Significance
Artemisinins provide important therapeutic agents for treatment of malaria and have potential for use in other infections and in cancer. Their use is threatened by the potential for resistance to arise, hence understanding their mechanism of action and identifying genetic changes that alter sensitivity are important goals. Our findings suggest that mutations in novel targets can contribute to the emergence of parasites with increased tolerance to artemisinin treatment and that such mutations can confer a fitness advantage, even in the absence of a notable shift in EC50. Our findings also support the idea that inhibition of mitochondrial function may be an important target in Toxoplasma gondii, as previously suggest from studies in yeast and human cancer cells.
Keywords: drug resistance, mutation rate, evolution, genome amplification, mitochondrion
Abstract
Artemisinins are effective against a variety of parasites and provide the first line of treatment for malaria. Laboratory studies have identified several mechanisms for artemisinin resistance in Plasmodium falciparum, including mutations in Kelch13 that are associated with delayed clearance in some clinical isolates, although other mechanisms are likely involved. To explore other potential mechanisms of resistance in parasites, we took advantage of the genetic tractability of Toxoplasma gondii, a related parasite that shows moderate sensitivity to artemisinin. Resistant populations of T. gondii were selected by culture in increasing concentrations and whole-genome sequencing identified several nonconservative point mutations that emerged in the population and were fixed over time. Genome editing using CRISPR/Cas9 was used to introduce point mutations conferring amino acid changes in a serine protease homologous to DegP and a serine/threonine protein kinase of unknown function. Single and double mutations conferred a competitive advantage over wild-type parasites in the presence of drug, despite not changing EC50 values. Additionally, the evolved resistant lines showed dramatic amplification of the mitochondria genome, including genes encoding cytochrome b and cytochrome c oxidase I. Prior studies in yeast and mammalian tumor cells implicate the mitochondrion as a target of artemisinins, and treatment of wild-type parasites with high concentrations of drug decreased mitochondrial membrane potential, a phenotype that was stably altered in the resistant parasites. These findings extend the repertoire of mutations associated with artemisinin resistance and suggest that the mitochondrion may be an important target of inhibition of resistance in T. gondii.
Toxoplasma gondii is a widespread parasite of animals that also frequently causes zoonotic infections in people (1). Serological testing suggests that approximately one-third of the world’s human population is chronically infected (2), although such infections are typically well controlled by the immune response following mild clinical symptoms in the acute phase (3). Chronic infections are characterized by semidormant tissue cysts containing a stage of the life cycle called the bradyzoite, which divides asynchronously and sporadically (4). Current therapies, and induction of a potent immune response, are insufficient to clear these stages and instead they undergo slow turnover to sustain infection over time. Reemergence of chronic infection in patients that are immunocompromised results in serious disease (5, 6), especially when this occurs in the central nervous system where tissue cysts most often occur in neurons (7). Toxoplasmosis is also a threat due to risk of congenital infection, especially in resource-limited regions (8). More severe disease has been reported in some regions of South America, where infections are associated with ocular disease in otherwise healthy individuals (9). Treatment of toxoplasmosis is typically provided by antifolates using a combination of pyrimethamine-sulfadiazine or trimethoprim-sulfamethoxazole, although pyrimethamine has also been used in combination with clindamycin, azithromycin, and atovaquone (also used as monotherapy) (10). Because infections are not commonly transferred from human to human, with the exception of congenital infections that do not occur serially, emergence of drug resistance is rarely a problem, although several examples of clinical failure have been reported, for example in patients given atovaquone as a monotherapy (11, 12).
Combination therapies including artemisinin (ART) are the first line of treatment against malaria (13). Since their original discovery, a wide variety of analogs have been made to improve solubility and other drug-like properties, including artemether, artesunate, and dihydroartemsinsin (14). ARTs are fast acting and effective across multiple life cycle stages; however, they have short half-lives in vivo and hence must be coupled with longer-lasting partner drugs, which reduces the risk of resistance arising (13). ART derivatives share an endoperoxide bond that is essential for activity, as shown by the fact that the deoxyartemisinin derivative losses antimalarial activity (15). More recently, completely synthetic analogs containing a similar ozonide group have been developed and some of these have increased half-lives in vivo (16). ART analogs are also effective in blocking growth of T. gondii in vitro (17–19) and controlling infection in vivo in murine models of toxoplasmosis (20, 21). Although analogs show similar potency ranking, they are effective against T. gondii at ∼50-fold higher concentrations when compared to Plasmodium falciparum (19, 22). This major difference in sensitivity may result from the fact that P. falciparum digests hemoglobulin in the food vacuole, thus releasing heme in high concentrations (23). Although most of the heme is detoxified into a crystalline form called hemozoin, the presumption is that elevated reduced iron (Fe2+) is present in the food vacuole. Although the exact mechanism is unknown, it is thought that free Fe2+ activates the endoperoxide bond (24), resulting in formation of adducts to a variety of protein and lipid targets that result in parasite death (25, 26). There is no analogous process to hemoglobin digestion in T. gondii, which may thus explain its lower sensitivity to ART. Instead, it is possible that heme derived from sources such as the mitochondria plays an important role in activating ART in T. gondii, similar to yeast and tumor cell models, as discussed further below.
Treatment failure in P. falciparum patients treated with ART in Southeastast Asia is associated with delayed clearance, rather than a shift in EC50, and this phenotype is monitored using a ring-stage survival assay (RSA) (27). Laboratory selection of ART-resistant P. falciparum lines lead to identification of mutations in a Kelch13 (K13) ortholog, which can confer resistance when expressed in a wild-type background, and similar mutations are found in high abundance in Southeast Asia (28, 29). Several mechanisms have been proposed to explain the role of K13 in ART resistance, including an enhanced cell stress response leading to up-regulation of the unfolded protein response, decreased ubiquitination, and increased cell survival (30). Consistent with this model, inhibitors of the proteasome synergize with ART (31, 32). However, other studies have reported the development of delayed clearance or dormancy phenotypes in P. falciparum-infected patients without associated K13 mutations (33, 34). Furthermore, genetic crosses between parasite lines bearing sensitive vs. resistant K13 mutations demonstrated a correlation with the RSA assay in vitro but not with clearance of blood infection following treatment of infected Aotus monkeys, suggesting other mechanisms are responsible in vivo (35). It has also been proposed that ART inhibits phosphoinositol 3-kinase (PI3K), and that K13 mutations that confer resistance lead to increased PI3K activity and increased inositol triphosphate and membrane vesicles that may confer protection to ring stages (36). In a separate study of in vitro selection for evolved resistance, mutations were identified in a number of genes, including the actin-binding protein coronin, changes that were shown to confer ART resistance (37). In summary, it seems evident that a number of distinct mechanisms can lead to decreased sensitivity of P. falciparum to ART, thus complicating tracking of the development of resistance in the field.
ART derivatives have also been promoted as potential treatment for cancer as they are active against a wide variety of tumor cells in vitro and a number of specialized analogs have been synthesized (38). The activities of the common ART derivatives against tumor cell lines are in the low micromolar range, comparable to T. gondii but much higher than what is typically observed against P. falciparum. The mechanism of action of ART against tumor cells is also uncertain, but it has been linked to oxidative stress, DNA damage, decreased cell replication, and activation of cell death pathways, including apoptosis (39, 40). Studies in Baker’s yeast (Saccharomyces cerevisiae) implicate the mitochondria as a target of ART, and consistent with this model, growth on fermentable carbon sources results in insensitivity as the mitochondria is dispensable under these conditions (41). Treatment of yeast with ART results in mitochondrial membrane depolarization, leading to the suggestion that ART is activated by the electron transport system, thus leading to oxidative stress and membrane damage (42). Inhibition of mitochondrial function in tumor cells treated with ART derivatives has also been shown to promote cell death due to apoptosis (43). In human tumor cells, it is proposed that mitochondrial heme activates the endoperoxide group of ART and interaction with the electron transport chain generates reactive oxygen species leading to cell death (44).
Here we sought to use the versatility of T. gondii as a genetic system to explore potential mechanisms of resistance to ART. Previous studies have shown that ART resistance can be selected in chemically mutagenized T. gondii parasites (45), although the molecular mechanism for the modest shift in EC50 observed in these resistant clones has not been defined. Chemical mutation introduces numerous changes in the genome, confounding analysis of those that might confer resistance. Consequently, we used a natural evolution process to select for mutations that would confer increased survival in ART, while monitoring the background mutation rate in the absence of selection in parallel. Whole-genome sequencing identified several single nucleotide polymorphisms (SNPs) that arose in the resistant populations and reached fixation over time. Reverse genetic approaches based on CRISPR/Cas9 gene editing were used to show that these mutations confer increased survival, despite the fact that they do not alter EC50 values. These findings define molecular changes associated with ART resistance in T. gondii, which may be informative for understanding mechanism of action of this important class of compounds.
Results
Estimating the Background Mutation Rate in T. gondii.
Prior to selecting for resistant parasites, we wanted first to establish the background mutation rate of T. gondii during a continuous in vitro passage under standard laboratory conditions. Because lines that have been passed for years in the laboratory are likely a mixture of genotypes that contain both private and shared SNPs, as well as potential copy number variations (CNVs), we first cloned the starting population by limiting dilution on monolayers of human foreskin fibroblasts (HFF cells). We obtained clones from the common laboratory RH strain, referred to as 5D and 12B lines, and grew them by sequential passage in HFF cells for the duration of 365 d, subculturing them every 2 to 3 d (Fig. 1A). During in vitro passage in this format, the total population parasite size of a given culture expands to ∼2 × 107 over a time frame of 2 to 3 d. The population is then reduced by 1:10 or 1:20 when a fraction of the growing culture is used to inoculate a new flask of previously uninfected HFF cells. At the time points of 182 and 365 d, separate subclones from the growing populations were isolated by limiting dilution, and these were named with the prefix of the parental line followed by a clone designation (Fig. 1A). Genomic DNA was extracted from all 12 of these clones and subjected to whole-genome sequencing using Illumina technology. In parallel we also sequenced the genomic DNA of the parental strains. The short reads obtained by Illumina sequencing were mapped with high stringency to the reference genome of the ME49 strain of T. gondii (https://toxodb.org/toxo/) using a specially developed pipeline (Materials and Methods). We then compared the frequency of reads that identified SNPs between the reference parental clones (i.e., 5D and 12B) from the subclones that were obtained from these lines following passage. Overall the number of mutations detected in the clones were quite low, with a range from 0 to 8 SNPs seen in each clone (Datasets S1 and S2).
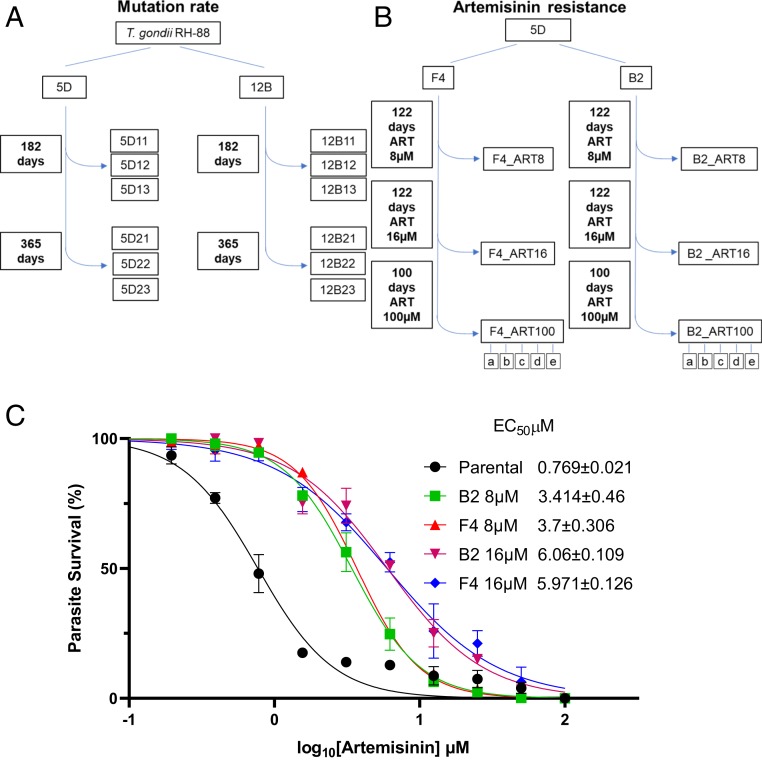
Fig. 1.
Generation of ART-resistant parasite populations. (A) Schematic for growth of parasites used to determine the background mutation rate. Two initial RH88 strain clones (5D and 12B) were sequentially passed in T25 flasks containing HFF monolayers every 48 h. The clones were subcloned by limiting dilution at 182- and 365-d time points. The genomes of the parental strain clones (5D and 12B) and the various subclones were sequenced using Illumina technology. (B) Schematic for selection of ART-resistant parasites. The parasite clone (5D) was further subcloned into 2 lines (F4 and B2) and subjected to 8 μM ART during sequential in vitro passages. The ART concentration was increased to 16 μM and 100 μM at the intervals shown. Populations were frozen for further analysis at the 8- and 16-μM levels and clones (a–e) were made after passage in 100 μM. (C) Dose–response curves for inhibition of T. gondii parental and ART adapted B2 and F4 lines growth in response to increasing concentration of ART. Parasite was monitored by measurement of LDH release from host cells as a consequence of rupture. Data presented as percent LDH release (% LDH) normalized to growth in naïve (untreated) HFF cells. Shown are 3 biological replicates each with technical replicates (n = 9) ±SE. EC50 values were determined using non‐linear regression analysis as a sigmoidal dose–response curve with variable slope. The EC50 data are presented as the average of 3 biological replicates (i.e., separate EC50 titrations), each with 3 technical replicates.
Analysis of the frequency of SNPs was used to derive approximate rates of mutation based on the assumption that the parasite doubles approximately every 6 h (4 generations per day) (46) and that the composite genome is roughly 6.5 × 106 bp (47). We calculated the rates for mutation in individual clones assuming that the mutants arose spontaneously, and that the rates of mutation and growth were constant during the course of the experiment (Datasets S1 and S2). Based on these calculations, the average rates over the course of the 365-d culture period was 6.62 × 10−11 mutations per base pair per generation for the 5D clones and 4.71 × 10−11 mutations per base pair per generation for the 12B clones. We also estimated the composite mutation rate for both sets of clones by treating the frequency of SNPs as mutants in a classic fluctuation analysis yielding a combined rate of 5.79 × 10−11 (Dataset S1). Since both methods give highly similar rates, we used the latter value of 5.8 × 10−11 for subsequent analysis.
In comparing the types of mutations found in each of the lines, there was approximately an equal number of variants found in coding and noncoding regions (19 coding vs. 13 noncoding in 5D and 10 coding vs. 13 noncoding in 12B). However, within coding regions, there was a marked increase in nonsynonymous mutations vs. synonymous mutations with a nonsynonymous/synonymous (dN/dS) ratio of 2.8 for all of the clones derived from 5D and dN/dS ratio of 9 for all of the clones derived from 12B. These high dN/dS ratios suggest strong selective pressure during the culture, although these clones were grown under standard laboratory conditions. In addition, there were some mutations that had apparently reached fixation as they were common to multiple clones from a particular line. For example, in clones from the 12B line, there was a missense mutation in a hypothetical protein that was found in all 6 clones (Dataset S1). Similarly, in clones from line 5D, there were 2 frameshift mutations in hypothetical proteins, 1 missense mutation in a hypothetical protein, and 1 intron mutation that occurred in multiple clones (Dataset S2).
Establishment of ART-Resistant Mutants of T. gondii.
Based on the background mutation rate, we designed an experimental protocol using sequential in vitro culture of relatively large parasite populations in combination with stepwise increases in drug concentrations to derive parasites with elevated ART tolerance. To initiate this trial, we recloned the 5D line to derive 2 parental lines, designated as B2 and F4 (Fig. 1B), and cultured them in HFF cells supplemented with 8 µM ART as a starting point for selecting parasite populations (Fig. 1A). Although growth was initially delayed, as evident from the time required to lyse the host cell monolayer, after 122 d of sequential passage at 8 µM ART both lineages exhibited reproducible growth and rapidly lead to lysis of the monolayer within 2 to 3 d of inoculation. At this time point, we cryopreserved each line for future study; these lines are referred to as F4_ART8 and B2_ART8 (Fig. 1B). We then increased the ART concentration by 2-fold to 16 µM and continued to passage both lines with drug until attaining the phenotype of reproducible growth and infectivity, which took ∼120 d. At this time point, we cryopreserved each line for future study; these lines are referred to as F4_ART16 and B2_ART816 (Fig. 1B). We then increased the concentration of ART to 100 µM and passaged the lines for an additional 100 d, after which they showed normal growth. At this stage, we generated subclones from each of the F4 and B2 lines by limiting dilution (Fig. 1B).
Identification of Candidate Genes Associated with ART Resistance.
To determine the extent of resistance that had developed during selection, we tested the 8-µM and 16-µM populations from both the F4 and B2 lines across a dilution series of ART and calculated their EC50 for growth inhibition. The evolved populations showed an ∼3- and ∼6-fold increase in EC50 in the 8-µM and 16-µM resistant populations, respectively (Fig. 1C). To analyze genetic changes that might have conferred enhanced ART resistance, genomic DNA from all 3 of the selected populations (i.e., 8, 16, and 100 µM from both clones) was extracted and subjected to whole-genome sequencing. Mapping of the reads to the reference genome was used to identify changes from the parental lines that showed altered alleles due to nonsynonymous mutations in the selected populations. Several of the changes were only detected in 1 of the 2 selected lines. For example, mutations were identified in 2 candidate genes in selected lines derived only from the B2 line and in 4 candidate genes derived only from the F4 line (Table 1). The majority of these changes were also only found at the highest level of selection (i.e., 100 µM ART), where they were present with allele frequencies of >90% (Table 1). However, because they were not common to both lines, we did not pursue them further, although it is possible that they contributed to the observed ART-resistance phenotypes observed here.
Table 1.
Mutations found in candidate genes identified by whole-genome sequencing
| Line | Gene ID* | Annotation | Frequency, % (ART 8 μM) | Frequency, % (ART 16 μM) | Frequency, % (ART 100 μM) | Coding region change | Amino acid change |
| B2 | TGME49_290840 | DegP2-serine protease | 68.08 | 98.61 | 100 | 2417G > A | Gly806Glu |
| B2 | TGME49_239420 | Ark1-protein kinase | 70.58 | 100 | 100 | 821G > T | Cys274Phe |
| B2 | TGME49_214110 | Protein yipf5 | 0 | 97.78 | 100 | 578A > C | Glu193Ala |
| B2 | TGME49_310700 | Serine/threonine phosphatase PP1 | 0 | 0 | 100 | 43C > G | Leu15Val |
| F4 | TGME49_290840 | DegP2-serine protease | 50 | 84 | 100 | 2461G > C | Glu821Gln |
| F4 | TGME49_239420 | Ark1-protein kinase | 4.1 | 90 | 100 | 820T > C | Cys274Arg |
| F4 | TGME49_263580 | Bromodomain-containing protein | 0 | 0 | 100 | 1291C > A | Gln431Lys |
| F4 | TGME49_286030 | Hypothetical protein | 6.25 | 100 | 100 | 1018T > G | Ser340Ala |
| F4 | TGME49_264030 | Aminotransferase | 0 | 0 | 94.29 | 1553G > C | Arg518Pro |
| F4 | TGME49_202900 | Zinc finger (CCCH type) motif-containing protein | 0 | 0 | 100 | 2083T > C | Trp695Arg |
| F4 | TGME49_314482 | Hypothetical protein | 0 | 0 | 100 | 305C > G | Ala102Gly |
In addition, we detected nonsynonymous changes in 2 genes that were common to the ART-resistant populations derived from both lines. The first of these was in the gene TGME49_290840, which is annotated in ToxoDB as a serine protease (Table 1). It has high similarity to DegP in P. falciparum (SI Appendix, Fig. S1) and is referred to here as a DegP2 ortholog. The DegP2 ortholog studied here is different from a previously named DegP-like serine protease that is localized in the rhoptry of T. gondii (48). We consider TGME49_290840 to be the more likely mitochondrial DegP2 ortholog based on the presence of a DegP-htrA superfamily motif and detection in the mitochondrial proteome (49). Moreover, a recent study in T. gondii reported that this gene is localized to the mitochondrion and it can be deleted without major loss of fitness (50). The T. gondii DegP2 ortholog was mutated in a different position in each of the F4 and B2 lines, although both nonconservative mutations resided in the PDZ2 domain (DegP2-F4-E821Q and DegP2-B2-G806E) (Table 1). The frequency of the mutated DegP2 alleles was 50% and 68.08% in F4 and B2 8-µM populations, respectively, and these rose to 84% and 98.61% in the 16-µM populations, respectively (Table 1). The second gene identified in both clones is TGME49_239420, which is annotated in ToxoDB as a serine/threonine protein kinase with similarity to calmodulin-dependent and myosin light-chain kinases. Because its function is unknown in T. gondii, it is referred to here as ART resistance kinase 1 (Ark1). The mutations in Ark1 occurred at the same site, which resides in the hinge region of the ATP binding pocket, with a different nonconservative substitution in each lineage (Ark1- F4-C274R and Ark1-B2-C274F). Mutations appeared in Ark1 in 70.58% of the reads in the B2 8-µM population but 4.1% mutant allele reads in the 8-µM population from the F4 line (Table 1). However, in the 16-µM populations Ark1 mutated alleles reached 90% and 100% abundance in F4 and B2 16-µM populations, respectively (Table 1). The frequency of mutations in both genes increased to 100% in the 100-µM ART-selected populations from both the B2 and F4 lines (Table 1).
Validation of the DegP and Ark1 Mutations in Conferring ART Tolerance.
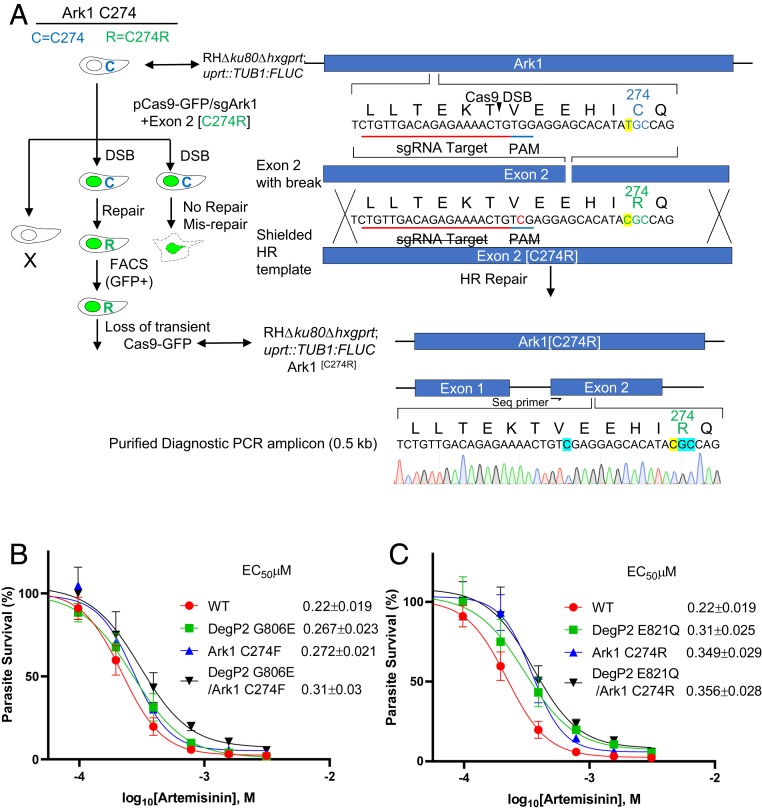
To establish a functional link between the identified mutations and ART resistance, we used a CRISPR/Cas9-based markerless genome-editing strategy to introduce nonsynonymous SNPs into the endogenous loci of the wild-type parental line (Fig. 2A) (51). Two pairs of mutations were created to simulate mutations identified in the B2 and F4 resistant lines, respectively. Each set consisted of clones with single mutants in DegP2, single mutants in Ark1, and double mutants of both DegP2 and Ark1. Initially, we combined the DegP2 G806E and Ark1 C274F mutations seen in the F4 line, either alone or together. We then constructed similar mutants based on the DegP2 E821Q and Ark1 C274R mutations seen in B2. The point mutations were introduced into a wild-type RH ∆ku80 line expressing firefly luciferase (FLUC), so that we could use a luciferase-based growth assay to monitor growth more precisely. The correct mutations were confirmed by restriction enzyme digestion followed by Sanger sequencing of an amplicon flanking the mutated residue. Surprisingly, introduction of any single- or double-point mutation combinations from either lineage resulted only in a slight increase in EC50, ranging from 1.2- to 1.6-fold (Fig. 2 B and C). Hence, introduction of the single- or double-point mutations seen in the original resistant populations does not recapitulate their phenotypes shown in Fig. 1C. One explanation for these results could be the existence of additional mutations that are needed to confer the full level of ART resistance seen in the original populations. However, there were no other mutations found in common (Table 1). Additionally, when we analyzed the genomic sequences of B2 and F4 lines grown at 100 µM ART and their respective clones, we also did not detect any additional novel mutations in coding regions that were common to both lineages (Table 1 and Datasets S3 and S4). It is also evident from analysis of the SNPs in these selected lines that the background rate of mutation is 2- to 3-fold higher, and a number of these are found in a majority of clones, albeit typically not in coding regions (Datasets S3 and S4). Analysis of the mutation frequency in these clones, including those mutations in DegP2 or Ark1, gave a rate of 1.39 × 10−10 for the B2 clones (Dataset S3) and 9.77 × 10−11 for the F4 clones (Dataset S4).
Fig. 2.
Effect of ART on growth of wild-type T. gondii and engineered lines containing point mutations in candidate genes. (A) Schematic illustration of the markerless CRISPR/Cas9 genome-editing strategy used to introduce mutations into the T. gondii chromosome for candidate genes. The scheme depicts introduction of the C274R mutation into the Ark1 gene. The wild-type allele is C274, while the mutant is C274R. CRISPR DSB, targeted Cas9 double-stranded break; HR, homologous recombination. Transformants were isolated by FACS for GFP expression followed by single-cell cloning and confirmation of the editing by diagnostic PCR and sequencing. (B and C) A dose–response curves for inhibition of T. gondii wild-type and point mutant tachyzoites growth in response to increasing concentration of ART. Parasite clones were inoculated in 96-well plates containing HFF cells and growth was monitored by measurement of luciferase activity after 72 h of incubation with different concentrations ART. Data presented as percent relative light units (% RLU) normalized to growth in naïve (untreated) HFF cells. Shown are 3 biological replicates each with technical replicates (n = 9) mean ± SE. EC50 values were determined using nonlinear regression analysis as a sigmoidal dose–response curve with variable slope. The EC50 data are presented as the average of 3 biological replicates (i.e., separate EC50 titrations), each with 3 technical replicates.
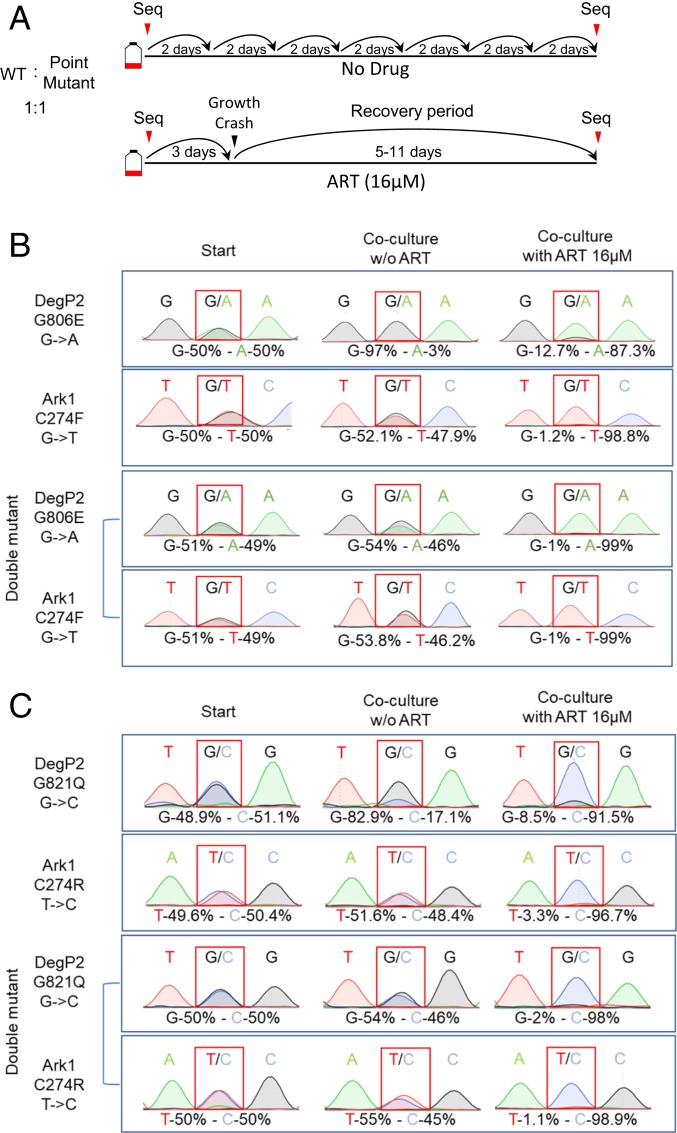
To further characterize the role of these mutations in ART resistance, we performed a growth competition assay in vitro under pressure of ART. In these assays an equal number of wild-type and mutant parasites were cocultured in the presence of 16 µM ART or in vehicle DMSO as a control. When grown in the presence of ART, the mixture of parasites exhibited a slight delay before lysing the monolayer at day 3 vs. the normal day 2 time period. When these parasites were passed onto a fresh HFF monolayer in the presence of ART, they exhibited a growth crash, with variable times of recovery that were dependent on the strains present in culture. Cultures containing DegP2 mutant parasites were the slowest, with a recovery time of ∼11 d. Cultures containing Ark1 mutants exhibited shorter recovery times, ranging between 6 and 8 d. The double mutants in both lineages consistently showed the fastest recovery time, ranging between 5 and 6 d.
To analyze the proportion of the wild-type vs. mutant strains at the initial inoculation and after the recovery from the ART-induced growth crash, genomic DNA was extracted at both time points and the respective loci were PCR-amplified and analyzed by Sanger sequencing (Fig. 3A). Remarkably, all single-point mutants and their double-mutant combinations reproducibly outcompeted the wild-type parental strain by the end of the culture period in the presence of ART (Fig. 3 B and C). Of note, while both DegP2 point mutants showed fitness defect when cocultured with the wild-type strain under control conditions, they were superior to the wild-type in the presence of ART (Fig. 3 B and C). Furthermore, this growth defect was rescued in the double mutant containing Ark1 mutations in addition to DegP2 mutations (Fig. 3 B and C).
Fig. 3.
Growth competition assays between wild-type and the point mutant strains. (A) Schematic for growth competition assay. Parental and point-mutant parasites were inoculated at 1:1 ratio (5 × 105 parasites of each strain) into T25 flasks of confluent HFFs with or without ART 16 μM. After natural egress, parasites were passed by a dilution of 1:10 to 1:20 into a fresh T25 flasks containing confluent HFF cells. Parallel cultures without drug treatment were serially passaged throughout the duration of the experiment. Toxoplasma genomic DNA was extracted at the starting time point and after the recovery from the ART induced growth crash. The mutated loci were amplified by PCR and analyzed by Sanger sequencing. DNA sequence traces were used to estimate the relative proportions of sequence variants for wild-type and mutant alleles. (B and C) DNA sequence traces results for analysis of the ratio of wild-type and the point mutants. Two different competition assays are shown: (B) DegP2 G806E or Ark1 C274F vs. the combined double mutant and DegP2 G821Q or (C) Ark1 C274R vs. the combined double mutant.
T. gondii ART-Resistant Parasites Show Amplification of the Mitochondrial Genome.
The resistance to ART in P. falciparum has been previously shown to be associated with increased pfmdr1 copy number (52). Hence, we considered the possibility that CNVs within the evolved resistant populations could explain the inability of the single- and double-point mutations to perfectly phenocopy the elevated ART resistance. To examine the genome for CNV, we mapped the Illumina reads to the whole-genome assembly containing the chromosomes of T. gondii and examined the read depth as a proxy for copy number. Our initial assessment of CNV within the resistant populations uncovered 4 genes with small regions of elevated read depth in the 100 µM ART-treated clones (SI Appendix, Fig. S2). The amplified regions were quite short, consisting of just 200 to 300 bp, and all 4 repeats resided in introns of genes with no known association with drug resistance (SI Appendix, Fig. S2). These regions were also elevated in the parental B2 and F4 lines, as compared to the rest of the 1X gene, but the read depth increased dramatically in the 100 µM ART-treated clones (SI Appendix, Fig. S2). Interestingly, all 4 repeats had top BLAST hits annotated as cytochrome b (COB) or cytochrome c oxidase (COX1), genes that are normally encoded in the mitochondria. The current T. gondii genome assembly does not include the mitochondrial genome sequence, as its assembly has been complicated by the presence of many short repeats derived from COX1 and COB (53). These genes are normally mitochondrially encoded, but many mtDNA fragments have been amplified and transferred to the nuclear genome of T. gondii as scattered imperfect copies that are often flanked by short-repeat elements (53). Therefore, we realigned the Illumina reads from the control and the resistant population genomes to the genome assembly, but this time included an independently sequenced portion of the mtDNA spanning COB and COX1 (54) and nonassembled supercontigs that likely also contain sequences from the mitochondrial genome. This analysis revealed an amplification of contigs that likely constitute the mtDNA, including genes encoding COX1, COX3, and COB (Fig. 4 and Dataset S5). The degree of amplification was very dramatic and proportional to the resistance level of the parasites, with ART 100-μM populations and clonal genomes showing the highest level of amplification being above that of ART 8-μM and ART 16-μM populations (Fig. 4 and Dataset S5). Furthermore, we confirmed the mtDNA amplification as shown by qRT-PCR (SI Appendix, Fig. S3). Analysis of the mitochondrially encoded genes identified only a few SNPs or short indels in a minority of reads (5 to 10% overall) without any pattern to ART treatment, indicating that the amplifications are not pseudogenes, truncation, or rearrangements, but rather represent increased copies of the intact genome.
Fig. 4.
CNV in the mtDNA in ART-resistant strains. (A) CNV for wild-type B2 parental and ART-resistant lines derived from this parent. Illumina reads were mapped across mtDNA genes COB (GenBank: JX473253.1, 1 to 1,080 bp), COX1 (GenBank: JX473253.1, 1,117 to 2,607 bp), and COX3 (supercontig KE138885). The B2 line was either untreated (blue) or treated with 8 µM (purple), 16 µM (orange), or 100 µM (red) ART. The individual 5 clones “a” through “e” from the 100-µM ART level are shown (green). Copy number estimates are based on the read depth per base pair normalized to 1X across the respective genome. (B) CNV for wild-type F4 parental and ART-resistant lines derived from this parent. Illumina reads were mapped across mtDNA genes COB (GenBank: JX473253.1, 1 to 1,080 bp), COX1 (GenBank: JX473253.1, 1,117 to 2,607 bp), and COX3 (supercontig KE138885). The B2 line was either untreated (blue) or treated with 8 µM (purple), 16 µM (orange), or 100 µM (red) ART. The individual 5 clones “a” through “e” from the 100-µM ART level are shown (green). Copy number estimates are based on the read depth per base pair normalized to 1X across the respective genome.
ART Affects T. gondii Mitochondrial Physiology.
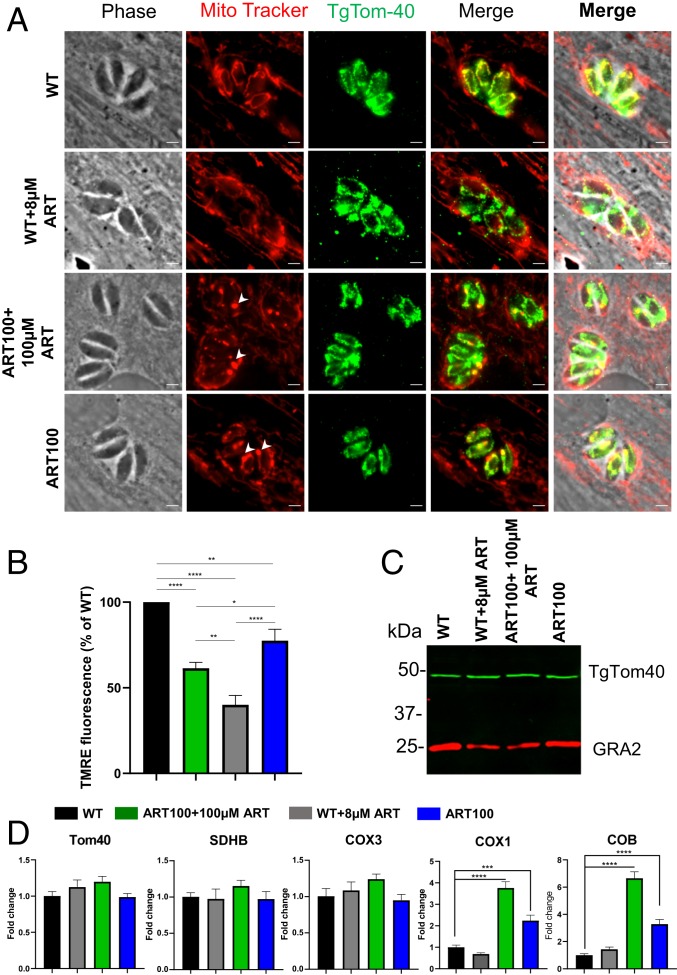
To examine the effect of ART on the mitochondrion, intracellular wild-type parasites were treated for 24 h with 8 µM ART or vehicle, as a negative control (Fig. 5). Due to the fact that the ART-resistant parasite populations were kept under constant drug pressure throughout the duration of this work, we also examined the effect of ART removal for 24 h (Fig. 5). Following the various treatments, cultures were stained with MitoTracker, fixed, and further stained for a previously characterized mitochondrial protein TgTom40 (55). Vehicle-treated wild-type parasites showed brightly fluorescent mitochondria with a characteristic annular morphology evident for both MitoTracker and TgTom40 staining (Fig. 5A). However, treatment of wild-type parasites for 24 h with 8 µM ART resulted in loss of bright MitoTracker staining indicative of a perturbed ΔΨm. In contrast, treatment of wild-type parasites with ART did not alter the staining pattern or the intensity of TgTom40 (Fig. 5A). We then investigated the ART100-resistant population in the presence of 100 µM ART, conditions under which they grow normally. MitoTracker staining revealed a highly atypical pattern consisting of a few bright foci, varying in size with a corresponding reduction in fluorescence intensity in other mitochondrial areas that were still readily stained by TgTom40 (Fig. 5A). Due to the reduced MitoTracker staining in most regions, there was limited overlap with TgTom40 staining (Fig. 5A). Culture of the ART100 population for 24 h in the absence of drug resulted in an increase in MitoTracker signal throughout the parasite mitochondria with the bright foci remaining intact (Fig. 5A). The differences above were uniformly seen within 95% of the vacuoles formed by the ART 100-resistant population. Interestingly, the engineered point mutant strains stained with MitoTracker showed similar staining with a few bright foci varying in size accompanied by reduced fluorescence intensity in other mitochondrial areas (SI Appendix, Figs. S4 and S5).
Fig. 5.
Effects of treatment with ART on Toxoplasma mitochondrial morphology and physiology. (A) HFF monolayers were infected with wild-type or ART100 mutant populations of T. gondii and treated with vehicle or ART for 24 h. Prior to fixation, monolayers were labeled for 30 min at 37 °C with 200 nM MitoTracker. Cells were visualized by fluorescence microscopy. Phase, MitoTracker (red), TgTom40 (green), and merged images are shown. (Scale bars, 2 µM.) (B) TMRE assay for mitochondrial membrane potential. HFF monolayers were infected and treated similarly to A. Data are means ± SEM of n = 3 independent biological experiments done with internal duplicates. Statistical analysis was performed using 1-way ANOVA with Tukey multiple comparisons test (*P < 0.05, **P < 0.005, ****P < 0.0001) compared to the untreated wild-type. (C) Immunoblot of lysates from wild-type and ART100 parasites treated with ART or vehicle for 24 h. Blots were probed with rabbit anti-TgTom40 and anti-rabbit IgG IRDye 800RD (green), mouse anti-GRA2 (used as a reference), and anti-mouse IgG IRDye 680CW (red). Combined channels of the membrane scan are shown. (D) Real-time PCR showing fold-induction of mRNA transcripts in wild-type or ART100 parasites treated with ART or vehicle for 24 h. Comparative cycle threshold values were used to evaluate the fold-change in transcripts using ACT1 as an internal transcript control. Data are plotted as fold-change ±SEM compared with wild-type vehicle-treated cells from at least 3 independent experiments per gene. Statistical analysis was performed using 1-way ANOVA with Dunnett multiple comparisons test (***P < 0.001, ****P < 0.0001) compared to the untreated wild-type.
To quantify the effects of ART treatment on ΔΨm we utilized tetramethylrhodamine, ethyl ester (TMRE), which accumulates in active mitochondria due to their relative negative charge. Following infection of HFF cells, the monolayers were labeled with TMRE the parasites were mechanically lysed and used for spectrophotometry in a microtiter plate. Consistent with the observations made with Mitotracker, ART treatment of wild-type parasites reduced parasite ΔΨm by 60% (Fig. 5B). Furthermore, the ART100 population grown in the presence of 100 μM ART showed significantly higher ΔΨm values compared to those of ART treated wild-type parasites (Fig. 5B). Removal of the drug from the ART100 for the 24-h population lead to a significant increase in ΔΨm, although it did not fully return to wild-type levels (Fig. 5B).
We next asked whether the amplification of the mitochondrial genome leads to a concomitant increase in nuclear-encoded mitochondrial components at both the mRNA and protein levels. Neither the addition of ART to wild-type parasites nor removal of the drug from the ART100-resistant population led to a change in TgTom40 levels when compared to that of the untreated wild-type cells (Fig. 5C). Nuclear-encoded mitochondrial genes (TOM40 and succinate dehydrogenase [ubiquinone] iron-sulfur subunit succinate dehydrogenase SDHB) showed similar transcript levels in wild-type parasites upon addition of ART, and in ART100 populations in the presence or absence of drug (Fig. 5D). In contrast, the ART100 population showed significantly higher transcript levels for the mitochondrially encoded COX1 and COB genes, although not COX3, when compared to wild-type parasites (Fig. 5D). Transcript levels for these genes were also responsive to ART removal, resulting in a significant drop (Fig. 5D), consistent with the change in ΔΨm in ART100 population grown without the drug (Fig. 5B).
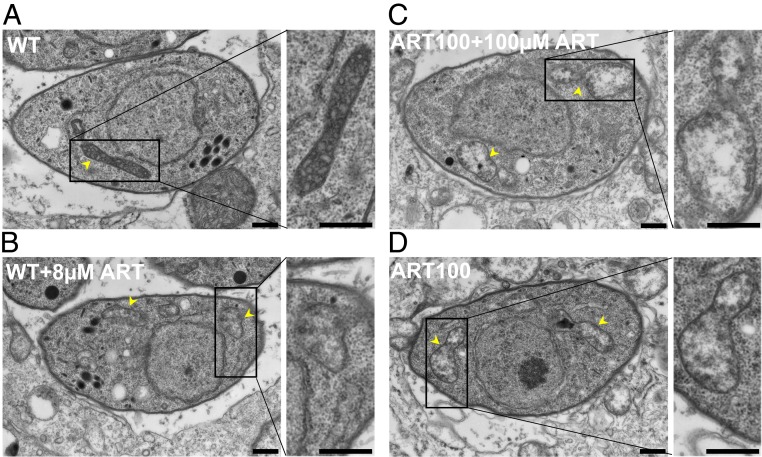
Finally, we characterized ART effects on the ultrastructure of mitochondria by transmission electron microscopy. Mitochondria of wild-type parasites treated with vehicle showed a typical dense matrix with multiple clearly defined cristae (Fig. 6A). Treatment of wild-type parasites with 8 µM ART for 24 h resulted in expansion of the matrix, loss of density, and a reduced number of cristae (Fig. 6B). The mitochondrial ultrastructure of the ART100-resistant population grown in the presence of 100 µM ART presented further reduction in matrix density with only few visible cristae (Fig. 6C). Removal of the drug from the ART100 population for 24 h led to partial increase in matrix density and restoration in cristae numbers (Fig. 6D). The observed ultrastructural alterations in the ART100-resistant population and ART-treated wild-type parasites were detected in the vast majority of the images (90%). All parasites contained a single mitochondrion with a typical elongated tubular morphology with several lobes (the apparent difference in Fig. 6 is due to different numbers of cross-sections obtained by thin section and not a true reflection of copy number).
Fig. 6.
ART effects on T. gondii mitochondria ultrastructure. Ultrastructural changes in parasites following treatment with ART. (A) Electron micrographs of wild-type parasites treated with vehicle (24 h), (B) wild-type treated with 8 µM ART for 24 h, (C) ART100-resistant population with 100 µM ART, (D) ART100-resistant population grown without ART for 24 h. Arrowheads indicate parasite mitochondria. (Scale bars, 500 nm.)
Discussion
We took advantage of flexible in vitro culture systems and genetic tools available in T. gondii to evolve lines that were resistant to ART. In parallel, we determined the background mutation rate after long-term passage in the absence of intentional selection. Surprisingly, spontaneous mutations showed a strong bias for nonsynonymous changes that in some cases were present at a very high frequency among separate clones, suggesting strong positive selection during normal culture. Passage in ART led to enhanced resistance at the population level and whole-genome sequencing of the ART-resistant lines identified several nonconserved SNPs in the coding sequences of DegP2, a serine protease, and Ark1, a serine/threonine kinase of unknown function. We engineered these point mutations into a wild-type background using CRISPR/Cas9 and confirmed that they confer a competitive advantage in the presence of ART, despite not altering the EC50 value for growth inhibition. In contrast, the ART-resistant lines, which had a shift in EC50, also showed up-regulation of heme-containing cytochromes encoded in the mitochondrial genome, consistent with previous studies suggesting a mitochondrial target for ART. Collectively, our studies define mechanisms of mutation that lead to ART resistance, including point mutations in specific genes, and alterations in mitochondrial function.
Analysis of the frequency of mutations in clones of T. gondii that were grown under standard laboratory conditions led to several surprising findings. First, the rate of apparent mutation in T. gondii was relatively low, at ∼5.8 × 10−11 mutations per base pair per generation, which is ∼30-fold lower than that estimated for P. falciparum (1.7 × 10−9) (56). Second, we observed a high level of nonsynonymous relative to synonymous mutations in coding regions, indicating that strong selection is operating under these conditions. Previous studies examining SNPs between different lineages of the RH strain that were passaged for an unknown number of doublings in different laboratories also reported elevated dN/dS ratios that ranged from 2 to 10 (57). Studies conducted on the model organism Escherichia coli have shown that strong selective pressures exist even under homogeneous conditions and that spontaneous mutations often contribute to fitness advantage and reach fixation (58). In the present study, we did not evaluate the fitness of the mutations that reached fixation in nonselected lines, although the high dN/dS ratio is more consistent with improved growth characteristics vs. potential bottlenecks during passage.
To test the importance of mutations in DegP2 and Ark1, we engineered point mutations into a wild-type background using CRISPR/Cas9. Somewhat surprisingly, these introduced point mutations did not shift the EC50 to a similar extent, as seen in the original evolved resistant clones. However, both point mutations did enhance fitness over the wild-type background in a competition assay. This outcome is similar to the phenotype seen in P. falciparum K13 mutations, where “resistant” lines show improved survival in the RSA but do not exhibit a shift in the EC50 (27). The differences in EC50 shift between the evolved lines and the engineered clones may relate to additional background mutations in each of the lines or differences in transcription that were not examined here. In this regard, it would be informative to revert these mutations in the evolved resistant mutants to determine to what extent they are required for the enhanced resistance. Consistent with previous studies in yeast (41) and tumor cells (43, 44), our findings also support the mitochondrion as a target of ART. We observed both a decrease in membrane potential and altered morphology in wild-type parasites treated with ART, in transgenic parasites expressing point mutants associated with increased tolerance, and in resistant populations that arose after long-term selection. Further studies are required to resolve the relative roles of mutations in individual genes vs. amplification of the mitochondrial genome in mediating ART resistance in T. gondii.
The mechanisms by which point mutations in DegP2 and Ark1 confer enhanced survival to ART in T. gondii are unclear, as previous mutations have not been observed in similar targets in other systems. The mutations seen in T. gondii Ark1 occur in the hinge region of the kinase, a critical spot for ATP binding, suggesting they might affect catalytic activity. The substrates of Ark1 are presently unknown and its role in the biology of the parasite has not previously been explored, although it has a fitness score in a genome-wide CRISPR screen that would suggest it is essential (−3.77) (59). DegP2 is a member of the high-temperature requirement A family (HtrA) and it is a periplasmic protease in E. coli (60). In bacteria, DegP is involved in protective stress responses to aid survival at high temperatures (60), a phenotype also reported in yeast (61). Htr orthologs in eukaryotes are typically found in the mitochondria where they function in quality control (62). Given the role of DegP in stress responses, it may play a similar to K13 in ART resistance in malaria, where mutations in K13 propeller domains have been linked to altered stress responses (30).
Our findings provide support for several previously suggested mechanisms of ART resistance and also make several new predictions about the frequency and consequences of mutation in T. gondii. First, we identify point mutations that confer enhanced survival in the presence of ART without shifting the EC50 in a classic resistance phenotype. Second, similar to P. falciparum, the mechanism of resistance to ART in T. gondii appears to be multigenic and may have a link to altered stress responses. Third, ART may inhibit mitochondrial function in T. gondii and alterations in targets there may thus lead to resistance, as previously suggested in yeast and tumor cells. Fourth, although the low intrinsic mutation rate in T. gondii may limit the potential for drug resistance to arise in the clinic, our finding that mutational drift occurs readily in the absence of purposefully applied selection predicts that mutations that may alter phenotypes are likely to accumulate during normal laboratory passage. Collectively, these processes may also drive evolution and affect the occurrence of drug resistance and other biological traits in vivo.
Materials and Methods
Reagents and Antibodies.
All chemicals were obtained from Sigma or Thermo Fisher Scientific unless otherwise stated. MitoTracker Red CMXRos was purchased from Life Technologies. Secondary antibodies used for immunofluorescence were conjugated to Alexa Fluor 488, Alexa Fluor 568, or Alexa Fluor 350 and were purchased from Invitrogen. The following primary antibodies were used: Rabbit polyclonal anti-TgTom40 antibody (a generous gift of Giel Van Dooren, Australian National University, Canberra, ACT, Australia) (55), and mouse monoclonal GRA2 (63). ART (Sigma) was prepared as 250-mM stock in 100% DMSO and stored at −80 °C until use. ART was further diluted in DMEM to achieve the required final concentration with DMSO final concentration of 0.1%.
Parasite and Cell Culture.
The type I T. gondii strain RH-88, transgenic lines reported previously RHΔhxgprtΔku80 (64), or developed here are listed in SI Appendix, Table S1. T. gondii tachyzoites were maintained in HFF monolayers cultured and purified, as described previously (51). All strains and host cell lines were determined to be mycoplasma negative using the e-Myco plus kit (Intron Biotechnology).
Parasite Transfection.
Following natural egress, freshly harvested parasites were transfected with plasmids, using protocols previously described (65). In brief, ∼2 × 107 extracellular parasites were resuspended in 370 μL cytomix buffer were mixed with ∼30 μL purified plasmid or amplicon DNA in a 4-mm gap BTX cuvette and electroporated using a BTX ECM 830 electroporator (Harvard Apparatus) using the following parameters: 1,700 V, 176-μs pulse length, 2 pulses, 100-ms interval between pulses. Transgenic parasites were isolated using a FACSAria II (BD Biosciences) FACS on the basis of Cas9-GFP fluorescence or by outgrowth under selection with pyrimethamine (3 µM) or 5-fluorodeoxyuracil (10 µM), as needed. Stable clones were isolated by limiting dilution on HFF monolayers grown in 96-well plates.
Plasmid Construction and Genome Editing.
All CRISPR/Cas9 plasmids used in this study were derived from the single-guide RNA (sgRNA) plasmid pSAG1:CAS9-GFP, U6:sgUPRT (66) by Q5 site-directed mutagenesis (New England Biolabs) to alter the 20-nt sgRNA sequence, as described previously (67). Primers for plasmids are listed in SI Appendix, Table S2. Separate sgRNA plasmids were made for editing mutations in DegP2 (i.e., pSAG1:CAS9-GFP, U6:sgDegP2 G806E; pSAG1:CAS9-GFP, U6:sgDegP2 [G821Q]) and Ark1 (i.e., pSAG1:CAS9-GFP, U6:sgArk1). To introduce point mutations into endogenous genes, we used a markerless genome-editing strategy: CRISPR/Cas9 combined with a repair template (SI Appendix, Tables S2 and S3).
Generation of FLUC-Tagged Strain, Rhluc.
To tag the RH ΔKU80 strain with FLUC, the reporter plasmid pUPRT::Floxed DHFR-TS*, TUB1:firefly luciferase plasmid (68) was targeted to the UPRT gene in T. gondii by cotransfection with the CRISPR plasmid pSAG1:CAS9, U6:sgUPRT. Parasites were sequentially selected in pyrimethamine (1.0 μM) followed by 5-fluorodeoxyracil (FUDR, 10 μM) and independent clones isolated by limiting dilution.
Immunodetection Methods.
Immunofluorescence microscopy experiments were performed as previously described (69). For staining of intracellular T. gondii, HFF monolayers grown on glass coverslips with MitoTracker Red CMXRos (Thermo), and medium was replaced with prewarmed DMEM containing MitoTracker at a concentration of 200 nM. After 30 min of incubation at 37 °C, cells were washed and then fixed prior to immunofluorescence staining. Western blotting experiments were performed as previously described (69).
TMRE Mitochondrial Membrane Potential Assay.
To quantify the T. gondii mitochondrial membrane potential, HFFs were infected at a multiplicity of infection of 5:1; 24 h postinfection the media was changed for DMEM media containing TMRE at a concentration of 500 nM. After 30 min of incubation at 37 °C, cells were washed 3 times with IC buffer (142 mm KCl, 5 mm NaCl, 1 mm MgCl2, 5.6 mm d-glucose, 2 mm EGTA, 25 mm Hepes, pH 7.4) and the parasites were mechanically released by scraping and syringe passage. The parasites were centrifuged followed by resuspension in IC buffer. Parasites were transferred in triplicate into to 96-well µCLEAR black plates (Greiner Bio International) at 100 µL per well. The wells were analyzed at Ex/Em = 549/575 nm with Cytation3 (Biotek). Mechanically released wild-type parasites without TMRE treatment were used for the estimation of the background signal.
Transmission Electron Microscopy.
For ultrastructural analyses, samples were fixed in 2% paraformaldehyde-2.5% glutaraldehyde (Polysciences) in 100 mM sodium cacodylate buffer, pH 7.2, for 2 h at room temperature and then overnight at 4 °C. Samples were washed in sodium cacodylate buffer at room temperature and postinfection fixed in 1% osmium tetroxide (Polysciences) for 1 h. Samples were then rinsed extensively in distilled water (dH2O) prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella) for 1 h. Following several rinses in dH2O, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella). Sections of 95 nm were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL) equipped with an AMT 8-megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques) as part of the Microbiology Imaging Facility, Washington University in St. Louis.
Real-Time PCR.
Samples were lysed, and RNA was extracted using the Qiagen RNeasy minikit per the manufacturer’s instructions. cDNA was prepared using the Bio-Rad iScript cDNA synthesis kit per the manufacturer’s instructions. Real-time PCR of all of the genes was performed using Clontech SYBR Advantage qPCR premix per the manufacturer’s instructions. Data acquisition was done in QuantStudio3 (Applied Biosystems) and analyzed in QuantStudio design and analysis software (Applied Biosystems). Primers are listed in SI Appendix, Table S2. Comparative threshold cycle (CT) values were used to evaluate fold-change in transcripts using actin as an internal transcript control.
In Vitro Growth Assays.
Lactate dehydrogenase release.
To assess ART sensitivity of the original resistant mutant clones, growth was monitored over a 10-point dose–response curve based on lactate dehydrogenase (LDH) release from the HFF monolayer. Briefly, parasites (100 μL per well) were added to HFF cells grown in a 96-well plate that contained 100 μL of 2× ART concentration (to achieve 1× final ART concentration in 200 μL total well volume containing 0.1% DMSO) and allowed to replicate for 48 h. Parasite growth was quantified by measuring LDH release from host cells as a consequence of rupture, as described previously (70). Supernatants were collected after 48 h and LDH measured using the CytoTox 96 assay (Promega) according to the manufacturer's instructions. Values were normalized to total lysis (100%) and uninfected (0%).
Luciferase assay.
To assess the ART sensitivity, T. gondii of the engineered point mutant strains, growth was monitored over a 10-point dose–response curve using luciferase activity. Briefly, luciferase-expressing parasites (100 μL per well) were added to HFF cells grown in a 96-well plate that contained 100 μL of 2× ART concentration (to achieve 1× final ART concentration in 200 μL total well volume containing 0.1% DMSO) and allowed to replicate for 72 h prior to measuring luciferase activity. Briefly, culture medium was aspirated and replaced with 40 μL of 1× passive lysis buffer (1× PLB, Promega, E1531) and incubated for 5 min at room temperature. Luciferase activity was measured on a Cytation 3 (BioTek) multimode plate imager using the following protocol: Inject 100 μL of luciferase assay reagent, shake 1 s, and read 10 s postinjection.
Data were analyzed using Prism (GraphPad) to determine EC50 values by plotting normalized, log‐transformed data (x axis), using nonlinear regression analysis as a sigmoidal dose–response curve with variable slope. The EC50 data are presented as the average of 3 biological replicates (i.e., separate EC50 titrations) each with 3 technical replicates (i.e., separate wells).
Growth Competition Assays.
Parental and point-mutant parasites were inoculated at 1:1 ratio into T25 flasks of confluent HFFs with or without ART (16 μM). Following growth for several days and natural egress, cultures were passed into a fresh T25 flask at a dilution of 1;10 or 1:20. Parasite cultures growing in the presence of ART exhibited growth crash upon second passage. Parallel cultures of the mixture of wild-type and mutant lines were serially passaged with no drug treatment. Parasite genomic DNA was extracted at the starting time point and after the recovery from the ART-induced growth crash. The target loci were amplified by PCR and analyzed by Sanger sequencing. QSVanalyser (71) was used for the analysis of DNA sequence traces for estimation of the relative proportions of the wild-type and mutant variants.
Genome Sequence Analysis, Variants, and Mutation Rates.
Whole-genome sequencing was performed for parental and resistant clones using Illumina technology, single-nucleotide variants, and CNVs we identified using methods detailed in SI Appendix. Mutation rates were determined based on the frequency of observed mutations compared to total doublings in the resistant lines or using bz-rates (http://www.lcqb.upmc.fr/bzrates) (72) based on equations that have been described previously (73), as described in SI Appendix.
Data Availability Statement.
Sequencing reads were deposited in the Sequence Read Archive repository at the National Center for Biotechnology Information, SRA accession no. PRJNA575881 (74).
Supplementary Material
Acknowledgments
We thank Sebastian Lourido and Jim Ajioka for insightful discussions; Sebastian Lourido for sharing unpublished data; Giel Van Dooren for providing TOM40 antibody; Jennifer Bark for technical assistance; and Wandy Beatty, Microbiology Imaging Facility, for assistance with electron microscopy. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for assistance with genomic sequencing. The Center is partially supported by National Cancer Institute Cancer Center Support Grant P30 CA91842 to the Siteman Cancer Center and by Institute for Clinical and Translational Science/Clinical and Translational Science Award Grant UL1TR002345 from the National Center for Research Resources, a component of the National Institutes of Health, and NIH Roadmap for Medical Research. This work was also supported by a Grant AI118426 from the NIH (to L.D.S.). A.R. was partially supported by a Berg Postdoctoral Fellowship from the Department of Molecular Microbiology at Washington University in St. Louis. M.R.L. was supported in part by Ruth L. Kirschstein Institutional National Research Award T32 GM008666 from the National Institute for General Medical Sciences. This publication is solely the responsibility of the authors and does not necessarily represent the official view of National Center for Research Resources or NIH.
Footnotes
The authors declare no competing interest.
Data deposition: Sequencing reads were deposited in the NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA575881).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914732116/-/DCSupplemental.
References
- 1.Dubey J. P., Toxoplasmosis of Animals and Humans (CRC Press, Boca Raton, 2010), p. 313. [Google Scholar]
- 2.Pappas G., Roussos N., Falagas M. E., Toxoplasmosis snapshots: Global status of Toxoplasma gondii seroprevalence and implications for pregnancy and congenital toxoplasmosis. Int. J. Parasitol. 39, 1385–1394 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Montoya J. G., Liesenfeld O., Toxoplasmosis. Lancet 363, 1965–1976 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Watts E., et al. , Novel approaches reveal that Toxoplasma gondii bradyzoites within tissue cysts are dynamic and replicating entities in vivo. MBio 6, e01155-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Israelski D. M., Remington J. S., Toxoplasmosis in the non-AIDS immunocompromised host. Curr. Clin. Top. Infect. Dis. 13, 322–356 (1993). [PubMed] [Google Scholar]
- 6.Luft B. J., Remington J. S., Toxoplasmic encephalitis in AIDS. Clin. Infect. Dis. 15, 211–222 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Cabral C. M., et al. , Neurons are the primary target cell for the brain-tropic intracellular parasite Toxoplasma gondii. PLoS Pathog. 12, e1005447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torgerson P. R., Mastroiacovo P., The global burden of congenital toxoplasmosis: A systematic review. Bull. World Health Organ. 91, 501–508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glasner P. D., et al. , An unusually high prevalence of ocular toxoplasmosis in southern Brazil. Am. J. Ophthalmol. 114, 136–144 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Dunay I. R., Gajurel K., Dhakal R., Liesenfeld O., Montoya J. G., Treatment of toxoplasmosis: Historical perspective, animal models, and current clinical practice. Clin. Microbiol. Rev. 31, e00057-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gajurel K., Gomez C. A., Dhakal R., Vogel H., Montoya J. G., Failure of primary atovaquone prophylaxis for prevention of toxoplasmosis in hematopoietic cell transplant recipients. Transpl. Infect. Dis. 18, 446–452 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Baatz H., Mirshahi A., Puchta J., Gümbel H., Hattenbach L. O., Reactivation of Toxoplasma retinochoroiditis under atovaquone therapy in an immunocompetent patient. Ocul. Immunol. Inflamm. 14, 185–187 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Eastman R. T., Fidock D. A., Artemisinin-based combination therapies: A vital tool in efforts to eliminate malaria. Nat. Rev. Microbiol. 7, 864–874 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller L. E., Roepe P. D., Artemisinin-based antimalarial drug therapy: Molecular pharmacology and evolving resistance. Trop. Med. Infect. Dis. 4, E89 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser M., et al. , Peroxide bond-dependent antiplasmodial specificity of artemisinin and OZ277 (RBx11160). Antimicrob. Agents Chemother. 51, 2991–2993 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannangelo C., Fowkes F. J. I., Simpson J. A., Charman S. A., Creek D. J., Ozonide antimalarial activity in the context of artemisinin-resistant malaria. Trends Parasitol. 35, 529–543 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Hencken C. P., et al. , Thiazole, oxadiazole, and carboxamide derivatives of artemisinin are highly selective and potent inhibitors of Toxoplasma gondii. J. Med. Chem. 53, 3594–3601 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones-Brando L., D’Angelo J., Posner G. H., Yolken R., In vitro inhibition of Toxoplasma gondii by four new derivatives of artemisinin. Antimicrob. Agents Chemother. 50, 4206–4208 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarciron M. E., Saccharin C., Petavy A. F., Peyron F., Effects of artesunate, dihydroartemisinin, and an artesunate-dihydroartemisinin combination against Toxoplasma gondii. Am. J. Trop. Med. Hyg. 62, 73–76 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Dunay I. R., Chan W. C., Haynes R. K., Sibley L. D., Artemisone and artemiside control acute and reactivated toxoplasmosis in a murine model. Antimicrob. Agents Chemother. 53, 4450–4456 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schultz T. L., et al. , A thiazole derivative of artemisinin moderately reduces Toxoplasma gondii cyst burden in infected mice. J. Parasitol. 100, 516–521 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Holfels E., McAuley J., Mack D., Milhous W. K., McLeod R., In vitro effects of artemisinin ether, cycloguanil hydrochloride (alone and in combination with sulfadiazine), quinine sulfate, mefloquine, primaquine phosphate, trifluoperazine hydrochloride, and verapamil on Toxoplasma gondii. Antimicrob. Agents Chemother. 38, 1392–1396 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sigala P. A., Goldberg D. E., The peculiarities and paradoxes of Plasmodium heme metabolism. Annu. Rev. Microbiol. 68, 259–278 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Klonis N., et al. , Altered temporal response of malaria parasites determines differential sensitivity to artemisinin. Proc. Natl. Acad. Sci. U.S.A. 110, 5157–5162 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ismail H. M., et al. , Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc. Natl. Acad. Sci. U.S.A. 113, 2080–2085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J., et al. , Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun. 6, 10111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witkowski B., et al. , Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: In-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 13, 1043–1049 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ariey F., et al. , A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straimer J., et al. , Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347, 428–431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tilley L., Straimer J., Gnädig N. F., Ralph S. A., Fidock D. A., Artemisinin action and resistance in Plasmodium falciparum. Trends Parasitol. 32, 682–696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bridgford J. L., et al. , Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nat. Commun. 9, 3801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dogovski C., et al. , Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol. 13, e1002132 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breglio K. F., Rahman R. S., Sa J. M., Roberts D. J., Wellems T. E., Kelch mutations in Plasmodium falciparum protein K13 do not modulate dormancy after artemisinin exposure and sorbitol selection in vitro. Antimicrob. Agents Chemother. 62, e02256-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutherland C. J., et al. , pfk13-independent treatment failure in four imported cases of Plasmodium falciparum malaria treated with artemether-lumefantrine in the United Kingdom. Antimicrob. Agents Chemother. 61, e02382-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sá J. M., et al. , Artemisinin resistance phenotypes and K13 inheritance in a Plasmodium falciparum cross and Aotus model. Proc. Natl. Acad. Sci. U.S.A. 115, 12513–12518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mbengue A., et al. , A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 520, 683–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demas A. R., et al. , Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc. Natl. Acad. Sci. U.S.A. 115, 12799–12804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu X., Cao J., Huang G., Zhao Q., Shen J., Biological activities of artemisinin derivatives beyond malaria. Curr. Top. Med. Chem. 19, 205–222 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Efferth T., Cancer combination therapies with artemisinin-type drugs. Biochem. Pharmacol. 139, 56–70 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Efferth T., From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin. Cancer Biol. 46, 65–83 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Li W., et al. , Yeast model uncovers dual roles of mitochondria in action of artemisinin. PLoS Genet. 1, e36 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., et al. , Artemisinin directly targets malarial mitochondria through its specific mitochondrial activation. PLoS One 5, e9582 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu M., Sun L., Zhou J., Yang J., Dihydroartemisinin induces apoptosis in colorectal cancer cells through the mitochondria-dependent pathway. Tumour Biol. 35, 5307–5314 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Mercer A. E., Copple I. M., Maggs J. L., O’Neill P. M., Park B. K., The role of heme and the mitochondrion in the chemical and molecular mechanisms of mammalian cell death induced by the artemisinin antimalarials. J. Biol. Chem. 286, 987–996 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagamune K., Moreno S. N. J., Sibley L. D., Artemisinin-resistant mutants of Toxoplasma gondii have altered calcium homeostasis. Antimicrob. Agents Chemother. 51, 3816–3823 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radke J. R., et al. , Defining the cell cycle for the tachyzoite stage of Toxoplasma gondii. Mol. Biochem. Parasitol. 115, 165–175 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Lorenzi H., et al. , Local admixture of amplified and diversified secreted pathogenesis determinants shapes mosaic Toxoplasma gondii genomes. Nat. Commun. 7, 10147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lentini G., et al. , Characterization of Toxoplasma DegP, a rhoptry serine protease crucial for lethal infection in mice. PLoS One 12, e0189556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seidi A., et al. , Elucidating the mitochondrial proteome of Toxoplasma gondii reveals the presence of a divergent cytochrome c oxidase. eLife 7, e38131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harding C. R., et al. , Genetic screens reveal a central role for heme biosynthesis in artemisinin susceptibility. bioRxiv . 10.1101/746974 (28 August 2019). [DOI] [PMC free article] [PubMed]
- 51.Brown K. M., Long S., Sibley L. D., Plasma membrane association by N-acylation governs PKG function in Toxoplasma gondii. MBio 8, e00375-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui L., et al. , Mechanisms of in vitro resistance to dihydroartemisinin in Plasmodium falciparum. Mol. Microbiol. 86, 111–128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ossorio P. N., Sibley L. D., Boothroyd J. C., Mitochondrial-like DNA sequences flanked by direct and inverted repeats in the nuclear genome of Toxoplasma gondii. J. Mol. Biol. 222, 525–536 (1991). [DOI] [PubMed] [Google Scholar]
- 54.Gjerde B., Characterisation of full-length mitochondrial copies and partial nuclear copies (numts) of the cytochrome b and cytochrome c oxidase subunit I genes of Toxoplasma gondii, Neospora caninum, Hammondia heydorni and Hammondia triffittae (Apicomplexa: Sarcocystidae). Parasitol. Res. 112, 1493–1511 (2013). [DOI] [PubMed] [Google Scholar]
- 55.van Dooren G. G., Yeoh L. M., Striepen B., McFadden G. I., The import of proteins into the mitochondrion of Toxoplasma gondii. J. Biol. Chem. 291, 19335–19350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bopp S. E., et al. , Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 9, e1003293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farrell A., et al. , Whole genome profiling of spontaneous and chemically induced mutations in Toxoplasma gondii. BMC Genomics 15, 354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Good B. H., McDonald M. J., Barrick J. E., Lenski R. E., Desai M. M., The dynamics of molecular evolution over 60,000 generations. Nature 551, 45–50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sidik S. M., et al. , A genome-wide CRISPR screen in Toxoplasma identifies essential apicomplexan genes. Cell 166, 1423–1435.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pallen M. J., Wren B. W., The HtrA family of serine proteases. Mol. Microbiol. 26, 209–221 (1997). [DOI] [PubMed] [Google Scholar]
- 61.Padmanabhan N., et al. , The yeast HtrA orthologue Ynm3 is a protease with chaperone activity that aids survival under heat stress. Mol. Biol. Cell 20, 68–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vande Walle L., Lamkanfi M., Vandenabeele P., The mitochondrial serine protease HtrA2/Omi: An overview. Cell Death Differ. 15, 453–460 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Charif H., Darcy F., Torpier G., Cesbron-Delauw M. F., Capron A., Toxoplasma gondii: Characterization and localization of antigens secreted from tachyzoites. Exp. Parasitol. 71, 114–124 (1990). [DOI] [PubMed] [Google Scholar]
- 64.Huynh M. H., Carruthers V. B., Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell 8, 530–539 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soldati D., Boothroyd J. C., Transient transfection and expression in the obligate intracellular parasite Toxoplasma gondii. Science 260, 349–352 (1993). [DOI] [PubMed] [Google Scholar]
- 66.Shen B., Brown K. M., Lee T. D., Sibley L. D., Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio 5, e01114-e14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Long S., et al. , Calmodulin-like proteins localized to the conoid regulate motility and cell invasion by Toxoplasma gondii. PLoS Pathog. 13, e1006379 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hopper A. T., et al. , Discovery of selective Toxoplasma gondii dihydrofolate reductase inhibitors for the treatment of toxoplasmosis. J. Med. Chem. 62, 1562–1576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown K. M., Sibley L. D., Essential cGMP signaling in Toxoplasma is initiated by a hybrid P-type ATPase-guanylate cyclase. Cell Host Microbe 24, 804–816.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lourido S., Tang K., Sibley L. D., Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J. 31, 4524–4534 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carr I. M., et al. , Inferring relative proportions of DNA variants from sequencing electropherograms. Bioinformatics 25, 3244–3250 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Gillet-Markowska A., Louvel G., Fischer G., bz-rates: A web tool to estimate mutation rates from fluctuation analysis. G3 (Bethesda) 5, 2323–2327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosche W. A., Foster P. L., Determining mutation rates in bacterial populations. Methods 20, 4–17 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosenberg A., Whole genome sequencing of Toxoplasma gondii parasite resistant to artemisinin. Sequence read Archive (SRA) Bioproject PRJNA575881. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA575881/. Deposited 4 October 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads were deposited in the Sequence Read Archive repository at the National Center for Biotechnology Information, SRA accession no. PRJNA575881 (74).