Significance
The stem cell niche is the in vivo microenvironment in which stem cells both reside and receive stimuli that determine their fate. Intestinal Paneth cells produce an arsenal of molecules involved in numerous biological processes, including critical support of the intestinal stem cell niche via the expression of niche ligands, growth factors, and cytokines. The relevance of the niche function of Paneth cells has been challenged, however. Here we show that specific, acute ablation of Paneth cells in mice does not affect the stem cell population, because alternative niche cells derive enteroendocrine and tuft cells. These cells serve as an alternative source of Notch signals, which are essential for intestinal stem cell maintenance.
Keywords: intestine, Paneth cells, stem cells, Notch
Abstract
Cycling intestinal Lgr5+ stem cells are intermingled with their terminally differentiated Paneth cell daughters at crypt bottoms. Paneth cells provide multiple secreted (e.g., Wnt, EGF) as well as surface-bound (Notch ligand) niche signals. Here we show that ablation of Paneth cells in mice, using a diphtheria toxin receptor gene inserted into the P-lysozyme locus, does not affect the maintenance of Lgr5+ stem cells. Flow cytometry, single-cell sequencing, and histological analysis showed that the ablated Paneth cells are replaced by enteroendocrine and tuft cells. As these cells physically occupy Paneth cell positions between Lgr5 stem cells, they serve as an alternative source of Notch signals, which are essential for Lgr5+ stem cell maintenance. Our combined in vivo results underscore the adaptive flexibility of the intestine in maintaining normal tissue homeostasis.
A stem cell niche is a unique microenvironment composed of specialized cells that provide the necessary repertoire of growth factors and physical interactions to maintain stem cells and control their behavior to safeguard proper tissue homeostasis. A relatively simple architecture combined with extraordinarily fast self-renewal makes the small intestine a unique model for studying adult tissue stem cells and their niches (1). To maintain intestinal tissue homeostasis, the balance between intestinal stem cell self-renewal and differentiation must be carefully regulated. To do so, various signaling pathways (e.g., Wnt, Notch, Hippo, and BMP signaling pathways) act in concert on the cycling Lgr5+ stem cell population, quiescent reserve stem cells, and niche cells (2–7).
The Lgr5+ stem cells are found at the base of the crypts of Lieberkühn, tiny invaginations that line the mucosal surface (8, 9). The symmetrically dividing Lgr5+ stem cells give rise to proliferating progenitors (transit-amplifying [TA] cells) that subsequently differentiate into 5 principal epithelial cell types: goblet cells, enteroendocrine cells, tuft cells, enterocytes, and Paneth cells (10, 11). Cellular differentiation takes place during migration from the crypts onto the flanks of the villi. It takes only 4 to 5 d for the cells to reach the villus tip, where they undergo apoptosis and exfoliate into the lumen of the intestine. Long-lived Paneth cells escape this upward flow (12); they migrate downward to settle at the crypt bottoms, where they can persist for weeks, with the oldest Paneth cells residing at the very base of the crypt (13). Paneth cells are removed from the crypt bottom by cellular fragmentation and phagocytosis from infiltrating macrophages.
Paneth cells contain granules rich in antimicrobial peptides (e.g., lysozymes, α-defensins/cryptdins) and immune modulators (14, 15). Paneth cell-derived antimicrobial peptides protect the host from enteric pathogens, help shape the composition of the colonizing microbiota, and act as a safeguard from bacterial translocation across the epithelium (16–18). The Paneth cell–stem cell interaction also plays a central role in response to the nutritional status of an organism. Paneth cells serve as sensors for nutritional status and enhance stem cell function in response to calorie restriction (19, 20). Moreover, Paneth cell dysfunction has been implicated in a subset of patients with Crohn's disease (21).
Paneth cells are in intimate connection with the Lgr5+ stem cells at the crypt base of the small intestine, small intestinal tumors, and intestinal-derived organoid cultures, suggestive of a functional interaction (8, 22–24). Indeed, coculturing of sorted stem cells with Paneth cells dramatically improves organoid formation (24). Moreover, Paneth cells are the source of multiple stem cell growth factors (e.g., Wnt3, Egf, Tgf-α) and express Notch-ligands (Dll4 and Dll1), essential signals for stem cell maintenance in culture (24). In addition, the Lgr5 stem cells present in intestinal organoids, which consist only of epithelial cells, depend on the presence of Paneth cells or their crucial niche signal Wnt3 (25), while drug-induced blocking of the Notch pathway in intestinal organoids results in the complete loss of proliferating stem cells (22).
The Wnt and Notch signaling pathways are essential for the maintenance of intestinal Lgr5+ stem cell in vivo (2, 26–29). The Wnt effector Tcf4 has a vital role during homeostasis of the adult mouse intestine (27). However, the Wnt ligand Wnt3, which is produced by Paneth cells, is dispensable for the maintenance of intestinal stem cells in mice, most likely due to the secretion of Wnt ligands (Wnt2B) by the mesenchymal cells surrounding the crypt base (25, 30). In contrast, the Notch signaling pathway can be activated only by cells that are physical neighbors. The intestinal epithelial-specific deletion of Dll1/Dll4 or Notch1/Notch2 unequivocally demonstrates that Notch signaling is essential for the preservation of Lgr5+ stem cells in vivo (28, 29). Importantly, these data also show that mesenchymal cells cannot compensate for the loss of Notch ligands of the intestinal epithelial cells.
The combined data suggest that Paneth cells, in addition to, for example, smooth muscle cells, fibroblasts, and intestinal subepithelial myofibroblasts present in the mesenchyme surrounding crypts, constitute the niche for the intestinal Lgr5+ stem cells. Therefore, we have proposed that Paneth cells serve as multifunctional guardians of intestinal stem cells (15, 24).
The relevance of the niche function of Paneth cells in vivo has been challenged, however. Conditional deletion of Math1/ATOH1, a target gene of Notch/Hes1-mediated repression, results in the complete elimination of all secretory cells, including the Paneth cells in normal intestinal tissue (31, 32) (SI Appendix, Fig. S1 A–F), as well as in Lgr5-derived adenomatous polyposis coli (APC)-deficient intestinal tumors (31) (SI Appendix, Fig. S1 G and H). Careful histological analysis showed that intestinal stem cells were still present and proliferated in the complete absence of Paneth cells. This apparent contradiction may be explained by the notion that Math1−/− stem cells are resistant to the loss of Notch signals normally provided by Paneth cells (31). Indeed, we have demonstrated that the loss of intestinal stem cells by their direct conversion into goblet cells on pharmacologically Notch inhibition requires Math1 (33).
Here we carefully analyzed the in vivo role of Paneth cells in the intestinal crypt stem cell niche via the generation and analysis of several newly generated Paneth cell-specific knockin (KI) mice.
A “Toolbox” of Genetically Modified KI Mice to Identify, Characterize, and Manipulate Paneth Cells
The mouse genome contains 2 closely linked genes encoding lysozyme isoforms, whose natural substrate is the bacterial cell wall peptidoglycan (34). One of these genes (mLys or Lys2) is expressed mainly in macrophages, while the other gene (pLys or Lys1) is specifically expressed in the intestinal Paneth cells. To be able to visualize, isolate, and/or manipulate Paneth cells, we generated a “toolbox” of 3 independent Paneth cell-specific genetically modified KI mouse lines: pLysdsRED, pLysCreErt2, and pLysDTR (SI Appendix, Fig. S2 A–C).
pLysdsRED KI Mice
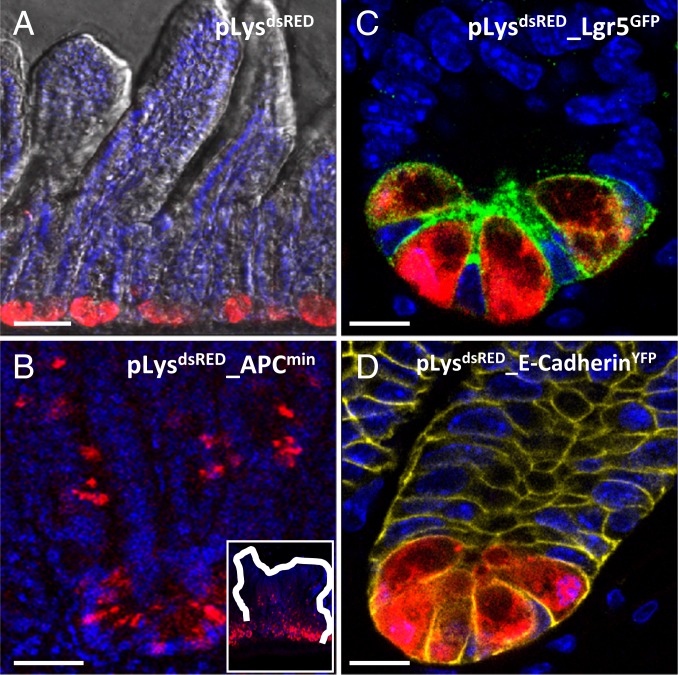
The pLysdsRED KI mice express the fluorescent dsRED marker under the specific regulatory sequences of the pLys gene. Confocal imaging of the pLysdsRED KI mice and the pLysdsRED_APCmin KI mice showed strong dsRED expression specifically in all Paneth cells present in healthy intestinal tissue (Fig. 1A), as well as in intestinal APCmin tumors (Fig. 1B). This Paneth cell-specific expression was further visualized by the analysis of pLysdsRED_Lgr5-GFP and pLysdsRED_E-cadherin-YFP compound KI mice (8, 35). Confocal imaging of the pLysdsRED_Lgr5-GFP mice showed that GFP+ Lgr5+ intestinal stem cells, containing typical large nuclei, were intermingled at the crypt base region with the terminally differentiated granulated dsRED+ Paneth cells (Fig. 1C). Analysis of the pLysdsRED_E-cadherin-YFP KI mice revealed specific membranous YFP staining of the cell adhesion molecule E-cadherin, expressed on all epithelial cells of the crypt, and the dsRED+-specific expression of Paneth cells (Fig. 1D).
Fig. 1.
Paneth cell-specific expression of fluorescent dsRed in pLysdsRED KI mice. (A) Confocal imaging of intestinal sections derived from pLysdsRED KI mice showing Paneth cell-specific dsRED expression. (B) Confocal imaging of intestinal sections derived from pLysdsRED_APCmin mice showing Paneth cell-specific dsRED expression in APCmin tumors. (Inset) Complete tumor. (C) Confocal imaging of Paneth cell-specific dsRED expression in combination with Lgr5+ stem cell-specific GFP expression in the intestine derived from the pLysdsRED_Lgr5-GFP compound mice. (D) Confocal imaging for Paneth cell-specific dsRED expression and E-cadherin–specific YFP membrane expression in the intestine derived from the pLysdsRED_E-cadherin-YFP compound mice. (Scale bars: 100 µm in A, 50 µm in B, 20 µm in C and D.)
Our combined analysis showed Paneth cell-specific dsRED expression in the pLysdsRED KI mice, demonstrating that these mice can be used to visualize, isolate, and characterize this unique cell type.
pLysCreErt2 KI Mice
pLysCreErt2 KI mice should express a tamoxifen-inducible version of the Cre enzyme specifically in all Paneth cells. To monitor the efficiency and specificity of Cre-mediated recombination, we analyzed pLysCreErt2_RosaLSL-LacZ KI compound mice. On tamoxifen administration, the Cre-mediated removal of a LoxP-STOP-LoxP (LSL) roadblock of the RosaLSL-LacZ KI reporter mice should result in the expression of β-galactosidase in a time-controlled and Paneth cell-specific fashion. The small intestines of adult pLysCreErt2_RosaLSL-LacZ mice and their littermate controls were histologically analyzed on day 10 after Cre induction. Expression of the β-galactosidase reporter was indeed explicitly restricted to the Paneth cells of the pLysCreErt2_RosaLSL-LacZ mice; however, up to 12% of the Paneth cells showed expression of this LacZ reporter gene (SI Appendix, Fig. S3 A and A′, Inset).
The inefficient targeting of the Paneth cell population resulted in the incomplete killing of Paneth cells via diphtheria toxin fragment A (DTA) expression on removal of the LSL cassette in the pLysCreErt2_RosaLSL-DTA compound mice (36). This inadequacy precluded us from using this compound mouse to analyze the role of Paneth cells as niche cells for intestinal stem cells in the crypt of Lieberkühn.
Of note, this pLysCreErt2 KI line has been used to successfully study the role of Paneth cells in the plasticity of the intestinal epithelium in response to inflammation (37) and their role in cancer initiation (SI Appendix, Fig. S3 B and C). Indeed, while Lgr5+ stem cells efficiently formed tumors on APC deletion and K-Ras activation (38) (SI Appendix, Fig. S3B), tumor formation is absent when these genes are specifically deleted or activated in Paneth cells during normal tissue homeostasis (SI Appendix, Fig. S3C).
pLysDTR KI Mice
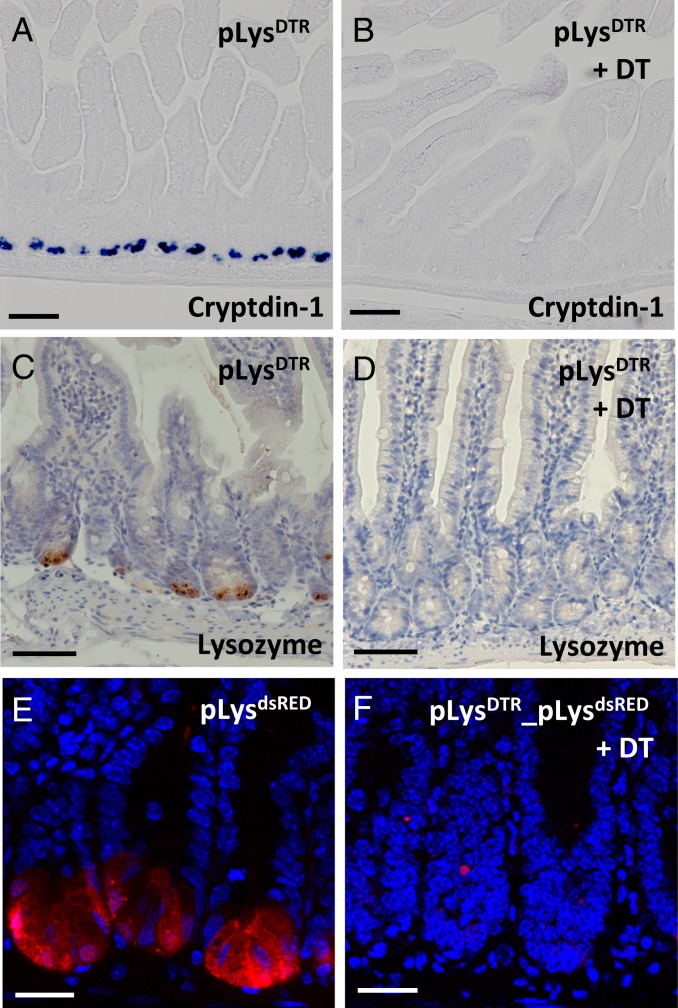
To increase the efficiency of Paneth cell ablation, we subsequently generated pLysDTR KI mice. In these mice, on the intraperitoneal (i.p.) injection of diphtheria toxin (DT), the DT receptor (DTR)-expressing Paneth cells should be specifically killed. After i.p. injection of DT for 6 consecutive days, histological analysis showed that virtually all Paneth cells were ablated in the duodenum and jejunum, as demonstrated by the absence of Paneth cell-specific markers, such as cryptdin-1 (Fig. 2B vs. Fig. 2A and SI Appendix, Fig. S5B vs. SI Appendix, Fig. S5A) and lysozyme (Fig. 2D vs. Fig. 2C). However, some cryptdin-1+ Paneth cells (average of 1 cell per crypt) were still present in the ileum (SI Appendix, Fig. S5D vs. SI Appendix, Fig. S5C). The efficient deletion of Paneth cells in the proximal regions of the intestine was further confirmed by the absence of dsRED+ Paneth cells on DT administration for 6 consecutive days in the pLysdsRED_pLysDTR KI mice (Fig. 2F vs. Fig. 2E). The efficient deletion of Paneth cells in the jejunum derived from the DT-treated pLysDTR KI mice at different time points (days 1 to 5) on daily DT injection via caspase-3 and lysozyme staining (SI Appendix, Fig. S4 A–J).
Fig. 2.
Efficient deletion of Paneth cells in the pLysDTR KI mice on DT administration. In situ hybridization of cryptdin-1 (A and B), lysozyme-specific immunostaining (C and D), and confocal imaging for dsRED expression (E and F) on jejunum sections from the untreated pLysDTR KI (A and C) and pLysdsRED (E) control mice and the pLysDTR KI (B and D) and pLysDTR_ pLysdsRED (F) mice DT-treated for 6 consecutive days, at 16 h after the last DT injection. The analysis shows the absence of Paneth cells in the intestines of DT-treated pLysDTR mice (B and D) and pLysDTR_pLysdsRED mice (F), in contrast to the controls (A, C, and E). (Scale bars: 100 µm in A–D; 50 µm in E and F.)
Ablation of Paneth Cells Does Not Affect Lgr5+ Stem Cells
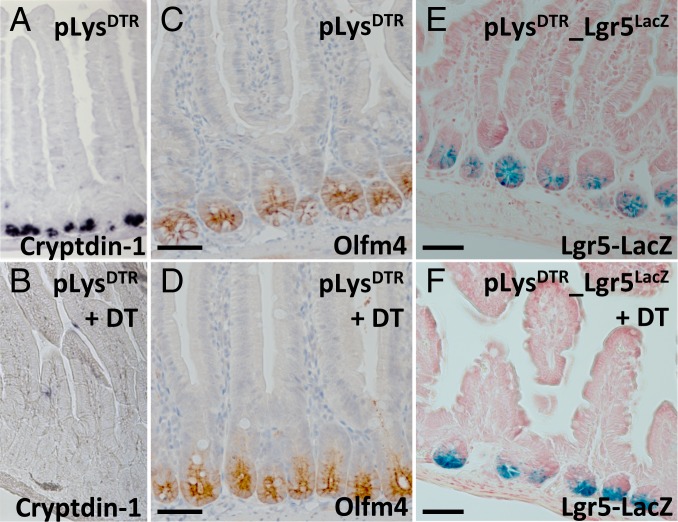
We then checked whether Lgr5+ stem cells survived in the absence of Paneth cells via histological analysis of the isolated duodenum/jejunum derived from pLysDTR KI mice that were treated with DT for 6 consecutive days. Of note, since DT injection beyond 6 d results in discomfort, we could not assess the long-term effects of Paneth cell ablation in vivo. We marked intestinal stem cells using an antibody directed against the Notch target Olfm4, a robust marker for human and mouse intestinal stem cells (39). While virtually all Paneth cells were killed on the i.p. injection of DT (Fig. 3B vs. Fig. 3A), Olfm4+ intestinal stem cells were not affected (Fig. 3D vs. Fig. 3C).
Fig. 3.
Ablation of Paneth cells does not affect intestinal stem cells. In situ hybridization of cryptdin-1+ Paneth cells (A and B), immunostaining of Ofm4+ stem cells (C and D), and Lgr5+ stem cell-specific LacZ expression (E and F) on jejunum sections derived from the indicated control mice (A, C, and D) and pLysDTR KI (B and D) and pLysDTR_Lgr5LacZ KI (F) mice DT-treated for 6 consecutive days, at 16 h after the last DT injection. This analysis shows the presence of normal numbers of intestinal stem cells in the absence of Paneth cells in the DT-treated pLysDTR (B and D) and pLysDTR_Lgr5LacZ KI mice (F), in contrast to the untreated control mice (A, C, and E). (Scale bars: 100 µm.)
To further demonstrate the presence of intestinal stem cells in the absence of Paneth cells, we analyzed the pLysDTR_Lgr5LacZ compound KI mice. The Lgr5LacZ allele faithfully recapitulates Lgr5 expression (8). These mice received DT for 6 consecutive days. At 16 h after the last DT administration, we found normal numbers of LacZ+ stem cells in pLysDTR_Lgr5LacZ KI mice (Fig. 3F) compared with untreated pLysDTR_Lgr5LacZ KI (Fig. 3E) or DT-treated control Lgr5LacZ mice.
Moreover, an in vivo BrdU pulse-labeling study showed that the number and location of proliferating (stem) cells at 2 and 24 h after i.p. BrdU administration are comparable in the pLysDTR KI mice treated with DT for 5 consecutive days (SI Appendix, Fig. S6 C and D), DT-treated wild-type mice, and untreated pLysDTR KI control mice (SI Appendix, Fig. S6 A and B).
Therefore, we concluded that the ablation of Paneth cells does not affect either the maintenance or proliferation of Lgr5+ stem cells or the proliferation of the pool of TA cells present in the intestinal crypt of Lieberkühn.
Notch Signaling Remains Active on Paneth Cell Ablation
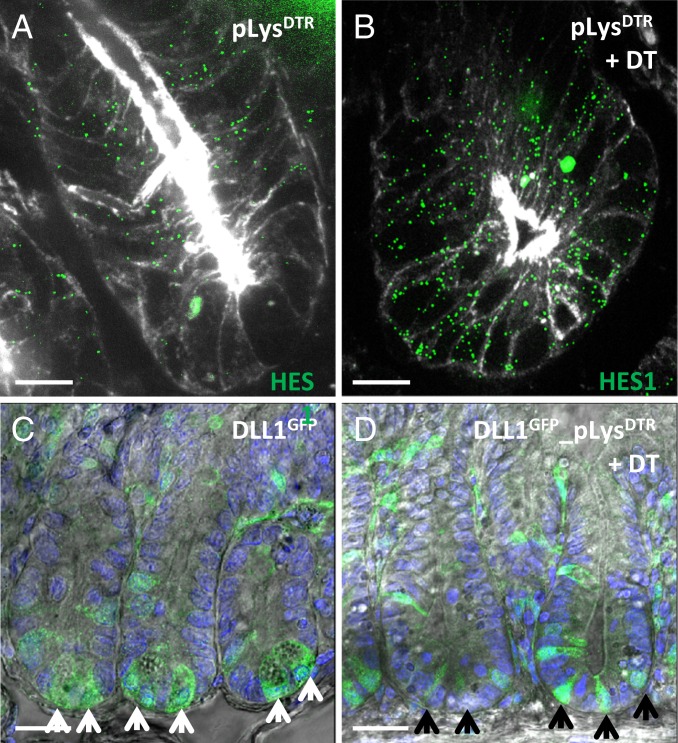
Paneth cells express the Notch ligands Dll1 and Dll4 (24). The Notch signaling pathway can only be activated by cells that are physical neighbors. The ablation of Notch signaling specifically within the intestinal epithelium results in loss of proliferating crypt (stem) cells owing to their conversion into postmitotic secretory cells (2, 28, 29, 33). Importantly, mesenchymal cells surrounding the crypt of Lieberkühn cannot compensate for the loss of Notch ligands of the epithelial cells. Therefore, we checked whether the Notch signaling pathway is indeed still active on Paneth cell depletion. Single molecule fluorescence in situ hybridization (smFISH analysis) using the Notch target gene Hes1 (40) as a specific probe showed that the Notch signaling pathway remained active after Paneth cell ablation on DT treatment for 6 consecutive days (Fig. 4B vs. Fig. 4A). Similarly, the Notch target Olfm4 (41) remained expressed in the Lgr5+ stem cells after the elimination of Paneth cells (Fig. 3D).
Fig. 4.
Active Notch signaling on Paneth cell ablation. (A and B) smFISH analysis of Notch target gene Hes1 on jejunum sections from untreated pLysDTR KI control mice (A) and pLysDTR KI mice DT-treated for 6 consecutive days (B), at 16 h after the last DT injection. This analysis shows, via Hes1 expression, that the Notch signaling pathway remained active on successful Paneth cell ablation (B vs. A). (C and D) Confocal microscopy imaging of jejunum sections from Dll1GFP-CreErt2 (C) and Dll1GFP-CreErt2_pLysDTR KI (D) mice DT-treated for 6 consecutive days, at 16 h after the last DT injection showing the absence of granulated Paneth cells on DT administration (D), which were present in the control mice (C, white arrows). Nongranulated GFP+/Dll1+ cells can be observed between the stem cells in the absence of Paneth cells (D, black arrows). (Scale bars: 20 µm in A and B; 50 µm in C and D.)
To determine whether the Notch ligands were still present, we took advantage of GFP expression regulated by the Dll1 promoter in the Dll1GFP-CreErt2 KI mice along with DTR expression in the pLysDTR KI mice (42). Confocal analysis of the intestine derived from the Dll1GFP-CreErt2 mice showed the presence of granulated Paneth cells (Fig. 4C, white arrows). However, similar analysis of the intestine derived from the Dll1GFP-CreErt2_pLysDTR KI mice confirmed the absence of granulated Paneth cells on DT administration for 6 consecutive days (Fig. 4D vs. Fig. 4C), while nongranulated, lysozyme, and cryptdin-negative cells—observed between the stem cells on Paneth cell ablation—were GFP+ (Fig. 4D, black arrows), that is, they expressed the Dll1 ligand.
The presence of Dll1+ non-Paneth cells adjacent to the Notch+ stem cells explained why the Notch signaling pathway remained active in stem cells on Paneth cell depletion. Active Notch signaling appeared to be the result of the presence of an alternative Notch ligand-expressing cell that served as an alternative source of Notch signaling for intestinal stem cells on Paneth cell ablation.
Single-Cell Sequencing to Identify the Alternative Niche Cells
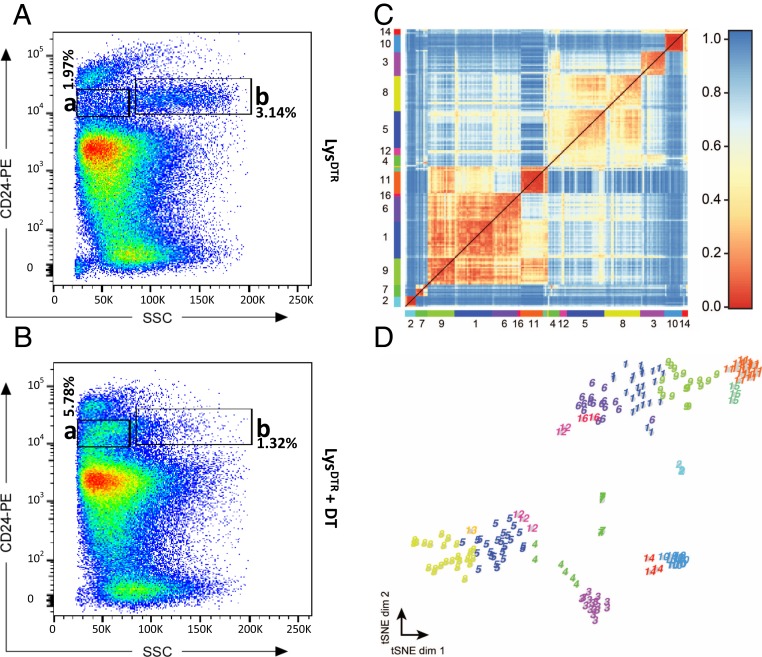
To further characterize this alternative niche cell population, we analyzed isolated crypt cells derived from the pLysDTR mice DT-treated for 6 consecutive days (Fig. 5B) and from the untreated control mice (Fig. 5A) via flow cytometry. This fluorescence-activated cell sorting (FACS) analysis was based on CD24 expression and side scatter intensity. CD24 discerns individual crypt cell types (24). This comparative analysis showed the expected strong reduction on DT administration of the Paneth cells containing gated cell populations (24) (Fig. 5A, gate b [3.14%] vs. Fig. 5B gate b [1.32%]), while (an)other cell population(s) was/were strongly increased (Fig. 5A, gate a [1.97%] vs. Fig. 5B, gate a [5.78%]).
Fig. 5.
Identification of the alternative niche cell on Paneth cell ablation via single-cell sequencing. FACS analysis, based on side-scatter intensity and CD24 expression, on isolated crypt cells (pooled from 3 mice) derived from the intestines of untreated control mice (A) and pLysDTR mice treated with DT for 6 consecutive days (B). This analysis shows a reduction in the Paneth cell population on DT administration (A, gate b [3.14%] vs. B, gate b [1.32%]), while (an)other cell population(s) was/were strongly increased (A, gate a [1.97%] vs. B, gate a [5.78%]). (C) Single-cell sequencing. The heat map shows the transcriptome similarities between cells measured by Pearson’s correlation coefficient after clustering with the RaceID2 algorithm. Colors and corresponding numbers along the axes represent the 16 identified clusters; only the numbers of the largest clusters are shown for clarity. (D) t-SNE map representation of transcriptome similarities. RaceID2 clusters are color-coded and numbered as in C.
Single-cell mRNA sequencing has emerged as a powerful method to simultaneously measure the cell-to-cell expression of thousands of genes and has the potential to enable the unbiased discovery of cell types and their corresponding marker genes. Therefore, we aimed to identify the alternative niche cells that emerge on Paneth cell ablation via single-cell sequencing. We applied a modified version of the CEL-seq method incorporating unique molecular identifiers to count transcripts (43, 44). We sequenced 192 randomly selected cells derived from gate a of the DT treated pLysDTR (Fig. 5B) and 96 cells derived from gate a of the untreated control mice (Fig. 5A). Of note, we used several regions of the intestine, including the regions with incomplete Paneth cell ablation. After quantifying transcript expression in all cells, we normalized by downsampling to a minimum number of 3,000 transcripts and discarded all cells with fewer than 3,000 transcripts. To reduce noise, we discarded genes that were not expressed with at least 5 transcripts in a cell in the dataset. Applying these filtering steps yielded 188 cells (116 derived from DT treated pLysDTR plus 72 from control mice) with a total of 839 expressed genes. To systematically screen for classes of cells with similar transcriptomes, we used k-medoids clustering and outlier analysis by RaceID2 (44). Via this approach, we identified 16 independent clusters (Fig. 5 C and D and Dataset S1).
The comparison between single cells derived from the DT-treated mice (Fig. 5B, gate a) and those derived from untreated mice (Fig. 5A, gate a) showed the presence of 5 clusters with increased cell numbers (clusters 1, 2, 4, 8, and 12). These cluster groups represented very early Paneth cells (cluster 1; low expression of Lyz1, Spink4, MMP7, and some cryptdins), tuft cells (cluster 8; expression of Dclk1, TRMP5, Sox9, SpiB, and Gfi1b), and subpopulations of enteroendocrine cells (cluster 2: high expression of ghrelin and somatostatin; cluster 4: high expression of chromogranin B, secretin, and ghrelin; and cluster 12: high expression of ghrelin) (Dataset S1 and SI Appendix, Table S1). The presence of these newly emerging clusters containing enteroendocrine cells and tuft cells suggests that these cell types might represent the alternative niche cell on Paneth cell ablation.
Tuft Cells and Enteroendocrine Cells Act as Novel Niche Cells on Paneth Cell Ablation
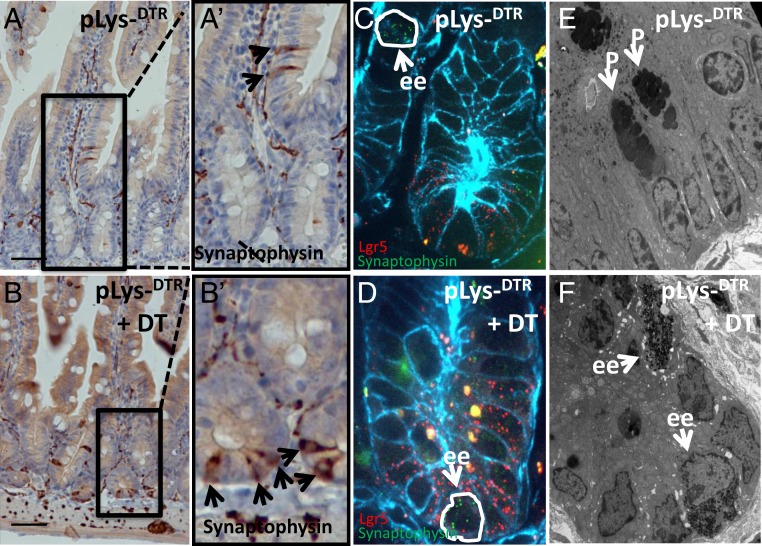
We next investigated, via histological analysis, whether tuft and/or enteroendocrine cells indeed replaced the ablated Paneth cells. We stained sections of intestines derived from the pLysDTR KI mice DT-treated for 6 consecutive days and from untreated pLysDTR KI mice (control) with antibodies directed against Dcamkl1 (a marker for Tuft cells) or synaptophysin (a general marker for enteroendocrine cells). Paneth cell ablation was successful (SI Appendix, Fig. S7B vs. SI Appendix, Fig. S7A). Synaptophysin and Dcamkl1 staining of intestinal sections derived from the control mice revealed enteroendocrine cells (Fig. 6 A and A′, Inset) and tuft cells (SI Appendix, Fig. S7C) in the TA zone of the crypt and on the villi, but not intermingled with the Lgr5+ stem cells at the bottom of the crypt. However, in the for 6 d DT-treated pLysDTR KI mice, synaptophysin+ enteroendocrine (Fig. 6 B and B′, Inset) and the Dcamkl1+ tuft cells (SI Appendix, Fig. S7) were also found between stem cells at the bottom of the crypt. Of note, we couldn’t detect any PAS+ goblet cell at the bottom of the crypt in the DT treated LysDTR mice.
Fig. 6.
Enteroendocrine cells support Lgr5+ crypt stem cells on Paneth cell ablation. Immunostaining (A, A′, B, and B′), smFISH (C and D), and TEM analysis (E and F) to detect synaptophysin+ enteroendocrine cells in jejunum sections from pLysDTR KI mice DT-treated for 6 consecutive days (B, D, and F) and untreated pLysDTR KI mice (A, C, and E) at 16 h after the last DT injection. The synaptophysin+ enteroendocrine cells could be detected in the TA zone of the crypt and on the villi (A, A′, and C), while in the DT-treated pLysDTR KI mice, the synaptophysin+ enteroendocrine cells were intermingled with Lgr5+ intestinal stem cells at the bottom of the crypt (arrows in B, B′, and D). Quantification revealed the presence of 0.89 enteroendocrine cells per crypt in a single section (counted: 600 crypts per mouse; n = 3). In contrast to the untreated control pLysDTR KI mice (E, white arrows marking Paneth cells [P]), TEM analysis confirmed the presence of enteroendocrine cells (ee) (F, white arrows) at the bottom of the crypt after DT-mediated Paneth cell ablation. (Scale bars: 100 µm.)
Next, we used smFISH, in situ hybridization method at single-cell resolution, to detect the localization of enteroendocrine cells in control mice and upon DT mediated Paneth cell ablation (45). Expression analysis of the enteroendocrine marker gene synaptophysin revealed the presence of enteroendocrine cells in the TA region (Fig. 6C) of the crypt and on villi in the intestines of the DT-treated and nontreated control pLys-DTR mice. However, enteroendocrine cells were also present at the bottom of the crypts, between Lgr5+ stem cells, on ablation of Paneth cells in the pLys-DTR mice (Fig. 6D).
The results of the histological and smFISH analyses were further confirmed by transmission electron microscopy (TEM) analysis, which revealed the presence of Paneth cells at the crypt bottoms of control mice (Fig. 6E), while Paneth cells were replaced by enteroendocrine cells in the LysDTR mice on DT administration (Fig. 6F). The combined analysis showed that Paneth cell ablation was followed by the formation of new niche cells (enteroendocrine cells and tuft cells) that physically occupy Paneth cell positions between Lgr5 stem cells. These Dll1+ cells serve as an alternative source of Notch signals that are essential for Lgr5+ stem cell maintenance.
In the present study, we show that specific, acute ablation of Paneth cells in mice does not affect the Lgr5+ stem cell population. Similar observations have been published by others and have been interpreted to mean that epithelial cells play no essential role in the crypt niche (31, 32). This interpretation is difficult to understand, however, given that Lgr5 stem cells are crucially dependent on Notch signals, which can be generated only by direct contact with neighboring cells. Of note, the basal lamina of the intestinal epithelium precludes the possibility that Notch signals can emanate from mesenchymal cells located in the subepithelium. We show here that an adaptation of the intestine occurs on Paneth cell loss, and an alternative niche is derived that represents a subpopulation of the secretory lineage, that is, the enteroendocrine and Tuft cells. These cells carry Dll1 and thus act as a source of Notch signals to maintain the Lgr5+ stem cells.
Previous studies have revealed that loss of Lgr5 stem cells in normal crypts is countered by the recruitment of more differentiated cells back into the stem cell pool (42, 46–49). In cancer, similar processes appear to play out; the destruction of Lgr5 cancer stem cells in primary intestinal tumors of murine or human origin results in their rapid replacement owing to the plasticity of differentiated daughter cells (50, 51). This analysis reveals yet another level of plasticity within the intestinal epithelium. We found that targeting the niche cells (i.e., Paneth cells) similarly triggers their rapid replacement by different types of secretory cells. These observations lead to the sobering prediction that in cancer, both stem cells and niche cells are replaceable, complicating therapies based on the cancer stem cell paradigm.
Materials and Methods
Mice.
All mouse experiments were conducted under a project license granted by the Central Animal Testing Committee of the Dutch government and approved by the Royal Netherlands Academy of Arts and Sciences–Hubrecht Institute Animal Welfare Body. pLysdsRED, pLysCreErt2, and pLysDTR mice were backcrossed with C57Bl6 mice for at least 5 generations. All other mice lines have been described elsewhere (8, 23, 35, 42). Both male and female mice were used for all experiments. Details on the experimental procedures, treatment regimens, and DT and tamoxifen dosages injected are provided in SI Appendix, Materials and Methods.
Immunohistochemistry, Single-Molecule In Situ Hybridization, and In Situ Hybridization.
Histological analysis of intestinal sections was performed as described previously and described in detail in SI Appendix, Materials and Methods.
TEM Analysis.
As described previously (8), 1.5-cm pieces of intestine were fixed in Karnovsky’s fixative (2% paraformaldehyde, 2.5% glutaraldehyde, 0.1 M sodium cacodylate, 2.5 mM CaCl2, and 5 mM MgCl2, pH 7.4) overnight at room temperature. The samples were embedded in Epon resin and examined with a Tecnai T12 Spirit transmission electron microscope equipped with an Eagle 4k × 4k CCD camera (Thermo Fisher Scientific).
X-Gal Staining.
To determine the pattern of Cre-mediated recombination of the RosaLSL-LacZ reporter locus on day 10 after tamoxifen-induced Cre induction, X-gal staining was performed on the isolated intestine as described previously (27) and explained in detail in SI Appendix, Materials and Methods.
Vibratome Sectioning and Confocal Imaging.
Isolated intestinal tissues were washed with PBS and fixed for 30 min in a 4% formaldehyde solution at room temperature. Fixed intestines were washed in PBS, embedded in 4% UltraPure low melting point agarose (Invitrogen), and vibratome-sectioned (Microm HM 650 V; Thermo Fisher Scientific) at 100 µm. The 100-µm sections were mounted in Vectashield hard set mounting medium with DAPI (Vector Laboratories) and analyzed within 24 h for dsRed expression with a confocal microscope. GFP signals were enhanced by incubation of sections after 2 h of permeabilization in PBS containing 1% BSA, 1% DMSO, and 0.2% Triton X-100 (PBDT) at room temperature, followed by overnight incubation at 4 °C with rabbit anti-GFP (1:500; Invitrogen) in PBDT. After four 15-min washes in PBDT, sections were incubated for 4 h in goat anti-rabbit-488 (1:500) in PBDT. After 4 more 15-min washes in PBDT, sections were incubated for 20 min with 4 µg/mL DAPI, then mounted in Vectashield. Images of intestinal sections were acquired with a Leica SP5 confocal microscope.
Flow Cytometry.
Crypts pooled from 3 mice per experimental group (pLysDTR and control mice treated with DT for 6 d) were isolated using a previously described protocol (24). Crypts were incubated in PBS containing 5 mM EDTA for 45 min. Dissociated cells were centrifuged for 5 min at 1,500 rpm, taken up in 5 mL of PBS, and filtered through a 70-µM EASYstrainer (Greiner Bio-One). After centrifugation, the cells were collected in 5 mL of TrypLE (Thermo Fisher Scientific) containing 2 U/µL of DNase I (Sigma-Aldrich). Cells were incubated at 37 °C for a maximum of 45 min, pipetted up and down, and checked every 10 min. On centrifugation, pellets of single cells were washed in Advanced DMEM/F12 (AdDMEM; Thermo Fisher Scientific) and then incubated in AdDMEM with rat anti-mouse CD24-PE (1:200; BioLegend) or rat isotype control (BioLegend) for 30 min on ice. Washed cells were resuspended in 800 µL of AdDMEM containing 2 U/µL DNase I and, after the addition of 2.5 µg/mL DAPI (Life Technologies) and filtration by a Falcon blue-cap strainer (Corning), analyzed and/or isolated with a MoFlo high-speed cell sorter (Dako Cytomation).
CEL-Seq Library Preparation.
The protocol for this step was as described previously (45). FACS-sorted cells were processed using the previously described CEL-seq technique with the following modifications (42). A 4-bp random barcode was inserted in the primer as a unique molecular identifier (UMI) between the cell-specific barcode and the poly T stretch. Dried RNA, prepared from single cells by TRIzol extraction with 2 µg of glycogen (Life Technologies), was resuspended in 5 ng/µL primer solution, denatured at 70 °C for 2 min, and quickly chilled on ice, followed by the addition of First-Strand synthesis mix (Invitrogen). Libraries were sequenced with the Illumina NextSeq 500 sequencing system using 75-bp paired-end sequencing.
Data Analysis.
Data analysis was performed as described previously (44). In brief, paired-end reads from the Illumina sequencing were aligned to the human transcriptome with Burrows–Wheeler Alignment. The 3′ mate contains the barcode and cell identity information, while the 5′ mate was mapped to gene models. Reads that mapped to multiple locations were discarded. Duplicate reads that had identical combinations of library, cellular, and molecular barcodes and were mapped to the same gene were removed. Transcript counts were then adjusted to the expected number of molecules based on counts, 264 possible UMIs, and Poisson counting statistics (52).
Data Availability.
All data generated or analyzed during this study are included in this published article and its SI Appendix.
Supplementary Material
Acknowledgments
We thank Stefan van der Elst for assisting with the FACS sorting experiments and Harry Begthel for performing some of the histological stainings.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801888117/-/DCSupplemental.
References
- 1.Tan D. W., Barker N., Intestinal stem cells and their defining niche. Curr. Top. Dev. Biol. 107, 77–107 (2014). [DOI] [PubMed] [Google Scholar]
- 2.van Es J. H., et al. , Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Koch U., Lehal R., Radtke F., Stem cells living with a Notch. Development 140, 689–704 (2013). [DOI] [PubMed] [Google Scholar]
- 4.He X. C., et al. , BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 36, 1117–1121 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Gregorieff A., Liu Y., Inanlou M. R., Khomchuk Y., Wrana J. L., Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Nusse R., Clevers H., Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Buczacki S. J., et al. , Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature 495, 65–69 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Barker N., et al. , Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Bjerknes M., Cheng H., Gastrointestinal stem cells, II: Intestinal stem cells. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G381–G387 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Schepers A. G., Vries R., van den Born M., van de Wetering M., Clevers H., Lgr5 intestinal stem cells have high telomerase activity and randomly segregate their chromosomes. EMBO J. 30, 1104–1109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKernan D. P., Egan L. J., The intestinal epithelial cell cycle: Uncovering its “cryptic” nature. Curr. Opin. Gastroenterol. 31, 124–129 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Ireland H., Houghton C., Howard L., Winton D. J., Cellular inheritance of a Cre-activated reporter gene to determine Paneth cell longevity in the murine small intestine. Dev. Dyn. 233, 1332–1336 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Bjerknes M., Cheng H., The stem-cell zone of the small intestinal epithelium, I: Evidence from Paneth cells in the adult mouse. Am. J. Anat. 160, 51–63 (1981). [DOI] [PubMed] [Google Scholar]
- 14.Ouellette A. J., Paneth cells and innate mucosal immunity. Curr. Opin. Gastroenterol. 26, 547–553 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Clevers H. C., Bevins C. L., Paneth cells: Maestros of the small intestinal crypts. Annu. Rev. Physiol. 75, 289–311 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Salzman N. H., Ghosh D., Huttner K. M., Paterson Y., Bevins C. L., Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422, 522–526 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Vaishnava S., Behrendt C. L., Ismail A. S., Eckmann L., Hooper L. V., Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U.S.A. 105, 20858–20863 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzman N. H., et al. , Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–83 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yilmaz O. H., et al. , mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igarashi M., Guarente L., mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell 166, 436–450 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Stappenbeck T. S., McGovern D. P. B., Paneth cell alterations in the development and phenotype of Crohn’s disease. Gastroenterology 152, 322–326 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato T., et al. , Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Schepers A. G., et al. , Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337, 730–735 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Sato T., et al. , Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415–418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farin H. F., Van Es J. H., Clevers H., Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology 143, 1518–1529.e7 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Korinek V., et al. , Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 19, 379–383 (1998). [DOI] [PubMed] [Google Scholar]
- 27.van Es J. H., et al. , A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Mol. Cell. Biol. 32, 1918–1927 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riccio O., et al. , Loss of intestinal crypt progenitor cells owing to inactivation of both Notch1 and Notch2 is accompanied by derepression of CDK inhibitors p27Kip1 and p57Kip2. EMBO Rep. 9, 377–383 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellegrinet L., et al. , Dll1- and Dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230–1240.e7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aoki R., et al. , Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell. Mol. Gastroenterol. Hepatol. 2, 175–188 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durand A., et al. , Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc. Natl. Acad. Sci. U.S.A. 109, 8965–8970 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim T. H., Escudero S., Shivdasani R. A., Intact function of Lgr5 receptor-expressing intestinal stem cells in the absence of Paneth cells. Proc. Natl. Acad. Sci. U.S.A. 109, 3932–3937 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Es J. H., de Geest N., van de Born M., Clevers H., Hassan B. A., Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat. Commun. 1, 18 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cross M., Renkawitz R., Repetitive sequence involvement in the duplication and divergence of mouse lysozyme genes. EMBO J. 9, 1283–1288 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snippert H. J., et al. , Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology 136, 2187–2194.e1 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Ivanova A., et al. , In vivo genetic ablation by Cre-mediated expression of diphtheria toxin fragment A. Genesis 43, 129–135 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth S., et al. , Paneth cells in intestinal homeostasis and tissue injury. PLoS One 7, e38965 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snippert H. J., Schepers A. G., van Es J. H., Simons B. D., Clevers H., Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 15, 62–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Flier L. G., Haegebarth A., Stange D. E., van de Wetering M., Clevers H., OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology 137, 15–17 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Jensen J., et al. , Control of endodermal endocrine development by Hes-1. Nat. Genet. 24, 36–44 (2000). [DOI] [PubMed] [Google Scholar]
- 41.VanDussen K. L., et al. , Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 139, 488–497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Es J. H., et al. , Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 14, 1099–1104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grün D., et al. , Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Grün D., et al. , De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19, 266–277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itzkovitz S., et al. , Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat. Cell Biol. 14, 106–114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jadhav U., et al. , Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell 21, 65–77.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tetteh P. W., et al. , Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203–213 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Yan K. S., et al. , Intestinal enteroendocrine lineage cells possess homeostatic and injury-inducible stem cell activity. Cell Stem Cell 21, 78–90.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu S., et al. , Paneth cell multipotency induced by Notch activation following injury. Cell Stem Cell 23, 46–59.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Sousa e Melo F., et al. , A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 543, 676–680 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Shimokawa M., et al. , Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 545, 187–192 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Grün D., Kester L., van Oudenaarden A., Validation of noise models for single-cell transcriptomics. Nat. Methods 11, 637–640 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its SI Appendix.