Significance
A challenge with the available synthetic viruses used for the treatment of genetic disorders is that they originate from wild-type viruses. These viruses benefit from infecting as many cells as possible in the body, while therapies should most often target a particular cell type, for example, dopamine neurons in the brain. In this paper, we present a technique for developing targeted synthetic viruses suitable for clinical therapy. We show that such viruses can be designed to target human dopamine neurons in vivo and to transport along the connective pathways in the brain, allowing for unprecedented accuracy of the therapy.
Keywords: vector evolution, gene therapy, barcoding, retrograde transport, dopamine
Abstract
Adeno-associated virus (AAV) capsid modification enables the generation of recombinant vectors with tailored properties and tropism. Most approaches to date depend on random screening, enrichment, and serendipity. The approach explored here, called BRAVE (barcoded rational AAV vector evolution), enables efficient selection of engineered capsid structures on a large scale using only a single screening round in vivo. The approach stands in contrast to previous methods that require multiple generations of enrichment. With the BRAVE approach, each virus particle displays a peptide, derived from a protein, of known function on the AAV capsid surface, and a unique molecular barcode in the packaged genome. The sequencing of RNA-expressed barcodes from a single-generation in vivo screen allows the mapping of putative binding sequences from hundreds of proteins simultaneously. Using the BRAVE approach and hidden Markov model-based clustering, we present 25 synthetic capsid variants with refined properties, such as retrograde axonal transport in specific subtypes of neurons, as shown for both rodent and human dopaminergic neurons.
The recombinant adeno-associated virus (AAV) vector has become the de facto standard for in vivo gene transfer (1). In clinical gene therapy, AAV has been proven to be safe and result in stable gene expression over many years (2, 3). However, with few exceptions, specificity and efficacy are still issues hindering some critical applications and have delayed broad clinical adoption in other areas (4). Engineering of the AAV capsid has been conducted over the last 2 decades to address these shortcomings and to generate capsids with altered tropism and function (5–15). AAV capsid engineering can broadly be divided into directed evolution and rational design. In directed evolution, random processes are utilized such as capsid gene shuffling of available serotypes (9, 16, 17), random peptide insertion into a known site of the AAV capsid (10, 14, 18–21), or phage display (22, 23). This approach, however, involves a selection process that requires multiple generations of screening to identify functional capsids (6–8), which excludes the use of tools such as single-cell or in situ sequencing. Due to the random nature of this process, it is also inherently unreproducible, and the resulting capsid variants provide little mechanistic insights into the molecular targets engaged. The alternative approach is rational design, where a priori knowledge is utilized to design fewer capsid variants and through systematic evaluation refine the capsid structure to achieve the desired function. Examples of this include the disruption of native cellular binding motifs and insertion of high-affinity ligands in the Cap gene, and through the use of structural and evolutionary shared sequences infer putative ancestral variants (24–29). What unifies the rational design approaches is that the resulting tested variants are counted in the tens or hundreds compared with the thousands to millions of capsids assessed using a directed evolution approach. While AAV capsid engineering has been around for over 25 y, few have broadly replaced wild-type variants in preclinical applications, and none has to date reached late-stage clinical trials despite distinct intellectual property incentives. The reason is that the improved features and functions have seldom transferred from the screening host species (most often mouse) to humans or the nonhuman primate (30).
To improve on this, we have developed a method for capsid engineering named barcoded rational AAV vector evolution (BRAVE), which encompasses all of the benefits of rational design (18, 26, 31–35) while maintaining the broad screening diversity permitted by directed evolution. The key to this method is a viral library production approach where each virus particle displays a protein-derived peptide on the surface, which is linked to a unique barcode in the packaged genome (36). Through hidden Markov model-based clustering (37), we were able to identify consensus motifs for neuronal cell type-specific retrograde transport and expression in the brain. The BRAVE approach enables the selection of functional capsid structures using only a single-generation screening. Furthermore, it can be used to map, with high resolution, the putative binding sequences of large protein libraries. Through the use of peptide sequences derived from proteins with a known mechanism in the human CNS, we believe that this approach provides significantly higher chances of success in clinical translation. Here, we have explored and validated this approach in the rodent brain as well as in human embryonic stem cell-derived dopaminergic (DA) neurons in vitro and in the brain of rats with human-derived DA transplants in vivo.
Results
In this, the first application of BRAVE screening, we utilized peptides derived from proteins with documented synapse interaction. The aim was to develop AAV capsid variants that are efficiently taken up and transported retrogradely in neurons in vivo. This combinatorial method enabled the generation and simultaneous functional mapping of close to 4 million unique AAV virion variants in parallel, both in vitro and in vivo. In a single round of screening, we selected 25 de novo capsid variants, all of which could be packaged into functional viruses with the capacity to become retrogradely transported in vivo and efficiently transduce neurons in vitro. We further characterized a capsid variant expressing highly efficient retrograde transport in both rat and human (stem cell-derived) DA neurons in vivo, and a pan-neuronal retrogradely transported AAV which we went on to use in an experiment aimed to elucidate the function of a subset of neurons in the basolateral nucleus of the amygdala projecting to the rostromedial striatum.
Design of Barcoded AAVs.
To link capsid structure to an in vivo expressed molecular barcode, we generated a refined AAV production plasmid. The barcode is inserted into the 3′ untranslated region (UTR) of GFP of a gutted self-complementary AAV genome (38, 39) and the AAV2 Rep/Cap genes are expressed from the same plasmid, but outside the inverted terminal repeats (ITRs), creating a linkage between capsid structure and barcode. Here, peptides are displayed at N587 of the VP1 capsid protein (18) (SI Appendix, Fig. S1). The insertion mutates the wild-type (WT) AAV2 heparan sulfate proteoglycan-binding motif (32). The AAV produced from this plasmid without an inserted peptide is hereafter referred to as MNMnull.
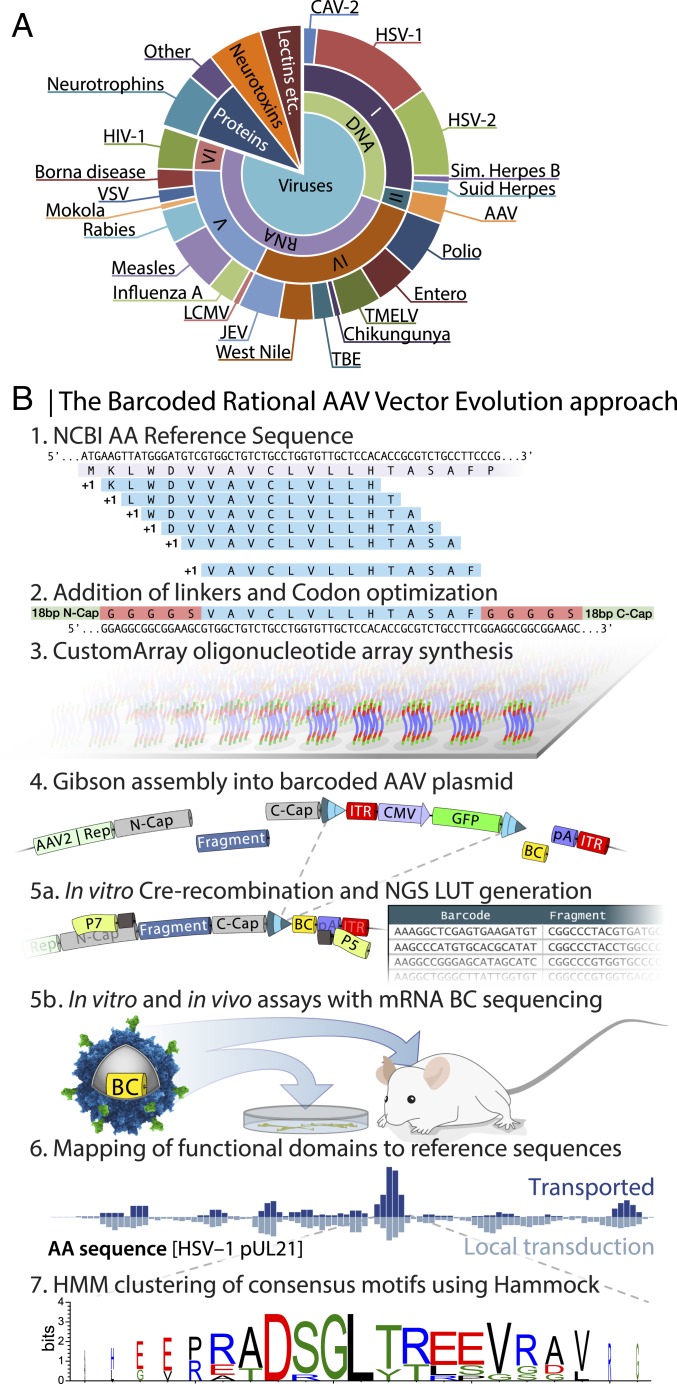
With the aim of targeting neurons at their terminals, we selected 131 proteins based on their documented affinity to synapses (Fig. 1A and SI Appendix, Fig. S2). We digested their amino acid (aa) sequences computationally into overlapping 14- or 22-aa-long polypeptides, and 3 alternative linkers were added to the 14-aa polypeptides (Fig. 1B, 1 and 2). The resulting 92,358 oligonucleotides were synthesized in parallel on a gene chip array (Fig. 1B, 3). They were then assembled into the backbone plasmid to allow for packaging of replication-deficient AAV particles, where the peptide is displayed on the capsid surface and a 20-bp random molecular barcode is included as part of the genome (Fig. 1B, 4 and SI Appendix, Fig. S1). In parallel, the same plasmid library was utilized to generate a look-up table (LUT), linking the random barcodes to the respective peptide (36) (see Fig. 1B, 5a and SI Appendix, Fig. S3 and Methods for details). The resulting library contained 3,934,570 unique combinations of peptide and barcode. On average, 50 barcodes point to the same peptide, but only 1 peptide is represented per barcode (40). This oversampling is essential for noise filtration and mapping of oligonucleotide array-induced mutations (see SI Appendix, Methods on LUT generation for details).
Fig. 1.
Barcoded rational AAV vector evolution screening approach. (A) Pie chart displaying the selected 131 proteins with documented affinity to synapses. (B) Schematics of the BRAVE procedure. 1: NCBI reference amino acid sequences were computationally digested into 14-aa (or 22-aa) -long polypeptides with a 1-aa shifting sliding window. 2: Three alternative linkers were added to the 14-aa polypeptides, for example, a rigid linker with 5 alanine residues (14aaA5). 3: Codon-optimized sequences (92,358) were synthesized in parallel on an oligonucleotide array. 4: The pool of oligonucleotides was assembled into an AAV production backbone with cis-acting AAV2 Rep/Cap and ITR-flanking CMV-GFP. A 20-bp random molecular barcode (BC) was simultaneously inserted into the 3′ UTR of the GFP gene. 5a: Using a fraction of the plasmid preparation, exposed to Cre-recombinase in vitro, a look-up table was generated linking the BC to the peptide. 5b: Using the same plasmid pool from step 4, an AAV batch was produced (MNMlib), and multiple parallel screening experiments were performed both in vitro and in vivo followed by BC sequencing from mRNA. 6: Through the combination of the sequenced barcodes and the LUT, transduction efficacy can be mapped back to each of the original 131 proteins, here exemplified by the aa sequence of HSV-1 pUL21. The height of each bar represents the relative abundance of that amino acid, normalized to the total library and summed from all peptides covering the region. 7: Consensus motifs can be determined using the Hammock, hidden Markov model-based clustering approach.
To ensure that each virion is assembled using only 1 mutated capsid protein variant and that the corresponding barcode is packaged inside, the AAV library plasmid was supplied at a 100- or 1,000-fold dilution compared with standard production (10, 41), resulting in 30 or 3 copies per cell (SI Appendix, Fig. S3), respectively. Using the AAV vector library, we performed multiple parallel screening experiments in vitro and in vivo, followed by the sequencing of barcodes expressed in the mRNA. Efficacy could be mapped back to the original 131 proteins with the help of the LUT and consensus motifs determined using the Hammock method (Fig. 1B, 6 and 7). The Hammock method utilizes a hidden Markov model approach to find underlying patterns in the complete dataset and cluster together peptides that share sequence homology.
Validation of the BRAVE Screening Principle.
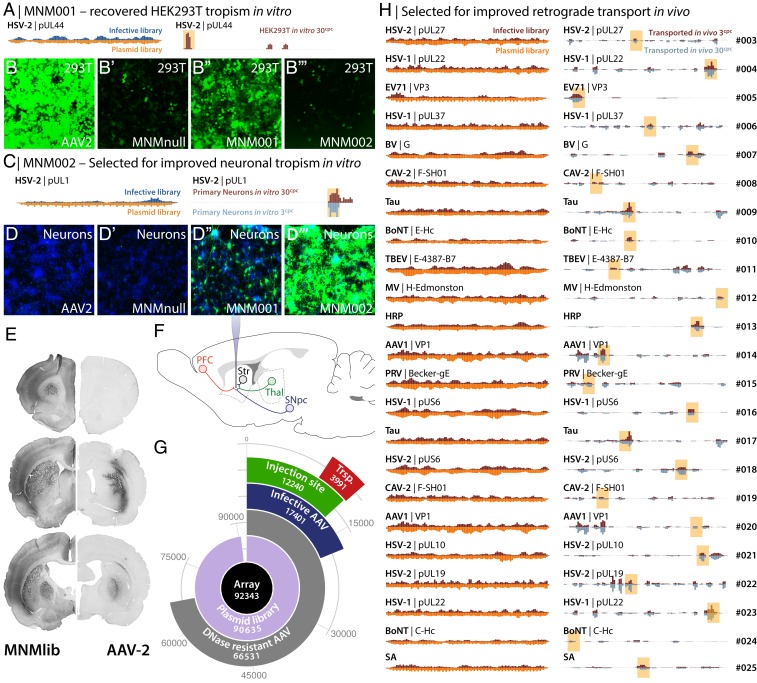
As an initial proof of concept, we utilized the BRAVE technology to screen for the reintroduction of tropism for HEK293T cells in vitro (Fig. 2 A–B″′), which was lost when the heparin sulfate proteoglycan (HS)-binding motif was removed in the MNMnull capsid (Fig. 2 B and B′). In the screening of the 4 million uniquely barcoded capsid variants, we found several regions from the 131 included proteins that conferred a significantly improved infectivity over the parent MNMnull capsid structure (SI Appendix, Fig. S4 and Dataset S1). One peptide from herpes simplex virus 2 (HSV-2) surface protein pUL44 was selected, and a first capsid variant was generated (MNM001) (Fig. 2A). This capsid indeed displayed a recovered tropism to the HEK293T cells (Fig. 2B″). Through a second BRAVE screen in primary cortical neurons, we identified several peptides clustering over a C-terminal region of the HSV-2 pUL1 protein (Fig. 2C). From these data, we generated an AAV capsid (MNM002) which improved the infectivity of primary neurons in culture dramatically compared with both the AAV2-WT and the MNMnull vector (Fig. 2 D–D″′, SI Appendix, Fig. S4, and Dataset S1). Together, these data provide compelling evidence of the efficiency of the BRAVE screening approach and indications that the peptides can be utilized more broadly than the initial protein inclusion criteria (synaptic association) would imply.
Fig. 2.
Assessment of improved in vitro infectivity and retrograde transport in the brain using BRAVE screening. (A) Infectivity mapping of HSV-2 pUL44 peptides is displayed comparing infective peptides (Top Left) plotted against total plasmid library (Bottom Left) and compared with peptides infecting HEK293T cells in vitro (Right). (B–B″′) The MNMnull capsid disrupts this binding (B′), compared to AAV2-WT (B). The first capsid generated from single-generation BRAVE screening in HEK293T, MNM001, displayed a significantly recovered tropism (B″), which is not observed in a capsid screened for in primary neurons (B″′). (C) Infectivity mapping of HSV-2 pUL1 peptides for introduced tropism to primary neurons. (D–D″′) In a second experiment, we used the BRAVE technology to improve the infectivity of primary cortical rat neurons in vitro. Both AAV2-WT and the MNMnull vector display very poor infectivity of primary neurons (D and D′) and the MNM001 displays some improvement (D″). BRAVE screening in primary neurons identified several peptides clustering over a C-terminal region of the HSV-2 pUL1 protein, which improved the infectivity of primary neurons in culture dramatically (D″′). (E) In vivo expression pattern of GFP after injection of a 30-cpc (copies per cell) library into the rat forebrain compared with an AAV2-WT vector at the same titer. (F) Connectivity diagram showing the injection site (striatum; Str) and connected neuronal populations frontal cortex (PFC), thalamus (Thal), and substantia nigra (SNpc) utilized for BRAVE screening in vivo for retrograde transport; 2.5 × 1010 or 4.4 × 108 vector genomes of the AAV library with capsid concentration of 30 or 3 cpc, respectively (5-µL total volume), were injected into the striatum (“3 cpc” and “30 cpc” refer to copies per cell of the capsid/genome plasmid utilized at the AAV production stage). (G) Polar plot showing the absolute quantities of the unique peptides recovered at each step of the BRAVE assay. (H) From the in vivo screening for improved retrograde transport capacity, we selected 23 peptides from 20 proteins that all were represented by multiple barcodes and found in multiple animals (Dataset S1). Twenty-three of the 25 de novo AAV capsid structures allowed for higher than or at par with AAV2-WT packaging efficacy (SI Appendix, Fig. S1), all with retrograde transportability (SI Appendix, Fig. S5).
Identification of Capsid Variants for Retrograde Axonal Transport.
In an experiment aimed to identify individual AAV capsid variants with efficient retrograde transport in neurons in vivo, we injected the AAV library (MNMlib) into the forebrain of adult rats. Compared with the standard AAV2-WT vector, the inserted peptides confer a striking change in the transduction pattern with both a broader spread of transduction and retrograde transport to the connecting afferent regions (Fig. 2E). Eight weeks after injection, total RNA was extracted from the injection site, and 3 connected areas (Fig. 2F) and the transcribed barcodes were sequenced (Dataset S1). Analysis of the unique peptides (identified using the barcodes and the LUT) throughout the BRAVE pipeline provides unique insights into the efficacy of the process (Fig. 2G). More than 98% of the designed oligonucleotides (array; black circle) were successfully cloned into the plasmid library and barcoded (purple ring) and 72% of the peptides permitted complete assembly of the AAV (as assessed by DNase treatment; gray ring). Barcodes recovered at the dissected regions reveal that ∼13% of the inserted peptides promoted efficient transduction in the brain (green ring segment) and ∼4% promoted retrograde transport in neurons (red ring segment). Pooling all experiments, including in vitro experiments, revealed that at least 19% of the peptides could promote infectivity in the cell types tested (dark blue ring segment). The large fraction, compared with what is observed in random peptide approaches, provides evidence that the rational design, even at this relatively high throughput, provides a significant advantage in the generation of functional capsids compared with a random approach. Even without any consideration of unsuitable peptide sequences for capsid insertion, only the introduction of a premature stop codon would remove 54 to 68% of all capsids with a random inserted peptide at this length.
Validation of Capsid Variants.
From this comprehensive in vivo BRAVE screening, we selected 23 peptides from 20 proteins that all were represented by multiple barcodes and found in multiple animals (Fig. 2H, SI Appendix, Fig. S5, and Dataset S1). Twenty-one of the 23 de novo AAV capsid structures found in vivo (and the 2 from the in vitro screening) allowed for higher than or on par with AAV2-WT packaging efficacy (SI Appendix, Fig. S1). Of note is that this experiment suggests that the insertion of 16 aa (14-aa peptide + 2 alanine [A] linkers) is approaching the upper limit of insertion into this site in the AAV capsid, as only 1 out of the 23 selected peptides was 24 aa in length (14 + 2 G4S/A5 linkers or 22 aa + 2 A linkers). Moreover, in the infective library, the 24-aa sequences were heavily underrepresented despite making up more than half of the total diversity in the peptide library (see Dataset S1 for details). To confirm the predictive nature of the single-generation BRAVE screen, we injected the 23 individually produced capsid variants into the rat striatum, one by one, and assessed retrograde transport to afferent regions (SI Appendix, Fig. S5). All 21 capsids selected for this capacity showed drastically improved transport compared with the parent AAV, while the 2 selected in vitro showed no improvement. Using a hidden Markov model (HMM)-based clustering (37), of all peptides with retrograde transportability, we could determine the putative consensus motif for each of the 23 capsids, providing a foundation for directed optimization (Dataset S1).
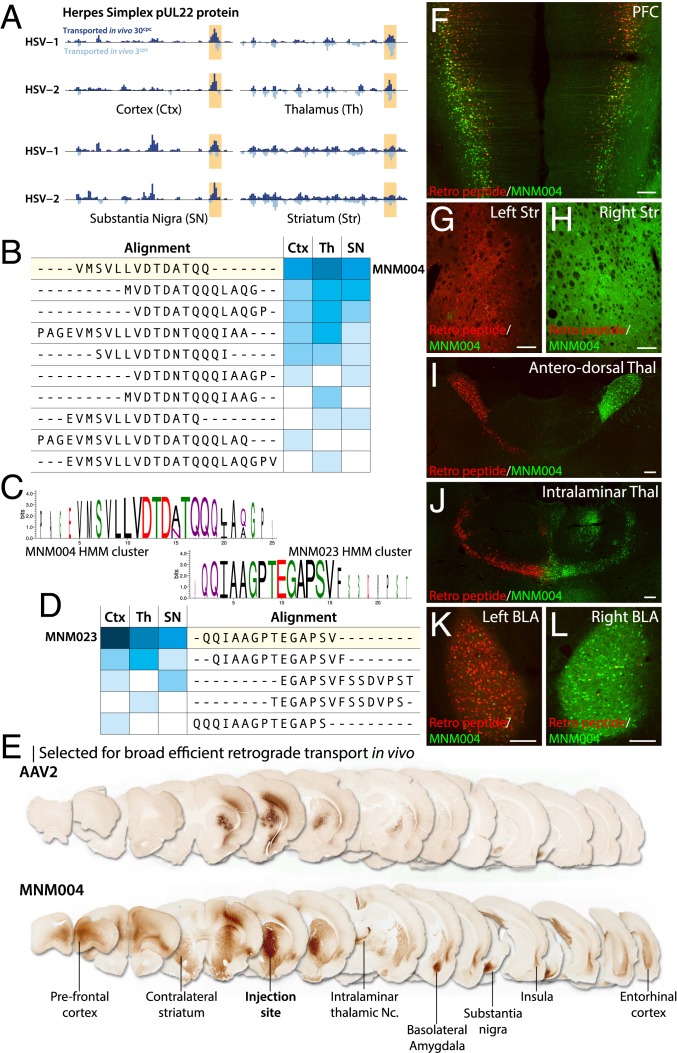
In the BRAVE design, we can compare the function of peptides across multiple regions of the brain and also between animals (Dataset S1). This data aggregation allows us to map domains of proteins that promote specific transport or uptake abilities. Exemplified by HSV-derived pUL22 protein, peptides throughout the protein can drive uptake at the injection site in the striatum (Fig. 3A). However, only peptide sequences from the C-terminal region of the protein displayed reproducible transport to all afferent brain structures: the cortex, thalamus, and substantia nigra (Fig. 3 A and B). HMM clustering of all peptides recovered at these sites revealed 2 overlapping consensus motifs (Fig. 3 B–D). These were generated into the MNM004 (VMSVLLVDTDATQQ) and MNM023 (QQIAAGPTEGAPSV) (Fig. 3C) capsid structures, respectively, with both displaying similar transport patterns in vivo (SI Appendix, Fig. S5 E and X). The MNM004 capsid promoted efficient retrograde transport to all afferent regions as far back as the medial entorhinal cortex (Fig. 3E and Movie S1). The parent AAV2-WT capsid, by contrast, promoted efficient transduction at the site of injection but very little retrograde transport of the vector (Fig. 3E). While this manuscript was being finalized, another peptide, LADQDYTKTA (Retro peptide), was published promoting strong retrograde transport when displayed in the same location on the AAV2 capsid surface (AAV2-Retro) (13). The paper also reports 2 additional point mutants in the capsid but without any data showing their influence on the transport. To allow the comparison between the peptides those were here omitted. When compared in vivo, the 2 vectors displayed very similar retrograde transport properties, with MNM004 showing equal or higher transport efficacy (Fig. 3 F–L and SI Appendix, Fig. S6).
Fig. 3.
Characterization of the MNM004 capsid for retrograde axonal transport in the rat brain. (A) A C-terminal region of the HSV pUL22 protein (highlighted) displayed reproducible transport to all afferent brain areas (cortex, thalamus, and substantia nigra) while not showing the same bias at the injection site (striatum). As this domain of the pUL22 protein is highly conserved between HSV strains (HSV-1 and HSV-2), both are visualized for comparison. (B–D) HMM clustering of all peptides displaying these properties revealed 2 overlapping consensus motifs (C). Those were generated into the MNM004 (B) and MNM023 (D) AAV capsid variants. (E) In vivo comparison between MNM004 and the parent AAV2 after unilateral striatal injection. Both vectors express GFP, and sections were developed using brown DAB (3, 3-diaminobenzidine)-peroxidase reaction (see Movie S1 for a 3D visualization). (F–L) Same animal comparison between MNM004 and AAV2 capsid with the previously published LADQDYTKTA peptide (AAV2-Retro). AAV2-Retro expressing mCherry was injected into the left striatum (G) while MNM004 expressing GFP was injected into right striatum (H) at matched titers. Monitored afferent regions include the prefrontal cortex (F), anterodorsal (I) and intralaminar thalamic nuclei (J), and basolateral amygdala (K and L) (see SI Appendix, Fig. S6 for additional information). (Scale bars, 200 µm.)
A Capsid Variant for the Retrograde Infectivity of Dopamine Neurons.
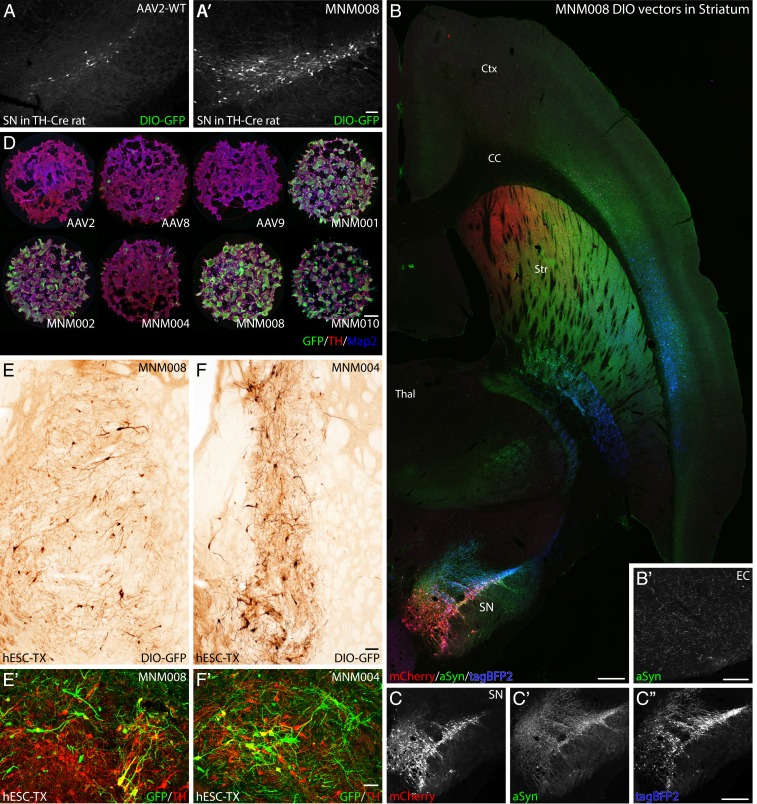
In the final component of the BRAVE screening, we aimed to develop a AAV capsid variant with efficient retrograde transport to dopamine neurons of the substantia nigra from injections into the striatal target region. In this screening, we identified 2 domains of the canine adenovirus (CAV-2) capsid protein [an often-used but immunogenic vector for targeting of DA neurons from their terminals in vivo (42)]. Using a Cre-inducible AAV genome (CMV-loxP-GFP) injected into the striatum of TH-Cre knockin rats, we found that the CAV-2–based MNM008 improved the retrograde infectivity of nigral neurons dramatically compared with the parent AAV2, from only a few scattered neurons transduced by AAV2-WT to the vast majority of the substantia nigra pars compacta neurons targeted by MNM008 (Fig. 4 A and A′). Next, we utilized this capsid to revisit a long-standing question of nigrostriatal innervation topography. The rodent striatum is functionally divided along each axis: the mediolateral, dorsoventral, and rostrocaudal (43). This topography is maintained in the striatal afferents from the forebrain neocortex, amygdala, and thalamic nuclei (43). Here we utilized 3 genes with available high-specificity antibodies, mCherry, TagBFP2, and alpha-synuclein (labeled red, blue, and green, respectively, in Fig. 4B). The latter was chosen due to the protein’s prominent localization to axon terminals and its excellent axonal transport properties in the nigrostriatal system. With this approach, we could confirm that the rostrocaudal topography of the projections to the striatum is linked to a mediolateral topography of the substantia nigra (Fig. 4 B–C″). Also, this experiment showed abundant collaterals from the dopamine neurons projecting to the central striatum reaching the entire ipsilateral neocortex, including as far back as the entorhinal cortex (Fig. 4B′).
Fig. 4.
Validation of AAV capsids infecting rat and human dopaminergic neurons in vivo and in vitro. (A and A′) In vivo comparison between the CAV-2–derived MNM008 and AAV2-WT assessing infectivity of dopamine neurons from their terminals in the striatum. Both vectors express Cre-inducible GFP (double-floxed inverted orientation, DIO-GFP) and were injected into the striatum of TH-Cre knockin rats. (B and C″) Mapping of nigral afferent topography to the striatum using 3 Cre-inducible genes (mCherry, red; alpha-synuclein, green; tagBFP2, blue) packaged in separate MNM008 vectors and injected into the striatum along the rostrocaudal axis (B), as illustrated in a horizontal section including both striatum (Str) and substantia nigra pars compacta (SN) imaged using a laser-scanning confocal microscope. (C–C″) The nigro-striatal topology is visible in the transgene expression pattern in the SN. (B′) transduced DA-neurons send collaterals as far back as the entorhinal cortex (EC). (D) Assessment of retained neuronal tropism in human embryonic stem cell-derived DA neuroblasts in vitro. (E and F) Assessment of retrograde infectivity in humanized rats. These animals first received an hESC-derived DA-rich neuronal transplant (expressing Cre) into the striatum. Six months later, the MNM008 or MNM004 vectors (expressing DIO-GFP) were injected into the frontal cortex. hESC-derived neurons in the graft were efficiently labeled by both vectors (E and F) and the vast majority expressed the DA neuron marker tyrosine hydroxylase (TH) determined using confocal microscopy (E′ and F′). (Scale bars: in A′ represents 100 µm in A and A′; in B represents 500 µm in B, C, and C″; in B′ represents 100 µm; in D represents 100 µm; in E represents 50 µm in E and F; and in E′ represents 50 µm in E′ and F′.)
A significant drawback of de novo capsid design using animal models has been the lack of predictability with regard to the translation to human cells. The BRAVE approach offers an exciting possibility to solve this problem through the use of naturally occurring peptides from viruses and proteins with known function in the human brain. To establish that the properties of MNM008 were not limited to rodent DA neurons, we performed an experiment on DA neurons derived from Cre-expressing human embryonic stem cells (hESCs) both in vitro and in vivo after transplantation into immunocompromised rats (44) (Fig. 4 B–D). Six months after transplantation, when the grafted neurons have grown to establish axonal projections with their usual forebrain target areas, the MNM008 vector was injected into the frontal cortex of the animals. MNM008 was efficiently transported back to the transplanted neurons innervating this region (Fig. 4E), with the majority of these cells being dopaminergic (Fig. 4E′).
Interestingly, MNM004, which does not have this ability in the rat brain, showed similar potency in the transplanted human neurons (Fig. 4 F and F′). Using the same in vitro hESC differentiation protocol, we confirmed that the de novo capsid variants (MNM002, 008, and 010) which displayed high tropism on primary rodent neurons also showed much higher tropism than the wild-type AAV variants (Fig. 4D and SI Appendix, Fig. S8) in human neurons in vitro. Of note is that the MNM004 capsid variant, so efficient in vivo, was not at all suitable for in vitro transduction (Fig. 4D).
Mapping of Tau-Derived Peptides for Neuronal Uptake of AAV.
The BRAVE approach provides a unique possibility to map protein function systematically. We therefore utilized this approach to display peptides from endogenous proteins involved in the pathogenesis of Alzheimer’s disease, amyloid precursor protein (APP) and microtubule-associated protein Tau, to provide insights into the mechanism underlying the proposed cell-to-cell communication of these proteins in the diseased brain (45, 46). In the mapping of APP, we found 2 regions that conferred retrograde transport, 1 in the soluble APP N-terminal region and 1 in the amyloid beta region (SI Appendix, Fig. S7). The functional properties of peptides originating from Tau were even more striking. In this protein, a central region conveyed very efficient retrograde transport (Fig. 5A, SI Appendix, Fig. S7, and Dataset S1). Two capsid structures were generated from this region, MNM009 and MNM017. Both capsids promoted retrograde transport in vivo (SI Appendix, Fig. S5), but MNM017 also displayed additional noteworthy properties. MNM017 infected both primary rat neurons and glial fibrillary acidic protein (GFAP)-positive primary rat glial cells in vitro with very high efficacy (Fig. 5B), including human primary astrocytes (Fig. 5 C–C″). Using this property, we then performed a displacement experiment comparing the MNM017 with the neurotrophic MNM002 capsid, which is not generated from a Tau-derived peptide (Fig. 5D). Three groups of primary neuron populations were pretreated with different recombinant Tau variants (T44, T39, and K18). The T44 variant had no apparent effect on the MNM017-to-MNM002 ratio of infectivity, while K18 enhanced the infectivity of MNM017 and the T39 variant efficiently blocked the infectivity compared with MNM002 (Fig. 5D). This outcome suggests that the Tau peptide is utilizing a receptor on the neurons that also has a binding activity of full-length human Tau protein.
Fig. 5.
AAV variants displaying peptides from the Alzheimer’s disease-related protein Tau allow competition with recombinant Tau uptake in astrocytes. (A) Peptides originating from a central region (highlighted) in the microtubule-associated protein Tau were found to convey both a very efficient retrograde transport and local infectivity in the rat brain. Two capsid variants were generated from this region, MNM009 and MNM017. (B and B′) In vitro assessment of 2 de novo capsid variants, the Tau-unrelated variant MNM002 and the Tau-derived MNM017 variant, in the transduction of primary GFAP+ rat astrocytes. (C and C′) In vitro transduction of human primary glia using MNM002 and MNM017. (D) Recombinant Tau displacement experiment in vitro of MNM017 compared with the MNM002 capsid, which is not generated from a Tau-related peptide. Primary cortical neurons were first treated with different recombinant Tau species at varying concentrations and then infected by MNM017(GFP) and MNM002(mCherry) simultaneously. (Scale bars: in B′, Upper represents 200 µm in B, Upper and B′, Upper; in B′, Lower represents 50 µm in B, Lower and B′, Lower; and in C′ represents 500 µm in C and C′.)
Basolateral Amygdala Regulates Anxiety Behavior via the Rostromedial Striatum.
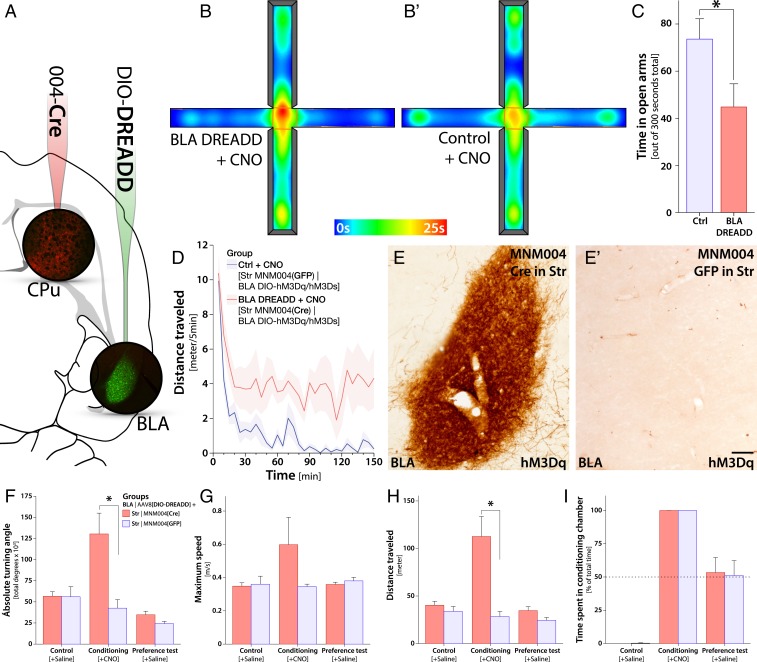
The BRAVE-generated capsid variants provide a powerful tool for brain connectivity studies. We utilized the MNM004 capsid variant to answer an outstanding question regarding the functional contribution of the afferents from the basolateral amygdala (BLA) to the rostromedial striatum. The experiment was conducted using a retrogradely induced chemogenetics (designer receptor exclusively activated by designer drugs, DREADDs) approach, using an injection of an MNM004-Cre vector in the rostromedial striatum combined with an injection of a Cre-inducible, activating DREADD vector in the BLA (Fig. 6A). After selective induction of activity of the BLA neurons projecting to the rostromedial striatum, induced by a systemic injection of the activating ligand clozapine-N-oxide (CNO), we found a striking fear and anxiety phenotype (n = 10) (Fig. 6 B and C) compared with control animals (n = 10) (where the Cre gene was replaced with GFP; Student’s 2-tailed t test, P < 0.05). This stands in stark contrast to the function of the BLA projections to the nucleus accumbens shown to promote positive stimuli (47). This increased anxiety phenotype was accompanied by significant hypermobility (Fig. 6D and SI Appendix, Fig. S9) and a fear phenotype including excessive digging, severe sweating, and episodes of freezing (Movie S2). Postmortem immunohistochemistry confirmed a highly selective DREADD expression in the BLA (Fig. 6 E and E′).
Fig. 6.
Selective activation of basolateral amygdala projections to the rostromedial striatum induced using the MNM004 capsid drives fear and anxiety phenotypes. (A) Experimental injection paradigm for functional assessment of the afferents from the basolateral amygdala to the rostromedial striatum (CPu). We injected the Cre-expressing MNM004 vector bilaterally into the CPu and a Cre-inducible (DIO) chemogenetic (DREADD) vector into the BLA bilaterally into wild-type rats (n = 20). Eight weeks later the animals received an s.c. injection of the DREADD-activating ligand clozapine-N-oxide. (B and C) Assessment of the fear and anxiety phenotype using the elevated plus maze, where the animals spent significantly less time in the open arms (n = 10) (B) compared with control animals (n = 10) (B′ and C), where the Cre gene was replaced with GFP. (D) The increased anxiety phenotype was accompanied by significant hypermobility in the open-field arena and a fear phenotype including excessive digging, severe sweating, and episodes of freezing (Movie S2). (E and E′) Visualization of selective DREADD expression in the BLA using immunohistochemistry together with brown DAB-peroxidase precipitation reaction. (F–I) After selective induction of the activity of the BLA neurons projecting to the dorsal striatum using the DREADD ligand CNO, we found a striking fear and anxiety phenotype in the animals with excessive defecation, sweating, digging, and freezing (Movie S2). This behavior was accompanied by an increase in both ipsilateral and contralateral rotation (F), high-speed rushes (G), and significantly elevated mobility (H). However, this did not result in any conditioning, as seen in the preference test performed the next day (I). Both animals in the active group and the control group spent equal time in both chambers with no signs of conditioned place aversion. (Scale bar, 50 µm in E and E′.) The asterisk in C, F, and H represents P < 0.05 in Student’s 2-tailed t test, corrected for multiple comparisons in F and H using Bonferroni correction. Error bars in C and F–I display the standard error of the mean (SEM).
Discussion
The approach to AAV capsid engineering described here provides distinct advantages over currently used methods with respect to both efficiency and speed. Compared with previous methods for the generation of AAV variants (5–14, 16–20, 24–27), the BRAVE screening approach is unique in that it allows for a systematic mapping of capsid motifs already after a single round of screening and enables the identification of capsid variants with efficient production yield and desired in vivo properties. Moreover, the highly efficient in vivo screening used here makes it possible to perform parallel and reproducible assessments in diverse tissues and cell types or different animal species. The method has several advantageous features. The use of a barcode in the packaged genome allows for removal of the Cap gene which allows for the utilization of a self-complementary AAV genome with swift and robust expression, and the expression of the barcode in the viral mRNA allows for detection of single virions transported to the target region without relying on adenoviruses for in vivo recovery. The efficient recovery of barcodes from the virally expressed mRNA also provides a guarantee that we only screen for capsid variants that have successfully undergone all critical steps of the AAV infectivity, namely cell attachment, internalization, lysosomal escape, and nuclear entry. This readout modality is of fundamental importance, as some versions of capsid engineering may result in capsid sequestration on the surface of the cell which could block the internalization and thus lead to false readouts. The BRAVE approach ensures that such capsid variants are filtered out. The barcodes from the injected library could be extracted from DNA as well. This would provide interesting data on which capsid variants can get stuck in the tissue (before or after internalization and uncoating) but not lead to successful transgene expression. However, as the primary purpose of this study was to design functional capsids driving transgene expression, we have chosen against that added step.
In a large pooled screen, the capsid variants can compete with each other or endogenous ligands for the same receptors. However, we believe that the mRNA readout and intrasample comparison in the same animal are key to resolving this. The end result would be that only the most potent receptor-binding variants (capable of successful infection) would result in the expression of an mRNA barcode. One such competition with an endogenous ligand is the finding of an uptake mechanism competing with the Tau protein.
An additional advantage of this method is the use of a stable bacterial library together with oversampling of barcodes, which ensures very efficient error control in the in vivo screening, since the identification of the vector-linked peptide transcript is based solely on unique barcode counts (not mRNA expression level) as well as the number of animals displaying the same readout for each individual peptide. This readout provides unprecedented accuracy and removes the need for multiple generations of screening. In future studies, the LUT and the associated in vivo data can be reused to further improve on target selection and filter out off-target infectivity. Furthermore, the single-generation BRAVE screening has the potential to be used together with both single-cell and in situ sequencing methods (48). This application is the main reason behind expressing the barcode from a polymerase II (PolII) promoter (cytomegalovirus; CMV) in a polyadenylated transcript (required for current in situ/single-cell sequencing technologies) and not a stronger PolIII promoter, successfully used for barcoding of AAV elsewhere (49).
The BRAVE approach depends on 2 fundamental advances: the tailored massively parallel array synthesis of DNA sequences and the generation of a look-up table linking a genetic barcode to the capsid structure. These techniques provide significant advantages to the screening approach such as the single-generation selection, potential mapping of changes over the entire capsid structure, and scanning of sequences derived from proteins with known function, but they also confer some limitations. The principal limitation is the comparatively small size and diversity of the libraries compared with libraries generated through processes such as capsid shuffling, degenerate primers, or error-prone PCR. However, this limitation is outweighed by the power of the rational design, which ensures the inclusion of only potentially functional sequences in the library without sequence repetitions or bias. While this approach cannot accommodate the full theoretical diversity of an entirely random approach, the continuous increase in synthesis size (now in the 106 range) together with a decrease in cost will further broaden the potential applications.
It is important to realize that the principle behind the BRAVE approach is not limited to the screening of capsid variants carrying modifications restricted to a single, small domain, such as the HS-binding motif used in this study (36, 50). It is possible that the BRAVE screening approach can be used where rational changes are introduced at any site of the 3 capsid proteins. In this case, where the long-read sequencing would be required to map the changes throughout the Cap gene, the implementation is greatly facilitated by the efficiency of the barcoding paradigm, allowing for this costly sequencing to be conducted only at the generation of the look-up table, not when assessing the function in vivo. However, the feasibility of such an approach remains to be shown.
In the presented studies, microdissection was used to separate afferent brain regions and allowed for the identification of transported variants, for example, to the DA neurons of the SN. However, the current library can potentially, without modification, be used together with FACS, single-cell/single-nucleus RNA-seq (sequencing) paradigms (e.g., from 10× genomics), or spatial sequencing [e.g., Slide-seq (51)]. This screen could allow for direct discrimination of capsids with specific, cell type-specific infectivity from those with high ubiquitous tropism. Such methods could further reduce the off-target infectivity, improve potency, and make possible the development of vector capsids with high selectivity and tropism of cellular subtypes.
In this first validation of the technique, we focused on sequences derived from proteins with known synaptic interactions. This approach was remarkably efficient, allowing us in a single round of screening to generate 25 AAV vector variants with the property of being efficiently transported retrogradely in diverse neuronal populations in the brain. Five of these were explored further in vitro and in vivo for their usefulness as research tools for the study of neuronal connectivity and function in the rodent brain. The HSV-derived peptide sequence used in MNM004 and the CAV-2–derived peptide sequence used in MNM008 provided these vectors with radically improved retrograde transport properties relative to the parent AAV2 vector, on par with or superior to previously published AAV variants. The properties of these AAV variants make them highly attractive as tools for studies of functional connectivity in the brain, as illustrated by the experiment performed on the neurons connecting the basolateral amygdala and the rostromedial striatum, where we used the MNM004 vector as a retrogradely induced chemogenetic tool. Moreover, the remarkable properties of MNM009 and MNM017, which express Tau-derived peptide sequences, indicate that the infectivity of these vectors depends on their ability to bind to a receptor that has binding activity for the human Tau protein, and thus possibly involved in the cell-to-cell spread of this disease-causing protein.
In summary, the BRAVE screening approach described here provides a tool for the design of AAV vector variants with tailored in vivo properties and tropism based on a highly efficient method for screening and selection of engineered capsid structures. This strategy, which can be applied in vivo in both rodents and primates, as well as in human cells in vitro, opens up the design and development of synthetic AAV vectors expressing capsid structures with unique properties and broad potential for clinical applications and brain connectivity studies. The results will provide possibilities to broaden the AAV toolbox for the exploration of vectors for use in future clinical gene therapies.
Data and Materials Availability.
The datasets supporting the conclusions of this article are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) with accession no. PRJNA473475. The R-based workflow is publicly available as a Git repository at https://bitbucket.org/MNM-LU/aav-library and as a Docker image: Bjorklund/aavlib:v0.2 (https://hub.docker.com/r/bjorklund/aavlib). The complete bioinformatics output and annotated code can be found in Dataset S1.
Supplementary Material
Acknowledgments
We thank the staff at the National Genomics Infrastructure of SciLifeLab, Sweden, and UCLA Clinical Microarray Core, United States, for expert assistance in the sequencing performed using the Illumina NextSeq technology. We also thank Anna Hammarberg for assistance and much appreciated help with in vitro fluorescence imaging. pscAAV-GFP was a gift from John T. Gray (Addgene plasmid 32396), pHGTI-adeno was a gift from Julie Tordo, and recombinant Tau was a gift from Virginia Lee and Alexander Crowe. This work was supported by Parkinson’s Disease Foundation International Research Grant PDF-IRG-1303, the Swedish Research Council (K2014-79X-22510-01-1 and ÄR-MH-2016-01997 starting grant), Swedish Parkinson Foundation, Swedish Alzheimer Foundation, Crafoord Foundation, The Bagadilico Linnaeus Consortium, Multipark, Schyberg Foundation, Thuring Foundation, Kocks Foundation, Åke Wiberg Foundation, Åhlén Foundation, Magnus Bergvall Foundation, Tore Nilsson Foundation, The Swedish Neuro Foundation, O. E. and Edla Johanssons Foundation, and Lars Hierta Foundation. M.P. is a New York Stem Cell Foundation Robertson Investigator. T.B. is supported by an associate senior lectureship from the Bente Rexed Foundation.
Footnotes
Competing interest statement: M.D. and T.B. are authors of a patent application covering components of the presented study.
This article is a PNAS Direct Submission.
Data deposition: Datasets supporting the conclusions of this article have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (accession no. PRJNA473475). The R-based workflow is publicly available as a Git repository at https://bitbucket.org/MNM-LU/aav-library and as a Docker image: Bjorklund/aavlib:v0.2.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910061116/-/DCSupplemental.
References
- 1.Hocquemiller M., Giersch L., Audrain M., Parker S., Cartier N., Adeno-associated virus-based gene therapy for CNS diseases. Hum. Gene Ther. 27, 478–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deverman B. E., Ravina B. M., Bankiewicz K. S., Paul S. M., Sah D. W. Y., Gene therapy for neurological disorders: Progress and prospects. Nat. Rev. Drug Discov. 17, 641–659 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Russell S., et al. , Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: A randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colella P., Ronzitti G., Mingozzi F., Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 8, 87–104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray S. J., et al. , Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood-brain barrier (BBB). Mol. Ther. 18, 570–578 (2010). Correction in: Mol. Ther.18, 1054 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan K. Y., et al. , Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deverman B. E., et al. , Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 34, 204–209 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojala D. S., et al. , In vivo selection of a computationally designed SCHEMA AAV library yields a novel variant for infection of adult neural stem cells in the SVZ. Mol. Ther. 26, 304–319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grimm D., et al. , In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 82, 5887–5911 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maheshri N., Koerber J. T., Kaspar B. K., Schaffer D. V., Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 24, 198–204 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Müller O. J., et al. , Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 21, 1040–1046 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Yang L., et al. , A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc. Natl. Acad. Sci. U.S.A. 106, 3946–3951 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tervo D. G., et al. , A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Körbelin J., et al. , Pulmonary targeting of adeno-associated viral vectors by next-generation sequencing-guided screening of random capsid displayed peptide libraries. Mol. Ther. 24, 1050–1061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsic D., Méndez-Gómez H. R., Zolotukhin S., High-accuracy biodistribution analysis of adeno-associated virus variants by double barcode sequencing. Mol. Ther. Methods Clin. Dev. 2, 15041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., et al. , Engineering and selection of shuffled AAV genomes: A new strategy for producing targeted biological nanoparticles. Mol. Ther. 16, 1252–1260 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Asuri P., et al. , Directed evolution of adeno-associated virus for enhanced gene delivery and gene targeting in human pluripotent stem cells. Mol. Ther. 20, 329–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girod A., et al. , Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 5, 1052–1056 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Wu P., et al. , Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74, 8635–8647 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalkara D., et al. , In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci. Transl. Med. 5, 189ra76 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Rabinowitz J. E., Xiao W., Samulski R. J., Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology 265, 274–285 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Chen Y. H., Chang M., Davidson B. L., Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat. Med. 15, 1215–1218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicklin S. A., et al. , Efficient and selective AAV2-mediated gene transfer directed to human vascular endothelial cells. Mol. Ther. 4, 174–181 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Tordo J., et al. , A novel adeno-associated virus capsid with enhanced neurotropism corrects a lysosomal transmembrane enzyme deficiency. Brain 141, 2014–2031 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zinn E., et al. , In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep. 12, 1056–1068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Münch R. C., et al. , Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol. Ther. 21, 109–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanaan N. M., et al. , Rationally engineered AAV capsids improve transduction and volumetric spread in the CNS. Mol. Ther. Nucleic Acids 8, 184–197 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grifman M., et al. , Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol. Ther. 3, 964–975 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Yang Q., et al. , Development of novel cell surface CD34-targeted recombinant adenoassociated virus vectors for gene therapy. Hum. Gene Ther. 9, 1929–1937 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Hordeaux J., et al. , The neurotropic properties of AAV-PHP.B are limited to C57BL/6J mice. Mol. Ther. 26, 664–668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adachi K., Enoki T., Kawano Y., Veraz M., Nakai H., Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat. Commun. 5, 3075 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opie S. R., Warrington K. H. Jr, Agbandje-McKenna M., Zolotukhin S., Muzyczka N., Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77, 6995–7006 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perabo L., et al. , Heparan sulfate proteoglycan binding properties of adeno-associated virus retargeting mutants and consequences for their in vivo tropism. J. Virol. 80, 7265–7269 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ried M. U., Girod A., Leike K., Büning H., Hallek M., Adeno-associated virus capsids displaying immunoglobulin-binding domains permit antibody-mediated vector retargeting to specific cell surface receptors. J. Virol. 76, 4559–4566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albright B. H., et al. , Mapping the structural determinants required for AAVrh.10 transport across the blood-brain barrier. Mol. Ther. 26, 510–523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidsson M., et al. , A novel process of viral vector barcoding and library preparation enables high-diversity library generation and recombination-free paired-end sequencing. Sci. Rep. 6, 37563 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krejci A., Hupp T. R., Lexa M., Vojtesek B., Muller P., Hammock: A hidden Markov model-based peptide clustering algorithm to identify protein-interaction consensus motifs in large datasets. Bioinformatics 32, 9–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCarty D. M., et al. , Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 10, 2112–2118 (2003). [DOI] [PubMed] [Google Scholar]
- 39.McCarty D. M., Monahan P. E., Samulski R. J., Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 8, 1248–1254 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Davidsson M., et al. , BRAVE AAV library for in vivo screening of retrograde transport in the CNS. National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA). https://www.ncbi.nlm.nih.gov/bioproject/PRJNA473475. Deposited 29 May 2018.
- 41.Jordan M., Schallhorn A., Wurm F. M., Transfecting mammalian cells: Optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24, 596–601 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hnasko T. S., et al. , Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc. Natl. Acad. Sci. U.S.A. 103, 8858–8863 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voorn P., Vanderschuren L. J., Groenewegen H. J., Robbins T. W., Pennartz C. M., Putting a spin on the dorsal-ventral divide of the striatum. Trends Neurosci. 27, 468–474 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Nolbrant S., Heuer A., Parmar M., Kirkeby A., Generation of high-purity human ventral midbrain dopaminergic progenitors for in vitro maturation and intracerebral transplantation. Nat. Protoc. 12, 1962–1979 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Brettschneider J., Del Tredici K., Lee V. M., Trojanowski J. Q., Spreading of pathology in neurodegenerative diseases: A focus on human studies. Nat. Rev. Neurosci. 16, 109–120 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dujardin S., et al. , Neuron-to-neuron wild-type Tau protein transfer through a trans-synaptic mechanism: Relevance to sporadic tauopathies. Acta Neuropathol. Commun. 2, 14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Correia S. S., McGrath A. G., Lee A., Graybiel A. M., Goosens K. A., Amygdala-ventral striatum circuit activation decreases long-term fear. eLife 5, e12669 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ståhl P. L., et al. , Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Xu M., et al. , High-throughput quantification of in vivo adeno-associated virus transduction with barcoded non-coding RNAs. Hum. Gene Ther. 30, 946–956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Davidsson M., et al. , Molecular barcoding of viral vectors enables mapping and optimization of mRNA trans-splicing. RNA 24, 673–687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriques S. G., et al. , Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.