Significance
PDAC epithelial (E) and quasi-mesenchymal (QM) subtypes can be modulated by different therapies demonstrating the plasticity of PDAC cells that contributes to the heterogeneity of PDAC tumors and their intrinsic resistance to a broad spectrum of therapies. Understanding and monitoring the fluidity of PDAC E and QM states are critical to the development of improved clinical trial design to target these different subpopulations.
Keywords: pancreatic ductal adenocarcinoma, molecular subtypes, Vitamin D
Abstract
Transcriptional profiling has defined pancreatic ductal adenocarcinoma (PDAC) into distinct subtypes with the majority being classical epithelial (E) or quasi-mesenchymal (QM). Despite clear differences in clinical behavior, growing evidence indicates these subtypes exist on a continuum with features of both subtypes present and suggestive of interconverting cell states. Here, we investigated the impact of different therapies being evaluated in PDAC on the phenotypic spectrum of the E/QM state. We demonstrate using RNA-sequencing and RNA-in situ hybridization (RNA-ISH) that FOLFIRINOX combination chemotherapy induces a common shift of both E and QM PDAC toward a more QM state in cell lines and patient tumors. In contrast, Vitamin D, another drug under clinical investigation in PDAC, induces distinct transcriptional responses in each PDAC subtype, with augmentation of the baseline E and QM state. Importantly, this translates to functional changes that increase metastatic propensity in QM PDAC, but decrease dissemination in E PDAC in vivo models. These data exemplify the importance of both the initial E/QM subtype and the plasticity of E/QM states in PDAC in influencing response to therapy, which highlights their relevance in guiding clinical trials.
A series of genomic and transcriptomic studies have demonstrated that human pancreatic ductal adenocarcinoma (PDAC) is not a homogeneous cancer, but is composed of subtypes with different functional behaviors in preclinical models and survival differences in clinical studies (1–3), suggesting that the efficacy of drugs may not be uniform across patients. The 2 major molecular subtypes consistently found are the classical epithelial (E) and the quasi-mesenchymal (QM) subtypes, the latter being also known as the basal-like or squamous subtype. Although there has been consistent analysis across datasets indicating poor prognosis of QM compared to E tumors, the predictive value of these subtypes for treatment response remains to be determined. Combination chemotherapy (FOLFIRINOX and Gemcitabine+nab-paclitaxel) remains the primary first line treatment for PDAC, and initial work in patient-derived organoids (4), retrospective correlative analysis (5), and a prospective clinical trial (6) suggest that these subtypes are linked with response to cytotoxic chemotherapy. In parallel work, Vitamin D (Vit D) has been shown in preclinical mouse models to enhance the delivery of chemotherapy by reprogramming cancer-associated fibroblasts into a more quiescent state that leads to increased survival of mice (7). This has led to the initiation of a number of PDAC clinical trials evaluating Vit D in combination with chemotherapy and immunotherapy. However, the impact of Vit D in the setting of E and QM PDAC subtypes has not been fully characterized.
Although a model of E and QM subtypes to dichotomize PDAC tumor behavior affords the simplicity of use in biomarker and functional studies, there has been increasing evidence of epithelial to mesenchymal transition (EMT) transcriptional plasticity of PDAC cells in model systems (8–10) as well as evidence from single-cell RNA-sequencing (RNA-seq) studies (11, 12). This suggests that the E and QM states are on a continuum that can result in interconverting cell types. Therefore, we aim to determine the effects of the cytotoxic combination chemotherapy and Vit D on PDAC cell E and QM states using patient-derived cell lines and translate these results to clinical specimens to better understand the dynamics and clinical implications for therapy for these subtypes in pancreatic cancer.
Results
PDAC Cell Lines Are on a Continuum of E and QM States.
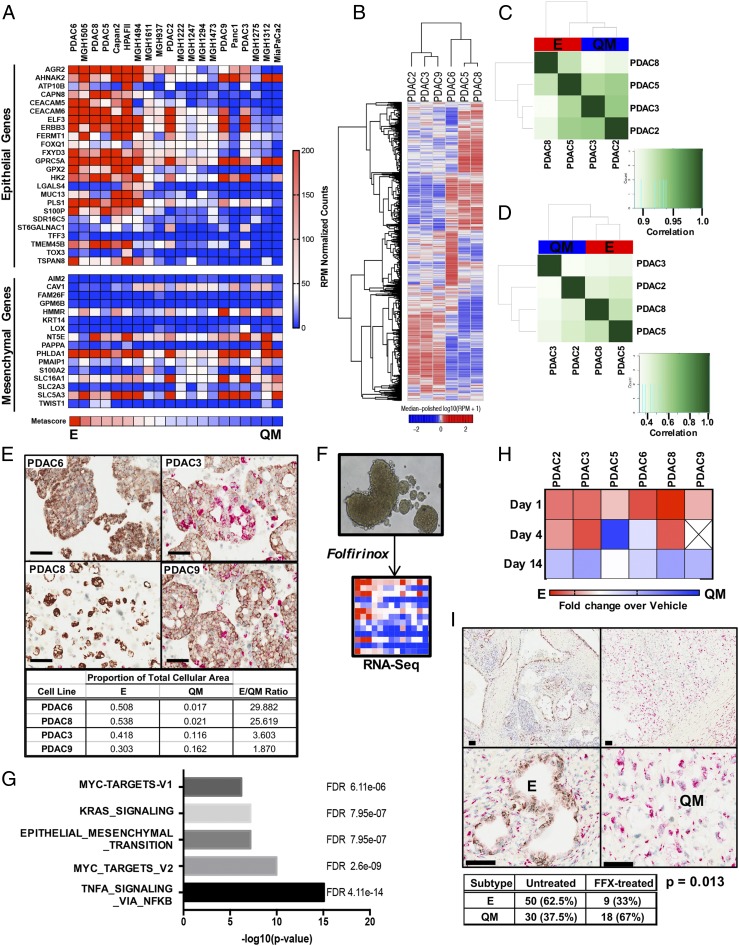
The established “PDAssigner” gene signature (2) was used to calculate an E versus QM metascore for a panel of PDAC cell lines, including both commonly utilized, commercially available and patient-derived cell lines, and demonstrated a diverse spectrum of molecular phenotypes (Fig. 1A). In addition, we assessed the Vitamin D receptor (VDR) expression levels across several of these cell lines for subsequent analyses, given that prior work has found lack of VDR expression in some PDAC lines (13). Our findings are consistent with prior data, revealing VDR expression in the commercially available E cell lines (Capan2, HPAFII) but not in the QM cell lines (MiaPaca2, Panc1) (SI Appendix, Fig. S1A). In contrast, we found VDR expression in all 15 patient-derived cell lines that was higher or comparable to that in a patient-derived cancer-associated fibroblasts (CAF) cell line (CAF1), and VDR protein was detectable in the 6 cell lines generated in our laboratory (14) (SI Appendix, Fig. S1B). Using this data, we selected 6 patient-derived and previously characterized (14) VDR-expressing cell lines, 3 with an E phenotype (PDAC5, PDAC6, PDAC8) and 3 with a QM phenotype (PDAC2, PDAC3, PDAC9) for molecular and functional studies. Unsupervised clustering of most variant genes across these 6 representative cell lines (Fig. 1B) again showed separation of the E and QM cell lines independent of the PDAssigner, indicating that these cell lines are transcriptionally distinct. We then wanted to determine if these subtype differences can be attributed to chromatin accessibility of genes using Assay for Transposase-Accessible Chromatin Sequencing (ATAC-seq) (Fig. 1 C and D). Although the QM (PDAC2, PDAC3) and E (PDAC5, PDAC8) lines clustered with each other with respect to relative openness of chromatin across the genome (Fig. 1C), there was a high level of similarity of global ATAC-seq profiles between cell lines (>0.9 correlation scores). However, when looking specifically at chromatin openness at gene promoters (Fig. 1D), QM and E cell lines again tended to cluster based on subtype, but greater differences were observed between these cell lines. This suggested that global chromatin accessibility is not the major driver of phenotypic heterogeneity in PDAC cell subtypes, but there may be some differences in specific gene promoters that can lead to distinct transcriptional response between QM and E subtypes.
Fig. 1.
PDAC exists on a continuum of epithelial to quasi-mesenchymal gene expression, and cytotoxic chemotherapy shifts toward a mesenchymal phenotype. (A) Expression heatmap to determine an E to QM metascore for each PDAC cell line using a validated gene signature (2). (B) Unsupervised clustering of RNA-seq data from 6 patient-derived PDAC cell lines. [Scale bar, log10(RPM+1).] (C and D) Correlation heatmap of ATAC-seq samples using chromatin accessibility peaks across the genome (C) and at promoter regions (D). (E) Representative images of PDAC cell lines stained with hematoxylin and by RNA-ISH for epithelial (brown) and mesenchymal (red) markers. Quantification of relative E vs. QM expression using the Halo image analysis platform shown below. (F) Schematic of experimental system in which tumor spheroids are grown in 3D culture conditions and treated with a single dose of FOLFIRINOX (FFX). RNA is extracted on day 14 for RNA-seq. (G) Top 5 enriched gene sets procured by computing overlaps between genes induced by FFX treatment across all 6 PDAC cell lines and Hallmark gene sets. (H) Heatmap depicting the relative change in E/QM metascore over time following FFX exposure in each PDAC cell line. Expressed as metascore fold change over untreated. (I) Representative images of PDAC tumors resected following neoadjuvant FFX chemotherapy and stained by RNA-ISH for E (brown) and QM (red) markers. (Scale bars, 100 μm.) Table depicts quantification of E and QM cases in the untreated cohort and the neoadjuvant FFX-treated cohort. P value calculated using Fisher’s exact 2-tailed test.
To understand the single-cell heterogeneity of PDAC cells, we utilized dual-color RNA in situ hybridization (RNA-ISH) for markers of EMT that has been previously used in human PDAC, colon cancer, and breast cancers (15, 16). Probes to these EMT markers were applied to representative E (PDAC6, PDAC8) and QM (PDAC3, PDAC9) cell lines revealing the presence of both E and QM PDAC cells in all cell lines, but with a higher proportion of QM cells in the PDAC3 and PDAC9 lines (Fig. 1E). This single-cell analysis indicates that the E and QM states exist as a heterogeneous mix of cells spanning a continuum of E to QM molecular phenotypes within cell lines rather than as a homogeneous and static state.
PDAC Cell Lines Enriched to QM State Following FOLFIRINOX Treatment Irrespective of Subtype.
Given the known survival differences between patients with E and QM tumors (1–3), we tested the effect of systemic therapies on the E/QM status of PDAC cell lines. E and QM PDAC cell lines were grown as tumorspheres in 3-dimensional (3D) culture and treated with a single dose of FOLFIRINOX or dimethyl sulfoxide (DMSO) vehicle control and then harvested at various time points for viability assays and RNA-seq (Fig. 1F). While all tested PDAC lines were sensitive to FOLFIRNOX, most E cell lines showed the highest sensitivity (SI Appendix, Fig. S1 C and D). RNA-seq analysis of surviving PDAC tumorspheres on day 14 following FOLFIRINOX treatment revealed up-regulation of many genes associated with a more QM state and the Hallmark Epithelial-Mesenchymal Transition Pathway was in the top 5 enriched pathways by gene set enrichment analysis (GSEA) (Fig. 1G and SI Appendix, Fig. S1E). To determine how the E vs. QM phenotype changes in each individual cell line over time after FOLFIRNOX exposure, we calculated an E/QM metascore at days 1, 4, and 14 following treatment. In all cell lines, the surviving cells had a more QM phenotype by day 14 compared with the starting population in each line (Fig. 1H), confirming the shift toward the QM end of the spectrum with FOLFIRINOX exposure.
In order to determine if tumors are selected for QM cells following treatment with systemic chemotherapy in patients, we performed dual-color RNA-ISH with probes to E and QM markers on resected PDAC tumors from a cohort of patients who underwent neoadjuvant FOLFIRINOX chemotherapy prior to surgical resection. This method was validated on PDAC cell lines with known E/QM molecular phenotypes determined by the Collisson PDAssigner, revealing a high correlation between E/QM status as determined by RNA-seq and RNA-ISH (Fig. 1E). In patient samples, tumors spanning the E to QM continuum were observed, with tumors displaying only E staining, both E and QM staining, or only QM staining (Fig. 1I). Scoring revealed that resected FOLFIRINOX-treated tumors were 66% (49/74) QM and 34% (25/74) E compared to 38% (46/121) QM vs. 62% (75/121) E in an untreated cohort of PDAC (Fisher’s exact P value 0.0001). Comparison with QM subtype frequency in resected PDAC tumors as determined by RNA expression analysis by others [Collisson 20/66, 30% (17); Moffitt QM, 36/125, 29% (3); Bailey 25/96, 26% 9 (1); see also ref. 16] also supports an enrichment of the QM subtype in post-FOLFIRINOX human PDAC primary tumors. Taken together, these data establish that systemic chemotherapy can alter the proportion of E and QM tumor cells, shifting a patient’s tumor toward an overall QM state.
Vitamin D Modulates Discrete Transcriptional Targets in PDAC Subtypes.
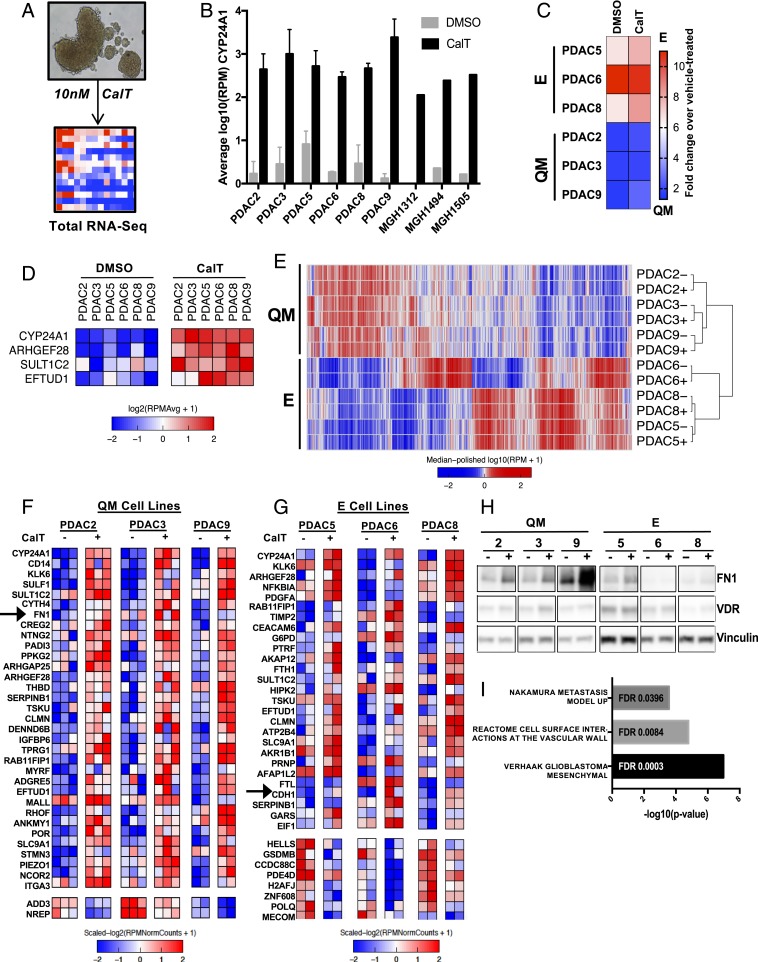
Vit D analogs are currently being evaluated in combination with chemotherapy and immunotherapy in patients with resectable and metastatic PDAC given its beneficial effect on CAFs in PDAC stroma in preclinical models (7). Therefore, we next sought to determine if Vit D also alters the E/QM phenotype of PDAC cell lines and the overall effects of VDR activation in each subtype. With the presence of VDR established, the effects of VDR activation in PDAC tumor cells were determined in patient-derived PDAC cell lines grown as tumorspheres in serum-free media—thereby avoiding undefined levels of Vit D species potentially present in serum—and exposed to 10 nM calcitriol (CalT) for 5 d (Fig. 2A). Each PDAC cell line displayed robust induction of CYP24A1, a Vit D 24-hydroxylase enzyme containing a Vit D response element (VDRE) and known to be a direct transcriptional target of VDR (18), in response to CalT (Fig. 2B), confirming VDR activation. Notably, induction of CYP24A1 was correlated with basal VDR expression, as commercially available cell lines without VDR (MiaPaCa2, Panc1) did not demonstrate significant induction of CYP24A1 in response to CalT (SI Appendix, Fig. S2A). Total RNA-seq was then performed on PDAC tumorspheres treated with CalT to determine transcriptional changes induced by VDR activation. An E/QM metascore calculated before and after exposure to Vit D for each cell line revealed general preservation of the initial overall E or QM molecular phenotype of each line (Fig. 2C). CalT exposure resulted in an enhancement in the epithelial nature of E lines, but did not appreciably shift QM lines more toward an E or QM phenotype.
Fig. 2.
E/QM status of PDAC dictates transcriptional response to VDR activation. (A) Schematic of experimental system in which tumor spheroids are grown in 3D culture conditions and dosed with 10 nM CalT or DMSO control on day 0 and day 2. Spheroids are harvested on day 5, and RNA is extracted for total RNA-seq. (B) CYP24A1 expression in a selection of PDAC cell-line spheroids following 5 d of CalT treatment compared with vehicle control as determined by RNA-seq, expressed as log10 reads per million (RPM). Error bars indicate SD. (C) Relative change in E/QM status of each PDAC cell line following 5 d of exposure to CalT expressed as fold change in metascore over vehicle-treated. (D) Expression heatmap of genes differentially expressed upon CalT treatment in all 6 PDAC cell lines. Data represents 2 to 3 independent experiments for each cell line. (E) Unsupervised clustering of RNA-seq data from 6 patient-derived PDAC cell lines at baseline (−) and after treatment with CalT (+). [Scale bar, log10(RPM+1).] (F and G) Expression heatmaps of differentially expressed genes following treatment of QM (F) and E (G) PDAC spheroids with CalT as determined by RNA-seq. Columns represent individual biological replicates. (H) Western blot demonstrating protein levels of FN1 and VDR in PDAC spheroids following treatment of PDAC spheroids with CalT for 5 d. (I) Enriched gene sets of interest from GSEA computing overlaps between CalT-induced genes in QM cell lines and hallmark, curated, and oncogenic signatures gene sets.
We then asked what the overall transcriptional changes are in response to Vit D treatment. While only 4 genes were commonly up-regulated in all 6 cell lines after exposure to CalT (Fig. 2D), including CYP24A1 as expected, each cell line independently exhibited significant global alterations in gene expression (SI Appendix, Fig. S2B). Unsupervised clustering of the 6 PDAC cell lines demonstrated variation in baseline gene expression by subtype that was not altered by CalT (Fig. 2E), supporting heterogeneous and cell context-dependent CalT-induced changes in PDAC cells. In order to determine if this was due to alterations in the chromatin state of VDRE-containing promoters, we performed ATAC-seq on PDAC cell lines at baseline and after exposure to 5 d of CalT. While PDAC cell lines tended to cluster by molecular subtype, the overall openness of all promoters was not altered by Vit D therapy (SI Appendix, Fig. S2C). Interestingly, when this analysis was confined to VDRE-containing promoters, we found that the baseline chromatin state at these promoters was more similar between PDAC cell lines of the same subtype (SI Appendix, Fig. S2D), consistent with the clustering by subtype seen in promoter regions (Fig. 1D) and suggesting a possibility of a common transcriptional response to VDR activation by subtype.
As expected, accounting for subtype-associated differences by performing differential expression analysis in cell lines within each subtype revealed many more differentially expressed genes from CalT treatment (Fig. 2 F and G) compared with analyzing all cell lines as similar samples (Fig. 2D). CYP24A1 remained the most differentially expressed gene regardless of molecular subtype. Notably, CalT increased the canonical epithelial gene E-cadherin (CDH1) in E cell lines and the established mesenchymal gene fibronectin-1 (FN1) in QM cell lines. Western blot analysis confirmed a specific increase in FN1 protein levels exclusively in QM type CalT-treated PDAC cells (Fig. 2H). Given that FN1 expression in tumor cells is linked to tumor invasiveness, EMT (19), metastasis, and poor clinical outcomes (5, 20–22), GSEA was performed to determine if CalT treatment induces transcriptional programs related to EMT or metastasis in QM tumor cells. Indeed, GSEA revealed enrichment of gene sets related to the mesenchymal phenotype in glioblastoma multiforme, which is reminiscent of EMT and is linked to shorter survival, disease progression, and chemoresistance (23, 24), as well as up-regulated genes in highly metastatic PDAC tumors (25) and cell-surface interactions with blood vessels (Fig. 2I). Taken together, these results reveal a divergent transcriptional response to CalT in PDAC with further induction of key subtype-associated genes in cell lines of each molecular subset. Moreover, they suggest the potential for a more aggressive or metastatic phenotype in QM PDAC cells after CalT treatment.
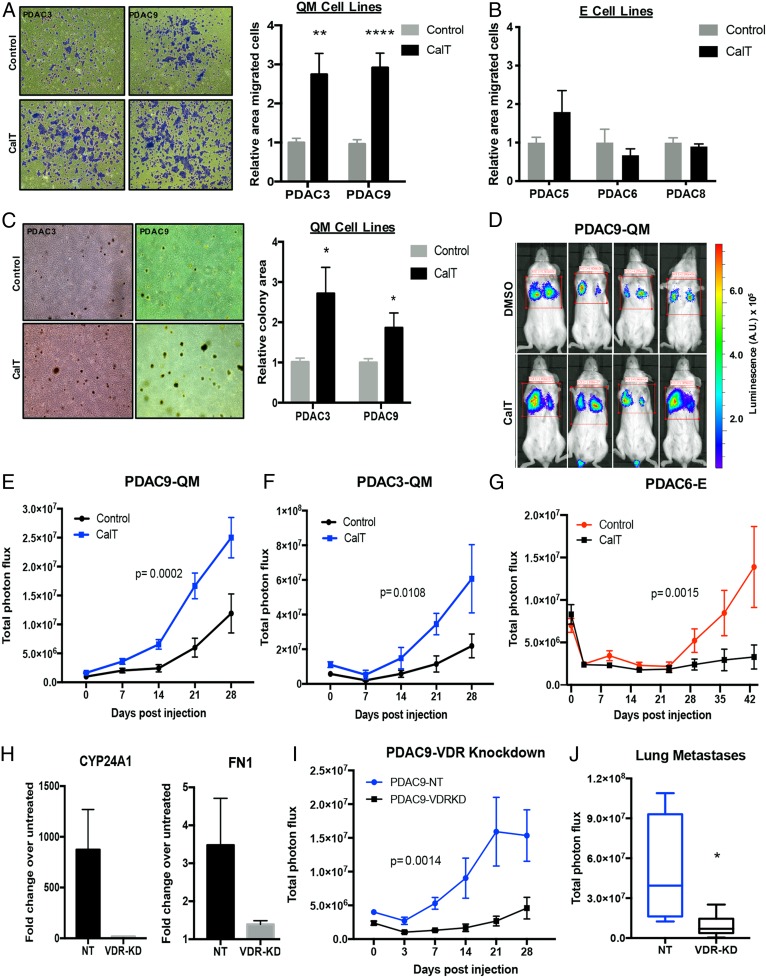
To test this possibility, the functional consequences of VDR activation in E and QM PDAC cells were investigated. Following treatment with CalT, dissociated tumorspheres from QM cell lines demonstrated increased migratory capacity in vitro (Fig. 3A), whereas no effect was detected in tumorspheres from E lines (Fig. 3B). Furthermore, tumor cells from intact CalT-treated PDAC3 (QM) tumorspheres had higher transwell migration capability, likely representing both single-cell and collective cluster motility (SI Appendix, Fig. S3A). In addition, QM but not E tumorspheres treated with CalT and then dissociated into single cells demonstrated increased colony formation under nonadherent growth conditions (Fig. 3C and SI Appendix, Fig. S3B), indicating enhanced anchorage-independent cell growth. These effects were not due to enhanced in vitro proliferation rates of QM PDAC cell lines in response to Vit D (SI Appendix, Fig. S3C). Together, these data reveal that activation of VDR specifically in QM PDAC cells enhances the ability to migrate and grow in anchorage-independent conditions in vitro, which may translate to increased metastatic propensity in vivo.
Fig. 3.
Vitamin D-induced VDR activation in QM PDAC cells increases anchorage-independent cell growth, in vitro migration, and metastasis formation in vivo. (A) Representative images (magnification, 4×) showing migration of pretreated QM PDAC tumor cells across 8-μm filters after 16 h following fixation and staining with crystal violet. Quantification of area covered by stained cells in 3 independent experiments (n = 3 to 5 per experiment) is shown for each cell line. **P < 0.01; ****P < 0.001. (B) Quantification of migration of E PDAC spheres using the same experimental design. (C) Representative images (magnification, 4×) of colonies grown in soft agar from single-cell suspension of QM PDAC cell-line tumorspheres pretreated with CalT for 5 d. Quantification of total colony area in 5 independent experiments (n = 3 per experiment) is shown. *P < 0.05. (D) Representative bioluminescent images of day 21 lung metastases in mice generated by injecting PDAC9 cells dissociated from spheroids pretreated with CalT or vehicle control into the tail vein of untreated mice. Scale representing photon flux in luminescence (A.U.). (E) Quantification of metastatic tumor burden in mice following tail-vein injection of PDAC9-dissociated tumorspheres pretreated with CalT or vehicle control. (F) Quantification of metastatic tumor burden in mice following tail-vein injection of QM PDAC3 dissociated tumorspheres pretreated with CalT or vehicle control. (G) Quantification of metastatic tumor burden in mice following tail-vein injection of E PDAC6 dissociated tumorspheres pretreated with CalT or vehicle control. (H) Induction of CYP24A1 and FN1 expression in response to CalT treatment in control (NT) and VDR-knockdown (VDR-KD) cell lines. Error bars represent SD. (I) Quantification of metastatic tumor burden in mice following tail-vein injection of VDR knockdown (PDAC9-VDRKD) or control PDAC9 (PDAC9-NT) dissociated tumorspheres pretreated with CalT. (J) Quantitation of metastases in explanted lungs from mice receiving VDR knockdown (PDAC9-VDRKD) or control PDAC9 (PDAC9-NT) dissociated tumorspheres pretreated with CalT on day of sacrifice. *P < 0.05. For E, G, and I, metastatic signal was determined by total photon flux in bioluminescent imaging performed weekly (P values determined by 2-way ANOVA).
Consistent with this hypothesis, direct intravascular inoculation of cells from dissociated QM tumorspheres pretreated with CalT into the tail vein of NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice demonstrated a significant increase in the rate and number of lung metastases compared with control-treated dissociated tumorspheres from 2 different cell lines (Fig. 3 D–F). Metastatic signal in explanted lungs at the time of sacrifice also revealed significantly higher metastatic burden in the group that received CalT-pretreated tumor cells (SI Appendix, Fig. S3D). In contrast, CalT-treated tumorspheres from PDAC6, an E PDAC line, generated fewer lung metastases in vivo compared with control-treated E cells (Fig. 3G and SI Appendix, Fig. S3E). To determine if the increase in development of lung metastases following CalT treatment in QM cells was dependent on VDR activation, we used short hairpin RNA (shRNA)-mediated VDR knockdown in PDAC9 tumorspheres (SI Appendix, Fig. S3F). Knockdown of VDR resulted in loss of induction of CYP24A1 and FN1 by CalT as expected (Fig. 3H). Indeed, the increased development of metastases observed with CalT pretreatment of QM tumorspheres was abolished in tumorspheres with low expression of VDR (Fig. 3 I and J), confirming that VDR activation is mediating these effects. Thus, CalT enhances migration and anchorage-independent growth in vitro and increases metastatic potential in vivo specifically in QM tumor cells, indicating that VDR activation may drive more aggressive behavior in this subset of PDAC.
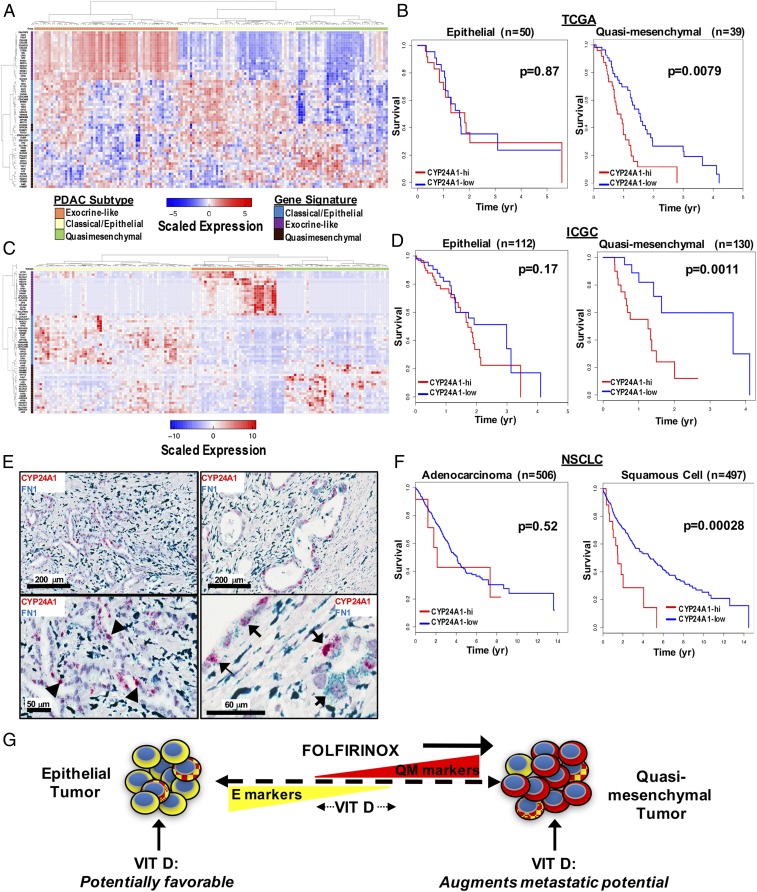
We next looked for evidence linking Vit D signaling in PDAC patients with QM tumors to clinical outcomes within The Cancer Genome Atlas (TCGA) database of surgically resected PDAC tumors. RNA-seq data from these tumors was used to determine CYP24A1 expression as a measure of Vit D signaling in tumor cells. On a population level, there was slightly higher, although nonsignificant, expression of CYP24A1 in pancreatic tumors (SI Appendix, Fig. S4A). We then compared overall survival of patients with pancreas tumors demonstrating high versus low CYP24A1 expression. Overall, there was no significant difference in survival when all tumors were analyzed irrespective of subtype (SI Appendix, Fig. S4B). Given the subtype-dependent effects of VDR activation in PDAC cells lines, we applied the PDAssigner gene signature (2) to the TCGA samples to separate them into E, QM, and exocrine-like tumors (Fig. 4A). Within the E and exocrine subtypes, expression of CYP24A1 did not correlate with changes in survival (Fig. 4B and SI Appendix, Fig. S4C). However, there was a striking association between high CYP24A1 expression and shorter overall survival in tumors of the QM subtype (Fig. 4B). Extension of this analysis to the International Cancer Genome Consortium (ICGC) microarray dataset of untreated and resected PDAC tumors (26) (Fig. 4C) again demonstrated an association between high CYP24A1 expression and shorter overall survival in patients with QM, but not E, tumors (Fig. 4D). We utilized RNA-ISH to determine the localization of CYP24A1 expression in human PDAC tumors, and indeed we found clear CYP24A1 expression in PDAC tumor cells but not in stromal cells (Fig. 4 E, Left). Furthermore, while FN1 expression was seen in both tumor and stromal cells, a subpopulation of tumor cells coexpressed CYP24A1 and FN1 (Fig. 4 E, Right). Therefore, the CYP24A1 expression detected in the TCGA and ICGC analyses is likely to reflect expression in tumor cells rather than in stroma. These results imply that patients harboring QM tumors with evidence of active VDR signaling have worse clinical outcomes, which is consistent with our data demonstrating the VDR-dependent functional gains in metastatic propensity in QM PDAC cells.
Fig. 4.
High CYP24A1 expression in specific subtypes of human cancers is associated with shorter overall survival. (A) Expression heatmap depicting classification of human PDAC tumor samples from the TCGA database into 3 clinical subtypes based on validated gene signatures (2). (B) Kaplan–Meier survival curves for high (red) vs. low (blue) CYP24A1 expression in tumors within the E and QM subtypes. (C) Expression heatmap depicting classification of human PDAC tumor samples from the ICGC database into 3 clinical subtypes based on validated gene signatures (2). (D) Kaplan–Meier survival curves for high (red) vs. low (blue) CYP24A1 expression in tumors within the E and QM subtypes from the ICGC dataset. (E) Representative images of human PDAC tumors with dual-color RNA-ISH stained for CYP24A1 (red) and FN1 (blue) showing expression of CYP24A1 in tumor cells (black arrowheads, Bottom Left). Higher power image showing coexpression of CYP24A1 and FN1 in a subset of tumor cells (black arrows, Bottom Right). (F) Kaplan–Meier curves for high (red) vs. low (blue) CYP24A1 expression in adenocarcinoma (Left) and squamous cell (Right) NSCLC tumors. (G) Model depicting the heterogeneous and plastic molecular phenotype of PDAC tumors in terms of E vs. QM subtypes and the effects of FOLFIRINOX (FFX) chemotherapy and Vit D therapy on the position of a tumor on the continuum.
This analysis was applied to other TCGA datasets to determine if the differential effect of VDR activation by tumor subtype exists within other cancers. There was no difference in survival associated with the level of CYP24A1 expression in any subtypes of breast cancer (luminal A, luminal B, basal and triple negative), esophageal cancer (epithelial and squamous cell), and head and neck squamous cell carcinoma (SI Appendix, Fig. S4 D–F). However, similar to the findings seen among PDAC tumors, nonsmall cell lung cancer (NSCLC) tumors of the squamous subtype demonstrated a marked survival disadvantage with high CYP24A1 expression while there was no difference in overall survival with CYP24A1 expression levels in adenocarcinomas (Fig. 4F). These unanticipated results support the concept of cellular context-dependent effects of VDR activation in tumor cells, which may have important clinical consequences.
Discussion
Overall, these studies provide definitive evidence that E and QM states of PDAC cells are fluid and can be modulated by therapies that are currently standard of care or under clinical investigation in PDAC patients (Fig. 4G). Genomic DNA-based clinical assays have provided opportunities for targeted therapies in a minority of PDAC patients, but we have shown the importance of RNA-based molecular subtyping in clinical practice. Recent work with single-marker GATA6 (6) or an EMT dual-color RNA-ISH (16) established the ability to identify E and QM PDAC subtypes with a histological stain. Although an RNA-seq–based approach can give an overall E/QM metascore for a tumor sample, an RNA-ISH–based approach affords the ability to detect E and QM marker expression in situ, allowing quantification of the proportion of each phenotype, and takes into account the heterogeneity in a particular sample. In addition, RNA-ISH is already used as a clinical biomarker platform and can be automated on the same machines that perform immunohistochemistry in core pathology laboratories. The assay is performed on standard archived formalin-fixed paraffin-embedded tissues and is completed within 24 h of assay initiation, which is critical for a diagnostic to be deployed in the clinical realm. Therefore, we have developed a dual-color RNA-ISH assay comprising a mixture of E markers and QM markers which was initially tested on PDAC cell lines (Fig. 1E) and validated against a cohort of resected PDAC tumors from untreated and neoadjuvant FOLFIRINOX-treated patients from our institution (Fig. 1I). These findings, combined with previous work by others demonstrating the inherent molecular and functional differences in E and QM PDAC subtypes, may account for differences in response to combination chemotherapy as well as explain the many failed investigational drug clinical trials, due to the lack of evaluating response in the context of PDAC subtypes.
Here, we have shown that the combination chemotherapy FOLFIRINOX results in a consistent shift toward the QM state for all PDAC cell lines and a general intrinsic chemoresistance profile seen in QM compared to E cell lines. This is consistent with initial reports demonstrating improved progression-free survival in patients with E classical subtype tumors treated with FOLFIRINOX (6). Our findings build upon this work and suggest that the shift of E and QM PDAC cells toward a QM state following FOLFIRINOX is likely transcriptionally mediated and linked to response rather than simple selection of discrete QM and E populations, given that both QM and E cell lines increase QM transcriptional profiles. However, a component of selection for preexisting QM cells within a cell line or tumor cannot be excluded.
In addition, we have identified a robust transcriptional response to VDR activation in patient-derived PDAC cell lines and discovered that the cancer cell-specific transcriptional program initiated by VDR activation is discordant between the 2 major subtypes of PDAC. In contrast to the universal shift toward QM state with chemotherapy, CalT exposure resulted in transcriptional changes that augmented the E phenotype of cell lines with a baseline E subtype, but did not appreciably shift QM lines to a more E state. Although the E/QM phenotype was not dramatically altered, we noted strong up-regulation of FN1, which is well known to be associated with the QM phenotype, and increased invasion and migration by CalT specifically in the QM cell lines. Functionally, this translated to increased anchorage-independent cell growth, migration, and development of lung metastasis in animal models when QM cells were treated with CalT, but decreased lung metastasis in the same animal models with exposure of E cells to CalT. Altogether, this raises the possibility that, while Vit D analogs could have favorable effects in E tumors, the same agents may not be effective or have unfavorable consequences in patients with QM tumors, although this requires further investigation. The poor survival outcomes of patients with QM tumors demonstrating high tumor CYP24A1 expression, which is not seen in E tumors, support this possibility and suggest that Vit D supplementation could be ineffective or may even result in disease progression and metastasis in these patients. Although CYP24A1 expression in human tumors may not be solely dependent on VDR activation, the requirement of VDR for CalT-induced metastasis in xenograft models argues that activation of VDR may indeed account for the poor prognosis in CYP24A1-high tumors. Our findings also highlight the need for further investigation into the mechanisms of, and the role of, the tumor microenvironment in PDAC subtypes and VDR activation given previous work showing a beneficial effect of Vit D analogs on stromal remodeling (7). The heterogeneous abundance of stromal CAFs and their distinct interactions with PDAC tumor cells (11) likely influence the effects of Vit D in human tumors, thus meriting further study with better coculture models that have been recently described (27). Taken together, these data provide an additional example of the need to stratify patients based on PDAC subtypes and stromal CAF composition to best evaluate the effects of drug therapy, in this case Vit D supplementation, in PDAC patients. In addition, our discovery of the ability of chemotherapy and CalT to shift the position of a PDAC tumor on the E to QM spectrum argues for the need to evaluate subtype changes in response to specific lines of therapy before considering next steps in treatment.
This collective work has demonstrated the importance of PDAC cell plasticity in response to drugs being used in the clinic and mirrors similar findings in lung, prostate, and breast cancers. The etiology of plasticity of QM and E cells is likely multifactorial, including changes in GATA6 transcription factor activity (28) and alterations in response to TGF-β (11, 28, 29). Further investigation into the mechanism of E and QM fluidity will be necessary to understand these changes in the context of therapy and reveal new therapeutic targets that can specifically modulate this plasticity. We have demonstrated the ability to spatially assess the proportion of E and QM cells in human tumors with an RNA-ISH assay, which is automated and can be deployed in clinical practice, representing a major advance in molecular subtyping of PDAC. Looking forward, a blood-based assay to determine the transcriptional profile of PDAC tumors at different time points in disease evolution and therapy is needed. Although this is unlikely to be achieved by circulating tumor DNA methods, it can potentially be assessed using other “liquid biopsy” technologies including circulating tumor cells or extracellular vesicles. Overall, this work supports the critical need to develop clinical grade assays for distinguishing E and QM PDAC that can finally bring RNA-based molecular subtyping to the field and change the landscape for current and future trials.
Experimental Methods
Cell Lines and Drug Treatments.
PDAC2, PDAC3, PDAC5, PDAC6, PDAC8, and PDAC9 were generated from metastatic ascites fluid of pancreatic adenocarcinoma patients at the Massachusetts General Hospital (MGH) under a discarded tissue protocol in accordance with the MGH IRB protocol 2011P001236 as previously described (14). CAF1 was derived from PDAC tumor tissue collected in accordance with the MGH IRB Protocol 2003P001289 as previously described (14). All samples were de-identified prior to use in the current study. Cell lines MGH927–1611 were generous gifts from the laboratory of A. Liss, Massachusetts General Hospital, Boston. Capan2 (HTB-80), HPAFII (CRL-1997), MiaPaCa2 (CRL-1420), and Panc1 (CRL-1469) were purchased from the American Type Culture Collection. For all experiments, cell lines were grown as tumorspheres under nonadherent conditions in 3D media [serum-free RPMI supplemented with 20 μL/mL B27 (Invitrogen/Life Technologies), 20 ng/mL epidermal growth factor (Invitrogen/Life Technologies), 20 ng/mL basic fibroblast growth factor (Invitrogen/Life Technologies), and 1% Pen/Strep (Gibco/Life Technologies)]. For shRNA experiments, PDAC9 cells were infected with pLKO-based lentiviral particles expressing either scrambled shRNA or shRNA targeting human FN1 (5′-GGCCTCCAGTTCGTGTGAATGATCTCGAGATCATTCACACGAACTGGAGGTTTTT-3′). Cells were selected 48 h after infection with puromycin (1 μg/mL) for 7 d. Knockdown was confirmed by performing qRT-PCR for VDR RNA expression on RNA isolated from nontarget control and VDR shRNA-transduced cells. Components of FOLFIRINOX including 5-fluorouracil (14416), oxaliplatin (13106), and irinotecan (14180) were purchased from Cayman Chemicals. Drugs were combined in a ratio similar to that given to patients, with 1× defined as 34.4 μm 5-fluououracil, 0.4 μm irinotecan, and 0.32 μm oxaliplatin. CalT (1α,25-dihydroxyvitaminD3) was purchased through Sigma-Aldrich (17936).
RNA-seq.
Cells (2 × 105 per well) were transferred to 6-well ultra-low attachment culture dishes in 3D media to allow tumorsphere formation. Calcitriol at 10 nM or DMSO vehicle control was added on day 0 and redosed 48 or 72 h later. Tumorspheres were collected on day 5. RNA was extracted using the miRNEasy Mini Kit (Qiagen) including the optional on-column DNase treatment (Qiagen). RNA quality was analyzed using the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). To generate libraries for total RNA-seq, the Illumina Smarter Stranded Total RNA-seq kit v2 (catalog #634413) was used according to the manufacturer’s instructions. Pooled libraries were sequenced on an Illumina Next. 500 sequencer.
Raw Illumina reads were quality-filtered as follows. First, ends of the reads were trimmed to remove N’s and bases with quality less than 20. After that, the quality scores of the remaining bases were sorted, and the quality at the 20th percentile was computed. If the quality at the 20th percentile was less than 15, the whole read was discarded. Also, reads shorter than 40 bases after trimming were discarded. If at least one of the reads in the pair failed the quality check and had to be discarded, we discarded the mate as well.
Quality filtered reads were mapped to the human genome (gencode annotation, build 38) and to repbase elements (release 20) using STAR aligner (24). Aligned reads were assigned to genes using the featureCounts function of the Rsubread (25) package using the external Ensembl annotation. This produced the raw read counts for each gene.
Mapping and counting of the reads was done in 2 stages. First, reads were mapped to the human genome, and the counts were determined using the Gencode annotation and the annotation derived from the repeatmasker output. After that, the reads which were not assigned to any feature in either Gencode and repeatmasker annotation were realigned to the repeat consensus sequence (repbase). Counts obtained from repeatmasker and repbase were added together.
The downstream analysis was carried out in R statistical programming language (26). In order to obtain an insight into the data, we selected the 100, 500, 1,000, and 2,000 most variant genes and performed a hierarchical clustering for all samples based on the expression of these genes until the clustering did not change anymore. The PDAC cell lines clustered according to their subtype with and without CalT treatment. Replicates for each cell line were averaged across each treatment. Following the clustering, the data were divided into replicates for each subtype to be compared for differential expression in response to CalT. We used the DE2 (27) package in R for the differential expression analysis. The heatmaps were plotted using the heatmap.2 function in the gplots (28) package in R.
qRT-PCR.
RNA was isolated using the miRNEasy Mini Kit (Qiagen) according to the manufacturer’s instructions including the optional on-column DNase treatment (Qiagen). qRT-PCR was conducted on the Dnase-treated RNA using the KAPA SYBR-Fast Onestep qRT-PCR kit according to the manufacturer’s instructions. Primers used include the following: GAPDH—Fw 5′ GGAGCGAGATCCCTC CAA AAT 3′, Rv 5′ GGCTGTTGTCATACTTCTCAT GG 3′; CYP24A1—Fw 5′ CATCATGGCCATCAA AACAAT 3′, Rv 5′ GCAGCTCGACTGGAGTGAC 3′; FN1—Fw 5′ GCAGCCTGCATCTGAGTACA 3′, Rv 5′ GGTGGAATAGAGCTCCCAG 3′; and VDR—Fw 5′ ACTTGCATGAGGAGGAGCAT 3′, Rv 5′ TCGGCTAGCTTCTGGATCAT 3′. Reactions were run on a Thermofisher (Quant Studio 3) thermocycler. All qPCR assays were conducted in triplicate.
Protein Analysis.
CalT- or vehicle-treated PDAC cells were washed twice in phosphate-buffered saline (PBS) and then resuspended in lysis buffer composed of radioimmunoprecipitation assay buffer (Thermo Scientific Product #89900) with 1% protease inhibitor (Thermo Scientific Product #1862209) and 1% phosphatase inhibitor (Thermo Scientific Product #1862495). Lysates were generated by incubating cells in lysis buffer on ice for 40 min with intermittent vortexing and then centrifuging at 16,100 × g for 20 min. Protein concentrations were determined using Bradford reagent (Bio-Rad). Proteins were resolved on Bio-Rad 15-well 4 to 15% gradient gel with the inclusion of Bio-Rad precision plus protein dual-color standards and transferred to nitrocellulose membrane. Membranes were blocked and probed with the following primary antibodies: CYP24A1 (Abcam, Reference #ab96691), FN1 (Abcam, Reference #ab2413), VDR (Cell Signaling Technology Reference #4499S), and loading control Histone H3 (Cell Signaling Technology Reference #12550S). CYP24A1, FN1, and VDR were applied at 1:1,000, and Histone H3 was applied at 1:2,000. Anti-rabbit IgG was used at 1:2,000 as a secondary antibody. Chemiluminescence was detected with Syngene Chemiluminescence and Fluorescence systems using the software Gene tools from Syngene.
GSEA.
GSEA was performed to investigate the dataset overlap with annotated gene sets in the Molecular Signature Database (MSigDB). All genes differentially expressed in QM PDAC cell lines after exposure to CalT were used to identify matching gene sets from MSigDB. Enrichment was based on the number of overlapping genes, the P value determining the probability of association with a given gene set, and a false discovery rate q-value.
Soft Agar Assays.
To evaluate anchorage-independent cell growth following treatment of tumorspheres with CalT in serum-free media, tumorspheres were first dissociated into single cells. Spheres were collected and washed once with PBS and then washed with a solution of 2 mM ethylenediaminetetraacetic acid + 0.5% bovine serum albumin. Tumorspheres were then resuspended in 0.05% warmed trypsin and incubated at room temperature for 10 min. Tumorspheres were then mechanically dissociated by passing through 21-gauge and then 25-gauge needles connected to 3-mL syringes. Dissociation of spheres into single cells was confirmed by light microscopy before proceeding. Following dissociation, 1.5 × 104 cells were added to 2× Dulbecco’s Modified Eagle Media (DMEM) + 10% fetal bovine serum (FBS) containing 0.3% agarose and plated on top of a base containing DMEM and 0.5% agarose in 12-well dishes. After the cell layer was set, DMEM + 10% FBS was added to cover each well and was replenished weekly. Colony area was quantified using ImageJ after ∼5 wk of incubation.
Migration Assays.
Tumorspheres were treated with CalT and dissociated to single-cell suspensions as described above. Cells were seeded onto 8-μm pore Transwell membranes (Corning) in serum-free growth media at 80,000 cells/well in a 24-well plate containing complete growth media (10% FBS) in the bottom chamber. After 16 h at 37 °C, cells on top of the chamber were stripped off with cotton swabs and cells on the bottom of the chamber were fixed in 4% paraformaldehyde and stained with Crystal Violet. Low-power images were acquired and the total crystal violet-stained area was quantitated using ImageJ and averaged across 3 images per well.
Tail-Vein Injection and IVIS Imaging.
The animal protocol was approved by the MGH Subcommittee on Research Animal Care (protocol 2014N000321). Eight- to 12-wk-old female NSG mice were purchased through Jackson Labs. PDAC cells (1 × 105) expressing luciferase were injected in 100 μl of sterile PBS into the lateral tail veins of the mice. Tumor formation in the lungs was monitored weekly by bioluminescence using IVIS Lumina II (Caliper Life Science). Mice were euthanized after 4 to 8 wk and lungs were imaged ex vivo.
Cancer Subtype Classification.
Processed RNA-seq expression data for pancreatic tumors were collected from TCGA (PAAD) (28, 29) and ICGC (PAAD-US, PACA-AU, PACA-CA) (30). Using gene sets published by Collison et al. (2), we classified each of the pancreatic samples as epithelial or quasi-mesenchymal. Briefly, we hierarchically clustered the samples based on the correlation between samples across the Collison tumor subtype gene sets on per-gene centered and scaled expression data [using the R (32) scale function]. Clustered samples associated with increased expression of a specific subtype gene set were classified as the appropriate tumor subtype.
Survival Analysis.
The association of CYP24A1 expression with respect to survival data was analyzed for several tumor types. Clinical and RNA-seq expression data were collected for the following tumor types, pancreas (PAAD) (28), breast (BRCA) (33), lung [LUSC (34), LUAD (35)], esophageal (ESCA) (28), and head-neck (HNSC) (36) from TCGA. Data for pancreas were collected from ICGC (PAAD-US, PACA-AU, PACA-CA). Samples were stratified between high and low CYP24A1 expression using a threshold defined by the Ostu method of minimizing the within-class variance. Kaplan–Meier curves were generated using a 5-y survival cutoff for pancreas and esophageal tumor datasets. A 15-y survival cutoff was selected for breast, head-neck, and lung tumor datasets. All analyses and plots were generated using R (32).
RNA-ISH.
For the detection of CYP24A1 and FN1 RNA levels, RNA-ISH was performed using RNAscope 2.5 Duplex technology using the manual platform (catalog no. 322430). Formalin-fixed paraffin-embedded tissue sections of cell pellets, human tissue microarrays (TMAs), and human tumor biopsies were cut in 5-µm sections on slides and were then deparaffinized and pretreated to allow unmasking and RNA probe accessibility. This was done by first baking the slides at 60 °C for 1 h followed by treatment with Histoclear and 100% ethanol. The samples were then exposed to hydrogen peroxide for 10 min at room temperature followed by a target retrieval solution for 15 min at 99 °C, and finally to protease enzyme for 30 min at 40 °C to allow probe accessibility.
The hybridization step was performed at 40 °C for about 2 to 3 h with CYP24A1 and FN1 probes to allow the target-specific probes to hybridize to target messenger RNA (mRNA). This was followed by a series of signal amplification steps: Amp 1 was added to bind to the target-specific probe, followed by Amp 2 to Amp 10, which were subsequently added to bind each other one after the other. Next, type-specific label probes conjugated to alkaline phosphates were added to bind to the Amps, thus completing the branched DNA tree and providing signal amplification due to this cascade of hybridizations.
The signal was visualized by sequential addition of Red substrate which binds to Amp 6 and Green substrate which binds to Amp 10, producing Red and Green precipitates (dots, Fig. 4E). The target mRNAs were then visualized using standard brightfield microscopy with CYP24A1 signal as Red and FN1 signal as Green.
For detection of epithelial and quasi-mesenchymal marker mRNA expression, RNA-ISH was performed on cell pellets and human TMAs and tumor biopsy samples using RNAscope 2.5 Duplex technology with the automation platform (catalog no. 322440) on the Bond RX automated immunohistochemistry and ISH staining system using BDZ 6.0 software (Leica Biosystems). The panel for E markers includes CDH1, EPCAM, KRT5, KRT7, KRT8, and KRT19 and the panel for QM markers includes CDH2, SERPINE1, and FN1. The antigen retrieval, probe hybridization, and signal amplification methods were similar to the manual staining described above. Positive ISH staining was defined as brown dots for E staining and red dots for QM staining.
Slides were imaged using a Leica Aperio CS-O slide scanning microscope at 40× magnification. To determine the amount of E versus QM expression, images were quantified using Halo software by Indica Labs. The color components for cell nuclei (blue, hematoxylin), RNA-ISH epithelial cells (brown, CDH1/EPCAM/KRT5/KRT7/KRT8/KRT19), and RNA-ISH quasi-mesenchymal cells (red, CDH2/SERPINE1/FN1) were extracted using color deconvolution. The hematoxylin, E, and QM areas (in units of micron squared) were quantified within representative regions that were annotated by a trained pathologist. The fractional E area was defined as the total E area divided by the total cellular area. The total cell area was defined as the sum of the hematoxylin, E, and QM areas. The fractional QM area was calculated in the same manner.
ATAC-seq.
ATAC-seq was performed based on the protocol published previously (30). Briefly, cells were trypsinized and washed with PBS, and 50,000 cells were pelleted and lysed in ATAC resuspension buffer containing 0.1% Nonidet P-40, 0.1% Tween-20, and 0.01% digitonin on ice for 10 min. Nuclei were then resuspended in a 50-μL transposition reaction mixture containing 25 μL 2× Tagment DNA buffer (Illumina Nextera DNA Library Prep Kit), 2.5 μL transposase (Illumina Nextera DNA Library Prep Kit), 16.5 μL PBS, 0.5 μL 1% digitonin, 0.5 μL 10% Tween-20, 5 μL H2O at 37 °C for 30 min in a thermomixer with 1,000 × g mixing. Transposed fragments were further amplified and purified before pooling together and sequencing on the Hi X platform.
Paired-end reads were mapped using BWA (31) to the human genome (hg19). Quality control, mitochondrial reads removal, and duplicate reads removal were performed using the R package ATAC-seqQC (32). To account for the expected shift from insertion of Tn5 transposase, alignments (bam files) were shifted using deepTools-alignmentsieve with parameter–ATACshift (33). ATAC-seq peak regions of each individual sample were called using MACS2 with parameters –nomodel (34). Blacklisted regions were excluded from called peaks (https://www.encodeproject.org/files/ENCFF001TDO/). Annotation of peaks to obtain exclusively peaks located in promoter regions (<3 kb from transcriptional start site) was performed using the R package ChIPseeker (35). Peaks called for each sample were used to draw correlation heatmaps of relative openness of chromatin using the R package DiffBind (36).
Data Availability Statement.
All sequencing data have been uploaded to the National Center for Biotechnology (NCBI) Gene Expression Omnibus and are accessible through GEO Series accession number GSE141116 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141116). Any data that is not found in the NCBI will be made available upon request from the corresponding author.
Quantification and Statistical Analyses.
For all experiments, P values were calculated using PRISM7 graphpad paired t test or 2-way ANOVA, unless otherwise noted (*<0.05, **<0.01, ***<0.001, ****<0.0001).
Supplementary Material
Acknowledgments
We thank Emily Silva and Danielle Bestoso for administrative support and Drs. Joseph Franses, Julia Philipp, and Mihir Rajurkar for helpful scientific discussions. This work was supported by the Warshaw Institute for Pancreatic Cancer Research (R.L.P.); NSF Grant PHY-1549535 (D.T.T., P.J.O., J.A.D.); Stand Up To Cancer (SU2C) and Lustgarten Foundation (D.T.T., P.J.O., J.A.D., T.S.H., D.P.R.); NIH grant 2R01CA129933 (D.A.H.), Howard Hughes Medical Institute (D.A.H.); National Foundation for Cancer Research (D.A.H.); and Advanced Cell Diagnostics/Biotechne (D.T.T., A.N., A.C., A.S.K., V.D.).
Footnotes
Competing interest statement: D.T.T. has received consulting fees from EMD Millipore-Sigma, Ventana-Roche, Foundation Medicine, Inc., and Merrimack Pharmaceuticals. D.T.T. is a founder and has equity in PanTher Therapeutics, ROME Therapeutics, and TellBio Inc., which are not related to this work. D.A.H. is founder and has equity in TellBio Inc., which is not related to this work. D.T.T.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. D.T.T. and V.D. receive sponsored research support from ACD-Biotechne that was performed for some parts of this work. V.D. receives sponsored research support from Agios. M.S.L. and E.A.C. are coauthors on a 2016 paper.
Data deposition: The data reported in this paper have been deposited in the National Center for Biotechnology Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE141116).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914915116/-/DCSupplemental.
References
- 1.Bailey P., et al. ; Australian Pancreatic Cancer Genome Initiative , Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Collisson E. A., et al. , Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moffitt R. A., et al. , Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 47, 1168–1178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiriac H., et al. , Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 8, 1112–1129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinelli P., et al. , GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 66, 1665–1676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aung K. L., et al. , Genomics-driven precision medicine for advanced pancreatic cancer: Early results from the COMPASS trial. Clin. Cancer Res. 24, 1344–1354 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman M. H., et al. , Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 159, 80–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., et al. , ATDC induces an invasive switch in KRAS-induced pancreatic tumorigenesis. Genes Dev. 29, 171–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aiello N. M., et al. , EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell 45, 681–695.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reichert M., et al. , Regulation of epithelial plasticity determines metastatic organotropism in pancreatic cancer. Dev. Cell 45, 696–711.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ligorio M., et al. , Stromal microenvironment shapes the intratumoral architecture of pancreatic cancer. Cell 178, 160–175.e27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting D. T., et al. , Single-cell RNA sequencing identifies extracellular matrix gene expression by pancreatic circulating tumor cells. Cell Rep. 8, 1905–1918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung H. C., et al. , Efficacy and safety of pembrolizumab in previously treated advanced cervical cancer: Results from the phase II KEYNOTE-158 study. J. Clin. Oncol. 37, 1470–1478 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Indolfi L., et al. , A tunable delivery platform to provide local chemotherapy for pancreatic ductal adenocarcinoma. Biomaterials 93, 71–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu M., et al. , Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahadevan K. K., et al. , Quasimesenchymal phenotype predicts systemic metastasis in pancreatic ductal adenocarcinoma. Mod. Pathol. 32, 844–854 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collisson E. A., Bailey P., Chang D. K., Biankin A. V., Molecular subtypes of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 16, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Haussler M. R., et al. , The nuclear vitamin D receptor: Biological and molecular regulatory properties revealed. J. Bone Miner. Res. 13, 325–349 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Lamouille S., Xu J., Derynck R., Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bae Y. K., et al. , Fibronectin expression in carcinoma cells correlates with tumor aggressiveness and poor clinical outcome in patients with invasive breast cancer. Hum. Pathol. 44, 2028–2037 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Wang J. P., Hielscher A., Fibronectin: How its aberrant expression in tumors may improve therapeutic targeting. J. Cancer 8, 674–682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu M., et al. , RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature 487, 510–513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olmez I., et al. , Targeting the mesenchymal subtype in glioblastoma and other cancers via inhibition of diacylglycerol kinase alpha. Neuro-oncol. 20, 192–202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips H. S., et al. , Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Nakamura T., Fidler I. J., Coombes K. R., Gene expression profile of metastatic human pancreatic cancer cells depends on the organ microenvironment. Cancer Res. 67, 139–148 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Nones K., et al. ; APGI , Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int. J. Cancer 135, 1110–1118 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Biffi G., et al. , IL-1-induced JAK/STAT signaling is antagonized by TGF-beta to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 9, 282–301 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellenrieder V., et al. , Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 61, 4222–4228 (2001). [PubMed] [Google Scholar]
- 29.Huang P. H., et al. , TGFβ promotes mesenchymal phenotype of pancreatic cancer cells, in part, through epigenetic activation of VAV1. Oncogene 36, 2202–2214 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Corces M. R., et al. , An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou J., et al. , ATACseqQC: A bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics 19, 169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramírez F., Dündar F., Diehl S., Grüning B. A., Manke T., deepTools: A flexible platform for exploring deep-uencing data. Nucleic Acids Res. 42, W187–W191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., et al. , Model-based analysis of ChIP- (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Stark R., Brown G., DiffBind: Differential binding analysis of ChIP-Seq peak data. http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf. Accessed 15 August 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data have been uploaded to the National Center for Biotechnology (NCBI) Gene Expression Omnibus and are accessible through GEO Series accession number GSE141116 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141116). Any data that is not found in the NCBI will be made available upon request from the corresponding author.