Significance
This study demonstrates a therapeutic option using induced pluripotent stem cells (iPSCs), gene editing, and tissue engineering techniques for the development of a long-lasting treatment that will result in the permanent closure of nonhealing wounds in dystrophic epidermolysis bullosa (DEB), especially for recessive DEB (RDEB). Here, we demonstrated that the combination of iPSCs, gene editing, and tissue engineering allows us to perform efficient gene correction by homology-directed repair with the CRISPR-Cas9 gRNA ribonucleoprotein complex system, differentiate the gene-corrected RDEB patient iPSCs into skin cells, and generate human skin equivalents (HSEs) that show skin integrity and type VII collagen restoration after grafting them onto nude mice. The results from this study serve as a foundation to translate this treatment into the clinic.
Keywords: type VII collagen, recessive dystrophic epidermolysis bullosa, CRISPR/Cas9 gene editing, iPSCs
Abstract
Recessive dystrophic epidermolysis bullosa (RDEB) is a severe inherited skin disorder caused by mutations in the COL7A1 gene encoding type VII collagen (C7). The spectrum of severity depends on the type of mutation in the COL7A1 gene. C7 is the major constituent of anchoring fibrils (AFs) at the basement membrane zone (BMZ). Patients with RDEB lack functional C7 and have severely impaired dermal–epidermal stability, resulting in extensive blistering and open wounds on the skin that greatly affect the patient’s quality of life. There are currently no therapies approved for the treatment of RDEB. Here, we demonstrated the correction of mutations in exon 19 (c.2470insG) and exon 32 (c.3948insT) in the COL7A1 gene through homology-directed repair (HDR). We used the clustered regulatory interspaced short palindromic repeats (CRISPR) Cas9-gRNAs system to modify induced pluripotent stem cells (iPSCs) derived from patients with RDEB in both the heterozygous and homozygous states. Three-dimensional human skin equivalents (HSEs) were generated from gene-corrected iPSCs, differentiated into keratinocytes (KCs) and fibroblasts (FBs), and grafted onto immunodeficient mice, which showed normal expression of C7 at the BMZ as well as restored AFs 2 mo postgrafting. Safety assessment for potential off-target Cas9 cleavage activity did not reveal any unintended nuclease activity. Our findings represent a crucial advance for clinical applications of innovative autologous stem cell-based therapies for RDEB.
Dystrophic epidermolysis bullosa (DEB) is a rare genetic skin fragility disorder characterized by blistered skin and mucosa that can be inherited in either a dominant (DDEB) or recessive (RDEB) manner. DEB is caused by mutations in the COL7A1 gene that encodes type VII collagen (C7), a crucial protein that forms anchoring fibrils (AFs) that stabilize dermal–epidermal adhesion at the basement membrane zone (BMZ) (1). There is currently no specific treatment available for DEB. Therefore, correction of COL7A1 mutations would be highly beneficial for affected patients and opens treatment avenues for induced pluripotent stem cell (iPSC)-based therapy.
Current strategies have shown limited potential for treating DEB. For example, gene therapy has been attempted by using a self-inactivating lentivirus or retrovirus to deliver the COL7A1 cDNA into keratinocytes (KCs) and/or fibroblasts (FBs) as a method of direct transplantation of the epithelium or skin (2). However, significant obstacles have delayed the safe use of these strategies for clinical applications. Recent studies have highlighted the potential use of the CRISPR/Cas9 technology in combination with iPSCs as a potential therapy to treat genetic diseases (3, 4). Gene correction techniques rely on the generation of a sequence-specific double-strand break (DSB) in the DNA. The DSB is subsequently repaired by homology-directed repair (HDR) or nonhomologous end-joining. HDR results in the permanent correction of a chromosomal mutation, the gain of function of the defective gene, and the reversal of the disease phenotype (5). Recently, the CRISPR/Cas9 endonuclease system has been shown to be an effective strategy to precisely repair a genomic mutation in human iPSCs derived from patients, using a plasmid strategy (6, 7). We previously knocked out the c.8068del17insGA-mutated allele that causes DDEB, using either CRISPR/Cas9 or TALENs constructs in RDEB patient-derived iPSCs (8).
Recently, a new system based on the Cas9 protein and chemically modified synthetic guide RNA (sgRNA) was developed. Direct use of the Cas9 protein and chemically modified synthetic sgRNA as a ribonucleoprotein (RNP) complex avoids prolonged Cas9 expression in the cells because of its short half-life. This strongly decreases the DNA off-target cleavage, and the modified sgRNA is improved in editing efficiency (9). Therefore, if the correction method is efficient and safe, it can serve as a foundation for adapting this strategy toward clinical use.
iPSCs are the product of reprogramming somatic cells to an embryonic cell-like state (10). iPSCs have unlimited proliferation potential, and their pluripotency allows them to differentiate into different cell lineages for therapeutic applications. Patient-derived iPSCs can be easily corrected using gene-editing tools. Rapid development in this field has produced several new methods in generating integration-free and feeder-free iPSCs, which have potential for therapeutic applications (11). Our group has already demonstrated that iPSCs derived from patients with DEB with subsequent gene correction can be differentiated into KCs and FBs, and can thereby offer potential clinical applications (8, 12–14).
The major challenge for the clinical application of iPSCs is the utilization of animal-derived products and undefined factors during in vitro establishment and expansion of the cell lines. To reduce the risk for graft rejection and immunologic reactions, optimization and standardization of a fully defined animal-free culture for iPSCs is necessary. Elimination of all animal components during derivation and long-term culture of iPSCs is required for future applications of iPSCs in clinical cell therapy. Therefore, developing a new second-generation protocol based on xeno-free (no animal products) culture conditions is needed.
Here, we show the feasibility of precisely correcting COL7A1 mutations using CRISPR/Cas9-mediated HDR in iPSCs through plasmid-based and RNP-based methods. In addition, we provide a preclinical demonstration that grafting 3D HSEs made from gene-corrected patient-derived iPSCs differentiated to keratinocytes (c-iKCs) and fibroblasts (c-iFBs) can restore C7 expression. We also found AF formation at the dermal–epidermal junction 2 mo after grafting.
Results
Generation and Characterization of RDEB Patient-Specific iPSCs.
In our study, we derived iPSCs from primary FBs isolated from the foreskin of healthy individuals and the dermis of 2 patients with RDEB. The first patient harbors a single homozygous frameshift mutation in exon 19 (c.2470insG) (15), and the second bears a compound heterozygous frameshift mutation in exons 19 and 32 (c.2470insG/c.3948insT) (16). The iPSCs were generated by electroporation of integration-free episomal vectors encoding the 4 reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) under feeder-free conditions with an efficiency of 5% (SI Appendix, Fig. S1A). The morphology of all iPSC lines appeared similar to that of human ESCs (small cell shape, a large nuclear-to-cytoplasmic ratio, prominent nucleoli, and tight/flat colonies with rounded edges). The WT-iPSC colonies, as well as RDEB patients’ mutant and corrected iPSC-derived colonies, were identified by the expression of several stem cell markers including OCT4, SOX2, Nanog, and TRA1–81 (SI Appendix, Fig. S1B). We also demonstrated by RT-PCR that additional pluripotent markers were expressed in both normal (WT iPSCs) and RDEB patient iPSCs (SI Appendix, Fig. S1C). Next, we examined the pluripotency of iPSCs by staining for the expression of germ layer-specific markers in embryoid body formation, which have been shown to promote spontaneous differentiation of iPSCs. Our analysis of the embryoid bodies demonstrated that the WT and 2 of our RDEB patient-derived iPSC lines were capable of differentiating into all 3 germ layers in vitro (SI Appendix, Fig. S2A). The RDEB patient-corrected iPSCs and the iPSC-derived KCs and FBs grew at similar rates to WT iPSCs and maintained normal karyotypes (46, XY) (SI Appendix, Fig. S2B). Moreover, we were able to confirm the in vivo differentiation capabilities of our iPSCs by teratoma formation upon intradermal injection of RDEB patient iPSCs into nude mice (SI Appendix, Fig. S2C). We did not detect any differences in the characteristics of normal and RDEB patient-derived iPSCs. Collectively, our findings indicate that the iPSCs we generated closely resembled human ESCs and other reported iPSCs in all respects, suggesting successful generation of iPSCs not only from healthy control individuals but also from 2 patients with RDEB for the first time under feeder-free conditions (8, 12, 17).
Biallelic Gene Correction of the Homozygous Mutations in the COL7A1 Gene, Using a Plasmid Based-CRISPR/Cas9 Approach.
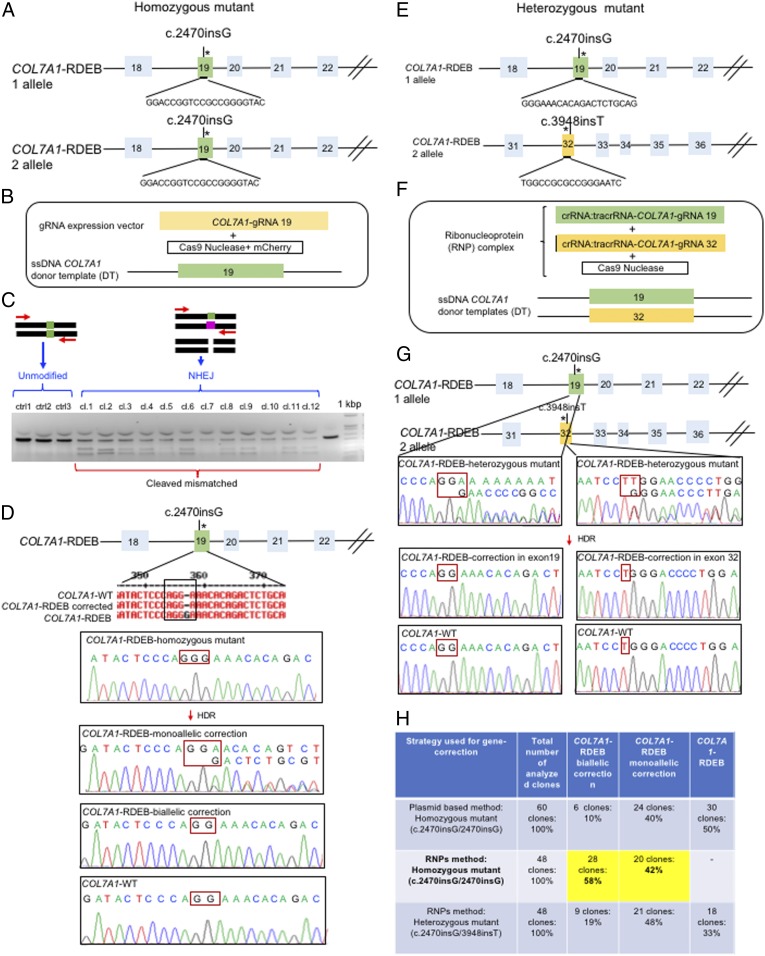
First, we performed CRISPR-mediated genome editing for the correction of the homozygous frameshift mutation in exon 19 in the COL7A1 gene in iPSCs, using a plasmid-based approach to evaluate the efficacy of gene correction in iPSC-derived cells. The selected guide RNAs (gRNAs) were designed using deskgen software (https://www.deskgen.com/landing/#/), following the rule 5′-GN (18–20) NGG-3′, and selected for the absence of off-target effects (Fig. 1A). The specific gRNA sequence for COL7A1 was used as a scaffold to generate a 100-bp fully double-stranded DNA fragment that was incorporated into the gRNA expression vector (SI Appendix, Fig. S4) (18). Next, we electroporated 3 components into single iPSCs: a 178-bp single-strand DNA (ssDNA) as the donor template, plasmids encoding a high-fidelity CRISPR/Cas9 nuclease together with an mCherry on the same plasmid, and a plasmid containing the gRNA (Fig. 1B). The ability of our selected gRNA to induce a site-specific DSB was demonstrated by nonhomologous end joining at the targeted region, and then analyzed by selecting iPSC colonies growing from single iPSCs through FACS without drug selection, using the T7 endonuclease I (T7E1) assay (Fig. 1C). T7E1 cleaves heteroduplexes formed by the hybridization of mutant and wild-type sequences or 2 different mutant sequences, resulting in multiple cleavage products (Fig. 1C). Approximately 80% of the selected clones displayed Cas9 activity, and direct sequencing of the PCR product generated around the targeted region showed that 10% of the clones had undergone biallelic correction and 40% of clones had undergone monoallelic correction of the COL7A1 mutation in exon 19 (Fig. 1 D and H). Moreover, sequence analysis indicated that the target sequence areas were free of off-target mutations (SI Appendix, Figs. S4 and S5).
Fig. 1.
Evaluation of CRISPR/Cas9 gene-correction efficiency using plasmid- and protein-based methods in iPS cells. (A) Schematic representation of the CRISPR target site for the homozygous (c.2470insG) mutation in exon 19 of COL7A. (B) Components used for plasmid-based gene-correction strategy. (C) T7E1 assay after gene editing targeting exon 19 of the COL7A1 gene shows that the Cas9-mCherry nuclease had high cleavage efficiency, demonstrated by the cleavage products of genome editing. (D) Sanger sequencing confirmed various genotypes from targeted colonies in COL7A1-RDEB homozygous mutant iPSCs. (E) Schematic representation of CRISPR target site for heterozygous (c.2470insG/c.3948insT) mutations in exon 19 of COL7A. (F) Components used for protein-based gene-correction strategy. (G) Sanger sequencing confirmed various genotypes from targeted colonies in COL7A1-RDEB heterozygous mutant iPSCs. (H) Summary of the detected genotypes and efficacy after CRISPR/Cas9 gene editing.
Delivery of the Dual sgRNA-Guided Cas9 Nuclease as a Ribonucleoprotein Complex Increases Biallelic Gene Correction in COL7A1.
To improve efficiency, we next examined the feasibility of CRISPR/Cas9-mediated correction, using the Cas9 protein and a chemically modified synthetic gRNA as a RNP complex, rather than as a plasmid. Because the Cas9 RNP degrades quickly after transfection into cells, delivery of the nuclease in this form is a method used to reduce off-target effects and prevent unwanted integration caused by the use of plasmid DNA (9, 19, 20). The Cas9 protein, the synthetic gRNAs (Fig. 1E), and a single-stranded DNA donor template were electroporated into single iPSCs, as described earlier (Fig. 1F). Having done this, we achieved 58% biallelic and 42% monoallelic correction for the homozygous mutation (c.2470insG) in exon 19 of COL7A1, and 19% biallelic and 48% monoallelic correction for the heterozygous mutation (c.2470insG/c.3948insT) in exons 19 and 32 of COL7A1, respectively (Fig. 1 G and H). This strategy creates a “scarless” phenotype without leaving a residual footprint. These include a remaining loxP site after removal of the resistance gene for positive clonal selection or silence, decoy mutations preventing prolonged Cas9 activity, and repair of endogenous mutations with a very high efficiency in single iPSCs without evidence of off-target activity (SI Appendix, Fig. S5).
Directed Differentiation of iPSCs to iKCs under Feeder-Free Conditions.
We next defined a feeder-free and xeno-free differentiation protocol to generate KCs from our iPSCs in vitro. On the basis of our previous studies, we used daily administration of 1 μM retinoic acid to promote ectodermal fate and 10 ng/μL bone morphogenetic protein 4 (BMP4) to block neural fate for 6 d when differentiating iPSCs to KCs (SI Appendix, Fig. S6A) (12). To determine whether the efficiency of differentiation is time-dependent, the iPS-KCs were first kept in culture without passaging for 12, 30, or 60 d. Next, they were subjected to in vitro and ex vivo functional analyses. Only the iPS-KCs that matured for 60 d (iPS 60-KCs) closely resembled normal human KCs (SI Appendix, Fig. S6 B and C). Approximately 96% of iPSC-derived KCs (iKCs) expressed keratin 14, and 50% of the 96% had high levels of p63 expression, showing that this population represents mature KCs (SI Appendix, Fig. S6D). To test the potential of these cells to differentiate ex vivo, the iPS 60-KCs were used to produce HSEs. The HSEs generated from the iPS 60-KCs resemble the architecture of normal skin, as demonstrated by H&E staining with stratified epidermis expressing keratin 14, keratin 10, and loricrin (SI Appendix, Fig. S6E). Taken together, we established a protocol for KC differentiation from iPSCs that we used to derive iKCs from RDEB patient-derived iPSCs in this study. Our data demonstrated that iPSCs can be used to generate iKCs that can then be differentiated into functional cells after 60 d of maturation in vitro. Moreover, we used feeder-free and xeno-free conditions to develop this protocol, thereby improving options for translating stem cell therapies for DEB.
Directed Differentiation of iPSCs to iFBs under Feeder-Free Conditions.
To generate full-thickness HSEs, 2 cellular skin compartments are needed. On the basis of our previous study, we developed a feeder-free differentiation protocol to differentiate FBs from iPSCs (13). We first formed embryoid bodies from iPSCs for differentiation and then used medium supplemented with ascorbic acid and TGFβ2 for 10 d to facilitate mesodermal differentiation, generating FBs from iPSCs (SI Appendix, Fig. S7A). These cells were cultured with regular DMEM without any supplements. After 31 d, we observed FB-like spindle-shaped cells growing from the attached embryoid bodies, and relatively uniform populations of FB-like cells were obtained after several passages (SI Appendix, Fig. S7A). We next examined the characteristics of iPSC-derived cells to define their FB-like properties. The analysis of markers indicative of mesodermal and FB induction, such as COL I, COL III, and Vimentin, were expressed in almost all the WT iPSC-differentiated FBs and COL7A1-RDEB mutant and biallelic-corrected iFB cells (SI Appendix, Fig. S7B). In addition, we compared the expression of FB-associated CD surface markers, CD10, CD44, CD73, and CD90, among normal human FBs and iPSC-derived FBs by FACS (SI Appendix, Fig. S7C). These markers were found in greater than 90% of iPSC-derived FBs with expression patterns consistent with those of normal human FBs.
Functional CRISPR-Based Correction of COL7A1 in Gene-Edited RDEB iPSC-Derived iFBs.
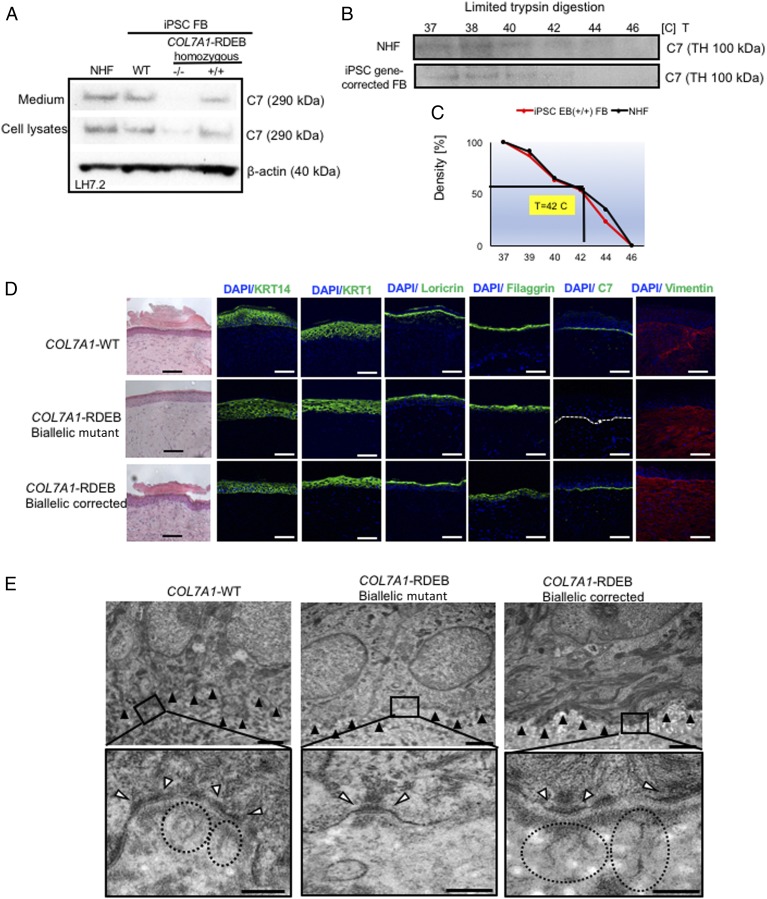
Next, we investigated the potential of c-iFBs to restore functional C7 by Western blot analysis. We assessed C7 expression and secretion levels in corrected RDEB patients’ c-iFBs vs iFBs derived from normal iPSCs and normal human FBs. LH7.2 antibody, specific to human C7, detected the intracellular C7 protein expression and the C7 secretion into the culture in gene-corrected iPSC-derived FBs vs normal human FBs and WT iPSC-derived FBs (Fig. 2A). In addition, the retained stability of triple helical conformation of C7 produced by c-iFBs was shown by the limited trypsin digestion assay, as previously reported (Fig. 2 B and C) (21).
Fig. 2.
Functional verification of gene-corrected RDEB patient iPSC-derived FBs and HSEs. (A) Western blot (WB) analysis assessing type VII collagen (C7) protein expression and secretion in normal human FBs, wild type iPSC-derived FBs (iPSC WT FB), COL7A1-RDEB homozygous mutant (−/−), and corrected (+/+) iPSCs differentiated to FBs. Conditioned medium was collected and directly probed for C7, using polyclonal rabbit LH7.2 antibody (kindly provided by Dr. A. Nystrom). (B) Thermal stability of C7 was analyzed and quantified by (C) limited trypsin digestion of medium from normal human FBs and iPSC gene-corrected RDEB-derived FBs at increasing temperatures, as previously reported. Triple helices (TH) of C7 were analyzed by immunoblot, using polyclonal rabbit NC2 antibody against C7 (kindly provided by Dr. A. Nystrom). (D) Generation of 3D HSEs using gene-corrected RDEB patient iPSC-derived KCs and FBs, which were then grafted onto nude mice are histologically comparable to those generated using iPSC WT KCs/FBs, 2 mo postgrafting. H&E staining revealed normal epidermal and dermal morphology. C7 deposition is demonstrated by immunofluorescence (IF) staining (green signal) 2 mo after grafting, using LH7.2 antibody. Additional IF staining was performed for keratin 14, keratin 10, loricrin, filaggrin, and vimentin on corrected, mutant, and WT xenografts. (E) Transmission electron microscopy was performed on positive iPSC WT KCs/FBs skin grafts, negative COL7A1-RDEB homozygous mutant and COL7A1-RDEB gene-corrected KCs/FBs skin grafts, and gene-corrected RDEB skin grafts, 2 mo postgrafting. (Top) Lower-magnification pictures; black boxes indicate where the higher magnification pictures were taken from. (Scale bar, 1 μm.) (Bottom) High-magnification pictures with an emphasis on the anchoring fibrils (AF), indicated by the dotted circles and white arrowheads point to the hemidesmosomes. (Scale bar, 200 nm.) The BMZ is indicated by black arrowheads.
Collagen VII Deposition and AF Formation Are Restored in 3D HSEs Generated Using Gene-Edited RDEB Patient-Derived KCs and FBs.
To test the functionality of gene-corrected differentiated iPSCs ex vivo, our aim was to build 3D HSEs composed exclusively of iPSC-derived iFBs and iKCs from gene-corrected cells and show their potential to re-express C7 and form AFs compared with HSEs made from iPSCs of healthy control individuals or uncorrected RDEB iPSCs (13). The 3D skin, derived from gene-corrected c-iKCs and c-iFBs, was then grafted onto immunodeficient mice and analyzed 2 mo postgrafting. Expression of C7, epidermal differentiation markers such as keratin 14, keratin 10, loricrin, filaggrin, and vimentin is shown by immunofluorescence staining on xenografts made of biallelic corrected iPSC-derived RDEB KCs and FBs compared with WT and mutant iPSC-derived xenografts 2 mo postgrafting. The iPSC-derived corrected xenografts expressed C7 and fully resemble the WT skin, demonstrating that the gene correction restored the protein function in iPSC-derived iKCs and iFBs. In addition, transmission electron microscopy analysis showed formation of AFs at the BMZ (Fig. 2E). Moreover, we provided safety data to demonstrate that 9 mo postgrafting, the mice grafted with gene-corrected HSEs made from iPSC-derived KCs and FBs did not develop tumors (SI Appendix, Fig. S8). Taken together, this experiment shows that iPSC reprogramming, differentiation, and CRISPR genome editing approaches are safe and nontumorigenic.
Discussion
RDEB caused by single gene mutations in the COL7A1 locus has serious effects on the patient’s quality of life. There are currently no therapies approved for the treatment of RDEB. iPSC technology combined with permanent gene correction gives a foundation for the potential treatment of RDEB, using patient-derived stem cells. To our knowledge, RDEB-derived iPSC correction using CRISPR/Cas9-mediated HDR to repair the COL7A1 mutation in iPSCs without drug selection has not yet been reported. This study shows that grafting gene-corrected HSEs made of gene-corrected iPSC-derived iKCs and iFBs restores C7 expression, functional AF formation, and dermal–epidermal adherence in our preclinical xenograft model. This proof-of-concept study serves as evidence that CRISPR/Cas9 technology has the therapeutic potential to improve RDEB and is an important step toward clinical implementation.
Until now, COL7A1 editing in RDEB through DSB-mediated HDR using the plasmid-based CRISPR/Cas9 system has been demonstrated only in primary FBs and KCs, and has used antibiotic selection to enrich for gene-edited cells (22, 23). However, primary cells have limited growth potential to undergo large-scale expansion and endure genetic manipulation. In addition, the random insertion of plasmid DNA into the host genome causes safety concerns. Fortunately, the cell limitations can be circumvented by using iPSCs, which can be reprogrammed from different cell sources to produce large numbers of desired cell types (24). Moreover, the risks of plasmid DNA can be minimized by using a protein-based RNP delivery system. iPSCs have been successfully reprogrammed from EB patient cells by several groups (8, 22, 25–27, 29, 30), but the seamless gene correction with the CRISPR RNP delivery system without drug selection has not yet been performed. We optimized our correction strategy by moving from a plasmid-based system to a protein-based RNP delivery system. A FACS approach was applied to select the targeted cells without drug selection, and the gene correction efficiency in positive iPSC colonies was increased from 10% to 58% for biallelic correction.
Naturally, this iPSC/RNP strategy carries some risks. iPSC reprogramming can cause genetic instability (31), and RNPs can result in off-target effects. However, in this study, we used a high-fidelity variant of Cas9 that reduced nonspecific DNA contact while retaining on-target activities similar to wild-type SpCas9. Previous studies demonstrate that the vast majority of off-target mutations induced by wild-type SpCas9 were not detected with high-fidelity SpCas9 (32–35). These results suggested us that the high-fidelity SpCas9 can be used as an alternative to wild-type SpCas9, especially for therapeutic applications. Moreover, our sequencing analysis did not detect any off-target mutations in the 5 top-predicted off-target sites. In addition, no tumors were observed on the xenograft mouse model 9 mo postgrafting with gene-corrected HSEs. This experiment shows that iPSC reprogramming, differentiation, and CRISPR genome-editing approaches are safe and nontumorigenic. However, it is possible that there were some undesired CRISPR-generated mutations that were not detected in the present study. For future clinical studies, tests such as oncopanel screenings and/or whole-genome sequencing could be performed on iPSC lines to detect any potential deleterious alterations within the genome at different stages of the manufacturing process; for example, after reprogramming, CRISPR/Cas9 gene editing, or differentiation into KCs and FBs (36).
Stem cell culture systems that rely on undefined animal-derived components introduce variability to the cultures and complicate their therapeutic application. The derivation of human iPSCs and the development of methods to produce iPSCs, combined with their potential to treat human diseases, have accelerated the drive to develop xeno-free (without animal products), chemically defined culture systems. Here, we developed xeno-free culture systems that were successful in supporting the undifferentiated growth of human iPSCs. In addition, we developed methods for xeno-free subculture, cryopreservation, and the differentiation of iPSCs to establish protocols for clinical applications. With this iPSC reprogramming protocol, we can achieve a 5% reprogramming efficiency, which is an improvement over previously published protocol (8).
Efficient differentiation of iPSCs to somatic cells is a crucial step in successful stem cell therapy. Our laboratory previously published iPSC differentiation protocols to iKCs and iFBs for research purposes (12, 13). Here, we developed second-generation differentiation protocols for iKCs and iFBs that will serve as a foundation for the development of therapeutic applications in the future. We demonstrated that iKCs require 60 d postdifferentiation to mature into iKCs expressing keratin 14 and p63 markers. Expression of both markers are needed to achieve 50% differentiation efficiency with reduced variability and better cell quality. For our iFB differentiation protocol without feeder culture, the efficiency of deriving unique FB populations increased from 10% to 50%. All iFBs derived with the protocol expressed all characteristic mesodermal and dermal markers.
Taken together, we demonstrated the feasibility of an iPSC-based gene correction strategy for the treatment of patients with RDEB. Our approach shows a highly efficient and safe method for gene correction, high-fidelity Cas9 (SpyFiCas9) nucleases with no detectable genome-wide off-target effects, a xeno-free, chemically defined culture system that supports pluripotent self-renewal and directed differentiation of iPSCs to somatic cells, and feasibility toward developing autologous therapies for RDEB, such as skin-replacement therapy and local treatment of difficult wounds. The data generated from this study will be used as a foundation for future applications and in preparation for clinical investigation.
Materials and Methods
Ethics Statement.
Informed consent was obtained from all subjects, and approval for this study was provided by the Institutional Review Board of Columbia University in accordance with the Declaration of Helsinki.
Isolation and Culture of FBs.
FBs were grown in DMEM (Gibco) supplemented with 10% FBS (Gibco) and 1% penicillin-streptomycin (Invitrogen). The cells were grown at 37 °C and kept at 5% CO2.
Generation and Characterization of Feeder-Free and Integration-Free iPSCs.
Integration-free iPSCs from fibroblasts were generated using transfection of an integration-free episomal plasmid, as described previously (8). The iPSCs were cultured with iPSCs E8 media supplemented with RevitaCell Supplement (Gibco) for the first 24 h after passaging.
Trilineage Differentiation of iPSCs.
To confirm the in vitro differentiation capacity of iPSCs into all 3 germ layers (ectoderm, mesoderm, and endoderm), a commercially available STEMdiff Trilineage Differentiation Kit was used (StemCell Technologies) following the manufacturer’s protocol.
RNA, DNA Isolation, PCR, and RT-PCR Conditions.
Total RNA was isolated from feeder-free generated iPSCs, using the Qiagen RNeasy Mini Kit, and cDNA was synthesized from RNA, using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions.
CRISPR/Cas9 Gene Editing.
For gene editing, 2 × 106 single, patient-derived iPSCs harboring either the homozygous c.2470insG mutation in exon 19 of COL7A or the heterozygous c.2470insG mutation in exon 19 and c.3948insT mutation in exon 32 of COL7A1 gene were electroporated with 15 sgRNA, 15 μg SpyFi Cas9, 10 μg ssDNA repair donor template, and a plasmid encoding GFP using Amaxa Nucleofector 2 Device (Lonza) transfection system with Nucleofector Kit for human stem cells. The iPSCs were then recovered in E8 medium supplemented with RevitaCell Supplement for 48 h after electroporation. Forty-eight hours posttransfection, the GFP-positive iPSCs were collected via FACS, plated at a low density in 6-well plates, and cultured for 10 d until iPSC colonies were ready to be picked.
Assessment of CRISPR/Cas9 Activity.
T7 endonuclease was used according to the manufacturer’s directions (New England Biolabs).
Off-Target Analysis.
The top 5 nonoverlapping predicted off-target sites were predicted using CRISPR design tool (https://zlab.bio/guide-design-resources).
Feeder-Free Direct Differentiation of iPSCs to KCs.
iPSC monolayer cultures were incubated in E8 medium supplemented with 1 μM all-trans retinoic acid (Sigma) and 10 ng/mL BMP4 (R&D Systems) for 7 d, as previously described (12).
Feeder-Free Direct Differentiation of iPSC to FBs.
To generate feeder-free iPSC-derived FBs, the embryoid bodies were formed, harvested, and plated onto 0.1% gelatin-coated tissue culture dishes and cultured in DMEM with 20% FBS and 0.3 mM ascorbic acid (Sigma-Aldrich) for 10 d. The medium was then switched to DMEM with 20% FBS for 10 d and continued with 10% FBS until the cells became FBs.
Engraftment of iPSC Derived 3D HSEs on Mice.
Next, 0.8 cm skin was removed from the dorsal anterior-posterior midline surface of 8- to 10-wk-old NU(NCR)-Fon1nu nude mice (Charles River, Wilmington, MA), using the pinch-cutting technique. The 3D HSEs were placed on the recipient and secured with 4 sutures (7-0 Nylon) in a simple interrupted pattern around the edge of the graft. To protect the HSE, we used a method described for the establishment of a humanized skin (37, 38), whereby the skin removed from the mouse was freeze-thawed 3 times by placing in DMEM and then inserting into liquid nitrogen before warming in a beaker of water. This decellularized and devitalized the skin, which was placed back over the skin construct on the mouse.
FACS Analysis, Western Blot Analysis, Trypsin Assay, Immunostaining and Imaging, H&E Staining, Transmission Electron Microscopy.
For details see Supplementary data in the SI Appendix online.
All data discussed in the paper are available in the SI Appendix.
Supplementary Material
Acknowledgments
We thank Emily Chang, Jade Hung, Ming Zhang, Wangyong Zeng, Avina Rami, and Emily Holliday for technical assistance. This study was supported by the Epidermolysis Bullosa Research Partnership grant and the Mandl grant from Columbia University (to J.J.). We thank Dr. Brigitte Sallee for her assistance with the human subject protocol for collecting biopsy samples from patients with EB. We are also grateful to Dr. Barbara Corneo and Alejandro Garcia Diaz from the Stem Cell Core Facility at Columbia University Medical Center for their technical advice about gene-editing strategies using the plasmid-based method.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See Commentary on page 26147.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907081116/-/DCSupplemental.
References
- 1.Hovnanian A., et al. , Characterization of 18 new mutations in COL7A1 in recessive dystrophic epidermolysis bullosa provides evidence for distinct molecular mechanisms underlying defective anchoring fibril formation. Am. J. Hum. Genet. 61, 599–610 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rashidghamat E., McGrath J. A., Novel and emerging therapies in the treatment of recessive dystrophic epidermolysis bullosa. Intractable Rare Dis. Res. 6, 6–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim E. J., Kang K. H., Ju J. H., CRISPR-Cas9: A promising tool for gene editing on induced pluripotent stem cells. Korean J. Intern. Med. 32, 42–61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox D. B., Platt R. J., Zhang F., Therapeutic genome editing: Prospects and challenges. Nat. Med. 21, 121–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreno A. M., Mali P., Therapeutic genome engineering via CRISPR-Cas systems. Wiley Interdiscip. Rev. Syst. Biol. Med. 9, e1380 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Byrne S. M., Mali P., Church G. M., Genome editing in human stem cells. Methods Enzymol. 546, 119–138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shinkuma S., Guo Z., Christiano A. M., Site-specific genome editing for correction of induced pluripotent stem cells derived from dominant dystrophic epidermolysis bullosa. Proc. Natl. Acad. Sci. U.S.A. 113, 5676–5681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S., Kim D., Cho S. W., Kim J., Kim J. S., Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi K., Okita K., Nakagawa M., Yamanaka S., Induction of pluripotent stem cells from fibroblast cultures. Nat. Protoc. 2, 3081–3089 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Kogut I., et al. , High-efficiency RNA-based reprogramming of human primary fibroblasts. Nat. Commun. 9, 745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh M., Kiuru M., Cairo M. S., Christiano A. M., Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc. Natl. Acad. Sci. U.S.A. 108, 8797–8802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itoh M., et al. , Generation of 3D skin equivalents fully reconstituted from human induced pluripotent stem cells (iPSCs). PLoS One 8, e77673 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenzel D., et al. , Genetically corrected iPSCs as cell therapy for recessive dystrophic epidermolysis bullosa. Sci. Transl. Med. 6, 264ra165 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Mellerio J. E., et al. , A recurrent frameshift mutation in exon 19 of the type VII collagen gene (COL7A1) in Mexican patients with recessive dystrophic epidermolysis bullosa. Exp. Dermatol. 8, 22–29 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Salas-Alanis J. C., et al. , Frameshift mutations in the type VII collagen gene (COL7A1) in five Mexican cousins with recessive dystrophic epidermolysis bullosa. Br. J. Dermatol. 138, 852–858 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., et al. , One-step biallelic and scarless correction of a β-Thalassemia mutation in patient-specific iPSCs without drug selection. Mol. Ther. Nucleic Acids 6, 57–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L., Mali P., Kim-Kiselak C., Church G., CRISPR-Cas-mediated targeted genome editing in human cells. Methods Mol. Biol. 1114, 245–267 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Zuris J. A., et al. , Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schumann K., et al. , Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. U.S.A. 112, 10437–10442 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacków J., et al. , Gene-corrected fibroblast therapy for recessive dystrophic epidermolysis bullosa using a self-inactivating COL7A1 Retroviral vector. J. Invest. Dermatol. 136, 1346–1354 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Webber B. R., et al. , CRISPR/Cas9-based genetic correction for recessive dystrophic epidermolysis bullosa. NPJ Regen. Med. 1, 16014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hainzl S., et al. , COL7A1 editing via CRISPR/Cas9 in recessive dystrophic epidermolysis bullosa. Mol. Ther. 25, 2573–2584 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z., et al. , Building a microphysiological skin model from induced pluripotent stem cells. Stem Cell Res. Ther. 4 (suppl. 1), S2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itoh M., Kawagoe S., Tamai K., Okano H. J., Nakagawa H., Integration-free T cell-derived human induced pluripotent stem cells (iPSCs) from a patient with recessive dystrophic epidermolysis bullosa (RDEB) carrying two compound heterozygous mutations in the COL7A1 gene. Stem Cell Res. 17, 32–35 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Matsumura W., et al. , Establishment of integration-free induced pluripotent stem cells from human recessive dystrophic epidermolysis bullosa keratinocytes. J. Dermatol. Sci. 89, 263–271 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Sebastiano V., et al. , Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci. Transl. Med. 6, 264ra163 (2014). Erratum in: Sci. Transl. Med. 6, 267er8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tolar J., et al. , Patient-specific naturally gene-reverted induced pluripotent stem cells in recessive dystrophic epidermolysis bullosa. J. Invest. Dermatol. 134, 1246–1254 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tolar J., et al. , Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. J. Invest. Dermatol. 131, 848–856 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umegaki-Arao N., et al. , Induced pluripotent stem cells from human revertant keratinocytes for the treatment of epidermolysis bullosa. Sci. Transl. Med. 6, 264ra164 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Laurent L. C., et al. , Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 8, 106–118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinstiver B. P., et al. , High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slaymaker I. M., et al. , Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J. S., et al. , Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vakulskas C. A., et al. , A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24, 1216–1224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman A. M., et al. , An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abaci H. E., et al. , Human skin constructs with spatially controlled vasculature using primary and iPSC-derived endothelial cells. Adv. Healthc. Mater. 5, 1800–1807 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koffler J., et al. , Improved vascular organization enhances functional integration of engineered skeletal muscle grafts. Proc. Natl. Acad. Sci. U.S.A. 108, 14789–14794 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.