into two main types according to a patient’s left ventricle ejection fraction (i.e. percentage of blood that comes out of the heart with each contraction), which can be reduced or preserved and is associated with differences in systemic and local mechanisms, risk factors, natural history and treatment options (14). Another example is the field of oncology. Diseases such as breast cancer used to be seen as the same condition across different patient groups. It is now known that breast cancer is a heterogeneous condition with varied molecular pathophysiology, such as the presence/absence of biomarkers including hormone-receptors and the HER2 protein (15). Patients are now treated and have their prognosis estimated according to these biomarkers which delineate disease subtypes with particular behaviors.
In knee OA, for example, clinically distinct subtypes exist such as medial and lateral tibiofemoral OA and patellofemoral knee OA, and potentially many others. However, the understanding of how this affects treatment decisions and prevention strategies is still in its infancy. Identifying specific OA phenotypes and endotypes can inform both prognosis and guide therapeutic development for this prevalent disease, with the potential of positively impacting patient care. This review summarizes the latest progress on phenotyping/endotyping OA research and includes a discussion on novel methodologies for phenotyping based on machine learning and big data.
APPROACHES FOR OA PHENOTYPING
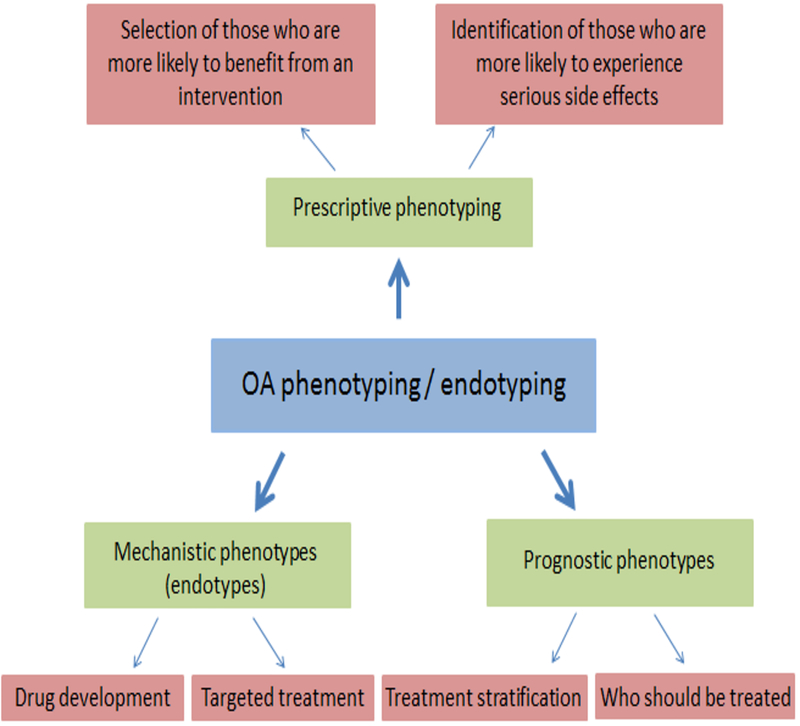
A phenotype can be understood as the composite observable characteristics of an individual that result from genetic combined with environmental factors. Subgroups of patients that have similar clinically observable characteristics are considered to represent a phenotype. Division of patients into discrete subgroups or subtypes is sometimes referred to as stratification. Prognostic phenotyping is the identification of subgroups that are more likely, within a specified period of time, to reach a specific outcome of interest (e.g., disease progression defined by deterioration in joint structural features and worsening pain) (Figure 1). Prescriptive phenotyping aims to define subgroups more likely to respond to a specific intervention with an outcome of interest (e.g., improved pain or function). Identifying subgroups by prognostic and prescriptive phenotyping (e.g., using prediction models) is necessary to meet the goals of precision medicine, defined as “treatments targeted to the needs of individual patients on the basis of genetic, biomarker, phenotypic, or psychosocial characteristics that distinguish a given patient from other patients with similar clinical presentations”(16).
Figure 1.
Examples of different types of phenotypes and endotypes and their potential uses.
Another concept that has emerged from phenotyping studies in chronic conditions such as asthma is the term “endotype”(17). Unlike phenotypes, which are based on clinical characteristics that are not necessarily connected to an established pathophysiologic mechanism of disease, an endotype “is a subtype of disease defined functionally and pathologically by a molecular mechanism”(17). It is important to note that a given OA phenotype (e.g. medial tibiofemoral OA) may be common to multiple endotypes (i.e., different mechanisms leading to the same manifestation). The importance of identifying endotypes for targeted treatment has gained much attention particularly from the point of view of drug discovery, where identifying the right target is key for success.
How to best subset OA into phenotypes and endotypes and whether certain subsets are of any clinical value is an important issue that has not yet been fully addressed but is under active investigation. A commentary was written in 2009 (18) by three notable figures in OA research in response to an article suggesting that “primary” or idiopathic OA could be divided into subsets consisting of genetic defects, menopause-associated estrogen deficiency, and aging (19). The authors of the commentary, Ken Brandt, Paul Dieppe, and Eric Radin, eloquently argued that dividing OA into primary and secondary subsets is not useful since “all OA is secondary” and that any attempt to subset OA had to take into account the fact that OA is largely a condition driven by the response to mechanical stress on the joint. They suggested that subsetting OA should be done on the basis of the mechanical abnormalities responsible for OA in a group of individuals that could include joint trauma, neuromuscular factors that affect the ability to absorb loading, congenital or developmental anatomic abnormalities causing joint incongruities or postinfectious. They also noted that it will be important to consider the large number of confounding variables involved.
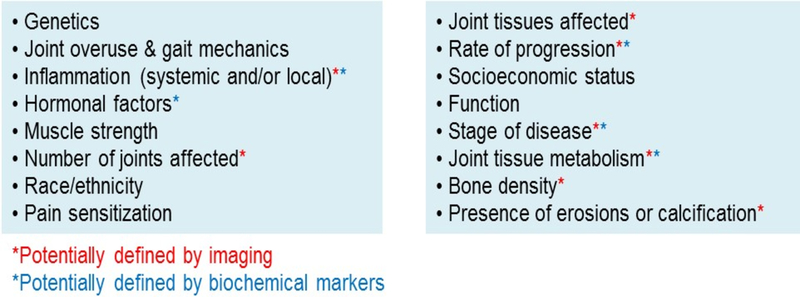
Like other chronic heterogeneous conditions, there are multiple genetic and environmental factors that increase the risk of developing the joint changes characteristic of OA, clinical manifestations of pain and loss of function, and progression to end-stage disease (20, 21). There is a critical need to accurately define the various factors that could contribute to phenotypes and subgroups of OA from a large number of potentially important variables (Figure 2). It is becoming clear that simply defining OA phenotypes based on risk factors (for example posttraumatic OA, obesity-related OA, age-related OA, postmenopausal OA, genetic OA, mechanical OA) is far too simplistic. Many individuals have more than one risk factor and, as already noted in the case of mechanics, there are shared mechanisms among risk factors with mechanical factors likely contributing to all OA. For phenotyping to be successful, datasets with a diverse set of variables and well-defined outcomes are needed. This may include various socio-demographic factors and clinical, imaging, and biochemical marker measurements in addition to mechanical measures. In some cases, genetic and “omic” data (transcriptomic, proteomic, metabolomic, microbiomic), or data from histologic analysis of tissue samples may be needed that will allow for more precise phenotyping.
Figure 2.
Factors and manifestations that could contribute to OA phenotypes/subgroups.
Various types of analysis can be used to define phenotypes using clinical and biological data, which are discussed below in the “Big data/ machine learning for OA phenotyping” section. Latent class analysis was used to cluster clinical and imaging data from the Osteoarthritis Initiative (OAI) database and four clusters were identified that represented mild OA, mild OA with areas of denuded bone that the authors called “classical OA”, and two severe OA groups of “aggressive OA” with larger areas of denuded bone and a high prevalence of progression with one of the two latter groups exhibiting more lateral involvement (22). Variables that were significantly different among clusters included BMI, alignment, and history of trauma which again emphasizes the importance of mechanics in OA.
A recent systematic review of OA phenotypes published in 2016 examined the current evidence for groups of variables that would distinguish OA phenotypes (23). This review identified six phenotypes from 24 published studies that included: 1) chronic pain phenotype with central sensitization; 2) inflammatory phenotype; 3) metabolic syndrome phenotype; 4) bone and cartilage metabolism phenotype; 5) mechanical (malalignment) phenotype; and 6) minimal joint disease phenotype. It is not clear how meaningful these phenotypes might be clinically or if they could be used for attempts at stratifying patients for clinical trials. A “metabolic syndrome phenotype” may simply represent individuals with obesity and OA. The association between metabolic syndrome and OA has not been well established and, after controlling for BMI, the correlations between OA and features of the metabolic syndrome are not significant (24). As mentioned above, all OA has a mechanical component and so it is unlikely to represent a distinct phenotype and all OA has involvement of bone and cartilage also making it unlikely to represent a distinct phenotype.
There has been much interest in defining an inflammatory phenotype or endotype in OA as well as a bone phenotype. Methodologies that can be used to better define an inflammatory phenotype include imaging (e.g., ultrasound or MRI) to detect synovitis (25, 26), blood levels of high sensitivity CRP and IL-6 (27–29), soluble macrophage markers (CD14 and CD163 in the synovial fluid and CD163 in serum) (30) and transcriptome data from peripheral blood leukocytes (31). Defining a bone phenotype would likely include imaging of osteophytes and subchondral sclerosis using radiographs and bone marrow lesions detected by MRI. However, since these are common features of OA, it is difficult to envision how they would define a distinct bone phenotype. Biomarkers of bone turnover such as urinary CTX-1, urinary NTX-1, serum PINP, and serum osteocalcin could be used to provide additional data on bone turnover (32, 33). Attempts to utilize inflammatory and bone phenotypes for targeted therapy, as well as the use of additional biomarkers, will be discussed below.
CURRENT STATE OF OA PHENOTYPING
Endotypes/mechanistic subgroups
As discussed above, endotypes are disease subtypes resulting from differences in specific pathobiological mechanisms. Research so far has suggested the existence of a few possible endotypes that will be discussed in this review. However, there may be many others that are not included herewith. In this regard, pre-clinical studies are key to understand how different etiologies such as post-traumatic and age-related OA may be associated with differences in disease pathophysiology and expression.
A structural endotype related to ageing or cell senescence has been suggested by a few studies, primarily in pre-clinical models (34, 35). Not only greater OA severity but differences in gene expression and pathways represented by these genes have been observed in older mice compared to younger mice in an injury-induced OA model (36), suggesting that the same OA model may result in different phenotypes depending on age. Defining a senescence endotype in human OA may be important as drugs for treatment of OA called senolytics are being developed that target and kill senescent cells in the joint (37).
There is an active interest in defining an inflammatory endotype that would have a shared mechanism related to a specific cytokine that could be targeted for therapy. Given the role for cytokines in OA this could be of value in advancing OA treatment as well. Supporting the existence of an endotype with increased inflammatory characteristics, Attur et al showed the existence of two subgroups of knee OA patients with different gene expression profiles in peripheral blood leukocytes. In this study, clinical and structural outcomes were worse in the subgroup with higher production of IL-1β compared to the subgroup with lower levels (31). However, whether targeting specific anti-cytokine therapies to an inflammatory OA phenotype would be useful remains unproven. A recent phase 2 trial of an anti-IL1α/β antibody lutikizumab in patients with knee OA and synovitis detected by MRI showed limited improvement in pain scores and no change in synovitis in the treated group compared to placebo controls (38). This does not necessarily mean that targeting the inflammatory phenotype will not be successful but rather that inhibition of IL-1 may not be the right target. Other potential endotypes include ones associated with metabolic factors (39, 40) and hormonal dysregulation (41, 42).
Pain endotypes have also been investigated. Individuals with or at risk of knee OA who displayed greater features of sensitization such as pressure pain sensitivity and temporal summation experienced worse clinical outcomes in cross-sectional studies (43–45) and were more likely to develop incident persistent pain after two years in a recent longitudinal analysis (46, 47). Presence of psychological characteristics such as pain catastrophizing has additionally been shown to negatively influence pain outcomes (48). Identifying individuals with those characteristics and tailoring pain therapies to each patient’s needs may be vital to achieve better clinical outcomes.
Prognostic subgroups
Heterogeneity in long-term OA outcomes has been highlighted by a number of recent studies using trajectory analysis. This statistical methodology uses a data-driven approach to identify clusters of people following different trajectories in a given outcome over time. Using trajectory analysis, we have recently shown that a minority of knee OA individuals (around 1 in 10) experience medial cartilage thickness loss assessed on MRI over 2 years (49), which is consistent with other studies using radiographs for outcome assessment (50, 51). This subgroup had greater odds of experiencing concurrent pain progression and requiring total knee replacement. Importantly, this subgroup could be identified with relatively good accuracy by a set of baseline clinical and disease characteristics. Other studies have also defined prediction models to identify knee OA progression (52–54) and incident knee OA with fast progression (55); however, there were different definitions of progression in these studies, such as an increase in Kellgren Lawrence grade (KLG) (52) and a reduction in medial joint space on radiographs (54). It should be highlighted that prediction models in most of these studies used characteristics that can be easily obtained through the clinical history, physical examination or radiographic assessment to facilitate their use in clinical practice or research using clinical datasets.
Other subgroups have been defined according to trajectories of clinical progression (56–59). Clear differences in prognosis have been shown, with the majority of individuals following a stable course over several years with mild to moderate symptoms, while others experience a more severe disease course with persistent intense pain and disability or significant decline over a few years. We have previously summarized these studies as well as the baseline characteristics that more frequently predicted worse trajectory outcomes (60). These included high BMI, lower education, more severe symptoms and radiographic disease at baseline, psychological factors (use of passive coping strategies and depression) and presence of other comorbidities including concomitant hip pain (60).
Defining individuals most likely to develop OA and the subgroup of individuals with OA most likely to exhibit structural progression within a selected time frame (rapid progressors) for both observational studies and clinical trials is extremely important. Latent class analysis of MRI variables collected at baseline from individuals without radiographic OA in the Multicenter Osteoarthritis Study, including cartilage damage, bone marrow lesions, meniscal tears, meniscal extrusion, synovitis, and effusion, was used to determine the odds of developing incident radiographic OA in 4 subgroups (61). As might be expected, those in the subgroups with more severe lesions were at greater risk than those with mild lesions. Radiographic criteria for progressor vs non-progressors by measurement of change in joint space width on plain films includes the OARSI-OMERACT criteria (62). These criteria were used to select progressors and non-progressors for a urine metabolomics study of participants in the Intensive Diet and Exercise for Arthritis (IDEA) trial that analyzed the metabolomics data using OPLS-DA, a commonly employed method for analysis of such high dimensional multicollinear data (63). OPLS-DA distinguished the metabolite profile of radiographic progressors from non-progressors and found a panel of metabolites that associated with radiographic progression (63).
Biochemical markers, measured in serum, plasma, urine or synovial fluid, represent measures that can provide important information for phenotyping. To date no single marker has been found to be sufficient for diagnosis or prognosis in OA. A study from the Foundation for the NIH (FNIH) OA biomarkers Consortium, examined 18 biomarkers measured at baseline, 12 and 24 months in 194 participants from the OAI study (64). Eight catabolic biomarkers (urine (u) c-terminal crosslinked telopeptide type II collagen (uCTXII), uC2C-Human Urine Sandwich Assay (HUSA), type I collagen cross-linked N-telopeptide (uNTXI), c-terminal crosslinked telopeptide of type I collagen α and β (uCTX1α and uCTXIβ, respectively), serum (s) hyaluronic acid (sHA) and c-terminal crosslinked telopeptide type II collagen (sCTXI)) and one anabolic marker, N-terminal pro-peptide of collagen IIA (sPIIANP), were shown to be the best predictors of pain and radiographic progression over 48 months. These and newer biomarkers under development may be useful to define phenotypes most likely when used in combination with other data such as imaging and clinical data.
Treatment response subgroups / prescriptive phenotyping
There is an increasing understanding and awareness that optimal effects of OA treatments might be attained by personalizing care according to clinically relevant characteristics which is a goal of precision medicine. Clinical trials can be used to investigate variations in the extent patients improve or fail to improve with a given treatment, while observational studies cannot differentiate the effect of the intervention from the disease natural history without an appropriate control. Most analyses investigating subgroup effects of interventions in OA have been post-hoc and exploratory. For example, there is a great interest to identify subgroups of patients in whom biomechanical interventions may be more effective, which has been highlighted as a research priority by the National Institute for Health Care and Excellence (65). It has been shown, in an underpowered analysis from a clinical trial, that unloading shoes alleviate knee pain in a significantly greater extent in patients with more severe structural disease (KLG 3 or 4) than those with milder disease (KLG 2) compared to conventional shoes, and that this may be mediated by a decrease in peak knee adduction moment (66). Baseline disease severity, represented by higher baseline knee pain scores (at least 70 in a 0–100 scale), was also significantly associated with a greater response to intra-articular glucocorticoid injection after up to 4 weeks compared to placebo injection (67).
There is also interest in determining if a bone phenotype exists in OA that might be responsive to agents that act on bone such as the antiresorptive bisphosphonates. Selecting OA patients with bone marrow lesions in a clinical trial of the bisphosphonate zoledronic acid resulted in reduction in pain and the size of the bone marrow lesions in the treatment group compared to the placebo group over 6 months but not over 3 or 12 months (68). An individual patient data meta-analysis is underway to investigate whether particular patient subgroups are more likely to benefit from bisphosphonates than others (69).
Trajectory analysis has been used to investigate heterogeneous response following exposure to interventions (70). Lee et al found four patterns of response to exercise over 12 weeks among knee OA patients, with over 70% of patients displaying early improvement while a minority experienced delayed improvement (15%) or no improvement (10%). Individuals with poorer physical and psychosocial status at baseline were more likely to follow an unfavorable trajectory. These findings highlight that early treatment is vital to reduce pain and disability and suggest that appropriate patient stratification may be needed to triage patients according to their likelihood of improvement with a given intervention. In other words, it would be helpful for clinicians to be able to differentiate patients who are likely to improve with safer and cheaper interventions, such as exercise and diet, from those who may need additional interventions (likely more complex and expensive) or a higher level of care. Currently, new OA models of care, characterized by a multi-disciplinary and holistic approach to personalize the therapeutic plan, have been implemented in several countries (71). At present, a decision support tool to identify persons with OA who would benefit the most from those programs and who would be best targeted by specific interventions is lacking.
Prescriptive phenotyping can also be used to identify individuals more likely to experience serious side effects from interventions. A subgroup of patients receiving anti-NGF experience more rapid progression of their structural changes which is a serious concern that could limit its use unless those at higher risk of this adverse effect can be identified before treatment is initiated (72). A recent exploratory study attempted to do just that using a panel of serum biomarkers in an attempt to phenotype participants in the tanezumab trials who were most at risk of developing a rapidly progressive OA phenotype (73).
BIG DATA/ MACHINE LEARNING FOR OA PHENOTYPING
Data sets in medicine are becoming ever larger, and in order to utilize these vast amounts of data, researchers need to look beyond traditional statistical methodology. Enter machine learning, where a computer learns from a variety of examples, eventually “learning” to classify new information based on these inputs (74). There are many names for machine learning methodologies, including artificial intelligence, artificial neural networks, deep learning, support vector machines, decision trees, etc. Machine learning algorithms can be supervised, unsupervised, or somewhere in between.
Only recently have these methodologies begun to impact the world of OA research (75). A variety of machine learning methodologies initially utilized primarily for image analysis in OA (76–78), are being increasingly applied to large datasets, often including detailed imaging, biochemical biomarker, and genetic/genomic information, for the purpose of identifying important OA subgroups (79–82). Of course, no method is a panacea, and analysis of large datasets can also generate seemingly meaningful results which are in fact spurious artifacts. As datasets become ever larger, and incorporate more complex objects, it becomes increasingly important to link confirmatory analysis with the scientific discovery process, while incorporating study design and subject area expertise.
Unsupervised learning
There are many alternative approaches to the validation and discovery of novel phenotypes (e.g., latent class methods, Bayesian approaches, factor analysis, etc.). A variety of tools are available for exploring clusters within data, all of which have positive and negative aspects, particularly in the high dimension and relatively low sample size setting (83). For example, cluster analysis (i.e. data segmentation) attempts to group items into clusters, such that items within a cluster are more closely related than those in different clusters. Once identified, these clusters are frequently arranged into a hierarchy to represent similarity among clusters. Principal components analysis and related methods attempt to reduce dimensionality in a dataset, thus providing more interpretable clusters, similar to the goals of latent class analysis (a subset of structural equation modeling) and factor analysis. Regardless of methods used, clusters can be readily identified in large datasets; the central challenge is to determine whether identified clusters are actually important in such unsupervised analyses. SigClust is an example of a novel method for hypothesis testing of clusters in high dimensions (84, 85), which can be used in conjunction with subject area expertise to determine whether identified clusters are likely important.
Supervised learning
In contrast to data-driven clustering, supervised machine learning methods are based in hypotheses, but take advantage of the computer’s ability to utilize high dimensional data, beyond what is often possible using traditional statistical methods. One approach, described by J.S. Marron and colleagues, is Object Oriented Data Analysis (OODA (86, 87)). OODA attempts to understand the data structure, determine appropriate data objects, and choose an appropriate analysis for the situation. Whereas in a typical analysis, the experimental unit may be a number or a set of numbers (i.e., a vector), OODA allows assessment of more complicated data objects, such as images or large multivariable datasets with repeated observations. Through consideration of all of the variables related to a given observation as a single data object, potential biases related to variable selection are alleviated, while providing a more complete picture using all available data.
Some of the OODA methods developed by Marron and colleagues include Distance Weighted Discrimination (DWD) and the Direction-Projection-Permutation (DiProPerm) test, which can be utilized in the setting where the classes are known, for example when validating previously hypothesized phenotypes, or comparing progressors and non-progressors. DWD is a linear discriminant analysis method allowing maximal separation of data points by class (88), which has been utilized in OA (78, 81), and is particularly suited to cases where the dimension of the data vector exceeds the number of samples (i.e., a large number of measurements relative to the sample size). The difference between two distributions obtained using DWD can then be tested for statistical significance using the DiProPerm test (89). DiProPerm ensures statistical specificity of the hypothesis test for two previously defined populations by first finding a separating direction (e.g. DWD), then projecting the data and using a one dimensional summary of the separation (e.g. the difference of the means). Statistical significance is obtained by a permutation approach, where the class labels are randomly shuffled and the DWD direction and projections are recomputed, giving a null distribution whose quantiles are used to compute p-values. This powerful machine learning method treats the overall vector of features as a single data object, so there is no requirement for adjustment for multiple comparisons.
Examples of applied machine learning in OA
Our group has applied DWD and DiProPerm to the publicly available OA Initiative FNIH Biomarkers Consortium dataset to assess differences between non-progressors and progressors by both radiographic (rKOA) and pain criteria (81). We found that, among 597 observations and 73 variables, the grouped baseline MRI variables contributed more (z score range 10.28–11.62) to the difference between progressors and non-progressors than did demographic and clinical variables (z score =1.47) or biochemical markers (z score =2.43). In addition, specific baseline variables (Western Ontario and McMaster Universities Osteoarthritis Index [WOMAC] pain, sPIIANP, and lateral meniscal extrusion) were higher among non-progressors, while uCTX-II, bone marrow lesions, and osteophyte number were higher among progressors; features that might inform phenotypes. In support of the validity of our methods, the published FNIH biomarkers study (64) also found sPIIANP was higher in non-progressors and uCTX-II was the strongest biomarker predictor of progression in the same dataset. The consistency of the results is reassuring, but notably, we were able to identify all of these associations in a single machine learning based analysis, rather than numerous studies focused on one or only a few features.
Strengths and limitations
Machine learning methodologies provide several advantages, including the ability to treat all available data as a single data object in high dimensional space, thereby obviating the need to adjust for multiple comparisons in some cases. All variables, even when there are hundreds or thousands of variables, can be considered together, reducing bias related to variable selection. A single analytic model can simultaneously identify a number of key variables or contributors to outcomes of interest. Limitations include the inherent limitations of the dataset (the analysis is limited to what is available), issues of generalizability which requires both internal and external validation, and the need for further analyses to determine the importance of the associated variables. These approaches, as currently described, are akin to genome-wide association study (GWAS) in that they provide several variables (i.e., single-nucleotide polymorphisms (SNPs)) of interest, but further “functional” assessments are needed to confirm and characterize the importance of those variables to the disease process both in the discovery cohort and in external populations.
FUTURE DIRECTIONS
The field of OA phenotyping has evolved significantly, with multiple studies aiming to identify phenotypes based on different OA aspects (e.g. clinical, structural, laboratory and aetiologic phenotypes) and employing different methodologies. A framework to guide future research is underway and will hopefully help to optimize efforts in this field. As highlighted in this review, efforts in OA phenotype research should focus on one or more of three main goals: identify those individuals at higher risk of progression; identify those more likely to benefit from a given existing treatment; and identify specific pathological processes (i.e. disease mechanisms representing specific endotypes) for targeted treatment, potentially with new agents/treatment strategies. Ultimately, all approaches aim to achieve improved clinical outcomes for individual OA patients. In addition, there is a widely recognized discordance between structural involvement and symptomatic disease in OA, and phenotypes are likely to be more helpful if defined according to a specific perspective (i.e. structural damage and pain mechanisms) to inform treatment strategies. There is a need to identify those individuals in whom structural and clinical progression are coupled and those in whom they are dissociated.
Research investigating phenotypes in other joints such as the hip, hand, foot and spine has lagged far behind that of knee OA and should also be the focus of future studies. Different OA phenotypes are likely to exist in joints with different pathophysiological drivers (knee vs. thumb base vs. hip), although risk factors that are common to more than one joint may be implicated in a phenotype of multiple joint OA (90).
Most evidence on the presence of phenotypes/endotypes so far come from single or few studies and lack validation. Further research is needed to validate previous findings and to assess their implications for clinically important outcomes and clinical trial design. In this regard, the availability of high-quality data, ideally longitudinal and from different populations, is key for OA phenotyping research. Efforts to combine datasets from existing OA cohorts/previous clinical trials are likely to be helpful by providing larger datasets for the identification and validation of phenotypes. In addition, imaging and/or laboratory biomarkers may be useful to define clinically relevant phenotypes. Nonetheless, to increase the uptake of proposed phenotypes and translation into practice, phenotypes should be recognizable using easy to obtain patient data (either clinical, imaging or laboratory).
REFERENCES
- 1.Cross M, Smith E, Hoy D, Nolte S, Ackerman I, Fransen M, et al. The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study. Annals of the rheumatic diseases. 2014;73(7):1323–30. [DOI] [PubMed] [Google Scholar]
- 2.Hunter DJ, Schofield D, Callander E. The individual and socioeconomic impact of osteoarthritis. Nature reviews Rheumatology. 2014;10(7):437–41. [DOI] [PubMed] [Google Scholar]
- 3.Bannuru RR, Osani MC, Vaysbrot EE, Arden NK, Bennell K, Bierma-Zeinstra SMA, et al. OARSI guidelines for the non-surgical management of knee, hip, and polyarticular osteoarthritis. Osteoarthritis and cartilage. 2019. July 3 pii: S1063–4584(19)31116–1. doi: 10.1016/j.joca.2019.06.011. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 4.Weinstein AM, Rome BN, Reichmann WM, Collins JE, Burbine SA, Thornhill TS, et al. Estimating the burden of total knee replacement in the United States. The Journal of bone and joint surgery American volume. 2013;95(5):385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierma-Zeinstra SM, Verhagen AP. Osteoarthritis subpopulations and implications for clinical trial design. Arthritis research & therapy. 2011;13(2):213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felson DT. Identifying different osteoarthritis phenotypes through epidemiology. Osteoarthritis and cartilage. 2010;18(5):601–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis and rheumatism. 2012;64(6):1697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neogi T. The epidemiology and impact of pain in osteoarthritis. Osteoarthritis and cartilage. 2013;21(9):1145–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lane NE, Schnitzer TJ, Birbara CA, Mokhtarani M, Shelton DL, Smith MD, et al. Tanezumab for the treatment of pain from osteoarthritis of the knee. The New England journal of medicine. 2010;363(16):1521–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnitzer TJ, Easton R, Pang S, Levinson DJ, Pixton G, Viktrup L, et al. Effect of Tanezumab on Joint Pain, Physical Function, and Patient Global Assessment of Osteoarthritis Among Patients With Osteoarthritis of the Hip or Knee: A Randomized Clinical Trial. JAMA : the journal of the American Medical Association. 2019;322(1):37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karsdal MA, Michaelis M, Ladel C, Siebuhr AS, Bihlet AR, Andersen JR, et al. Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: lessons learned from failures and opportunities for the future. Osteoarthritis Cartilage. 2016;24:2013–21. [DOI] [PubMed] [Google Scholar]
- 12.Mirza S, Benzo R. Chronic Obstructive Pulmonary Disease Phenotypes: Implications for Care. Mayo Clinic proceedings. 2017;92(7):1104–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samson R, Jaiswal A, Ennezat PV, Cassidy M, Le Jemtel TH. Clinical Phenotypes in Heart Failure With Preserved Ejection Fraction. Journal of the American Heart Association. 2016;5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah SJ, Kitzman DW, Borlaug BA, van Heerebeek L, Zile MR, Kass DA, et al. Phenotype-Specific Treatment of Heart Failure With Preserved Ejection Fraction: A Multiorgan Roadmap. Circulation. 2016;134(1):73–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waks AG, Winer EP. Breast Cancer Treatment: A Review. JAMA. 2019;321(3):288–300. [DOI] [PubMed] [Google Scholar]
- 16.Jameson JL, Longo DL. Precision medicine--personalized, problematic, and promising. The New England journal of medicine. 2015;372(23):2229–34. [DOI] [PubMed] [Google Scholar]
- 17.Anderson GP. Endotyping asthma: new insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet. 2008;372(9643):1107–19. [DOI] [PubMed] [Google Scholar]
- 18.Brandt KD, Dieppe P, Radin EL. Commentary: is it useful to subset “primary” osteoarthritis? A critique based on evidence regarding the etiopathogenesis of osteoarthritis. Seminars in arthritis and rheumatism. 2009;39(2):81–95. [DOI] [PubMed] [Google Scholar]
- 19.Herrero-Beaumont G, Roman-Blas JA, Castaneda S, Jimenez SA. Primary osteoarthritis no longer primary: three subsets with distinct etiological, clinical, and therapeutic characteristics. Seminars in arthritis and rheumatism. 2009;39(2):71–80. [DOI] [PubMed] [Google Scholar]
- 20.Blagojevic M, Jinks C, Jeffery A, Jordan KP. Risk factors for onset of osteoarthritis of the knee in older adults: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2010;18(1):24–33. [DOI] [PubMed] [Google Scholar]
- 21.Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best Pract Res Clin Rheumatol. 2014;28(1):5–15. [DOI] [PubMed] [Google Scholar]
- 22.Waarsing JH, Bierma-Zeinstra SM, Weinans H. Distinct subtypes of knee osteoarthritis: data from the Osteoarthritis Initiative. Rheumatology (Oxford). 2015;54(9):1650–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dell’Isola A, Allan R, Smith SL, Marreiros SS, Steultjens M. Identification of clinical phenotypes in knee osteoarthritis: a systematic review of the literature. BMC musculoskeletal disorders. 2016;17(1):425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berenbaum F, Wallace IJ, Lieberman DE, Felson DT. Modern-day environmental factors in the pathogenesis of osteoarthritis. Nature reviews Rheumatology. 2018;14(11):674–81. [DOI] [PubMed] [Google Scholar]
- 25.Conaghan PG, Tennant A, Peterfy CG, Woodworth T, Stevens R, Guermazi A, et al. Examining a whole-organ magnetic resonance imaging scoring system for osteoarthritis of the knee using Rasch analysis. Osteoarthritis Cartilage. 2006;14 Suppl A:A116–21. [DOI] [PubMed] [Google Scholar]
- 26.Felson DT, Niu J, Neogi T, Goggins J, Nevitt MC, Roemer F, et al. Synovitis and the risk of knee osteoarthritis: the MOST Study. Osteoarthritis Cartilage. 2016;24:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller GD, Nicklas BJ, Loeser RF. Inflammatory biomarkers and physical function in older, obese adults with knee pain and self-reported osteoarthritis after intensive weight-loss therapy. Journal of the American Geriatrics Society. 2008;56(4):644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messier SP, Mihalko SL, Legault C, Miller GD, Nicklas BJ, DeVita P, et al. Effects of intensive diet and exercise on knee joint loads, inflammation, and clinical outcomes among overweight and obese adults with knee osteoarthritis: the IDEA randomized clinical trial. JAMA : the journal of the American Medical Association. 2013;310(12):1263–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beavers KM, Beavers DP, Newman JJ, Anderson AM, Loeser RF jr., Nicklas BJ, et al. Effects of total and regional fat loss on plasma CRP and IL-6 in overweight and obese, older adults with knee osteoarthritis. Osteoarthritis Cartilage. 2015;23(2):249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daghestani HN, Pieper CF, Kraus VB. Soluble macrophage biomarkers indicate inflammatory phenotypes in patients with knee osteoarthritis. Arthritis & rheumatology. 2015;67(4):956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Attur M, Belitskaya-Levy I, Oh C, Krasnokutsky S, Greenberg J, Samuels J, et al. Increased interleukin-1beta gene expression in peripheral blood leukocytes is associated with increased pain and predicts risk for progression of symptomatic knee osteoarthritis. Arthritis and rheumatism. 2011;63(7):1908–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huebner JL, Bay-Jensen AC, Huffman KM, He Y, Leeming DJ, McDaniel GE, et al. Alpha C-telopeptide of type I collagen is associated with subchondral bone turnover and predicts progression of joint space narrowing and osteophytes in osteoarthritis. Arthritis & rheumatology. 2014;66(9):2440–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Spil WE, Nair SC, Kinds MB, Emans PJ, Hilberdink WK, Welsing PM, et al. Systemic biochemical markers of joint metabolism and inflammation in relation to radiographic parameters and pain of the knee: data from CHECK, a cohort of early-osteoarthritis subjects. Osteoarthritis Cartilage. 2015;23(1):48–56. [DOI] [PubMed] [Google Scholar]
- 34.Collins JA, Diekman BO, Loeser RF. Targeting aging for disease modification in osteoarthritis. Curr Opin Rheumatol. 2018;30(1):101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon OH, Kim C, Laberge RM, Demaria M, Rathod S, Vasserot AP, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 2017;23(6):775–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loeser RF, Olex AL, McNulty MA, Carlson CS, Callahan MF, Ferguson CM, et al. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis and rheumatism. 2012;64(3):705–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeon OH, David N, Campisi J, Elisseeff JH. Senescent cells and osteoarthritis: a painful connection. J Clin Invest. 2018;128(4):1229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleischmann RM, Bliddal H, Blanco FJ, Schnitzer TJ, Peterfy C, Chen S, et al. A Phase II Trial of Lutikizumab, an Anti-Interleukin-1alpha/beta Dual Variable Domain Immunoglobulin, in Knee Osteoarthritis Patients With Synovitis. Arthritis & rheumatology. 2019;71(7):1056–69. [DOI] [PubMed] [Google Scholar]
- 39.Roze RH, Bierma-Zeinstra SM, Agricola R, Oei EH, Waarsing JH. Differences in MRI features between two different osteoarthritis subpopulations: data from the Osteoarthritis Initiative. Osteoarthritis and cartilage. 2016;24(5):822–6. [DOI] [PubMed] [Google Scholar]
- 40.Mobasheri A, Rayman MP, Gualillo O, Sellam J, van der Kraan P, Fearon U. The role of metabolism in the pathogenesis of osteoarthritis. Nature reviews Rheumatology. 2017;13(5):302–11. [DOI] [PubMed] [Google Scholar]
- 41.Sniekers YH, Weinans H, Bierma-Zeinstra SM, van Leeuwen JP, van Osch GJ. Animal models for osteoarthritis: the effect of ovariectomy and estrogen treatment - a systematic approach. Osteoarthritis and cartilage. 2008;16(5):533–41. [DOI] [PubMed] [Google Scholar]
- 42.Tanamas SK, Wijethilake P, Wluka AE, Davies-Tuck ML, Urquhart DM, Wang Y, et al. Sex hormones and structural changes in osteoarthritis: a systematic review. Maturitas. 2011;69(2):141–56. [DOI] [PubMed] [Google Scholar]
- 43.Cardoso JS, Riley JL 3rd, Glover T, Sibille KT, Bartley EJ, Goodin BR, et al. Experimental pain phenotyping in community-dwelling individuals with knee osteoarthritis. Pain. 2016;157(9):2104–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frey-Law LA, Bohr NL, Sluka KA, Herr K, Clark CR, Noiseux NO, et al. Pain sensitivity profiles in patients with advanced knee osteoarthritis. Pain. 2016;157(9):1988–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osgood E, Trudeau JJ, Eaton TA, Jensen MP, Gammaitoni A, Simon LS, et al. Development of a bedside pain assessment kit for the classification of patients with osteoarthritis. Rheumatology international. 2015;35(6):1005–13. [DOI] [PubMed] [Google Scholar]
- 46.Carlesso LC, Segal NA, Frey-Law L, Zhang Y, Na L, Nevitt M, et al. Pain Susceptibility Phenotypes in Those Free of Knee Pain With or at Risk of Knee Osteoarthritis: The Multicenter Osteoarthritis Study. Arthritis & rheumatology. 2019;71(4):542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlesso LC, Neogi T. Identifying Pain Susceptibility Phenotypes in Knee Osteoarthritis. Clinical and Experimental Rheumatology. 2019. (this supplement). [PMC free article] [PubMed] [Google Scholar]
- 48.Egsgaard LL, Eskehave TN, Bay-Jensen AC, Hoeck HC, Arendt-Nielsen L. Identifying specific profiles in patients with different degrees of painful knee osteoarthritis based on serological biochemical and mechanistic pain biomarkers: a diagnostic approach based on cluster analysis. Pain. 2015;156(1):96–107. [DOI] [PubMed] [Google Scholar]
- 49.Deveza LA, Downie A, Tamez-Pena JG, Eckstein F, Van Spil WE, Hunter DJ. Trajectories of femorotibial cartilage thickness among persons with or at risk of knee osteoarthritis: development of a prediction model to identify progressors. Osteoarthritis and cartilage. 2019;27(2):257–65. [DOI] [PubMed] [Google Scholar]
- 50.Bartlett SJ, Ling SM, Mayo NE, Scott SC, Bingham CO 3rd. Identifying common trajectories of joint space narrowing over two years in knee osteoarthritis. Arthritis care & research. 2011;63(12):1722–8. [DOI] [PubMed] [Google Scholar]
- 51.Collins JE KJ, Losina E . Identifying Rapid Structural Disease Progression in Knee Osteoarthritis [abstract]. Arthritis Rheumatol. 2017; 69 (suppl 10). http://acrabstracts.org/abstract/identifying-rapid-structural-disease-progression-in-knee-osteoarthritis/. Accessed June 20, 2019. [Google Scholar]
- 52.Zhang W, McWilliams DF, Ingham SL, Doherty SA, Muthuri S, Muir KR, et al. Nottingham knee osteoarthritis risk prediction models. Annals of the rheumatic diseases. 2011;70(9):1599–604. [DOI] [PubMed] [Google Scholar]
- 53.Liukkonen MK, Mononen ME, Klets O, Arokoski JP, Saarakkala S, Korhonen RK. Simulation of Subject-Specific Progression of Knee Osteoarthritis and Comparison to Experimental Follow-up Data: Data from the Osteoarthritis Initiative. Scientific reports. 2017;7(1):9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LaValley MP, Lo GH, Price LL, Driban JB, Eaton CB, McAlindon TE. Development of a clinical prediction algorithm for knee osteoarthritis structural progression in a cohort study: value of adding measurement of subchondral bone density. Arthritis Res Ther. 2017;19(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riddle DL, Stratford PW, Perera RA. The incident tibiofemoral osteoarthritis with rapid progression phenotype: development and validation of a prognostic prediction rule. Osteoarthritis Cartilage. 2016;24(12):2100–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins JE, Katz JN, Dervan EE, Losina E. Trajectories and risk profiles of pain in persons with radiographic, symptomatic knee osteoarthritis: data from the osteoarthritis initiative. Osteoarthritis and cartilage. 2014;22(5):622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bastick AN, Wesseling J, Damen J, Verkleij SP, Emans PJ, Bindels PJ, et al. Defining knee pain trajectories in early symptomatic knee osteoarthritis in primary care: 5-year results from a nationwide prospective cohort study (CHECK). The British journal of general practice : the journal of the Royal College of General Practitioners. 2016;66(642):e32–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holla JF, van der Leeden M, Heymans MW, Roorda LD, Bierma-Zeinstra SM, Boers M, et al. Three trajectories of activity limitations in early symptomatic knee osteoarthritis: a 5-year follow-up study. Annals of the rheumatic diseases. 2014;73(7):1369–75. [DOI] [PubMed] [Google Scholar]
- 59.Wesseling J, Bastick AN, ten Wolde S, Kloppenburg M, Lafeber FP, Bierma-Zeinstra SM, et al. Identifying Trajectories of Pain Severity in Early Symptomatic Knee Osteoarthritis: A 5-year Followup of the Cohort Hip and Cohort Knee (CHECK) Study. The Journal of rheumatology. 2015;42(8):1470–7. [DOI] [PubMed] [Google Scholar]
- 60.Deveza LA, Melo L, Yamato TP, Mills K, Ravi V, Hunter DJ. Knee osteoarthritis phenotypes and their relevance for outcomes: a systematic review. Osteoarthritis and cartilage. 2017;25(12):1926–41. [DOI] [PubMed] [Google Scholar]
- 61.Niu J, Felson DT, Neogi T, Nevitt MC, Guermazi A, Roemer F, et al. Patterns of Coexisting Lesions Detected on Magnetic Resonance Imaging and Relationship to Incident Knee Osteoarthritis: The Multicenter Osteoarthritis Study. Arthritis & rheumatology. 2015;67(12):3158–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ornetti P, Brandt K, Hellio-Le Graverand MP, Hochberg M, Hunter DJ, Kloppenburg M, et al. OARSI-OMERACT definition of relevant radiological progression in hip/knee osteoarthritis. Osteoarthritis Cartilage. 2009;17(7):856–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loeser RF, Pathmasiri W, Sumner SJ, McRitchie S, Beavers D, Saxena P, et al. Association of urinary metabolites with radiographic progression of knee osteoarthritis in overweight and obese adults: an exploratory study. Osteoarthritis Cartilage. 2016;24(8):1479–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kraus VB, Collins JE, Hargrove D, Losina E, Nevitt M, Katz JN, et al. Predictive validity of biochemical biomarkers in knee osteoarthritis: data from the FNIH OA Biomarkers Consortium. Annals of the rheumatic diseases. 2017;76(1):186–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Osteoarthritis: Care and Management in Adults. National Institute for Health and Clinical Excellence: Guidance. London: 2014. [PubMed] [Google Scholar]
- 66.Paterson KL, Kasza J, Bennell KL, Wrigley TV, Metcalf BR, Campbell PK, et al. Moderators and mediators of effects of unloading shoes on knee pain in people with knee osteoarthritis: an exploratory analysis of the SHARK randomised controlled trial. Osteoarthritis and cartilage. 2018;26(2):227–35. [DOI] [PubMed] [Google Scholar]
- 67.van Middelkoop M, Arden NK, Atchia I, Birrell F, Chao J, Rezende MU, et al. The OA Trial Bank: meta-analysis of individual patient data from knee and hip osteoarthritis trials show that patients with severe pain exhibit greater benefit from intra-articular glucocorticoids. Osteoarthritis and cartilage. 2016;24(7):1143–52. [DOI] [PubMed] [Google Scholar]
- 68.Laslett LL, Dore DA, Quinn SJ, Boon P, Ryan E, Winzenberg TM, et al. Zoledronic acid reduces knee pain and bone marrow lesions over 1 year: a randomised controlled trial. Ann Rheum Dis. 2012;71(8):1322–8. [DOI] [PubMed] [Google Scholar]
- 69.Deveza LA, Bierma-Zeinstra SMA, van Spil WE, Oo WM, Saragiotto BT, Neogi T, et al. Efficacy of bisphosphonates in specific knee osteoarthritis subpopulations: protocol for an OA Trial Bank systematic review and individual patient data meta-analysis. BMJ open. 2018;8(12):e023889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee AC, Harvey WF, Han X, Price LL, Driban JB, Bannuru RR, et al. Pain and functional trajectories in symptomatic knee osteoarthritis over up to 12 weeks of exercise exposure. Osteoarthritis and cartilage. 2018;26(4):501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Allen KD, Choong PF, Davis AM, Dowsey MM, Dziedzic KS, Emery C, et al. Osteoarthritis: Models for appropriate care across the disease continuum. Best practice & research Clinical rheumatology. 2016;30(3):503–35. [DOI] [PubMed] [Google Scholar]
- 72.Hochberg MC. Serious joint-related adverse events in randomized controlled trials of anti-nerve growth factor monoclonal antibodies. Osteoarthritis and cartilage. 2015;23 Suppl 1:S18–21. [DOI] [PubMed] [Google Scholar]
- 73.Karsdal MA, Verburg KM, West CR, Bay-Jensen AC, Keller DS, Arends R. Serological biomarker profiles of rapidly progressive osteoarthritis in tanezumab-treated patients. Osteoarthritis Cartilage. 2019;27(3):484–92. [DOI] [PubMed] [Google Scholar]
- 74.Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. The New England journal of medicine. 2019;380(14):1347–58. [DOI] [PubMed] [Google Scholar]
- 75.Jamshidi A, Pelletier JP, Martel-Pelletier J. Machine-learning-based patient-specific prediction models for knee osteoarthritis. Nature reviews Rheumatology. 2019;15(1):49–60. [DOI] [PubMed] [Google Scholar]
- 76.Du Y, Almajalid R, Shan J, Zhang M. A Novel Method to Predict Knee Osteoarthritis Progression on MRI Using Machine Learning Methods. IEEE Trans Nanobioscience. 2018;17(3):228–36. [DOI] [PubMed] [Google Scholar]
- 77.Huang C, Shan L, Charles HC, Wirth W, Niethammer M, Zhu H. Diseased Region Detection of Longitudinal Knee Magnetic Resonance Imaging Data. IEEE transactions on medical imaging. 2015;34(9):1914–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.An H, Marron JS, Schwartz TA, Renner JB, Liu F, Lynch JA, et al. Novel statistical methodology reveals that hip shape is associated with incident radiographic hip osteoarthritis among African American women. Osteoarthritis Cartilage. 2016;24(4):640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lazzarini N, Runhaar J, Bay-Jensen AC, Thudium CS, Bierma-Zeinstra SMA, Henrotin Y, et al. A machine learning approach for the identification of new biomarkers for knee osteoarthritis development in overweight and obese women. Osteoarthritis and cartilage / OARS, Osteoarthritis Research Society. 2017;25(12):2014–21. [DOI] [PubMed] [Google Scholar]
- 80.Hu T, Oksanen K, Zhang W, Randell E, Furey A, Sun G, et al. An evolutionary learning and network approach to identifying key metabolites for osteoarthritis. PLoS Comput Biol. 2018;14(3):e1005986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nelson AE, Fang F, Arbeeva L, Cleveland RJ, Schwartz TA, Callahan LF, et al. A machine learning approach to knee osteoarthritis phenotyping: data from the FNIH Biomarkers Consortium. Osteoarthritis and Cartilage. 2019;27(7):994–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mobasheri A, van Spil WE, Budd E, Uzieliene I, Bernotiene E, Bay-Jensen AC, et al. Molecular taxonomy of osteoarthritis for patient stratification, disease management and drug development: biochemical markers associated with emerging clinical phenotypes and molecular endotypes. Current opinion in rheumatology. 2019;31(1):80–9. [DOI] [PubMed] [Google Scholar]
- 83.Hastie T, Friedman JH, Tibshirani R. The elements of statistical learning : data mining, inference, and prediction Second edition, corrected 7th printing. ed. 1 online resource (xxii, 745pages) p. [Google Scholar]
- 84.Liu Y, Hayes DN, Nobel A, Marron JS. Statistical Significance of Clustering for High-Dimension, Low-Sample Size Data. J Am Stat Assoc. 2008;103(483):1281–93. [Google Scholar]
- 85.Huang HW, Liu YF, Yuan M, Marron JS. Statistical Significance of Clustering Using Soft Thresholding. J Comput Graph Stat. 2015;24(4):975–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Marron JS. Object oriented data analysis: Sets of trees. Ann Stat. 2007;35(5):1849–73. [Google Scholar]
- 87.Marron JS, Alonso AM. Overview of object oriented data analysis. Biometrical journal Biometrische Zeitschrift. 2014;56(5):732–53. [DOI] [PubMed] [Google Scholar]
- 88.Marron JS, Todd MJ, Ahn J. Distance-weighted discrimination. J Am Stat Assoc. 2007;102(480):1267–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wei S, Lee C, Wichers L, Marron JS. Direction-Projection-Permutation for High-Dimensional Hypothesis Tests. J Comput Graph Stat. 2016;25(2):549–69. [Google Scholar]
- 90.Gullo TR, Golightly YM, Cleveland RJ, Renner JB, Callahan LF, Jordan JM, et al. Defining multiple joint osteoarthritis, its frequency and impact in a community-based cohort. Seminars in arthritis and rheumatism. 2019;48(6):950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]