Significance
Understanding the molecular response of plants to drought is critical to efforts to improve agricultural yields under increasingly frequent droughts. We grew 2 cultivars of the naturally drought-tolerant food crop sorghum in the field under drought stress. We sequenced the mRNA from weekly samples of these plants, resulting in a molecular profile of drought response over the growing season. We find molecular differences in the 2 cultivars that help explain their differing tolerances to drought and evidence of a disruption in the plant’s symbiosis with arbuscular mycorrhizal fungi. Our findings are of practical importance for agricultural breeding programs, while the resulting data are a resource for the plant and microbial communities for studying the dynamics of drought response.
Keywords: drought, RNA-Seq, S. bicolor, arbuscular mycorrhizal fungi
Abstract
Drought is the most important environmental stress limiting crop yields. The C4 cereal sorghum [Sorghum bicolor (L.) Moench] is a critical food, forage, and emerging bioenergy crop that is notably drought-tolerant. We conducted a large-scale field experiment, imposing preflowering and postflowering drought stress on 2 genotypes of sorghum across a tightly resolved time series, from plant emergence to postanthesis, resulting in a dataset of nearly 400 transcriptomes. We observed a fast and global transcriptomic response in leaf and root tissues with clear temporal patterns, including modulation of well-known drought pathways. We also identified genotypic differences in core photosynthesis and reactive oxygen species scavenging pathways, highlighting possible mechanisms of drought tolerance and of the delayed senescence, characteristic of the stay-green phenotype. Finally, we discovered a large-scale depletion in the expression of genes critical to arbuscular mycorrhizal (AM) symbiosis, with a corresponding drop in AM fungal mass in the plants’ roots.
Drought dramatically impacts crop growth and productivity, causing losses estimated at $2.9 billion annually (1–5). While the physiological responses of plants to drought have been extensively studied in model organisms under controlled laboratory or greenhouse conditions, results from these studies typically translate poorly to field-grown plants that are often distantly related to model organisms. The collection of molecular information gathered from crops grown under field conditions will likely contribute to a more complete understanding of the complex interactions between environmental factors and drought responses that crops encounter in the field during the growing season. Thus, a better mechanistic understanding of drought responses in the field is needed to develop effective drought-mitigation strategies.
Sorghum [Sorghum bicolor (L.) Moench], one of the 5 major cereal crops in use today worldwide, is an ideal organism for studying genomic responses to drought, due to its innate drought tolerance, small sequenced diploid genome, extensive germplasm, and C4-photosynthetic pathway (6). While drought-tolerant, the phenotypic impact of drought on sorghum can be impactful, depending on the developmental stage at which it occurs. Drought occurring before flowering (preflowering drought), when panicle development takes place, typically results in leaf rolling and uncharacteristic leaf erectness (to reduce heat load and transpiration), leaf bleaching, delayed flowering, and poor panicle exsertion and development (7). The most commonly used metrics to quantify a plant’s tolerance to preflowering drought are related to floral development and are measured after the termination of drought. Preflowering drought-recovery mechanisms are thus of special interest to fully understand these processes (7). Postflowering drought, in contrast, causes premature leaf senescence and stalk lodging. Drought-tolerant cultivars of sorghum are typically characterized as tolerant to either preflowering or postflowering drought, with known cultivars demonstrating strong drought tolerance to only 1 of these 2 drought stresses (7). A subset of postflowering drought-tolerant genotypes have the capacity to maintain high levels of chlorophyll in severe postflowering drought stress and are thus known as stay-green genotypes (7–9). Of critical importance, BTx642 has the added feature of being a “functional stay-green” variety because it also continues to fill grain at levels comparable to nondroughted controls under severe postflowering drought, implying that the totality of photosynthesis continues to function under these severe postflowering drought conditions (10). Previous studies have identified numerous quantitative trait loci (QTLs) associated with this stay-green phenotype, and recombinant inbred lines containing those QTLs showed modified canopy development, root growth, and different rates of water usage and uptake (10). Understanding the exact molecular underpinnings of the stay-green varieties may serve as a robust resource for improving drought tolerance in other crop plants.
The full genome sequencing and resequencing of several S. bicolor cultivars (11, 12) have enabled a number of transcriptome surveys of drought responses in sorghum. However, these surveys have been restricted to greenhouse studies (i.e., not in field conditions) using early time points (13–18), and all but 1 of these have taken samples exclusively from leaves (14–18). Several studies profile responses to various abiotic stresses (13–18), including drought (13–15, 18), and biotic stimuli (17). Taken together, these studies confirm that drought elicits a broad, multifaceted response, notably featuring coordination between phytohormone signaling pathways, reduction of photosynthetic gene expression, and alteration in expression of genes involved in stress-induced leaf senescence (19, 20). While these studies provide insight into the broader processes involved in environmental stresses in sorghum, they are limited by the type of tissue assayed (primarily leaf), as well as the duration of treatment (2 mo, which is before manifestation of postflowering stay-green phenotype).
Here, we provide a field-based, temporal transcriptomic study of sorghum. We profiled almost 400 transcriptomes sampled weekly from leaves and roots of 2 genotypes (the preflowering drought-tolerant RTx430 and the stay-green type BTx642), under control watering conditions and 2 different drought regimes. This transcriptomic study also follows the recovery from preflowering drought, by reintroducing water after anthesis, and the effects of postflowering drought, by removing water after anthesis.
Our weekly time series reveals that sorghum detects and adapts to preflowering and postflowering drought stresses within the 1-wk resolution of our study, ultimately influencing the gene expression of over 40% of the genome. This transcriptomic response includes genes involved in critical functions, such as biotic defense and abiotic stress responses, and photosynthesis. We demonstrate that the stay-green genotype, BTx642, maintains high levels of photosynthetic gene expression during postflowering drought, compared to RTx430. We highlight, as part of the drought-tolerance response, a potential role for reactive oxygen species (ROS) scavenging and glutathione S-transferase (GST) activity, which is associated with a previously identified stay-green QTL.
Finally, these data are part of a larger drought project, EPICON (Epigenetic Control of Drought Response in S. bicolor), funded by the US Department of Energy (DOE, Grant DESC0014081), designed to study the molecular responses of sorghum to agronomically relevant drought conditions. As part of this effort, the root and rhizosphere microbiomes were simultaneously sampled, and analysis of bacterial and fungal communities have been published (21, 22). Our mRNA-sequencing (mRNA-Seq) data combined with these microbial data result in an unprecedented fine-grain study of the drought effects on sorghum and associated microbial communities in the field. In particular, the results of ref. 22 demonstrated that arbuscular mycorrhizal (AM) fungi show remarkably strong succession with sorghum development. Our study of the transcriptome of the roots of these plants further shows that drought precipitates a coordinated reduction in the transcription of genes important to AM symbiosis, a drop that we show to be coincidental with a loss of total AM fungal biomass under drought. When drought is relieved, the preflowering drought-tolerant genotype, RTx430, is able to resume this critical symbiosis more quickly than the postflowering tolerant variety, BTx642.
This transcriptomic study of field-grown sorghum and, paired with soil, rhizosphere, and root microbiomes, represents an unparalleled resource for studying drought tolerance in agronomically relevant settings.
Results and Discussion
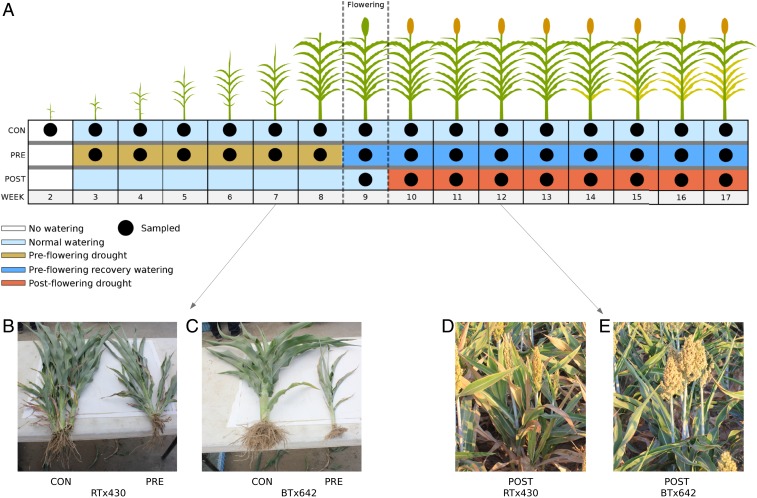
We conducted a large-scale field experiment exposing sorghum plants to 3 distinct watering schemes over a 17-wk period. The control condition consisted of irrigation once per week, 5 d before sampling, starting with week 3 and continuing throughout the experiment. Preflowering drought was imposed by providing no irrigation prior to flowering (weeks 3 to 8), then resuming watering in subsequent weeks (drought recovery; weeks 9 to 17). Postflowering drought was implemented by halting irrigation after the weekly watering applied before week 9 (flowering), resulting in postflowering drought exposure for samples in weeks 10 to 17 (SI Appendix, Methods and Materials and Fig. 1). Roots and leaves were sampled weekly, starting in week 2 and continuing through week 17. We conducted this experiment on 2 sorghum genotypes: RTx430, a genotype tolerant to preflowering drought and sensitive to postflowering drought; and BTx642, a stay-green genotype, tolerant to postflowering drought (having the ability to delay leaf senescence) and sensitive to preflowering drought (23, 24). Both of these varieties of sorghum are photoperiod-insensitive and do not exhibit drought escape (SI Appendix, Table S6), defined as flowering earlier under drought. Further, in sorghum, drought tolerance is defined as demonstrating high yields under drought stress, which both of these varieties have been shown to do under their respective drought stresses (7, 25– 27). Cumulatively, these points ensure that the plants exhibit drought tolerance. At each weekly sampling, we took 3 independent, biological replicates, each consisting of 10 plants.
Fig. 1.
Experimental design. (A) Schematic overview of the experimental design for control (CON), preflowering (PRE), and postflowering (POST) drought. Black dots represent whether plants were sampled for the specified treatment/week, and the color of the boxes reflects the irrigation status for the plants (light blue, watered; brown, preflowering drought; dark blue, watered, preflowering recovery; red, postflowering drought); no plots were irrigated prior to week 3 (white boxes). All samples marked as “watered” were irrigated 5 d prior to sample collection. Samples from week 3 of preflowering drought and week 10 of postflowering drought are considered the 1st samples of drought-exposed plants from the 2 drought regimes (i.e., the 1st samples experiencing different watering regimes from control; Materials and Methods). (B and C) Photos of side-by-side comparisons of control (CON; left) and preflowering droughted (PRE; right) plants at week 7 for RTx430 (B) and BTx642 (C). (D and E) Field picture at week 12 after 3 wk of postflowering drought of RTx430 (D) and BTx642 (E), showing delayed senescence in this stay-green variety.
Paired-end RNA-Seq was performed on 198 root and 198 leaf samples (396 samples total) (28). Each sample was independently aligned, processed for quality control, and normalized (SI Appendix, Methods and Materials). To identify genes affected by drought, we first modeled the expression of each gene as a function across time and selected genes that exhibited a significant deviation in expression between control versus either drought treatment. We then clustered these genes based on their expression model, independently across tissues (root and leaf) and treatments (preflowering and postflowering drought stress) (SI Appendix, Methods and Materials). Within each of the 60 clusters obtained, we assessed the biological functions of the component genes by performing gene-enrichment analysis to identify overrepresented networks from Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology terms (SI Appendix, Cluster Summaries).
Overall, our analysis showed a massive transcriptional response to drought. From all comparisons performed, we identified a cumulative total of 10,727 genes (44% of expressed genes) significantly affected by drought stress under agriculturally relevant conditions, consistent with the scale of gene-expression changes observed in laboratory and greenhouse studies (15, 18) (SI Appendix, Table S1). In analyzing the time-dependent transcriptional response to drought, both treatments resulted in a large-scale response within 1 wk of drought exposure (week 3 for preflowering drought and week 10 for postflowering drought), with roughly 10% of all expressed genes showing a change in the 1st wk of drought exposure (SI Appendix, Table S2). These expression differences occurred several weeks before the plants exhibited physical signs of stress (SI Appendix, Figs. S2 and S3).
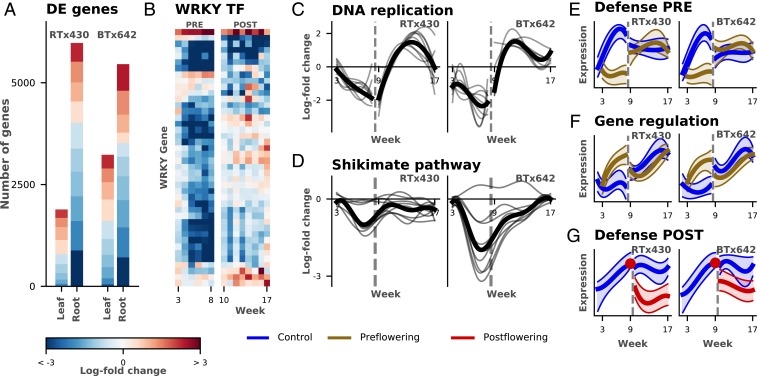
Plants recovering from preflowering drought also exhibited dramatic expression changes in both root and leaf samples. Of the genes affected during preflowering drought, 75% reverted to expression levels similar to that of well-watered control plants within 1 wk of watering resumption. However, a large subset of genes (2,424 genes) were still differentially expressed following the resumption of watering. This included a large number of genes from the DNA-replication pathway (in particular, homologs to MCM 2, 3, 4, 6, and 7) that were strongly down-regulated in leaf tissue in preflowering and postflowering drought and in the root under postflowering drought stress, but were overexpressed (relative to control) during preflowering drought recovery (Fig. 2C and SI Appendix, Figs. S7 and S8). Taken together, these results highlight the global and temporally complex nature of drought and drought recovery in field-grown sorghum.
Fig. 2.
Analysis of temporal transcriptional changes. (A) Bar plot of significantly differentially expressed (DE) genes with a log2-fold difference in expression of at least 2 between drought and watered conditions in preflowering drought (see SI Appendix, Fig. S4 for postflowering drought). The bar height corresponds to the number of genes found DE; each bar is broken down to indicate the number of genes falling in different categories of log-fold change via the color scale (key shown at bottom). (B) Heatmap of average log2-fold change per week between droughted and watered plants for WRKY transcription factors (TF) shows the specificity of preflowering (Left) and postflowering (Right) drought responses in roots (color scale key at bottom). (C and D) Log-fold change between droughted and watered plants (y axis) estimated per-gene as a smoothed function over time (x axis). The dark line corresponds to the average. C demonstrates overexpression during recovery in 9 genes in the DNA-replication pathway, while D shows the genotype difference in drought response in the 9 genes of the shikimate pathway and the overall stronger down-regulation in BTx642. (E–G) Illustrative examples of temporal patterns found via clustering of the genes (see SI Appendix, Cluster Summaries for all clusters). The scaled expression of the cluster average (y axis) is plotted against the week (x axis), for each of the droughted and watered conditions along with the 20 to 80 percentile bands. (E) Root expression remains constant under preflowering drought (PRE), while developmentally changing in the watered condition. (F) Preflowering drought and watered conditions show temporal changes. (G) Postflowering drought (POST) shows initial drop, while expression in watered plants remains constant.
Roots Show Greater Transcriptional Disruption than Leaves.
While both leaf and root samples showed a widespread response to drought, we observed that root samples exhibited both a larger number of differentially expressed genes, as well as a larger absolute change in expression of those genes, compared to leaf samples (Fig. 2A and SI Appendix, Fig. S4 and Table S1). This difference in expression between root and leaf tissue is rapidly apparent; in the 1st wk of drought exposure, twice as many genes were differentially expressed in roots than in leaves (SI Appendix, Table S2). Under both drought treatments, the tolerant genotype (RTx430 for preflowering and BTx642 for postflowering) had fewer genes differentially expressed in the leaf after a week of drought exposure, while there was not a large difference in the number of transcriptional changes between the 2 genotypes in the root. These observations suggest that while the root is more substantially affected overall by drought (possibly due to its role in detection and absorption of water), genotype-specific adaptations to preflowering and postflowering drought stress may be primarily determined by attempts to maintain normal activity in leaves.
We observed more down-regulated than up-regulated transcripts in roots in response to drought (Fig. 2A). We found enrichment of several functional categories among these down-regulated genes—notably, a large number of plant defense pathways (e.g., response to fungi and pathogens) (SI Appendix, Figs. S11–S13). Among these pathways, we observed that expression of the majority of WRKY transcription factors (known to regulate, for example, pathogen-defense responses) was suppressed in leaf and root tissue (29) (Fig. 2B and SI Appendix, Figs. S5 and S6), while jasmonic and salicylic acid-responsive genes were sharply down-regulated in expression in roots (SI Appendix, Figs. S9 and S10). This observation further supports observed phenomena that responsiveness of plants to the biotic environment is affected by drought (30).
Preflowering Drought Causes More Complex Temporal Changes than Postflowering Drought.
Most genes that we identified as responsive to postflowering drought exhibited an expression pattern of rapid change in the 1st wk of drought exposure, followed by little variation in expression in later weeks, relative to control-treated plants (Fig. 2G). This expression pattern differed from that seen for genes responsive to preflowering drought, which showed a variety of drought-specific temporal patterns. For example, some clusters in preflowering drought showed relatively constant gene expression under drought, in contrast to the well-watered samples (Fig. 2E), while genes in other clusters demonstrated expression patterns that varied in both drought and control (Fig. 2F). This may reflect greater developmental plasticity (and a more varied suite of drought-tolerance strategies) prior to flowering, or the comparative severity of responses to postflowering drought versus preflowering drought. Despite the large-scale differences in gene-expression patterns between the 2 drought conditions, we found many genes that responded in the same direction to both preflowering and postflowering drought (leaf: 41%; root: 30%). Of the genes that showed patterns that differed in the 2 drought conditions, the most common pattern was for the gene to show strong effects in 1 drought condition and only small or nonexisting effects in the other condition (root: 2,639 genes; leaf: 1,155 genes). For example, in the root samples, the WRKY transcription factors, highlighted earlier as largely suppressed under drought, were particularly sensitive to preflowering, but less so to postflowering, drought stress (Fig. 2B). A smaller subset of genes actually showed opposite regulatory patterns under the 2 conditions (51 in leaf and 235 in root; SI Appendix, Table S3).
Large Differences between Genotypes Found in Many Transcriptional Pathways.
To better understand differences in drought-adaptation strategies, we compared expression patterns between genotypes. From this analysis, we identified 3,977 genes (16% of expressed genes) that were significantly affected by drought in a genotype-specific way (SI Appendix, Methods and Materials and Table S4). Approximately 25% of these genes were differentially expressed in only 1 genotype. Other genes had similar qualitative responses in both genotypes, but differed in the magnitude of that response (depending on the drought regime and sample condition, between 30 and 50% of the 3,977 genes were found to be differentially expressed; SI Appendix, Table S4). Interestingly, a small subset of the genotype-dependent, drought-responsive genes were regulated in opposing directions between the 2 genotypes (SI Appendix, Table S4). Lastly, the 2 genotypes exhibited strong differences in their expression during preflowering drought recovery, during which 50% of those 3,977 genes respond differently.
Among the annotated functions of genotype-specific, drought-responsive genes, we observed a high representation of genes within the shikimate pathway, which were strongly down-regulated during preflowering in drought-sensitive BTx642, but not in the tolerant RTx430 (Fig. 2D). Aromatic amino acids produced by this pathway are essential precursors for a wide range of secondary metabolites that are important for plant growth, including auxin and lignin. We also observed that 479 genes were not expressed in 1 of the 2 genotypes (SI Appendix, Table S4), including homologs of the WAK kinase family proteins, CYP and GST homologs, heat shock proteins, and dehydration-responsive element-binding factors.
Data from our transcriptomic study are also a resource for exploration of the developmental differences in the genotypes under normal watering conditions, and our analysis demonstrated that these 2 genotypes show large constitutive transcriptomic changes, with 22% of all genes differentially expressed with at least a log-fold change of 2 between the 2 genotypes. Constitutive differences between the genotypes govern the potential of the plant to respond to drought and could potentially be important in understanding differential responses to drought.
Stay-Green Phenotype Is Associated with Differences in Photosynthesis and ROS Scavenging.
Preflowering and postflowering drought influences the expression of many genes involved in photosynthesis (31). We observed that photosynthetic genes (especially light-harvesting complex [LHC] genes) shared a similar preflowering drought response in leaf tissue in both genotypes: down-regulated or unchanged expression during the drought weeks, but sharply up-regulated expression during drought recovery (SI Appendix, Fig. S14). The preflowering drought-tolerant genotype, RTx430, showed a greater increase than BTx642 in photosynthetic gene expression during recovery, a pattern paralleled by total chlorophyll levels (Fig. 3D). The up-regulation of photosynthesis genes during recovery may represent a compensatory response to drought-induced developmental delay. More generally, the data suggest that RTx430 not only exhibits greater tolerance during preflowering drought, but also a more rapid recovery when watering was resumed.
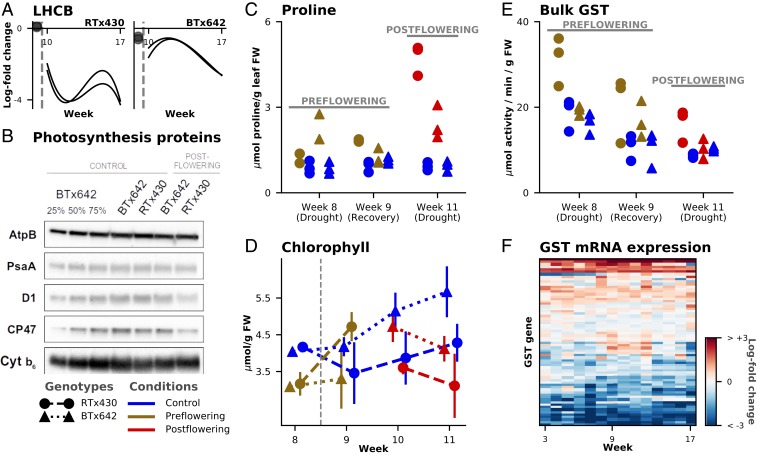
Fig. 3.
Stay-green phenotype, photosynthesis, and drought. (A) Two LHC PSII genes (LHCBs) (Sobic.003G209800 and Sobic.003G209900) show stronger down-regulation in RTx430 than BTx642 under postflowering drought stress. (B) Immunoblot analysis of representative subunits of photosynthetic complexes PSI (PsaA), PSII (D1 and CP47), and cytochrome b6f complex (Cyt b6) under control and 3 wk postflowering drought (week 11) with ATP synthase (AtpB) as a loading control (additional replicate is in SI Appendix, Fig. S15). (C) Proline levels determined on fresh weight (FW) leaf samples via a ninhydrin spectrophotometric assay for both drought conditions and genotypes. (D) Average value of 3 HPLC assays of total chlorophyll levels (y axis) plotted against week (x axis) showing constitutive differences between BTx642 and RTx430 after flowering and a sharp increase in chlorophyll levels under preflowering drought of RTx430; bars indicate ±SD intervals. (E) Bulk GST enzymatic activity in leaf extracts (y axis) at 3 timepoints (x axis). (F) Heatmap of centered log2 difference between the constitutive gene-expression values of RTx430 and BTx642 for the 68 expressed GST genes.
Under postflowering drought, we observed a significant down-regulation of homologs of photosystem II (PSII) LHCB genes, Sobic.003G209800 and Sobic.003G209900, with a stronger down-regulation in RTx430 than BTx642 (Fig. 3A). Similarly, at the protein level in RTx430, we observed reduced levels in 2 core PSII subunits, D1 (PsbA) and CP47 (PsbB), to 25 to 50% of the levels observed in the well-watered control (Fig. 3B). In contrast, D1 and CP47 protein levels were unchanged during postflowering drought in BTx642, relative to the well-watered control. Thus, BTx642 retains a greater capacity to participate in photochemistry at PSII in postflowering drought.
Limiting excess accumulation of ROS is a vital component of drought tolerance (32). Proline biosynthesis is a key regulator of ROS-dependent processes and osmoprotection. We observed constitutively higher mRNA expression of the rate-limiting enzyme of proline biosynthesis, P5CS2 (Sobic.003G356000; SI Appendix, Fig. S18), in BTx642 compared to RTx430, consistent with the results of a greenhouse study of 45-d-old sorghum plants (16). We also observed similar increases in P5CS2 mRNA expression under drought conditions for both genotypes. While our gene-expression results supported higher capacity for proline biosynthesis in BTx642, we observed lower levels of drought-induced proline accumulation in BTx642 relative to RTx430 (Fig. 3C), and we did not see a constitutive difference in proline abundance (16). This indicates a reduced demand for proline as an osmoprotectant and regulator of ROS in field-grown BTx642 in postflowering drought, despite an apparent higher capacity for proline biosynthesis.
GSTs are also key regulators of ROS in plant cells (18, 33, 34), and expression of GSTs is induced by drought (18). We observed both constitutive and drought-specific expression differences between the 2 genotypes in multiple individual GST genes, despite similar levels of total GST mRNA expression between the genotypes in both control and drought conditions. One such gene, GST29 (Sobic.003G264400), was strongly up-regulated in both preflowering and postflowering drought in BTx642, but was not differentially expressed in RTx430 (SI Appendix, Fig. S16). GST29 is located near a known stay-green QTL (STG2) on chromosome 3 (27) and led us to speculate that the specific roles of individual GSTs in ROS scavenging may result in genotypic differences in drought tolerance. To further explore genotype-specific GST redox-scavenging activity, we measured total GST enzymatic activity in leaf extracts sampled during weeks 8, 9, and 11 (representing preflowering drought, recovery, and postflowering drought, respectively). We detected similar levels of bulk GST enzymatic activity in both genotypes under control conditions, but observed that both preflowering and postflowering drought led to significantly greater increases in GST activity in RTx430 compared to BTx642 (Fig. 3E). Taken together, the lower proline levels, lower bulk GST enzymatic activity, elevated LHCB gene expression, and elevated PSII protein levels in BTx642 relative to RTx430 during postflowering drought all point toward lower levels of perceived drought-induced ROS stress in BTx642 (Fig. 3).
In addition to constitutive up-regulation of GST29, we observed genotype-specific differences in 2 other pathways that might enable a higher capacity for the control of ROS levels in BTx642. First, a chloroplast-targeted ferredoxin FD3 (Sobic.003G364400; SI Appendix, Fig. S17), located close to STG1, was constitutively up-regulated in BTx642, consistent with an increase in the efficiency of ROS scavenging in chloroplasts (35). Second, we found that BTx642 maintained constitutively higher mitochondrial alternative oxidase (AOX) capacity (SI Appendix, Fig. S19), a feature that prevents excess ROS accumulation in plant mitochondria (36) and that might be important for drought tolerance (37). From fresh field-sampled leaves, we found that AOX-mediated electron transport capacity was significantly higher in BTx642 leaf tissue relative to RTx430 (SI Appendix, Fig. S19) (false discovery rate-adjusted P value ), despite similar dark respiration rates. Thus, we hypothesized that BTx642’s postflowering drought tolerance may be due to enhanced redox balancing in the chloroplast and mitochondria. Given the observed relative expression and protein abundance of core photosynthetic machinery, BTx642 is better at retaining the proteins necessary to sustain photochemistry under postflowering drought, likely contributing to its classification as a functional stay-green variety. This work provides an attractive dataset to identify candidate genes that improve productivity under drought in other agronomically relevant crops.
Symbiosis with AM Fungi Is Disrupted in Drought.
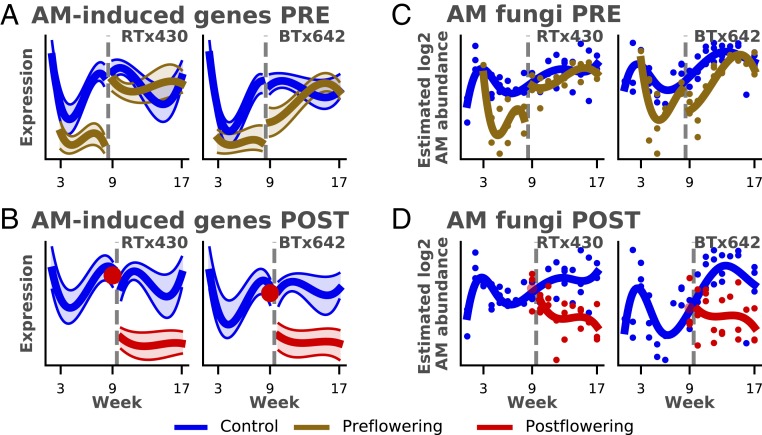
In addition to the abiotic stress responses detailed above, we investigated whether drought affects important biotic interactions, particularly the symbiosis between sorghum and AM fungi. Central to this relationship is the exchange of photosynthetic carbon for phosphorus and other mineral nutrients delivered by the AM fungi (38–40). AM fungi can also alter the plants’ stomatal conductance (38, 41), all of which collectively enhance plant drought tolerance (42). Among the clusters of drought-responsive genes in our dataset, 2 root clusters were identified as highly enriched (with a 69 to 75% overlap) for genes previously identified as markers of AM fungus colonization of the plant (43) (Fig. 4 A and B and SI Appendix, Table S9).
Fig. 4.
Symbiosis of AM fungi and sorghum under drought. (A and B) Average scaled gene expression of genes in the cluster highly overlapping in AM-induced genes (y axis) plotted against week (x axis) for preflowering drought (PRE; A) and postflowering drought (POST; B). (C and D) AM fungal abundance estimates (y axis; Materials and Methods) plotted against week (x axis) for preflowering drought (C) and postflowering drought (D) show the correlation of AM fungal abundance with the gene expression of the AM-induced genes. Gray dashed vertical line indicates the respective watering changes for the 2 drought regimes (Fig. 1).
Under normal watering conditions, these genes appeared to be developmentally regulated, with an initial drop in expression in week 3, followed by a gradual increase throughout the preflowering period until flowering, at which point the expression of these genes plateaued. Under both drought conditions, however, we saw only low expression levels of these AM-associated genes relative to control. In postflowering drought, we saw a drop to low expression levels in the 1st wk of drought exposure relative to control, while in preflowering drought, the effect of drought manifested as a failure to increase the expression of these AM-related genes, resulting in a large difference in expression after prolonged drought. These results suggest that drought diminishes the symbiotic relationship between host plant and AM fungi. Previous results have shown AM fungal biomass decreased by drought in a semiarid steppe (44). This prompted us to estimate the absolute abundance of AM fungi in the roots of our samples under drought by combining the published relative abundance of AM fungi via internal transcribed spacer (ITS) amplicon-sequencing data (22) with additional qPCR of total fungal ribosomal RNA (SI Appendix, Methods and Materials). Our estimates showed a drop in AM fungal levels during preflowering and postflowering drought (Fig. 4 C and D) that corresponded to decreased mRNA expression observed in the AM symbiosis-induced genes. Taken together, these results strongly indicate that drought leads to a reduction of both AM fungal abundance and activity of this vital symbiotic interaction. The AM symbiosis-induced genes include genes related to transport of minerals and photosynthate, biotic defense, and root development (43). In particular, the exchange of photosynthetic carbon for phosphorus (Pi) and other mineral nutrients delivered by the AM fungi is central to AM symbiosis. Among the AM symbiosis-induced genes are genes potentially related to this exchange, such as 4 Pi transporter genes (Sobic.003G243400, Sobic.006G026800, Sobic.001G502000 and Sobic.006G026900), and 12 homologs to senescence-associated gene 12 (SAG12), which all demonstrate this pattern of reduced expression under both drought conditions. This decreased association between plants and AM fungi under drought suggests a complex dynamical interaction, possibly related to reduced availability of photosynthate, decreased demand for phosphorus, or AM fungal mass decreasing due to severe water deficit.
Moreover, these AM symbiosis-induced genes showed very different responses between the 2 genotypes to rewatering after preflowering drought. The preflowering drought-tolerant genotype, RTx430, resumed normal gene-expression levels immediately, while BTx642 exhibited a slower recovery over a period of several weeks. Our estimates of AM fungal abundance suggested a corresponding delay in the recovery of AM fungal symbiosis in the root of BTx642 following preflowering drought, but were not conclusive. The difference in gene-expression recovery for these genes represented the largest genotype-specific drought response for a single functional category of genes that we identified in our data and indicates that AM symbiosis may explain some of the differences seen between the 2 genotypes during preflowering drought recovery. AM fungal colonization is known to increase photosynthetic efficiency and chlorophyll levels (45), so the greater increase in photosynthetic activity seen in RTx430 during the recovery period may potentially be linked to faster reestablishment of AM fungal symbiosis. To our knowledge, the different resilience of AM fungi to drought stress has not been seen before. This genotypic difference in drought recovery further points toward a complex dynamical system between plants and AM fungi.
Immediate Up-Regulation of Stress Response under Drought, with Delayed Recovery after Rewatering.
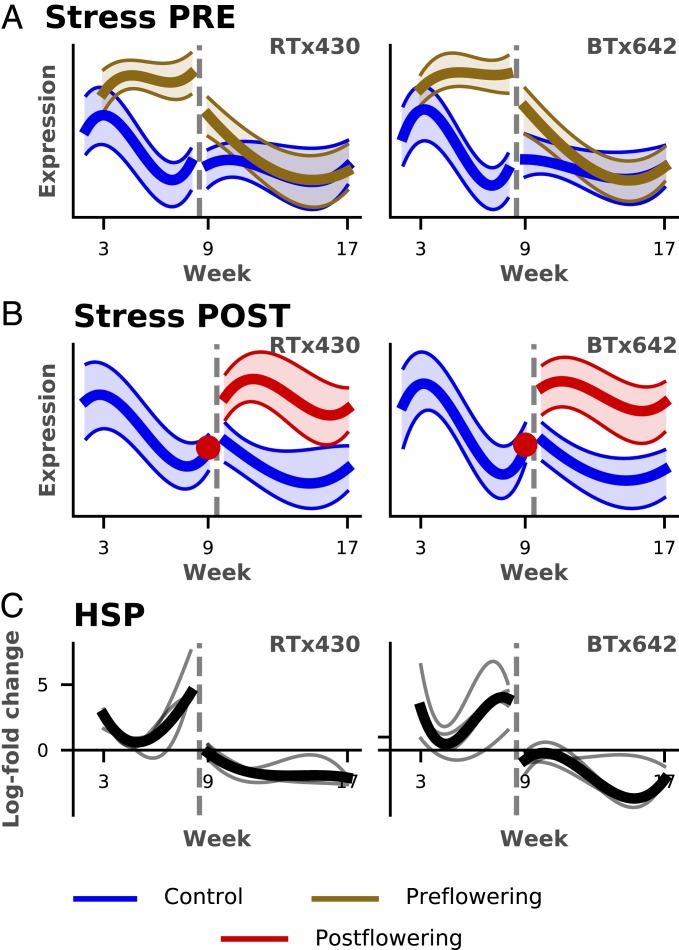
In addition to clusters associated with biotic stress responses and metabolism described above, 9 of the expression clusters had high representation for genes known to be involved in responses to abiotic stress (Fig. 5A and SI Appendix, Cluster Summaries). Common putative functions for these genes included heat shock (e.g., HSF and chaperonins), response to abscisic acid (ABA), response to ROS/oxidative stress, and programmed cell death (SI Appendix, Cluster Summaries). Genes within these stress clusters exhibited dramatic responses to both preflowering and postflowering drought in both leaf and root tissue. Stress cluster genes either remained highly expressed or were sharply up-regulated in plants experiencing preflowering and postflowering drought, respectively, despite sharply declining or remaining low in normally watered plants. In both cases, changes in expression of these genes were apparent in the 1st wk of drought exposure. Similar to the AM cluster genes, many abiotic stress cluster genes also showed a delay in returning to expression levels seen in control plants during the preflowering drought-recovery period, though this delay was seen in both genotypes. Other stress-related genes, including 4 putative heat shock proteins (Fig. 5B), were down-regulated (compared to control plants) during recovery, despite being strongly up-regulated during preflowering drought weeks.
Fig. 5.
Temporal expression of known stress signaling and response pathways. (A and B) The scaled expression of the cluster average (y axis) is plotted against the week (x axis) for 2 clusters in the root found to be enriched in stress responders under preflowering (PRE; A) and postflowering (POST; B) drought, showing strong up-regulation under both drought treatments and slow recovery after resumption of water in preflowering drought (A) (see SI Appendix, Cluster Summaries for all stress-enriched clusters) (C) Log-fold change (y axis) shown as a smooth function of time for 4 heat shock proteins (HSP) demonstrating strong up-regulation under drought, then down-regulation during recovery.
Apart from these stress cluster genes, we found many other genes belonging to known stress-response pathways (e.g., ABA signal transduction and oxidative stress) strongly down-regulated by preflowering and postflowering drought. Peroxidase family homologs, produced in response to oxidative stress (46), were predominant among these negative responders. This group also included genes sharing homology to DREB1 transcription factors [known to respond to low-temperature and drought stress (47, 48)] and PYR6 (an ABA receptor). We also found that HAI2 and HAI3 homologs, which inhibit ABA signaling, and CYP707As, involved in ABA catabolism, were strongly overexpressed. Up-regulation of these latter genes likely buffers ABA signaling to prevent an excessive response (20, 49). The mixed transcriptional response of known stress responders indicates that a robust drought response is already apparent in the 1st wk of drought exposure, resulting in complex feedback dynamics of abiotic stress-response pathways.
While our discussion has primarily focused on the genes that have homology to known biological functions, a large number of differentially expressed genes are completely uncharacterized. Of these, 3,923 are unannotated, while 1,870 have no homologs to Arabidopsis (SI Appendix, Table S5). This is due to the relatively poor level of annotation of the sorghum genome: 42.9% of the transcriptome of sorghum is not annotated. These genes have exciting potential for trait-engineering efforts in sorghum, as well as other (more drought-prone) species. In addition, better characterization of their involvement in drought will ultimately improve our understanding of how drought tolerance is manifested in plants.
Conclusion
We have described a comprehensive transcriptomics study, assaying the molecular underpinnings of drought responses in field-grown sorghum plants. This work comprises roughly 400 samples of leaf and root tissues under well-watered conditions, preflowering and postflowering drought stress, and a preflowering drought-recovery period for 2 genotypes with different drought responses. Overall, our data demonstrate that sorghum rapidly detects and adapts to drought stress. This response involves massive changes in the transcriptome, impacting more than 40% of all expressed genes encompassing a wide variety of molecular mechanisms.
Weekly sampling of droughted sorghum plants revealed dynamic properties of many transcriptional pathways. We highlighted differences between genotypes with respect to how quickly they recover from preflowering drought after rewatering, in particular, genes involved in photosynthesis. We have also observed constitutive differences between genotypes that may impact how prepared the plant is to respond to preflowering and postflowering drought, an element reported among ecotypes of other plants (50, 51). Finally, we described important interorganismal interactions between sorghum and its associated fungi that appear to be impacted by drought, reducing AM fungal symbiosis.
The data generated in this study provide many impactful avenues for future research into the development of sorghum and other crops, as well as biotic and metabolic responses to drought. The value of these data are magnified when considering insights noted from the companion measurements of microbial community composition (21, 22). The scale and scope of these data provide an unprecedented resource to deeply explore the molecular mechanisms of drought tolerance and its interplay with the plants’ biotic environment.
Materials and Methods
Leaf and root samples of 2 sorghum genotypes exposed to preflowering and postflowering drought stress were collected from field-grown plants in Parlier, California. Field setup and sample collection, transcriptomic profiling, protein extraction and Western blotting, spectrophometric metabolite and enzyme assays, leaf respiration measurements, high-performance liquid chromatography analysis, and computational analysis were performed as described in SI Appendix, Methods and Materials.
Data Access
The sequences and processed data files have been submitted to the NCBI Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE128441. Additional data are available in SI Appendix and at https://www.stat.berkeley.edu/∼epicon/publications/rnaseq/.
Supplementary Material
Acknowledgments
We thank Ciera Martinez, Diya Das, Nicholas Karavolias, and Alexandra Paxton for insightful discussions and advice on the manuscript; and Barbara Alonso for designing and creating Fig. 1. This research was funded in part by DOE Grant DE-SC0014081 awarded to P.G.L., D.C.D., J.D., R.H., E.P., J.T., A.V., and J.V.; the Gordon and Betty Moore Foundation Grant GBMF3834 and Alfred P. Sloan Foundation Grant 2013-10-27 to the University of California, Berkeley (N.V.); the ENS-CFM Data Science Chair (E.P.); the Office of Science (BER), DOE, DE-SC0012460 (M.J.H.); the US Cooperative Extension Service through the Division of Agriculture and Natural Resources of the University of California (P.G.L., J.D., and R.H.); the Berkeley Fellowship and the NSF Graduate Research Fellowship Program Grant DGE 1752814 (D.P.). K.K.N. is an investigator of the Howard Hughes Medical Institute. Work conducted by the US DOE Joint Genome Institute was supported by the Office of Science of the DOE under Contract no. DE-AC02-595-05CH11231. Work conducted at the Environmental Molecular Sciences Laboratory, a DOE Office of Science User Facility, was sponsored by the Office of Biological and Environmental Research.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE128441).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907500116/-/DCSupplemental.
References
- 1.Intergovernmental Panel on Climate Change IPCC 2014: Climate change 2014: Synthesis report. Contribution of Working Groups I, II and III to the fifth assessment report of the Intergovernmental Panel on Climate Change (Intergovernmental Panel on Climate Change, Geneva, 2014). [Google Scholar]
- 2.Barnabas B., Jager K., Feher A., The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 31, 11–38 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Fahad S., et al. , Crop production under drought and heat stress: Plant responses and management options. Front. Plant Sci. 8, 1–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lesk C., Rowhani P., Ramankutty N., Influence of extreme weather disasters on global crop production. Nature 529, 84–87 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Food and Agriculture Organization of the United Nations 2017: The impact of disasters and crises on agriculture and food security (Food and Agriculture Organization of the United Nations, Rome, 2018). [Google Scholar]
- 6.Mace E. S., et al. , Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 4, 2320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenow D. T., Quisenberry J. E., Wendt C. W., Clark L. E., Drought Tolerant Sorghum and Cotton Germplasm (Elsevier B.V., Amsterdam, 1983), vol. 12. [Google Scholar]
- 8.Xu W., Rosenow D. T., Nguyen H., Stay green trait in grain sorghum: Relationship between visual rating and leaf chlorophyll concentration. Plant Breed. 119, 365–367 (2008). [Google Scholar]
- 9.Kusaba M., Tanaka A., Tanaka R., Stay-green plants: What do they tell us about the molecular mechanism of leaf senescence. Photosyn. Res. 117, 221–234 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Borrell A. K., et al. , Stay-green alleles individually enhance grain yield in sorghum under drought by modifying canopy development and water uptake patterns. New Phytol. 203, 817–830 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Paterson A. H., et al. , The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009). [DOI] [PubMed] [Google Scholar]
- 12.McCormick R. F., et al. , The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93, 338–354. [DOI] [PubMed] [Google Scholar]
- 13.Buchanan C. D., et al. , Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Mol. Biol. 58, 699–720 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Dugas D. V., et al. , Functional annotation of the transcriptome of Sorghum bicolor in response to osmotic stress and abscisic acid. BMC Genom. 12, 514 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson S. M., et al. , Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genom. 15, 456 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson S. M., Cummins I., Lim F. L., Slabas A. R., Knight M. R., Transcriptomic analysis comparing stay-green and senescent Sorghum bicolor lines identifies a role for proline biosynthesis in the stay-green trait. J. Exp. Bot. 66, 7061–7073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yazawa T., Kawahigashi H., Matsumoto T., Mizuno H., Simultaneous transcriptome analysis of Sorghum and Bipolaris sorghicola by using RNA-seq in combination with de novo transcriptome assembly. PLoS One 8, e62460 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fracasso A., Trindade L. M., Amaducci S., Drought stress tolerance strategies revealed by RNA-Seq in two sorghum genotypes with contrasting WUE. BMC Plant Biol. 16, 115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubois M., Van den Broeck L., Inze D., The pivotal role of ethylene in plant growth. Trends Plant Sci. 23, 311–323 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daszkowska-Golec A., The Role of Abscisic Acid in Drought Stress: How ABA Helps Plants to Cope with Drought Stress (Springer International Publishing, Cham, Switzerland, 2016), pp. 123–151. [Google Scholar]
- 21.Xu L., et al. , Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, E4284–E4293 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao C., et al. , Strong succession in arbuscular mycorrhizal fungal communities. ISME J. 13, 214–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith R. H., Bhaskaran S., Miller F. R., Screening for drought tolerance in sorghum using cell culture. In Vitro Cellular amp; Dev. Biol. 21, 541–545 (1985). [Google Scholar]
- 24.Thomas H., Howarth C. J., Five ways to stay green. J. Exp. Bot. 51, 329–337 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Burke J., Franks C. D., Burow G., Xin Z., Selection system for the stay-green drought tolerance trait in sorghum germplasm. Agronomy J. 102, 1118–1122 (2010). [Google Scholar]
- 26.Matt Sowder C., Tarpley L., Vietor D., Miller F. R., Leaf photoassimilation and partitioning in stress-tolerant sorghum. Crop Sci. 37, 833–838 (1997). [Google Scholar]
- 27.Harris K., et al. , Sorghum stay-green QTL individually reduce post-flowering drought-induced leaf senescence. J. Exp. Bot. 58, 327–338 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Varoquaux N., et al. , Lifecycle transcriptomics of field-droughted sorghum reveals rapid biotic and metabolic responses. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128441. Deposited 18 March 2019. [DOI] [PMC free article] [PubMed]
- 29.Pandey S. P., Somssich I. E., The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bostock R. M., Pye M. F., Roubtsova T. V., Predisposition in plant disease: Exploiting the nexus in abiotic and biotic stress perception and response. Annu. Rev. Phytopathol. 52, 517–549 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Majeed Zargar S., et al. , Impact of drought on photosynthesis: Molecular perspective. Plant Gene 11, 154–159 (2017). [Google Scholar]
- 32.Suzuki N., Koussevitzky S., Mittler R., Miller G., ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Ding N., et al. , Identification and analysis of glutathione S-transferase gene family in sweet potato reveal divergent GST-mediated networks in aboveground and underground tissues in response to abiotic stresses. BMC Plant Biol. 17, 225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das K., Roychoudhury A., Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2, 53 (2014). [Google Scholar]
- 35.Lin Y. H., et al. , Overexpression of ferredoxin, PETF, enhances tolerance to heat stress in Chlamydomonas reinhardtii. Int. J. Mol. Sci. 14, 20913–20929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxwell D. P., Wang Y., McIntosh L., The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. U.S.A. 96, 8271–8276 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahal K., Vanlerberghe G. C., Alternative oxidase respiration maintains both mitochondrial and chloroplast function during drought. New Phytol. 213, 560–571 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Neumann E., George E., Colonisation with the arbuscular mycorrhizal fungus Glomus mosseae (Nicol. & Gerd.) enhanced phosphorus uptake from dry soil in Sorghum bicolor (L.). Plant Soil 261, 245–255 (2004). [Google Scholar]
- 39.Smith S. E., Mycorrhizal fungi. CRC Crit. Rev. Microbiol. 3, 275–313 (1974). [DOI] [PubMed] [Google Scholar]
- 40.MacLean A. M., Bravo A., Harrison M. J., Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 29, 2319–2335 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Augé R. M., Toler H. D., Saxton A. M., Arbuscular mycorrhizal symbiosis alters stomatal conductance of host plants more under drought than under amply watered conditions: A meta-analysis. Mycorrhiza 25, 13–24 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Augé R. M., Water relations, drought and vesicular-arbuscular mycorrhizal symbiosis. Mycorrhiza 11, 3–42 (2001). [Google Scholar]
- 43.Watts-Williams S. J., et al. , Diverse Sorghum bicolor accessions show marked variation in growth and transcriptional responses to arbuscular mycorrhizal fungi. Plant Cell Environ. 42, 1758–1774 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Gao C., et al. , Increased precipitation, rather than warming, exerts a strong influence on arbuscular mycorrhizal fungal community in a semiarid steppe ecosystem. Botany 94, 459–469 (2016). [Google Scholar]
- 45.Tyagi J., Sultan E., Mishra A., Kumari M., Pudake R. N., The Impact of AMF Symbiosis in Alleviating Drought Tolerance in Field Crops (Springer International Publishing, Cham, Switzerland, 2017), pp. 211–234. [Google Scholar]
- 46.Lazzarotto F., Turchetto-Zolet A. C., Margis-Pinheiro M., Revisiting the non-animal peroxidase superfamily. Trends Plant Sci. 20, 807–813 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Liu Q., et al. , Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srivasta A., Mehta S., Lindlof A., Bhargava S., Over-represented promoter motifs in abiotic stress-induced DREB genes of rice and sorghum and their probable role in regulation of gene expression. Plant Signal. Behav. 5, 775–784 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu S., Ligang C., Liping Z., Diqiu Y., Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J. Biosci. 35, 459–471 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Juenger T. E., et al. , Exploring genetic and expression differences between physiologically extreme ecotypes: Comparative genomic hybridization and gene expression studies of kas-1 and Tsu-1 accessions of Arabidopsis thaliana. Plant Cell Environ. 33, 1268–1284 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Juenger T. E. Natural variation and genetic constraints on drought tolerance. Curr. Opin. Plant Biol. 16, 274–281 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.