Significance
Bioluminescence is generated by luciferases that oxidize a specific luciferin. The enzymes involved in the synthesis of the luciferin from widespread cellular metabolites have so far been identified for only 2 bioluminescence systems, those of bacteria and fungi. In these cases, the complete reaction cascade is genetically encodable, meaning that heterologous expression of the corresponding genes can potentially produce autonomous bioluminescence in cell types other than the bacterial or fungal host cells. However, the light levels achieved in mammalian cells so far are not sufficient for single-cell applications. Here we present autonomous bioluminescence images of single mammalian cells by coexpression of the genes encoding the 6 enzymes from the bacterial bioluminescence system.
Keywords: bioluminescence, luciferase, lux, microscopy
Abstract
Bioluminescence-based imaging of living cells has become an important tool in biological and medical research. However, many bioluminescence imaging applications are limited by the requirement of an externally provided luciferin substrate and the low bioluminescence signal which restricts the sensitivity and spatiotemporal resolution. The bacterial bioluminescence system is fully genetically encodable and hence produces autonomous bioluminescence without an external luciferin, but its brightness in cell types other than bacteria has, so far, not been sufficient for imaging single cells. We coexpressed codon-optimized forms of the bacterial luxCDABE and frp genes from multiple plasmids in different mammalian cell lines. Our approach produces high luminescence levels that are comparable to firefly luciferase, thus enabling autonomous bioluminescence microscopy of mammalian cells.
Bioluminescence is an enzyme-catalyzed process by which living cells can produce light. It requires a luciferin substrate that is oxidized by a luciferase, a reaction during which the bioluminescence light is emitted. In addition to the luciferase, several other enzymes participate in the process of bioluminescence by synthesizing the luciferin from cellular metabolites and by converting it back into its reduced form. For most bioluminescence systems, these proteins have not fully been identified so far, with the exception of the bacterial (1–5) and the recently solved fungal systems (6). Knowledge of all enzymes participating in the reaction cascade means that they can, in principle, be heterologously expressed in any cell type in order to produce autonomous bioluminescence for imaging applications. This circumvents the problems associated with external luciferin supply, such as elevated background signal due to nonenzymatic luciferin oxidation in solution (7) or decay of the signal over time upon luciferin consumption (8, 9).
Of particular interest for numerous applications in medical research (10) is the bioluminescence imaging of mammalian cells. However, neither the bacterial nor the fungal bioluminescence systems are widely used for this purpose. In the case of fungal bioluminescence, autonomous bioluminescence imaging of mammalian cells has not been demonstrated so far. One obstacle for this is that fungal luciferase loses its activity at temperatures above 30 °C (6). The bacterial bioluminescence system has been implemented in mammalian cells (11, 12), but the resulting bioluminescence is much dimmer than that produced by other luciferases which require an external luciferin (13). The low brightness of bacterial bioluminescence in mammalian cells impedes, particularly, single-cell imaging applications due to insufficient contrast.
Many scientists in the field have contributed to elucidating the biochemistry of bacterial bioluminescence, which provides the basis for the present work (for reviews, see, e.g., refs. 14–18). The light emitter in bacterial bioluminescence is assumed to be FMN-C4a-hydroxide (19, 20) which is formed in its excited state during the bioluminescence reaction and emits a photon upon return to its ground state. During this reaction, reduced flavin mononucleotide (FMNH2) is oxidized to flavin mononucleotide (FMN), and a long-chain aliphatic aldehyde (RCHO) is oxidized to the corresponding carboxylic acid (RCOOH),
| [1] |
Both products are recycled under consumption of cellular energy. This is performed by a flavin reductase which reduces the oxidized FMN to FMNH2 as well as by the fatty acid reductase complex that converts the acid back into the aldehyde,
| [2] |
| [3] |
The involved enzymes are coded by the lux operon. The genes luxA and luxB encode the luciferase subunits, whereas luxC, luxD, and luxE encode the subunits of the fatty acid reductase complex. FMNH2 is either produced by the FMN reductase coded by luxG or by other cellular enzymes.
Although none of the required substrates is exclusively found in bacteria and hence bacterial bioluminescence should, in principle, be functional in other cell types as well, expression of the lux genes in eukaryotic cells has only been moderately successful so far. It has been demonstrated that codon optimization of the lux genes improves their expression in mammalian cells (11, 12, 21), yet the achieved brightness has been comparatively low (12, 13), hampering the prospects for single-cell visualization by microscopy. In addition to variations in codon usage between different cell types, the requirement for simultaneous multigene expression of all Lux proteins at high levels poses further challenges for the use in mammalian cells. Here, we demonstrate that codon-optimized (co) lux genes expressed from individual plasmids in an optimized ratio produce bright autonomous bioluminescence in different mammalian cell lines. The resulting signal is comparable to that of firefly luciferase and enables imaging of single cells over extended time periods. Moreover, it allows the observation of toxic compounds effects and starvation on cell viability and metabolic activity.
Results
Expression of Lux Proteins in Mammalian Cells.
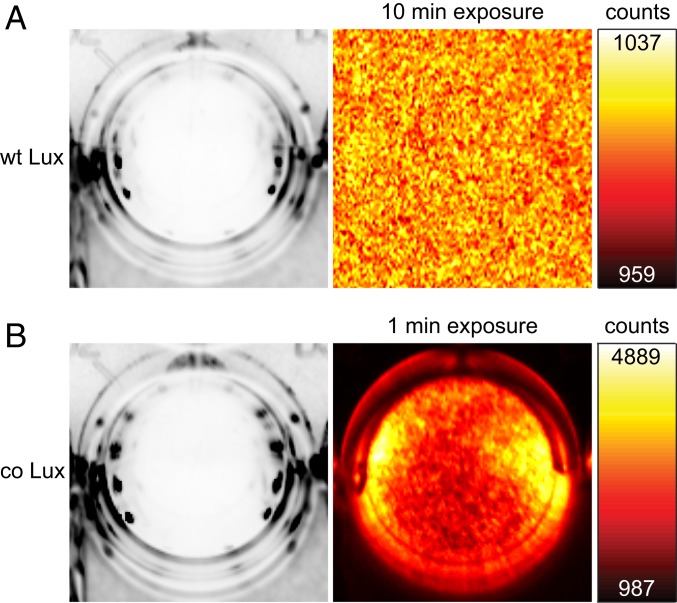
Since the wild-type lux genes have been shown to be poorly expressed in mammalian cells in their native form (11), we generated co versions of the luxCDABE genes from Photorhabdus luminescens and the NADPH (nicotinamide adenine dinucleotide phosphate)-flavin oxidoreductase gene frp from Vibrio campbellii using the codon adaptation tool JCat (http://www.jcat.de) (22). The resulting genes (SI Appendix, Fig. S1) were synthesized, cloned separately into the expression vector pcDNA3.1(+), and cotransfected in equal amounts into HEK 293 cells. While expression of the wild-type genes did not result in detectable bioluminescence levels, the optimized genes produced high signal (Fig. 1). Assuming that a signal 3-fold higher than the SD of the background (∼8 counts per pixel for 10-min exposure time) would have been readily detectable, comparison of the camera counts between wild-type (no signal) and co lux (∼3,000 counts per pixel for 1-min exposure time; Fig. 1) indicated that the gene optimization improved the light output by at least 3 orders of magnitude. This demonstrates that the Lux proteins are functional in mammalian cells, but that their expression is highly dependent on the codon usage or possibly other factors determined by the nucleotide sequence. Variation of the ratios of the transfected plasmids while keeping the total amount of DNA constant indicated that the brightness was increased by 66% if a 3-fold molar amount of the fatty acid reductase components luxC, luxD, and luxE was used (Fig. 2). Therefore, synthesis of the aldehyde seems to be the most rate-limiting step of the overall reaction cascade, rather than the luciferase-catalyzed bioluminescence reaction itself.
Fig. 1.
Bioluminescence of wild-type Lux and co Lux. HEK 293 cells were transfected with (A) wild-type lux and frp genes without codon optimization or (B) the co lux genes in a ratio of luxA:luxB:luxC:luxD:luxE:frp = 1:1:3:3:3:1. Cells were imaged in a 24-well plate with an Amersham Imager 600. (Left) White-light images of the respective well with a monolayer of cells, and (Right) bioluminescence signal after the indicated exposure time. The color maps were scaled to the minimum and maximum camera counts per pixel of the bioluminescence images.
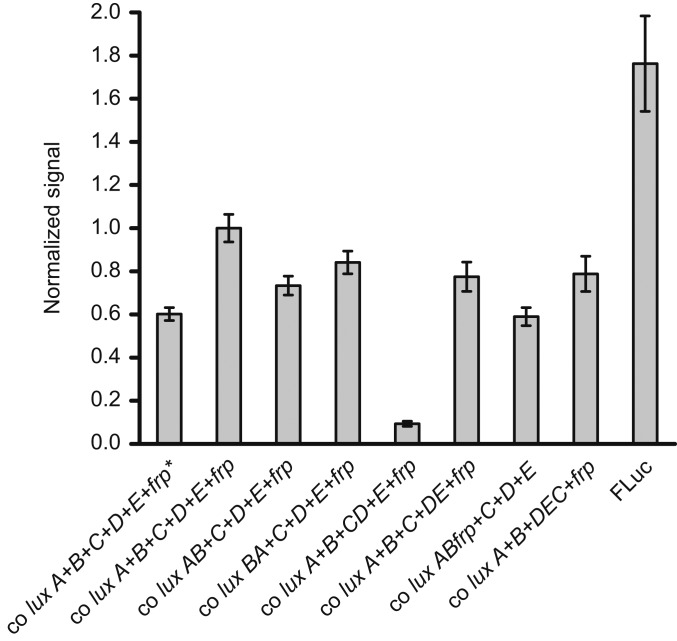
Fig. 2.
Bioluminescence of HEK 293 cells expressing different combinations of co lux genes or FLuc from pcDNA3.1(+). Cells were grown in 24-well plates, transfected with the same total amount of DNA, and imaged with an Amersham Imager 600. Asterisk (*) indicates transfection of all plasmids in equal amounts. Otherwise, plasmids encoding LuxA, LuxB, LuxC, LuxD, LuxE, and Frp were transfected in a ratio of 1:1:3:3:3:1. Plasmids encoding 2 or 3 genes were transfected in 2- and 3-fold amounts, respectively. The following abbreviations are used: luxAB, luxA-P2A-luxB; luxBA, luxB-P2A-luxA; luxCD, luxC-P2A-luxD; luxDE, luxD-P2A-luxE; luxABfrp, luxA-P2A-luxB-T2A-frp; luxDEC, luxD-P2A-luxE-T2A-luxC. Plus (+) signs indicate expression from separate plasmids. For imaging of FLuc, 150 µg/mL d-luciferin was added to the medium. Error bars represent SD of the signal of 5 separate wells.
Since simultaneous cotransfection of 6 different plasmids reduces the transfectable amount for each gene, we tested whether the brightness can be increased by reducing the number of plasmids and expressing more than one gene from the same vector. For this purpose, we linked several genes by viral 2A sequences, as this approach has been reported to increase the transfection efficiency and brightness (12). The 2A sequence impairs the formation of a peptide bond during translation and thus results in the synthesis of 2 separate proteins, with the major part of the peptide remaining attached to the C terminus of the first protein (P2A: ATNFSLLKQAGDVEENPG↓P, T2A: EGRGSLLTCGDVEENPG↓P) (23). Although the transfected amount of the combined genes was accordingly increased, none of the tested combinations showed an increase in bioluminescence (Fig. 2). This might be caused by a lower transfection efficiency, decreased translation rates, or impaired function of the Lux proteins due to the remaining 2A fragments. Consequently, maximum brightness was achieved when all 6 proteins were expressed from separate plasmids (Fig. 2).
We compared the brightness of co Lux to the widely used firefly luciferase (FLuc). Although FLuc was transfected in a 12-fold molar amount compared to the luxAB luciferase subunits, its brightness was only 76% higher (Fig. 2). Hence, bacterial luciferase is functional in mammalian cells when efficiently expressed and produces signal comparable to firefly luciferase if cellular production of the substrates is high enough.
Since bioluminescence systems based on the exogenous luciferin coelenterazine often suffer from background signal due to autooxidation of coelenterazine in solution, we compared its background to the co Lux system. For this purpose, we measured the signal from nontransfected cells with 5 µM coelenterazine and from cells transfected with the FMNH2-producing co frp (SI Appendix, Fig. S2). Whereas frp expression resulted in no detectable bioluminescence emission, nonenzymatic coelenterazine oxidation produced clear signal. Therefore, the low background of the co Lux system allows its bioluminescence detection with a higher specificity than coelenterazine-based systems.
As co versions of the lux genes have been previously expressed in mammalian cells (11, 12), we compared the brightness of our co Lux system to pCMVLux (12). pCMVLux is a humanized lux expression plasmid that contains all 6 luxCDABE and frp genes separated by 2A sequences and produces the highest levels of autonomous bioluminescence in mammalian cells reported so far. Expression of pCMVLux in HEK 293 cells was not detectable with our bioluminescence imager at 37 °C (SI Appendix, Fig. S3). Weak signal (on average, ∼20 counts above background in 10 min) was only detectable after the cells had cooled down to room temperature, but the overall brightness was still ∼3 orders of magnitude lower than with our co Lux system (SI Appendix, Fig. S3 and Fig. 1). In order to find out the main reasons for this large difference, we first compared the codon adaptation indices (CAIs) of our co lux genes and the humanized genes from pCMVLux (referred to as hlux) and the nonoptimized wild-type genes (SI Appendix, Table S1). While the low CAIs of the nonoptimized genes (average 0.13) may account for poor expression and consequently nondetectable light levels, the differences of the CAIs of the hlux and co lux genes are smaller (average 0.74 and 0.95, respectively). Therefore, differences in codon usage may not be the only factor that contributes to the decreased brightness from the hlux genes. To test the influence of the 2A sequences contained in pCMVLux on the bioluminescence levels, we cloned the individual hlux genes without the 2A sequences into pcDNA3.1(+), analogous to our co lux genes. Cotransfection of the hlux pcDNA3.1(+) plasmids in HEK 293 cells produced detectable bioluminescence, but only 1 to 2% of that from the co lux pcDNA3.1(+) plasmids (SI Appendix, Fig. S4). This suggests that the 2A peptides are one important factor that reduces expression or activity of the Lux proteins, in accordance with the results from our co lux 2A constructs (Fig. 2). In addition, we combined single hlux pcDNA3.1(+) plasmids with the remaining co lux pcDNA3.1(+) plasmids to test the influence of the individual hlux genes on bioluminescence levels (SI Appendix, Fig. S4). All hlux combinations exhibited decreased brightness compared to co lux, with the brightness lowest for hfrp. While the observed differences may partly be attributed to the lower CAIs of the hlux genes, other reasons may be responsible for the strongly decreased light emission with the hfrp gene. The most likely explanation is a lower functionality of the protein in mammalian cells due to multiple differences in the protein sequence, since a different host strain was chosen for hfrp (Vibrio harveyi instead of V. campbellii for co frp). Overall, our co Lux system provides an improvement in brightness of at least 3 orders of magnitude compared to previous approaches of lux gene expression in mammalian cells.
Test of Toxic Effects Caused by the Expression of co lux.
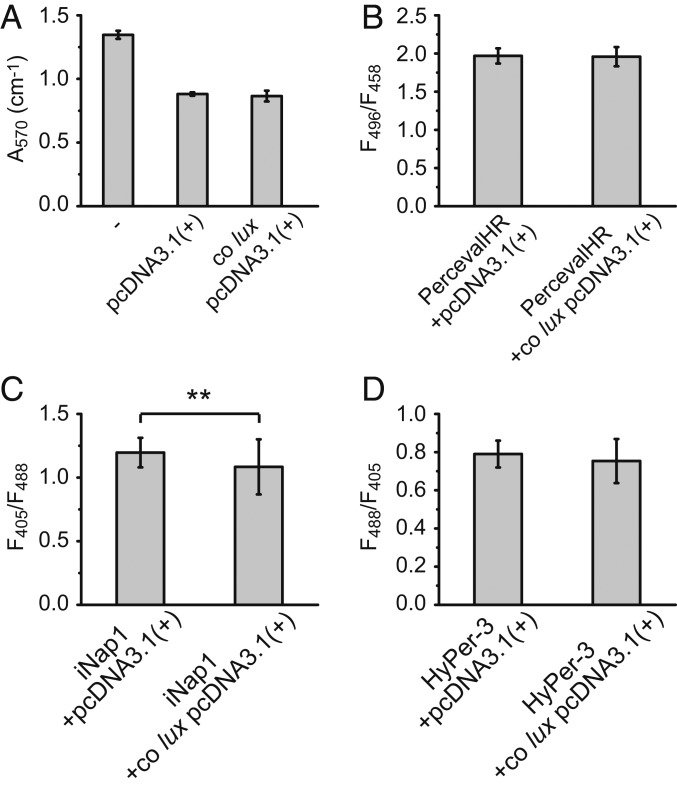
Since the bacterial bioluminescence reaction requires a long-chain aldehyde substrate that is potentially cytotoxic, we tested whether the expression of co lux has adverse cellular effects. First, we performed an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay based on the biochemical reduction of the tetrazolium dye MTT by NAD(P)H. Decreased cell growth and viability result in lower formation of the reduced formazan, which can be detected as a lower absorption. HeLa cells transfected with co lux or the empty pcDNA3.1(+) vector using Lipofectamine 2000 did not show significant differences (P > 0.5) in MTT reduction (Fig. 3A). However, lysates of untransfected cells exhibited a 53% higher absorption, indicating that the only toxicity observed is caused by the transfection reagent rather than expression of the co lux genes.
Fig. 3.
Co lux toxicity control measurements. HeLa cells were cotransfected with co lux genes in pcDNA3.1(+) or the empty vector, and a fluorescent sensor where indicated. (A) Absorption of cell lysates from MTT assay with untransfected cells (−) for comparison. Error bars represent SD of 3 separate coverslips. (B–D) Comparison of (B) ATP, (C) NADPH, and (D) H2O2 levels by ratiometric fluorescence imaging of the sensors (B) PercevalHR, (C) iNap1, and (D) HyPer-3 using the indicated excitation wavelengths. Error bars represent SD of 50 cells. ** represents a P value of <0.01 as calculated by a 2-tailed Student’s t test.
Since toxic effects may involve more subtle cellular changes due to depletion of ATP (adenosine triphosphate) or NADPH or increased production of hydrogen peroxide due to reaction of the luciferase-bound FMNH2 with molecular oxygen (17, 24), we compared their cellular levels with and without co lux expression. For this purpose, we cotransfected HeLa cells with a ratiometric fluorescent sensor for ATP, NADPH, or H2O2 and imaged them by fluorescence microscopy (SI Appendix, Fig. S5). For imaging of ATP, we used the sensor PercevalHR which measures the cellular ATP-to-ADP (adenosine diphosphate) ratio. ATP and ADP binding increases its fluorescence excited at 500 nm and 420 nm, respectively; there is a fixed isosbestic point at ∼455 nm (25). NADPH concentration was imaged with the sensor iNap1, which increases its fluorescence excited at 420 nm and decreases its fluorescence excited at 485 nm upon NADPH binding (26). H2O2 levels were compared using the sensor HyPer-3 which increases its fluorescence ratio F500/F420 upon excitation at 500 and 420 nm (27). No significant change of the fluorescence ratios between cells transfected with co lux or empty pcDNA3.1(+) vector was observed for ATP and H2O2 (P > 0.05; Fig. 3 B and D). In the case of NADPH, we found a small (9%) but significant (P < 0.01) change in the iNap fluorescence ratio (Fig. 3C). The reason for this might be increased NADPH consumption for the synthesis of the aldehyde or FMNH2.
Overall, no major toxic effects of co lux expression were observed. Aldehyde toxicity might not be observed because its concentration is too low. The reason for this could be relatively low aldehyde production by the fatty acid reductase or fast back-conversion of the aldehyde into the acid by the bioluminescence reaction if the activity of all Lux proteins is well balanced.
Single-Cell Bioluminescence Imaging.
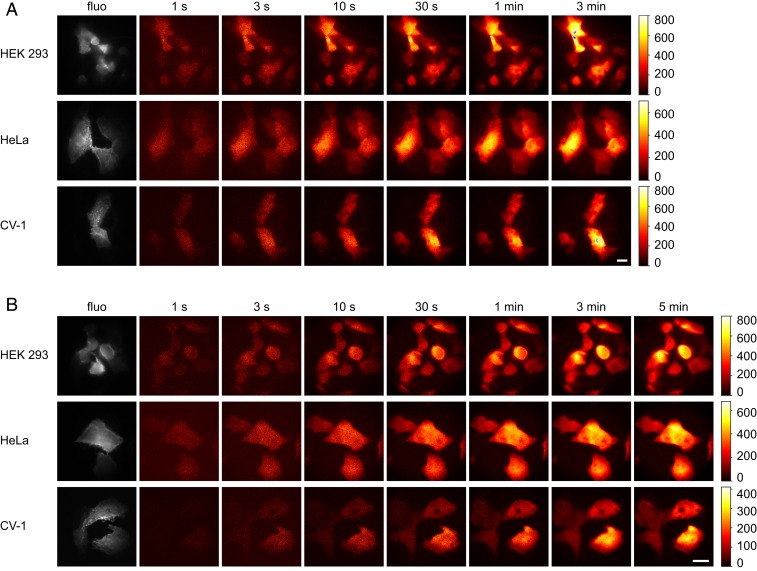
Next, we investigated whether the light produced by co lux in mammalian cells is sufficient to detect single cells with a custom-built bioluminescence microscope. HEK, HeLa, and CV-1 cells were transfected with the co lux genes and imaged using different exposure times (Fig. 4). Bioluminescence was already detectable after only 1 s in all cell lines tested, and the signal-to-noise ratio increased with prolonged exposure times, as expected. A calibration of the camera demonstrated that several hundred thousand and, in some cases, more than a million photons per minute per cell were detected.
Fig. 4.
Bioluminescence images of different cell lines expressing the co lux genes. Images were taken with the indicated exposure times using a (A) 60× or (B) 100× objective lens. The color map was scaled to the minimum and maximum pixel values of each image. For the (A) 3-min and (B) 5-min images, the color bars represent the number of detected photons per pixel. Blue pixels represent saturation of the camera. Fluorescence images (fluo) of cotransfected lifeact-EYFP excited at 491 nm are shown in gray. (Scale bars, 20 µm.)
Since cotransfection of 6 co lux plasmids is required to generate bioluminescence, which might result in a low number of cells expressing all components in sufficient amounts, we determined the fraction of bioluminescent cells. For this purpose, we stained co lux-transfected cells with the fluorescent DNA-binding dye Hoechst 33342. This membrane-permeable dye labels the nuclei of living cells and was used to determine the total cell number. Concomitant bioluminescence imaging indicated that ∼30% of the cells exhibited detectable bioluminescence emission during the 2-min exposure time and therefore expressed all 6 genes in sufficient amounts for single-cell imaging (SI Appendix, Fig. S6).
We compared the brightness of co lux to FLuc and pCMVLux on the single-cell level. In line with the measurements on whole wells of cells (Fig. 1 and SI Appendix, Fig. S3), FLuc produced similar levels of bioluminescence as co lux, whereas no signal from pCMVLux was detectable (SI Appendix, Fig. S7).
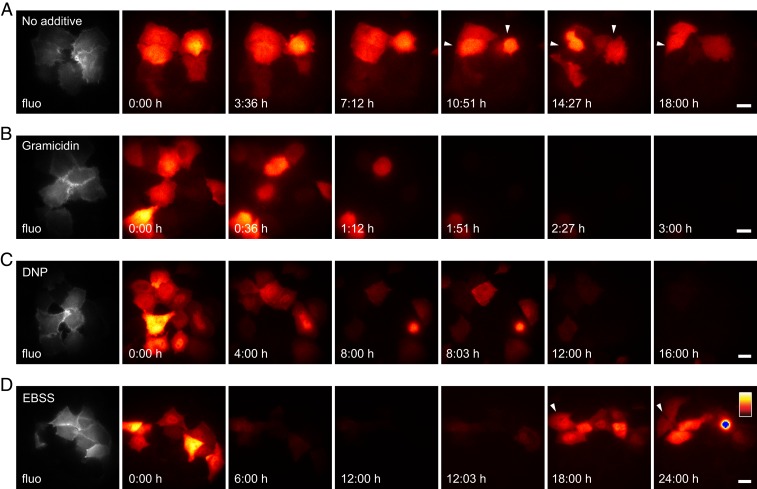
Subsequently, we imaged co lux-expressing cells for longer timespans. First, imaging was performed in cell culture medium without additives (Fig. 5A and Movie S1). Some cells divided, whereas other cells lost their bioluminescence signal, possibly due to cell death resulting from toxicity of the transfection reagent. Since ATP and NADPH are required for sustained bioluminescence, the bioluminescence signal provides information about the metabolic state and cellular viability. As a proof-of-principle application, we tested the influence of the pore-forming toxin gramicidin on co lux-expressing cells (Fig. 5B and Movie S2). Gramicidin is a peptide antibiotic mainly against Gram-positive bacteria that also affects the ion permeability of eukaryotic cells. It forms ion channels in the cell membrane and thereby disturbs the electrochemical gradient, which finally leads to cell death. This became apparent as rounding of the cells and complete loss of bioluminescence within a few hours, confirming its toxicity to eukaryotic cells. The second substance tested was 2,4-dinitrophenol (DNP) that acts as a proton ionophore and thus decouples the proton gradient across the mitochondrial membrane. As a result, ATP synthesis by oxidative phosphorylation is impaired, and the energy of the proton gradient is instead lost as heat. DNP has therefore been used as a drug against obesity, but, due to its high toxicity and its severe and sometimes fatal side effects, its use is no longer authorized. Up to now, no efficient antidote against DNP poisoning is known. Cells treated with DNP maintained their original shape, except for some cases where cells rounded immediately before cell death (Fig. 5C and Movie S3), but they moved less on the coverslip than untreated cells (Movie S1). In addition, the signal steadily decreased over time (SI Appendix, Fig. S8A). A sudden increase in bioluminescence was observed when DNP was removed, but the signal then continued decreasing (Fig. 5C, SI Appendix, Fig. S8A, and Movie S3). Therefore, the effect of DNP does not seem to be fully reversible, probably either due to its limited water solubility that impedes its washing out or due to causing irreversible cellular damage. Cellular accumulation of DNP might also be responsible for cases of death after repeated intake of sublethal DNP doses (28).
Fig. 5.
Long-term bioluminescence measurements of HeLa cells expressing co lux. Cells were imaged in cell culture medium with (A) no additive, (B) gramicidin (25 µg/mL), or (C) DNP (25 µg/mL) or (D) in EBSS without glucose. In C, cells were washed 5 times with EBSS after 8 h, and imaging was continued in cell culture medium without additives. In D, cells were washed 10 times with EBSS before starting the measurement, and EBSS was replaced with cell culture medium after 12 h. Bioluminescence images were taken with a 60× objective lens and exposure times of (A, C, and D) 3 min or (B) 2 min. The color map was scaled to the minimum and maximum pixel values of each image series. Blue pixels represent saturation. Arrows indicate dividing cells. Fluorescence images (fluo) of cotransfected lifeact-EYFP excited at 491 nm are shown in gray. Complete time series are shown in Movies S1–S4. (Scale bars, 20 µm.)
Since the energy for the synthesis of ATP and NADPH and thus the generation of bioluminescence is obtained from nutrients in the medium, the bioluminescence signal should decrease upon nutrient deprivation. This effect was investigated by imaging the cells in Earle’s Balanced Salt Solution (EBSS) without glucose, instead of cell culture medium (Fig. 5D and Movie S4). The signal decreased rapidly during the first ∼30 min and then continued decreasing at a lower rate to almost zero after 12 h (SI Appendix, Fig. S8B). When the EBSS was replaced by cell culture medium, the signal increased immediately and rose further over the next hours (Fig. 5D, SI Appendix, Fig. S8B, and Movie S4). Although several cells died during the 24-h observation time, other cells survived and divided, demonstrating that some cells are able to recover after the starvation period. The energy source of the prolonged bioluminescence which was still detectable in many cells after 12 h in EBSS is not known. Energy might be derived from remnants of nutrients from the cell culture medium, from intracellularly stored nutrients, or from uptake of components of the extracellular matrix (29).
As we previously reported for Escherichia coli cells (30), the bioluminescence signal from a few HeLa cells fluctuated before cell death (Movies S3 and S4). This “blinking” occurred less frequently than in kanamycin-treated E. coli cells where it was shown to be accompanied by changes in the cellular ATP concentration (30). The cause of the blinking is unknown, but might involve similar processes affecting the energy metabolism during cell death in both cell types.
Discussion and Conclusion
We demonstrated autonomous bioluminescence imaging of single mammalian cells by coexpression of 6 co genes from the bacterial bioluminescence system. Although the brightness of bacterial bioluminescence has so far been considered to be much lower than that of other luciferases requiring an external luciferin, we found that co Lux produced autonomous bioluminescence at levels similar to firefly luciferase. Codon optimization was found to be crucial for high expression in mammalian cells, although we cannot rule out that features of the nucleotide sequence other than the codon usage contribute to enhanced expression (e.g., due to formation of messenger RNA secondary structures). In addition, several other factors affect the expression or function of the Lux proteins and may explain the previously observed decreased performance of bacterial bioluminescence in mammalian cells: 1) choice of the specific LuxCDABE and flavin reductase amino acid sequences, as the proteins from different bacterial species may differ in their stability at 37 °C and their expression, maturation, or activity under the conditions in mammalian cells, which we observed in particular for the flavin reductase; 2) the presence of fusion partners at the N and C termini of the Lux proteins, as even the small 2A peptides can strongly reduce the bioluminescence signal; 3) the ratio at which the 6 genes are transfected, since aldehyde synthesis seems to be the most rate-limiting step of the overall process, and expression of all co lux genes from individual plasmids allows freely adjusting the protein ratios; and 4) choice of the expression vector and distance of the lux genes from the promoter.
Although the aldehyde produced by the Lux system may be cytotoxic for eukaryotic cells (31), we did not observe significant effects on cell number or metabolic activity 24 h posttransfection, which is in line with other reports (11, 12, 32). Furthermore, co lux-expressing cells maintained their physiological shape and their ability to divide. The only significant effect of co lux expression was found to be a small decrease of NADPH levels which possibly results from elevated NADPH consumption during synthesis of the aldehyde or FMNH2. It is not clear whether this represents already the metabolic limit for the light levels achievable by autonomous bioluminescence due to its high energy demand, or whether the brightness could be further increased by optimization of the involved proteins and their expression levels in mammalian cells.
Because of its dependence on metabolic energy, bioluminescence generated by the co lux genes can be used to investigate metabolic changes in mammalian cells, for example, during the cell cycle phases, during nutrient deprivation, or in response to toxic compounds. On the other hand, the generation of bioluminescence requires substantial amounts of metabolic energy, which may itself influence cellular processes. Bioluminescence measurements with co Lux can easily be performed with cell monolayers using a commercial luminescence imager. Owing to the high bioluminescence signal, they are also feasible on the single-cell level, which allows the observation of individual differences between cells and events such as fluctuations in brightness, cell division, and cell death. Therefore, co Lux holds promise as an easy-to-use reporter system for numerous applications in biological and medical research. Although fluorescence-based methods remain superior for imaging of cells at high speed and resolution due to their much higher signals, further improvements of the co Lux system will extend its scope of application in single-cell imaging.
Materials, Methods, and Data Availability
Details of plasmid generation, cell culture, bioluminescence imaging, and toxicity tests are described in SI Appendix, SI Materials and Methods. In brief, co lux and frp genes were cloned separately into the vector pcDNA3.1(+) and cotransfected into mammalian cell lines. Bioluminescence imaging was performed with a custom microscope containing a cooled electron-multiplying charge-coupled device camera and a 491-nm excitation laser for fluorescence measurements.
The co lux pcDNA3.1(+) plasmids are available from Addgene (www.addgene.org, plasmid numbers 129085 to 129090). All data are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank Rainer Pick and Dr. Ellen Rothermel for technical assistance. We thank Dr. Steven Ripp (490 BioTech) for kindly providing the pCMVLux plasmid, and Prof. Yi Yang (East China University of Science and Technology) for the iNap1 pcDNA3.1/Hygro(+) plasmid.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913616116/-/DCSupplemental.
References
- 1.Cormier M. J., Kuwabara S., Some observations on the properties of crystalline bacterial luciferase. Photochem. Photobiol. 4, 1217–1225 (1965). [Google Scholar]
- 2.Puget K., Michelson A. M., Studies in bioluminescence. VII. Bacterial NADH:flavin mononucleotide oxidoreductase. Biochimie 54, 1197–1204 (1972). [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez A., Riendeau D., Meighen E., Purification of the acyl coenzyme A reductase component from a complex responsible for the reduction of fatty acids in bioluminescent bacteria. Properties and acyltransferase activity. J. Biol. Chem. 258, 5233–5237 (1983). [PubMed] [Google Scholar]
- 4.Rodriguez A., Wall L., Riendeau D., Meighen E., Fatty acid acylation of proteins in bioluminescent bacteria. Biochemistry 22, 5604–5611 (1983). [Google Scholar]
- 5.Carey L. M., Rodriguez A., Meighen E., Generation of fatty acids by an acyl esterase in the bioluminescent system of Photobacterium phosphoreum. J. Biol. Chem. 259, 10216–10221 (1984). [PubMed] [Google Scholar]
- 6.Kotlobay A. A., et al. , Genetically encodable bioluminescent system from fungi. Proc. Natl. Acad. Sci. U.S.A. 115, 12728–12732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao H., et al. , Characterization of coelenterazine analogs for measurements of Renilla luciferase activity in live cells and living animals. Mol. Imaging 3, 43–54 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Bhaumik S., Gambhir S. S., Optical imaging of Renilla luciferase reporter gene expression in living mice. Proc. Natl. Acad. Sci. U.S.A. 99, 377–382 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inoue Y., et al. , Gaussia luciferase for bioluminescence tumor monitoring in comparison with firefly luciferase. Mol. Imaging 10, 377–385 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Sharifian S., Homaei A., Hemmati R., B Luwor R., Khajeh K., The emerging use of bioluminescence in medical research. Biomed. Pharmacother. 101, 74–86 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Close D. M., et al. , Autonomous bioluminescent expression of the bacterial luciferase gene cassette (lux) in a mammalian cell line. PLoS One 5, e12441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu T., Ripp S., Sayler G. S., Close D. M., Expression of a humanized viral 2A-mediated lux operon efficiently generates autonomous bioluminescence in human cells. PLoS One 9, e96347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Close D. M., et al. , Comparison of human optimized bacterial luciferase, firefly luciferase, and green fluorescent protein for continuous imaging of cell culture and animal models. J. Biomed. Opt. 16, 047003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson T., Hastings J. W., Bioluminescence. Annu. Rev. Cell Dev. Biol. 14, 197–230 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Haddock S. H., Moline M. A., Case J. F., Bioluminescence in the sea. Annu. Rev. Mar. Sci. 2, 443–493 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Dunlap P., “Biochemistry and genetics of bacterial bioluminescence” in Bioluminescence: Fundamentals and Applications in Biotechnology–Volume 1, Thouand G., Marks R., Eds. (Advances in Biochemical Engineering/Biotechnology, Springer, Berlin, Germany, 2014), vol. 144, pp. 37–64. [DOI] [PubMed] [Google Scholar]
- 17.Tinikul R., Chaiyen P., “Structure, mechanism, and mutation of bacterial luciferase” in Bioluminescence: Fundamentals and Applications in Biotechnology–Volume 3, Thouand G., Marks R., Eds. (Advances in Biochemical Engineering/Biotechnology, Springer, Berlin, Germany, 2014), vol. 154, pp. 47–74. [DOI] [PubMed] [Google Scholar]
- 18.Brodl E., Winkler A., Macheroux P., Molecular mechanisms of bacterial bioluminescence. Comput. Struct. Biotechnol. J. 16, 551–564 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurfürst M., Ghisla S., Hastings J. W., Characterization and postulated structure of the primary emitter in the bacterial luciferase reaction. Proc. Natl. Acad. Sci. U.S.A. 81, 2990–2994 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurfürst M., Macheroux P., Ghisla S., Hastings J. W., Isolation and characterization of the transient, luciferase-bound flavin-4a-hydroxide in the bacterial luciferase reaction. Biochim. Biophys. Acta 924, 104–110 (1987). [Google Scholar]
- 21.Patterson S. S., Dionisi H. M., Gupta R. K., Sayler G. S., Codon optimization of bacterial luciferase (lux) for expression in mammalian cells. J. Ind. Microbiol. Biotechnol. 32, 115–123 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Grote A., et al. , JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33, W526–W531 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szymczak-Workman A. L., Vignali K. M., Vignali D. A., Design and construction of 2A peptide-linked multicistronic vectors. Cold Spring Harb. Protoc. 2012, 199–204 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Chaiyen P., Fraaije M. W., Mattevi A., The enigmatic reaction of flavins with oxygen. Trends Biochem. Sci. 37, 373–380 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Tantama M., Martínez-François J. R., Mongeon R., Yellen G., Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 4, 2550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao R., et al. , Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat. Methods 14, 720–728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bilan D. S., et al. , HyPer-3: A genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem. Biol. 8, 535–542 (2013). [DOI] [PubMed] [Google Scholar]
- 28.McFee R. B., Caraccio T. R., McGuigan M. A., Reynolds S. A., Bellanger P., Dying to be thin: A dinitrophenol related fatality. Vet. Hum. Toxicol. 46, 251–254 (2004). [PubMed] [Google Scholar]
- 29.Muranen T., et al. , Starved epithelial cells uptake extracellular matrix for survival. Nat. Commun. 8, 13989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregor C., Gwosch K. C., Sahl S. J., Hell S. W., Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging. Proc. Natl. Acad. Sci. U.S.A. 115, 962–967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollis R. P., et al. , Toxicity of the bacterial luciferase substrate, n-decyl aldehyde, to Saccharomyces cerevisiae and Caenorhabditis elegans. FEBS Lett. 506, 140–142 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Gupta R. K., Patterson S. S., Ripp S., Simpson M. L., Sayler G. S., Expression of the Photorhabdus luminescens lux genes (luxA, B, C, D, and E) in Saccharomyces cerevisiae. FEMS Yeast Res. 4, 305–313 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.