Abstract
Spinal Muscular Atrophy with Respiratory Distress type 1 (SMARD1) is an autosomal recessive disease that develops early during infancy. The gene responsible for disease development is immunoglobulin helicase μ-binding protein 2 (IGHMBP2). IGHMBP2 is a ubiquitously expressed gene but its mutation results in the loss of alpha-motor neurons and subsequent muscle atrophy initially of distal muscles. The current SMARD1 mouse model arose from a spontaneous mutation originally referred to as neuromuscular degeneration (nmd). The nmd mice have the C57BL/6 genetic background and contain an A to G mutation in intron 4 of the endogenous Ighmbp2 gene. This mutation causes aberrant splicing, resulting in only 20–25% of full-length functional protein. Several congenital conditions including hydrocephalus are common in the C57BL/6 background, consistent with our previous observations when developing a gene therapy approach for SMARD1. Additionally, a modifier allele exists on chromosome 13 in nmd mice that can partially suppress the phenotype, resulting in some animals that have extended life spans (up to 200 days). To eliminate the intrinsic complications of the C57BL/6 background and the variation in survival due to the genetic modifier effect, we created a new SMARD1 mouse model that contains the same intron 4 mutation in Ighmbp2 as nmd mice but is now on a FVB congenic background. FVB-nmd are consistently more severe than the original nmd mice with respect to survival, weigh and motor function. The relatively short life span (18–21 days) of FVB-nmd mice allows us to monitor therapeutic efficacy for a variety of novel therapeutics in a timely manner without the complication of the small percentage of longer-lived animals that were observed in our colony of nmd mice.
Keywords: SMARD1, IGHMBP2, FVB, nmd, C57BL/6
1. Introduction
Spinal Muscular Atrophy with Respiratory Distress type 1 (SMARD1) is an autosomal recessive disease affecting infants [1]. The most common clinical symptoms include early diaphragm paralysis and subsequent respiratory failure, as well as distal lower muscle weakness progressing to proximal muscles [1–3]. SMARD1 is an aggressive disease and mostly fatal as the children suffering from this disease are paralyzed and require continuous artificial ventilation. The gene responsible for disease development is immunoglobulin helicase μ-binding protein 2 (IGHMBP2) located on chromosome 11q13.3 [4, 5]. IGHMBP2 is a ubiquitously expressed gene but its mutation results in the loss of alpha-motor neurons and consequent muscle atrophy [6, 7]. IGHMBP2 protein is involved in many cellular activities including transcription regulation [8, 9], RNA maturation and translation [10, 11], and immunoglobulin-class switching [5], but the exact loss of function that leads to motor neuron loss and respiratory complications is unknown. Although genetically and clinically distinct, proximal Spinal Muscular Atrophy (5q-linked SMA) is also caused by the loss of a ubiquitously expressed gene (SMN) that is intricately involved in several aspects of RNA metabolism [12]. Robust animal models were important in the development of the recent clinical successes in the SMA field, including the approval of Spinraza [13–15] and ZOLGENSMA [16, 17]. The current SMARD1 mouse model contains a previously characterized mutation in the endogenous Ighmbp2 gene of C57BL/6 mice referred to nmd mice [18]. To push SMARD1 therapeutic development, additional models are needed to rapidly evaluate the potential of IGHMBP2-dependent and –independent therapeutics.
We and others utilized a similar viral-based gene replacement strategy to improve the severe nmd phenotype using AAV9-IGHMBP2 [19–21]. Based on these results, AAV9-IGHMBP2 significantly improved the nmd phenotype, with intracerebroventricular delivery providing better rescuing activity than systemic delivery (IV) [19, 20]. These studies revealed that repeated administration via ICV resulted in a significant increase in hydrocephalus and premature death in homozygous and heterozygous (unaffected) nmd mice [19]. Interestingly, none of the side effects occurred in SMNΔ7 mice following multiple ICV injections of AAV9-IGHMBP2. It is important to note that the background of the nmd mice is C57BL/6 and the background of the SMNΔ7 mice is FVB. In addition to hydrocephalus (a genetically prone condition associated with C57BL/6), a modifier allele exists in chromosome 13 of the nmd mice that can suppresses its phenotype, leading to unusually long life span (up to 200 days) in some untreated animals [18]. Collectively, these complications limited our ability to conclude the survival results in a timely fashion or perform a straightforward dose escalation and time course study in nmd mice.
Currently there are no effective treatments for SMARD1. To move any therapeutic to clinic, an animal model that provides a consistent and robust phenotype is essential such that important disease phenotypes (life span, weight gain, motor function and quantifiable repair at the cellular level) can be evaluated. These parameters can vary depending on the genetic background and the sensitivity of the mice model to high dose, delivery route, and time of delivery. To achieve this goal, we have created a new SMARD1 mouse model on FVB genetic background. The severe and consistent phenotype of the FVB-nmd mice makes this model a balanced and stable context to test and measure the impact of a variety of therapeutic approaches.
2. Materials and methods
2.1. Animals
All experimental procedures were approved by the University of Missouri’s Institutional Animal Care and Use Committee and were performed according to the guidelines set forth in the Guide for the Use and Care of Laboratory Animals. FVB/NJ embryo donor females (3 weeks of age) and stud males (10 weeks of age) were purchased from Jackson Laboratory. CD1 surrogate females (8 weeks of age) were purchased from Charles River. Mice were housed in ventilated cages (Thoren) and kept on a 14:10 light cycle. Food and water were available ad libitum.
2.2. sgRNA design and synthesis
FVB/NJ mouse genomic sequence for immunoglobulin μ-binding protein 2 (Ighmbp2) was obtained from Ensembl.org. The CRISPR RGEN Tools website maintained by the Center for Genome Engineering Institute (Korea) was used to calculate off target scores and design a sgRNA near the desired mutation position in Ighmbp2 intron 4. After designing, the sgRNA was ordered as a chemically-modified synthetic sgRNA through Synthego. The chemical modifications were 2’-O-methyl analogs and 3’ phosphorothioate internucleotide linkages at the first three 5’ and 3’ terminal RNA residues.
2.3. Repair template design and synthesis
A repair template was designed to introduce the desired A to G substitution at the 23rd bp in Ighmbp2 intron 4. The repair template was chemically synthesized by IDT as a 200 bp single-stranded DNA oligo (ssODN). The ssODN was complementary to the non-target strand and contained symmetrical homology arms. A silent mutation was designed in the repair template to prevent the CRISPR sgRNA/Cas9 RNP complex from cleaving after the desired point mutation was introduced.
2.4. Microinjection and embryo transfer
A microinjection mix containing a final concentration of 3.0 μM sgRNA, 2.0 μM enhanced specificity Cas9 protein (Sigma), and 1.6 μM ssODN was made immediately prior to microinjections. Briefly, CRISPR sgRNA/Cas9 RNP complexes were form by gently mixing the sgRNA and Cas9 protein together and incubating at room temperature for 10 minutes. After RNP formation, the ssODN repair template was added to the mixture. Pronuclear injected zygotes were surgically transferred to pseudopregnant surrogate females the same day as microinjections.
2.5. Western blot
Western blot with transfected HEK293 cells were conducted as described previously [22, 23]. Spinal cord tissue harvested from postnatal day 14 animals were immediately placed in liquid nitrogen postharvest, and kept in −80° C until protein extraction. Hundred micrograms of tissue were homogenized in 100 μl of Jurkat Lysis Buffer (JLB) buffer and equal amounts of protein were loaded onto 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis gel as previously described [19, 20, 24, 25]. Blots were incubated overnight at 4° C with rabbit IGHMBP2 antibody (Invitrogen, Carlsbad, CA) diluted to 1:1,000 and horseradish peroxidase anti-rabbit IgG (Jackson-Immunoresearch) was used as secondary antibody. Blot was stripped with 30% H2O2/PBS (1:1) and re-probed with Mouse Tubulin (Sigma, St. Louis, MO) as loading control. Blots were visualized by chemiluminescence and images were captured by UVP ChemStudio imaging system.
2.6. Muscle histology
14 days old FVB-nmd, “HET” in FVB background, and original nmd mice were transcardially perfused, gastrocnemius muscles were harvested after 24 hours, 16-μm cross-sections were prepared from gastrocnemius and immunostained with laminin. A blinded quantification assessments of muscle fibers were performed using image J as previously described [26].
2.7. Neuromuscular junction Immunostaining
Teased muscle fibers from gastrocnemius of 14 days old mice were labeled with α-BTX (Invitrogen) for AChRs, anti-neurofilament (Abcam), and antivesicular acetylcholine transporter (VAChT; synaptic system) antibodies for nerve terminals. Fluorescently labeled NMJs were observed with confocal microscopy and quantified as previously described [20, 27].
2.8. Statistical analysis
The statistical significance in comparing three experimental groups (FVB-nmd, “HET”, and original nmd) was calculated using one-way analysis of variance and Bonferroni multiple comparison post hoc test. Analyses were performed with GraphPad Prism software; error bars represent means ± standard error of the mean. Significance in survival was determined with log-ranked (Mantel-Cox) test.
3. Results
3.1. Crisper/Cas9-mediated generation of a FVB-nmd model
The nmd mouse model contains a single A to G transition mutation 23 bp into Ighmbp2 intron 4, resulting in abnormal splicing of ~75–80% of the Ighmbp2 transcripts [18]. As the nmd model has a well-characterized neurodegenerative phenotype, our objective was to generate the same mutation in the congenic FVB background. Using a CRISPR/Cas9, the intron 4 A to G transition was specifically recreated in Ighmbp2 of FVB mice. The correct mutation was confirmed by sequencing of the tail genomic DNA as well as demonstrating that additional mutations were not present in the surrounding sequences (data not shown). Heterozygous (HET) breeding pairs were used to obtain the homozygous FVB-nmd pups.
3.2. FVB-nmd mice show a severe phenotype compared to nmd mice
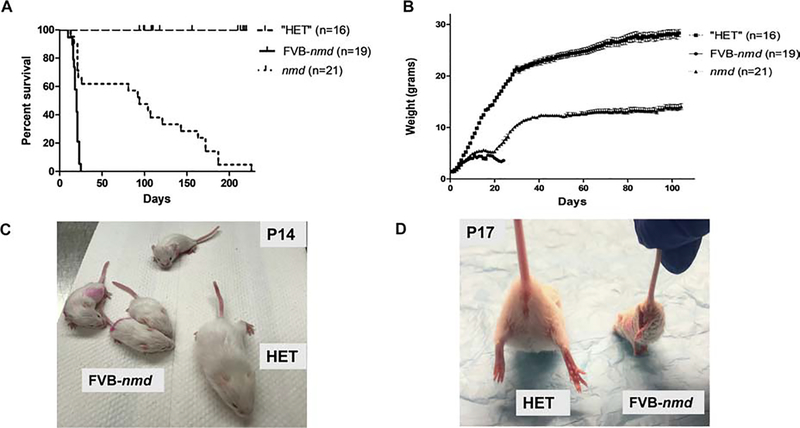
We monitored the life span and weight of the FVB-nmd newborns compared to the unaffected heterozygous (HET) littermates and the original nmd mice. FVB-nmd pups demonstrated a consistently more severe phenotype compared to the nmd mice as the majority of FVB-nmd mice died by post-natal day (P) 20 and only 2/19 pups survived to P23 (Figure 1A). In contrast, the average life span of the original nmd mice is 94 days and only 36% died by P21–22 (Figure 1A). Consistent with the life span data, FVB-nmd mice initially gained weight but stopped gaining for several days and then declined rapidly. In contrast, the nmd mice that survive beyond P22 continued to gain weight, with an average total body mass of greater than 10 g by P40 (Figure 1B). Similar to nmd mice, the FVB-nmd mice develop symptoms by the second week of age, which is reflected in the significant difference in body weight of the FVB-nmd and nmd mice compared to unaffected mice (Figure 1B, C). The weight of the FVB-nmd mice fluctuated between 3–4 grams but only 2 out of 19 pups reached 5 grams while nmd mice can easily reach above 10 grams (Figure 1B, C). A hallmark of the nmd model is the quantifiable contracture in the hind limbs as measured by the distance between the legs while positioned upside down from the tail [19, 20] The hind limb phenotype of the FVB-nmd mice is more severe than the nmd mice as they have lost the ability to splay their hind limbs at P17. The representative images of the FVB-nmd pups and the hind limb phenotype are demonstrated side by side to the “HET” littermate (Figure 1C and 1D).
Figure 1. The phenotype of the FVB-nmd mice is significantly more severe than nmd mice.
(A) Survival of homozygous FVB-nmd was compared to the “HET” littermates and to homozygous nmd mice. Survival was determined by Kaplan-Meier curves, and the P value was determined by the log-rank (Mantel-Cox) test. Median survival of FVB-nmd mice (n=19) was 20 days compared to 94 days in homozygous nmd mice (n=21) (P<0.0001). (B) Weight assessment of FVB-nmd compared to “HET” littermates and nmd mice. The average weight of FVB-nmd mice is 3.6 ± 0.18 g compared to 10.75 ± 0.35 g in nmd mice (two-tailed t test P=0.0009) and 21.50 ± 0.72 g in “HET” littermates (two-tailed t test P<0.0001). (C) Four FVB-nmd pups and one “HET” littermate are photographed side by side. (D) Suspended from the tail, all of the FVB-nmd pups at P17 show severe hindlimb contracture compared with age-matched “HET” littermate.
3.3. Severe pathology in the FVB-nmd gastrocnemius muscle
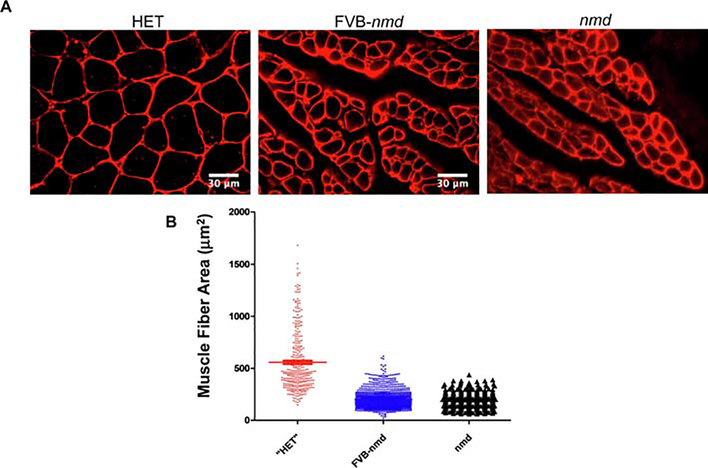
To determine the level of the skeletal muscle pathology in FVB-nmd mice, gastrocnemius muscle fiber cross-sectional area was determined. Gastrocnemius tissue collected from P14 FVB-nmd (n=6), unaffected “HET” littermates in FVB background (n=3), and original nmd (n=3) were examined by laminin immunostaining (Figure 2). A blinded, quantitative assessment of muscle pathology revealed that the gastrocnemius cross sectional fiber area was significantly reduced in the FVB-nmd mice compared to the unaffected littermates (Figure 2A, B). On the other hand, muscle fiber area in FVB- nmd gastrocnemius was statistically larger than original nmd mice (Figure 2A, B). These results correlate the reduced gastrocnemius fiber area in FVB-nmd to a reduced ability to splay their hind limbs (Figure 1D).
Figure 2. Gastrocnemius muscle fibers are severely affected n FVB-nmd mice.
(A) Laminin-stained cross sections of gastrocnemius muscles of FVB-nmd mice (n=5) were examined at P14 and compared with those of the age-matched “HET” littermates (n=3) and original nmd mice (n=3). (B) The area size of muscle fibers in FVB-nmd mice was significantly reduced in comparison with the “HET” littermate (one-way ANOVA P<0.0001) but statistically larger than the original nmd (one-way ANOVA P<0.01). The average fiber area of FVB-nmd gastrocnemius is 202.8 ± 2.58 μm2 compared to 557.9 ± 16.53 μm2 in “HET” and 182.4 ± 2.90 μm2 in original nmd. Error bars represent mean ± SEM.
3.4. Ighmbp2 protein is significantly reduced in FVB-nmd spinal cord
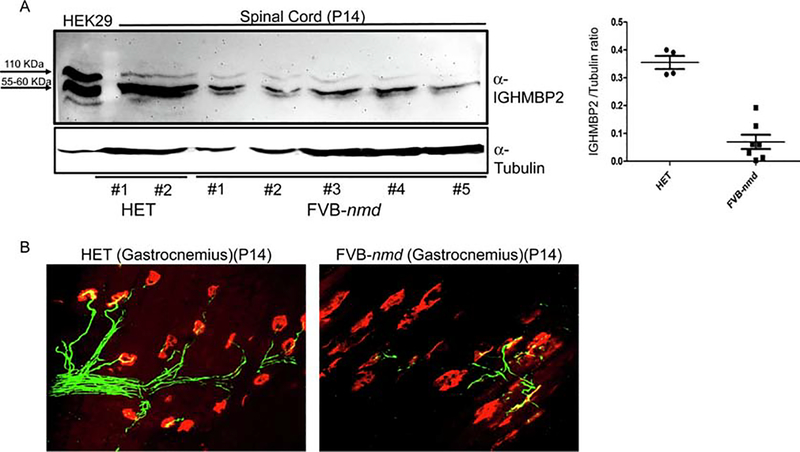
To examine Ighmbp2 protein expression in the FVB-nmd mice, western blot analysis was performed using spinal cord extracts from FVB-nmd and “HET” littermates at P14. To verify the size of the endogenous Ighmbp2 protein in FVB-nmd, HEK293 cells were transfected with the AAV9-IGHMBP2 plasmid and used as size control. Western blot revealed the predicted Ighmbp2 product of ~110 kDa in “HET” spinal cord extracts. As anticipated, Ighmbp2 was significantly reduced in FVB-nmd spinal cord extract compared to unaffected “HET” animals (Figure 3A), associating the SMARD1 phenotype to decreased Ighmbp2 protein in the central nervous system. Reduced expression level of Ighmbp2 in the spinal cord extracts of the original nmd mice at P14 and P30 have been previously reported [19]. An additional prominent product of the western blot was a 55–60 kDa protein that was detected in all of the samples. This isoform has been observed in nmd tissue extracts previously and it has been suggested to originate from alternative splicing or an additional start codon [7].
Figure 3. Ighmbp2 protein levels in the spinal cord and the gastrocnemius NMJ of the FVB-nmd mice are severely affected compared to “HET”.
(A) Western blot of the spinal cord extracts of the FVB-nmd at P14 (n=7) demonstrates a considerable decrease in Ighmbp2 protein levels compared to the age-matched “HET” littermates (n=4) (two-tailed t test P<0.0001). (B) NMJs from gastrocnemius muscle of FVB-nmd mice at P14 compared to those of age-matched “HET’ littermates. Teased fibers were labeled with α-BTX for acetylcholine receptors (ACHRs), anti-neurofilament, and anti-vesicular acetylcholine transporter (VAChT) for nerve terminals. NMJs of the FVB-nmd are mostly denervated as opposed to the “HET” that shows no sign of denervation.
3.5. Severe NMJ pathology in FVB-nmd skeletal muscle
Based upon the reduced fiber area in the gastrocnemius and decreased Ighmbp2 protein in the spinal cord extracts, we predicted that neuromuscular junctions (NMJs) innervating the gastrocnemius would also exhibit a severe phenotype in FVB-nmd mice. In a blinded assessment of NMJ pathology from the gastrocnemius, NMJs from FVB-nmd exhibited the hallmark pattern of neurodegeneration, including nearly 90% denervated and fragmented endplates (Figure 3B). In contrast, NMJs from unaffected “HET” control tissues showed no detectable denervation or motor endplate fragmentation (Figure 3B). Collectively, the analysis of the NMJ phenotype of FVB-nmd mice is consistent with the gastrocnemius muscle pathology and hind limb phenotype, providing evidence of a more severe neurodegenerative phenotype in the FVB congenic background compared to the C57BL/6 background.
4. Discussion
Developing a portfolio of disease-appropriate models is important from a biological perspective as well as in the development of novel therapeutics. The current SMARD1 mouse model, the nmd mice, has been an important tool to further our understanding of SMARD1 pathology and has allowed the development and testing of AAV9-IGHBMP2 vector. To determine if a more severe and consistent phenotype could be generated in a different genetic background, we engineered the nmd allele in a congenic FVB background. This resulted in a model that retained the important phenotypic features of the original nmd mice, but led to a more severe model with a shorter life span. While a shorter life span is not always advantageous, from the perspective of therapeutic development, it provides an important disease context to be able to quickly assess the efficacy of novel therapeutics with respect to life span and the impact upon the disease phenotype. All FVB-nmd mice died by P23 providing an excellent disease context to assess potential therapeutics in a timely manner while still retaining pre-symptomatic and symptomatic disease stages. This model is also suitable for quantifying small but measurable improvements in testing IGHMBP2-dependent or -independent therapeutics and genetic modifiers.
One of the complications we observed previously in the delivery of a gene therapy vector to the nmd mice was that they were prone to hydrocephalus [19]. While unrelated to disease, this complicated the survival and cellular analysis and made further studies, such as a time course study to determine the therapeutic window, technically challenging. By changing the genetic background, we believe that this FVB-nmd model will provide an important addition to the research portfolio for SMARD1 investigators from a biological perspective but also as a means to analyze potential therapeutics. This model will provide an important context to provide supporting evidence regarding optimal dosing/delivery, temporal requirements, and the identification of potential side effects of increasing IGHMBP2 protein via gene therapy or other therapeutic interventions focused on increasing IGHMBP2 levels in SMARD1 model mice.
Novel severe mouse model of spinal muscular atrophy with respiratory distress
Severe pathology in skeletal muscle and neuromuscular junctions correlates with Ighmbp2 levels.
New context ideal for therapeutic development.
Acknowledgements
We would like to thank members of the Lorson lab who contributed to the early stages of this project and to animal colony maintenance. This work is supported by NIH/NINDS grant R21NS093175 (to C.L.L. and M.S.), R21NS109762 (to CLL), and the Sims’ Fund to Cure SMARD (to M.S.). CES is supported by NIH Training Grant (T32 GM008396).
Footnotes
Conflicts of interest
CLL. is the co-founder and Chief Scientific Officer of Shift Pharmaceuticals.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Grohmann K, Varon R, Stolz P, et al. , Infantile spinal muscular atrophy with respiratory distress type 1 (SMARD1), Ann Neurol., 54 (2003) pp. 719–724. [DOI] [PubMed] [Google Scholar]
- [2].Rudnik-Schoneborn S, Stolz P, Varon R, et al. , Long-term observations of patients with infantile spinal muscular atrophy with respiratory distress type 1 (SMARD1), Neuropediatrics., 35 (2004) pp. 174–182. [DOI] [PubMed] [Google Scholar]
- [3].Grohmann K, Wienker TF, Saar K, et al. , Diaphragmatic spinal muscular atrophy with respiratory distress is heterogeneous, and one form Is linked to chromosome 11q13-q21, Am J Hum Genet., 65 (1999) pp. 1459–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Viollet L, Barois A, Rebeiz JG, et al. , Mapping of autosomal recessive chronic distal spinal muscular atrophy to chromosome 11q13, Ann Neurol., 51 (2002) pp. 585–592. [DOI] [PubMed] [Google Scholar]
- [5].Fukita Y, Mizuta TR, Shirozu M, et al. , The human S mu bp-2, a DNA-binding protein specific to the single-stranded guanine-rich sequence related to the immunoglobulin mu chain switch region, J Biol Chem., 268 (1993) pp. 17463–17470. [PubMed] [Google Scholar]
- [6].Grohmann K, Schuelke M, Diers A, et al. , Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1, Nat Genet., 29 (2001) pp. 75–77. [DOI] [PubMed] [Google Scholar]
- [7].Grohmann K, Rossoll W, Kobsar I, et al. , Characterization of Ighmbp2 in motor neurons and implications for the pathomechanism in a mouse model of human spinal muscular atrophy with respiratory distress type 1 (SMARD1), Hum Mol Genet., 13 (2004) pp. 2031–2042. [DOI] [PubMed] [Google Scholar]
- [8].Chen NN, Kerr D, Chang CF, et al. , Evidence for regulation of transcription and replication of the human neurotropic virus JCV genome by the human S(mu)bp-2 protein in glial cells, Gene., 185 (1997) pp. 55–62. [DOI] [PubMed] [Google Scholar]
- [9].Miao M, Chan SL, Fletcher GL, et al. , The rat ortholog of the presumptive flounder antifreeze enhancer-binding protein is a helicase domain-containing protein, Eur J Biochem., 267 (2000) pp. 7237–7246. [DOI] [PubMed] [Google Scholar]
- [10].de Planell-Saguer M, Schroeder DG, Rodicio MC, et al. , Biochemical and genetic evidence for a role of IGHMBP2 in the translational machinery, Hum Mol Genet., 18 (2009) pp. 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guenther UP, Handoko L, Varon R, et al. , Clinical variability in distal spinal muscular atrophy type 1 (DSMA1): determination of steady-state IGHMBP2 protein levels in five patients with infantile and juvenile disease, J Mol Med (Berl). 87 (2009) pp. 31–41. [DOI] [PubMed] [Google Scholar]
- [12].Lorson CL, Rindt H, Shababi M, Spinal muscular atrophy: mechanisms and therapeutic strategies, Hum Mol Genet., 19 (2010) pp. R111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bennett CF, Krainer AR, Cleveland DW, Antisense Oligonucleotide Therapies for Neurodegenerative Diseases, Annu Rev Neurosci, 42 (2019) pp. 385–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Darras BT, Chiriboga CA, Iannaccone ST, et al. , Nusinersen in later-onset spinal muscular atrophy: Long-term results from the phase 1/2 studies, Neurology, 92 (2019) pp. e2492–e2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Montes J, Dunaway Young S, Mazzone ES, et al. , Nusinersen Improves Walking Distance and Reduces Fatigue in Later-Onset SMA, Muscle Nerve, (2019) pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Keeler AM, Flotte TR, Recombinant Adeno-Associated Virus Gene Therapy in Light of Luxturna (and Zolgensma and Glybera): Where Are We, and How Did We Get Here?, Annu Rev Virol, (2019) pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Saffari A, Weiler M, Hoffmann GF, et al. , [Gene therapies for neuromuscular diseases], Nervenarzt, (2019) pp. [DOI] [PubMed] [Google Scholar]
- [18].Cox GA, Mahaffey CL, Frankel WN, Identification of the mouse neuromuscular degeneration gene and mapping of a second site suppressor allele, Neuron., 21 (1998) pp. 1327–1337. [DOI] [PubMed] [Google Scholar]
- [19].Shababi M, Feng Z, Villalon E, et al. , Rescue of a Mouse Model of Spinal Muscular Atrophy With Respiratory Distress Type 1 by AAV9-IGHMBP2 Is Dose Dependent, Mol Ther, 24 (2016) pp. 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shababi M, Villalon E, Kaifer KA, et al. , A Direct Comparison of IV and ICV Delivery Methods for Gene Replacement Therapy in a Mouse Model of SMARD1, Mol Ther Methods Clin Dev, 10 (2018) pp. 348–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nizzardo M, Simone C, Rizzo F, et al. , Gene therapy rescues disease phenotype in a spinal muscular atrophy with respiratory distress type 1 (SMARD1) mouse model, Science Advances., 1 (2015) pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Baughan T, Shababi M, Coady TH, et al. , Stimulating full-length SMN2 expression by delivering bifunctional RNAs via a viral vector, Mol Ther., 14 (2006) pp. 54–62. [DOI] [PubMed] [Google Scholar]
- [23].Coady TH, Shababi M, Tullis GE, et al. , Restoration of SMN function: delivery of a trans-splicing RNA re-directs SMN2 pre-mRNA splicing, Mol Ther., 15 (2007) pp. 1471–1478. [DOI] [PubMed] [Google Scholar]
- [24].Shababi M, Habibi J, Ma L, et al. , Partial restoration of cardio-vascular defects in a rescued severe model of spinal muscular atrophy, J Mol Cell Cardiol, 52 (2012) pp. 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shababi M, Lorson CL, Optimization of SMN trans-splicing through the analysis of SMN introns, J Mol Neurosci., 46 (2012) pp. 459–469. [DOI] [PubMed] [Google Scholar]
- [26].Kaifer KA, Villalon E, Osman EY, et al. , Plastin-3 extends survival and reduces severity in mouse models of spinal muscular atrophy, JCI Insight, 2 (2017) pp. e89970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Villalon E, Shababi M, Kline R, et al. , Selective vulnerability in neuronal populations in nmd/SMARD1 mice, Hum Mol Genet, 27 (2018) pp. 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]