Significance
Influenza virus and some other human pathogens evolve to evade herd immunity and, hence, to reinfect previously immune individuals. Individual humoral immune responses also evolve, through somatic mutation and selection in a process known as affinity maturation. We describe a detailed molecular “case history” of this coevolution (to our knowledge, the most thorough such study so far) through structural and biophysical analysis of a lineage of human antibodies directed at the influenza hemagglutinin receptor binding site. We show how affinity maturation in this lineage has maintained adaptability (“breadth” of neutralizing activity) by generating a branched and, hence, diversifying phylogeny. The mechanisms underlying affinity maturation appear to ensure such diversification while also enhancing affinity of the most potent resulting antibodies.
Keywords: influenza virus, antibody, B cell, vaccine, X-ray crystallography
Abstract
Affinity maturation of the B cell antigen receptor (BCR) is a conserved and crucial component of the adaptive immune response. BCR lineages, inferred from paired heavy- and light-chain sequences of rearranged Ig genes from multiple descendants of the same naive B cell precursor (the lineages’ unmutated common ancestor, “UCA”), make it possible to reconstruct the underlying somatic evolutionary history. We present here an extensive structural and biophysical analysis of a lineage of BCRs directed against the receptor binding site (RBS) of subtype H1 influenza virus hemagglutinin (HA). The lineage includes 8 antibodies detected directly by sequencing, 3 in 1 principal branch and 5 in the other. When bound to HA, the heavy-chain third complementarity determining region (HCDR3) fits with an invariant pose into the RBS, but in each of the 2 branches, the rest of the Fab reorients specifically, from its position in the HA-bound UCA, about a hinge at the base of HCDR3. New contacts generated by the reorientation compensate for contacts lost as the H1 HA mutated during the time between the donor’s initial exposure and the vaccination that preceded sampling. Our data indicate that a “pluripotent” naive response differentiated, in each branch, into 1 of its possible alternatives. This property of naive BCRs and persistence of multiple branches of their progeny lineages can offer broader protection from evolving pathogens than can a single, linear pathway of somatic mutation.
Affinity maturation drives B cells to evolve somatically toward expression of higher-affinity B cell antigen receptors (BCRs) (1). Following initial antigen exposure, cycles of rapid B cell proliferation, somatic hypermutation, and affinity-based selection can lead to reduction of the dissociation constant for the antigen–BCR interaction by several orders of magnitude (2, 3). This “Darwinian” competition takes place in germinal centers (GCs), specialized compartments within peripheral lymphoid tissues (e.g., lymph nodes, spleen) (4).
Single-cell sequencing of paired heavy-chain and light-chain B cell cDNA provides, in favorable cases, enough data to reconstruct clonal lineages, all members of which descend from the same naive progenitor (5). The root of the phylogenetic tree derived from experimentally determined sequences of multiple BCRs in a sample is a good estimate for the sequence of the unmutated common ancestor (UCA) of those BCRs—i.e., the sequence of the antigen BCR expressed on the naive B cell that gave rise to the lineage (6). The properties of the intermediates thus report on the history of affinity maturation during cycles of mutation, division, and selection in cells of the lineage.
Affinity is not the only property likely to be important for effective host defense, however. Especially when the antigens themselves evolve in response to human herd immunity—as does influenza virus, for example—adaptability to respond to mutationally altered antigen may be equally relevant, once a certain affinity threshold has been reached (1, 7). Evidence that the GC output covers a range of antibody affinities suggests that the affinity maturation regime has evolved to match such requirements (8).
We have described in previous work from these laboratories a variety of lineages for human antibodies that interact directly with the receptor-binding site on influenza hemagglutinin (HA) (9–13). One important conclusion, which generalizes to antibodies that recognize other HA epitopes, is that the donor’s initial exposure to influenza (likely by infection, in all of the subjects we have studied so far) conditions all subsequent responses, in which immunological memory of the first encounter dominates (12–14). Later exposures, through infection or vaccination, can update that memory, by selecting for mutations that enhance complementarity to the evolved epitope; in general, the selected variants still bind the original, “imprinting” HA, although sometimes with low affinity (14).
We describe here the results of a detailed structural and biophysical analysis of a particular RBS-directed antibody lineage (designated 652), which includes 8 antibodies detected directly by sequencing, 3 in 1 main branch of the lineage and 5 in the other (14, 15). We have determined a total of 6 structures of lineage-member Fabs bound with HA heads and 6 structures of free Fabs. In all of the structures of HA-bound Fabs, the heavy-chain third complementarity determining loop (HCDR3) of the antibody fits into the RBS with an essentially invariant pose. In products of the primary response in the 652 lineage, the “body” of the Fab appears to have at least 3 possible orientations with respect to the HCDR3 loop (and, hence, with respect to HA). By the time the subject was vaccinated, accumulated mutations in circulating HAs and, hence, in the vaccine strain had eliminated several of the specific HCDR3 interactions; affinity maturation had compensated by introducing mutations outside that loop, creating contacts from other CDRs and selecting a distinct alternative orientation for each of the 2 branches. In other words, a “pluripotent” naive response differentiated in each branch into 1 of its potential alternatives. We discuss these observations in view of their relevance for understanding the coevolution of virus and B cell antigen receptor and for design of next-generation influenza vaccines.
Results
Donor TIV01 and Clonal Antibody Lineage 652.
Donor TIV01 was born between 1989 and 1990 and in 2008 received a trivalent seasonal influenza vaccine containing H1N1 components from A/Solomon Islands/3/2006(H1N1) (SI-2006). Antibody sequences were determined from plasmacytes drawn 2 wk after the 2008 immunization (15). At least 2 antigenic exposures have driven affinity maturation of the identified BCRs/antibodies: an inferred primary exposure (presumably by infection) to an H1 virus that circulated during the donor’s early childhood (i.e., early 1990s) and a vaccination that the donor received in 2008 containing SI-2006 as its H1 component. We have found no clear evidence of intervening H1 exposures (14).
Antibody sequences defining clonal lineage 652 (Fig. 1A and SI Appendix, Fig. S1 A and B) were among those determined from donor TIV01 (15). We have reported previously that the computationally inferred, naive BCR, the UCA of the lineage, binds HAs from the early-to-mid 1990s, but not those from the late 1990s onward, while H2227 binds more broadly to seasonal H1 HAs, including H1 SI-2006 (14). Breadth therefore emerged through affinity maturation.
Fig. 1.
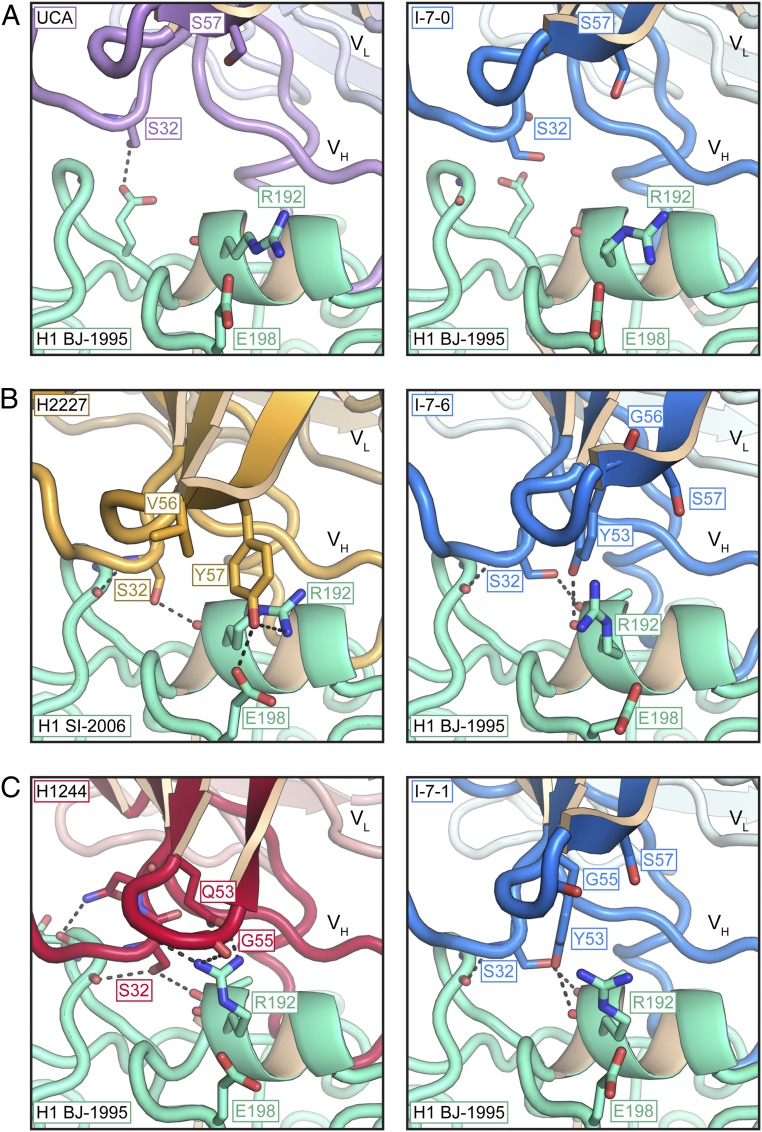
Clonal antibody lineage 652 engages the HA receptor binding site. (A) Phylogram of lineage 652 including its UCA and inferred evolutionary intermediates (denoted with an “I-”). Its 2 branches are named for their last common ancestor, I-1 and I-6, respectively. Antibodies for which we determined structures with bound HA are in boldface and colored to correspond to their colors in later structures. (B) Structure of the UCA Fab bound with the HA head domain of A/Beijing/262/1995 (H1N1) (H1 BJ-1995). (C) Contacts of the 652 UCA HCDR3 with the RBS. Key interactions are shown in sticks. Sites in HA that did not change between 1995 and 2006 are labeled with a white background. Sites where contacts were lost as influenza viruses drifted are labeled with a black background. UCA residues are labeled in purple. In B and C, the Fab light chain is in blue, the heavy chain in violet, and the HA head in green.
Lineage 652 comprises a single inferred UCA and its clonal descendants: 7 inferred evolutionary intermediates and 8 antibodies directly isolated from plasmacytes (Fig. 1A). Each has an HCDR3 23 amino acid residues in length, formed by recombination of IGHV4-4*02, IGHD2-2*01, and IGHJ6*03. The IGHD2-2*01 gene supplies a germ-line–encoded pair of cysteine residues predicted to form a disulfide linkage within HCDR3. All members of the inferred lineage share I-7 as their last common ancestor. The remaining antibodies occupy 2 phylogenetically distinct branches, I-1 and I-6 (named for their last common intermediates) (Fig. 1A).
Structures.
We determined the structure of the Fab from lineage 652 UCA bound with the HA head domain of A/Beijing/262/1995 (H1N1) (H1 BJ-1995) (Fig. 1B), 3 structures of I-7 (the first intermediate), and the structure of Fab H1244 (in the I-1 branch of the lineage), all bound with the same H1 BJ-1995 head domain and the structure of Fab H2227 (in the I-6 branch of the lineage) bound with the HA head domain of H1 SI-2006 (SI Appendix, Fig. S2). We also determined structures of the free Fabs of the UCA, H1244, and H2227 (SI Appendix, Fig. S2). In all 3 cases, the variable module of the free Fab superposed within experimental error on the corresponding module of the bound Fab, except for some side-chain rotamer differences (SI Appendix, Fig. S2).
The loop in HCDR3 from residue 102 through residue 113 is effectively a small, internally stabilized subdomain, delimited by the hydrogen bond from the amide of Cys103 to the carbonyl of Tyr-112 (Fig. 1C). A chain of van der Waals contacts, a set of internal hydrogen bonds, and a disulfide linking Cys103 with Cys108 determine its conformation. This loop inserts into the RBS nearly identically in all of the HA-bound complexes. Most of the residues between 103 and 112 participate in HA contacts. The dipeptide Pro109-Asp110 recapitulates many of the sialic acid receptor interactions (Fig. 1C), and the disulfide recapitulates another, by packing against Ile194.
UCA and I-7.
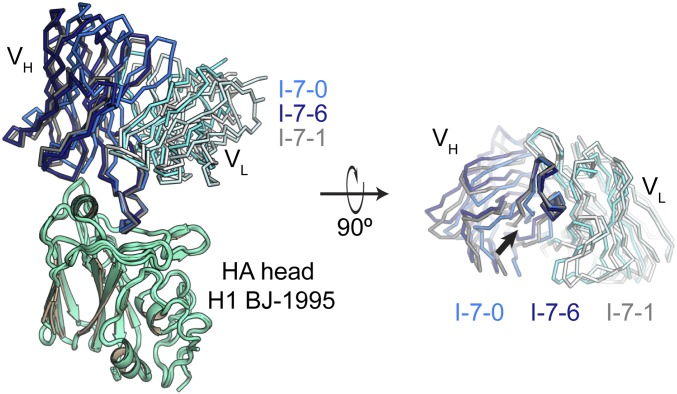
Intermediate I-7 is the last common intermediate between the 2 major branches of lineage 652 (Fig. 1A). In the 3 structures of this Fab fragment bound with the HA head domain of H1 BJ-1995, the pose of the HCDR3 loop in the RBS was essentially invariant, but in each of the structures, the rest of the Fab had a distinctive pitch with respect to HA (Fig. 2 and SI Appendix, Fig. S3). The 3 orientations, achieved by tilting and rotating about a fulcrum at the base of the HCDR3, corresponded approximately to those of the HA-bound UCA, H1244, and H2227 Fab fragments, and created different HA contacts between HCDR1 and HCDR2 and HA residues in the 156–159 loop and the 190 helix (Figs. 2 and 3). We therefore refer to these 3 confomers as I-7-0, I-7-1, and I-7-6, respectively.
Fig. 2.
Structures of the HA-bound I-7 intermediate. We determined 3 structures of I-7 bound with the HA head domain of H1 BJ-1995 (green); they are shown superposed on HA. With reference to HA, the Fab is pitched differently in each. We call these 3 conformations I-7-0, I-7-6, and I-7-1 (Left). The color of I-7-0 matches the colors used throughout; I-7-6 and I-7-1 are colored differently for clarity in this figure. Fab heavy chains are shown in bolder colors; light chains are shown in weaker colors. The right has been rotated about the x axis by 90° degrees, and the HA head has been removed to view the Fab at the HA interface. An arrow points to the base of HCDR3, which projects toward the viewer. The individual structures are compared in SI Appendix, Fig. S3.
Fig. 3.
Distinct orientations of lineage 652 Fabs with respect to HA. Structures of UCA (colored in shades of purple), H2227 (colored shades of orange), and H1244 (colored in shades of red) HA complexes, each aligned on HA (colored green) to a I-7-0, I-7-6, and I-7-1 (colored in shades of blue) Fab–HA complex with similar pitch. (A) UCA and I-7-0. (B) H2227 and I-7-6. (C) H1244 and I-7-1. Heavy chains are shown in stronger colors; light chains in weaker colors.
Most of the contacts between the HCDR3 loop and the rest of the Fv module are with LCDR1. A hydrogen bond from the phenolic -OH group of light-chain Tyr30 to the carbonyl oxygen of Cys103 is present in all of the structures and appears to help determine the pivot point. The rings of light-chain Tyr31 and heavy-chain Tyr114 are in van der Waals contact in all 3 configurations but would slide against each other in the transitions.
The substitutions that became fixed between the UCA and I-7 (Ser55Gly, Ser104Thr, Thr106Ala, all in the heavy chain) are not at positions likely to have imparted flexibility to the junction between HCDR3 and the rest of the heavy-chain variable domain. We suggest that, like I-7, the UCA could explore multiple conformations, with different principal members of this ensemble becoming fixed in each of the 2 principal lineage branches. Although none of the identified BCRs in our set of sequences had a structure like that of I-7-0, retention of that conformation in a branch not represented among the cells analyzed is in principle a possibility.
Affinity-matured antibodies.
Between 1990, the approximate year of the donor’s first exposure, and 2006, the year of the H1 strain used in the vaccine he received, evolution of the viral HA introduced substitutions that altered contacts at the periphery of the RBS with lineage 652 antibodies (Fig. 1C and SI Appendix, Fig. S1C). In particular, the mutation Glu156Gly eliminated a hcTyr112-Glu156 polar hydrogen bond; Ser186Pro eliminated a side-chain hydrogen bond with hcSer105 and required a small displacement at the tip of HCDR3; and Arg189Gly removed a participant in an extended H-bond network. Moreover, deletion of Lys133a (H3N2 numbering) in strains after 1995 likely eliminated yet another contact (Fig. 1C). Loss of these interactions would have reduced the affinities of the UCA and I-7. Affinity for SI-2006 and other post-1995 isolates would therefore have been acquired through somatic hypermutation and selection for new contacts from other CDR loops, enabled by reorientation of the Fab “body” with respect to the HCDR3 loop, as shown in Fig. 3.
Affinities.
To determine the consequences of the structural changes just described, we measured by biolayer interferometry (BLI) the affinities of the UCA, I-7, I-6, I-3, H2227, H1228, and H1244 Fabs for monomer HA heads of seasonal and pandemic H1N1 viruses (Fig. 4). I-7 and I-6 differ from the UCA at only 3 and 5 positions, respectively, in the heavy chain and at none in the light chain (SI Appendix, Fig. S1 A and B). These intermediates could therefore represent antibodies encoded in the genomes of memory B cells generated during the primary immune response, with an overall 4-fold gain in affinity between UCA and I-6. The transition from I-6 to I-3, the common ancestor of H2227 and H1228, likely includes the consequences of a waning primary response and a recall during secondary exposure. While we cannot rule out the possibility that I-3 (or I-3–like BCRs) were present during the primary response, it is more likely that I-3 arose during the secondary response. I-3 has an additional 6 changes in the heavy chain and 4 in the light chain—the 5% overall frequency of differences from the presumed germ-line precursor is twice the generally observed upper limit for memory cells from a primary response. Moreover, I-3 binds not only HA heads from strains circulating in the early 1990s but also all strains we tested from 1986 to 2008 (Fig. 4). The donor, who received the seasonal vaccine in 2008, reported no previous vaccination. Selection for the mutations present in I-3 could therefore have been by the vaccine immunogen (HA A/Solomon Islands/03/2006) or by a somewhat earlier exposure. Affinity profiles of I-3 and Fab H2227 are nearly identical, and they differ at only 2 positions in the heavy chain. We infer that I-3 and H2227 were probably responses to the same exposure—either the 2008 vaccination or an H1 infection between the late 1990s and 2008.
Fig. 4.
Lineage 652 affinity maturation. Affinities of Fab fragments from selected lineage members for HA heads of H1 isolates. Viruses isolated between 1990 and 2009 circulated during the donor’s lifetime. HA structures determined in this study are in bold. Coloring according to the key indicates the apparent equilibrium dissociation constant (KD), measured by biolayer interferometry.
The affinity profiles of H1228 and H1244—representing the lower part of the I-6 branch and the I-1 branch, respectively—are essentially the same as those of I-3 and H2227 (Fig. 4). Although the mutational events between I-7 and members of each of those branches are distinct, they generated similar breadth and similar incremental affinities. Under selection in germinal centers, divergent outcomes from the same starting point converged on essentially identical phenotypes. Moreover, as we have noted previously for imprinted responses, all of the isolated antibodies in this lineage retained high affinity for the HA of the initial exposure.
The Two Major Branches of the 652 Lineage.
In each of the 2 observed branches, pivoting of the heavy chain about the base of the HCDR3 loop defines a distinct pitch for the Fab with respect to HA (Fig. 3). The changes have negligible effects on HCDR3-RBS contacts (rms Cα displacements, with respect to the HA RBS, for residues 103–112 are 0.31 and 0.26 Å for H1224 and H2227, respectively), while generating HA contacts with HCDR1 and HCDR2 (Fig. 3). In both lineages, HCDR2 approaches the 190 helix, and HCDR1 projects between the C-terminal end of that helix and the 156–159 loop. Conformational adjustments in HCDR2 generate interactions, distinct for each branch, with residues in the 190 helix (Fig. 5).
Fig. 5.
Evolution of additional HA contacts. Selected contacts between HCDR1, HCDR2, and HA are shown as sticks. (A) UCA (Left) and I-7-0 (Right). (B) H2227 (Left) and I-7-6 (Right). (C) H1244 (Left) and I-7-1 (Right). Heavy chains in bolder colors in foreground; light chains in weaker colors in background; HA, in green.
Two positions in HCDR2 have branch-specific amino acid substitutions for all antibodies and intermediates in that branch. At position 53, a tyrosine in the UCA (and I-7) has mutated to glutamine in the I-1 branch and to serine in the I-6 branch; at position 57, a serine has mutated to glycine in I-1 and to tyrosine in I-6. Both Gln53 in the I-1 branch and Tyr57 in the I-6 branch interact with Arg192 of HA. The former is part of a hydrogen-bonding network that includes the carbonyls of residues 32, 54, and 55; the latter is part of a triple interaction with Arg192 and Glu198 (Fig. 5).
Because the germ-line residues at these positions in I-7 cannot participate in either of these branch-specific interaction networks, contacts with HA in the I-7-1 and I-7-6 complexes resemble each other, rather than those made by antibodies in the branch to which they correspond (Fig. 5). The conformation of HCDR2 in all 3 I-7 structures is similar to that of HCDR2 in the UCA.
We also examined potential intermediate events in the I-6 branch. Capacity to bind post-1995 isolates occurred between I-6 and I-3. One substitution in HCDR2, Gly56Val, occurred in this interval. We produced a mutant I-6, designated I-6V, bearing the substitution. Introduction of Val produced detectable binding to H1 SI-2006 and to HAs isolated between 1995 and 2006 (Fig. 4). The range of HAs (“breadth”) to which I-6V binds is comparable to the ranges for I-3 and H2227, although in some instances the affinity of I-6V for HA is lower. We determined structures for the I-6 and I-6V Fabs (SI Appendix, Fig. S2). The configuration of HCDR1 and HCDR2 in I-6 are nearly identical to those in the UCA and I-7. In I-6V, HCDR2 has a conformation that is similar to its conformation in H2227. The Gly at position 56 in the germ-line sequence has a positive φ angle. Accommodating Val requires a polypeptide backbone with a negative φ angle. The outcome redirects the side chain of hcTyr57, with the critical contacts shown (SI Appendix, Fig. S2). The Gly to Glu substitution in the I-5 branch presumably has a similar effect.
Discussion
Somatic hypermutation and selection of antibody variants appear to have at least 2 roles in generating a robust B cell immune response, both illustrated by events in development of the 652 lineage. One is the classical process of affinity maturation, which molds an antibody to fit more perfectly against an antigen. The other is the generation of affinity-independent diversity (1). The latter outcome may be particularly important for protection against pathogens such as influenza virus that mutate rapidly enough to reinfect a previously exposed individual.
The 3 structures of the I-7 intermediate bound with HA head capture 3 distinct modes of HA engagement. They correspond to the structure of the UCA and foreshadow those observed on the tips of both lineage branches. Substitutions distinguishing I-7 from the UCA are not at positions likely to impart or preclude conformational plasticity. Thus, the germ-line antibody (the UCA of the lineage) and early intermediates could probably bind HA in several modes. Mutation and selection in germinal centers would then have directed subsequent differentiation of the lineage toward at least 2 of the conformations represented in the I-7 ensemble.
Two principal conformational events, produced by mutation and selection, allowed lineage members to bind post-1995 H1 HAs. One was pivoting of the Fab with respect to contacts in the RBS; the other was local reconfiguration of HCDR2. Together, these changes created new contacts outside the RBS and presumably compensated for the loss of HCDR3 contacts with HAs from H1 strains after 1995. Despite somewhat different modes in the 2 branches for each of these conformational adjustments, the binding profiles of H1224, H1128, and H2227 for HAs from various historical influenza virus strains are essentially the same (Fig. 4). This phenotypic convergence is presumably the result of selection by the same series of antigens. We noticed the same phenomenon is our earlier characterization of the CH65-CH67 lineage, from the same donor (TIV01) (10).
Structural pluripotency of the germ-line precursor has at least 2 potential advantages. One is the capacity, as seen here, to evolve toward the same functional result along more than 1 mutational pathway, increasing the probability of reaching an outcome with substantially increased fitness. The other is an enhanced capacity to resist escape, as the alternative HCDR1 and HCDR2 contacts in the I-1 and I-6 branches imply that some potential HA resistance mutations to antibodies from 1 branch might still be sensitive to antibodies from the other. Presence of cells producing antibodies from divergent branches like these would thus offer broader protection than cells from only a single, linear evolutionary trajectory.
The information available in 1 or a few lineages of the kind we have studied here restricts the extent to which we can safely infer broad features of B cell evolution in this individual’s response. We have shown with some confidence, from comparison of 6 RBS-directed linages, that early exposure to an H1 strain similar to A/Massachusetts/1/1990 imprinted the response we have studied here, but we have no information about other influenza virus infections between that time and the time of vaccination in 2008 (14). Nonetheless, the relatively large increment in extent of somatic mutations between I-6 and I-3 and between I-7 and I-1 and the convergence of phenotype in the 2 branches strongly suggest that the BCRs from which we inferred the 652 lineage resulted from recall of B cell memory from the primary exposure, either by the 2008 vaccination itself or by infection with a strain circulating within a few years of 2006. In the former case, updating of the recalled memory by affinity maturation following reentry into germinal centers would have yielded the plasmablast/cytes from which the sequences were derived. In the latter case, updating would have occurred during the presumptive infection, yielding high-affinity memory cells that would have differentiated directly into plasmacytes when recalled by the 2008 vaccine.
Methods
Cell Lines.
Human embryonic kidney (HEK) 293F cells were maintained at 37 °C with 5% CO2 in FreeStyle 293 Expression Medium (Thermo Fisher Scientific) supplemented with penicillin and streptomycin. HEK 293T cells were maintained at 37 °C with 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, penicillin, and streptomycin. High Five Cells (BTI-TN-5B1-4) (Trichoplusia ni) were maintained at 28 °C in EX-CELL 405 medium (Sigma) supplemented with penicillin and streptomycin.
Recombinant Fab Expression and Purification.
Synthetic heavy-chain and light-chain variable domain genes were cloned into a modified pVRC8400 expression vector, as previously described (10). Fab fragments were produced either by polyethylenimine facilitated, transient transfection of 293F cells that were maintained in FreeStyle 293 Expression Medium or by Lipofectamine 2000 (Thermo Fisher Scientific) facilitated, transient transfection of 293T cells. Transfection complexes were prepared in Opti-MEM and added to cells. For 293T transfections, Opti-MEM was exchanged for FreeStyle 293 Expression Medium for expression 4 h after transfection. Supernatants were harvested 4–5 d after transfection and clarified by low-speed centrifugation. Fabs were purified by passage over cobalt-nitrilotriacetic acid (Co-NTA) agarose (Clontech) followed by gel filtration chromatography on Superdex 200 column (GE Healthcare) in 10 mM Tris⋅HCl, 150 mM NaCl at pH 7.5 (buffer A).
Recombinant HA Expression and Purification.
All recombinant HA constructs were expressed by infection of insect cells with recombinant baculovirus as previously described (10, 12, 13, 16). In brief, synthetic DNAs corresponding to the globular HA-head domain were subcloned into a pFastBac vector modified to encode a C-terminal rhinovirus 3C protease site and a 6xHis tag.
Supernatant from recombinant baculovirus infected High Five Cells was harvested 72 h after infection and clarified by centrifugation. Proteins were purified by adsorption to Co-NTA agarose resin, followed by a wash in buffer A, elution in buffer A plus 350 mM imidazole (pH 8), and gel filtration chromatography on a Superdex 200 column in buffer A.
For binding and crystallographic studies, the 6xHis tag was removed from HA heads by treatment with PreScission protease (MolBioTech; Thermo Fisher Scientific) and the protein repurified on Co-NTA agarose followed by gel filtration chromatography on Superdex 200 in buffer A to remove the protease, tag, and uncleaved protein.
BLI.
Binding of Fabs with HA heads was analyzed by BLI (BLItz: forteBIO; Pall); all measurements were in buffer A at room temperature. Purified Fab was immobilized on a Ni-NTA biosensor, and HA heads (with tag removed by PreScission protease cleavage) were titrated to obtain rate constants and affinities. KD values were obtained by applying a 1:1 binding isotherm using vendor-supplied software (“Advanced Kinetics” program).
Crystallization.
All proteins were prepared in buffer A for crystallographic experiments. Fab fragments of UCA, I-6, and I-6V were concentrated to a final concentration of ∼12–15 mg/mL. Fab fragments of H2227 and the HA head domain of A/Solomon Islands/03/2006(H1N1) were coconcentrated and stoichiometric 1:1 complexes were purified by gel filtration chromatography on Superdex 200 and concentrated to a final concentration of ∼12 mg/mL. Crystals of an unbound form H2227 were also obtained from this preparation. Fab fragments of UCA and the HA head domain of A/Beijing/262/1995 (H1N1) were coconcentrated in molar ratio of 1:2 to a final concentration of ∼20 mg/mL. Crystals from all other Fab–HA head complexes were obtained by coconcentrating the Fab fragment and HA head domain of A/Beijing/262/1995 (H1N1) in a ∼1–1.3 molar ratio to a final concentration of ∼20 mg/mL. Crystals of unbound I-7 and H1244 Fab fragments were obtained with similarly prepared samples using the HA head of A/Solomon Islands/03/2006. All crystals were grown in hanging drops over a reservoir of the conditions detailed in SI Appendix, Table S1. Crystals were cryprotected in the well solution for crystals obtained in PEG 400 conditions with concentrations ≥30%. Other crystals were cryprotected with glycerol at concentrations of 12–25% in cryoprotectant buffers that were 20% more concentrated than the well solution. These solutions were added directly to the drop, harvested, and flash cooled in liquid nitrogen.
Structure Determination and Refinement.
We recorded diffraction data at the Advanced Light Source (ALS) on beamline 8.2.2 and the Advanced Photon Source on NE-CAT beamlines 24-ID-C and 24-ID-E. Data were processed with XDS (17). Molecular replacement was carried out with PHASER (18). I-6 was phased using a 3-component search with the CHCL from PDB: ID code 4YK4; VH from PDB: ID code 4YK4; and VL from PDB: ID code 4QHK. Three I-6 components CHCL, VH, and VL were used as the Fab components of all other structures. The HA head PDB: ID code 4YK4 was used to phase the H2227 and UCA Fab–HA complexes. The HA head from the UCA–HA head complex was used to phase all other structures that used the A/Beijing/262/1995(H1N1) HA head domain. We carried out refinement calculations with PHENIX (19) and model modifications, with COOT. Refinement of atomic positions and B factors was followed by translation-liberation-screw (TLS) parameterization and placement of water molecules (if warranted). Final coordinates were validated with the MolProbity server (20). Data collection and refinement statistics are in SI Appendix, Table S2. Figures were made with PyMOL (Schrödinger).
Data and Software Availability.
V(D)J sequences for all members of lineage 652 are available at GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession nos. MN178246–MN178253 and MN181516–MN181523. Coordinates and diffraction data have been deposited at the Protein Data Bank (PDB) (ID codes 6Q0E, 6Q0H, 6Q0I, 6Q0L, 6Q0O, 6Q18, 6Q19, 6Q1A, 6Q1E, 6Q1G, 6Q1J, and 6Q1K).
Supplementary Material
Acknowledgments
We thank Garnett Kelsoe (Duke), Simon Jenni (Harvard Medical School), Yoana N. Dimitrova (Harvard Medical School and Genetech), and Lindsey Robinson-McCarthy (Harvard Medical School) for advice, discussion, and instruction; Thomas Kepler (Boston University), for interest and support; and Allan Parelli (Boston Children’s Hospital) for technical assistance. Affinity measurements were carried out in the Harvard Medical School Center for Macromolecular Interactions, directed by Dr. Kelly Arnett. X-ray diffraction data were recorded at beamline 8.2.2 (operated by the Berkeley Center for Structural Biology and the Howard Hughes Medical Institute) at the Advanced Light Source (Lawrence Berkeley Laboratory). We thank beamline staff members for advice and assistance in data collection. The work was supported by NIH Grants P01 AI089618 (to S.C.H.) and U19 AI117892 (subcontract from Boston University to S.C.H.). S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no competing interest.
Data deposition: V(D)J sequences for all members of lineage 652 are available at GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession nos. MN178246–MN178253 and MN181516–MN181523. Coordinates and diffraction data have been deposited at the Protein Data Bank (PDB) (ID codes 6Q0E, 6Q0H, 6Q0I, 6Q0L, 6Q0O, 6Q18, 6Q19, 6Q1A, 6Q1E, 6Q1G, 6Q1J, and 6Q1K).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915620116/-/DCSupplemental.
References
- 1.Eisen H. N., Affinity enhancement of antibodies: How low-affinity antibodies produced early in immune responses are followed by high-affinity antibodies later and in memory B-cell responses. Cancer Immunol. Res. 2, 381–392 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Eisen H. N., Siskind G. W., Variations in affinities of antibodies during the immune response. Biochemistry 3, 996–1008 (1964). [DOI] [PubMed] [Google Scholar]
- 3.McKean D., et al. , Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 81, 3180–3184 (1984). [PubMed] [Google Scholar]
- 4.Jacob J., Kelsoe G., Rajewsky K., Weiss U., Intraclonal generation of antibody mutants in germinal centres. Nature 354, 389–392 (1991). [DOI] [PubMed] [Google Scholar]
- 5.Haynes B. F., Kelsoe G., Harrison S. C., Kepler T. B., B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat. Biotechnol. 30, 423–433 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kepler T. B., Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000 Res. 2, 103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garces F., et al. , Affinity maturation of a potent family of HIV antibodies is primarily focused on accommodating or avoiding glycans. Immunity 43, 1053–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuraoka M., et al. , Complex antigens drive permissive clonal selection in germinal centers. Immunity 44, 542–552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whittle J. R., et al. , Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U.S.A. 108, 14216–14221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt A. G., et al. , Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. U.S.A. 110, 264–269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt A. G., et al. , Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raymond D. D., et al. , Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat. Med. 22, 1465–1469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy K. R., et al. , Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity 48, 174–184.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt A. G., et al. , Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell Rep. 13, 2842–2850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moody M. A., et al. , H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One 6, e25797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H., et al. , Key mutations stabilize antigen-binding conformation during affinity maturation of a broadly neutralizing influenza antibody lineage. Proteins 83, 771–780 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.