Abstract
In this issue of Cell Chemical Biology, Sakin et al. (2017) investigate the nanoscale behavior of the HIV-1 envelope (Env) glycoprotein complex by using genetic code expansion, bioorthogonal amino acids, synthetic dyes, and click chemistry. This minimally invasive approach allows the measurement of native Env cellular distribution and dynamics.
The HIV-1 envelope (Env) glycoprotein complex plays an essential role in the virus replication cycle by binding to receptor and co-receptor on the surface of target cells and catalyzing a membrane fusion reaction between the viral lipid envelope and cellular membranes. This fusion reaction delivers the viral core into the cytoplasm to initiate the infection process. Virion incorporation of the Env glycoprotein complex, which comprises a hetero-trimer of surface (gp120) and transmembrane (gp41) subunits, is therefore a crucial step in virus replication (Freed, 2015). Although the structure and function of Env glycoproteins during virus entry are now fairly well understood, the mechanism by which Env traffics to the plasma membrane, clusters at the site of virus particle assembly, localizes to the virological synapse, and incorporates into virions remains to be fully elucidated.
A number of recent studies have used advanced fluorescence microscopy techniques to probe the behavior of viral proteins in infected cells and virus particles. These studies have used a variety of methods for protein visualization (re-viewed in Sakin et al., 2016). Many studies have engineered in-frame fusions between proteins of interest and auto-fluorescent proteins such as green fluorescent protein (GFP) (Tsien, 1998). Self-labeling enzymes, for example those found in SNAP and Halo tags, can be fused to proteins of interest and subsequently labeled with fluorescent dyes. Small peptide tags that serve as binding sites for compounds that fluoresce upon binding can be introduced into proteins in a minimally disruptive fashion. An example of such a peptide tag is the tetracysteine motif (Cys-Cys-X-X-Cys-Cys), which binds the biarsenical dyes FlAsH and ReAsH (Adams et al., 2002). Click chemistry reactions, such as Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC), can be used to label proteins and nucleic acids (Link and Tirrell, 2003).
While application of these methods has provided a wealth of information about the function of both viral and cellular proteins, these approaches are subject to a variety of limitations. Auto-fluorescent proteins are large (~25 kDa) and thus can disrupt protein structure and function, have relatively long folding times, and are less photo-stable than currently available fluorescent dyes. Self-labeling enzymes are also quite large (~18–34 kDa). Peptide tags, such as tetracysteine, are only a few amino acids in length, but labeling is constrained by a limited number of available dyes and by high background (non-specific) staining. Immunolabeling with fluorescently tagged antibodies or Fab fragments can also disrupt protein dynamics, and the length of the antibody molecule can reduce the accuracy of protein localization. The CuAAC method is very efficient in vitro or with fixed cells but its use is problematic in living samples due to the cytotoxicity of copper. Minimally invasive fluorescent labeling of Env would enhance our understanding of Env biology within its native environment.
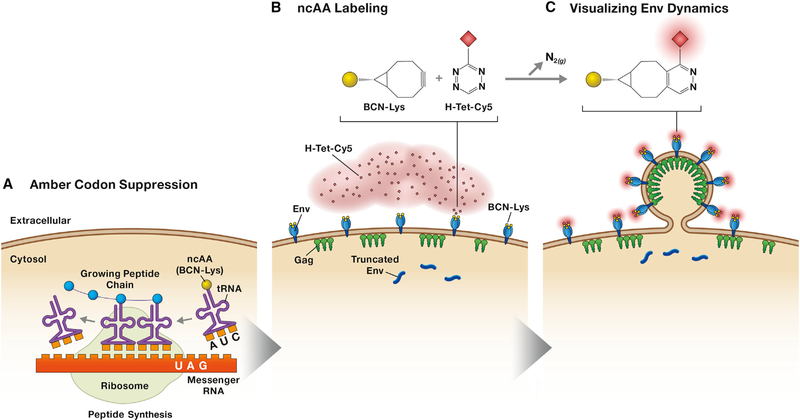
Recently, a new method for protein labeling based on genetic code expansion and click chemistry has been developed (Nikić et al., 2014; Plass et al., 2012) (Figure 1). This technique overcomes the limitations of the protein labeling techniques mentioned above. Genetically encodable, non-canonical amino acids (ncAAs) and bioorthogonal, site-specific labeling via click chemistry now allow for single-molecule analysis in living cells. This technique entails the use of site-directed mutagenesis to introduce an amber (UAG) stop codon into the protein of interest. Co-expression of this engineered protein with an aminoacyl tRNA synthase/tRNA pair orthogonal to the host in the presence of ncAA directs the incorporation of the ncAA at the amber stop codon and allows translation to continue through the amber stop codon, rather than terminating translation. This process is known as amber stop codon suppression. Because the ncAA used in this system bears a ring-strained alkyne or alkene, it reacts with azide- or tetrazine-containing dyes in a click- labeling reaction via strain-promoted alkyne-azide cycloaddition (SPAAC) or strain-promoted inverse-electron-demand Diels-Alder cycloaddition (SPIEDAC, also known as SPDAC) reactions. This approach allows the rapid and efficient grafting of fluorophores onto the protein of interest with minimal disruption to the protein coding region. Several studies have used this labeling system to interrogate the behavior of some viral proteins (Nikić et al., 2014), but it had not been applied to an HIV protein.
Figure 1. A Site-Specific, Covalent Labeling Strategy for the HIV-1 Env Glycoprotein Complex Expressed on the Plasma Membrane of Living Cells.
(A) Translational machinery orthogonal to the host machinery is introduced into the target cell in the presence of ncAA (in this example the ncAA is BCN-Lys) to suppress amber stop codons. The tRNA synthase is engineered to specifically aminoacylate tRNACUA with the ncAA, resulting in incorporation of the ncAA into the growing polypeptide chain wherever there is an amber stop codon.
(B) Translation in suppressed cells results in a mixture of full-length, ncAA-labeled, plasma membrane-associated Env and truncated, cytoplasmic Env. Supplementation of cellular media with H-Tet-Cy5 or H-Tet-KK114 (for fluorescence recovery after photobleaching or STED analysis, respectively) results in a click labeling reaction between ncAA and tetrazine to produce a covalently labeled Env trimer on membrane-associated, full-length Env trimers.
(c) Env mobility and association with Gag-GFP can then be measured.
In this issue of Cell Chemical Biology, Sakin and colleagues developed a site-specific, covalent labeling strategy for the HIV-1 Env glycoprotein complex expressed on the plasma membrane of living cells (Sakin et al., 2017). An ncAA residue was introduced by amber stop codon suppression in the gp120 exposed variable region 4 (V4) following amino acid 407 of the Env protein, resulting in ncAA incorporation at the crown of the Env trimer facing out into the extracellular space. After protein expression and membrane localization of the engineered Env trimer, Env-specific click labeling with a tetrazine-conjugated fluorophore was carried out in culture via a rapid SPIEDAC reaction. This strategy enabled the authors to measure Env mobility and plasma membrane distribution without the disruptive incorporation of fusion proteins (e.g., GFP) or other peptide tags. Due to the modularity of this technique, any probe fused to tetrazine can covalently modify protein-incorporated ncAAs, making the strategy very adaptable to super-resolution methods including stimulated emission depletion (STED), super-resolution optical fluctuation imaging, photo-activated localization, and stochastic optical reconstruction microscopy.
Sakin et al. co-transfected HEK293T cells with three constructs encoding the key components for this reaction: (1) Env containing the amber stop codon at the labeling site, (2) a pyrrolysyl-tRNA synthase along with the cognate tRNA (tRNApyl/pylRSAF) specific for amber stop codon recognition, and (3) a dominant-negative eukaryotic tRNA release factor, eRF1 (E55D) (Sakin et al., 2017). This approach results in competition between endogenous translation machinery and the bioorthogonal machinery for the engineered Env sequence (Figure 1A). Therefore, two Env products are formed: a truncated Env that terminates at the amber stop codon (amino acid 407) and the full-length, engineered protein (which the authors show is functional) containing an incorporated clickable ncAA (Figure 1B). The ncAA utilized by Sakin et al. was endo Bicyclo [6.1.0] non-yne-L-Lysine (BCN-Lys), which was previously shown to efficiently react with H-tetrazine (H-Tet) (Nikić et al., 2014). Supplementation of the cellular media for 10 min with H-Tet-Cy5 or H-Tet-KK114 results in a click-labeling reaction between BCN-Lys and H-Tet. The quick and specific nature of this reaction enabled measurement of Env mobility on the cell surface by live-cell imaging. HIV-1 particles were generated by the addition of eGFP-tagged Gag to the system, allowing the visualization of Env accumulation at sites of particle budding by STED nanoscopy (Figure 1C). In this study, the efficiency of amber suppression was low, and could be only partially rescued by co-expression of eRF1 (E55D), which competes with the endogenous eRF1 to increase the incorporation of the ncAA at amber stop codons without increasing read-through at ochre (UAA) or opal (UGA) stop codons (Schmied et al., 2014). Direct visualization of virion-associated Env could therefore not be achieved, limiting the analysis to Env trimers on the plasma membrane. Despite this limitation, the preliminary results of the study confirm previous Env mobility measurements on the plasma membrane and localization of Env to distinctive clusters at nascent viral budding sites (Muranyi et al., 2013; Roy et al., 2013).
The strategy developed by Sakin and colleagues expands the possibilities for further study of Env trafficking and incorporation into virus particles. Improvements in labeling efficiencies could be achieved by engineering a single vector that expresses all the necessary components for amber codon suppression and HIV protein expression, or by creating cell clones that stably express the components of the system. Minimally invasive labeling of single amino acid residues on proteins in intact living cells allows tracking of labeled proteins in their natural environment. Click chemistry is therefore a powerful tool for studying the cell biology of HIV replication beyond Env-related questions. SPIEDAC reactions are the most efficient representative of the in vivo bioorthogonal chemical reactions to date, and reagents are continuously being developed and improved. For example, cell-permeable dyes are now available and may be coupled with the membrane-impermeable cell labeling strategy described here. Membrane-associated protein topography and cellular trafficking could be studied by introducing two amber stop codons in any given protein and labeling extracellular portions with one (membrane-impermeable) dye and intracellular regions with another (membrane-perme- able) dye, or, similarly, differentially labeling intracellular and surface-exposed populations of a protein of interest. If the efficiency of amber suppression could be substantially increased, multiple amber codons could be introduced into one protein for amplification of the fluorophore signal. Protein-protein interactions could also be studied by dual ncAA incorporation and click chemistry labeling to enable Förster resonance energy transfer (FRET) analysis (Lee et al., 2016). Imaging studies of viral transmission by cell-to-cell transfer, nuclear import of viral complexes, and viral membrane protein recycling are also amenable to this new technology. In short, the tools developed by Muller, Lemke, and colleagues are likely to find broad application in the study of HIV and other viruses.
REFERENCES
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, and Tsien RY (2002). J. Am. Chem. Soc. 124, 6063–6076. [DOI] [PubMed] [Google Scholar]
- Freed EO (2015). Nat. Rev. Microbiol. 13, 484–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TC, Kang M, Kim CH, Schultz PG, Chapman E, and Deniz AA (2016). ChemBio-Chem 17, 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link AJ, and Tirrell DA (2003). J. Am. Chem. Soc. 125, 11164–11165. [DOI] [PubMed] [Google Scholar]
- Muranyi W, Malkusch S, Müller, Heilemann M, and Kräusslich HG (2013). PLoS Pathog. 9, e1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikić I, Plass T, Schraidt O, Szymaiński J, Briggs JA, Schultz C, and Lemke EA (2014). Angew. Chem. Int. Ed. 53, 2245–2249. [DOI] [PubMed] [Google Scholar]
- Plass T, Milles S, Koehler C, Szymański J, Mueller R, Wiessler M, Schultz C, and Lemke EA (2012). Angew. Chem. Int. Ed. 51, 4166–4170. [DOI] [PubMed] [Google Scholar]
- Roy NH, Chan J, Lambele M, and Thali M (2013). J. Virol. 87, 7516–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakin V, Paci G, Lemke EA, and Müller (2016). FEBS Lett. 590, 1896–1914. [DOI] [PubMed] [Google Scholar]
- Sakin V, Hanne J, Dunder J, Anders-Össwein M, Laketa V, Nikić I, Krässlich H-G, Lemke EA, and Müller B (2017). Cell Chem. Biol. 24, this issue, 635–645. [DOI] [PubMed] [Google Scholar]
- Schmied WH, Elsässer SJ, Uttamapinant C, and Chin JW (2014). J. Am. Chem. Soc. 136, 15577–15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY (1998). Annu. Rev. Biochem. 67, 509–544. [DOI] [PubMed] [Google Scholar]