Significance
The origin of transmissible BSE in cattle remains unestablished. Sheep scrapie is a potential source of this known zoonotic. Here we investigated the capacity of sheep scrapie to propagate in bovine PrP transgenic mice. Unexpectedly, transmission of atypical but not classical scrapie in bovine PrP mice resulted in propagation of classical BSE prions. Detection of prion seeding activity by in vitro protein misfolding cyclic amplification demonstrated BSE prions in the original atypical scrapie isolates. BSE prion seeding activity was also detected in ovine PrP mice inoculated with limiting dilutions of atypical scrapie. Our data demonstrate that classical BSE prions can emerge during intra- and interspecies passage of atypical scrapie and provide an unprecedented insight into the evolution of mammalian prions.
Keywords: prion, atypical scrapie, c-BSE
Abstract
Atypical/Nor98 scrapie (AS) is a prion disease of small ruminants. Currently there are no efficient measures to control this form of prion disease, and, importantly, the zoonotic potential and the risk that AS might represent for other farmed animal species remains largely unknown. In this study, we investigated the capacity of AS to propagate in bovine PrP transgenic mice. Unexpectedly, the transmission of AS isolates originating from 5 different European countries to bovine PrP mice resulted in the propagation of the classical BSE (c-BSE) agent. Detection of prion seeding activity in vitro by protein misfolding cyclic amplification (PMCA) demonstrated that low levels of the c-BSE agent were present in the original AS isolates. C-BSE prion seeding activity was also detected in brain tissue of ovine PrP mice inoculated with limiting dilutions (endpoint titration) of ovine AS isolates. These results are consistent with the emergence and replication of c-BSE prions during the in vivo propagation of AS isolates in the natural host. These data also indicate that c-BSE prions, a known zonotic agent in humans, can emerge as a dominant prion strain during passage of AS between different species. These findings provide an unprecedented insight into the evolution of mammalian prion strain properties triggered by intra- and interspecies passage. From a public health perspective, the presence of c-BSE in AS isolates suggest that cattle exposure to small ruminant tissues and products could lead to new occurrences of c-BSE.
Transmissible spongiform encephalopathies (TSEs), or prion diseases, are fatal neurodegenerative disorders that affect a large spectrum of mammalian species. These conditions include scrapie in small ruminants, classical bovine spongiform encephalopathy (c-BSE) in cattle, and sporadic Creutzfeldt–Jakob disease (sCJD) or variant CJD (vCJD) in humans.
The fundamental event in prion propagation is the conversion of the normal cellular prion protein (PrPC) into an abnormal disease-associated isoform (PrPSc) in tissues of infected individuals. PrPC is completely degraded by digestion with proteinase K (PK), whereas PrPSc is N-terminally truncated, resulting in a PK-resistant core termed PrPres (1). According to the prion concept, PrPSc is the principal, if not sole, component of the transmissible prion agent (2), and PrPres is a disease marker for prion diseases (1, 3). Particular biochemical properties of PrPSc, such as detergent solubility, PK resistance, and electromobility evidenced by Western blot can be used to distinguish between different prion agents or strains (4, 5).
Intraspecies transmission of prion disease between individuals is typically quite efficient. In contrast, interspecies transmission of prions can be unpredictable, with apparent failure of disease transmission on many occasions. In other cases, clinical prion disease may not be evident but, rather, there is the presence of subclinical infection (6). When interspecies prion transmission does occur, the propagating agent can remain identical to the original prion strain or can display different biological properties compared to the original inoculum (7, 8). This complex set of outcomes for interspecies prion challenge are collectively referred to as the transmission barrier phenomenon.
After identification of the gene encoding PrP, it was soon discovered that differences in amino acid sequence between host PrPC and donor PrPSc constitutes the principal determinant of the transmission barrier. For example, the resistance of wild-type mice to clinical prion disease induced by hamster scrapie is abrogated by transgenic expression of hamster PrPC in mice (9, 10). As a consequence, mice genetically engineered to express particular species forms of PrP sequence, in the absence of endogenous mouse PrP, have emerged as relevant models to experimentally characterize the outcome of prion strain transmission between species (11). It is also now well established that strain properties have a significant impact on the ability of prions to cross the species barrier. For instance, human vCJD can be transmitted readily to conventional mice, but it is extremely difficult for sCJD to propagate in the same mouse lines (12, 13). Furthermore, the amino acid sequence of PrPSc influences the efficacy of interspecies prion transmissions, since studies in human PrP transgenic mouse models indicate that the human species barrier is more permeable to sheep-passaged BSE compared to its cattle counterpart (14).
From a public health perspective, the transmission barrier phenomenon and its capacity to limit the interspecies propagation of prion disease has long been considered as an effective protection of humans against animal TSEs (15). However, in 1996, the new human prion disease vCJD was observed in people in the United Kingdom. Multiple lines of evidence indicated that vCJD was the likely consequence of dietary exposure of humans to the agent responsible for c-BSE in cattle, an epizootic prion disease that has spread in bovine hosts through the recycling of prion-contaminated animal carcasses in the animal food chain (16). Since the emergence of vCJD, considerable efforts have been deployed to characterize not only the zoonotic potential of animal prions but also their capacity to propagate in farmed animal species.
Atypical/Nor98 scrapie (AS) probably represents the largest geographically spread known animal prion disease. Since its original discovery in 1998 in Norway, AS has been identified in most EU member states, in Asia, and in North and South America (17). AS has also been detected in Australia and New Zealand, 2 countries that were believed to be free of animal TSEs (18, 19). Retrospective studies carried out in archived animal tissues identified an AS case in a sheep that died in 1972 in the United Kingdom, demonstrating that the disease has been present in small ruminant populations for many decades (20).
Bioassay in ovine PrP transgenic mice provided evidence that AS comprised a single prion strain (21–23). The AS prion strain was associated with a multiband PrPres signature that contrasted with those normally observed in small ruminant TSE cases (24). Since there is no statistical difference in the apparent prevalence of atypical scrapie between sheep flocks in general and those flocks where a positive case had been identified, atypical scrapie is considered by many as a noncontagious prion disease that arises sporadically in sheep and goats (25). However, atypical scrapie can be experimentally transmitted via the oral route in small ruminants, resulting in a similar clinicopathological phenotype to that observed in natural cases (26). Consequently, the origin of atypical scrapie (spontaneous disorder versus acquired disease) remains an open question.
In this study, we used mice transgenic for bovine PrP (tgBov mouse line) to characterize the capacity of sheep AS isolates to cross the bovine transmission barrier. Unexpectedly, the TSE agent that propagated in tgBov mice was indistinguishable from the prion strain that was responsible for the c-BSE epizootic in cattle. In addition, our sensitive detection of c-BSE by in vitro PMCA methodology indicated that this bovine prion strain was present as a minor prion strain in the original sheep AS isolates, and that AS prion strain replication in an ovine PrP host was accompanied by the generation of c-BSE prions. Collectively, these data provide compelling evidence for the emergence and the propagation of zoonotic mammalian prions during intra- and interhost transmission of the AS prion strain.
Results
A panel of 8 atypical scrapie (AS) cases collected from sheep and goats in 5 different European countries was obtained (Table 1). All of the AS isolates displayed a multiband abnormal PrP (PrPres) Western blot profile that was considered to be specific for small ruminant AS (SI Appendix, Fig. S1). This panel of AS isolates was transmitted to the VRQ ovine PrP transgenic mice (tg338). The transmission properties such as incubation period (SI Appendix, Table S1), vacuolar lesion profile (Fig. 1A), and PrPres Western blot profile in the brain (SI Appendix, Fig. S1) of the propagated AS isolates observed after 2 or 3 iterative passages in tg338 were similar and were the same as previously reported for AS passage in tg338 mice (21–23).
Table 1.
Inoculation of atypical scrapie isolates in bovine PrP (tgBov) expressing mice
| Isolates | TgBov | |||||||
| First passage | Second passage | Third passage | ||||||
| Identifiant | Origin | Genotype | Positive mice | Incubation, mean ± SD | Positive mice | Incubation, mean ± SD | Positive mice | Incubation, mean ± SD |
| AS 1 | Fr | ARQ*/ARQ | 1/6 | 533 | 3/5 | 317 ± 63 | 7/7 | 235 ± 16 |
| AS 2 | Sp | ARR/ARQ | 0/6 | >650 | 7/9 | 354 ± 26 | 5/5 | 273 ± 5 |
| AS 3 | Sp | ARQ/ARH | 0/6 | >650 | 2/11 | 504, 525‡ | 12/12 | 269 ± 13 |
| AS 4 | Nor | ARQ*/ARQ* | 3/4 | 395 ± 44 | 6/6 | 230 ± 17 | 6/6 | 271 ± 18 |
| AS 5 | Sp | ARQ/ARQ | 0/6 | >650 | 0/4 | >650 | NA | — |
| AS 6 | Sp | ARQ/ARH | 0/6 | >650 | 1/4 | >650‡ | NA | — |
| AS 7† | It | ARQ/AHQ | 0/6 | >650 | 1/7 | 424 | 6/6 | 286 ± 14 |
| AS 8 | Po | ARQ/ARQ | 1/5 | 439 | 5/5 | 297 ± 14 | 6/6 | 250 ± 4 |
| PS42 | Fr | VRQ/VRQ | 0/6 | >650 | 0/6 | >650 | 0/6 | >650 |
| Ovine c-BSE | Fr | ARQ/ARQ | 6/6 | 254 ± 19 | 6/6 | 234 ± 12 | 6/6 | 232 ± 6 |
| Cattle c-BSE | Fr | — | 6/6 | 295 ± 12 | 6/6 | 265 ± 35 | 6/6 | 243 ± 7 |
Transgenic mice that express the bovine PrP (tgBov) were inoculated intracerebrally (6 to 12 mice, 20 µL per mouse) with 8 sheep or goat (†) atypical scrapie (AS) isolates originating from 5 different countries, France (Fr), Spain (Sp), Norway (Nor), Italy (It) or Portugal (Po). The AS affected animals displayed a different PRNP genotype at codons 136, 154, and 171. Some also displayed a F/L dimorphism at codon 141 (*). Cattle classical BSE (c-BSE), ovine c-BSE (first passage of cattle c-BSE in an ARQ/ARQ sheep by the intracerebral route), and classical scrapie (PS42) isolates were inoculated in the same mouse model. After first and second passages, clinically affected or asymptomatic mice that had lived for more than 500 d post inoculation were pooled and used for subsequent passage in the same line. Mice were considered positive when abnormal PrP deposition was detected in the brain. Incubation periods (in days) are shown as mean ± SD except when less than 50% of the mice were found to be positive. In that case, the incubation periods of the positive mice are individually presented. NA, not available. Cattle c-BSE transmission in tgBov data from ref. 51. PS42 transmission data from ref. 15.
Abnormal PrP positive and found dead animals (without symptoms).
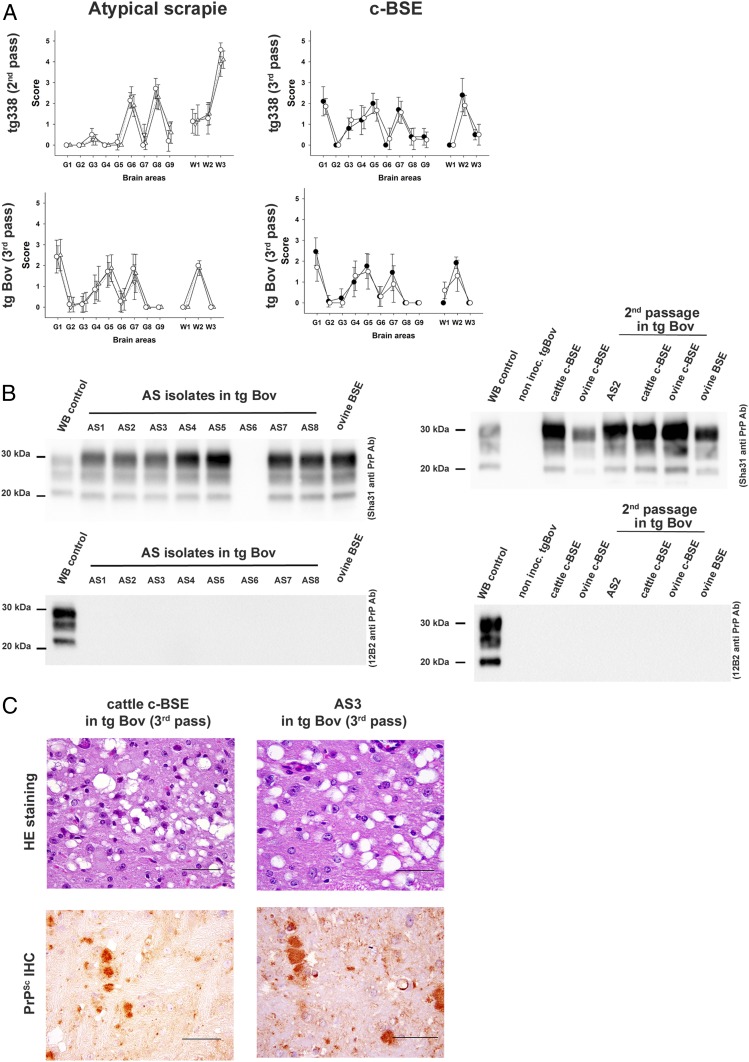
Fig. 1.
Brain lesion profile and PrPres Western blot profiles in tgBov and tg338 mice inoculated with atypical/Nor98 scrapie (AS) or c-BSE. Groups of mice (n ≥ 6) that express either ovine VRQ PrP (tg338 mice) or bovine PrP (tgBov mice) were intracerebrally challenged with atypical scrapie isolates (AS) or an ovine classical BSE isolate (c-BSE). (A) After 2 or 3 iterative passages in each mouse line, a standard lesion profile was established by scoring the vacuolar changes observed in predefined brain areas. In c-BSE graphs, ●, ovine c-BSE; ○, cattle c-BSE. In AS graphs, ○, AS 1; △, AS 2; ▽, AS 3. (B) The accumulation of PK-resistant PrP (PrPres) in the brain of tgBov mice was established by Western blot using anti-PrP monoclonal antibodies Sha31 (epitope 145-YEDRYYRE-152) and/or 12B2 (epitope 89-WGQGG-93). The same Western blot PrPres control (classical scrapie isolate) was used on all of the gels labeled as WB control. Cattle c-BSE and an ovine c-BSE (original isolate and isolate passaged in tgBov) were included as controls. (C) Vacuolar lesions (thalamus level, conventional histology; hematoxylin-eosin) and abnormal PrP deposition (mesencephalon: tegmentum, immunohistochemistry using 6H4 anti-PrP antibody, epitope 147-DYEDRYYRE-155) in tgBov mice inoculated with AS3 and cattle BSE. (Scale bars: vacuolar lesions, 25 µm; abnormal PrP deposition, 50 µm.)
The panel of 8 AS isolates was individually serially transmitted (2 or 3 iterative passages) in bovine PrP transgenic mice (tgBov; Table 1). On first passage, signs of clinical prion disease were observed in a low proportion of inoculated tgBov mice. PrPres was detected by Western blot in the brains of clinically affected mice and in some mice that displayed no apparent clinical signs of prion disease when euthanized at the end of their life expectancy. No PrPres accumulation was observed in tgBov mice after inoculation with several of the AS isolates (AS 2, AS 3, AS 5, AS 6, and AS 7), as was the case with classical scrapie PS42 (Table 1).
Second passage of the AS isolates in tgBov mice was performed using either first-passage PrPres-positive brains or pooled PrPres-negative brains as inoculum. During this process, 7 out of the 8 AS isolates caused the occurrence of clinical prion disease in a proportion of animals in each group inoculated. On third passage (available for AS 1, 2, 3, 4, and 7), 100% attack rates and mean incubation periods that ranged between 235 and 286 dpi were recorded (Table 1).
At each passage stage, a 3-band PrPres Western blot profile characterized by a prominent di-glycosylated PrP band was observed in the brains of the clinically positive tgBov mice (Fig. 1B). Strikingly, the lesion profile (Fig. 1A), the PrPres Western blot profile (Fig. 1B), and the histopathological lesions (Fig. 1C) in the brains of AS inoculated tgBov mice were identical to those observed for transmission of c-BSE (sheep or cattle origin) to tgBov mice. Importantly, the inoculation of one classical scrapie isolate (PS42) to the same mouse models resulted in the occurrence of a 100% attack rate for prion disease with a short incubation period in tg338 mice (SI Appendix, Table S1), but no clinical disease or PrPres accumulation in the brains of tgBov (Table 1).
In order to further characterize the nature of the TSE agent that propagated in tgBov mice inoculated with the AS isolates, prions obtained after second passage (isolate AS 2) or third passage (isolate AS 3) in this mouse line were transmitted (2 iterative passages) to VRQ (tg338) and ARQ (tgARQ) ovine PrP transgenic mice (Table 2). The incubation periods (Table 2 and SI Appendix, Table S1), the lesion profile (Fig. 2A), and the PrPres Western blot profile in the brain (Fig. 2B) of tg338 mice inoculated with tgBov-adapted AS isolates clearly differed from those observed in the same mouse line inoculated with the original AS isolates.
Table 2.
Inoculation of atypical scrapie adapted in tgBov and c-BSE isolates in ovine PrP expressing mouse models (tg338 and tgARQ)
| Tg338 | TgARQ | ||||||||
| Isolates | First passage | Second passage | First passage | Second passage | |||||
| Identifiant | Origin | Positive mice | Incubation, mean ± SD | Positive mice | Incubation, mean ± SD | Positive mice | Incubation, mean ± SD | Positive mice | Incubation, mean± SD |
| AS 2 | Second pass in TgBov | 6/6* | >650 | 6/6 | 617 ± 75 | 6/6 | 350 ± 9 | 6/6 | 260 ± 3 |
| AS 3 | Third pass in TgBov | 6/6* | >650 | 6/6 | 672 ± 83 | 6/6 | 354 ± 21 | 6/6 | 257 ± 2 |
| Ovine c-BSE | Second pass in TgBov | 6/6* | >750 | 6/6 | 653 ± 32 | 6/6 | 270 ± 12 | 6/6 | 259 ± 4 |
| Cattle c-BSE | Cattle | 6/6* | >700 | 6/6 | 682 ± 52 | 6/6 | 321 ± 16 | 6/6 | 263 ± 7 |
Transgenic mice that express the VRQ (tg338) or ARQ (tgARQ) variants of ovine PrP were inoculated intracerebrally (6 mice, 20 µL per mouse) with atypical scrapie isolates or ovine c-BSE isolate that had previously been adapted in tgBov (2 iterative passages). Cattle BSE was also included as control. After first passage, clinically affected or asymptomatic mice that had lived for more than 500 d post inoculation were pooled and used for second passage in the same line. Mice were considered positive when abnormal PrP deposition was detected in the brain. Incubation periods (in days) are shown as mean ± SD.
Abnormal PrP positive and found dead animals (without symptoms).
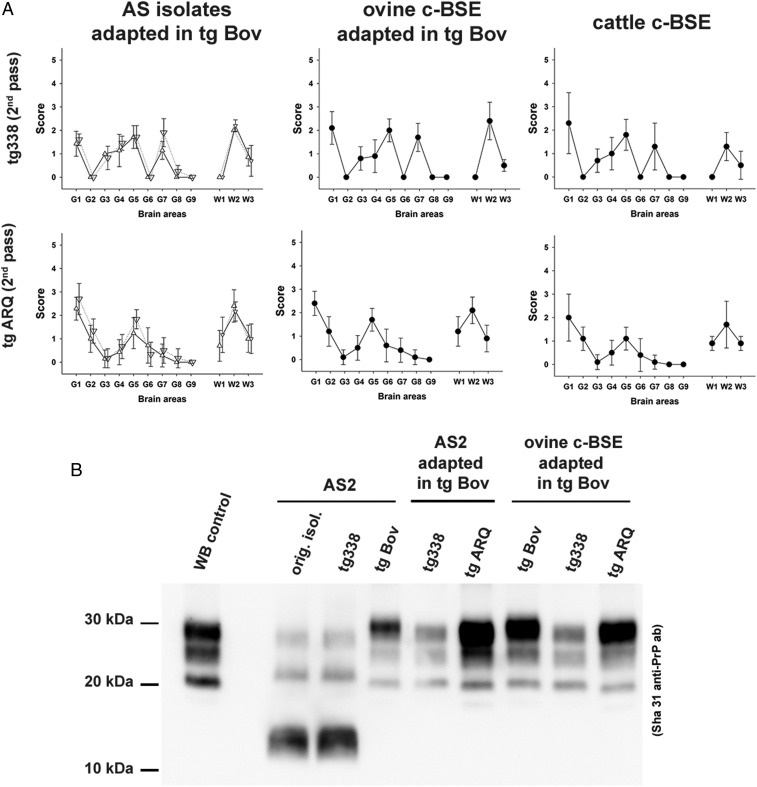
Fig. 2.
Brain lesion profile and PrPres Western blot in the brain of tgARQ and tg338 mice inoculated with AS scrapie adapted in tgBov. Groups of mice (n ≥ 6) that express ovine VRQ PrP (tg338 mice) or ovine ARQ PrP (tgARQ mice) were intracerebrally challenged with atypical scrapie isolates (AS) or an ovine c-BSE isolate that had previously been adapted (2 iterative passages) in tgBov mice. (A) After 2 iterative passages in each mouse line, a standard lesion profile was established by scoring the vacuolar changes observed in predefined brain areas. In AS graphs, △, AS 2; ▽, AS 3. (B) The Western blot profile of PK-resistant PrP (PrPres) in the original AS isolates and in the brain of inoculated mice was established by Western blot using anti PrP monoclonal antibody Sha31 (epitope 145-YEDRYYRE-152). The same Western blot PrPres control (classical scrapie isolate) was used on all of the gels labeled as WB control.
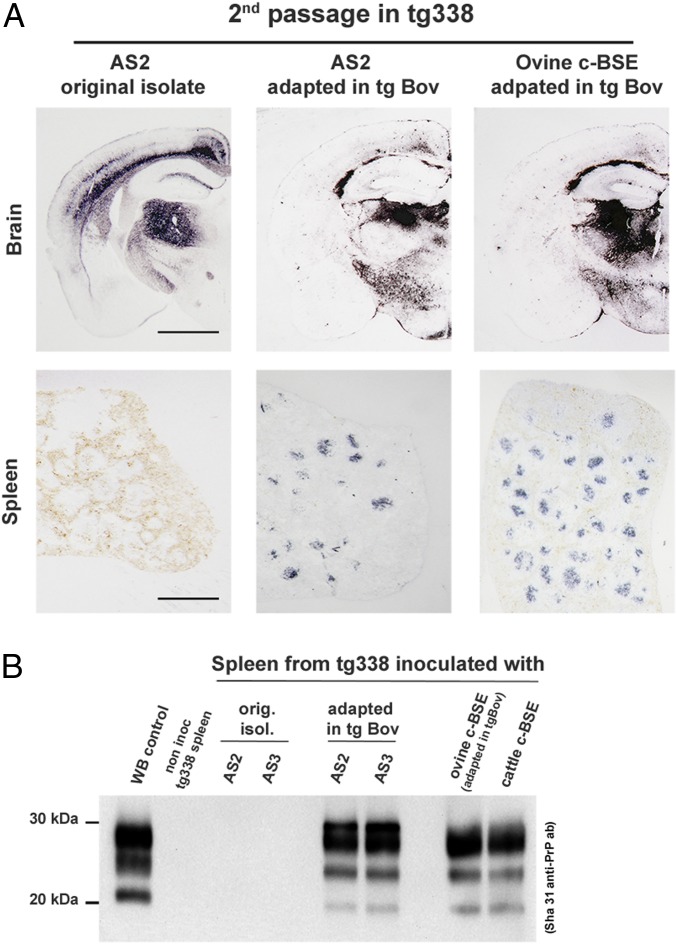
No PrPres deposition could be detected in the spleen of tg338 mice inoculated with the original AS isolates (Fig. 3A). Conversely, transmission in tg338 of tgBov-adapted AS isolates was associated with a PrPres accumulation in the spleen, as was transmission of ovine and cattle c-BSE (Fig. 3A). PrPres WB profile in the spleen of tg338 mice that were inoculated with c-BSE and AS isolates passaged in tgBov were identical (Fig. 3B).
Fig. 3.
Brain and spleen PrPres accumulation in tg338 mice inoculated with AS scrapie adapted in tgBov. Groups of mice (n ≥ 6) that express ovine VRQ PrP (tg338 mice) were intracerebrally challenged with atypical scrapie isolates (AS) and AS that had previously been adapted (2 iterative passages) in tgBov mice. In parallel, cattle c-BSE isolate and ovine BSE isolate (adapted in tgBov) were transmitted (2 iterative passages) in tg338 mice. (A) In tg338 mice (second passage), the PrPres distribution pattern in the brain (thalamic coronal section) and in the spleen was established by paraffin-embedded tissue blot using anti-PrP monoclonal antibody Sha31 (epitope 145-YEDRYYRE-152). (Scale bars: brain, 160 µm; spleen, 100 µm.) (B) The Western blot profile of PK-resistant PrP (PrPres) in the spleen of tg338 mice (second passage) inoculated with AS isolates and AS isolates passaged in tgBov and c-BSE (cattle and ovine origin) was established using anti-PrP monoclonal antibody Sha31.
Transmission of tgBov-adapted AS isolates in both tg338 and tgARQ mice resulted in prion incubation periods (Table 2), brain vacuolar lesion profiles (Fig. 2A), PrPres Western blot profile patterns (Fig. 2B), and PrPres distribution patterns in the brain (Fig. 3A) that were similar to c-BSE passaged in tg338 and tgARQ mice. Collectively, these results demonstrate beyond reasonable doubt that our transmission of AS in a bovine PrP host resulted in the propagation of the c-BSE agent. Since the bioassays reported here were performed in 3 independent institutes (located in France and Spain) that used inoculum prepared by 5 distinct laboratories, we exclude the possibility of a cross-contamination of the original AS isolates by the c-BSE agent.
Two hypotheses could explain the emergence of the c-BSE agent in tgBov mice after their inoculation with AS isolates. First, c-BSE prions could be present at a low level in the original AS isolates. The high sensitivity of tgBov mice for detection of the c-BSE agent could allow this potentially low level of bovine prions to be identified during passage of the original AS isolates in the bovine PrP host. Alternatively, the occurrence of c-BSE in AS-inoculated tgBov mice could result from a mutation of AS strain properties triggered by passage across the bovine transmission barrier for this particular ovine prion strain.
In order to explore the origin of the c-BSE agent observed in tgBov mice inoculated with AS, we employed in vitro protein misfolding cyclic amplification (PMCA), a methodology that mimics prion replication in vitro, but in an accelerated form, allowing amplification of minute amounts of PrPSc and prion infectivity (27). In PMCA, a PrPC-containing substrate is combined with a seed that contains PrPSc. Following repeated cycles of incubation and sonication, the amount of PrPSc increases.
PMCA has been previously reported to amplify the c-BSE agent with great efficacy using either tgARQ or tgBov mouse brain homogenate as substrate (28). Using this protocol, 2 (tgARQ substrate) or 3 (tgBov substrate) amplification rounds were sufficient to reach the detection limit for c-BSE prion seeding activity (SI Appendix, Fig. S2). The level of prion infectivity and prion seeding activity of a reference sheep-passaged c-BSE isolate were endpoint-titrated by both bioassays in tgBov mice and PMCA, respectively (SI Appendix, Table S2). The infectious prion titer of the sheep-passaged c-BSE isolate was ∼107.2 LD50/g IC in tgBov mice. The prion seeding titer (SA50) was estimated to be ∼1011.1 SA50/g using tgARQ mouse tissue as substrate and 1011.05 SA50/g using tgBov mouse tissue as substrate. Considering the fact that mice were inoculated using 20 µL of sample and the PMCA reactions were seeded using 5 µL of the same sample, the PMCA can be considered to be about 1,500 fold more sensitive than the bioassay in tgBov. This also means that 1 c-BSE LD50 in tgBov mice corresponds to ∼1,500 SA50 assessed by PMCA.
In addition to its high sensitivity, in vitro PMCA can reproduce, at least partly, the transmission barrier phenomenon observed during the in vivo prion bioassay (29). Therefore, amplification of prion seeding activity in AS isolates by PMCA using tgBov mouse tissue as substrate offered an opportunity to characterize the potential impact of the bovine transmission barrier on AS strain properties.
The AS isolates that were originally transmitted to tgBov mice (except AS8) and 18 additional AS isolates (originating from Norway, France, and Portugal) were subjected to PMCA (Table 3). Each AS isolate was used to seed reactions containing either bovine PrP or ovine ARQ PrP substrate (10 to 18 replicates per substrate). After amplification, PrPres was detected by Western blot in a low proportion of the reactions seeded, with 19 out of the 25 AS isolates for tgBov and tgARQ combined (Table 3). In most instances, a similar proportion of PrPres-positive PMCA reactions were observed when either bovine PrP or ovine ARQ PrP was used as substrate. However, in some cases (n = 3), a low number of PrPres-positive reactions were observed when bovine PrP was used as substrate (in the case of AS 10) or when ovine ARQ PrP was used as substrate (in the cases of AS 9 and AS 25). Whatever combination of AS isolate and substrate PrP was used, the PrPres Western blot profile in PMCA-positive reaction products and its reactivity with 12B2 antibody were indistinguishable from those observed for PMCA reaction products seeded with authentic ovine c-BSE prions (Fig. 4). No PrPres was observed in PMCA reactions that were unseeded (n = 120) or in those reactions seeded (n = 60) with prion-free sheep brain homogenate (representative samples shown in Fig. 4). It should be noted that the PrP amino sequence was 100% homologous between certain AS isolates (AS 5, AS 26) and the ovine PrP substrate (tgARQ) used in PMCA reactions. Therefore, in vitro amplification of c-BSE prions in PMCA reactions seeded with these AS isolates using ovine ARQ PrP as substrate cannot be a consequence of mutation of prion strain properties triggered by a transmission barrier.
Table 3.
Protein Misfolding Cyclic Amplification seeding activity in atypical scrapie isolates
| Isolates | PMCA positive reactions | |||
| Identifiant | Origin | Genotype | TgBov substrate | TgARQ substrate |
| AS 1 | Fr | ARQ*/ARQ | 3/12 | 5/12 |
| AS 2 | Sp | ARR/ARQ | 2/12 | 3/12 |
| AS 3 | Sp | ARQ/ARH | 3/12 | 4/18 |
| AS 4 | No | ARQ*/AFRQ | 12/12 | 9/12 |
| AS 5 | Sp | ARQ/ARQ | 4/12 | 3/12 |
| AS 6 | Sp | ARQ/ARH | 5/12 | 1/12 |
| AS 7 | It | ARQ/AHQ | 1/12 | 2/12 |
| AS 8 | Po | ARQ/ARQ | ND | ND |
| AS 9 | Nor | ARR/ARQ | 0/12 | 2/10 |
| AS 10 | — | ARQ*/AHQ | 1/12 | 0/10 |
| AS 11 | — | AHQ/ARQ | 3/12 | 7/12 |
| AS 12 | — | ARR/ARQ | 0/12 | 0/12 |
| AS 13 | — | ARR/AHQ | 0/12 | 0/12 |
| AS 14 | — | ARQ/AHQ | 1/12 | 1/12 |
| AS 15 | — | ARR/ARR | 3/12 | 1/12 |
| AS 16 | — | ARR/AHQ | 1/12 | 3/10 |
| AS 17 | — | ARQ*/AHQ | 1/12 | 1/10 |
| AS 18 | — | ARQ*/AHQ | 0/12 | 0/10 |
| AS 19 | Po | ARR/ARR | 0/12 | 0/12 |
| AS 20 | — | ARR/AHQ | 3/12 | 1/12 |
| AS 21 | — | ARR/ARR | 0/12 | 0/10 |
| AS 22 | — | ARQ*/AHQ | 0/12 | 0/12 |
| AS 23 | — | ARQ*/ARQ | 1/12 | 1/12 |
| AS 24 | — | ARQ*/ARQ | 2/12 | 4/12 |
| AS 25 | Fr | AHQ/AHQ | 0/12 | 1/12 |
| AS 26 | — | ARQ/ARQ | 1/12 | 1/12 |
| TSE free sheep | — | ARQ/ARQ | 0/12 | 0/12 |
| Unseeded | — | — | 0/120 | 0/120 |
Twenty-six AS scrapie cases (1/50 diluted 10% brain homogenates) originating from 5 different countries (France [Fr], Spain [Sp], Italy [It], Portugal [Po] and Norway [Nor]) were used to seed PMCA reactions (5 µL of seed per reaction). The AS affected animals displayed different Prnp genotypes at codons 136, 154, and 171. Two different PMCA substrates were used. The first one was prepared using brains from transgenic mice overexpressing the ARQ variant of the sheep prion protein (tgARQ). The second was prepared using brains from transgenic mice overexpressing the bovine prion protein (tgBov). For each isolate and substrate, 10 to 18 individual replicates were tested. Reactions were subjected to 3 amplification rounds. After each round, reaction products (1 volume) were mixed with fresh substrate (9 volumes) to seed the following round. PMCA reaction products (third amplification round) were analyzed by Western blot for the presence of PrPres. The number of PrPres Western blot positive reactions/total number of reactions are reported. Unseeded reactions and reactions seeded with brain homogenate prepared from a TSE-free sheep were included as specificity controls. ND, not done.
F/L dimorphism displayed at codon 141.
Fig. 4.
PrPres detection in PMCA reactions seeded with atypical/Nor98 scrapie isolates. Protein misfolding cyclic amplification (PMCA) reactions were seeded with atypical/Nor98 scrapie (AS) isolates (1/50 diluted 10% brain homogenate) that had been identified in 5 European countries (Table 3). PMCA reactions seeded with brain homogenate from a TSE-free sheep (originating from New Zealand) and unseeded PMCA reactions were included as specificity controls. PMCA substrate consisted of brain homogenate from either bovine PrP (tgBov) or ovine PrP (tgARQ) mice. PMCA reactions were subjected to 3 (tgARQ substrate) or 4 (tgBov substrate) amplification rounds, each comprising 96 cycles (10 s sonication, 14 min and 50 s incubation at 39.5 °C) in a Qsonica700 device. The PMCA reactions were analyzed by Western blot for the presence of abnormal PK-resistant PrP (PrPres) using anti-PrP monoclonal antibodies Sha31 (epitope 145-YEDRYYRE-152) and/or 12B2 (epitope 89-WGQGG-93). Each Western blot included a classical scrapie isolate (labeled as WB control) and an ovine c-BSE isolate as controls.
Taken together, the tgBov mouse bioassay and PMCA results strongly support the view that a low level of c-BSE prions was initially present in at least 21 out of the 26 AS isolates tested.
To further clarify the origin of the c-BSE agent detected in AS isolates, 2 of these isolates (AS 25 and AS 26) were endpoint-titrated in tg338 mice (1/10 dilution series, 6 or 7 tg338 mice inoculated per dilution). For both isolates, the last positive transmissions were observed in mice that received a 10−6 log10 dilution of the original 10% w/vol brain material (SI Appendix, Table S3). The brains of these endpoint-titration tg338 mice were subsequently subjected to PMCA. Irrespective of the substrate used for PMCA, either bovine or ovine ARQ PrP, PrPres was observed in a similar proportion of the PMCA reactions seeded with either the original AS isolates or AS isolates passaged in tg338 mice (Table 4). The PrPres Western blot profile observed in all of the PMCA-positive reactions was identical to that seen in reactions seeded with authentic c-BSE prions (Fig. 5). PMCA reactions seeded with brain homogenate prepared from age-matched noninoculated tg338 mice remained PrPres-negative (Table 4 and Fig. 5).
Table 4.
PMCA seeding activity in atypical scrapie passaged in tg338
| PMCA seeds | PrPres positive PMCA reactions | Seeding activity, SA50/mL | ||||
| Case | Origin | TgBov substrate | TgARQ substrate | TgBov substrate | TgARQ substrate | |
| AS 25 | Sheep | 0/12 | 1/12 | 0 (0–101.86)* | 101.40 | |
| First passage in tg338 (neat) | Mouse 1 | 2/12 | 1/12 | 101.72 | 101.40 | |
| Second passage in tg338 (neat) | Mouse 1 | 2/12 | 1/12 | 101.72 | 101.40 | |
| End-point titration in tg338 (10−6 dilution) | Mouse 1 | 2/12 | 3/12 | 101.72 | 101.92 | |
| Mouse 2 | 1/12 | 3/12 | 101.40 | 101.92 | ||
| Mouse 3 | 2/12 | 2/12 | 101.72 | 101.72 | ||
| AS 26 | Sheep | 1/12 | 1/12 | 101.40 | 101.40 | |
| First passage in tg338 (neat) | Mouse 1 | 2/12 | 0/12 | 101.72 | 0 (0–101.86) * | |
| Second passage in tg338 (neat) | Mouse 1 | 2/12 | 0/12 | 101.72 | 0 (0–101.86) * | |
| End-point titration in tg338 (10−6 dilution) | Mouse 1 | 2/12 | 1/12 | 101.72 | 101.40 | |
| Mouse 2 | 1/12 | 1/12 | 101.40 | 101.40 | ||
| Noninoculated tg338 | Mouse 1 | 0/12 | 0/12 | — | — | |
| Mouse 2 | 0/12 | 0/12 | — | — | ||
| Mouse 3 | 0/12 | 0/12 | — | — | ||
Two sheep atypical scrapie (AS) isolates were selected. The 10% w/vol brain homogenates were inoculated into tg338 mice (2 iterative passages). Groups of 6 tg338 mice were inoculated intracerebrally with 20 µL of serial 10-fold dilutions of the same homogenates. Transmission was observed in 3 (AS 25) and 2 (AS 26) mice inoculated with 10−6 brain homogenate. No transmission was observed at lower dilutions. PMCA reactions (12 replicates) were seeded with 1/50 diluted brain homogenate (10% w/vol) from 1) the original sheep, 2) the second passage tg338 mice (pool of brains), and 3) individual brain from positive tg338 in the endpoint titration experiment. Brain homogenates (10% w/vol) from age-matched, noninoculated tg338 mice were also used as seeds (1/50 diluted). Two different PMCA substrates were used. The first one was prepared using brains from transgenic mice overexpressing the ARQ variant of the sheep prion protein. The second was prepared using brains from transgenic mice overexpressing the bovine prion protein (tgBov). Reactions were subjected to up to 4 amplification rounds. After each round, reaction products (1 volume) were mixed with fresh substrate (9 volumes) to seed the following round. PMCA reaction products were analyzed by Western blot for the presence of PrPres. The number of PrPres Western blot-positive reactions/total number of reactions are reported. Seeding activity titers were estimated using the Spearman–Karber’s limiting dilution titration method (most likely value) or, when no positive reaction was observed, by the Poisson’s probabilistic model. Titers are given as the number of PMCA SA50 per milliliter of 10% brain homogenate.
Most likely value and IC 95% as described by Brown et al. (66).
Fig. 5.
PMCA seeding activity detection in 2 bioassay endpoint-titrated atypical/Nor98 isolates. Two atypical/Nor98 isolates, AS 25 and AS 26 (Table 3), were endpoint-titrated in tg338 mice (1/10 dilution series, 6 tg338 mice per dilution). For both isolates, the last positive transmissions were observed in mice that received a 10−6 dilution of the original 10% w/vol brain material (SI Appendix, Table S3). The original AS isolates and the brains of clinically affected mice inoculated with neat and 10−6 diluted isolates were used to seed PMCA reactions that either used tgARQ or tgBov as substrate. PMCA reactions seeded with age-matched inoculated tg338 mice and unseeded reactions were included as specificity controls. Reactions were subjected to 3 (tgARQ substrate) or 4 (tgBov substrate) amplification rounds, each comprising 96 cycles (10 s sonication, 14 min and 50 s incubation at 39.5 °C) in a Qsonica700 device. The PMCA reactions were analyzed by Western blot for the presence of abnormal PK-resistant PrP (PrPres) using the Sha31 (epitope 145-YEDRYYRE-152) and/or the 12B2 (89-WGQGG-93) anti-PrP antibodies. Each Western blot included a classical scrapie isolate (labeled as WB control) and an ovine c-BSE isolate as controls.
Considering the level of c-BSE seeding activity originally present in isolates AS 25 and AS 26 (less than 100 SA50/mL; Table 4), there is an extremely low level of probability that one of the 6 tg338 mice inoculated with 20 µL of a 10−6 diluted AS isolate (<2 10−6 SA50 per dose of inoculum) could be exposed to 1 infectious dose of c-BSE agent (1 c-BSE LD50 is ∼1,500 SA50). Consequently, the presence of c-BSE prion seeding activity in the brains of tg338 mice inoculated with a 10−6 log10 dilution of original AS isolate implies that a low titer of c-BSE prions was generated during the propagation of ovine AS prions in a host that expressed ovine PrP, namely tg338 mice.
Discussion
The mechanism(s) that lead to an alteration in the phenotype of prion strains as these transmissible entities undergo transmission between different host species remain uncertain. This is despite the identification that differences in amino acid sequence between host PrPC and donor PrPSc, together with prion strain identity, are principal determinants of the transmission barrier (9, 10). Based on the concept that conformation of PrPSc molecules/aggregates encode prion strain information (2, 4, 5, 30, 31), at least 2 nonexclusive hypotheses, “deformed templating” (32, 33) and the “conformational selection model” (32–35), have been proposed to explain the mutation of prion strains.
The deformed templating hypothesis postulates that a prion strain replicates as a clone of PrPSc molecules/aggregates. When confronted by a transmission barrier that does not allow clonal prion replication, the propagation process is modified so that “altered” PrPSc structural variants are generated in an attempt to convert the new host PrPC. While the majority of these presumably fail to replicate efficiently in the new host, variants eventually emerge that are successful and adapt to the new PrP environment through multiple trial-and-error replication events. In this deformed templating model, confrontation of the transmission barrier serves as the triggering event that initiates the generation of new prion variant(s) and as a filter for their selection (35).
The conformational selection model proposes that a prion strain naturally propagates in its host as an ensemble of PrPSc conformers dominated by a stable energetically favorable conformation responsible for the observed prion strain phenotype. Furthermore, this model predicts that the number of stable PrPSc conformers is limited for each PrP amino acid sequence, which would explain the existence of a finite number of stable prion strains that can propagate in a given species. It is further proposed that, during transmission of a prion strain to a new host, one of the less dominant PrPSc conformers of those present in the ensemble is selected with a resultant change, or mutation, in the properties of the newly propagating prion strain. In the conformational selection model, the transmission barrier acts simply as a selective filter for new prion variants, and ease of permeation of the barrier results from the extent of overlap of PrPSc conformers that exist between the interacting species (32, 33).
Our data reported here showed that c-BSE prions are present as a minor variant in natural isolates of ovine AS. In addition, transmission of ovine AS to bovine PrP mice demonstrated that c-BSE can emerge during these transmissions as the dominant prion strain. These results provide a cogent argument in favor of the conformational selection model as the mechanism for prion strain mutation during interspecies prion transmission. This would be expected to occur by selection of a preexisting PrPSc variant in AS isolates, one best suited to the new replicative environment. Within this conceptual framework, the occurrence of prion strain mutation is dependent upon the particular repertoire of PrPSc variants associated with distinct prion strains. This notion is supported by our observation that c-BSE prions emerged during serial transmission of ovine AS in tgBov mice but not from serial passage of classical scrapie in the same mouse line [Table 1 and Cassard et al. (15)].
The diversity of prion strains that exist in small ruminants remains undefined, although it is established that at least 5 different natural ovine prion strains exist, including AS (6, 36–39). According to the conformational selection model, each of these different ovine prion strains is associated with a unique and stable PrPSc conformer and a distinct set of minor variants. The tgBov mouse line has previously been reported to support the propagation of a variety of natural ovine prions, of which several displayed significantly shorter incubation periods than c-BSE (15). Strikingly, in our experiments, the diversity of prion variants in the AS isolates (7 different cases) revealed by the serial passage in tgBov was restricted to the c-BSE agent (Table 1). This consistent emergence of a single prion strain argues against the view that AS prion replication in sheep can randomly generate all of the existing stable PrPSc variants associated with a particular ovine PrPC amino acid sequence. Instead, our data support the view that individual prion strains are associated with a restricted repertoire of stable PrPSc variants in a given host. Whether AS is unique in its ability to generate c-BSE prion particles during its replication process remains to be established.
Classical BSE was first recognized in 1984 and 1985 as a novel prion disease affecting cattle in the United Kingdom (40). Epidemiological data clearly established that the number of cases of c-BSE was amplified by the recycling of infected animal carcasses into cattle feed in the form of meat and bone meal (MBM) (41). Since bovine prion disease had not been recognized in cattle prior to the c-BSE epizootic and the disease is apparently noncontagious between cattle, several hypotheses were proposed to explain its emergence. These range from the spontaneous occurrence of c-BSE in cattle to the passage and adaptation of a prion originating from another species (42, 43). Our studies here that show the presence of c-BSE prions in AS isolates, combined with the demonstrated presence of AS in the United Kingdom long before the appearance of the c-BSE epizootic in cattle, suggests that the recycling of AS cases in MBM might be a source of bovine prion disease (20). In addition to its potential role in the initial emergence of c-BSE in cattle, the presence of c-BSE prions in natural cases of AS has current and direct implications for both the continued risk of this ovine prion disease to other farmed animals and for human exposure risks. The distribution of AS cases are widespread across the world (17–19). A recent retrospective analysis of surveillance data collected over a period exceeding 10 y in the European Union (EU) concluded that the prevalence of detected AS cases has remained relatively stable in the different member states, with between 2 and 6 positive cases per 10,000 tested animals per year. This implies that a substantial number of AS-infected animals could enter either the animal or human food chain each year (44, 45), and each case represents a potential source of exposure to the c-BSE agent for farmed animals (MBM derived from rendered small ruminants) and human consumers (consumption of healthy slaughtered animals), respectively. The epidemiological features of AS within the EU is likely to reflect the situation of the disease in other countries that breed and maintain small ruminants.
In Europe, the c-BSE crisis and the emergence of vCJD resulted in the implementation of a strong and coherent policy (EU regulation 999/2001) aimed at control and eradication of this animal prion disease. The total feed ban on the use of MBM in animal feed and the systematic retrieval from the food chain of ruminant tissues that have the potential to contain high levels of prion infectivity, so-called Specified Risk Material (SRM) measures, were instrumental for control of c-BSE in cattle and prevention of dietary human exposure to these bovine prions (46, 47). As a side effect, these measures also strongly limited the exposure of farmed animals and human consumers to the other TSE agents circulating in farmed animal species, including AS.
With the decline of the c-BSE epizootic in cattle and the combined increase in pressure from industry, EU authorities have begun to consider discontinuing certain TSE control measures. The abrogation of the SRM measures for small ruminants and the partial reauthorization of the use of processed animal protein, formerly known as MBM, in animal feed are part of the EU authorities’ agenda. Our observation of the presence of the c-BSE agent in AS-infected small ruminants suggests that modification of the TSE control measures could result in an increased risk of exposure to c-BSE prions for both animals and humans. Whether or not this exposure will result in further c-BSE transmission in cattle and/or humans remains an open and important question.
Methods
Ethics Statement.
All animal experiments were performed in compliance with institutional and French national guidelines and in accordance with the European Directives 86/609/EEC and 2010/63/EU. In France, the animal experiments that are part of this study (national registration 01734.01) were approved by the local ENVT ethics committee. Experiments developed in CISA-INIA (Madrid, Spain) were approved by the Committee on the Ethics of Animal Experiments of the Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria and the General Directorate of the Madrid Community Government (permit numbers: CEEA 2009/004 and PROEX 228–16). Mouse inoculations were performed under anesthesia (isoflurane).
Experiments developed in IRTA-CReSA (Barcelona, Catalonia) involving animals were approved by the animal experimentation ethics committee of the Autonomous University of Barcelona (reference number 585–3487) in agreement with Article 28, sections (a), (b), (c), and (d) of the “Real Decreto 214/1997 de 30 de Julio,” European Directive 86/609/CEE, and the European Council Guidelines included in the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes.
Mice that displayed clinical signs were anesthetized with isoflurane before sacrifice using CO2 inhalation.
Atypical/Nor98 Scrapie Cases and Control Sheep.
Natural atypical scrapie (AS) cases identified through active or passive surveillance programs were selected according to their geographical origin (France, Spain, Italy, Norway, and Portugal) and PRNP genotypes (SI Appendix, Table S1 and Table 1). These cases have been originally classified as AS by TSE national reference laboratories in each country. All of the cases corresponded to sheep except the AS 7 case (goat).
In all cases, PrP genotype was checked by sequencing exon 3 of the PRNP gene as previously described (48). The polymorphisms at codons 136 (A/V), 154 (H/R), and 171 (R/Q/H), which have been demonstrated to strongly influence the susceptibility to TSE in sheep, are indicated (49). Additionally, the presence of a phenylalanine at codon 141 (F/L), which has been shown to impact on the susceptibility to atypical/Nor98 scrapie, are also indicated (Tables 1 and 4) (23, 48). Brain material collected in TSE-free Poll–Dorset sheep (APHA) was used as control (50).
c-BSE Isolates.
Cattle and ovine classical BSE (c-BSE) isolates were used as control. The cattle c-BSE isolate was a natural case originating from France. This isolate was used in previous studies aimed at the characterization of c-BSE strain properties through transmission to mice overexpressing the PrP sequence of various host species (51). The ovine c-BSE isolate was obtained by the intracerebral inoculation of the same cattle c-BSE isolate in ARQ/ARQ TSE-free sheep (first passage) as described in Andréoletti et al., 2004 (50).
Mouse Bioassays.
Bioassays were carried out using mice expressing bovine PrP (tgBov/tg110) (52, 53) and/or mice expressing ovine ARQ (tgARQ) (54) or VRQ (tg338) PrP (55).
Groups of 6- to 10-wk-old female mice (n ≥ 6) were anesthetized and inoculated with 20 µL of a 10% tissue homogenate in the right parietal lobe using a 25-gauge disposable hypodermic needle. Mice were observed daily, and their neurological status was assessed weekly. When clinically progressive TSE disease was evident, the animals were euthanized and their brains harvested. Half of the brain was fixed by immersion in 10% formol saline and the other half was frozen at −20 °C. Tissues from animals found dead were frozen (no formalin fixation). In animals where no clinical signs were observed, mice were killed at the end of their natural lifespan (650 to 750 d). In those cases, incubation periods reported in Tables 1 and 2 as >650 dpi corresponded to the survival time observed in at least 3 out of the 6 mice.
PMCA Amplification.
Brains from tgBov, tgARQ, and tg338 mice were used to prepare the PMCA substrates. PMCA was performed as previously described (28, 56). Briefly, PMCA reactions (50 µL final volume) were seeded with 5 µL of sample to be tested. PMCA reactions were then subjected to 3 amplification rounds, each comprising 96 cycles (10 s sonication, 14 min and 50 s incubation at 39.5 °C) in a Qsonica700 device. After each round, reaction products (1 volume) were mixed with fresh substrate (9 volumes) to seed the following round. The PMCA reaction products were analyzed by Western blot for the presence of PK-resistant PrP. Each WB line was loaded with the equivalent of 20 µL of PK-digested PMCA product. Each PMCA run included a reference ovine BSE sample (10% brain homogenate) as a control for the amplification efficiency. Unseeded controls (2 unseeded controls for 8 seeded reactions) were also included in each run.
Western Blot Detection of Abnormal PrP.
PK-resistant abnormal PrP (PrPres) extraction and Western blot were performed as previously described (57). Immunodetection was performed using 2 different PrP-specific monoclonal antibodies, Sha31 (1 µg/mL) (58) and 12B2 (4 µg/mL) (59), which recognize the amino acid sequences YEDRYYRE (145–152) and WGQGG (89–93), respectively (60).
Paraffin-Embedded Tissue Blot.
Paraffin-embedded brain tissue from inoculated mice was analyzed as previously described (61–63).
Lesion Profiling and Abnormal PrP Immunohistochemistry.
Vacuolar brain lesion profiles were established following the method described by Fraser et al. (64). In situ PrPSc immune labeling was performed as previously described (63) using 6H4 anti-PrP antibody (epitope: 147DYEDRYYRE155 of the bovine PrP concentration 3 µg/mL).
Infectious and Seeding Activity Titer Estimates.
A series of 1/10 dilutions of a reference 10% w/vol brainstem from an ovine-BSE (ARQ/ARQ) isolate and 2 AS isolates (AS 25 and AS 26) were prepared. Successive 1/10 dilutions of brain homogenate were inoculated intracerebrally (20 µL) into tgBov or tg338 mice (n = 6 per inoculum). Dilutions of the same c-BSE isolate were used to seed PMCA reactions that used brain tissue from either bovine PrP (tgBov mice) or ovine ARQ PrP (tgARQ mice) as substrate. Twelve individual replicates of each sample dilution were tested. Reactions were then subjected to 3 amplification rounds. PMCA reaction products (third amplification round) were analyzed by Western blot for the presence of PrPres. The titer of prion seeding activity was estimated by the Spearman–Kärber’s method (65).
For AS and AS passaged in tg338 isolates (10% brain homogenate), 1/50 diluted material was used to seed 12 individual reactions (tgBov or tgARQ substrates). After 3 amplification rounds, the number of PrPres Western blot-positive reactions per total number of reactions was established. These ratios were used to estimate seeding activity titers (SA/µL of 10% brain homogenate) by the limiting dilution titration method (application of Poisson’s probabilistic model) described by Brown et al. (66) or by Spearman–Kärber’s method. According to Fisher et al. (67) and as previously used for prion infectivity comparisons (68), 1 SA50 was considered to be equivalent to 0.693 SA.
Data Availability.
All data are available in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
This work was funded by Fonds Europeens de Developpement Regional Programme Operationnel de Cooperation Territoriale Espagne France Andorre TRANSPRION (EFA282/13) and REDPRION (EFA148/16); the UK Food Standards Agency Exploring permeability of the species barrier (M03043 and FS231051); the European Union through FP7 222887 “Priority;” the Spanish Ministerio de Economía y Competitividad (AGL2016-78054-R [AEI/FEDER, UE]); and Fundació La Marató de TV3 (201821-31). A.M.-M. was supported by a fellowship from the INIA (FPI-SGIT-2015-02), and P.A.-C. was supported by a fellowship from the Spanish Ministerio de Economía y Competitividad (BES-2010-040922).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915737116/-/DCSupplemental.
References
- 1.McKinley M. P., Bolton D. C., Prusiner S. B., A protease-resistant protein is a structural component of the scrapie prion. Cell 35, 57–62 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Prusiner S. B., Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 (1982). [DOI] [PubMed] [Google Scholar]
- 3.Race R., Raines A., Raymond G. J., Caughey B., Chesebro B., Long-term subclinical carrier state precedes scrapie replication and adaptation in a resistant species: Analogies to bovine spongiform encephalopathy and variant Creutzfeldt-Jakob disease in humans. J. Virol. 75, 10106–10112 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bessen R. A., Marsh R. F., Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 66, 2096–2101 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessen R. A., Marsh R. F., Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 68, 7859–7868 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Béringue V., Vilotte J. L., Laude H., Prion agent diversity and species barrier. Vet. Res. 39, 47 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Pattison I. H., Scrapie in the Welsh mountain breed of sheep and its experimental transmission to goats. Vet. Rec. 77, 1388–1390 (1965). [DOI] [PubMed] [Google Scholar]
- 8.Bruce M. E., Dickinson A. G., Biological evidence that scrapie agent has an independent genome. J. Gen. Virol. 68, 79–89 (1987). [DOI] [PubMed] [Google Scholar]
- 9.Kimberlin R. H., Walker C. A., Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J. Gen. Virol. 39, 487–496 (1978). [DOI] [PubMed] [Google Scholar]
- 10.Scott M., et al. , Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59, 847–857 (1989). [DOI] [PubMed] [Google Scholar]
- 11.EFSA Panel on Biological Hazards (BIOHAZ) , Joint scientific opinion on any possible epidemiological or molecular association between TSEs in animals and humans. EFSA J. 9, 1945 (2011). [Google Scholar]
- 12.Gibbs C. J. Jr, Gajdusek D. C., Experimental subacute spongiform virus encephalopathies in primates and other laboratory animals. Science 182, 67–68 (1973). [DOI] [PubMed] [Google Scholar]
- 13.Bruce M. E., et al. , Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature 389, 498–501 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Padilla D., et al. , Sheep and goat BSE propagate more efficiently than cattle BSE in human PrP transgenic mice. PLoS Pathog. 7, e1001319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cassard H., et al. , Evidence for zoonotic potential of ovine scrapie prions. Nat. Commun. 5, 5821 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hill A. F., et al. , The same prion strain causes vCJD and BSE. Nature 389, 448–450, 526 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Benestad S. L., Arsac J. N., Goldmann W., Nöremark M., Atypical/Nor98 scrapie: Properties of the agent, genetics, and epidemiology. Vet. Res. 39, 19 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Kittelberger R., et al. , Atypical scrapie/Nor98 in a sheep from New Zealand. J. Vet. Diagn. Invest. 22, 863–875 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Cook R. W., et al. , Atypical scrapie in Australia. Aust. Vet. J. 94, 452–455 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Chong A., et al. , Archival search for historical atypical scrapie in sheep reveals evidence for mixed infections. J. Gen. Virol. 96, 3165–3178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Dur A., et al. , A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc. Natl. Acad. Sci. U.S.A. 102, 16031–16036 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths P. C., et al. , Characterization of atypical scrapie cases from Great Britain in transgenic ovine PrP mice. J. Gen. Virol. 91, 2132–2138 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Andréoletti O., et al. , Atypical/Nor98 scrapie infectivity in sheep peripheral tissues. PLoS Pathog. 7, e1001285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benestad S. L., et al. , Cases of scrapie with unusual features in Norway and designation of a new type, Nor98. Vet. Rec. 153, 202–208 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Fediaevsky A., et al. , The prevalence of atypical scrapie in sheep from positive flocks is not higher than in the general sheep population in 11 European countries. BMC Vet. Res. 6, 9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons M. M., et al. , The natural atypical scrapie phenotype is preserved on experimental transmission and sub-passage in PRNP homologous sheep. BMC Vet. Res. 6, 14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saborio G. P., Permanne B., Soto C., Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Lacroux C., et al. , Preclinical detection of variant CJD and BSE prions in blood. PLoS Pathog. 10, e1004202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castilla J., et al. , Crossing the species barrier by PrP(Sc) replication in vitro generates unique infectious prions. Cell 134, 757–768 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bessen R. A., et al. , Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature 375, 698–700 (1995). [DOI] [PubMed] [Google Scholar]
- 31.Telling G. C., et al. , Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 274, 2079–2082 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Collinge J., Clarke A. R., A general model of prion strains and their pathogenicity. Science 318, 930–936 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Li J., Browning S., Mahal S. P., Oelschlegel A. M., Weissmann C., Darwinian evolution of prions in cell culture. Science 327, 869–872 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghaemmaghami S., et al. , Continuous quinacrine treatment results in the formation of drug-resistant prions. PLoS Pathog. 5, e1000673 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makarava N., Baskakov I. V., The evolution of transmissible prions: The role of deformed templating. PLoS Pathog. 9, e1003759 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Béringue V., et al. , A bovine prion acquires an epidemic bovine spongiform encephalopathy strain-like phenotype on interspecies transmission. J. Neurosci. 27, 6965–6971 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thackray A. M., Hopkins L., Klein M. A., Bujdoso R., Mouse-adapted ovine scrapie prion strains are characterized by different conformers of PrPSc. J. Virol. 81, 12119–12127 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thackray A. M., Lockey R., Beck K. E., Spiropoulos J., Bujdoso R., Evidence for co-infection of ovine prion strains in classical scrapie isolates. J. Comp. Pathol. 147, 316–329 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Tixador P., et al. , The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog. 6, e1000859 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells G. A., et al. , A novel progressive spongiform encephalopathy in cattle. Vet. Rec. 121, 419–420 (1987). [DOI] [PubMed] [Google Scholar]
- 41.Wilesmith J. W., Wells G. A., Cranwell M. P., Ryan J. B., Bovine spongiform encephalopathy: Epidemiological studies. Vet. Rec. 123, 638–644 (1988). [PubMed] [Google Scholar]
- 42.Eddy R. G., Origin of BSE. Vet. Rec. 137, 648 (1995). [PubMed] [Google Scholar]
- 43.Colchester A. C., Colchester N. T., The origin of bovine spongiform encephalopathy: The human prion disease hypothesis. Lancet 366, 856–861 (2005). [DOI] [PubMed] [Google Scholar]
- 44.EFSA Panel on Biological Hazards (BIOHAZ) , Scientific opinion on the scrapie situation in the EU after 10 years of monitoring and control in sheep and goats. EFSA J. 12, 3781 (2014). [Google Scholar]
- 45.EFSA Panel on Biological Hazards (BIOHAZ) , Scientific opinion on BSE/TSE infectivity in small ruminant tissues. EFSA J. 8, 1875 (2010). [Google Scholar]
- 46.Ducrot C., et al. , Modelling BSE trend over time in Europe, a risk assessment perspective. Eur. J. Epidemiol. 25, 411–419 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Adkin A., Webster V., Arnold M. E., Wells G. A., Matthews D., Estimating the impact on the food chain of changing bovine spongiform encephalopathy (BSE) control measures: The BSE control model. Prev. Vet. Med. 93, 170–182 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Arsac J. N., et al. , Similar biochemical signatures and prion protein genotypes in atypical scrapie and Nor98 cases, France and Norway. Emerg. Infect. Dis. 13, 58–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunter N., et al. , Natural scrapie in a closed flock of Cheviot sheep occurs only in specific PrP genotypes. Arch. Virol. 141, 809–824 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Andréoletti O., et al. , PrPSc accumulation in myocytes from sheep incubating natural scrapie. Nat. Med. 10, 591–593 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Torres J. M., et al. , Elements modulating the prion species barrier and its passage consequences. PLoS One 9, e89722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castilla J., et al. , Early detection of PrPres in BSE-infected bovine PrP transgenic mice. Arch. Virol. 148, 677–691 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Douet J. Y., et al. , Detection of infectivity in blood of persons with variant and sporadic Creutzfeldt-Jakob disease. Emerg. Infect. Dis. 20, 114–117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Groschup M. H., Buschmann A., Rodent models for prion diseases. Vet. Res. 39, 32 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Vilotte J. L., et al. , Markedly increased susceptibility to natural sheep scrapie of transgenic mice expressing ovine prp. J. Virol. 75, 5977–5984 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Douet J. Y., et al. , Distribution and quantitative estimates of variant Creutzfeldt-Jakob disease prions in tissues of clinical and asymptomatic patients. Emerg. Infect. Dis. 23, 946–956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huor A., et al. , Infectivity in bone marrow from sporadic CJD patients. J. Pathol. 243, 273–278 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Féraudet C., et al. , Screening of 145 anti-PrP monoclonal antibodies for their capacity to inhibit PrPSc replication in infected cells. J. Biol. Chem. 280, 11247–11258 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Langeveld J. P., et al. , Rapid and discriminatory diagnosis of scrapie and BSE in retro-pharyngeal lymph nodes of sheep. BMC Vet. Res. 2, 19 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Uro-Coste E., et al. , Beyond PrP9res) type 1/type 2 dichotomy in Creutzfeldt-Jakob disease. PLoS Pathog. 4, e1000029 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langevin C., Andréoletti O., Le Dur A., Laude H., Béringue V., Marked influence of the route of infection on prion strain apparent phenotype in a scrapie transgenic mouse model. Neurobiol. Dis. 41, 219–225 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Lacroux C., et al. , Dynamics and genetics of PrPSc placental accumulation in sheep. J. Gen. Virol. 88, 1056–1061 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Andreoletti O., et al. , Astrocytes accumulate 4-hydroxynonenal adducts in murine scrapie and human Creutzfeldt-Jakob disease. Neurobiol. Dis. 11, 386–393 (2002). [DOI] [PubMed] [Google Scholar]
- 64.Fraser H., Dickinson A. G., The sequential development of the brain lesion of scrapie in three strains of mice. J. Comp. Pathol. 78, 301–311 (1968). [DOI] [PubMed] [Google Scholar]
- 65.Markus R. A., Frank J., Groshen S., Azen S. P., An alternative approach to the optimal design of an LD50 bioassay. Stat. Med. 14, 841–852 (1995). [DOI] [PubMed] [Google Scholar]
- 66.Brown P., et al. , Further studies of blood infectivity in an experimental model of transmissible spongiform encephalopathy, with an explanation of why blood components do not transmit Creutzfeldt-Jakob disease in humans. Transfusion 39, 1169–1178 (1999). [DOI] [PubMed] [Google Scholar]
- 67.Fisher R. A., Uncertain inference. Proc. Am. Acad. Arts Sci. 71, 245–258 (1936). [Google Scholar]
- 68.Gregori L., et al. , Reduction in infectivity of endogenous transmissible spongiform encephalopathies present in blood by adsorption to selective affinity resins. Lancet 368, 2226–2230 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript and SI Appendix.