Summary
The axoneme of motile cilia is the largest macromolecular machine of eukaryotic cells. In humans, impaired axoneme function causes a range of ciliopathies. Axoneme assembly, structure, and motility requires a radially arranged set of doublet microtubules, each decorated in repeating patterns with non-tubulin components. We use single-particle cryo-electron microscopy to visualize and build an atomic model of the repeating structure of a native axonemal doublet microtubule, which reveals the identities, positions, repeat lengths, and interactions of 38 associated proteins including 33 microtubule inner proteins (MIPs). The structure demonstrates how these proteins establish the unique architecture of doublet microtubules, maintain coherent periodicities along the axoneme, and stabilize the microtubules against the repeated mechanical stress induced by ciliary motility. Our work elucidates the architectural principles that underpin the assembly of this large, repetitive eukaryotic structure, and provides a molecular basis for understanding the etiology of human ciliopathies.
Graphical Abstract
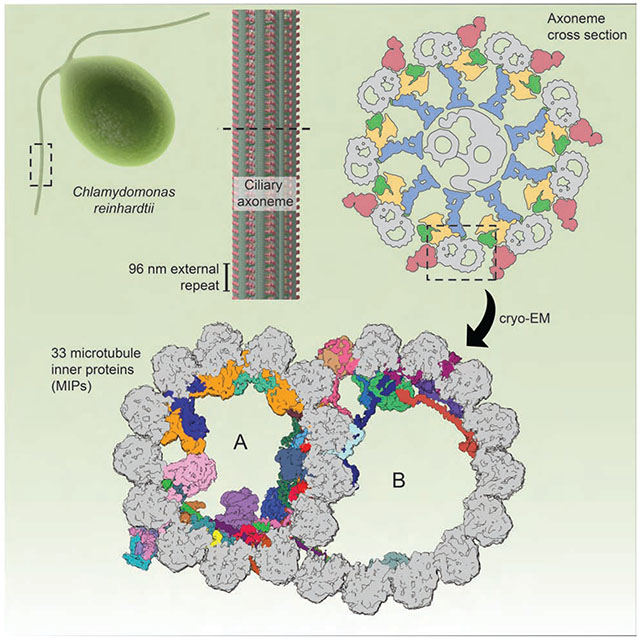
Visualizing axonemal microtubules and the proteins that decorate them, on the outside and inside, points to how the underlying periodic architecture supports cilia function.
Introduction
Cilia (also known as flagella) protrude from the surface of almost all eukaryotic cells and have many diverse roles in human physiology. Motile cilia are involved in locomotion, for example propelling spermatozoa and coordinating directional fluid flow in the embryonic node, respiratory tract, fallopian tube, and brain (Zhou and Roy, 2015). Immotile (primary) cilia are involved in sensory perception and developmental and homeostatic signaling pathways (Singla and Reiter, 2006). A large group of clinically and genetically heterogeneous human diseases, known as ciliopathies, are associated with cilia dysfunction, leading to symptoms including chronic respiratory infections, laterality abnormalities, and infertility (Mitchison and Valente, 2017; Reiter and Leroux, 2017).
The elongated shape of the cilium is supported by a microtubule-based structure, the “axoneme”, which spans almost the entire length of the cilium. The axoneme shows remarkable evolutionary conservation (Li et al., 2004), and much of our current understanding of cilia structure and function comes from work with the unicellular alga Chlamydomonas reinhardtii (Ishikawa, 2017). In most motile cilia, including those of C. reinhardtii and humans, the axoneme has a 9+2 architecture in which nine doublet microtubules surround a central pair of singlet microtubules (Figure 1A). Each doublet microtubule has a distinctive structure with a complete ring of 13 protofilaments (the A tubule) and an incomplete ring of 10 protofilaments (the B tubule). Each protofilament is formed from α/β-tubulin heterodimers stacked head-to-tail with a guanine nucleotide bound between each monomer (Nogales et al., 1999). Protofilaments have intrinsic polarity, so that the microtubule plus end lies at the ciliary tip and the minus end is anchored to the basal body, a modified centriole at the base of the cilium. The axonemal doublet microtubules are contiguous with the A and B tubules of the triplet microtubules of the basal body with the proximal-most region of the axoneme referred to as the transition zone (Reiter et al., 2012).
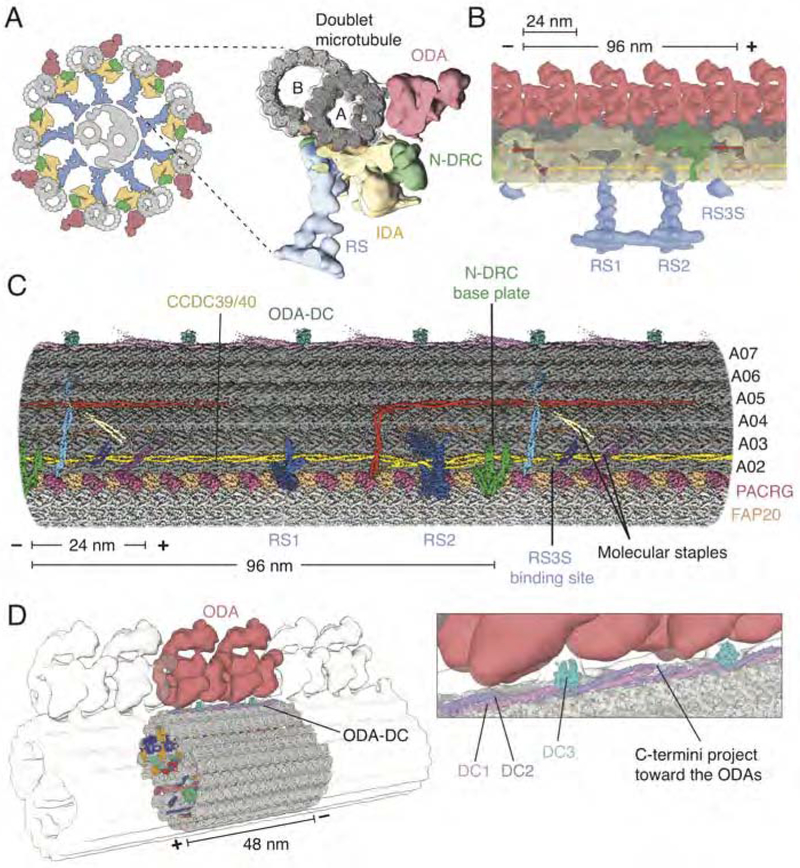
Figure 1. Structure of the 96-nm Repeat of the Doublet Microtubule and Relationship to the Axoneme.
(A) Left, schematic representation of the cross-section of the axoneme from C. reinhardtii showing nine doublet microtubules surrounding a central pair of singlet microtubules (grey). Attached to the doublet microtubules are the radial spokes (RS; blue), inner dynein arm (IDA; yellow), nexin-dynein regulatory complex (N-DRC; green) and outer dynein arm (ODA; red). Right, subtomogram average (EMD-6872) (Kubo et al., 2018) of the axoneme with our map of the 96-nm doublet microtubule repeat (grey) docked inside.
(B) Longitudinal view of the doublet microtubule docked into the subtomogram average of the axoneme (EMD-6872).
(C) Density map of the 96-nm repeat showing axonemal proteins decorating the external surface of the A tubule.
(D) The ODA-DC repeats every 24 nm and, based on fitting the structure into the subtomogram average (EMD-6872), is the main attachment point for the ODA. Inset, detail of the interaction between the ODA and ODA-DC.
In all panels, the minus (−) and plus (+) ends of the doublet microtubule are indicated at the ends of the scale bar.
See also Figures S1, S2, S3, S4 and Tables S1, S2, S3 and S4.
The doublet microtubules of the axoneme are tracks for intraflagellar transport (IFT), the molecular transport mechanism specific to cilia (Lechtreck, 2015; Nakayama and Katoh, 2018). IFT is necessary to build and maintain cilia, and to establish cilium-dependent signal transduction pathways. During IFT, large protein assemblies including cargo proteins and motor complexes associate into linear arrays known as IFT trains, which traverse in both directions along the microtubule tracks (Jordan et al., 2018). Anterograde transport occurs along the B tubules and retrograde transport occurs along the A tubules (Stepanek and Pigino, 2016). This spatial segregation avoids collisions between anterograde and retrograde trains.
Regularly spaced along the A tubule of the doublet microtubule, following multiples of the 8-nm repeat of the tubulin heterodimer, are large multi-subunit protein complexes known as the “axonemal complexes” (Figures 1A and 1B). These include the dynein arms that drive ciliary motility (Lin and Nicastro, 2018). The outer dynein arm (ODA) repeats every 24 nm (Nicastro et al., 2005) and seven different inner dynein arms (IDAs) occupy specific sites within the 96-nm repeat (Bui et al., 2008; Nicastro et al., 2006). The bending of the axoneme is regulated by the nexin-dynein regulatory complex (N-DRC), which connects adjacent doublet microtubules every 96 nm (Heuser et al., 2009) and regulates dynein activity (Huang et al., 1982), as well as by radial-spoke complexes that connect the doublet microtubules to the central pair of singlet microtubules. In C. reinhardtii each 96-nm axonemal repeat has two full-length radial spokes, RS1 and RS2, and a shorter complex, RS3S (Barber et al., 2012). The regular spacings of these axonemal complexes along the length of the doublet microtubule are essential for efficient and coordinated ciliary movements (Oda et al., 2014). Periodicity must be preserved both along the length of each doublet microtubule and collectively among all doublet microtubules of the axoneme. Genetic studies of C. reinhardtii motility mutants have identified a “molecular ruler” — a proposed coiled-coil heterodimer consisting of CCDC39 and CCDC40 — that spaces the axonemal complexes with 96 nm repeat (Lin et al., 2015; Oda et al., 2014). However, a single molecular ruler cannot account for the different periodicities that coexist in the axoneme.
In addition to the large axonemal complexes, many smaller proteins, of hitherto unknown identity and function, bind the external and luminal faces of doublet microtubules. Proteins on the luminal surface, known as microtubule inner proteins (MIPs) (Nicastro et al., 2006), are believed to stabilize the axoneme, especially to resist the mechanical stress associated with ciliary beating (Ichikawa et al., 2017; Owa et al., 2019). In contrast to the overall 96-nm repeat of the doublet microtubule exterior, the internal MIP architecture repeats every 48 nm but in coherent register with the 96-nm repeat (Nicastro et al., 2006; 2011).
We report the structure of a native axonemal doublet microtubule isolated from C. reinhardtii, highly decorated with associated proteins. Our high-resolution (3.4-3.6 Å) reconstructions unambiguously establish the identities, positions, repeat lengths, structures, and interactions of 38 of the associated proteins, creating a direct link between structure and genetic data. The structure demonstrates how axonemal proteins establish the unique architecture of doublet microtubules, limit the protofilament tracks that can be used for IFT, determine the different repeat lengths of the individual axonemal complexes, and maintain a coherent global periodicity along the axoneme.
Results
Exploiting the different periodicities of the complex, we have determined three-dimensional structures for the 96-nm repeating unit at 3.6 Å resolution and for the internal 48-nm repeating unit at 3.4 Å resolution by single-particle cryo-electron microscopy (cryo-EM) (Figures S1 and S2; Table S1). Docking our structures into subtomogram averages of the intact C. reinhardtii axoneme (Bui et al., 2012; Kubo et al., 2018) showed that many of the external axonemal proteins (including radial spokes and the baseplate of the N-DRC) and all discernable MIPs had been retained during purification (Figures 1 and S3).
The 96-nm external repeat structure
The positions of proteins bound to the external surface of the 96-nm repeat map, including several long coiled-coils, correspond with the locations of the axonemal complexes on the A tubule (Figures 1 and S3). The relative sparsity of proteins bound to the external surface of the B tubule may provide the axonemal dynein arms with unfettered access to the tubulin surface for the powerstroke that drives motility (Lin and Nicastro, 2018).
The longest coiled-coil is a 96-nm complex of CCDC39 and CCDC40 (CCDC39/40) that runs the entire length of the furrow between protofilaments A02 and A03 (Figure 1C, yellow). Periodic densities along the length of CCDC39/40 correspond to the bases of RS1, RS2, and the N-DRC (Figures 1A–1C and S3), consistent with the idea that CCDC39/40 is a molecular ruler that defines binding sites and periodicities of axonemal complexes (Lin et al., 2015; Oda et al., 2014). Binding across CCDC39/40 are coiled-coils that interact laterally with neighboring protofilaments (Figure 1C). The density does not yet allow identification of these proteins, which appear to act as “molecular staples” that fix the ruler in place. Two of them generate a unique binding site for RS3S (Figure S3C).
Interacting with the CCDC39/40 complex is a long, L-shaped coiled-coil that occupies the cleft between protofilaments A04 and A05 and turns roughly 90 degrees to bind laterally across protofilaments A02-A04 (Figure 1C, red). The positioning of this coiled-coil is compatible with a role in determining the periodic docking of the two-headed IDAs to protofilaments A03 and A04. Unlike CCDC39/40 and the filamentous MIPs (see later), this coiled-coil does not bind end-to-end and has a the 24-nm gap between repeats. This discontinuity suggests that its periodicity is not self-governed but depends on CCDC39/40.
Not all proteins bind the external surface with 96-nm periodicity. We identified that the ODA docking complex (ODA-DC) occupies the cleft between protofilaments A07-A08 and repeats every 24 nm (Figure 1D). As proposed, the ODA-DC consists of three proteins; DC1, DC2, and DC3 (Takada et al., 2002). Due to the local resolution of this region (Figure S2C), we built a partial atomic model of the heterotrimer (Figure S2D). The N-terminal halves of DC1 and DC2 form a microtubule-bound coiled-coil, while their poorly resolved C-terminal halves project from the surface of the doublet. DC3, which has a calmodulin-like domain, lies midway along the DC1/DC2 coiled-coil. Docking the model into the subtomogram average of the intact axoneme (Kubo et al., 2018) revealed that all three proteins contact the ODAs (Figure 1D).
The 48-nm internal repeat structure
In the 48-nm map of the internal repeat, we could unambiguously identify and build atomic models for 33 MIPs (Figure 2; Table S2). Based on their morphologies and locations, we divide MIPs into three classes; globular MIPs that protrude into the microtubule lumen, single-helix filamentous MIPs (fMIPs) (Ichikawa et al., 2017) that bind the clefts between protofilaments, and extended MIPs that generally lack secondary structure. Previously, MIPs had been numbered based on their low-resolution features in cryo-EM maps (Ichikawa et al., 2017; Maheshwari et al., 2015; Nicastro et al., 2006; 2011), but our analysis shows that the current MIP nomenclature failed to include all MIPs or distinguish the molecular boundaries between them. We therefore refer to MIPs by their protein names (Table S2).
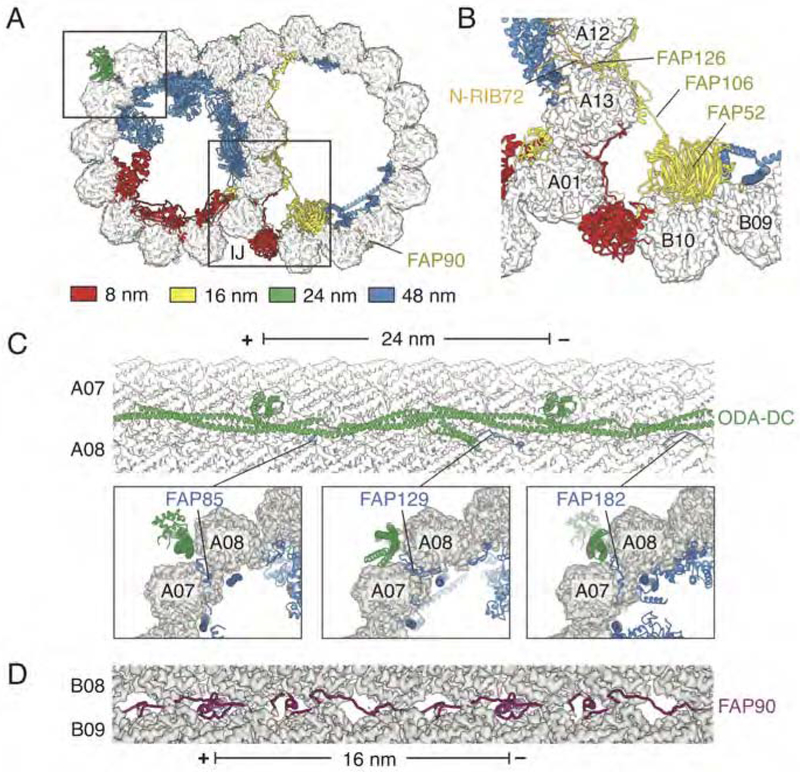
Figure 2. The 48-nm Internal Repeat Structure.
(A). Two slices through the doublet microtubule showing the microtubule inner proteins (MIPs), each uniquely colored. Protein labeling continues in panel B.
(B) Three longitudinal sections through the A tubule of the doublet microtubule. From left-to-right: the first section shows protofilaments A10-A01, the second section shows protofilaments A03-A08, and the third section shows protofilaments A06-B01. In all panels, the minus (−) and plus (+) ends of the doublet microtubule are indicated and the seam of the A tubule is marked with an asterisk.
Individual MIPs were identified in the reconstruction using a combination of prior information, automated fold recognition, and de novo tracing of the map density (STAR Methods and Data S1). For all proteins, the presence of well-defined sidechain densities allowed unambiguous protein assignment and accurate model building (Figures S2D–G and Data S1). As expected, all identified MIPs appear in the mass spectrometry analysis of the cryo-EM sample (Table S3).
Notably, two of the 33 detected MIPs were not previously annotated as ciliary proteins. We name these proteins RIB21 and RIB30 according to their molecular masses and their binding site on the “ribbon”, the three-protofilament arc (A11-A13) that partitions the lumens of the A and B tubules (Linck, 1976). The identification of proteins in the doublet-microtubule structure not previously associated with cilia indicates that the number of proteins in cilia may be higher than reported (Pazour et al., 2005).
To validate our protein identification strategy, we used single-particle cryo-EM to determine the structures of two mutants lacking either RIB72 or FAP166 to 3.9 and 4.0 Å resolution, respectively (Figure S4). RIB72 is a large, multidomain protein that spans seven protofilaments, whereas FAP166 was chosen as it is small and has the lowest exponentially modified protein abundance index score (emPAI; Ishihama et al., 2005) in our mass-spectrometry analysis (Table S3). In both cases, the expected loss of the corresponding protein densities from the reconstructions confirm the protein assignment. In the rib72 mutant, proteins FAP115, FAP222 and FAP252, which bind RIB72, are also lost (Figure S4). This observation is in agreement with a cryo-electron tomography study of Tetrahymena thermophila RIB72 knockout strains, which revealed simultaneous loss of several MIP densities (Stoddard et al., 2018), and a reported association between RIB72 and FAP252 in C. reinhardtii (Ikeda et al., 2007).
Due to averaging, our single-particle approach loses spatial information such as the radial asymmetry among the nine doublet microtubules (Hoops and Witman, 1983) and the longitudinal differences along the length of the axoneme (Bui et al., 2012). We observe only weak density for a “beak” projection in the B tubule (Figure S3E), which is reported to be restricted to doublets 1, 5 and 6 in vivo (Bui et al., 2012; Hoops and Witman, 1983). The beak density becomes more apparent after 3D classification of the local region (Figure S3F). We also lack additional copies of FAP182 (or an unknown MIP with similar size and position to FAP182) on protofilament A06 that are present in proximal but not distal sections of the doublet microtubule (Bui et al., 2012) (Figures S3G and S3H).
MIPs form interconnected networks of different periodicities
Proteins that associate with doublet microtubules follow five different periodicities; 8, 16, 24, 48, and 96 nm, which are multiples of the 8-nm length of a tubulin heterodimer. Proteins with 24 and 96 nm repeat length are specific to the external surface, whereas all fMIPs have 48-nm periodicity, and globular and extended MIPs have repeat lengths of 8, 16 or 48 nm (Table S2). MIPs that bind with 8-nm periodicity follow the repeat of the tubulin heterodimer. Other periodicities are determined by proteins that have a physical footprint larger than a single copy of the tubulin heterodimer. For example, the globular protein FAP52 establishes a repeat length of 16 nm, and all 48-nm fMIPs span six copies of the tubulin heterodimer. By breaking the intrinsic 8-nm periodicity of the tubulin heterodimer, these large proteins generate novel binding sites for other MIPs.
MIPs with similar periodicities cluster together in intricate protein-protein networks (Figure 3A). Proteins with 8-nm repeat length (FAP115, FAP252, and RIB72) occur predominantly on protofilaments A01-A05, whereas proteins with 16-nm repeat length cluster around the ribbon and the inner junction (where the A and B tubules join). The cluster of proteins with 16-nm repeat include six MTPs (FAP52, FAP90, FAP106, FAP126, FAP222, and FAP276) and the N-terminus of RFB72. RIB72 is unusual among MIPs as its other domains show 8-nm periodicity (Figure 3A). All MIPs with 16-nm repeat interact with each other in a network that crosses the junction between protofilaments A12 and A13, where FAP126 on the B tubule comes into direct contact with the N-terminus of RIB72 on the A tubule (Figure 3B). FAP106 links the interactions occurring on the ribbon of the A tubule with FAP52 and FAP276 on the B tubule. Proteins with 48-nm periodicity coincide with the locations of fMIPs on both tubules.
Figure 3. MTPs form Networks of Different Periodicities.
(A) Slice through the doublet microtubule showing non-tubulin components colored by periodicity. Boxes represent regions highlighted in panels B and C.
(B) The cluster of proteins around the inner junction (IJ) with 16-nm repeat length. The RIB72 N-terminal domain (N-RIB72) is shown in orange as although it follows 16-nm repeat, the rest of the protein has 8-nm periodicity.
(C) Top, longitudinal view of the outer dynein arm docking complex (ODA-DC) bound in the external cleft between protofilaments A07 and A08. MIPs FAP85, FAP129 and FAP182 protrude through the cleft of the microtubule and make contact with the ODA-DC. Bottom, cross-sections showing the interactions between 48-nm repeat MIPs and the 24-nm repeat ODA-DC.
(D) Longitudinal view showing the N-terminal half of FAP90 occupying the external cleft between protofilaments B08 and B09.
See also Figures S4 and S7.
MIPs modify the external surface and allow inside-outside communication
MIPs can indirectly affect the external surface of the tubulin lattice by either widening or constricting the angle between protofilaments (Ichikawa et al., 2017). We also notice a more direct effect. Many MIPs extend to the external surface of the doublet microtubule through openings in the lattice between neighboring protofilaments. For example, three MIPs with 48-nm periodicity (FAP85, FAP129, and FAP182) penetrate the cleft between protofilaments A07 and A08 and contact the 24-nm repeat ODA-DC (Figure 3C). Of these proteins, FAP129 interacts both with the fMIP FAP127 on the inside of the A07/A08 cleft and the coiled-coil subunits of the ODA-DC on the exterior. A physical link between the external 24-nm repeat with the luminal 48-nm repeat could ensure accurate alignment and explain why mutations in the mammalian equivalent of FAP127, MNS1, show defects in ODA-DC assembly (Ta-Shma et al., 2018; Zhou et al., 2012). Moreover, the observation that MIPs extend to the exterior and contact proteins bound there shows that MIPs can create novel binding sites that are recognized on the external surface.
Surface-exposed MIPs are also present at the seam of the A tubule (see later) and on the B tubule. The structure of the extended MIP, FAP90, on the B tubule almost belies its annotation as a MIP, as half of its length lies at the external surface (at the cleft between protofilaments B08 and B09) and half internally between protofilaments B09 and B10 (Figure 3D).
Maintenance of periodicity through end-to-end self-association
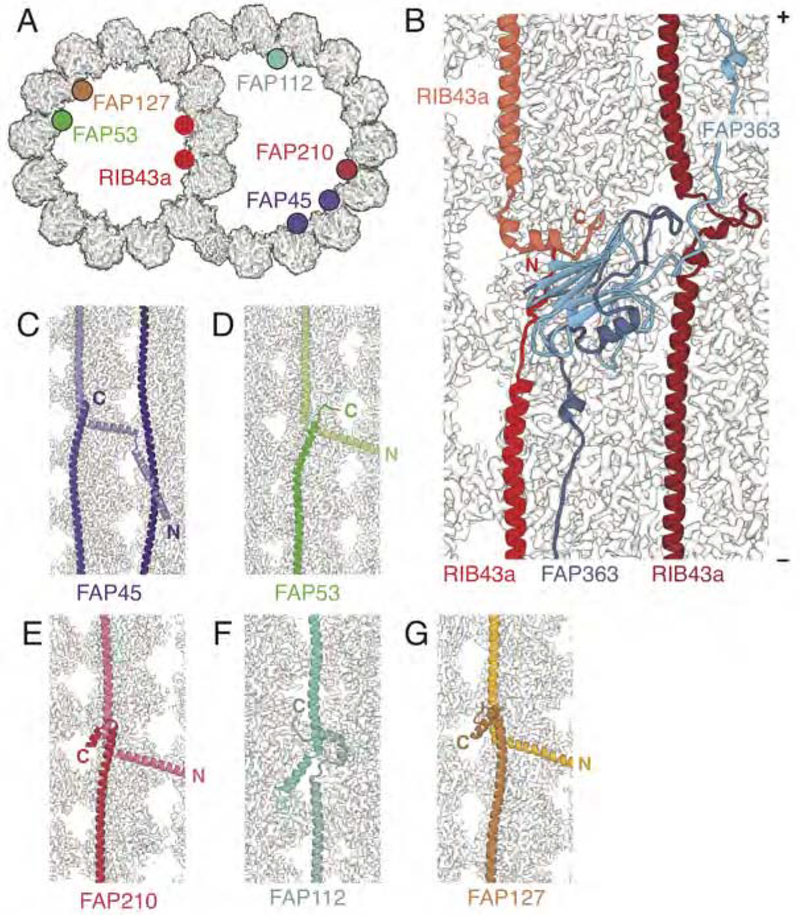
The external coiled-coils (CCDC39/40 and DC1/DC2) and the six internal fMIPs (RIB43a, FAP45, FAP53, FAP112, FAP127, FAP210) show end-to-end self-association (Figure 4). For example, RIB43a occupies the furrows between protofilaments A11-A12 and A12-A13, with an offset of 16 nm between the two fMIPs (Figure 4A). In each furrow, the N-terminus of RIB43a interacts with the C-terminus of the preceding RIB43a molecule to form an uninterrupted filament (Figure 4B). Bridging the two copies of RIB43a is FAP363, a non-filamentous MIP that also shows end-to-end self-association (Figures 2B and 4B). FAP363 has both a globular C-terminal HSP70-like substrate-binding domain and an extended N terminus. This N terminus acts like a HSP70 substrate and docks into the binding groove of the HSP70-like domain of the next copy of FAP363 (Figure 4B). All fMIPs follow the same end-to-end rule, although the interactions between termini differ greatly (Figure 4C–4G). End-to-end self-association provides a mechanism that allows axonemal proteins to maintain their own periodicities.
Figure 4. Maintenance of Longitudinal Periodicity through End-to-end Self-association.
(A) The positions of the fMIPs bound to the 48-nm doublet microtubule. There are two copies of RIB43a and FAP45.
(B) Overview of RIB43 and FAP363 bound to the ribbon showing end-to-end association.
(C-G) Details of the end-to-end self-association between molecules of (C) FAP45, (D) FAP53, (E) FAP210, (F) FAP112, and (G) FAP127. In all longitudinal sections the plus (+) end of the microtubule is facing upwards.
MIPs nonuniformly coat the luminal face
There are more MIPs inside the A tubule than the B tubule, with the highest density of MIPs occurring on the protofilaments of the ribbon (A11-A13) (Figure 2). Most MIPs form both lateral and longitudinal interactions with the tubulin protofilaments (Table S2), in contrast to classic microtubule-associated proteins such as tau (Kellogg et al., 2018), which form exclusively longitudinal interactions. Furthermore, many protofilaments bind multiple MIPs. For example, the ribbon features an interconnected network of 14 proteins (Figure 2B). The number and interconnectivity of MIPs correlates with resistance of protofilament segments to solubilization by Sarkosyl detergent in vitro, during which the ribbon is solubilized only after the sequential losses of the B and A tubules (Witman et al., 1972). Thus, networks of MIPs stabilize lateral and longitudinal interactions of the tubulin lattice to prevent depolymerization of the doublet microtubule.
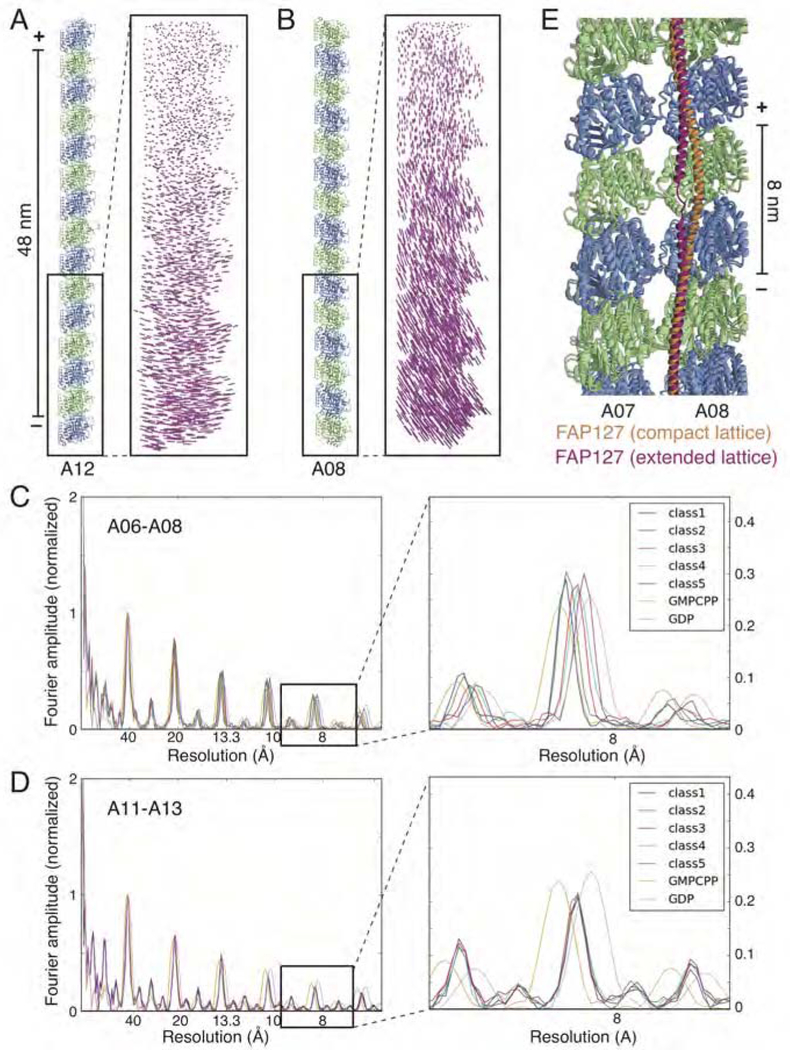
The nonuniform coverage of the luminal walls by MIPs also raises the possibility that MIPs confer variable flexibility within the tubulin lattice. Using focused three-dimensional classification (see STAR Methods), we detected structural variations of different regions of the doublet microtubule within the dataset (Figure 5). Besides a common lateral displacement (in the cross-section plane) (Figure 5A), reflecting the bending of the doublet microtubules that is apparent from the cryo-EM images, we found that the longitudinal spacing between tubulin heterodimers also varied between 82.1 Å and 83.5 Å in most protofilaments (Figure 5B–C). In general, we found greater variation in longitudinal spacings in the A tubule than in the B tubule. The exception was the MIP-dense ribbon, which showed almost no lattice extension/compaction (Figure 5A and 5D). A similar range of longitudinal spacings distinguishes the GDP- and GMPCPP-bound states of cytosolic singlet microtubules (Alushin et al., 2014; Zhang et al., 2015) (Figures 5C and 5D), but the variation of longitudinal spacings in the doublet microtubule is not related to the nucleotide state of the β-tubulin, as only GDP is present at the E site (Figure S5A). We also found that the helix of FAP127 underwent local straightening and unwinding when bound to a more extended tubulin lattice (Figure 5E). Structural plasticity of tubulin (Kueh and Mitchison, 2009) and corresponding structural changes in the associated proteins likely allow doublet microtubules to tolerate both tensile and compressive stresses during ciliary beating.
Figure 5. Tubulin Lattice Variation for Different Regions of the Doublet Microtubule.
(A) Left, protofilament A12 of the ribbon, with the right side facing the central axis of the A tubule. Right, vector display (Cα displacement for each residue of α- and β-tubulin) of protofilament A12 between the two most distinct classes (out of five) following 3D classification of protofilaments A11–13 shows a lateral movement in the cross-section plane.
(B) Left, protofilament A08, in the same orientation as (A). Right, vector display of protofilament A08 between the two most distinct classes from 3D classification of protofilaments A06-A08 shows a longitudinal extension/compaction in addition to the lateral movement as seen in (A).
(C) Layer line profile for the 5 different classes from 3D classification of protofilaments A06-A08. The magnified section shows variable longitudinal spacings of tubulin heterodimers among the 5 different classes, from 82.1 to 83.5 Å, which is comparable to the difference between cytosolic microtubules bound with GMPCPP (84 Å, gold, mimicking the GTP-bound state) and GDP (81.6 Å, pink).
(D) Layer line profile for the 5 different classes from 3D classification of protofilaments A11-A13 (the ribbon region). The magnified section shows that the longitudinal spacings of tubulin heterodimers among the 5 different classes have a consistent value around 82.6 Å.
(E) FAP127 adopts different conformations when bound to different lattice dimensions of protofilaments A07-A08. To accommodate the longer lattice dimensions, a helical region of FAP127 straightens and unwinds.
MIPs at the outer and inner junctions
MIPs at the junctions between A and B tubules help create and maintain the unique architecture of the doublet microtubule. At the outer junction, where the first protofilament of the B tubule (B01) attaches to protofilaments A10-A11, horizontal α-helices belonging to three MIPs (FAP127, FAP53, and FAP141) intersect the three-way junction (Figure 6A), linking together the A and B tubules. The external cleft of the outer junction is bound by an unidentified axonemal protein with 24-nm repeat, potentially providing additional stability (Figure 3A). The α-helices from the three junction-spanning MIPs also interact with the seam of the A tubule, which is between protofilaments A09-A10 (Maheshwari et al., 2015) (Figures 6A and S6). The seam is unusual among interprotofilament interactions as it features heterotypic tubulin contacts and therefore represents a unique site on the microtubule. This physical link between the seam of the A tubule and the outer junction suggests that the location of the seam determines the site of B-tubule nucleation.
Figure 6. MIPs at the Junctions between Tubules.
(A) Three slabs (labeled 1–3) through the doublet microtubule showing the locations of MIPs at the seam and outer junction (OJ). The OJ is penetrated by three MIPs; FAP127 (shown in slab 1), FAP53 (slab 2) and FAP141 (slab 3). The seam is recognized by a number of MIPs including the kinase FAP67. See also Figures S5 and S6.
(B) Alternating copies of FAP20 and PACRG connect the A and B tubules at the inner junction (IJ). One copy of PACRG adjacent to the nexin-dynein regulatory complex (N-DRC) is absent every 96 nm.
(C) Details of the interactions among FAP20, PACRG and the tubulin heterodimer viewed from the microtubule lumen. Ten residues of the typically disordered C-terminal tail of β-tubulin are resolved.
See also Figures S5 and S6.
Besides the three α-helical proteins, the seam is also recognized by five extended MIPs (FAP68, FAP95, FAP107, FAP143, and FAP161) that extend from the luminal side to the external side of the doublet microtubule (Figure S6A). These extended MIPs also interact with the outer junction-spanning MIPs (FAP127, FAP53, and FAP141), potentially helping to anchor them at the seam (Figures S6B–S6D). The seam is also the binding site of FAP67, a nucleoside diphosphate kinase that binds twice within the 48-nm repeat, in which each copy exists in a different conformation due to the influence of neighboring MIPs (Figures 6A and S5B–D). Thus, the seam of the A tubule is recognized by a variety of proteins. In contrast, no proteins that bind the seam of cytosolic microtubules have been identified so far.
At the inner junction, the A01 and B10 protofilaments are connected indirectly by a ladder of two alternating proteins; FAP20 and PACRG (Figure 6B), consistent with a recent cryo-ET study (Dymek et al., 2019). FAP20 has a β-sandwich fold; PACRG has helical armadillo repeats. The interactions of FAP20 and PACRG with protofilament B10 are enhanced by helical elements from FAP276 (Figure 6C). Interestingly, the FAP20/PACRG ladder is not strictly continuous, as a copy of PACRG close to the N-DRC baseplate is absent every 96 nm, forming an opening that was also observed by cryo-ET (Nicastro et al., 2011) (Figures 6B and S3D). A role for the FAP20-PACRG ladder in stabilizing the doublet microtubule architecture is consistent with the observation that Chlamydomonas mutants that lack FAP20 showed split or frayed axonemes (Yanagisawa et al., 2014).
FAP52, which has two 7-bladed β-propeller WD-40 domains, sits adjacent to the inner junction (Figure 6C). It has large footprint that spans three tubulin monomers (α/β/α) on two protofilaments (B09 and B10). The combined absence of FAP52 and FAP20 in Chlamydomonas results in a phenotype more severe than caused by the loss of FAP20 alone: the B tubules detach from A tubules and short paralyzed flagella assemble (Owa et al., 2019). These observations suggest that MIPs function synergistically to maintain the architecture of doublet microtubules.
MIPs bind sites of tubulin post-translational modifications
Axonemal microtubules undergo a range of post-translational modifications (PTMs) that accumulate as cilia assemble and mature and are unequally distributed over the surface of the doublet microtubule (Magiera et al., 2018; Wloga et al., 2017). In particular, the acidic C-terminal tails of α- and β-tubulin undergo polyglutamylation, polyglycylation, and tyrosination/detyrosination. Recent in vitro studies have linked these C-terminal tails to the inhibition of doublet microtubule formation leading to the proposition that MIPs sequester tubulin C-terminal tails at the outer junction to allow B-tubule nucleation (Schmidt-Cernohorska et al., 2019). However, our structure shows that the C-termini of tubulin including those at protofilaments A10 and A11 lack clear density. The absence of density may reflect either disorder or cleavage by subtilisin A, which we used to prepare doublet microtubules (see STAR Methods) and can cleave tubulin C-terminal tails (Sackett et al., 1985). We do, however, identify a role for the C-termini of β-tubulin in the closure of the B tubule at the inner junction. An additional ten residues of the C-terminal tail of β-tubulin on protofilament A01 binds the interface between FAP20 and PACRG (Figure 6C). Depletion of FAP20 in Drosophila melanogaster and human cell lines affects the extent and type of tubulin PTMs (Mendes Maia et al., 2014), suggesting that the positioning of axonemal proteins may help define which tubulin molecules are modified and the type of PTM by recruiting different enzymes. We also observe additional densities for the C-terminal tails of α- and β-tubulin on protofilament A12, which interact with FAP106 and an unidentified MIP.
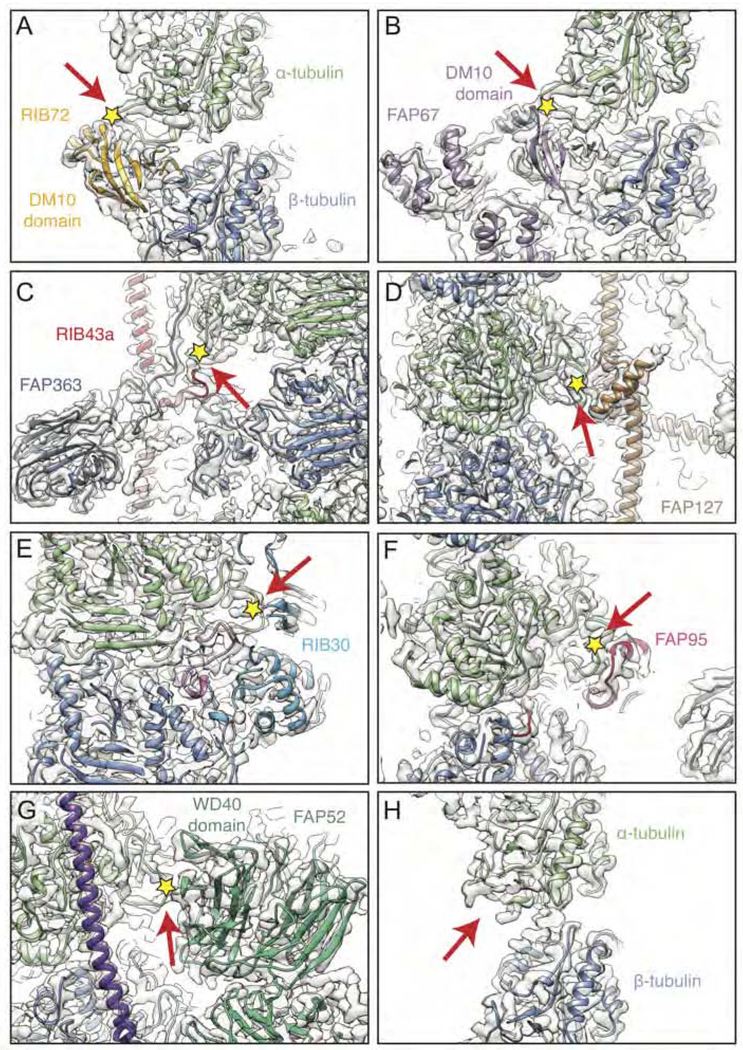
In Chlamydomonas, the majority of axonemal α-tubulin is acetylated on residue K40 within a loop facing the microtubule lumen (Greer et al., 1985; LeDizet and Piperno, 1987). The role of microtubule acetylation is unclear, but makes microtubules more resistant to mechanical stress in vitro (Xu et al., 2017). In all previously determined structures of microtubules, the αK40 loop is poorly resolved regardless of acetylation status (Eshun-Wilson et al., 2019; Howes et al., 2014; Zhang et al., 2015) indicating flexibility. In our structure around 40% of the αK40 loops in the A tubule and 10% in the B tubule are resolved in a wide range of conformations that interact with 13 different proteins (Figure 7). In particular, the αK40 loop is bound by the DM10 domain, which occurs in only two protein families (King, 2006), both of which are axonemal MIPs. The first DM10-containing MIP is the seam-binding kinase, FAP67 (Figure 7A), the second is RIB72, which has three sequential DM10 domains that recognize the αK40 loops on protofilaments A01, A03, and A05 (Figure 7B). In undecorated regions of the doublet microtubule, the αK40 loops are disordered (Figure 7H). At the current resolution of the map, we cannot distinguish the acetylation status of individual αK40 residues. Whether αK40 acetylation is required for MIP recruitment or prevented from occurring once shielded by MIPs remains to be tested.
Figure 7. Interactions between the K40 Loop of α-tubulin and MIPs.
(A-G) Interaction between the αK40 loop on (A) protofilament A01 and the DM10 domain of RIB72, (B) protofilament A09 and the DM10 domain of FAP67, (C) protofilament A13 and RIB43a and an extended region of FAP363, (D) protofilament A07 and FAP127, (E) protofilament A12 and RIB30, (F) protofilament A11 and FAP95, (G) protofilament B09 and FAP52.
(H) The αK40 loop is unresolved in undecorated regions, such as on protofilament A02. In all panels, the αK40 loop is indicated with a red arrow and the K40 residue with a yellow star.
Discussion
A model for the origin and maintenance of axonemal periodicity
More than half of the MIPs present in our structure are annotated in the proteomes of centrioles and basal bodies (Hamel et al., 2017; Keller et al., 2009; 2005) (Table S2). Furthermore, several of these (FAP67, RIB72 and the beak) appear to be visible in the subtomogram averages of basal bodies from Chlamydomonas (Li et al., 2012; 2019b) and mammalian cells (Greenan et al., 2018) (Figure S7). The presence of proteins shared by the basal body and doublet microtubules raises the tantalizing possibility that the periodicity of the MIP structure could be a continuation of the A and B tubules of the triplet microtubules. However, there are two caveats to this model. First, the basal body has a reported periodicity of 8 or 16 nm (Li et al., 2012; 2019b), which is shorter than the 48 nm internal repeat of doublet microtubules and, second, among our identified MIPs only PACRG and RIB72 were found in the proteome of isolated transition zones (Diener et al., 2015). Therefore, it is possible that the internal 48-nm repeat arises together with the external 96-nm repeat, which forms only after the transition zone. Further structural studies will be needed to examine the relationship between the repeat structure of MIPs in the basal body and the axoneme in detail.
During ciliogenesis, our structure suggests a precise and sequential sequence of assembly in which the external coiled-coils and luminal filamentous MIPs likely co-assemble with tubulin and promote their own periodicity by end-to-end self-association (Figure 4). They then define the periodicity of other proteins either by creating new binding sites or shielding others. Consistent with this model, genes encoding these proteins are among the first to be transcribed following deflagellation and new flagellar assembly, as might be expected if they are needed for the periodicity of other structures (Albee et al., 2013).
The external 96-nm repeat (or 24-nm repeat in the case of the ODAs) and internal 48-nm repeat can be self-maintained through faultless end-to-end binding of external coiled-coils and internal fMIPs, respectively. However, we also observe direct communication between the external 24-nm repeat of the ODAs and the internal 48-nm repeat through a set of surface-exposed MIPs near the ODA-DC binding site (Figure 3C) that reinforce the alignment between these two regions. Collectively, interconnected networks of different periodicities would reduce the likelihood of the overall periodicity slipping out of register and disrupting the coordination of ciliary movement.
Diverse MIP functions
Many MIPs have architectural roles, including specifying the position of the B tubule adjacent to the seam of the A tubule (FAP53, FAP127, and FAP141) and connecting the two tubules at the inner junction (the FAP20-PACRG filament). Others stabilize doublet microtubules against the repeated mechanical stress induced by motility, as shown experimentally for FAP45, FAP52 (Owa et al., 2019) and RIB72 (Stoddard et al., 2018). Beyond the architectural and stabilizing roles, two MIPs have folds consistent with enzymatic functions. FAP182 has a nucleotidyltransferase domain and FAP67 is a nucleoside diphosphate kinase that catalyzes transphosphorylation between ATP and GDP (Watanabe and Flavin, 1976). The presence of regular copies of FAP67 (Figure S5B) could keep local GTP-tubulin concentrations high during assembly to promote axonemal extension, since GTP is necessary for microtubule growth. In the mature cilium, high local GTP concentrations could promote microtubule self-repair and suppress catastrophe events (Schaedel et al., 2015).
We also note that EF-hand motifs are highly represented among MIPs; at least eight of the axonemal proteins contain them (Table S2). These EF-hand motifs bind directly to the microtubule surface (FAP85, FAP115, FAP161, FAP166, and RIB30) or to other proteins (DC3, RIB72, and FAP252). As EF-hand motifs undergo conformational changes upon calcium binding (Ikura, 1996) and calcium is sufficient to activate motility of isolated axonemes (Murakami and Eckert, 1972), we speculate that these EF-hand-containing proteins could respond to fluctuations in intraciliary calcium concentration. In particular, C. reinhardtii strains that lack the calmodulin-like DC3 subunit of the ODA-DC cannot swim backwards in response to photoshock, which is mediated by an increase in intraciliary calcium concentration (Tamm and Terasaki, 1994).
Implications for intraflagellar transport
Anterograde and retrograde IFT are mediated by different motors; ciliary kinesins power anterograde transport whereas dynein-2 powers retrograde transport (Lechtreck, 2015). Because the direction of transport is segregated between the two tubules of the doublet microtubule, the prevailing model is that the specific motors recognize the A and B tubules differently (Stepanek and Pigino, 2016). Not all protofilaments on the A-tubule are accessible to the motors. This is especially true for the A tubule, where the axonemal complexes and their docking apparatus spatially restrict dynein-2 to protofilaments A08-A10. There are fewer obvious restrictions on the protofilaments available for anterograde transport. However, the requirement for IFT trains to associate with the ciliary membrane during transport (Stepanek and Pigino, 2016) would constrain kinesin to the protofilaments of the B tubule near the outer junction (B01-04). Thus, the arc of protofilaments available for IFT include the seam of the A tubule and the outer junction. The unique geometries of these sites, and the proteins bound there, could specialize protofilaments for recognition by a particular motor either through direct interactions or by recruiting enzymes that modify the tubulin C-termini. Consistent with the second model, PTMs are known to be variably distributed on the A and B tubules (Wloga et al., 2017) and have been shown to influence the velocity and processivity of human kinesin-II in vitro (Sirajuddin et al., 2014).
Relevance to human cilia and ciliopathies
Our structure of the Chlamydomonas doublet microtubule provides a reference for interpreting prior and future genetic, biochemical, and physiological data from different cell types and for understanding the etiology of human ciliopathies. Based on the sequences of the identified axonemal proteins, we collated a table of their human orthologs with their associated disease phenotypes (Table S4). This table is unlikely to be exhaustive as bioinformatic identification of orthologs of the EF-hand-containing and extended MIPs is challenging and, ultimately, high-resolution structures of mammalian doublet microtubules will be needed for a full and detailed comparison. Nevertheless, the table reveals that external axonemal proteins and MIPs have distinct disease associations. Mutations within human orthologs of the external proteins are associated with primary cilia dyskinesia (PCD), a ciliopathy caused by defective ciliary motility and characterized by chronic respiratory infections and infertility (Noone et al., 2004). In contrast, mutations in human MTPs (including FAP52, FAP53, FAP67 and FAP106) are more often associated with laterality abnormalities such as situs inversus totalis and heterotaxy (which are also often associated with PCD). These laterality abnormalities originate in the failure of cilia-driven fluid flow within the embryonic node to correctly establish left-right asymmetry (Nonaka et al., 1998). The apparent vulnerability of nodal cilia to mutations in MTPs may be related to four key differences from other motile cilia; 1) nodal cilia have a 9+0 axonemal architecture and lack the central pair of singlet microtubules and radial spokes (Shinohara et al., 2015), 2) nodal cilia rotate in a clockwise direction rather than beat in a whip-like motion (Nonaka et al., 1998), 3) nodal cilia are tilted and physically strike the cell surface during rotation (Nonaka et al., 2005) and 4) the density of nodal cilia is low (Shinohara et al., 2012). The mechanical stress of the rotary motion coupled to a less stable axonemal ultrastructure (Shinohara et al., 2015) may place more importance on maintaining the integrity of the doublet microtubule.
In summary, we have determined the structure of the fully decorated native doublet microtubule and built an atomic model of its 48-nm repeat. The structure reveals the identities, binding sites, and periodicities of 38 proteins that bind the external and luminal faces of the doublet microtubules. The presence of MTPs at the seam of the A tubule, at sites of tubulin PTMs, and crossing microtubule walls, expands our knowledge of the repertoire of binding mechanisms used by microtubule-associated proteins learnt from cytosolic microtubules. The structure shows how these proteins, through end-to-end self-association, extensive interconnectivity and redundancy, contribute to establishing and maintaining the accurate register and coherent periodicities of components along the length of the cilium critical for ciliary function.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Rui Zhang (zhangrui@wustl.edu). All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
We used either wild-type Chlamydomonas reinhardtii cells (CC-125) or insertional mutants from the CLiP collection (Li et al., 2016; Zhang et al., 2014) as sources for doublet microtubules. These mutants include LMJ.RY0402.238057 (fap166), LMJ.RY0402.040835 (rib72-1), and LMJ.RY0402.153393 (rib72-2). The mutants were verified by PCR as described in the Method Details section.
Wild-type and mutant Chlamydomonas reinhardtii cells were spread onto 5 Tris-Acetate-Phosphate (TAP) agar plates at a starting density of 8×106 cells/plate and grown for 2 days at 25°C to reach a final density of 1.28×108 cells/plate. Cells from the 5 plates were resuspended in 80 mL of M-N/5 (Sager and Granick, 1954) medium and incubated at 25°C for 2 hrs to allow flagellar assembly.
METHOD DETAILS
RNA preparation
For RNA isolation, cells from two agar plates grown for 5 days were resuspended in 40 ml nitrogen-free medium (M-N/5) for 2 hrs at room temperature. The cells were then collected by centrifugation, and RNA extraction was performed with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s recommendation. Two micrograms of RNA were used in a reverse transcription reaction with SuperScript III (Invitrogen) with random primers as previously described (Lin et al., 2010).
Verification of the rib72 and fap166 mutant strains
Because the parental strain (CMJ030) of the insertion mutants obtained from the CLiP collection has altered swimming and cell wall defects, each strain was crossed to wild type CC-124 or CC-125 sequentially at least two times. The presence of the insertion was monitored by resistance to paromomycin and by PCR using primers that flank the insertion. Primers are listed in the Table S5. The insertions in strains fap166, rib72-1, and rib72-2 showed cosegregation of these traits in 48, 42, and 57 tetrads, respectively. The insertion in the FAP166 gene is in exon 5 (Figure S4A) and the insertions in the RIB72 gene are in exons 4 and 5 (Figure S4B). The fap166 mutant appears to be a null and has no mRNA (Figure S4C). Both rib72 mutants result in truncated mRNAs (Figure S4D).
To verify the absence of FAP166 in the mutant strain, we inserted a HA tag at the C-terminus of FAP166 but found no stable tagged protein in over 500 transformations by immunoblot. Similarly, we made two constructs of RIB72 with HA and mNeonGreen tags at the C- and N-termini but found no stable tagged protein in over 500 transformants by immunoblots.
Doublet purification
Axonemes from wild-type and mutant strains were isolated following a published protocol (Craige et al., 2013). Briefly, cells were deflagellated with 25 mM dibucaine (Sigma-Aldrich) in HMDS buffer (10 mM HEPES, 5 mM MgSO4, 1 mM DTT, 4% Sucrose, pH 7.4) and centrifugated at 1,800 × g for 5 min to remove cell bodies. The supernatant containing flagella was collected and laid over HMDS buffer containing 25% sucrose. After centrifugation at 2,400 × g for 10 min, the supernatant containing flagella was collected down to the sucrose interface. To remove membranes and matrix from flagella, NP-40 (USB Chemicals) was added to flagella to a final concentration of 1% and centrifuged at 30,000 × g for 20 min. The white pellet at the bottom of tube (representing the axonemes) was resuspended with HMDEKP buffer (30 mM HEPES, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA, 25 mM KCl, PH 7.4) containing 1x ProteaseArrest protease inhibitors (G-Biosciences). In the last step, axonemes (equal to 32 OD280) were mildly digested with 10 μg/ml subtilisin A (Sigma-Aldrich) on ice for 30 min in the presence of 2 mM ATP in a total reaction volume of 10 μl before plunge freezing. ProteaseArrest protease inhibitors were present in the buffer to minimize the effects of subtilisin digestion.
Mass spectrometry
Surplus sample from making the cryo-EM grids was spun down and sent for mass spectrometry analysis at the Keck Biotechnology Resource Laboratory at Yale University. The data are provided as Table S3.
Over 600 proteins were identified in the sample by mass spectrometry analysis. The proteins were ranked by the exponentially modified Protein Abundance Index (emPAI), which is a method of estimating protein abundances from peptide counts (Ishihama et al., 2005). Known contaminants and cytosolic proteins (for example human keratins and ribosomal proteins) were then removed from the ranked list. To further reduce the number of candidates on the list, we removed proteins that had previously been annotated as being components of known axonemal complexes. This included 54 components of the central pair of singlet microtubules (Zhao et al., 2019), 46 dynein components, 17 subunits of the radial spokes (Yang et al., 2006), 11 subunits of the nexin-dynein regulatory complex (Lin et al., 2011), 5 subunits of the IFT complexes (Cole et al., 1998), 4 basal body proteins, 3 subunits of the CSC (Heuser et al., 2012), 3 subunits of the tether-head complex (Urbanska et al., 2018), and the 3 subunits of the ODA-DC (Casey et al., 2003). The remaining proteins on the list were used to help identify MTPs in the cryo-EM map. All MTPs identified in our reconstruction appear in the first 104 proteins when ranked by emPAI. The remaining highly ranked proteins on this list may include unidentified MIPs (including components of the beak), proteins bound to the external surface of the 96-nm repeat (for example the molecular staples) or unknown components of other axonemal complexes.
Cryo-EM data collection
The samples were applied to Quantifoil or C-flat R1.2/1.3 copper grids mounted in a Vitrobot Mark IV (Thermo Fisher Scientific) operated at 4°C and 95% humidity. Following blotting for 4 seconds, the grids were plunge frozen in liquid ethane.
Data were collected over 5 cryo-EM sessions using a 300 keV Titan Krios microscope equipped with a Cs-corrector (Thermo Fisher Scientific) and a Bioquantum Energy Filter (Gatan) at the Center for Cellular Imaging (WUCCI) in Washington University in St. Louis (WUSTL). All data (8,314 micrographs in total) were collected using a K2 Summit direct electron detector (Gatan) in counting mode, with an exposure rate of 8.5 electrons/pixel/s on the detector camera. The images were recorded at a nominal magnification of 81,000×, corresponding to a calibrated pixel size of 1.403 Å. A total exposure time of 9 s, corresponding to a total dose of 38.9 electrons/Å2 on the specimen, was fractionated into 30 movie frames. A defocus range from −1.0 to −3.5 μm was set during data acquistion. The data were collected automatically using EPU software (Thermo Fisher Scientific). The data collection statistics for doublet microtubules isolated from wild type cells are listed in Table S1. Statistics for the mutant strains can be found associated with their EMDB entries.
Data processing
Each movie stack was motion corrected using MotionCor2 (Zheng et al., 2017). The contrast transfer function (CTF) parameters for each motion-corrected micrograph were estimated using Gctf (Zhang, 2016). Following a protocol for processing microtubule data (Zhang and Nogales, 2015; Zhang et al., 2015), microtubules (including intact doublet microtubules and over-digested singlet microtubules) were manually selected from the micrographs using the APPION image-processing suite (Lander et al., 2009). The selected microtubule segments were computationally cut into overlapping boxes (512 x 512 pixels) with an 8-nm non-overlapping region (step size) between adjacent boxes (corresponding to the length of a tubulin heterodimer). We refer to these boxed images as 8-nm microtubule particles. Initial alignment parameters and classification results for each 8-nm microtubule particle were obtained via multi-reference alignment in EMAN1 (Ludtke et al., 1999), using one doublet microtubule and one singlet microtubule as reference models. Particles corresponding to doublet microtubules were selected and subjected to further refinement in FREALIGN v9 (Grigorieff, 2007), using the doublet microtubule structure from Tetrahymena thermophila (EMD-8528) (Ichikawa et al., 2017) as an initial model. Based on the similarity scores between the raw particles and the reference model, microtubule particles that were misaligned by 4 nm (corresponding to the length of a tubulin monomer) were identified and shifted into register with the other 8-nm particles at this step.
To determine the register of each 8-nm microtubule particle in the context of the 48-nm repeat for most of the MIPs (Ichikawa et al., 2017), we first imported their alignment parameters from FREALIGN to RELION-3.0 (Zivanov et al., 2018) and did a refinement using Refine3D in RELION-3.0. Only a local search of alignment parameters was performed in this step. After the Refine3D step, local CTF parameters for each 8-nm particle were refined using CTF refinement in RELION-3.0. Next, we performed 3D classification of all the 8-nm microtubule particles while keeping the alignment parameters from the previous Refine3D run. To ensure the classification was driven by the MIP densities, we signal-subtracted the tubulin densities from the raw particle images using Particle subtraction in RELION-3.0, and performed 3D classification with a customized soft-edged binary mask covering protofilaments A08 to A13 (Figure S1C), which has many repetitive MIPs bound with 48-nm periodicity (Ichikawa et al., 2017; Nicastro et al., 2011). The 3D classification produces 6 major classes with roughly the same number of particles (Figure S1D). These 6 classes correspond to the 6 possible registers of the 8-nm particles, agreeing with previous reports that the overall MIP structure has a 48-nm repeat (Ichikawa et al., 2017; Nicastro et al., 2011).
To compute the structure of the doublet microtubule with 48-nm repeat, we arbitrarily selected 1 of the 6 classes from the previous 3D classification result, reverted the signal-subtracted particles to the original particles, which we refer to as 48-nm particles, and did a refinement with Refine3D in RELION-3.0. To improve the local resolution, we employed the strategy of “focused refinement” by restricting the refinement to only a small portion of the doublet microtubule structure each time. First, we used 10 cylindrical masks to divide the doublet microtubule into several subregions, each containing 2 or 3 protofilaments (Figure S1G). We further used short cylindrical masks to divide each subregion into three longitudinal sections, i.e. top, middle and bottom (Figure S1H). Using these methods, the resolution within the mask of each portion of the doublet microtubule is substantially improved. Finally, we stitched the reconstructions of all the 30 (10×3) portions together to generate a composite map for structural analysis and model building. In brief, the generation of the composite map involved two steps in Chimera (Pettersen et al., 2004): 1) aligning and resampling each of the 30 reconstructions to a common reference, which is the 48-nm map before focused refinement; 2) merging the resampled maps by taking the maximum density value at each voxel using the vop maximum command. We employed the same merging strategy to generate the composite half maps, which were used to estimate the overall resolution (Figure S2A) and the B-factor for map sharpening, using Post-processing in RELION-3.0. The same half maps were also used to compute local resolution (Figure S2C), using the blocres function in the Bsoft package (Cardone et al., 2013). To examine the structural variations of the tubulin lattice across different regions, 3D classification was performed for each of the 10 subregions shown in Figure S1G using Class3D in RELION-3.0, while keeping the alignment parameters from the previous Refine3D run.
To obtain the structure of the doublet microtubule with 96-nm repeat, we signal-subtracted the tubulin densities from the original 48-nm particle stack using Particle Subtraction in RELION-3.0. Next, we performed 3D classification with a customized soft-edged binary mask that covers protofilaments A02 to A06 (Figure S1E). This region has many structural features with 96-nm periodicity, including the bases of the radial spokes and N-DRC. The 3D classification produces 2 major classes with approximately equal number of particles, corresponding to the 2 possible registers of the 48-nm particles, each covering half of the 96-nm repeat (Figure S1F). As we were unable to directly compute a 96-nm map because the required particle box size exceeded the capacity of our computational resources, we reverted the signal-subtracted particles to the original particles and refined each half of the 96-nm repeat map separately with Refine3D in RELION-3.0. The 96-nm map was then obtained by joining the two reconstructions together. To improve the local resolution, we employed the same strategy of focused refinement and map merging as we did for the 48-nm map to generate composite maps for each half of the 96-nm repeat map. The nominal resolution of the two 48-nm repeat composite maps was estimated to be 3.6 Å using Post-processing in RELION-3.0 (Figure S2B).
The 48-nm repeat structures of the two mutants (fap166 and rib72) were calculated in the same way as the wild type.
Model building
The process of assignment and model building for all proteins is provided in Data S1. All modeling was performed with Coot (Emsley and Cowtan, 2004). During real-space refinement in Coot, torsion, planar peptide and Ramachandran restraints were used.
To begin modeling the structure of the Chlamydomonas doublet microtubule, we first placed the model of the cytosolic microtubule from Bos taurus (Zhang et al., 2015) into the density of the A tubule and mutated the residues to match the sequences of C. reinhardtii α-tubulin (Uniprot ID: P09204) and β-tubulin (Uniprot ID: P04690). The models were adjusted to fit the density using real-space refinement. Protofilaments from the A tubule were subsequently used to model the B tubule. The αK40 loop and acidic C-terminus were modeled de novo for each tubulin molecule if density was present.
Models for the DC1 and DC2 subunits of the ODA-DC were built by assigning residues to idealized helices positioned in the map using Coot. No density was observed for the C-terminal halves of either of these proteins. The DC3 subunit was modeled using a homology model generated from the iTASSER server (Zhang, 2008) as a template. The density for the CCDC39/40 coiled coil was insufficient to build an accurate atomic model. Therefore, the CCDC39/40 complex is not included in the final deposited model.
The positions of three MIPs (FAP20, FAP52, and PACRG) had been well predicted based on biochemical and cryo-ET experiments (Owa et al., 2019; Yanagisawa et al., 2014) and could be placed manually using homology models generated from the iTASSER server (Zhang, 2008). These homology models were then comprehensively rebuilt to fit the density in Coot. To identify other MIPs, we used different strategies depending on the class of MIP. The folds of large globular MIPs were identified by density-based fold recognition using the MOLREP-BALBES pipeline (Brown et al., 2015). In this approach, densities for individual MIPs were “cut” from the overall map to generate a library of map fragments each with a radius of 30 Å. Each unique domain present in the BALBES database was then fit to each map fragment using MOLREP and ranked on the basis of the contrast score. The fit to the density of the top 20 ranked hits were then inspected manually and the best solution (based on the visual consistency of the model to the map) was used to filter the mass spectrometry results. This approach identified the kinase domains of FAP67, the β-trefoil domain of FAP161, the DM10 domains of RIB72, the HSP70-like domain of FAP363, the transferase domain of FAP182, and multiple EF-hand motifs within the density. It should be noted that this process only identifies the most likely fold, definitive protein assignment in all cases was achieved based on sidechain density. In the case of FAP182, although the protein could be assigned based on the sidechain density of its extended C-terminus, the density for the transferase domain is less well resolved and is not included in the final deposited model. Filamentous MIPs were built de novo from poly(alanine) helices. Sequence was assigned to these poly(alanine) traces based on sidechain density and consistency with secondary structure predictions. The remaining MIPs were built de novo, starting from manually placed poly(alanine) helices. Sequences for the C. reinhardtii MIPs were obtained from UniProt and Phytozome, the plant comparative genomics portal (Table S2). A number of small proteins, or fragments of larger proteins, remain unassigned due to insufficient sidechain information to unambiguously identify them. These unassigned proteins are not included in the deposited atomic model, which only includes proteins that can be confidently assigned.
Refinement
The atomic model for the 48-nm repeat was refined against the 48-nm composite map using Phenix.real_space_refine v1.15 (Afonine et al., 2018). During refinement the resolution limit was set to 3.6 Å to limit overfitting. Secondary structure, local symmetry, Ramachandran and rotamer restraints were applied during refinement. As each refinement run took 130 hours, refinement was restricted to two rounds of refinement, each with three macrocycles. A round of manual model correction in Coot was performed between the rounds of refinement. The quality of the final model was analyzed by MolProbity (Chen et al., 2010). Model statistics can be found in Table S1 and Data S1.
Visualization
All figures were created using Chimera v.1.13 (Pettersen et al., 2004).
QUANTIFICATION AND STATISTICAL ANALYSIS
Resolution estimations of cryo-EM density maps are based on the 0.143 Fourier Shell Correlation (FSC) criterion (Rosenthal and Henderson, 2003). All statistical validation performed on the deposited models (PDB: 6U42) was done using the PHENIX package (Table S1).
DATA AND CODE AVAILABILITY
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/): EMD-20631, EMD-20632, EMD-20633, EMD-20634, EMD-20635, EMD-20636, EMD-20637. The model of the 48-nm repeat of the doublet microtubule has been deposited in the Protein Data Bank (PDB; https://www.rcsb.org/): 6U42.
Supplementary Material
Table S3. Proteins identified in the cryo-EM sample by mass spectrometry, Related to Figures 1 and 2. Provided as an Excel table. Proteins identified in the cryo-EM density map are highlighted in yellow.
Table S1. Statistics for data collection, data processing, model refinement and validation, Related to Figures 1 and 2.
Table S2. Identified proteins of the doublet microtubule, Related to Figures 1 and 2.
Table S4. Human orthologs of MIPs and their disease association, Related to Figures 1 and 2. Reports from other organisms are given if reports on human disease associations are not available.
Table S5. Primers used to assay the presence of the insertion in the C. reinhardtii mutants, Related to STAR Methods.
Figure S1. Data Collection and Processing, Related to Figures 1 and 2
(A) Section of a micrograph showing doublet microtubules with different orientations.
(B) Zoomed-in section of a micrograph highlighting radial spokes bound to a doublet microtubule.
(C) Following global alignment of 8-nm microtubule particles, tubulin densities were subtracted from the raw particle images and 3D classification performed with a binary mask covering protofilaments A08 to A13.
(D) Classification identifies six equally distributed classes (classes 1-6). The red and black dashed circles mark the two different copies of FAP67.
(E) To separate the two classes of 48-nm repeat that together form the 96-nm repeat, tubulin densities were subtracted from the raw particle images and 3D classification performed with a binary mask covering protofilaments A02 to A06.
(F) Classification identifies two equally distributed classes (classes 1-2) that together form the 96-nm repeat of the doublet microtubule. The black dashed circle marks the density of the N-DRC.
(G) Using a set of soft-edged cylindrical masks, the doublet microtubule was split into 10 overlapping subregions each containing 2 or 3 protofilaments.
(H) Each subregion shown in (G) was subdivided into 3 sections (top, middle, and bottom) and refined separately. The final composite map consists of 30 independently refined maps merged together.
Figure S2. Global and Local Resolution, Related to Figures 1 and 2
(A) Fourier shell correlation (FSC) curve calculated between masked independent half maps for the 48-nm repeat (blue). The nominal resolution was estimated as 3.4 Å using the FSC = 0.143 criterion (dashed line).
(B) Fourier shell correlation (FSC) curve calculated between masked independent half maps for the two 48-nm sections that combine to form the 96-nm repeat (red and black). The nominal resolution was estimated as 3.6 Å using the FSC = 0.143 criterion (dashed line).
(C) Composite map of the 48-nm repeat of the doublet microtubule colored by local resolution showing the cross-section midway through the 48-nm repeat (left) and a longitudinal section of the ribbon as viewed from the position marked in the cross-section. Most MIPs have sections of density at better than 4.0 Å resolution.
(D) Example of the density for an external coiled-coil (the central portion of the outer dynein arm docking complex; ODA-DC).
(E) Example of the density for two globular MIPs (FAP20 and PACRG).
(F) Example of the density for a fMIP (RIB43a) with landmark residues labeled.
(G) Example of the density for an extended MIP (FAP363) with landmark residues labeled.
Figure S3. Comparison with Subtomogram Averages, Related to Figure 1
(A) The map of the 96-nm repeat of the doublet microtubule docked into the subtomogram average of the C. reinhardtii axoneme (EMD-6872). Segmented densities for the radial spokes (RS1, RS2, and RS3S), the nexin-dynein regulatory complex (N-DRC), and the outer dynein arms (ODA) are shown. The inner dynein arms are excluded for clarity. The boxed regions represent the regions magnified in panels B-D.
(B) The positions of RS1 and RS2 on the doublet microtubule. The densities for the radial spokes in the doublet microtubule map are truncated due to the mask applied during processing.
(C) The binding site for RS3S is formed by the CCDC39/40 complex (yellow) and two unidentified “molecular staples” that cross CCDC39/40.
(D). Position of the N-DRC base-plate that co-purifies with the doublet microtubule. The position of N-DRC base-plate coincides with the “hole” in the PACRG-FAP20 filament.
(E) Schematic representation of the radial asymmetry of the C. reinhardtii axoneme. Doublet microtubules are labeled 1 to 9. Doublet microtubule 1 lacks the ODA (red). Doublet microtubules 1, 5, and 6 have an additional structure, known as the beak (marked with a purple asterisk), in the proximal and central regions of the axoneme.
(F) Map of the 48-nm doublet microtubule showing density for the beak structure on protofilaments B05-B06, which is at lower occupancy than the rest of the doublet microtubule but becomes more apparent after 3D classification of the local subregion. One of the classes with stronger density for the beak is shown in purple.
(G) The map of the doublet microtubule docked into the subtomogram average of doublet microtubule 2-8 in the distal/central region of the C. reinhardtii axoneme (EMD-2132) (Bui et al., 2012).
(H) The map of the doublet microtubule docked into the subtomogram average of doublet microtubule 2-9 in the proximal region of C. reinhardtii axoneme (EMD-2131) (Bui et al., 2012). Additional copies of FAP182 (or an unknown MTP with a similar size to FAP182) are present in the proximal region of the doublet microtubule that are not present in our cryo-EM map, even after 3D classification of the local region.
Figure S4. Characterization of MIP Mutants, Related to Figures 1–3 and Table S5.
(A) Diagram of the gene structure of FAP166 showing 5′UTR (purple), exons E1-E5 (green), and 3′UTR (light blue). Dark blue inverted triangle indicates the insertion site in exon E5.
(B) Diagram of the gene structure of RIB72 showing 5′UTR (purple), exons E1-E5 (orange), and 3′UTR (light blue). Dark blue inverted triangle indicates the insertion sites in exons E4 and E5.
(C) Reverse transcriptase PCR products of FAP166 using primers, listed in the Key Resources Table, that amplify between the sites indicated on the left. Lane a, wild-type (CC-125); b, wild type with no reverse transcriptase; c, fap166; d, fap166 with no reverse transcriptase; e, no template.
(D) Reverse transcriptase PCR products of RIB72 using primers, listed in the Key Resources Table, that amplify between the sites indicated on the left. Lane a, wild-type (CC-125); b, wild type with no reverse transcriptase; c, rib72-1; d, rib72-1 with no reverse transcriptase; e, rib72-2; f, rib72-2 with no reverse transcriptase.
(E) Slice of the density map of the 48-nm repeat of the doublet microtubule from wild-type C. reinhardtii colored by protein. Proteins absent in the fap166 and rib72 mutant structures are labeled.
(F) Slice of the density map of the 48-nm repeat of the doublet microtubule from the fap166 mutant. The absence of FAP166 is indicated by a dashed oval.
(G) Slice of the density map of the 48-nm repeat of the doublet microtubule from the rib72 mutant.
(H) View from the lumen of protofilaments A11-B01 of the doublet microtubule structure from wild-type C. reinhardtii.
(I) View from the lumen of protofilaments A11-B01 of the doublet microtubule structure from the fap166 mutant. The positions of the 2 missing copies of FAP166 are indicated by dashed ovals.
(J) View from the lumen of protofilaments A11-B01 of the doublet microtubule structure from the rib72 mutant.
(K) View from the lumen of protofilaments A01-A06 of the doublet microtubule structure from wild-type C. reinhardtii.
(L) View from the lumen of protofilaments A01-A06 of the doublet microtubule structure from the fap166 mutant. The position of a missing copy of FAP166 is indicated by a dashed oval.
(M) View from the lumen of protofilaments A01-A06 of the doublet microtubule structure from the rib72 mutant.
Figure S5. Nucleotide Status of Tubulin and a Putative Nucleotide Diphosphate Kinase, Related to Figures 2 and 6
(A) section of the ribbon showing density for nucleotides bound at the interfaces between tubulin monomers. The density unambiguously identifies GTP at the α-β interface within the tubulin heterodimer and GDP at the interface between dimers.
(B) Two copies of FAP67 occupy the luminal cleft between protofilaments A09 and A10 at the seam and repeat every 48 nm.
(C) Each copy of FAP67 has two putative nucleotide-binding domains, but only one of these is solvent accessible. The other interacts with the tubulin surface. In the first copy of FAP67, additional density can be seen in the nucleotide-binding pocket.
(D) Superposition of the second copy of FAP67 on the DM10 domain of the first copy of FAP67 reveals that the two copies of FAP67 have different conformations on the doublet microtubule. Panel D is related to panel C by a 90° rotation.
Figure S6. Seam-binding MIPs, Related to Figure 6
(A) Views from the lumen and exterior of the seam of the A tubule with associated MIPs. Five seam-binding MIPs (FAP68, FAP95, FAP107, FAP143, and FAP161) extend from the luminal side to the external side of the doublet microtubule. Three MIPs (FAP53, FAP127, and FAP141) protrude through the outer junction.
(B) Molecular interactions of FAP107 (dark orange), FAP161 (blue) and FAP127 (light orange) with the seam.
(C) Molecular interactions of FAP68 (olive), FAP143 (pink) and FAP53 (green) with the seam. FAP67 is not shown for clarity.
(D) Molecular interactions of FAP95 (red) and FAP141 (purple) with the seam.
Figure S7. Comparison of the Doublet Microtubule with Centrioles/Basal Bodies, Related to Figures 2 and 3
(A) The atomic model of the 48-nm repeat of the doublet microtubule docked into the subtomogram average of the triplet microtubule of the C. reinhardtii procentriole (EMD-9167). MIPs present in the doublet microtubule that have also been identified in mass-spectrometry studies of the C. reinhardtii centriole/basal body are labeled.
(B) The atomic model of the 48-nm repeat of the doublet microtubule docked into the subtomogram average of the triplet microtubule of the C. reinhardtii basal body (EMD-5252) (Li et al., 2012).
(C) The atomic model of the 48-nm repeat of the doublet microtubule docked into the subtomogram average of the triplet microtubules of the centriole of Chinese hamster ovary (CHO) cell line (EMD-7775) (Greenan et al., 2018).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| SuperScript III Reverse Transcriptase | Invitrogen | 18080085 |
| Dibucaine | Sigma | D0638 |
| ProteaseArrest protease inhibitor cocktail | G-Biosciences | 786-108 |
| Subtilisin A | Sigma | P5380 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | Qiagen | 74104 |
| Deposited Data | ||
| Composite cryo-EM density map of the 48-nm repeat of the doublet microtubule from wild-type Chlamydomonas reinhardtii | This paper | EMD-20631 |
| Composite cryo-EM density map of half 1 of the 96-nm repeat of the doublet microtubule from wild-type Chlamydomonas reinhardtii | This paper | EMD-20632 |
| Composite cryo-EM density map of half 2 of the 96-nm repeat of the doublet microtubule from wild-type Chlamydomonas reinhardtii | This paper | EMD-20633 |
| Composite cryo-EM density map of protofilaments A07-A08 (extended lattice) | This paper | EMD-20634 |
| Composite cryo-EM density map of protofilaments A07-A08 (compact lattice) | This paper | EMD-20635 |
| Composite cryo-EM density map of the 48-nm repeat of the doublet microtubule from Chlamydomonas reinhardtii rib72 mutant | This paper | EMD-20636 |
| Composite cryo-EM density map of the 48-nm repeat of the doublet microtubule from Chlamydomonas reinhardtii fap166 mutant | This paper | EMD-20637 |
| Atomic model of the 48-nm repeat of the doublet microtubule | This paper | PDB: 6U42 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| Chlamydomonas reinhardtii wild type cells (CC-125) | Chlamydomonas Resource Center | N/A |
| Chlamydomonas reinhardtii fap166 | (Li et al., 2016) | LMJ.RY0402.238057 |
| Chlamydomonas reinhardtii rib72-1 | (Li et al., 2016) | LMJ.RY0402.040835 |
| Chlamydomonas reinhardtii rib72-2 | (Li et al., 2016) | LMJ.RY0402.153393 |
| Oligonucleotides | ||
| Primers to assay mutants, see Table S5 | This paper | N/A |
| Recombinant DNA | ||
| Software and Algorithms | ||
| EPU version 1.9.0.30REL | ThermoFisher | https://www.fei.com/software/epu-automated-single-particles-software-for-life-sciences/ |
| MotionCor2 | (Zheng et al., 2017) | https://msg.ucsf.edu/software |
| Gctf v1.06 | (Zhang, 2016) | https://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| APPION | (Lander et al., 2009) | http://emg.nysbc.org/redmine/projects/appion/wiki/Appion_Home |
| EMAN1 | (Ludtke et al., 1999) | https://blake.bcm.edu/emanwiki/EMAN1 |
| FREALIGN v9 | (Grigorieff, 2007) | http://grigoriefflab.janelia.org/frealign |
| RELION-3.0 | (Zivanov et al., 2018) | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| Blocres | (Cardone et al., 2013) | https://lsbr.niams.nih.gov/bsoft/programs/blocres.html |
| Chimera v.1.13.1 | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| MOLREP v.11.6 | (Vagin and Teplyakov, 2010) | http://www.ccp4.ac.uk/html/molrep.html |
| Coot v0.8.9.1 | (Emsley and Cowtan, 2004) | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| phenix.real_space_refine v1.15.2-3472 | (Afonine et al., 2018) | https://www.phenix-online.org/ |
| phenix.molprobity v1.15.2-3472 | (Chen et al., 2010) | https://www.phenix-online.org/ |
| JPred4 | (Drozdetskiy et al., 2015) | http://www.compbio.dundee.ac.uk/jpred/ |
| DSSP | (Kabsch and Sander, 1983) | http://2struc.cryst.bbk.ac.uk/about/ |
| Other | ||
Cell highlights.
High-resolution cryo-EM structure of a native axonemal doublet microtubule
Atomic model for the 48-nm repeat structure includes 38 proteins
Coherent register between different periodicities via interconnected networks
Insights into diverse MIP functions and roles in ciliopathies
Acknowledgments
We thank Michael Rau and James Fitzpatrick for microscopy support, Jean Kanyo for mass spectrometry, Rui Ma for his contribution at the early stage of the project, Saikat Chowdhury for advice on generating composite maps, and John Cooper, Stephen Harrison, Huawen Lin, and Travis Walton for critical reading. Research in the Dutcher laboratory is supported by funds from NIGMS (R01GM032843). Research in the Brown laboratory is supported by the International Retinal Research Foundation, the E. Matilda Ziegler Foundation for the Blind, the Smith Family Foundation, and the Pew Charitable Trusts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes seven figures, four tables and one document.
Declaration of interests
The authors declare no competing interests.
References
- Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, and Adams PD (2018). Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albee AJ, Kwan AL, Lin H, Granas D, Stormo GD, and Dutcher SK (2013). Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3 (Bethesda) 3, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, and Nogales E (2014). High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 157, 1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony D, Becker-Heck A, Zariwala MA, Schmidts M, Onoufriadis A, Forouhan M, Wilson R, Taylor-Cox T, Dewar A, Jackson C, et al. (2013). Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum. Mutat 34, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin-Tse C, Halbritter J, Zariwala MA, Gilberti RM, Gee HY, Hellman N, Pathak N, Liu Y, Panizzi JR, Patel-King RS, et al. (2013). Zebrafish Ciliopathy Screen Plus Human Mutational Analysis Identifies C21orf59 and CCDC65 Defects as Causing Primary Ciliary Dyskinesia. Am. J. Hum. Genet 93, 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JN, Patterson C, de Nijs L, Durón RM, Nguyen V-H, Tanaka M, Medina MT, Jara-Prado A, Martínez-Juárez IE, Ochoa A, et al. (2017). EFHC1 variants in juvenile myoclonic epilepsy: reanalysis according to NHGRI and ACMG guidelines for assigning disease causality. Genet Med 19, 144–156. [DOI] [PubMed] [Google Scholar]
- Barber CF, Heuser T, Carbajal González BI, Botchkarev VV, and Nicastro D (2012). Three-dimensional structure of the radial spokes reveals heterogeneity and interactions with dyneins in Chlamydomonas flagella. Mol. Biol. Cell 23, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Heck A, Zohn IE, Okabe N, Pollock A, Lenhart KB, Sullivan-Brown J, McSheene J, Loges NT, Olbrich H, Haeffner K, et al. (2011). The coiled-coil domain containing protein CCDC40 is essential for motile cilia function and left-right axis formation. Nat. Genet 43, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchon S, Legendre M, Copin B, Duquesnoy P, Montantin G, Kott E, Dastot F, Jeanson L, Cachanado M, Rousseau A, et al. (2012). Delineation of CCDC39/CCDC40 mutation spectrum and associated phenotypes in primary ciliary dyskinesia. J. Med. Genet 49, 410–416. [DOI] [PubMed] [Google Scholar]
- Bragina EY, Babushkina NP, Garaeva AF, Rudko AA, Tsitrikov DY, Gomboeva DE, and Freidin MB (2019). Impact of the Polymorphism of the PACRG and CD80 Genes on the Development of the Different Stages of Tuberculosis Infection. Iran J Med Sci 44, 236–244. [PMC free article] [PubMed] [Google Scholar]
- Brown A, Long F, Nicholls RA, Toots J, Emsley P, and Murshudov G (2015). Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr 71, 136–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Mosley M, Montes-Berrueta D, Hou Y, Yang F, Scarbrough C, Witman GB, and Wirschell M (2017). Characterization of a new oda3 allele, oda3–6, defective in assembly of the outer dynein arm-docking complex in Chlamydomonas reinhardtii. PLoS ONE 12, e0173842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, and Ishikawa T (2008). Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J. Cell Biol 183, 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, and Ishikawa T (2012). Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol 198, 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardone G, Heymann JB, and Steven AC (2013). One number does not fit all: Mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol 183, 226–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey DM, Inaba K, Pazour GJ, Takada S, Wakabayashi K-I, Wilkerson CG, Kamiya R, and Witman GB (2003). DC3, the 21-kDa subunit of the outer dynein arm-docking complex (ODA-DC), is a novel EF-hand protein important for assembly of both the outer arm and the ODA-DC. Mol. Biol. Cell 14, 3650–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, and Rosenbaum JL (1998). Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol 141, 993–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craige B, Brown JM, and Witman GB (2013). Isolation of Chlamydomonas flagella. Curr Protoc Cell Biol Chapter 3, Unit3.41.1-Unit3.41.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SD, Rosenfeld M, Lee H-S, Ferkol TW, Sagel SD, Dell SD, Milla C, Pittman JE, Shapiro AJ, Sullivan KM, et al. (2019). Primary Ciliary Dyskinesia: Longitudinal Study of Lung Disease by Ultrastructure Defect and Genotype. Am. J. Respir. Crit. Care Med 199, 190–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Lupetti P, and Rosenbaum JL (2015). Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr. Biol 25, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, and Barton GJ (2015). JPred4: a protein secondary structure prediction server. Nucleic Acids Research 43, W389–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek EE, Lin J, Fu G, Porter M, Nicastro D, and Smith EF (2019). PACRG and FAP20 form the inner junction of axonemal doublet microtubules and regulate ciliary motility. Mol. Biol. Cell 30, 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Gegg M, Böttcher A, Burtscher I, Hasenoeder S, Van Campenhout C, Aichler M, Walch A, Grant SGN, and Lickert H (2014). Flattop regulates basal body docking and positioning in mono- and multiciliated cells. Elife 3, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremek M, Schoenmaker F, Zietkiewicz E, Pogorzelski A, Diehl S, Wijmenga C, and Witt M (2008). Sequence analysis of 21 genes located in the Kartagener syndrome linkage region on chromosome 15q. Eur. J. Hum. Genet 16, 688–695. [DOI] [PubMed] [Google Scholar]
- Ghandourah H, and Dell SD (2018). Severe disease due to CCDC40 gene variants and the perils of late diagnosis in primary ciliary dyskinesia. BMJ Case Rep 2018, bcr-2018–224964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenan GA, Keszthelyi B, Vale RD, and Agard DA (2018). Insights into centriole geometry revealed by cryotomography of doublet and triplet centrioles. Elife 7, 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer K, Maruta H, L’Hernault SW, and Rosenbaum JL (1985). Alpha-tubulin acetylase activity in isolated Chlamydomonas flagella. J. Cell Biol 101, 2081–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N (2007). FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol 157, 117–125. [DOI] [PubMed] [Google Scholar]
- Hamel V, Steib E, Hamelin R, Armand F, Borgers S, Flückiger I, Busso C, Olieric N, Sorzano C-OS, Steinmetz MO, et al. (2017). Identification of Chlamydomonas Central Core Centriolar Proteins Reveals a Role for Human WDR90 in Ciliogenesis. Curr. Biol 27, 2486–2498. e2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Dymek EE, Lin J, Smith EF, and Nicastro D (2012). The CSC connects three major axonemal complexes involved in dynein regulation. Mol. Biol. Cell 23, 3143–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, and Nicastro D (2009). The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J. Cell Biol 187, 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjeij R, Onoufriadis A, Watson CM, Slagle CE, Klena NT, Dougherty GW, Kurkowiak M, Loges NT, Diggle CP, Morante NFC, et al. (2014). CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am. J. Hum. Genet 95, 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ, and Witman GB (1983). Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol 97, 902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes SC, Alushin GM, Shida T, Nachury MV, and Nogales E (2014). Effects of tubulin acetylation and tubulin acetyltransferase binding on microtubule structure. Mol. Biol. Cell 25, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Ramanis Z, and Luck DJ (1982). Suppressor mutations in Chlamydomonas reveal a regulatory mechanism for Flagellar function. Cell 28, 115–124. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Liu D, Kastritis PL, Basu K, Hsu TC, Yang S, and Bui KH (2017). Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins. Nat Commun 8, 15035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Ikeda T, Morikawa K, and Kamiya R (2007). Axonemal localization of Chlamydomonas PACRG, a homologue of the human Parkin-coregulated gene product. Cell Motil. Cytoskeleton 64, 814–821. [DOI] [PubMed] [Google Scholar]
- Ikura M (1996). Calcium binding and conformational response in EF-hand proteins. Trends in Biochemical Sciences 21, 14–17. [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, and Mann M (2005). Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics 4, 1265–1272. [DOI] [PubMed] [Google Scholar]
- Ishikawa T (2017). Axoneme Structure from Motile Cilia. Cold Spring Harbor Perspectives in Biology 9, a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan MA, Diener DR, Stepanek L, and Pigino G (2018). The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol 20, 1250–1255. [DOI] [PubMed] [Google Scholar]
- Kabsch W, and Sander C (1983). Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Keller LC, Geimer S, Romijn E, Yates J, Zamora I, and Marshall WF (2009). Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell 20, 1150–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, and Marshall WF (2005). Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Current Biology 15, 1090–1098. [DOI] [PubMed] [Google Scholar]
- Kellogg EH, Hejab NMA, Poepsel S, Downing KH, DiMaio F, and Nogales E (2018). Near-atomic model of microtubule-tau interactions. Science 360, 1242–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM (2006). Axonemal protofilament ribbons, DM10 domains, and the link to juvenile myoclonic epilepsy. Cell Motil. Cytoskeleton 63, 245–253. [DOI] [PubMed] [Google Scholar]
- Knowles MR, Leigh MW, Ostrowski LE, Huang L, Carson JL, Hazucha MJ, Yin W, Berg JS, Davis SD, Dell SD, et al. (2013). Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am. J. Hum. Genet 92, 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Hou Y, Cochran DA, Witman GB, and Oda T (2018). A microtubule-dynein tethering complex regulates the axonemal inner dynein f (I1). Mol. Biol. Cell 29, 1060–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueh HY, and Mitchison TJ (2009). Structural plasticity in actin and tubulin polymer dynamics. Science 325, 960–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, et al. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol 166, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF (2015). IFT-Cargo Interactions and Protein Transport in Cilia. Trends in Biochemical Sciences 40, 765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, and Piperno G (1987). Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc. Natl. Acad. Sci. U.S.A 84, 5720–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, et al. (2004). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541–552. [DOI] [PubMed] [Google Scholar]
- Li P, He Y, Cai G, Xiao F, Yang J, Li Q, and Chen X (2019a). CCDC114 is mutated in patient with a complex phenotype combining primary ciliary dyskinesia, sensorineural deafness, and renal disease. Journal of Human Genetics 64, 39–48. [DOI] [PubMed] [Google Scholar]
- Li S, Fernandez J-J, Marshall WF, and Agard DA (2012). Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. Embo J. 31, 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fernandez J-J, Marshall WF, and Agard DA (2019b). Electron cryo-tomography provides insight into procentriole architecture and assembly mechanism. Elife 8, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, Gang SS, Blum SR, Ivanova N, Yue R, Robertson JM, Lefebvre PA, Fitz-Gibbon ST, et al. (2016). An Indexed, Mapped Mutant Library Enables Reverse Genetics Studies of Biological Processes in Chlamydomonas reinhardtii. Plant Cell 28, 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Kwan AL, and Dutcher SK (2010). Synthesizing and salvaging NAD: lessons learned from Chlamydomonas reinhardtii. PLoS Genet. 6, e1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zhang Z, Guo S, Chen F, Kessler JM, Wang YM, and Dutcher SK (2015). A NIMA-Related Kinase Suppresses the Flagellar Instability Associated with the Loss of Multiple Axonemal Structures. PLoS Genet. 11, e1005508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, and Nicastro D (2018). Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science 360, eaar1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Tritschler D, Song K, Barber CF, Cobb JS, Porter ME, and Nicastro D (2011). Building blocks of the nexin-dynein regulatory complex in Chlamydomonas flagella. J. Biol. Chem 286, 29175–29191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linck RW (1976). Flagellar doublet microtubules: fractionation of minor components and alpha-tubulin from specific regions of the A-tubule. J. Cell. Sci 20, 405–439. [DOI] [PubMed] [Google Scholar]
- Lorenzetti D, Bishop CE, and Justice MJ (2004). Deletion of the Parkin coregulated gene causes male sterility in the quaking(viable) mouse mutant. Proc. Natl. Acad. Sci. U.S.A 101, 8402–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, and Chiu W (1999). EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol 128, 82–97. [DOI] [PubMed] [Google Scholar]
- Maheshwari A, Obbineni JM, Bui KH, Shibata K, Toyoshima YY, and Ishikawa T (2015). α- and β-Tubulin Lattice of the Axonemal Microtubule Doublet and Binding Proteins Revealed by Single Particle Cryo-Electron Microscopy and Tomography. Structure 23, 1584–1595. [DOI] [PubMed] [Google Scholar]
- Mendes Maia T, Gogendeau D, Pennetier C, Janke C, and Basto R (2014). Bug22 influences cilium morphology and the post-translational modification of ciliary microtubules. Biol Open 3, 138–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng D, Cao M, Oda T, and Pan J (2014). The conserved ciliary protein Bug22 controls planar beating of Chlamydomonas flagella. J. Cell. Sci 127, 281–287. [DOI] [PubMed] [Google Scholar]
- Merveille A-C, Davis EE, Becker-Heck A, Legendre M, Amirav I, Bataille G, Belmont J, Beydon N, Billen F, Clément A, et al. (2011). CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet 43, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MT, Alcaïs A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, et al. (2004). Susceptibility to leprosy is associated with PARK2 and PACRG. Nature 427, 636–640. [DOI] [PubMed] [Google Scholar]
- Mitchison HM, and Valente EM (2017). Motile and non-motile cilia in human pathology: from function to phenotypes. J. Pathol 241, 294–309. [DOI] [PubMed] [Google Scholar]
- Mizuno K, Dymek EE, and Smith EF (2016). Microtubule binding protein PACRG plays a role in regulating specific ciliary dyneins during microtubule sliding. Cytoskeleton (Hoboken) 73, 703–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami A, and Eckert R (1972). Cilia: activation coupled to mechanical stimulation by calcium influx. Science 175, 1375–1377. [DOI] [PubMed] [Google Scholar]
- Nakayama K, and Katoh Y (2018). Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin-2 and dynein-2 motors. Journal of Biochemistry 163, 155–164. [DOI] [PubMed] [Google Scholar]
- Narasimhan V, Hjeij R, Vij S, Loges NT, Wallmeier J, Koerner-Rettberg C, Werner C, Thamilselvam SK, Boey A, Choksi SP, et al. (2015). Mutations in CCDC11, which encodes a coiled-coil containing ciliary protein, causes situs inversus due to dysmotility of monocilia in the left-right organizer. Hum. Mutat 36, 307–318. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Fu X, Heuser T, Tso A, Porter ME, and Linck RW (2011). Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc. Natl. Acad. Sci. U.S.A 108, E845–E853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, McIntosh JR, and Baumeister W (2005). 3D structure of eukaryotic flagella in a quiescent state revealed by cryo-electron tomography. Proc. Natl. Acad. Sci. U.S.A 102, 15889–15894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, and McIntosh JR (2006). The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313, 944–948. [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, and Downing KH (1999). High-resolution model of the microtubule. Cell 96, 79–88. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, and Hirokawa N (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829–837. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Yoshiba S, Watanabe D, Ikeuchi S, Goto T, Marshall WF, and Hamada H (2005). De novo formation of left-right asymmetry by posterior tilt of nodal cilia. PLoS Biol. 3, e268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noone PG, Leigh MW, Sannuti A, Minnix SL, Carson JL, Hazucha M, Zariwala MA, and Knowles MR (2004). Primary ciliary dyskinesia: diagnostic and phenotypic features. Am. J. Respir. Crit. Care Med 169, 459–467. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, and Kikkawa M (2014). Cilia and flagella. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science 346, 857–860. [DOI] [PubMed] [Google Scholar]
- Onoufriadis A, Paff T, Antony D, Shoemark A, Micha D, Kuyt B, Schmidts M, Petridi S, Dankert-Roelse JE, Haarman EG, et al. (2013). Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am. J. Hum. Genet 92, 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owa M, Uchihashi T, Yanagisawa H-A, Yamano T, Iguchi H, Fukuzawa H, Wakabayashi K-I, Ando T, and Kikkawa M (2019). Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat Commun 10, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, and Witman GB (2005). Proteomic analysis of a eukaryotic cilium. J. Cell Biol 170, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perles Z, Cinnamon Y, Ta-Shma A, Shaag A, Einbinder T, Rein AJJT, and Elpeleg O (2012). A human laterality disorder associated with recessive CCDC11 mutation. J. Med. Genet 49, 386–390. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Reish O, Aspit L, Zouella A, Roth Y, Polak-Charcon S, Baboushkin T, Benyamini L, Scheetz TE, Mussaffi H, Sheffield VC, et al. (2016). A Homozygous Nme7 Mutation Is Associated with Situs Inversus Totalis. Hum. Mutat 37, 727–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, and Leroux MR (2017). Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol 18, 533–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Blacque OE, and Leroux MR (2012). The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13, 608–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, and Henderson R (2003). Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol 333, 721–745. [DOI] [PubMed] [Google Scholar]
- Sackett DL, Bhattacharyya B, and Wolff J (1985). Tubulin subunit carboxyl termini determine polymerization efficiency. J. Biol. Chem 260, 43–45. [PubMed] [Google Scholar]
- Sager R, and Granick S (1954). Nutritional control of sexuality in Chlamydomonas reinhardi. J. Gen. Physiol 37, 729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedel L, John K, Gaillard J, Nachury MV, Blanchoin L, and Théry M (2015). Microtubules self-repair in response to mechanical stress. Nat Mater 14, 1156–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Cernohorska M, Zhernov I, Steib E, Le Guennec M, Achek R, Borgers S, Demurtas D, Mouawad L, Lansky Z, Hamel V, et al. (2019). Flagellar microtubule doublet assembly in vitro reveals a regulatory role of tubulin C-terminal tails. Science 363, 285–288. [DOI] [PubMed] [Google Scholar]
- Shinohara K, Chen D, Nishida T, Misaki K, Yonemura S, and Hamada H (2015). Absence of Radial Spokes in Mouse Node Cilia Is Required for Rotational Movement but Confers Ultrastructural Instability as a Trade-Off. Dev. Cell 35, 236–246. [DOI] [PubMed] [Google Scholar]
- Shinohara K, Kawasumi A, Takamatsu A, Yoshiba S, Botilde Y, Motoyama N, Reith W, Durand B, Shiratori H, and Hamada H (2012). Two rotating cilia in the node cavity are sufficient to break left-right symmetry in the mouse embryo. Nat Commun 3, 622. [DOI] [PubMed] [Google Scholar]
- Sigg MA, Menchen T, Lee C, Johnson J, Jungnickel MK, Choksi SP, Garcia G, Busengdal H, Dougherty GW, Pennekamp P, et al. (2017). Evolutionary Proteomics Uncovers Ancient Associations of Cilia with Signaling Pathways. Dev. Cell 43, 744–762. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla V, and Reiter JF (2006). The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science 313, 629–633. [DOI] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, and Vale RD (2014). Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nat Cell Biol 16, 335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanek L, and Pigino G (2016). Microtubule doublets are double-track railways for intraflagellar transport trains. Science 352, 721–724. [DOI] [PubMed] [Google Scholar]
- Stoddard D, Zhao Y, Bayless BA, Gui L, Louka P, Dave D, Suryawanshi S, Tomasi RF-X, Dupuis-Williams P, Baroud CN, et al. (2018). Tetrahymena RIB72A and RIB72B are microtubule inner proteins in the ciliary doublet microtubules. Mol. Biol. Cell 29, 2566–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subaran RL, Conte JM, Stewart WCL, and Greenberg DA (2015). Pathogenic EFHC1 mutations are tolerated in healthy individuals dependent on reported ancestry. Epilepsia 56, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Baek I-J, Kim YH, Gho YS, Oh SP, Lee YJ, and Lee H-W (2016). PIERCE1 is critical for specification of left-right asymmetry in mice. Sci Rep 6, 27932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta-Shma A, Hjeij R, Perles Z, Dougherty GW, Abu Zahira I, Letteboer SJF, Antony D, Darwish A, Mans DA, Spittler S, et al. (2018). Homozygous loss-of-function mutations in MNS1 cause laterality defects and likely male infertility. PLoS Genet. 14, e1007602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta-Shma A, Perles Z, Yaacov B, Werner M, Frumkin A, Rein AJJT, and Elpeleg O (2015). A human laterality disorder associated with a homozygous WDR16 deletion. Eur. J. Hum. Genet 23, 1262–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Wilkerson CG, Wakabayashi K-I, Kamiya R, and Witman GB (2002). The outer dynein arm-docking complex: composition and characterization of a subunit (oda1) necessary for outer arm assembly. Mol. Biol. Cell 13, 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm SL, and Terasaki M (1994). Visualization of calcium transients controlling orientation of ciliary beat. J. Cell Biol 125, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska P, Joachimiak E, Bazan R, Fu G, Poprzeczko M, Fabczak H, Nicastro D, and Wloga D (2018). Ciliary proteins Fap43 and Fap44 interact with each other and are essential for proper cilia and flagella beating. Cell. Mol. Life Sci 100, 2008–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadnais ML, Lin AM, and Gerton GL (2014). Mitochondrial fusion protein MFN2 interacts with the mitostatin-related protein MNS1 required for mouse sperm flagellar structure and function. Cilia 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A, and Teplyakov A (2010). Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr 66, 22–25. [DOI] [PubMed] [Google Scholar]
- Watanabe T, and Flavin M (1976). Nucleotide-metabolizing enzymes in Chlamydomonas flagella. J. Biol. Chem 251, 182–192. [PubMed] [Google Scholar]
- Wilson GR, Sim ML-J, Brody KM, Taylor JM, McLachlan RI, O’Bryan MK, Delatycki MB, and Lockhart PJ (2010a). Molecular analysis of the PArkin co-regulated gene and association with male infertility. Fertil. Steril 93, 2262–2268. [DOI] [PubMed] [Google Scholar]
- Wilson GR, Wang HX, Egan GF, Robinson PJ, Delatycki MB, O’Bryan MK, and Lockhart PJ (2010b). Deletion of the Parkin co-regulated gene causes defects in ependymal ciliary motility and hydrocephalus in the quakingviable mutant mouse. Hum. Mol. Genet 19, 1593–1602. [DOI] [PubMed] [Google Scholar]
- Witman GB, Carlson K, Berliner J, and Rosenbaum JL (1972). Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol 54, 507–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloga D, Joachimiak E, Louka P, and Gaertig J (2017). Posttranslational Modifications of Tubulin and Cilia. Cold Spring Harbor Perspectives in Biology 9, a028159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Théry M, and Nachury MV (2017). Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H-A, Mathis G, Oda T, Hirono M, Richey EA, Ishikawa H, Marshall WF, Kikkawa M, and Qin H (2014). FAP20 is an inner junction protein of doublet microtubules essential for both the planar asymmetrical waveform and stability of flagella in Chlamydomonas. Mol. Biol. Cell 25, 1472–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, et al. (2006). Radial spoke proteins of Chlamydomonas flagella. J. Cell. Sci 119, 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Patena W, Armbruster U, Gang SS, Blum SR, and Jonikas MC (2014). High-Throughput Genotyping of Green Algal Mutants Reveals Random Distribution of Mutagenic Insertion Sites and Endonucleolytic Cleavage of Transforming DNA. Plant Cell 26, 1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, and Nogales E (2015). A new protocol to accurately determine microtubule lattice seam location. J. Struct. Biol 192, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Alushin GM, Brown A, and Nogales E (2015). Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell 162, 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li D, Wei S, Guo T, Wang J, Luo H, Yang Y, and Tan Z (2019). Whole-exome sequencing identifies a novel CCDC151 mutation, c.325G>T (p.E109X), in a patient with primary ciliary dyskinesia and situs inversus. Journal of Human Genetics 64, 249–252. [DOI] [PubMed] [Google Scholar]
- Zhang Y (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Hou Y, Picariello T, Craige B, and Witman GB (2019). Proteome of the central apparatus of a ciliary axoneme. J. Cell Biol 218, 2051–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, and Roy S (2015). SnapShot: Motile Cilia. Cell 162, 224–224. e1. [DOI] [PubMed] [Google Scholar]
- Zhou J, Yang F, Leu NA, and Wang PJ (2012). MNS1 is essential for spermiogenesis and motile ciliary functions in mice. PLoS Genet. 8, e1002516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, and Scheres SH (2018). New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Proteins identified in the cryo-EM sample by mass spectrometry, Related to Figures 1 and 2. Provided as an Excel table. Proteins identified in the cryo-EM density map are highlighted in yellow.
Table S1. Statistics for data collection, data processing, model refinement and validation, Related to Figures 1 and 2.
Table S2. Identified proteins of the doublet microtubule, Related to Figures 1 and 2.
Table S4. Human orthologs of MIPs and their disease association, Related to Figures 1 and 2. Reports from other organisms are given if reports on human disease associations are not available.
Table S5. Primers used to assay the presence of the insertion in the C. reinhardtii mutants, Related to STAR Methods.
Figure S1. Data Collection and Processing, Related to Figures 1 and 2
(A) Section of a micrograph showing doublet microtubules with different orientations.
(B) Zoomed-in section of a micrograph highlighting radial spokes bound to a doublet microtubule.
(C) Following global alignment of 8-nm microtubule particles, tubulin densities were subtracted from the raw particle images and 3D classification performed with a binary mask covering protofilaments A08 to A13.
(D) Classification identifies six equally distributed classes (classes 1-6). The red and black dashed circles mark the two different copies of FAP67.
(E) To separate the two classes of 48-nm repeat that together form the 96-nm repeat, tubulin densities were subtracted from the raw particle images and 3D classification performed with a binary mask covering protofilaments A02 to A06.
(F) Classification identifies two equally distributed classes (classes 1-2) that together form the 96-nm repeat of the doublet microtubule. The black dashed circle marks the density of the N-DRC.
(G) Using a set of soft-edged cylindrical masks, the doublet microtubule was split into 10 overlapping subregions each containing 2 or 3 protofilaments.
(H) Each subregion shown in (G) was subdivided into 3 sections (top, middle, and bottom) and refined separately. The final composite map consists of 30 independently refined maps merged together.
Figure S2. Global and Local Resolution, Related to Figures 1 and 2
(A) Fourier shell correlation (FSC) curve calculated between masked independent half maps for the 48-nm repeat (blue). The nominal resolution was estimated as 3.4 Å using the FSC = 0.143 criterion (dashed line).
(B) Fourier shell correlation (FSC) curve calculated between masked independent half maps for the two 48-nm sections that combine to form the 96-nm repeat (red and black). The nominal resolution was estimated as 3.6 Å using the FSC = 0.143 criterion (dashed line).
(C) Composite map of the 48-nm repeat of the doublet microtubule colored by local resolution showing the cross-section midway through the 48-nm repeat (left) and a longitudinal section of the ribbon as viewed from the position marked in the cross-section. Most MIPs have sections of density at better than 4.0 Å resolution.
(D) Example of the density for an external coiled-coil (the central portion of the outer dynein arm docking complex; ODA-DC).
(E) Example of the density for two globular MIPs (FAP20 and PACRG).
(F) Example of the density for a fMIP (RIB43a) with landmark residues labeled.
(G) Example of the density for an extended MIP (FAP363) with landmark residues labeled.
Figure S3. Comparison with Subtomogram Averages, Related to Figure 1
(A) The map of the 96-nm repeat of the doublet microtubule docked into the subtomogram average of the C. reinhardtii axoneme (EMD-6872). Segmented densities for the radial spokes (RS1, RS2, and RS3S), the nexin-dynein regulatory complex (N-DRC), and the outer dynein arms (ODA) are shown. The inner dynein arms are excluded for clarity. The boxed regions represent the regions magnified in panels B-D.
(B) The positions of RS1 and RS2 on the doublet microtubule. The densities for the radial spokes in the doublet microtubule map are truncated due to the mask applied during processing.
(C) The binding site for RS3S is formed by the CCDC39/40 complex (yellow) and two unidentified “molecular staples” that cross CCDC39/40.
(D). Position of the N-DRC base-plate that co-purifies with the doublet microtubule. The position of N-DRC base-plate coincides with the “hole” in the PACRG-FAP20 filament.
(E) Schematic representation of the radial asymmetry of the C. reinhardtii axoneme. Doublet microtubules are labeled 1 to 9. Doublet microtubule 1 lacks the ODA (red). Doublet microtubules 1, 5, and 6 have an additional structure, known as the beak (marked with a purple asterisk), in the proximal and central regions of the axoneme.
(F) Map of the 48-nm doublet microtubule showing density for the beak structure on protofilaments B05-B06, which is at lower occupancy than the rest of the doublet microtubule but becomes more apparent after 3D classification of the local subregion. One of the classes with stronger density for the beak is shown in purple.
(G) The map of the doublet microtubule docked into the subtomogram average of doublet microtubule 2-8 in the distal/central region of the C. reinhardtii axoneme (EMD-2132) (Bui et al., 2012).
(H) The map of the doublet microtubule docked into the subtomogram average of doublet microtubule 2-9 in the proximal region of C. reinhardtii axoneme (EMD-2131) (Bui et al., 2012). Additional copies of FAP182 (or an unknown MTP with a similar size to FAP182) are present in the proximal region of the doublet microtubule that are not present in our cryo-EM map, even after 3D classification of the local region.
Figure S4. Characterization of MIP Mutants, Related to Figures 1–3 and Table S5.
(A) Diagram of the gene structure of FAP166 showing 5′UTR (purple), exons E1-E5 (green), and 3′UTR (light blue). Dark blue inverted triangle indicates the insertion site in exon E5.
(B) Diagram of the gene structure of RIB72 showing 5′UTR (purple), exons E1-E5 (orange), and 3′UTR (light blue). Dark blue inverted triangle indicates the insertion sites in exons E4 and E5.
(C) Reverse transcriptase PCR products of FAP166 using primers, listed in the Key Resources Table, that amplify between the sites indicated on the left. Lane a, wild-type (CC-125); b, wild type with no reverse transcriptase; c, fap166; d, fap166 with no reverse transcriptase; e, no template.
(D) Reverse transcriptase PCR products of RIB72 using primers, listed in the Key Resources Table, that amplify between the sites indicated on the left. Lane a, wild-type (CC-125); b, wild type with no reverse transcriptase; c, rib72-1; d, rib72-1 with no reverse transcriptase; e, rib72-2; f, rib72-2 with no reverse transcriptase.
(E) Slice of the density map of the 48-nm repeat of the doublet microtubule from wild-type C. reinhardtii colored by protein. Proteins absent in the fap166 and rib72 mutant structures are labeled.
(F) Slice of the density map of the 48-nm repeat of the doublet microtubule from the fap166 mutant. The absence of FAP166 is indicated by a dashed oval.
(G) Slice of the density map of the 48-nm repeat of the doublet microtubule from the rib72 mutant.
(H) View from the lumen of protofilaments A11-B01 of the doublet microtubule structure from wild-type C. reinhardtii.
(I) View from the lumen of protofilaments A11-B01 of the doublet microtubule structure from the fap166 mutant. The positions of the 2 missing copies of FAP166 are indicated by dashed ovals.
(J) View from the lumen of protofilaments A11-B01 of the doublet microtubule structure from the rib72 mutant.
(K) View from the lumen of protofilaments A01-A06 of the doublet microtubule structure from wild-type C. reinhardtii.
(L) View from the lumen of protofilaments A01-A06 of the doublet microtubule structure from the fap166 mutant. The position of a missing copy of FAP166 is indicated by a dashed oval.
(M) View from the lumen of protofilaments A01-A06 of the doublet microtubule structure from the rib72 mutant.
Figure S5. Nucleotide Status of Tubulin and a Putative Nucleotide Diphosphate Kinase, Related to Figures 2 and 6
(A) section of the ribbon showing density for nucleotides bound at the interfaces between tubulin monomers. The density unambiguously identifies GTP at the α-β interface within the tubulin heterodimer and GDP at the interface between dimers.
(B) Two copies of FAP67 occupy the luminal cleft between protofilaments A09 and A10 at the seam and repeat every 48 nm.
(C) Each copy of FAP67 has two putative nucleotide-binding domains, but only one of these is solvent accessible. The other interacts with the tubulin surface. In the first copy of FAP67, additional density can be seen in the nucleotide-binding pocket.
(D) Superposition of the second copy of FAP67 on the DM10 domain of the first copy of FAP67 reveals that the two copies of FAP67 have different conformations on the doublet microtubule. Panel D is related to panel C by a 90° rotation.
Figure S6. Seam-binding MIPs, Related to Figure 6
(A) Views from the lumen and exterior of the seam of the A tubule with associated MIPs. Five seam-binding MIPs (FAP68, FAP95, FAP107, FAP143, and FAP161) extend from the luminal side to the external side of the doublet microtubule. Three MIPs (FAP53, FAP127, and FAP141) protrude through the outer junction.
(B) Molecular interactions of FAP107 (dark orange), FAP161 (blue) and FAP127 (light orange) with the seam.
(C) Molecular interactions of FAP68 (olive), FAP143 (pink) and FAP53 (green) with the seam. FAP67 is not shown for clarity.
(D) Molecular interactions of FAP95 (red) and FAP141 (purple) with the seam.
Figure S7. Comparison of the Doublet Microtubule with Centrioles/Basal Bodies, Related to Figures 2 and 3
(A) The atomic model of the 48-nm repeat of the doublet microtubule docked into the subtomogram average of the triplet microtubule of the C. reinhardtii procentriole (EMD-9167). MIPs present in the doublet microtubule that have also been identified in mass-spectrometry studies of the C. reinhardtii centriole/basal body are labeled.
(B) The atomic model of the 48-nm repeat of the doublet microtubule docked into the subtomogram average of the triplet microtubule of the C. reinhardtii basal body (EMD-5252) (Li et al., 2012).
(C) The atomic model of the 48-nm repeat of the doublet microtubule docked into the subtomogram average of the triplet microtubules of the centriole of Chinese hamster ovary (CHO) cell line (EMD-7775) (Greenan et al., 2018).
Data Availability Statement
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB; https://www.ebi.ac.uk/pdbe/emdb/): EMD-20631, EMD-20632, EMD-20633, EMD-20634, EMD-20635, EMD-20636, EMD-20637. The model of the 48-nm repeat of the doublet microtubule has been deposited in the Protein Data Bank (PDB; https://www.rcsb.org/): 6U42.